Abstract
PURPOSE
Radiotherapy (RT)-induced lymphopenia (RIL) is commonly associated with adverse clinical outcomes in patients with cancer. Using machine learning techniques, a retrospective study was conducted for patients with esophageal cancer treated with proton and photon therapies to characterize the principal pretreatment clinical and radiation dosimetric risk factors of grade 4 RIL (G4RIL) as well as to establish G4RIL risk profiles.
METHODS
A single-institution retrospective data of 746 patients with esophageal cancer treated with photons (n = 500) and protons (n = 246) was reviewed. The primary end point of our study was G4RIL. Clustering techniques were applied to identify patient subpopulations with similar pretreatment clinical and radiation dosimetric characteristics. XGBoost was built on a training set (n = 499) to predict G4RIL risks. Predictive performance was assessed on the remaining n = 247 patients. SHapley Additive exPlanations were used to rank the importance of individual predictors. Counterfactual analyses compared patients' risk profiles assuming that they had switched modalities.
RESULTS
Baseline absolute lymphocyte count and volumes of lung and spleen receiving ≥ 15 and ≥ 5 Gy, respectively, were the most important G4RIL risk determinants. The model achieved sensitivitytesting-set 0.798 and specificitytesting-set 0.667 with an area under the receiver operating characteristics curve (AUCtesting-set) of 0.783. The G4RIL risk for an average patient receiving protons increased by 19% had the patient switched to photons. Reductions in G4RIL risk were maximized with proton therapy for patients with older age, lower baseline absolute lymphocyte count, and higher lung and heart dose.
CONCLUSION
G4RIL risk varies for individual patients with esophageal cancer and is modulated by radiotherapy dosimetric parameters. The framework for machine learning presented can be applied broadly to study risk determinants of other adverse events, providing the basis for adapting treatment strategies for mitigation.
INTRODUCTION
Lymphocytes, cells of the immune system, play a critical role in promoting systemic antitumor response. They are highly sensitive to ionizing radiation,1 and radiotherapy (RT) is an established causal determinant of immunotoxicity, that is, radiation-induced lymphopenia (RIL). Several studies have demonstrated associations between lymphopenia and adverse clinical outcomes.2-8 Development of severe RIL has been shown to be strongly associated with worse survival. Our own results have shown that grade 4 RIL (G4RIL) during concurrent chemoradiation therapy among patients with esophageal cancer is associated with worse 5-year survival (hazard ratio = 1.40; 95% CI, 1.04 to 1.89; P = .027) compared to those with grade 0-3.2 Despite its prevalence and negative consequences, RIL is often ignored in the current practice of RT as an unavoidable side effect.
CONTEXT
Key Objective
Characterize radiation-induced lymphopenia risk profiles for patients with esophageal cancer receiving proton and photon therapy.
Knowledge Generated
The risk of experiencing severe lymphopenia following radiotherapy (RT) varies for individual patients with esophageal cancer and is modulated by RT dosimetric parameters. Lymphopenia risk for an average patient receiving protons increased by 19% had the patient switched to photons. Reductions in risk were maximized with proton therapy for patients with older age, lower baseline absolute lymphocyte count, and higher lung and heart dose.
Relevance
The choice of treatment modality (protons v photons) may significantly alter the risk of severe lymphopenia for older patients and patients with low absolute lymphocyte count. Treatment determinants of severe lymphopenia risk identified in our study may be modifiable and adopted to further individualize RT planning.
Currently, RT planning is founded on population-based dose-volume (DV) response predictors that measure volumes of critical normal tissues receiving ≥ specified doses and ignores the heterogeneity in pretreatment clinical factors. Treatment planning based on population-averaged trends may reduce severe adverse events for some, but not all patients. Future innovations in dose distribution optimization require innovations in risk modeling to ascertain the principal risk determinants of immunotoxicity as well as calculate individualized risk predictions that can be used to identify patient subpopulations requiring mitigation. Previous studies have compared immunotoxicity risks between photon versus proton therapies1,3 as well as investigated the implications of immunotoxicity for cancer prognosis.4,9-12 RIL risk prediction models integrating dosimetric features with clinical factors have also been developed.13 Patient-specific lymphocyte loss kinetics were identified as important biomarkers for constraining radiation dose to immune organs at risk, for example, spleen, whole brain, or body as a whole.4,14 A model facilitating individualized predictions of lymphopenia risk integrating both patient-specific pretreatment clinical and treatment dosimetric factors has yet to be established.
