Abstract
PURPOSE
Typically stored as unstructured notes, surgical pathology reports contain data elements valuable to cancer research that require labor-intensive manual extraction. Although studies have described natural language processing (NLP) of surgical pathology reports to automate information extraction, efforts have focused on specific cancer subtypes rather than across multiple oncologic domains. To address this gap, we developed and evaluated an NLP method to extract tumor staging and diagnosis information across multiple cancer subtypes.
METHODS
The NLP pipeline was implemented on an open-source framework called Leo. We used a total of 555,681 surgical pathology reports of 329,076 patients to develop the pipeline and evaluated our approach on subsets of reports from patients with breast, prostate, colorectal, and randomly selected cancer subtypes.
RESULTS
Averaged across all four cancer subtypes, the NLP pipeline achieved an accuracy of 1.00 for International Classification of Diseases, Tenth Revision codes, 0.89 for T staging, 0.90 for N staging, and 0.97 for M staging. It achieved an F1 score of 1.00 for International Classification of Diseases, Tenth Revision codes, 0.88 for T staging, 0.90 for N staging, and 0.24 for M staging.
CONCLUSION
The NLP pipeline was developed to extract tumor staging and diagnosis information across multiple cancer subtypes to support the research enterprise in our institution. Although it was not possible to demonstrate generalizability of our NLP pipeline to other institutions, other institutions may find value in adopting a similar NLP approach—and reusing code available at GitHub—to support the oncology research enterprise with elements extracted from surgical pathology reports.
INTRODUCTION
Although electronic health record (EHR) systems are designed principally to facilitate billing and clinical care, they also contain real-world clinical data captured in both structured and unstructured formats that may be useful for research. Natural language processing (NLP) methods, a suite of techniques for rendering unstructured, free-text data amenable to computational analysis, have provided the opportunity to uncover insights about disease trajectories and other health outcomes that would otherwise require manual abstraction.1,2 With the advent of NLP techniques, there has been an influx of oncology studies applying such methods, as many of the data points critical for conducting population-level research in oncology remain scattered across disparate components of the EHR, often with limited or nonexistent structure. Differing groups have used diverse NLP techniques to extract oncology-specific clinical predictors and outcomes, with varying levels of success for particular tasks, including, but not limited to, case identification, staging, and outcome.2
CONTEXT
Key Objectives
To develop a natural language processing (NLP) method for surgical pathology reports extracting TNM staging scores and International Classification of Diseases, Tenth Revision codes regardless of the disease area.
Knowledge Generated
Extracting TNM staging scores substantially reduces the need for manual review of patients' charts to support several cancer investigations use cases, such as predictive modeling and patient stratification. We developed a method to enhance generalizability to other oncologic domains and support the research enterprise in our institution.
Relevance
The majority of the NLP methods developed to this date focused on only one disease area to extract a set of disease-specific concepts of interest, limiting the generalizability of those methods to other disease areas. Our NLP method demonstrated that extracting TNM staging is achievable within a large medical center with a broad range of oncology patients.
Surgical pathology reports are a particularly rich source of oncology-specific data elements and a potentially valuable target for NLP techniques. These reports often contain disease-agnostic data elements ranging from tumor classification to diagnostic indications of the patient. Some of these oncology-specific data elements, although only found within surgical pathology reports for neoplastic specimens, contain information critical for assessing disease severity at presentation, including the TNM staging score. Surgical pathology reports often contain the formal pathologic diagnosis, standardized with reference terminologies, such as SNOMED clinical terms, International Classification of Diseases, Ninth Revision (ICD-9), or International Classification of Diseases, Tenth Revision (ICD-10), often offering a more specific description of the diagnosis. Structured diagnoses provided by pathologists in a formal report may often display more specificity than diagnoses assigned by clinicians in the course of an office visit, specifically denoting laterality and/or morphology by their use of particular terms. For example, a patient diagnosed with an ICD-10 code of C74.9 (“malignant neoplasm of unspecified part of adrenal gland”) by an oncologist or primary care physician might see their disease coded as C74.12 (“malignant neoplasm of medulla of left adrenal gland”) in a surgical pathology report for a biopsy or adrenalectomy.
The College of American Pathologists has published several protocols3 promoting required data elements to be reported in pathology reports.4 Although these protocols have been crucial in harmonizing how pathologists report specific data elements within the past couple of years (eg, TNM staging), there are a large number of legacy pathology reports written before implementation of these protocols. When developing an NLP technique to extract data elements of interest, reporting styles before and after implementation of these protocols should be taken into account.
