Abstract
PURPOSE
Cloud computing has led to dramatic growth in the volume, variety, and velocity of cancer data. However, cloud platforms and services present new challenges for cancer research, particularly in understanding the practical tradeoffs between cloud performance, cost, and complexity. The goal of this study was to describe the practical challenges when using a cloud-based service to improve the cancer clinical trial matching process.
METHODS
We collected information for all interventional cancer clinical trials from ClinicalTrials.gov and used the Google Cloud Healthcare Natural Language Application Programming Interface (API) to analyze clinical trial Title and Eligibility Criteria text. An informatics pipeline leveraging interoperability standards summarized the distribution of cancer clinical trials, genes, laboratory tests, and medications extracted from cloud-based entity analysis.
RESULTS
There were a total of 38,851 cancer-related clinical trials found in this study, with the distribution of cancer categories extracted from Title text significantly different than in ClinicalTrials.gov (P < .001). Cloud-based entity analysis of clinical trial criteria identified a total of 949 genes, 1,782 laboratory tests, 2,086 medications, and 4,902 National Cancer Institute Thesaurus terms, with estimated detection accuracies ranging from 12.8% to 89.9%. A total of 77,702 API calls processed an estimated 167,179 text records, which took a total of 1,979 processing-minutes (33.0 processing-hours), or approximately 1.5 seconds per API call.
CONCLUSION
Current general-purpose cloud health care tools—like the Google service in this study—should not be used for automated clinical trial matching unless they can perform effective extraction and classification of the clinical, genetic, and medication concepts central to precision oncology research. A strong understanding of the practical aspects of cloud computing will help researchers effectively navigate the vast data ecosystems in cancer research.
INTRODUCTION
There has been dramatic growth in the volume, variety, and velocity of cancer data in the cloud. However, unlocking the full potential of cloud computing will require improving interoperability between highly diverse and heterogeneous cancer data sets. Artificial intelligence (AI), machine learning, and natural language processing (NLP) are just some of the technologies that could potentially meet these needs in oncology.1,2
CONTEXT
Key Objective
What are the practical issues that must be considered when implementing and applying cloud computing services to important cancer research use cases?
Knowledge Generated
Cloud health care services are capable of providing some of the core standardized data building blocks required for automated clinical trial matching. Investigators considering cloud computing should follow a process of categorizing cloud costs, assessing technical resources, and making project decisions that balance scientific, technical, and cloud expertise with their particular research needs.
Relevance
A strong understanding of the practical aspects of cloud computing can help researchers effectively navigate the vast data ecosystems in cancer research.
Cloud vendors have recently developed AI and NLP services to standardize and harmonize large volumes of unstructured health care text, including Amazon Web Services Comprehend Medical, Google Cloud Healthcare Natural Language (HNL), and Microsoft Azure Text Analytics for Health. In general, these cloud-based services receive large volumes of text through an application programming interface (API) and perform medical entity analyses, which return machine-readable collections of knowledge categories, relationships, and codes from relevant medical vocabularies.
However, cloud platforms and services also present new challenges for cancer research, particularly in understanding the practical tradeoffs between cloud performance, cost, and complexity. The Google Cloud HNL API, for example, could be used for cancer research since it analyzes text using relevant standards and terminologies, including the National Cancer Institute (NCI) Thesaurus (NCIt).3 To our knowledge, however, very few studies have investigated the practical aspects of using cloud vendor-specific services for cancer-related research.4-6
The goal of this study was to describe the practical challenges of leveraging a cloud-based service to improve the cancer clinical trial matching process. Specifically, we investigated the technical, performance, and cost issues encountered when using the Google Cloud HNL API to extract structured cancer types and eligibility criteria from cancer studies in ClinicalTrials.gov.