This retrospective review applies machine learning (ML) techniques to a cohort of 746 patients with esophageal cancer. The analysis identifies factors that contribute to the heterogeneity in lymphopenia risk observed in clinical practice. The ML model defines risk profiles for proton and photon modalities, identifying vulnerable patient subpopulations for which early preventive measures may be warranted to protect against immunotoxicities. Furthermore, counterfactual analyses elucidate differences in G4RIL risk between proton and photon therapies. Attributes of patients predicted to experience clinically significant differences in G4RIL risk between photon and proton therapy are summarized.
METHODS
Study Cohort
We evaluated a total of 860 patients with biopsy-proven esophageal cancer who received concurrent chemoradiotherapy (with or without surgery) at the University of Texas MD Anderson Cancer Center between January 2004 and November 2017. Exclusion criteria included prescribed radiation dose other than 50.4 Gy, RT modality other than proton therapy or intensity-modulated RT, split-course RT, missing baseline blood sample data (eg, absolute lymphocyte count [ALC]), < 3 weekly documented ALCs during the treatment course, simultaneous irradiation of a second primary tumor, tumor overall stage IV (or unknown), histologic diagnosis other than adenocarcinoma or squamous cell carcinoma, history of hematologic malignancy, and endomucosal resection before chemoradiotherapy.
The primary end point of the study was G4RIL (ALC < 0.2 k/µL). Candidate predictors consisted of 34 clinical pre-RT patient characteristics and 59 dosimetric and midtreatment variables, which represented a subset of the collected information that was sufficiently complete for statistical interrogation. The latter set included DV parameters in terms of percentage of volumes receiving ≥ specified dose, for example, volume of heart receiving ≥ 5 Gy, denoted as V5. Details of predictors are given in the Data Supplement. Patients missing any of these variables were excluded from the analyses. The final cohort evaluable for statistical analysis comprised 746 patients. This study was approved by the University of Texas MD Anderson Cancer Center Institutional Review Board with a waiver of informed consent. Table 1 summarizes the clinical and treatment characteristics of the analysis cohort.
TABLE 1.
Patient and Treatment Characteristics of the Study Cohort, N = 746

Unsupervised Learning and Supervised Learning
Unsupervised learning using the Hierarchical Density-Based Spatial Clustering of Applications with Noise15 and Uniform Manifold Approximation and Projection16-18 algorithms was applied to study patterns among RT treatment dosimetry and pretreatment clinical attributes (Fig 1). Sensitivity analyses were performed to ensure the robustness of clustering results.19,20 Comparisons of patient and treatment features by different subgroups (eg, clusters, RT types, and G4RIL status) were performed using chi-square tests for categorical variables and two-sample t-test (if two subgroups) or analysis of variance (if +2 subgroups) for continuous variables (Data Supplement). More details are provided in the Data Supplement.
FIG 1.

Illustration of the analytic framework. Data preprocessing used unsupervised learning to explore sources of treatment and clinical heterogeneity. Explorative analyses developed independent models for multiple subpopulations. The findings were used to create a composite model consolidating the principal determinants of G4RIL risk that were evident across subpopulations. BMI, body mass index; D1, D2 and D3, the first, second and third column (dimension) of the UMAP embedding; G4RIL, grade 4 radiotherapy-induced lymphopenia; HDBSCAN, hierarchical density-based spatial clustering of applications with noise; SHAP, SHapley Additive exPlanations; UMAP, Uniform Manifold Approximation and Projection.
Supervised learning based on the XGBoost,21 a scalable and accurate gradient-boosting algorithm, was applied to build G4RIL risk models (Fig 2). Exploratory analyses evaluated RT-modality–specific models within subpopulations identified through unsupervised learning. Each model was built upon a unique set of variables that were selected by the algorithm from a total of 94 candidate predictors (Data Supplement). Bayesian optimization22 was implemented to optimize each model's configuration. Because of the limited sample size of some of the subpopulations, the training and evaluation of predictive performance for these models used five-fold cross-validation with 10 repetitions.
