Significance
Our study, involving 1,873 patients and 36,370 healthy individuals, is an extensive genome-wide study of myasthenia gravis. Our genome-wide association and transcriptome-wide association analyses identified two signals, namely CHRNA1 and CHRNB1, encoding acetylcholine receptor subunits, which were replicated in an independent cohort obtained from the UK Biobank. Identifying these genes confirms the potential utility of using genetics to identify proteins that are the antigenic targets of autoantibodies. We confirmed that the genetic abnormalities underlying early-onset and late-onset myasthenia gravis are different. Our data offer a broader insight into the genetic architecture underlying the pathophysiology of myasthenia gravis.
Keywords: myasthenia gravis, genome-wide association study, genetic correlation, pathway analysis
Abstract
Myasthenia gravis is a chronic autoimmune disease characterized by autoantibody-mediated interference of signal transmission across the neuromuscular junction. We performed a genome-wide association study (GWAS) involving 1,873 patients diagnosed with acetylcholine receptor antibody-positive myasthenia gravis and 36,370 healthy individuals to identify disease-associated genetic risk loci. Replication of the discovered loci was attempted in an independent cohort from the UK Biobank. We also performed a transcriptome-wide association study (TWAS) using expression data from skeletal muscle, whole blood, and tibial nerve to test the effects of disease-associated polymorphisms on gene expression. We discovered two signals in the genes encoding acetylcholine receptor subunits that are the most common antigenic target of the autoantibodies: a GWAS signal within the cholinergic receptor nicotinic alpha 1 subunit (CHRNA1) gene and a TWAS association with the cholinergic receptor nicotinic beta 1 subunit (CHRNB1) gene in normal skeletal muscle. Two other loci were discovered on 10p14 and 11q21, and the previous association signals at PTPN22, HLA-DQA1/HLA-B, and TNFRSF11A were confirmed. Subgroup analyses demonstrate that early- and late-onset cases have different genetic risk factors. Genetic correlation analysis confirmed a genetic link between myasthenia gravis and other autoimmune diseases, such as hypothyroidism, rheumatoid arthritis, multiple sclerosis, and type 1 diabetes. Finally, we applied Priority Index analysis to identify potentially druggable genes/proteins and pathways. This study provides insight into the genetic architecture underlying myasthenia gravis and demonstrates that genetic factors within the loci encoding acetylcholine receptor subunits contribute to its pathogenesis.
Myasthenia gravis manifests as ocular, bulbar, and limb weakness with muscle fatigability (1, 2). In severe cases, respiratory muscles are affected, leading to acute respiratory failure that can be fatal (myasthenic crisis) (1). This archetypal neuroimmunological disease is relatively rare, affecting ∼77 persons per million of the population, though the actual rate may be rising due to population aging in Western societies (2). Epidemiological studies distinguish two incidence peaks, with the first occurring before 40 and predominantly affecting women (3). The second peak occurs at the age of 60 and is mainly observed in men (3).
The discovery of autoantibodies has been central to advancing our understanding of myasthenia gravis (4). Myasthenic patients have autoantibodies against proteins in the neuromuscular junction involving the nicotinic acetylcholine receptor, muscle-specific kinase, and lipoprotein receptor-related protein 4, among others (5). These antibodies are responsible for an autoimmune attack on the neuromuscular junction that interferes with neuromuscular transmission (6). Current treatment strategies focus on decreasing the antibody levels using immunosuppressive medications, such as corticosteroids and azathioprine, or plasma exchange (5).
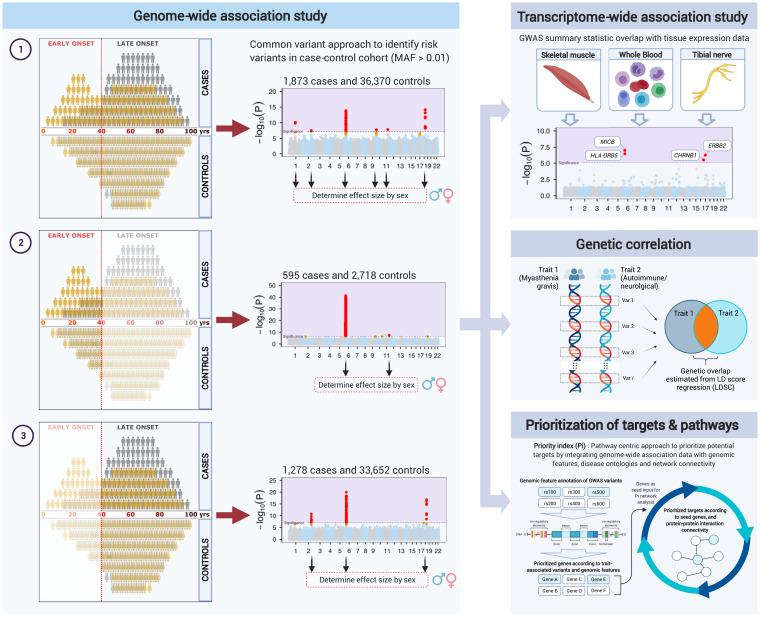
Despite their relevance to the pathogenesis of myasthenia gravis, the exact mechanisms by which autoantibodies develop and what nonimmunologic biology may modify disease onset and progression are poorly understood. More in-depth knowledge of this process may provide additional targets for drug development and guide therapeutic agent selection to treat the disease. Here, we performed a genome-wide association study (GWAS) and a transcriptome-wide association study (TWAS) to identify the genetic risks and candidate genes involved in disease etiology to address this knowledge gap (Fig. 1). This effort increased the sample size from 972 cases and 1,977 controls in the previous GWAS (7) to 1,873 cases and 36,370 controls, expanding our power to detect genetic risk factors associated with myasthenia gravis. We then used the genetic data to identify diseases linked to myasthenia gravis and prioritize genes and pathways that could be potentially targeted for disease-modifying therapy.