NLP techniques reported to date have primarily focused on extraction for specific disease areas, as opposed to surgical pathology reports writ large. Efforts focused on disease-specific areas, although useful in a particular subspecialty, hinder the potential impact of such methods to support the research enterprise within large medical centers with a broad range of oncology patients. To the best of our knowledge, the majority of studies that used NLP techniques to parse pathology reports focused on only one disease area to extract a set of disease-specific concepts of interest. Relevant work often focused on a specific disease area such as prostate,5,6 breast,7 lung,8-10 colorectal,11 or bladder12 cancer, except for two studies.13,14 The breadth of methods and models used in each study varied from using regular expressions5,10,12,14 to more complex machine learning techniques.6,7,9,13
Although the studies mentioned had relatively good performances, the disease-specific nature of the NLP techniques limits the algorithms' generalizability to other disease areas. We aimed to develop a technique to support our research enterprise and provide assistance to researchers interested in various fields and disease areas. In this study, we developed and evaluated a rule-based NLP method for surgical pathology reports to extract TNM staging scores and diagnosis codes regardless of the diagnosis.
METHODS
Setting
Weill Cornell Medicine (WCM), with more than 20 outpatient sites across New York City, is an academic medical center with approximately 1,000 attending physicians and 250,000 patient visits annually. WCM physicians hold admitting privileges at NewYork-Presbyterian Hospital. Since 2000, WCM physicians have used the EpicCare Ambulatory EHR system to document clinical care in the outpatient setting. This study was approved by the WCM Institutional Review Board.
Data Collection
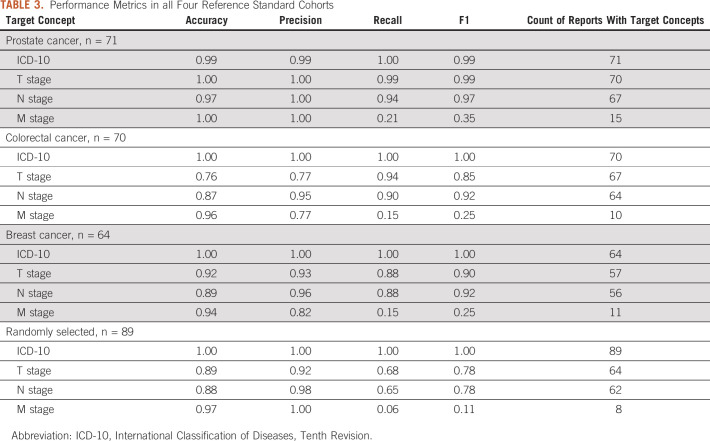
We identified a total of 555,681 surgical pathology reports of 329,076 patients from December 1, 2017, to February 12, 2020. These reports represent a broad array of specimen types, including both malignant and nonmalignant samples. The reports varied in the level of details and structure depending on the pathologic, anatomic, topological, and morphological characteristics of the sample submitted. Figure 1 provides an example of a surgical pathology report containing TNM staging and ICD-10 diagnosis codes. As shown in Figure 1, pathology reports at our institution often begin with hospital and laboratory information, followed by clinical assessment and diagnosis. The gross description and histological type of the tumor appear toward the end of each report.
FIG 1.
Mock-up pathology report that contains sections with pathologic staging and ICD-10 code information. h/o, history of ICD-10, International Classification of Diseases, Tenth Revision; PSA, prostate-specific antigen.
Leo NLP System
The NLP method was implemented on an open-source framework called Leo previously developed by the Department of Veterans Affairs.15 The Leo framework comprises services and libraries to facilitate rapid creation and deployment of Apache Unstructured Information Management Architecture—Asynchronous Scale-out annotators.16,17 Leo includes a client component for data input and output, a core component that facilitates local or external NLP annotations, and a service component that provides functionalities to build custom annotators and launch Unstructured Information Management Architecture—Asynchronous Scale-out services.