FIG 2.

G4RIL risk prediction by clinical stage and RT modality. Each panel demonstrates risk prediction for all patients in the subpopulation (left plot of each panel) as well as the prediction process for one randomly selected patient (right). Prediction performance is reported by the area under the ROC curve (AUC: mean [SD]) and misclassification rate (percentage of incorrectly classified cases; error rate: mean [SD]), respectively. Classification performance is reported from internal five-fold cross-validation with 10 repetitions. Numbers in the parentheses for selected patient provide the values of the corresponding predictors. The x-axes describe predicted probabilities of G4RIL. The y-axes rank G4RIL predictors by their importance. For example, on the right side of the top left panel, the selected proton patient is predicted to have approximately 50% probability of developing G4RIL based on their profile (BMI 34.8, baseline ALC 1.25 k/µL, heart V50 4.37%, spleen V50 9.51%, baseline eosinophil counts 0.1, total blood volume 6.45 L, and mean spleen dose approximately 20 Gy). The slope of the line segments indicates the extent of change in risk. This individual's immunotoxicity susceptibility is higher than the average estimated risk score (14.5%) of the subgroup. Vertical gray lines depict means of predicted G4RIL risk. Blue (red) lines delineate the patient's predicted risk that is below (above) the subpopulation average. ALC, absolute lymphocyte count; AUC, area under the receiver operating characteristics curve; BMI, body mass index; G4RIL, grade 4 radiotherapy-induced lymphopenia; NLR, neutrophil-lymphocyte ratio; ROC, receiver operating characteristic; RT, radiotherapy; SD, standard deviation.
Quantitative metrics, called the SHAP (SHapley Additive exPlanations) values,23 developed using game theory to quantify each variable's contributions to predictive models, were computed to inform variable selection and interpretations of variable importance across subpopulations. The SHAP values were also used to illustrate individualized patterns of prediction (Data Supplement). Performance for exploratory analyses is reported by misclassification error and area under the receiver operating characteristics curve (AUC).24
Counterfactual Analyses
Counterfactual prediction analyses were applied to the resulting models to elucidate the extent to which G4RIL risk varied by RT modality. Using each patient as her own synthetic (virtual) control, the probability of experiencing G4RIL following RT was predicted for each patient assuming she had received each modality. The difference in G4RIL risk was quantified as:
| (1) |
Differences in RIL risk were summarized using histograms. The proportion of patients experiencing higher and lower risk for each modality is reported.
Composite Model
After synthesizing the heterogeneity in G4RIL risk observed among clinical subpopulations, a final model combining all patients and modalities was developed. Final selection of nondosimetric predictors was guided by the synthesis of performance in our internal exploratory analyses and review of independent study.12 The model was trained using two third of the study cohort (n = 499) and evaluated using the remaining one third of patients (n = 247), which were held out of model fitting. Youden's index25 was used to select an optimal classification threshold for designating a patient as having elevated risk of G4RIL from model predictions. Model performance is reported for the test set in terms of accuracy, positive predictive value, specificity, sensitivity, and F1 score. All analyses were conducted in Python26 and R.27 Analysis scripts can be found on GitHub.28
RESULTS
A total of 285 (38.2%) of G4RIL cases were observed among the 746 patients evaluable for analyses (Table 1). The incidence of G4RIL was 22.0% among the 246 proton patients and 46.2% among the 500 photon patients (Data Supplement). The average (±standard deviation) baseline ALC, age, and body mass index (BMI) were 1.63 (±0.61) k/µL, 63.0 (±10.8) years, and 26.1 (±5.7) kg/m2, respectively. The majority of patients (84.5%) were males. All 746 patients received concurrent chemotherapy with cycles ranging from 1 to 7. Most patients (634 of 746, 85%) received five cycles of concurrent chemotherapy. A total of 215 patients received induction chemotherapy with cycles ranging from 1 to 8, with two cycles (125 of 215, 58.1%) as most predominant. Most commonly used concurrent and induction chemotherapy regimens were either oxaliplatin-based or taxane-based (Data Supplement). Tumors observed in this patient cohort were mostly stage cT3 (88.3%) and cN1-3 (67.7%) and located in the distal third of the esophagus (86.5%).