Fig. 1.
Analysis workflow. The flow diagram gives an overview of the analyses performed in this study. The Left panel depicts the GWAS of 1) all, 2) early-onset, and 3) late-onset myasthenia gravis. The Manhattan plots and variant and gene names do not correspond to the actual results. The Right-hand panels depict the advanced analytical approaches applied to the data, including TWAS (Right Top) to identify genes whose expression may mediate the risk of developing myasthenia gravis, genetic correlation (Right Middle) to identify diseases that overlap with myasthenia gravis, and Priority Index (Right Bottom) to identify gene targets and pathways that may be amenable to therapeutic intervention. The image was generated using BioRender.com.
Results
GWAS Implicates Loci in Myasthenia Gravis.
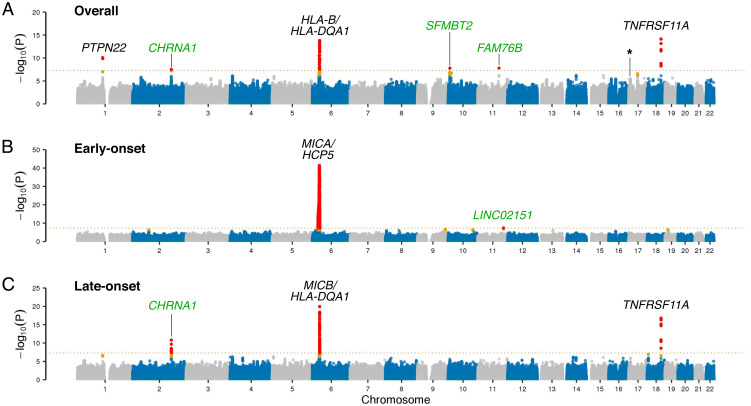
We performed a GWAS involving ∼24 million single nucleotide polymorphisms (SNPs) across a cohort of 1,873 patients diagnosed with myasthenia gravis and 36,370 healthy individuals. We focused on acetylcholine receptor antibody-positive cases, as these antibodies are found in ∼90% of patients with generalized myasthenia gravis. In addition to the previously reported signals in PTPN22, HLA-DQA1/HLA-B, and TNFRSF11A (7, 8), we observed association signals on chromosomes 2q31.1, 10p14, and 11q21 (see Fig. 2A for the Manhattan plot and Table 1 and SI Appendix, Table S1 for details of the associated SNPs). SI Appendix, Figs. S1 and S2 show the forest and regional association plots for each index variant.
Fig. 2.
GWAS in myasthenia gravis. The Top (A), Middle (B), and Bottom (C) panels show the Manhattan plots depicting the GWAS results of the overall discovery cohort (n = 1,873 myasthenia gravis cases and 36,370 control individuals), the early-onset cohort (n = 595 cases and 2,718 controls), and the late-onset cohort (n = 1,278 cases and 33,652 controls). The x-axis denotes the chromosomal position for the autosomes in hg38, and the y-axis indicates the association P values on a −log10 scale. Each dot represents a variant, where red dots denote variants that reached genome-wide significance and orange dots denote variants that are one log-fold lower than the significant threshold. A dashed line shows the conservative Bonferroni threshold for genome-wide significance (P = 5.0 × 10−8). Black font highlights known risk loci, while green font indicates association signals identified in this study not previously reported. The subsignificant hit on chromosome 17 corresponding to the CHRNB1 locus is marked with an asterisk.
Table 1.