METHODS
Identifying Cancer Studies From ClinicalTrials.gov
Reporting was guided by the Standards for Reporting Implementation Studies guidelines and checklist.7 ClinicalTrials.gov is a public database of registered clinical trials created to increase transparency through improved availability and accessibility of information for clinical studies.8-10 A recent version of the ClinicalTrials.gov database was downloaded in pipe-delimited format from the Clinical Trials Transformation Initiative.11 Clinical trials were included in the study if (1) the Conditions field contained one or more cancer terms commonly used by the ClinicalTrials.gov search engine (cancer, neoplasm, tumor, malignancy, oncology, oncologic, neoplasia, neoplastic syndrome, and neoplastic disease) and (2) had the interventional study type. Similar to prior studies, we focused on clinical trial Title and Eligibility Criteria text from the official_title and criteria fields, respectively.12,13
Preparing for Cloud-Based Medical Entity Analysis
The HNL API currently processes data requests on a per text record basis, with a single text record defined as having up to 1,000 unicode characters.14 Current pricing for entity analysis is $0.10 US dollars (USD) per text record, where the number of text records for a single request is calculated as the total characters in the current request divided by 1,000, rounded up to the nearest whole number, and limited to a maximum of 10,000 unicode characters.14 We estimated the number of words and text records for each clinical trial, and truncated the text of any submission request exceeding the character limit to meet cloud vendor–specific API requirements. Although the HNL API was available at no cost to all Google Cloud users during the study period, we still performed text record calculations and API cost estimates using currently documented rates.
API requests were collected into two main data sets, one for processing clinical trial Title text and one for Criteria text. Best practices for cloud pipeline development usually involve pilot testing with smaller files before working with larger data sets.15 As a result, the Title data set was split into four files: two smaller files with 2,500 clinical trials each (for preliminary runs), followed by larger files with 10,000 and 23,851 trials, respectively. The relatively larger Criteria data set was split into six files: two smaller files with 2,500 clinical trials each, three files with 10,000 trials each, and one with 3,851 trials. The Title data set was processed first, running its two smallest files in sequence and (if successful) running the remaining two files in parallel. The two smallest files of the Criteria data set were then run concurrently, followed by running the last four files in parallel as well. Script processing times for both data sets were tracked.
Cloud Health Care Service Input and Output
Configuring the cloud API involved creating a cloud project, enabling the Cloud Health Care and HNL APIs, and setting up appropriate application credentials. We used the default settings specified in the technical documentation to create API request calls, including the us-central1 region where the API is currently available.16 A simple shell script was written that read in the text input for each clinical trial, created and submitted an entity analysis request to the cloud API, and stored the returned JavaScript Object Notation (JSON)-formatted response in a text file.
Once the HNL API response was returned for each clinical trial, the JSON output was processed to extract relevant codes for analysis. This included NCIt codes under the PROBLEM medical knowledge category, Logical Observation Identifiers Names and Codes (LOINC) under the LABORATORY_DATA medical knowledge category, Human Genome Organization Gene Nomenclature Committee (HGNC) codes under the LABORATORY_DATA category, and RxNorm codes under the MEDICINE medical knowledge category.
Cancer Categorization Using NCIt
The NCIt provides a useful organized collection of cancer-related terms and relationships.1 To understand how well the cloud-based service extracted cancer site from clinical trial Title text, we created a mapping between NCIt codes and broad cancer categories guided by the cancer site classifications used in the NCI SEER Cancer Registration and Surveillance Modules: (1) bladder, (2) brain and central nervous system, (3) breast, (4) cervical and uterine, (5) colorectal, (6) head and neck, (7) kidney and ureter, (8) leukemia and lymphoma, (9) lung, (10) ovarian, fallopian, and peritoneal, (11) pancreatic and biliary, (12) prostate, (13) skin cancer and melanoma, (14) testicular, and (15) upper gastrointestinal tract.17,18
Using the current version of the NCIt Neoplasm Core Hierarchy Plus file from the NCI Enterprise Vocabulary Services, relevant NCIt terms and codes from the Neoplasm by Site section with a malignant neoplastic status flag were mapped to each category listed above.19 This allowed cloud service output to be compared with clinical trial counts per cancer category obtained by directly searching ClinicalTrials.gov using the same broad NCIt terms.