Exploratory Analysis
Unsupervised analyses defining interdependencies among clinical attributes and dosimetric parameters revealed that the database was predominantly composed of two subpopulations (Data Supplement), which were differentiated by a patient's clinical stage at diagnosis (I and II v III). In general, clinical stage is characterized according to tumor characteristics, for example, T stage and N stage (see the Data Supplement for the full comparison between the subpopulations). Sensitivity analyses demonstrated that results obtained from Hierarchical Density-based Spatial Clustering of Applications with Noise were consistent with consensus clustering and consistent among various configurations of hyperparameters. Separate exploratory prediction models (Data Supplement) were developed for subpopulations grouped by clinical stage (I-II: 266 patients; III: 480 patients) and RT modality.
As shown in Figure 2, baseline ALC was identified as an important feature among all subpopulations. The relative importance of other features tended to vary by stage and modality (Fig 2 and Data Supplement). For example, DV parameters heart V50 and spleen V50 were identified for proton clinical stage I and II patients, compared with lung V15 and spleen V5 for clinical stage III patients. Age was an important predictor for photon patients in both subpopulations but not for proton patients.
Photon patients demonstrated wider distribution of G4RIL risk, whereas proton patients tended toward lower risk. Approximately 14% of proton patients were predicted to have > 50% G4RIL risk compared with approximately 41% of photon patients (Data Supplement). Risk prediction was slightly more accurate among clinical stage I and II patients versus clinical stage III patients. A full assessment of model performance is provided in the Data Supplement. According to model predictions, proton patients experienced lower G4RIL risks when compared with photon patients across subpopulations (clinical stage I and II-photons: 40.8%; clinical stage I and II-protons: 14.6%; clinical stage III-photons: 49.4%; and clinical stage III-protons: 28.1%).
Counterfactual Analysis
Exploratory models were used to compare the G4RIL risk predicted for each patient assuming that they had received each modality (protons and photons). Figure 3 describes the distributions of absolute changes in risk computed, assuming each patient had switched RT modality. The predicted change in risk varies in magnitude and direction from patient to patient. In general, however, proton RT mitigates G4RIL risk for patients originally treated with photons. 83.7% of clinical stage I and II patients who received protons were predicted to have higher G4RIL risk if they had received photons. Similarly, 75.6% of clinical stage III patients who received protons would have experienced a higher G4RIL risk with photons. On average, a proton RT patient experienced a 19% increase in G4RIL risk if they had been treated with photon RT. By contrast, patients with clinical stage I and II receiving photon therapy experienced 27% risk reduction on average if they had been treated with proton therapy. The mean risk reduction if the RT modality had been switched from photons to protons for patients with clinical stage III was 10.4%.
FIG 3.
Histograms from counterfactual analyses depicting distributional shifts in G4RIL risk attributable to switching modality. The x-axis depicts absolute change in predicted G4RIL risk for the modalities compared. The y-axis reports the number of patients in each bin. G4RIL, grade 4 radiotherapy-induced lymphopenia.
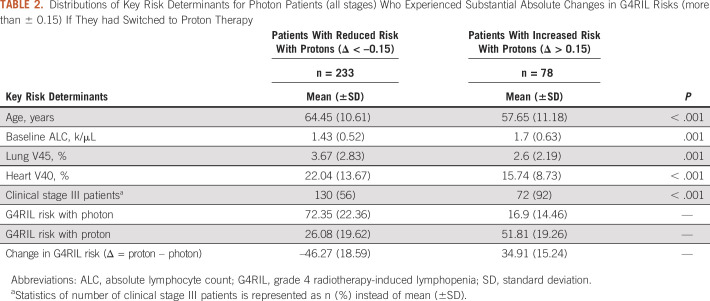
Let Δ denote the difference in predicted G4RIL risk between proton and photon therapy (Δ = proton risk – photon risk). Table 2 summarizes the distributions of risk determinants identified from counterfactual analysis as key features for differentiating between patients predicted to have substantial decreases (Δ < –0.15) versus substantial increases (Δ > 0.15) in G4RIL risk when switched from photon to proton therapy. Patients experiencing reduced risk with proton therapy tended to be older (64.45 [±10.61] v 57.65 [±11.18]; P < .001), with lower baseline ALC (1.43 [±0.52] v 1.7 [±0.63]; P = .001) and higher RT doses delivered to the lung and heart under photon planning.