Genome-wide significant association signals in the myasthenia gravis GWAS
| Discovery | Replication | Meta-analysis | |||||||||
| Chr | Position (SNPid) | Nearest gene | EA/OA | Cohort | EAF | OR (95% CI) | P | EAF | OR (95% CI) | P | P |
| 1 | 113834946 (rs2476601) | PTPN22 | A/G | All | 0.101/0.093 | 1.49 (1.32–1.67) | 7.95 × 10−11 | 0.141/0.107 | 1.36 (1.10–1.69) | 5.11 × 10−3 | 1.85 × 10−12 |
| M | 0.111/0.093 | 1.60 (1.38–1.86) | 6.50 × 10−10 | 0.135/0.107 | 1.30 (0.96–1.76) | 9.59 × 10−2 | 7.73 × 10−10 | ||||
| F | 0.089/0.093 | 1.34 (1.12–1.59) | 1.35 × 10−3 | 0.147/0.107 | 1.41 (1.05–1.90) | 2.38 × 10−2 | 1.15 × 10−4 | ||||
| 2 | 174764492 (rs35274388) | CHRNA1 | A/G | All | 0.057/0.036 | 1.57 (1.34–1.84) | 3.07 × 10−8 | 0.047/0.03 | 1.60 (1.11–2.31) | 1.17 × 10−2 | 1.18 × 10−9 |
| M | 0.063/0.036 | 1.77 (1.46–2.15) | 8.06 × 10−9 | 0.066/0.03 | 2.34 (1.52–3.60) | 1.13 × 10−4 | 7.19 × 10−12 | ||||
| F | 0.051/0.036 | 1.34 (1.06–1.71) | 1.48 × 10−2 | 0.026/0.03 | 0.86 (0.44–1.70) | 0.67 | 4.42 × 10−2 | ||||
| 6 | 31358836 (*rs9266277) | HLA-B | A/G | All | 0.361/0.347 | 1.29 (1.20–1.40) | 3.30 × 10−11 | 0.395/0.385 | 1.04 (0.89–1.22) | 0.61 | 6.70 × 10−10 |
| M | 0.323/0.347 | 1.09 (0.99–1.21) | 8.10 × 10−2 | 0.41/0.385 | 1.09 (0.89–1.35) | 0.40 | 6.64 × 10−2 | ||||
| F | 0.403/0.347 | 1.60 (1.44–1.78) | 7.15 × 10−18 | 0.379/0.385 | 0.98 (0.79–1.22) | 0.86 | 1.19 × 10−13 | ||||
| 6 | 32620936 (rs76815088) | HLA-DQA1 | C/T | All | 0.024/0.061 | 0.42 (0.34–0.52) | 1.58 × 10−14 | 0.028/0.052 | 0.54 (0.34–0.84) | 6.37 × 10−3 | 5.70 × 10−16 |
| M | 0.021/0.061 | 0.37 (0.27–0.50) | 5.55 × 10−10 | 0.03/0.052 | 0.58 (0.32–1.06) | 7.47 × 10−2 | 2.82 × 10−10 | ||||
| F | 0.027/0.061 | 0.53 (0.39–0.71) | 2.73 × 10−5 | 0.026/0.052 | 0.50 (0.26–0.96) | 3.89 × 10−2 | 3.25 × 10−6 | ||||
| 10 | 7410781 (rs2245569) | SFMBT2 | G/A | All | 0.277/0.213 | 1.27 (1.17–1.38) | 1.66 × 10−8 | 0.216/0.205 | 1.07 (0.89–1.28) | 0.49 | 5.58 × 10−8 |
| M | 0.278/0.213 | 1.28 (1.15–1.42) | 5.82 × 10−6 | 0.232/0.205 | 1.16 (0.91–1.49) | 0.23 | 4.08 × 10−6 | ||||
| F | 0.275/0.213 | 1.26 (1.12–1.41) | 7.73 × 10−5 | 0.199/0.205 | 0.97 (0.74–1.27) | 0.82 | 3.47 × 10−4 | ||||
| 11 | 95578258 (rs4409785) | FAM76B | C/T | All | 0.217/0.174 | 1.29 (1.18–1.41) | 1.54 × 10−8 | 0.171/0.172 | 1.00 (0.81–1.22) | 0.97 | 2.42 × 10−7 |
| M | 0.220/0.174 | 1.30 (1.16–1.45) | 6.88 × 10−6 | 0.188/0.172 | 1.11 (0.85–1.45) | 0.44 | 1.47 × 10−5 | ||||
| F | 0.213/0.174 | 1.27 (1.12–1.43) | 2.11 × 10−4 | 0.153/0.172 | 0.88 (0.65–1.18) | 0.39 | 2.59 × 10−3 | ||||
| 18 | 62342581 (rs4574025) | TNFRSF11A | C/T | All | 0.535/0.461 | 1.33 (1.24–1.43) | 7.08 × 10−15 | 0.492/0.474 | 1.09 (0.93–1.27) | 0.30 | 7.12 × 10−14 |
| M | 0.559/0.461 | 1.47 (1.33–1.61) | 2.27 × 10−15 | 0.526/0.474 | 1.23 (1.00–1.53) | 5.13 × 10−2 | 2.60 × 10−15 | ||||
| F | 0.508/0.461 | 1.20 (1.09–1.33) | 4.08 × 10−4 | 0.457/0.474 | 0.94 (0.76–1.17) | 0.59 | 4.06 × 10−3 | ||||
Significant hits were stratified by sex to determine the contribution of sex and are highlighted in bold. Positions were in build hg38; Chr, chromosome; EA/OA, effect allele/other allele; EAF, effect allele frequency in cases and controls; discovery, meta-analysis from the US and Italian cohorts (1,873 cases [989 male, 884 female] and 33,370 controls); replication, cohort from the UK Biobank where controls were age- and sex-matched (354 cases [181 male, 173 female] and 7,078 controls); *rs9266277 is an independent variant identified from conditional analysis on rs76815088. Results in the table are from the unconditional analysis. Discovery phase conditional statistics for rs9266277: OR (95% CI) = 1.28 (1.19–1.38), P = 1.92 × 10−10. The replication significance threshold was set at P = 0.1/7 = 0.0143 (Bonferroni correction for seven loci with the same effect direction).