Integrating Criteria Results on Genes, Laboratory Tests, and Medications
Cloud-based entity analysis of the Criteria text for cancer studies can provide valuable insight into the diverse characteristics being used to include or exclude patients from clinical trials. We therefore leveraged three established databases of standardized terms and codes to describe the distribution of genes (HGNC), medical laboratory tests (LOINC), and medications (RxNorm), which were extracted from clinical trial Eligibility Criteria.20-22
For genes, we downloaded the most recent version of the HGNC database in tab-separated values file format, and found the corresponding approved symbol and approved name by joining cloud output against HGNC ID.20 For laboratory tests, we downloaded recent versions of the Core LOINC Table (version 2.69) and LOINC Part File (version 5.1 beta), and matched cloud output codes to LOINC_NUM and LONG_COMMON_NAME or to PartNumber and PartName with COMPONENT PartType, respectively.21 For medications, we downloaded the RxNorm Full Monthly Release and used the pipe-delimited rxnconso.rrf file to map RxCUI codes returned from cloud-based entity analysis to the associated medication ingredients.22
Evaluation of Eligibility Criteria and Title Extraction
We randomly selected 50 clinical trials and reported the accuracy of Criteria gene, laboratory, and medication detection as the number of concept terms correctly identified by the cloud service compared with the total number of extracted concepts. Accuracy was similarly reported for the extraction and categorization of Titles for 50 randomly selected clinical trials.
Summary Statistics and Analytics
Summary statistics were collected for clinical trial characteristics as means with standard deviations and medians with interquartile range (IQR). Frequencies and percentages were calculated for categorical data involving clinical trials, genes, laboratory tests, and medications.
Differences in the distribution of clinical trial cancer type categorization between cloud-based entity analysis and direct search of ClinicalTrials.gov were evaluated using the chi-square test. A P value < .05 was considered significant for the analysis, which was performed using RStudio.
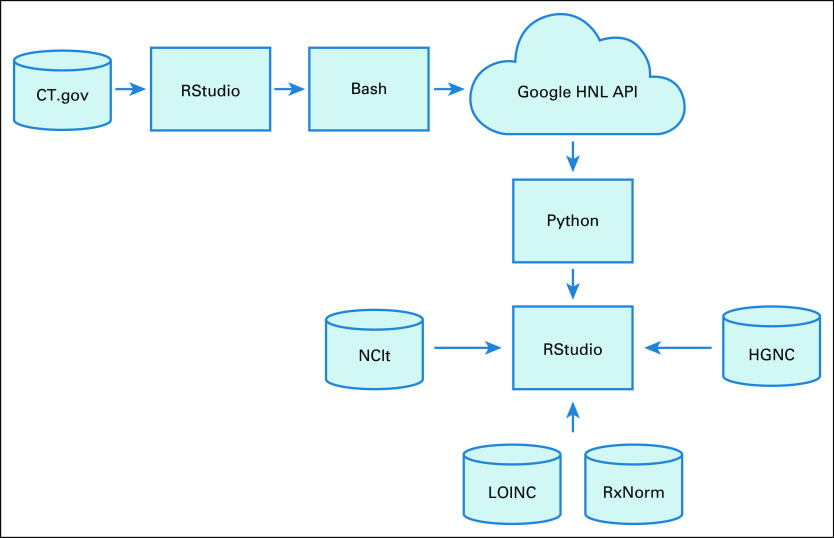
Several technologies were used to perform the steps in the informatics pipeline described above (Fig 1). An R (version 4.0.3) script written in RStudio (version 1.3.1093) performed preprocessing of the ClinicalTrials.gov database; postprocessing integration of the NCIt, HGNC, LOINC, and RxNorm databases; and all statistical analyses. A bash shell script submitted all requests to the Google Cloud HNL API, and a python script (version 3.8.5) in a Jupyter Notebook (version 6.1.4) processed the returned cloud JSON output.
FIG 1.
Informatics processing pipeline. CT.gov, ClinicalTrials.gov; Google HNL API, Google Healthcare Natural Language Application Programming Interface; HGNC, Human Genome Organization Gene Nomenclature Committee; LOINC, Logical Observation Identifiers Names and Codes; NCIt, National Cancer Institute Thesaurus.