NLP Method Development
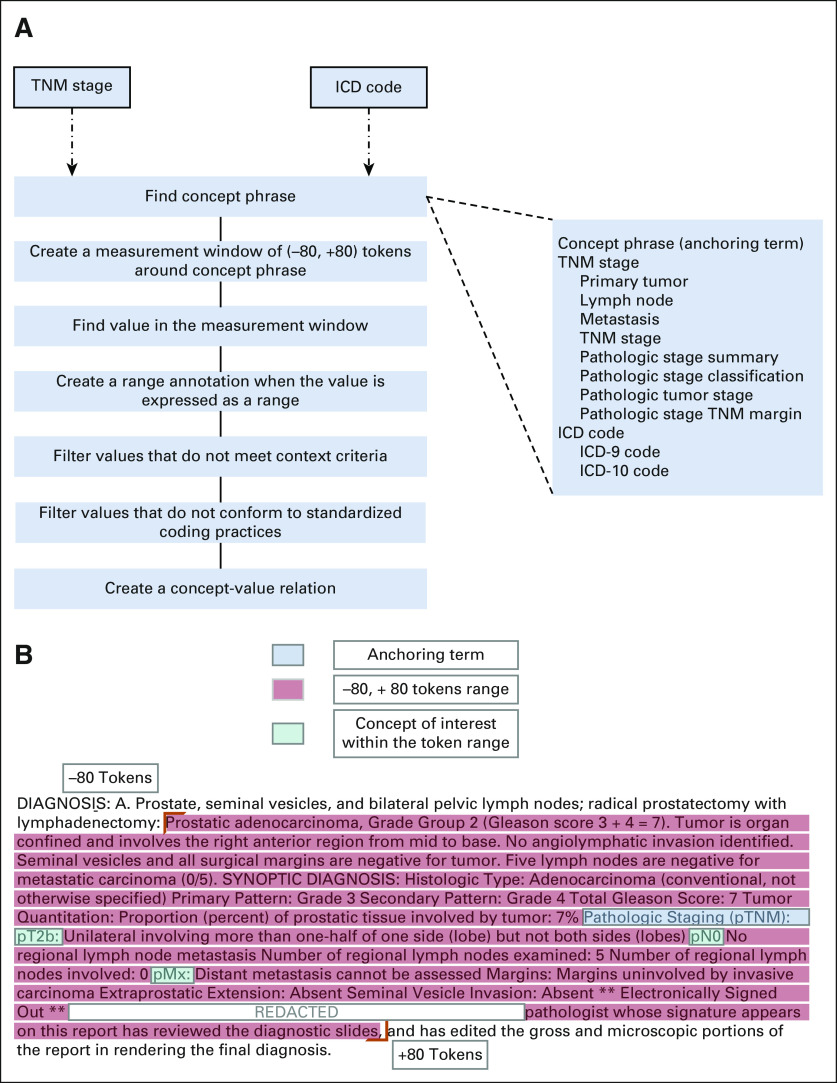
We previously used Leo NLP system15-17 to extract structured data elements from clinical free text, including Patient Health Questionnaire-9 scores18 and race and ethnicity.19 As shown in Figure 2A, we implemented a rule-based approach for data extraction following an iterative process focused on concept definition, context analysis, rule definition, system application, and manual review. For TNM staging and International Classification of Diseases codes, we used a similar approach to develop rules, working through the steps defined in Figure 2B. For each of the two concepts of interest, the first step was to define an anchoring term. For example, we defined patterns such as primary tumor or lymph nodes as potential anchoring terms for TNM staging data elements in the reports. Next, we used iterative context analysis to determine whether concepts were used in the appropriate context, depending on the concept of interest. We also defined, for each concept of interest, a window size of 80 tokens around the anchoring term, beyond which we would reject mentions of the concept of interest as irrelevant or referring to another concept. Of note, the NLP method was developed against all surgical pathology reports regardless of the diagnosis for all types of malignancies.
FIG 2.
(A) NLP logic implemented for extracting TNM staging and diagnosis codes from surgical pathology reports. (B) NLP logic implementation on a pathology report to extract TNM staging from a surgical pathology report. NLP, natural language processing; pTNM, Pathological Tumor-Node-Metastasis.
All concepts of interest were identified on the basis of predefined patterns. For example, we defined ICD-10 Clinical Modification concepts as a three- to seven-character code, starting with an alphabetic character and followed by one numeric character and up to five alphanumeric characters along with an optional decimal after the third character, if the code is longer than three digits. Similarly, each of the TNM staging patterns was developed by allowing only particular numbers or letters as suffixes and/or prefixes for each staging pattern. For instance, the allowed prefixes for the T component were p, rp, or yp, whereas the allowed suffix options were a, b, or c. Informal review of the corpus indicated that in instances with multiple mentions of the concepts of interest, the final mention of the concept contained the most recent or pertinent mention. Accordingly, we defined rules in such a way as to extract the final mention of each concept of interest, ignoring earlier mentions where present.