The locus on chromosome 2 lies in a promoter region of the cholinergic receptor nicotinic alpha 1 subunit (CHRNA1) gene (rs35274388, P = 3.07 × 10−8, odds ratio [OR] =1.57), which encodes a glycoprotein subunit of the nicotinic acetylcholine receptor on the forward strand, and in the noncoding transcript of AC010894.2, which is an antisense gene to CHRNA1. The two other signals were found within the first intron of the Scm-like with four mbt domains 2 (SFMBT2) gene on chromosome 10 (rs2245569, P = 1.66 × 10−8, OR = 1.27) and downstream of the family with sequence similarity 76 member B (FAM76B) gene on chromosome 11 (rs4409785, P = 1.54 × 10−8, OR =1.29). Within the locus on chromosome 6, we identified two association signals near the HLA-B gene (rs9266277, P = 3.31 × 10−11, OR = 1.29) and the HLA-DQA1 gene (rs76815088, P = 1.58 × 10−14, OR = 0.42). These variants were located ∼1.3 million base pairs apart, and conditional analysis confirmed that the loci were independent of each other (SI Appendix, Fig. S3). Fine-mapping at the HLA region showed that rs9266277 was highly correlated with amino acids 45 and 74 in the HLA-B gene, whereas rs76815088 was tagging the HLA-DRB1*13:01:02 and HLA-DQB1*06:03:02 alleles (SI Appendix, Figs. S4A and S5A). Only CHRNA1 showed association in the UK Biobank replication cohort (P = 0.012, OR = 1.60; see Table 1), likely reflecting the small number of cases and age distribution within this cohort (n = 354 cases and 7,058 controls). Variant rs2476601 (PTPN22, P = 5.11 × 10−3, OR = 1.36) was replicated in the UK Biobank dataset, but variants rs2245569 (SFMBT2, P = 0.487, OR = 1.07) and rs4409785 (FAM76B, P = 0.965, OR = 1.00) did not.
Early-Onset and Late-Onset Myasthenia Gravis.
Because of the age-dependent genetic heterogeneity known to exist in myasthenia gravis, we performed genome-wide association testing separately in early-onset cases (n = 595 cases and 2,718 controls, aged 40 y or younger) and late-onset cases (n = 1,278 cases and 33,652 controls). SI Appendix, Fig. S6 shows the age and sex distribution of the cohort. As in our previous GWAS of myasthenia gravis (3), we found distinct differences in the genetic architecture among these cohorts (Fig. 2 B and C). An association on chromosome 11q23.3 was also identified downstream of a long intergenic noncoding RNA, LINC02151 (rs73007767, P = 4.36 × 10−8, OR = 1.85). However, the genetic risk of early-onset (predominantly female) myasthenia gravis cases was driven primarily by the Major Histocompatibility Complex (MHC) on chromosome 6 (Fig. 2B and Table 2 and SI Appendix, Table S2) with the index variant tagging the HLA-B*08:01:02 and HLA-C*07:01:02 alleles (SI Appendix, Fig. S4B and S5B).
Table 2.
Genome-wide significant association signals in early-onset myasthenia gravis
| Chr | Position (SNPid) | Nearest gene | EA/OA | Cohort | EAF | OR (95%CI) | P |
| 6 | 31442744 (rs3093958) | HCP5/MICA | G/A | All | 0.245/0.094 | 4.49 (3.62–5.57) | 3.97 × 10−42 |
| M | 0.117/0.094 | 1.90 (1.28–2.82) | 1.47 × 10−3 | ||||
| F | 0.290/0.094 | 6.39 (5.01–8.14) | 7.46 × 10−51 | ||||
| 11 | 116158033 (rs73007767) | LINC02151 | G/T | All | 0.129/0.087 | 1.85 (1.48–2.30) | 4.36 × 10−8 |
| M | 0.133/0.087 | 1.80 (1.26–2.56) | 1.14 × 10−3 | ||||
| F | 0.128/0.087 | 1.81 (1.42–2.32) | 2.17 × 10−6 |
Positions are in build hg38; Chr, chromosome; EA/OA, effect allele/other allele; EAF, effect allele frequency in cases and controls; meta-analysis is based on early-onset myasthenia gravis cases from the US and Italian cohorts (595 cases and 2,718 controls).