RESULTS
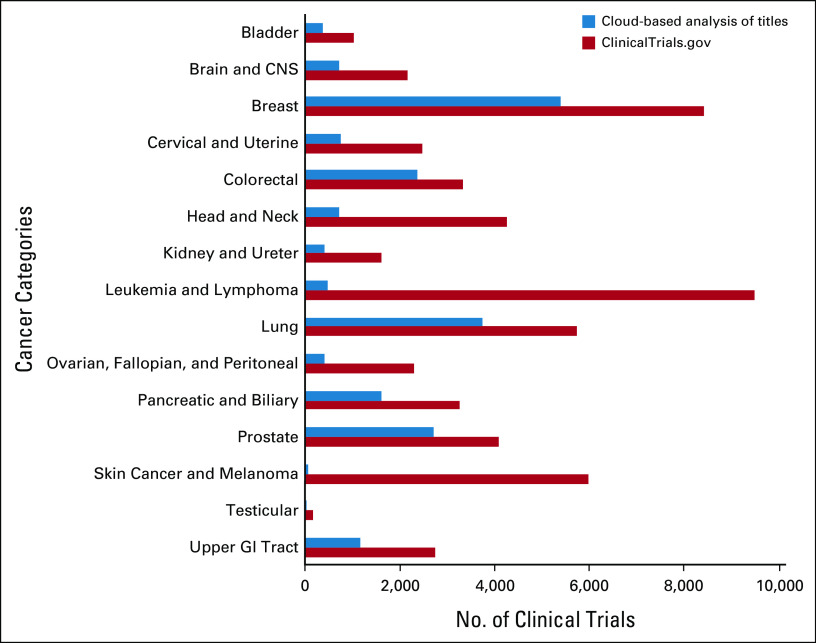
There were a total of 38,851 cancer-related clinical trials, with clinical trial Title text having a mean 21.9 ± 8.7 words (median 21, IQR 16-27 words) and 151.2 ± 59.7 characters (median 144, IQR 108-187 characters). Cancer clinical trial Criteria text had a mean of 352.3 ± 333.4 words (median 247, IQR 116-472 words) and 2,889.8 ± 2,647.7 characters (median 2,069, IQR 1,003-3,886 characters). There were 1,083 (2.8%) trials with Criteria text that exceeded 10,000 characters and were truncated during preprocessing to meet API requirements for the cloud service, after which Criteria text submitted for cloud-based analysis had a mean of 344.3 ± 304.1 words (median 247, IQR 116-472 words) and 2,824.0 ± 2,405.9 characters (median 2,069, IQR 1,003-3,886 characters). There were 479 (1.2%) cancer clinical trials without an entry in the Title field, and 6 (0.02%) without an entry in the Criteria field. The distribution of clinical trials by cancer category from cloud-based entity analysis of Title text (Fig 2) was significantly different than the general distribution in ClinicalTrials.gov (P < .001).
FIG 2.
Number of cancer clinical trials categorized by cloud-based entity analysis of titles compared with ClinicalTrials.gov.