TABLE 2.
Distributions of Key Risk Determinants for Photon Patients (all stages) Who Experienced Substantial Absolute Changes in G4RIL Risks (more than ± 0.15) If They had Switched to Proton Therapy
Final Model
A final prediction model combining patient subpopulations and modalities was created (Fig 4) from baseline ALC, lung V15, spleen V5, planning target volume (PTV), heart V45, age, and BMI. SHAP values were used to select the most important DV parameter for each organ (lung, spleen, and heart) identified in exploratory analyses.
FIG 4.
G4RIL risk for the combined model. The top panel displays G4RIL risk predictions for all patients (left plot of each panel) as well as one randomly selected patient (right). Numbers in the parentheses for the selected patient report values of the corresponding predictors. The model achieved an area under the ROC curve = 0.783 and misclassification error rate = 0.283 when applied to the test set. The x-axes report predicted probabilities of G4RIL. The population averaged G4RIL risk is depicted by a solid gray vertical line. The predicted individual G4RIL risk for the randomly selected patient is marked on the x-axis by the dashed line. Blue (red) lines delineate each patient's predicted risk that is below (above) the average. The y-axes rank G4RIL risk predictors ranked by their importance. The left lower panel displays the relative importance of variables for the combined prediction model. Each dot in the right lower panel indicates the SHAP value for an individual patient. Color specifies the feature value according to scale on the right. For example, high baseline ALC (red) tends to correspond to large negative SHAP values, which reduce G4RIL risk. As another example, increases in BMI tend to decrease in the G4RIL risk (red dots on the negative x-axis or blue dots on the positive x-axis). ALC, absolute lymphocyte count; AUC, area under the receiver operating characteristics curve; BMI, body mass index; G4RIL, grade 4 radiotherapy-induced lymphopenia; PTV, planning target volume; ROC, receiver operating characteristic; SHAP, SHapley Additive exPlanations.
The model achieved superior overall prediction performance (AUC: 0.783, error rate (accuracy): 0.283 (0.717), specificity: 0.667, sensitivity: 0.798, positive predictive value: 0.595, and F1-score: 0.682) than modality-specific models (Data Supplement). The optimal prediction threshold for defining a patient as high risk for G4RIL was 0.3 (Data Supplement), which was determined by Youden's index.25
The top left panel in Figure 4 depicts prediction trajectories for each individual patient. The top right panel shows the full risk calculation for a randomly selected patient. As shown in the figure, the patient is predicted to have approximately 10% risk of developing G4RIL according to their profile (baseline ALC: 1.94 k/µL, lung V15: 21.9%, spleen V5: 25.32%, PTV: 663.3, heart V45: 1.47%, age: 47, and BMI: 28.8); this individual's susceptibility to immunotoxicity was predicted to be lower than the average estimated risk score (approximately 37%, the gray vertical line) of the study cohort.
DISCUSSION
This article describes a framework for using ML to personalize prediction of RT-induced severe lymphopenia. Yielding true-positive and true-negative rates of 0.798 and 0.667, respectively, our study demonstrates that clinical and radiation dosimetry informatics can be combined to ascertain a patient's risk of immunotoxicity before receiving RT for esophageal cancer. Baseline ALC, lung V15, spleen V5, PTV, heart V45, age, and BMI were identified as the predominant determinants of G4RIL risk. The treatment dosimetric risk factors, in general, are modifiable through treatment plan optimization customized to each patient's risk profile. Furthermore, the choice of treatment modality (protons v photons) may significantly alter the risk. Counterfactual analyses defined attributes of patients for which risk of lymphopenia differed to a clinically significant extent between proton and photon therapy.