The MHC locus was also strongly associated with an increased risk of late-onset myasthenia gravis. However, the risk-associated variants were distinct from those observed among the early-onset cases (Table 3 and SI Appendix, Table S3). Furthermore, the PTPN22, CHRNA1, and TNFRSF11A loci were more prominently associated with disease in the late-onset myasthenia gravis cases than the early-onset cases (Table 3). These findings show that genetic factors other than those in the MHC play a prominent role in the occurrence of myasthenia gravis in older, predominantly male patients. A correlation plot comparing the effect sizes among early-onset and late-onset cases further confirmed their distinct patterns of genetic predispositions to myasthenia gravis (SI Appendix, Fig. S7).
Table 3.
Genome-wide significant association signals in late-onset myasthenia gravis
| Discovery | Replication | Meta-analysis | |||||||||
| Chr | Position (SNPid) | Nearest gene | EA/OA | Cohort | EA/OA | OR (95% CI) | P | EA/OA | OR (95% CI) | P | P |
| 2 | 174764492 (rs35274388) | CHRNA1 | A/G | All | 0.065/0.036 | 1.86 (1.56–2.23) | 1.53 × 10−11 | 0.049/0.03 | 1.69 (1.15–2.48) | 7.32 × 10−3 | 1.40 × 10−12 |
| M | 0.066/0.036 | 1.94 (1.58–2.40) | 6.00 × 10−10 | 0.071/0.03 | 2.56 (1.66–3.95) | 2.18 × 10−5 | 1.35 × 10−12 | ||||
| F | 0.062/0.036 | 1.73 (1.29–2.32) | 2.87 × 10−4 | 0.022/0.03 | 0.72 (0.32–1.64) | 0.44 | 1.02 × 10−3 | ||||
| 6 | 32603181 (*rs679242) | HLA-DRB1 | T/G | All | 0.143/0.162 | 1.13 (1.00–1.27) | 0.055 | 0.212/0.189 | 1.18 (0.97–1.45) | 0.11 | 1.87 × 10−2 |
| M | 0.143/0.162 | 1.12 (0.97–1.30) | 0.12 | 0.22/0.189 | 1.25 (0.95–1.64) | 0.10 | 3.68 × 10−2 | ||||
| F | 0.143/0.162 | 1.13 (0.93–1.37) | 0.23 | 0.204/0.189 | 1.11 (0.82–1.50) | 0.49 | 0.19 | ||||
| 6 | 32619290 (rs9271375) | HLA-DRB1/HLA-DQB1 | A/G | All | 0.387/0.481 | 0.66 (0.60–0.72) | 1.09 × 10−20 | 0.419/0.474 | 0.80 (0.68–0.95) | 9.52 × 10−3 | 1.79 × 10−19 |
| M | 0.361/0.481 | 0.60 (0.54–0.66) | 2.38 × 10−21 | 0.42/0.474 | 0.81 (0.65–1.01) | 0.06 | 5.20 × 10−20 | ||||
| F | 0.436/0.481 | 0.78 (0.68–0.90) | 5.81 × 10−4 | 0.418/0.474 | 0.79 (0.62–1.01) | 0.06 | 4.50 × 10−4 | ||||
| 18 | 62343215 (rs4369774) | TNFRSF11A | A/G | All | 0.560/0.461 | 1.45 (1.33–1.58) | 1.62 × 10−17 | 0.492/0.473 | 1.08 (0.92–1.27) | 0.37 | 6.45 × 10−19 |
| M | 0.569/0.461 | 1.54 (1.39–1.71) | 4.65 × 10−16 | 0.532/0.473 | 1.27 (1.02–1.58) | 0.03 | 8.98 × 10−19 | ||||
| F | 0.542/0.461 | 1.34 (1.16–1.54) | 3.78 × 10−5 | 0.444/0.473 | 0.88 (0.69–1.12) | 0.31 | 1.36 × 10−4 | ||||
Significant hits were stratified by sex to determine the contribution of sex and are highlighted in bold. Positions are in build hg38; EA/OA, effect allele/other allele; EAF, effect allele frequency in cases and controls; discovery, meta-analysis from the US and Italian cohorts (1,278 cases [835 male, 443 female] and 33,652 controls); replication, a cohort from the UK Biobank where controls were age- and sex-matched (308 cases [168 male, 140 female] and 7,056 controls); *rs679242 is an independent variant identified from conditional analysis on rs9271375. The results in the table are from the unconditioned analysis. Discovery phase conditional statistics for rs679242: OR (95% CI) = 1.72 (1.49–2.00), P = 4.42 × 10−13. The replication significance threshold was set at P = 0.1/3 = 0.0333 (Bonferroni correction for three loci identified on chromosomes 2, 6, and 18 with the same effect direction).