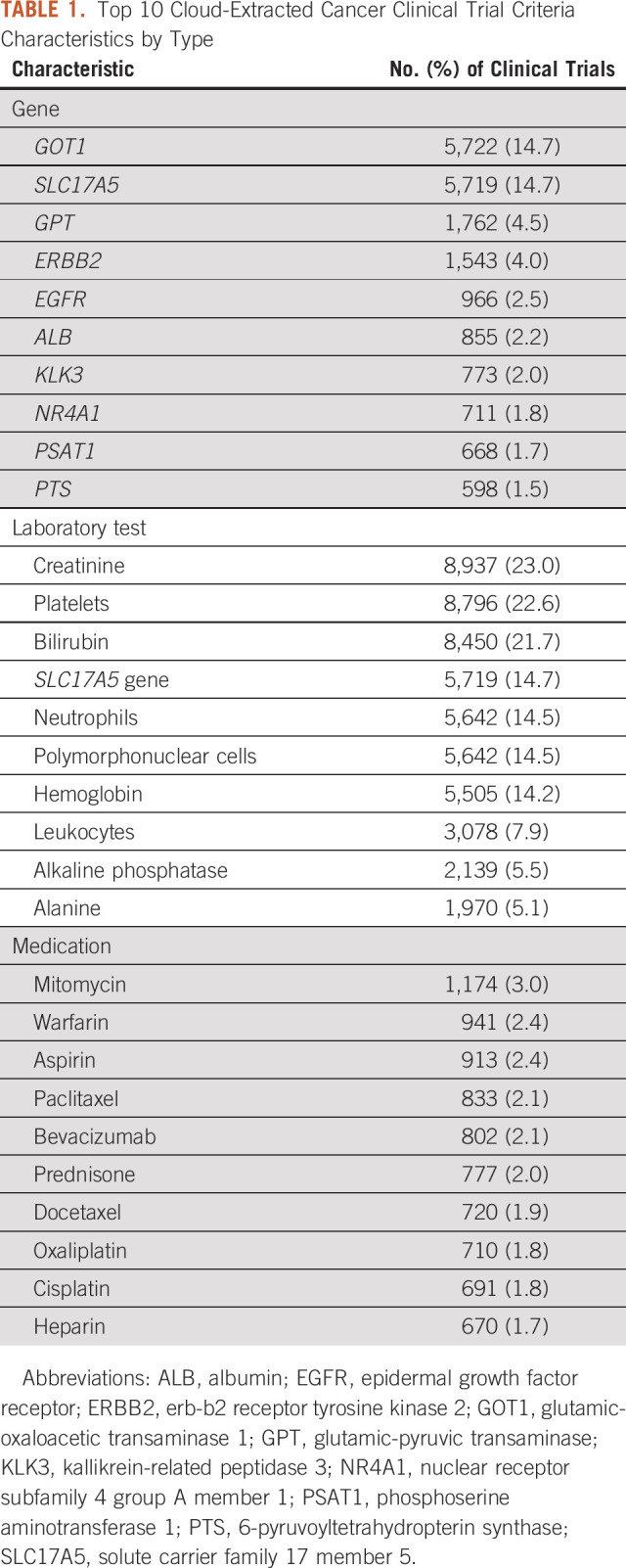
Cloud-based entity analysis of clinical trial Criteria extracted a total of 949 HGNC-coded genes, 1,782 LOINC-coded laboratory tests, 2,086 RxNorm-coded medications, and 4,902 NCIt terms. Details regarding the most frequently occurring genes, laboratory tests, and medications from cancer-related clinical trial Eligibility Criteria are summarized in Table 1.
TABLE 1.
Top 10 Cloud-Extracted Cancer Clinical Trial Criteria Characteristics by Type
Analysis of Eligibility Criteria for 50 randomly selected clinical trials identified 492 coded concepts, with an accuracy of 12.8% (14/109) for gene extraction, 64.5% (196/304) for laboratory test extraction, and 89.9% (71/79) for medication extraction. Title extraction of 50 randomly selected clinical trials demonstrated an accuracy of 90% (45/50), with commonly missed terms including metastatic medullary thyroid, advanced or metastatic solid tumors, and refractory cancer.
There were a total of 77,702 API calls made to the HNL API, which took a total of 1,979 processing-minutes (33.0 processing-hours), or approximately 1.5 seconds per API call. More specifically, analysis of clinical trial Titles took 1,006 processing-minutes (16.8 processing-hours) or roughly 1.6 seconds per API call, whereas clinical trial Criteria took 973 processing-minutes (16.2 processing-hours) or 1.5 seconds per API call. Partial parallelization of scripts for Title data resulted in 858 minutes (14.3 hours) of actual script execution time, whereas the increased parallelization of Criteria scripts yielded 313 minutes (5.2 hours) of script execution time.
For cloud-based entity analysis of clinical trial Titles, there were estimated to be 38,851 text records created for a price of $3,885.10 USD and 128,328 records for clinical trial Criteria for a price of $12,832.80 USD, giving a total of 167,179 records processed for cloud-based medical entity analysis at an estimated price of $16,717.90 USD.