The analysis methodology presented in this article stands in contrast to traditional statistical analyses that rely on population-averaging with two-sample summaries or linear trends estimated by the regression. Two-sample hypothesis testing assumes that patient risk is homogenous, whereas the regression assumes that the effects of individual predictors (including treatment) are consistent and linear for all patient subpopulations. These assumptions apply to treatment effects, which are assumed to persist with the same magnitude for all patients for conventional approaches. Authors have applied conventional statistical approaches to this database to estimate linear trends in lymphopenia risk and construct nomograms.12 By way of contrast, the ML algorithms applied in our study characterize complex patterns among the studied features without the need to assume linearity. For example, our analysis identified that BMI increases G4RIL risk for some patients, while reducing risk for other patients. By characterizing heterogeneity in patterns of risk predictors and risk profiles across subpopulations, this study builds upon previous research conducted for predicting RIL.13 The impact of interventions may also vary among patients and patient subpopulations, which can be elucidated with the methodology for counterfactual patient profiling implemented in our study. Exploratory analyses identified interdependencies among baseline G4RIL risk, RT dose distribution, and clinical stage. Tumor characteristics, which comprise components of clinical stage, were associated with PTV size, which determines the extent of irradiated volume and thus modulates G4RIL risk. A nearly linear relationship was observed between baseline ALC and G4RIL risk within the ALC interval of approximately 1.5-2.0 k/µL, whereas outside this range, G4RIL risk plateaued at high and low values (Data Supplement). Patients with baseline ALC values within this range should be the target of future G4RIL risk mitigation strategies.
In addition, nonmonotonic relationships were observed among dosimetric factors and the G4RIL risk. For example, we found that G4RIL risk increased rapidly as mean heart dose increased from 20 to 30 Gy. G4RIL risk remained unchanged, however, as mean heart dose decreased below 20 Gy or increased beyond 30 Gy (Data Supplement). Such data may be used to define patient-specific dosimetric constraints for the anatomical structures and incorporated into treatment plan optimization to mitigate G4RIL risk. Adapting dosimetric strategies may be insufficient for patients with baseline ALCs between 1.5 and 2.0 k/µL, however, necessitating pharmacologic interventions or pretreatment extraction, expansion, and reinfusion of lymphocytes.
Counterfactual analyses for photon patients, had they been treated with protons, suggest reductions in G4RIL risk were maximized for patients treated with photon plans that delivered high RT doses to the lung and heart as well as for older patients, and patients with lower baseline ALC. The integral dose delivered by proton therapy is on average approximately 60% lower than photon therapy.29 Thus, smaller volumes of immune organs at risk and circulating lymphocyte are irradiated. This is advantageous for vulnerable patients whose reservoir of ALCs may be insufficient to provide an adequate buffer to counter lymphocyte depletion induced over the course of RT.
A few limitations should be noted. The database arises from a single institution, which has the potential to limit its generalizability to centers that have adopted different models for RT treatment planning and selection. Implementation of the prediction algorithm in practice will require precise methods for data collection and preprocessing to ensure sufficient quality and reproducibility. Our subpopulation analyses (proton stage I and II in particular) were potentially affected by sample size. Thus, subpopulation analyses used internal cross-validation. Efforts to validate all models using external and independent databases are ongoing.
In conclusion, ML can be used to extract insights into G4RIL risk from complex patient informatics. Determinants of G4RIL risk identified in our study may be modifiable and adopted to further individualize RT planning. For patients predicted to be at high risk of G4RIL, dosimetric constraints can be applied to immune organs at risk, for example, heart and lungs, and be customized to that given patient's baseline clinical characteristics. Such constraints can be determined using the models discussed above and incorporated into the criteria of optimization of radiation dose distribution to mitigate the risk. We believe this framework is generalizable to other cancer sites and RT-induced toxicities. Future research endeavors will interrogate larger databases and additional sources of patient informatics. Moreover, although immune toxicity risk was the emphasis of this study, treatment selection requires evaluation of the potential trade-offs between toxicity mitigation and clinical efficacy. Methods for developing individualized utilities for treatment selection based on both types of outcomes are under development.