Reference Standard Creation
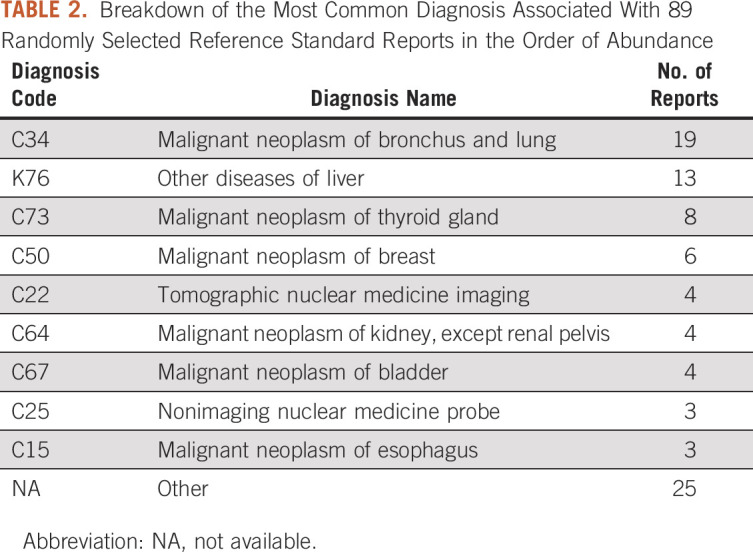
To evaluate the effectiveness of the NLP method, we identified a reference standard, which consisted of 294 surgical pathology reports generated during the period from December 1, 2017, to February 12, 2020. Because the NLP pipeline was developed regardless of diagnosis and against all subtypes, we decided to evaluate the pipeline against a randomly selected reference standard in addition to three different disease subtypes. Of the 294 reports in the reference standard, 71 were associated with prostate cancer, 70 were associated with colorectal cancer, 65 were associated with breast cancer, and 89 were associated with a random array of malignancies (Table 1). Each disease subtype in the reference standard was annotated by a pair of reviewers, with disagreements adjudicated by a third reviewer. We only included samples associated with a diagnosis of cancer in the reference standard, and we divided the reference standard into four subsets for patients with particular types of cancer. A total of 297 ICD-10, 399 T stage, 326 N stage, and 51 M stage target concepts were present across all 294 reports. All 294 reports in the reference standard contained at least one ICD-10 code, 258 of those had at least one mention of a T staging, 249 contained at least one N staging, and only 44 included at least one mention M staging classification. Reviewers were instructed to indicate the last mention of each target concept as the most pertinent whenever there were more than one available. Table 2 demonstrates the wide range of associated billing diagnoses in the randomly selected cohort. We then evaluated the interrater reliability for each subset of the reference standard by calculating Cohen's κ across all concepts of interest. Ideally, 10% of the overall corpus should be allocated to the reference standard creation; however, this may not be feasible in this study given the large corpus size.
TABLE 1.
Reference Standard Details and Assignments
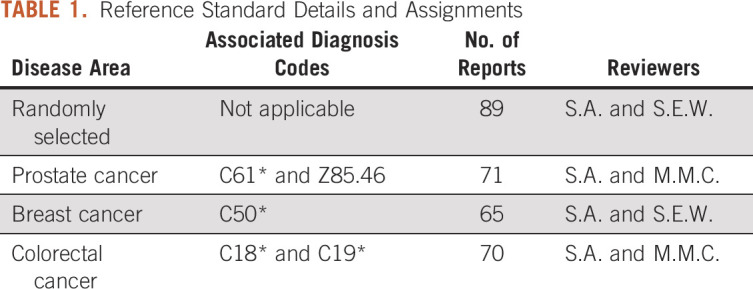
TABLE 2.
Breakdown of the Most Common Diagnosis Associated With 89 Randomly Selected Reference Standard Reports in the Order of Abundance
Evaluation of NLP Performance
For each of the 294 reports in the reference standard, we compared the results of manual annotation with the results of NLP output. For each concept of interest in reports, we classified performance into one of the following categories:
A true positive was defined as an instance when the output of the NLP method matched the reference standard.
A true negative was defined as an instance when both the NLP method and reference standard did not contain the concept of interest.