DISCUSSION
Technology-driven approaches to find cancer patient cohorts for clinical trials or recommend clinical studies to eligible patients with cancer are expected to streamline cancer clinical trial recruitment.5,9,10,12,13 Even from clinical trial Titles alone, the cloud service used in this study extracted meaningful information for some cancers (eg, breast, colorectal, and lung), whereas others (eg, hematopoietic and skin cancers) could still be refined further. Manual review of a random sample of Titles suggest cloud service extraction correctly handled studies with single or specific cancer types, but struggled when studies became more complex, described multiple cancers, or included broadly or vaguely defined cancer topographies. Integrating context-specific data from ClinicalTrials.gov (eg, Conditions field) or the NCI Clinical Trials Reporting Program would likely improve cloud entity analyses and facilitate cancer patient cohort matching from structured sources (eg, electronic health records).
The poor detection accuracies for tests (64.5%) and genes (12.8%), along with challenges differentiating these concepts, suggest that the current technology is not ready for automated clinical trial matching. Both EGFR and ERBB2 mutations, for example, are known to affect clinical responsiveness to different therapies for lung or breast cancer, and thus expected to be inclusion or exclusion criteria in many interventional clinical trials.23,24 Other genes (eg, GOT1 and GPT) on the list were more surprising and may have been extracted for other reasons. Investigating entity analysis output showed that AST and ALT were the most frequent Criteria strings associated with these genes. AST and ALT are both common laboratory tests to evaluate a patient's liver function, but they are also synonyms for the approved names of the glutamic oxaloacetic transaminase (GOT1) and glutamic-pyruvic transaminase (GPT) genes, respectively. In these situations, reporting LOINC codes for organ function tests would be more appropriate than HGNC codes, unless those genes were listed in Eligibility Criteria or the published literature supported their relevance in cancer clinical trials.
Similarly, although many of the extracted laboratory tests assessed important aspects of patient health (eg, hematologic, kidney, and liver function), the laboratory test for the solute carrier family 17 member 5 (SLC17A5) gene was unexpected and often occurred when the AST string was found in Criteria text. In this context, the cloud technology could be interpreting the AST liver function test as the AST gene (which is a known alias for SLC17A5). This may explain the unexpected appearances of SLC17A5 in the list of extracted genes as well as laboratory tests. Furthermore, manual review of a random sample of Criteria suggests that the cloud service needs better study context to more accurately distinguish genes and laboratory tests. For example, estimated glomerular filtration rate to measure kidney function was often miscoded as the EGFR gene, whereas tests for hepatitis B were instead coded for blood type B. Accurately extracting genes and laboratory tests from unstructured text while minimizing false positives remains an ongoing challenge, but could lead to novel decision support tools that accelerate precision medicine and precision oncology.12
Cloud computing allows researchers to store, manage, and analyze large volumes of data that would be difficult or impossible to do locally, but requires clearly understanding important tradeoffs when using cloud vendor–based tools. In our study, for example, the Criteria text of more than 1,000 cancer clinical trials had to be reduced to meet character limits set by the cloud API. Other options were considered (eg, splitting Criteria text into logical blocks), but would have introduced additional complexity, uncertainty, and costs to the project. Currently, most cancer studies leveraging cloud tools will likely require significant preprocessing of source data to meet cloud technical requirements, as well as robust filtering and refinement of cloud output to meet research needs.