Consistent with the pattern observed in the discovery cohort, the association signals at the HLA-DRB1/DQB1 tagging the HLA-DRB1*13:01:02 and HLA-DQB1*06:03:02 alleles (rs9271375, P = 9.52 × 10−3, OR = 0.80) and the CHRNA1 locus (rs35274388, P = 7.32 × 10−3, OR = 1.69) were independently replicated in the subset of the UK Biobank cohort consisting of 308 late-onset cases and 7,056 age- and sex-matched controls (Table 3). Of note, the risk conferred by rs35274388 (CHRNA1) was highest among men manifesting disease in late age in the replication dataset (P = 2.18 × 10−5, OR = 2.56; n= 168 male cases and 7,056 controls, Table 3). The secondary signal on chromosome 6 was also replicated upon conditioning on rs9271375 (rs679242, P = 2.98 × 10−4, OR = 1.61). This variant mapped to exon 2 of the HLA-DRB1 gene (SI Appendix, Figs. S4C and S5C).
TWAS Implicates Loci in Myasthenia Gravis.
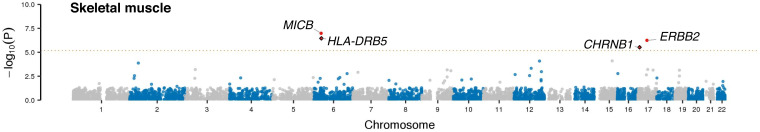
Next, we performed a TWAS of myasthenia gravis to identify additional genes relevant to the disease’s pathogenesis. This analytical approach correlates genotype and expression to phenotypic traits to identify the cis-genetic component of gene expression relevant to the trait (9). To do this, we integrated our myasthenia gravis GWAS results from the overall and subgroup analysis with the gene expression profile in normal skeletal muscle, peripheral nerve, and whole blood obtained from public Genotype-Tissue Expression (GTEx) datasets (10). We then performed association testing of the predicted gene expression and risk (9). This approach identified six genes whose differential expression was significantly associated with myasthenia gravis risk across tissues (Fig. 3, SI Appendix, Figs. S8 and S9, Table 4, and SI Appendix, Table S4). Notably, lower expression of the gene encoding another acetylcholine receptor subunit, namely the cholinergic receptor nicotinic beta 1 subunit (CHRNB1; rs4151121, P = 3.01 × 10−6, Z = −4.67) and epidermal growth factor receptor family of receptor tyrosine kinases (ERBB2, rs2102928, P = 5.63 × 10−7, Z = −5.00), was predicted to increase disease risk. We found that the rs4151121 variant within the CHRNB1 locus was a subsignificant hit in the GWAS of all myasthenia gravis patients (P = 1.13 × 10−5, OR = 1.18; see asterisk denoting hit on chromosome 17 in Fig. 2A). Colocalization analysis confirmed that the GWAS-associated variant and the variant driving the expression of CHRNB1 were the same (posterior probability H4 [PPH4] = 0.97, Table 4). In contrast, rs2102928 within ERBB2 had a small effect size (P = 0.031, OR = 1.03).
Fig. 3.
TWAS in myasthenia gravis. Manhattan plots depicting the TWAS results of the overall discovery cohort (n = 1,873 myasthenia gravis cases and 36,370 control individuals) using expression data for skeletal muscle obtained from GTEx. The x-axis denotes the chromosomal position for the autosomes in hg38, and the y-axis indicates the association P values on a −log10 scale. Each dot represents a variant, where red dots denote variants that reached genome-wide significance and permutation P values < 0.05. The significance threshold for skeletal muscle was 6.8 × 10−6 and is shown with the dashed line. The orange dots denote variants that are one log-fold lower than the significant threshold. The dots with a black diamond outline are variants with colocalization posterior prior probability H4 (PPH4) > 0.75.
Table 4.