Cong Zhu
Stock and Other Ownership Interests: Biogen, AbbVie, Regeneron, Hepion Pharmaceuticals, Galectin Therapeutics
Radhe Mohan
Stock and Other Ownership Interests: General Electric, Berkshire Hathaway
Steven H. Lin
Employment: MD Anderson Cancer Center
Honoraria: AstraZeneca/MedImmune, BeyondSpring Pharmaceuticals
Speakers' Bureau: AstraZeneca
Research Funding: BeyondSpring Pharmaceuticals, Nektar, STCube Pharmaceuticals Inc
Travel, Accommodations, Expenses: STCube Pharmaceuticals Inc, Elekta
Qianxia Wang
Employment: Philips (I)
Brian P. Hobbs
Stock and Other Ownership Interests: Presagia
Consulting or Advisory Role: STCube Pharmaceuticals Inc, Bayer
Research Funding: Amgen
Uncompensated Relationships: Presagia
No other potential conflicts of interest were reported.
DISCLAIMER
The content is solely the responsibility of the authors and does not necessarily represent the official views of the Cancer Prevention and Research Institute of Texas.
SUPPORT
U19 CA021239 and Cancer Center Support Grant P30 CA016672 from the National Cancer Institute. UTHealth Innovation for Cancer Prevention Research Training Program Predoctoral Fellowship (Cancer Prevention and Research Institute of Texas grant no. RP210042).
DATA SHARING STATEMENT
The data can be requested for research purposes in accordance with a material transfer agreement with MD Anderson Cancer Center.
AUTHOR CONTRIBUTIONS
Conception and design: Cong Zhu, Radhe Mohan, Steven H. Lin, Brian P. Hobbs
Financial support: Radhe Mohan, Steven H. Lin
Administrative support: Radhe Mohan
Provision of study materials or patients: Steven H. Lin
Collection and assembly of data: Cong Zhu, Radhe Mohan, Steven H. Lin, Brian P. Hobbs
Data analysis and interpretation: Cong Zhu, Goo Jun, Ashraf Yaseen, Xiaoqian Jiang, Qianxia Wang, Wenhua Cao, Brian P. Hobbs
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/cci/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Cong Zhu
Stock and Other Ownership Interests: Biogen, AbbVie, Regeneron, Hepion Pharmaceuticals, Galectin Therapeutics
Radhe Mohan
Stock and Other Ownership Interests: General Electric, Berkshire Hathaway
Steven H. Lin
Employment: MD Anderson Cancer Center
Honoraria: AstraZeneca/MedImmune, BeyondSpring Pharmaceuticals
Speakers' Bureau: AstraZeneca
Research Funding: BeyondSpring Pharmaceuticals, Nektar, STCube Pharmaceuticals Inc
Travel, Accommodations, Expenses: STCube Pharmaceuticals Inc, Elekta
Qianxia Wang
Employment: Philips (I)
Brian P. Hobbs
Stock and Other Ownership Interests: Presagia
Consulting or Advisory Role: STCube Pharmaceuticals Inc, Bayer
Research Funding: Amgen
Uncompensated Relationships: Presagia
No other potential conflicts of interest were reported.
REFERENCES
- 1.Shiraishi Y, Fang P, Xu C, et al. Severe lymphopenia during neoadjuvant chemoradiation for esophageal cancer: A propensity matched analysis of the relative risk of proton versus photon-based radiation therapy Radiother Oncol 128154–1602018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wei D, Cai X, Amy L, et al. The relationship of lymphocyte recovery and prognosis of esophageal cancer patients with severe radiation-induced lymphopenia after chemoradiation therapy Radiother Oncol 1339–152019 [DOI] [PubMed] [Google Scholar]
- 3.Routman DM, Garant A, Lester SC, et al. A comparison of grade 4 lymphopenia with proton versus photon radiation therapy for esophageal cancer Adv Radiat Oncol 463–692019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mohan R, Liu AY, Brown PD, et al. Proton therapy reduces the likelihood of high-grade radiation-induced lymphopenia in glioblastoma patients: Phase II randomized study of protons vs photons Neuro Oncol 23284–2942021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grossman SA, Ye X, Lesser G, et al. Immunosuppression in patients with high-grade gliomas treated with radiation and temozolomide Clin Cancer Res 175473–54802011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wild AT, Ye X, Ellsworth SG, et al. The association between chemoradiation-related lymphopenia and clinical outcomes in patients with locally advanced pancreatic adenocarcinoma. Am J Clin Oncol. 2015;38:259. doi: 10.1097/COC.0b013e3182940ff9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campian JL, Ye X, Brock M, et al. Treatment-related lymphopenia in patients with stage III non-small-cell lung cancer Cancer Invest 31183–1882013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davuluri R, Jiang W, Fang P, et al. Lymphocyte nadir and esophageal cancer survival outcomes after chemoradiation therapy Int J Radiat Oncol Biol Phys 99128–1352017 [DOI] [PubMed] [Google Scholar]