Assessing and optimizing cloud costs remains a complex challenge in cancer research. We thus identified four cloud cost categories that guided our decision making: cloud storage, cloud compute, cloud data transfer, and cloud services.25-30 In general, cloud storage involves the data being accessed for research, with costs directly related to (1) larger volumes of data stored or (2) shorter data retrieval times (latency).27,29,31 Cloud compute involves resources (eg, workflows, pipelines, platforms, and environments) to perform research analyses, with costs directly related to (1) virtual machine configuration, (2) compute usage, or (3) compute instance availability (eg, on-demand v pre-emptible or spot instances).25,27,32 Cloud data transfer can involve upload (ingress) or download (egress), as well as moving data between cloud resources located in different regions.26,27 Finally, the costs of vendor-specific cloud services can be as variable as the types of technologies involved (eg, manual or automated AI, machine learning, and NLP tools).4,14,26
If all our scripts were run from the cloud, performing API calls in parallel batches would have yielded lower costs in the cloud compute category since virtual machine usage would have been markedly shorter. Furthermore, if the HNL API had not been freely available to Google Cloud users during the study period, our estimates show that cloud services would have been the largest contributor of the four cost categories. Finally, while centralizing data, medical vocabularies, API calls, and analyses entirely in the cloud would have streamlined development considerably, we ultimately chose to use local storage and local compute resources with the cloud API service to minimize overall expenses. Investigators considering cloud computing will need to go through a similar process of categorizing cloud costs, assessing technical resources, and making project decisions that balance scientific, technical, and cloud expertise with their particular research needs.15
Although cloud-based API services contain useful building blocks for standardizing data, poor detection performance and other shortcomings limit its current value to the cancer research community. The following missing functionality should be added before the technology can be used for automated clinical trial matching: classifying coded concepts as inclusion or exclusion criteria, expanding cloud limits to handle clinical trial content, harmonizing genomic and laboratory results, and robust context annotations of extracted concepts for all cancer types, treatments, and relevant findings.1,5,10
This study had several limitations. First, because the cloud-based tool in this study was proprietary in nature, the traditional process of building, training, testing, and refining the technology was not possible.1 Second, the mapping of cloud entity analysis output to medical vocabularies may have been incomplete, since the API technical documentation did not describe which version of the medical vocabularies were used or how often they were updated; including this information would likely improve interoperability of the cloud service. Finally, complex cloud pricing models made it challenging to assess the cost effectiveness of different project pipelines. Additional studies are needed to clarify cloud computing costs across the diverse use cases in biomedical research.
In conclusion, current general-purpose cloud health care tools cannot be used for automated clinical trial matching without substantial informatics improvements to address the core aspects of cancer and precision oncology research. A strong understanding of the practical aspects of cloud computing will help researchers effectively navigate the vast data ecosystems in cancer research.
DISCLAIMER
This is a US Government work. There are no restrictions on its use.
SUPPORT
Supported by General Google Cloud Platform Cloud Credits via the NCI Cancer Research Data Commons Cloud Resources.
AUTHOR CONTRIBUTIONS
Conception and design: Jay G. Ronquillo
Collection and assembly of data: Jay G. Ronquillo
Data analysis and interpretation: All authors
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/cci/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
No potential conflicts of interest were reported.
REFERENCES
- 1.Yim WW, Yetisgen M, Harris WP, et al. Natural language processing in oncology review JAMA Oncol 2797–8042016 [DOI] [PubMed] [Google Scholar]
- 2.Kehl KL, Xu W, Lepisto E, et al. Natural language processing to ascertain cancer outcomes from medical oncologist notes JCO Clin Cancer Inf 4680–6902020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Google Cloud Healthcare natural language API. 2020. https://cloud.google.com/healthcare/docs/concepts/nlp
- 4. Zeng Y, Zhang J. A machine learning model for detecting invasive ductal carcinoma with Google Cloud AutoML Vision. Comput Biol Med. 2020;122:103861. doi: 10.1016/j.compbiomed.2020.103861. [DOI] [PubMed] [Google Scholar]