Causal genes associated with myasthenia gravis based on TWAS of skeletal muscle
| MAF | eQTL | TWAS | ||||||||
| Chr | ID | Cases | Cont. | Z | GWAS.Z | Z | P | PERM.P | PPH4 | |
| Overall | ||||||||||
| CHRNB1 | 17 | rs4151121 | 0.433 | 0.372 | −11.79 | 4.39 | −4.67 | 3.01 × 10−6 | 2.03 × 10−4 | 0.98 |
| ERBB2 | 17 | rs2102928 | 0.353 | 0.322 | −3.42 | 2.16 | −5.00 | 5.63 × 10−7 | 7.99 × 10−4 | 0.18 |
| Early-onset | ||||||||||
| CYP21A2 | 6 | rs3101018 | 0.225 | 0.089 | 4.55 | 12.72 | 10.90 | 7.32 × 10−28 | 3.76 × 10−3 | 0.98 |
| HLA-DMA | 6 | rs35404844 | 0.061 | 0.042 | 7.12 | 3.58 | 7.60 | 2.88 × 10−14 | 1.13 × 10−3 | 0.89 |
| Late-onset | ||||||||||
| C2 | 6 | rs1048709 | 0.180 | 0.180 | −3.97 | 4.51 | −6.80 | 1.04 × 10−11 | 1.23 × 10−3 | 0.71 |
| HLA-DRB5 | 6 | rs9271055 | 0.180 | 0.158 | 16.95 | 5.46 | 7.68 | 1.65 × 10−14 | 5.05 × 10−3 | 0.00 |
Chr, chromosome; ID, rs number of the best eQTL locus in the locus; MAF, minor allele frequency; cont., controls; eQTL Z, Z-score of the best eQTL in the locus; GWAS.Z, GWAS Z-score for the eQTL; PERM.P, TWAS permutation P values; PPH4, Bayesian derived posterior probability for colocalization of the eQTL and the GWAS variant.
Genetic Overlap of Myasthenia Gravis with Other Diseases.
Epidemiological studies have shown that myasthenic patients are more likely to develop other autoimmune diseases (11). An overlap with neurodegenerative diseases has also been suggested (12). For this reason, we investigated shared genetic risk between myasthenia gravis, nine autoimmune diseases, and four neurological diseases using Linkage Disequilibrium Score Regression modeling (13). We found evidence of genetic overlap with myasthenia gravis for all autoimmune diseases except systemic lupus erythematosus: rheumatoid arthritis (regression coefficient representing the degree of genetic correlation [rg] = 0.41, pFDR adjusted = 1.10 × 10−4), multiple sclerosis (rg = 0.18, pFDR adjusted = 0.04), type 1 diabetes (rg = 0.44, pFDR adjusted = 2.84 × 10−4), ulcerative colitis (rg = 0.20, pFDR adjusted = 0.05), and hypothyroidism (rg = 0.28, pFDR adjusted = 4.31 × 10−3). There was no significant correlation with any of the tested neurological disorders (SI Appendix, Table S5). Of note, the genetic correlation was present after excluding chromosome 6, confirming that the HLA locus was not the only source of the observed correlation (SI Appendix, Table S5). Different genetic overlaps were also observed for the early-onset and late-onset myasthenia gravis cohorts (SI Appendix, Table S5).
Identifying Targets and Pathways for Myasthenia Gravis Based on Genetic Data.
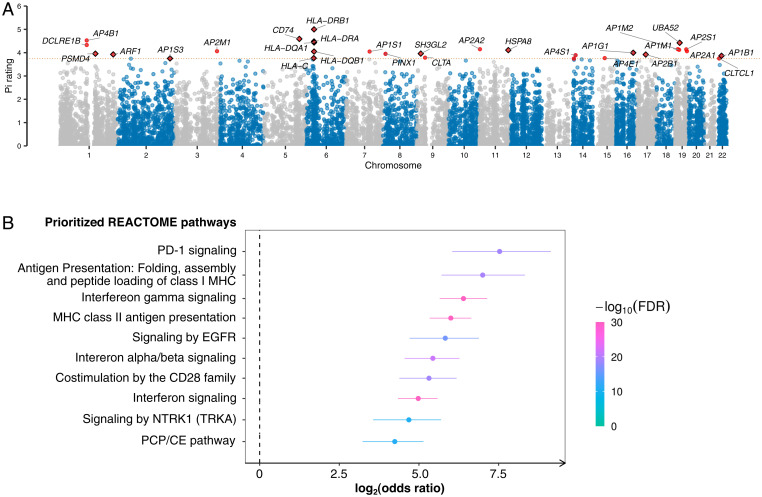
Finally, we performed a target prioritization analysis using the Priority Index pipeline to gain insights into the pathways involved in myasthenia gravis and identify potentially druggable targets (14). This analysis integrated the genetic information from our GWAS with functional genomic data and immune-related annotation data to nominate the most relevant genes (Fig. 4A). Enrichment analysis of the top 1% genes with the highest Priority Index score identified a set of enriched pathways involved in the adaptive immune response, cytokine signaling in the immune system, and receptor tyrosine kinase signaling (Fig. 4B).
Fig. 4.