- 9.Bigley AB, Spielmann G, LaVoy EC, et al. Can exercise-related improvements in immunity influence cancer prevention and prognosis in the elderly? Maturitas 7651–562013 [DOI] [PubMed] [Google Scholar]
- 10.Nagalla S, Chou JW, Willingham MC, et al. Interactions between immunity, proliferation and molecular subtype in breast cancer prognosis Genome Biol 141–82013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vesely MD, Kershaw MH, Schreiber RD, et al. Natural innate and adaptive immunity to cancer Annu Rev Immunol 29235–2712011 [DOI] [PubMed] [Google Scholar]
- 12.Van Rossum PS, Deng W, Routman DM, et al. Prediction of severe lymphopenia during chemoradiation therapy for esophageal cancer: Development and validation of a pretreatment nomogram Pract Radiat Oncol 10e16–262020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhu C, Lin SH, Jiang X, et al. A novel deep learning model using dosimetric and clinical information for grade 4 radiotherapy-induced lymphopenia prediction. Phys Med Biol. 2020;65:035014. doi: 10.1088/1361-6560/ab63b6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yalamanchali A, Zhang H, Huang KC, et al. Patient-specific lymphocyte loss kinetics as biomarker of spleen dose in patients undergoing radiation therapy for upper abdominal malignancies. Adv Radiat Oncol. 2021;6:100545. doi: 10.1016/j.adro.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McInnes L, Healy J, Astels S. HDBSCAN: Hierarchical density based clustering. J Open Source Softw. 2017;2:205. [Google Scholar]
- 16. McInnes L, Healy J, Melville J. UMAP: Uniform manifold approximation and projection for dimension reduction. 2018 arXiv: 1802.03426. [Google Scholar]
- 17.UMAP Documentation. https://github.com/lmcinnes/umap [Google Scholar]
- 18.Allaoui M, Kherfi ML, Cheriet A.Considerably improving clustering algorithms using UMAP dimensionality reduction technique: A comparative study. International Conference on Image and Signal Processing, Cham, Switzerland, Springer, June 4, 2020, pp 317-325.
- 19.Wilkerson MD, Hayes DN.ConsensusClusterPlus: A class discovery tool with confidence assessments and item tracking Bioinformatics 261572–15732010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Şenbabaoğlu Y, Michailidis G, Li JZ.Critical limitations of consensus clustering in class discovery Sci Rep 41–32014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen T, Guestrin C.XGBoost: A scalable tree boosting system. Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, August 2016, pp 785-794.
- 22.Nogueira F. Bayesian Optimization: Open Source Constrained Global Optimization Tool for Python. 2014. https://github.com/fmfn/BayesianOptimization [Google Scholar]
- 23. Lundberg S, Lee SI. A unified approach to interpreting model predictions. 2017 arXiv: 1705.07874. [Google Scholar]
- 24.Fawcett T.An introduction to ROC analysis Pattern Recognit Lett 27861–8742006 [Google Scholar]
- 25.Schisterman EF, Faraggi D, Reiser B, et al. Youden index and the optimal threshold for markers with mass at zero Stat Med 27297–3152008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Python Software Foundation . Python Language Reference, Version 3.7.1. 2021. http://www.python.org [Google Scholar]
- 27.R Core Team . R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2021. https://www.R-project.org/ [Google Scholar]
- 28.Zhu C. Analyses Pipeline, 2021 https://github.com/random-git/radiotherapy
- 29.Mitin T, Zietman AL.Promise and pitfalls of heavy-particle therapy J Clin Oncol 322855–28632014 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data can be requested for research purposes in accordance with a material transfer agreement with MD Anderson Cancer Center.