- 5.Beck JT, Rammage M, Jackson GP, et al. Artificial intelligence tool for optimizing eligibility screening for clinical trials in a large community cancer center JCO Clin Cancer Inf 450–592020 [DOI] [PubMed] [Google Scholar]
- 6.Carey VJ, Ramos M, Stubbs BJ, et al. Global alliance for genomics and health meets bioconductor: Toward reproducible and agile cancer genomics at cloud scale JCO Clin Cancer Inf 4472–4792020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pinnock H, Barwick M, Carpenter CR, et al. Standards for reporting implementation studies (StaRI) statement. BMJ. 2017;356:i6795. doi: 10.1136/bmj.i6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hudson KL, Collins FS.Sharing and reporting the results of clinical trials JAMA 313355–3562014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu DTY, Hanauer DA, Mei Q, et al. Assessing the readability of clinicaltrials.gov J Am Med Inf Assoc 23269–2752016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ni Y, Wright J, Perentesis J, et al. Increasing the efficiency of trial-patient matching: Automated clinical trial eligibility pre-screening for pediatric oncology patients. BMC Med Inf Decis Mak. 2015;15:28. doi: 10.1186/s12911-015-0149-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clinical Trials Transformation Initiative Improving public access to aggregate content of ClinicalTrials.gov. 2020. https://aact.ctti-clinicaltrials.org
- 12.Xu J, Lee HJ, Zeng J, et al. Extracting genetic alteration information for personalized cancer therapy from ClinicalTrials.gov J Am Med Inf Assoc 23750–7572016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tu SW, Peleg M, Carini S, et al. A practical method for transforming free-text eligibility criteria into computable criteria J Biomed Inf 44239–2502011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Google Cloud Healthcare natural language API pricing. 2020. https://cloud.google.com/healthcare/healthcare-natural-language-api-pricing
- 15. Bai J, Jhaney I, Wells J. Developing a reproducible microbiome data analysis pipeline using the Amazon Web Services Cloud for a cancer research group: Proof-of-concept study. J Med Internet Res. 2019;7:e14667. doi: 10.2196/14667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Google Cloud Using the healthcare natural language API. 2020. https://cloud.google.com/healthcare/docs/how-tos/nlp
- 17.National Cancer Institute SEER training modules: Site-specific modules. 2020. https://training.seer.cancer.gov/modules_site_spec.html
- 18.Jouhet V, Mougin F, Bréchat B, et al. Building a model for disease classification integration in oncology, an approach based on the National Cancer Institute thesaurus J Biomed Semant 81–122017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.National Cancer Institute Enterprise vocabulary services. 2017. https://evs.nci.nih.gov/ftp1/NCI_Thesaurus//Neoplasm/
- 20.HUGO Gene Nomenclature Committee: HGNC: The resource for approved human gene nomenclature, 2021 https://www.genenames.org
- 21.Regenstrief Institute LOINC from Regenstrief. 2020. https://loinc.org/
- 22.National Library of Medicine RxNorm files. 2020. https://www.nlm.nih.gov/research/umls/rxnorm/docs/rxnormfiles.html
- 23.National Comprehensive Cancer Network Non-small cell lung cancer. NCCN Clinical Practice Guideline Oncology. 2020. https://www.nccn.org/professionals/physician_gls/pdf/nscl.pdf
- 24.National Comprehensive Cancer Network Breast cancer. NCCN Clinical Practice Guideline Oncology. 2020. https://www.nccn.org/professionals/physician_gls/pdf/breast.pdf
- 25.Seven Bridges . Compute Costs. Seven Bridge Platform Document; 2020. https://docs.sevenbridges.com/docs/compute-costs [Google Scholar]
- 26.Amazon Web Services . How AWS Pricing Works: AWS Pricing Overview. AmazOn Web Services; 2020. http://media.amazonwebservices.com/AWS_Pricing_Overview.pdf [Google Scholar]
- 27.Hajian A. Understanding and Controlling Cloud Costs. Terra Support; 2020. https://support.terra.bio/hc/en-us/articles/360029748111 [Google Scholar]
- 28.Seven Bridges . Cloud Infrastructure Pricing. Seven Bridge Platform Document; 2020. https://docs.sevenbridges.com/docs/about-pricing [Google Scholar]
- 29.Seven Bridges . Storage Costs. Seven Bridge Platform Document; 2020. https://docs.sevenbridges.com/docs/storage-costs [Google Scholar]
- 30.Seven Bridges . Data Transfer Costs. Seven Bridge Platform Document; 2020.. https://docs.sevenbridges.com/docs/data-transfer-costs [Google Scholar]
- 31. Krumm N, Hoffman N. Practical estimation of cloud storage costs for clinical genomic data. Pr Lab Med. 2020;21:e00168. doi: 10.1016/j.plabm.2020.e00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yazar S, Gooden GEC, Mackey DA, et al. Benchmarking undedicated cloud computing providers for analysis of genomic datasets. PLoS One. 2014;9:E108490. doi: 10.1371/journal.pone.0108490. [DOI] [PMC free article] [PubMed] [Google Scholar]