A false positive was defined either as an instance when the NLP method's outcome did not match the reference standard, or an instance when the NLP method extracted a value, but the reference standard did not contain any value.
A false negative was defined as an instance when the NLP method failed to extract any concept of interest when one existed in the reference standard.
For each of the prostate, colon, breast, and random specimen groups described above, we created a confusion matrix and calculated precision, recall, and F1-score.
RESULTS
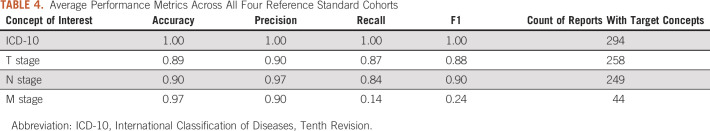
As described in Table 3, the NLP method extracted ICD-10 code concepts with the highest level of accuracy, with F1 scores ≥ 0.99 across all disease subtypes in the reference standard. Of note, all pathology reports in the reference standard contained at least one ICD-10 code concept. All manually reviewed notes were created during the period from the beginning of December 2017 to February 21, 2020, in which all health care organizations were mandated to use ICD-10 Clinical Modification. Owing to the timeframe we selected for our evaluation, none of the manually reviewed notes contained an ICD-9 code. Accordingly, we excluded the ICD-9 concepts from our evaluation. We observed the highest average Cohen's κ, 0.93, among the pairs of reviewers when annotating ICD-10 codes across four reference standards. The Cohen's κ was fairly consistent across other concepts of interest with 0.75, 0.87, and 0.73 for T, N, and M stages, respectively.
TABLE 3.
Performance Metrics in all Four Reference Standard Cohorts
The overall performance of the NLP method was the worst when evaluating M stage concepts, with the lowest F1 score of 0.11 in the randomly selected reference standard. M stage concepts also had the lowest data element availability across all reference standards with 15, 10, 11, and 8 notes with M stage present in each of the disease-specific subgroups. Finally, the NLP method's performance was comparable in extracting T and N stage concepts, with an average F1 score of 0.88 and 0.90 across all standard references for T and N stage concepts, respectively.
DISCUSSION
In this study, we developed and evaluated an NLP method to obtain staging and diagnosis information from surgical pathology reports regardless of the disease area. To the best of our knowledge, previous studies demonstrated the use of NLP to extract data elements specific to particular cancer subtypes. In contrast, our approach extracts data elements across multiple oncologic subtypes. The current approach may be valuable to other institutions seeking to unlock data in unstructured surgical pathology reports.
The NLP method performed particularly well in extracting ICD-10 code concepts, demonstrating an almost perfect F1 score (0.99 and above) across all disease subtypes in the reference standard. However, we observed substantially poorer performance in extracting M stage concepts across all reports. Despite high precision, this method displayed poor sensitivity. As shown in Table 4, only 44 reports (of 294) contained an M stage target concept. Since a pathology report contains only data derived from observations about a specific specimen, its dictator cannot make an accurate assessment of the M stage without access to full-body imaging data, such as a positron emission tomography scan. This is likely the reason for the relative paucity of M stage data in our gold standard reference set. Considering the low number of reports containing M staging in the reference standard, the false-negative cases were more heavily punished when evaluating M stage concepts.
TABLE 4.
Average Performance Metrics Across All Four Reference Standard Cohorts
Further manual review of false-positive samples from all reference standards indicated a systematic error for instances with several mentions of each concept, more specifically TNM stages. Each surgical pathology note may contain more than one TNM staging value for multiple biopsy samples. However, in our institution, the majority of surgical pathology reports only report on one specimen. In rare cases where there are multiple mentions of the target concept in one report, the NLP method is designed to extract the final mention of each concept of interest. However, when creating the reference standards, the annotators considered the last mentions of staging classifications as the most pertinent. Lexical variability of the repeating concepts may also contribute to an increased number of false positives. For instance, if the T stage concept is once indicated as pT4a in the pathologic stage classification section of the report, and once again indicated in the body of the report as T4a, the NLP method favors the last mention. This is a major limitation of this rule-based NLP system, more prominently in settings where one report contains information pertaining to more than one specimen.
One of the main limitations of NLP techniques developed and evaluated in only one institution is lack of external validation. Owing to the variability in how pathology reports are written at each institution, NLP pipelines may not perform similarly across all. This is a major limitation of this study, as we have not evaluated the performance of our NLP pipeline against pathology reports written at other institutions.
In contrast to our rule-based NLP method, other studies have used a number of machine learning (ML) techniques to extract structured information from pathology reports for particular cancer disease areas.6,7,9 Notably, the ML models reported in the literature have not substantially outperformed rule-based approaches. For example, Leyh-Bannurah et al6 developed a convolutional neural network to extract prostate-specific elements but needed to supplement the convolutional neural network with a set of regular expression rules to improve performance.
The majority of NLP techniques applied to surgical pathology reports to date have focused on a single disease area,5-12 limiting the generalizability of the methods to other disease areas. However, in a 2018 study, AAlAbdulsalam et al developed a rule-based system to extract and classify TNM staging information from multiple disease areas using data from a state cancer registry. Although AAlAbdulsalam et al14 evaluated performance using pathology reports for colon, lung, and prostate cancer, the corpus included only reports containing target concepts (eg, TNM staging). In contrast, our study included reports with and without target concepts, which enabled us to better evaluate the NLP method's precision, or how many selected items were relevant. In addition to the studies discussed, Savova et al13 presented DeepPhe software that enables automated extraction of phenotype information from EHRs of cancer patients regardless of the disease area. To the best of our knowledge, the authors have not evaluated the performance of this software, and the article cited here only details the NLP system and its challenges.
The main task of the present NLP method, identifying TNM stages and ICD-10 codes, may be relatively simple, with low lexical variability and similar note structure across all pathology reports. Despite the simplicity of this task, the importance of ready availability of such quantitative parameters remains important for research, quality improvement, and clinical care cannot be exaggerated—without accurate data on initial tumor staging, it is difficult to conduct observational research on treatment efficacy and safety. Although several diagnosis-specific data elements exist in surgical pathology reports, we sought to extract the most common shared elements regardless of the associated diagnosis to enhance the generalizability of the NLP method to support the research enterprise in our institution. Using a similar approach, we previously imputed the race and ethnicity of underrepresented patient populations to fill gaps in structured EHR data. Notably, those patients were older, more likely to be male, less likely to have commercial insurance, and more likely to have higher comorbidity, demonstrating the value of rule-based NLP methods to support the research enterprise.19 We plan to scale the current NLP pipeline to also capture disease-specific concepts of interest, such as Gleason scores indicated in surgical pathology reports.
Oncology research requires accurate, comprehensive, and high-quality data. The extraction of TNM staging values and ICD-10 codes, which clinical researchers have identified as of immediate use in patient stratification and predictive modeling, directly supports cancer investigations by substantially reducing the need for manual review of patients' charts. Although it was not possible to demonstrate generalizability of our NLP pipeline to other institutions, other institutions may find value in adopting a similar NLP approach—and reusing code20—to support the oncology research enterprise with elements extracted from surgical pathology reports.
Evan T. Sholle
Stock and Other Ownership Interests: Moderna Therapeutics
Jonathan E. Shoag
Research Funding: Bristol Myers Squibb Foundation
Jim C. Hu
Speakers' Bureau: Genomic Health, Intuitive Surgical
Travel, Accommodations, Expenses: Intuitive Surgical
No other potential conflicts of interest were reported.
SUPPORT
Supported by NewYork-Presbyterian Hospital (NYPH) and Weill Cornell Medical College (WCMC), including the Clinical and Translational Science Center (CTSC) (UL1 TR000457) and Joint Clinical Trials Office (JCTO).
AUTHOR CONTRIBUTIONS
Conception and design: Sajjad Abedian, Evan T. Sholle, Prakash M. Adekkanattu, Jonathan E. Shoag, Jim C. Hu, Thomas R. Campion
Financial support: Thomas R. Campion
Administrative support: Thomas R. Campion
Provision of study materials or patients: Thomas R. Campion
Collection and assembly of data: Sajjad Abedian, Evan T. Sholle, Prakash M. Adekkanattu, Marika M. Cusick, Jonathan E. Shoag, Thomas R. Campion
Data analysis and interpretation: Sajjad Abedian, Evan T. Sholle, Prakash M. Adekkanattu, Marika M. Cusick, Stephanie E. Weiner, Thomas R. Campion
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/cci/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Evan T. Sholle
Stock and Other Ownership Interests: Moderna Therapeutics
Jonathan E. Shoag
Research Funding: Bristol Myers Squibb Foundation
Jim C. Hu
Speakers' Bureau: Genomic Health, Intuitive Surgical
Travel, Accommodations, Expenses: Intuitive Surgical
No other potential conflicts of interest were reported.
REFERENCES
- 1.Griffon N, Charlet J, Darmoni SJ.Managing free text for secondary use of health data Yearb Med Inform 9167–1692014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yim W-W, Yetisgen M, Harris WP, et al. Natural language processing in oncology: A review JAMA Oncol 2797–8042016 [DOI] [PubMed] [Google Scholar]
- 3.Cancer Protocol Templates|College of American Pathologists. https://www.cap.org/protocols-and-guidelines/cancer-reporting-tools/cancer-protocol-templates [Google Scholar]
- 4.Renshaw AA, Mena-Allauca M, Gould EW, et al. Synoptic reporting: Evidence-based review and future directions JCO Clin Cancer Inform 21–92018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim BJ, Merchant M, Zheng C, et al. A natural language processing program effectively extracts key pathologic findings from radical prostatectomy reports J Endourol 281474–14782014 [DOI] [PubMed] [Google Scholar]
- 6.Leyh-Bannurah S-R, Tian Z, Karakiewicz PI, et al. Deep learning for natural language processing in urology: State-of-the-art automated extraction of detailed pathologic prostate cancer data from narratively written electronic health records JCO Clin Cancer Inform 21–92018 [DOI] [PubMed] [Google Scholar]
- 7.Yala A, Barzilay R, Salama L, et al. Using machine learning to parse breast pathology reports Breast Cancer Res Treat 161203–2112017 [DOI] [PubMed] [Google Scholar]
- 8.Nguyen AN, Lawley MJ, Hansen DP, et al. Symbolic rule-based classification of lung cancer stages from free-text pathology reports J Am Med Inform Assoc 17440–4452010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCowan I, Moore D, Fry M-J.Classification of cancer stage from free-text histology reports Conf Proc IEEE Eng Med Biol Soc 20065153–51562006 [DOI] [PubMed] [Google Scholar]
- 10.Warner JL, Levy MA, Neuss MN, et al. Recap: Feasibility and accuracy of extracting cancer stage information from narrative electronic health record data J Oncol Pract 12157–158; e1692016 [DOI] [PubMed] [Google Scholar]
- 11.Martinez D, Cavedon L, Pitson G. Stability of text mining techniques for identifying cancer staging. Louhi 2013, Sydney, Australia, February 11, 2013. [Google Scholar]
- 12.Glaser AP, Jordan BJ, Cohen J, et al. Automated extraction of grade, stage, and quality information from transurethral resection of bladder tumor pathology reports using natural language processing JCO Clin Cancer Inform 21–82018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Savova GK, Tseytlin E, Finan S, et al. DeepPhe: A natural language processing system for extracting cancer phenotypes from clinical records Cancer Res 77e115–e1182017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.AAlAbdulsalam AK, Garvin JH, Redd A, et al. Automated extraction and classification of cancer stage mentions from unstructured text fields in a central cancer registry AMIA Jt Summits Transl Sci Proc 201716–252018 [PMC free article] [PubMed] [Google Scholar]
- 15.Cornia R, Patterson OV, Ginter T, Duvall SL. Rapid NLP development with leo. AMIA Annu Symp Proc; 2014. p. 1356. [Google Scholar]
- 16.Ferrucci D, Lally A.UIMA: An architectural approach to unstructured information processing in the corporate research environment Nat Lang Eng 10327–3482004 [Google Scholar]
- 17.Soundrarajan BR, Ginter T, DuVall SL.An interface for rapid natural language processing development in UIMA. Proceedings of the 49th Annual Meeting of the Association for Computational Linguistics: Human Language Technologies: Systems Demonstrations, Portland, OR, June 19-24, 2011.
- 18.Adekkanattu P, Sholle ET, DeFerio J, et al. Ascertaining depression severity by extracting patient health questionnaire-9 (PHQ-9) scores from clinical notes AMIA Annu Symp Proc 2018147–1562018 [PMC free article] [PubMed] [Google Scholar]
- 19.Sholle ET, Pinheiro LC, Adekkanattu P, et al. Underserved populations with missing race ethnicity data differ significantly from those with structured race/ethnicity documentation J Am Med Inform Assoc 26722–7292019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weill Cornell Medicine Research Informatics. PathExtractor. https://github.com/wcmc-research-informatics/PathExtractor