Prioritization of immunological targets in myasthenia gravis based on Priority Index and druggability. (A) Autosomal genes were scored based on the gene-predictor matrix generated from the Priority Index pipeline. The x-axis denotes the chromosomal position in hg38, and the y-axis indicates the Priority Index rating on a scale of 0 to 5. Each dot represents a gene, and the red dots indicate the top 30 genes. Dots with a black diamond outline are druggable genes based on a Pocketome analysis. The red dashed line shows the threshold for the top 30 genes (scale > 3.9). (B) An enrichment analysis shows the prioritized target pathways in the immune and the signal transduction modules. The x-axis shows the REACTOME pathways that were significantly overrepresented by the top genes. The y-axis denotes the strength of the enrichment quantified by the OR on a log2 scale. The 95% CIs are represented by lines flanking each dot. Enrichment significance was measured by calculating the false discovery rate (FDR) from a one-sided Fisher’s exact test.
Discussion
We performed a GWAS in a large cohort of patients diagnosed with myasthenia gravis and attempted to replicate our findings in an independent cohort. Our data confirmed that early-onset and late-onset myasthenia gravis have distinct genetic bases. We also identified several susceptibility loci as having a role in the disease’s pathogenesis. Most interesting among these, we implicated genetic variants in two genes encoding nicotinic acetylcholine receptor subunits as relevant to the disease. We used our data to show a genetic overlap between myasthenia gravis and other autoimmune diseases, most notably thyroid abnormalities, rheumatoid arthritis, and multiple sclerosis. Building on those observations, we nominated genes and pathways that may contribute to the underlying disease pathobiology and are worthy of additional study as therapeutic targets.
The adult acetylcholine receptor is a complex heteropentamer consisting of alpha, beta, epsilon, and delta subunits (α2βεδ). Our GWAS and TWAS analyses identified association signals in the genes encoding two of these subunits (CHRNA1 and CHRNB1). These nicotinic acetylcholine receptor subunits are targets of autoantibodies in myasthenia gravis (15, 16). Furthermore, coding mutations in CHRNA1 and CHRNB1 are known causes of congenital myasthenia gravis, a condition characterized by a markedly diminished number of functioning acetylcholine receptors on the postsynaptic membrane (17). Based on what is already known about myasthenia gravis, there are several mechanisms by which the genetic signals identified in our study could lead to disease. For example, altered expression of CHRNA1 in the thymus or other immune cells may interfere with the development of immunological tolerance to the acetylcholine receptor (18) or cause intrathymic immunization (19).
Alternatively, the associated variants appear to reduce the expression of their respective acetylcholine receptor subunits. The disease-associated SNP that we identified on chromosome 2q31.1 is located within the CCAAT-Enhancer-Binding Protein-Beta transcription factor binding site of CHRNA1 (20) and within the antisense gene to CHRNA1 (AC010894.2) on the reverse strand, suggesting that it may decrease CHRNA1 expression. The allele showing subsignificant association with myasthenia gravis in CHRNB1 similarly reduces expression of that subunit. The reduced expression of these subunits could, in turn, decrease the number of functional acetylcholine receptors within the neuromuscular synapse, similar to what is observed with autoantibody-mediated myasthenia gravis and congenital myasthenic syndrome. Altered subunit composition may also alter how the immune system perceives the acetylcholine receptors on the neuromuscular synapse, leading to an abnormal response.
Our data will be a seeding point for future research into myasthenia gravis. This will include functional studies to confirm the molecular mechanisms by which CHRNA1 and CHRNB1 expression changes lead to disease. Our genomic data already provide clues to this mechanism; patients who were homozygous for the CHRNA1 risk alleles had a four times higher risk of developing myasthenia gravis than patients who carried only one risk allele (rs35274388, OR in homozygotes = 5.86 and OR in heterozygous = 1.45). A similar, though smaller, trend was observed for CHRNB1 (rs4151121, OR for homozygous carriers = 1.42 and OR in heterozygous carriers = 1.10). Skeletal muscle expression data from GTEx also showed that homozygous carriers of the rs4151121 risk allele had an 8-fold reduced expression compared to heterozygous carriers (https://gtexportal.org/home/snp/rs4151121). Existing autoimmune acquired mouse models of myasthenia gravis are unsuitable for modeling the implicated genetic loci, meaning that transgenic lines will have to be developed. Patient-orientated studies would require tissue samples collected from individuals diagnosed with myasthenia gravis, and our work argues in favor of establishing a public biobank to facilitate this work.
Identifying the ERBB2 gene in our TWAS analysis further supports the central importance of the acetylcholine receptor in the pathogenesis of myasthenia gravis. The encoded ERBB2 protein is highly expressed at the neuromuscular junction in skeletal muscle (21), where it regulates the expression of acetylcholine receptor subunits via MAP kinase activation (22). Synapses were less efficient in mutant mice with reduced Erbb2 muscle expression, and there were reduced levels of acetylcholine receptors at the motor endplates in these animals (4). The risk conferred by ERBB2 in our study appears to be mediated by its low expression, which would affect the formation of the functional acetylcholine receptor complex.
As with previous GWASes of myasthenia gravis (7, 8), our study identified the HLA locus as a susceptibility locus for this autoimmune disease. Despite the individual-level and population-level variability intrinsic to this locus, our results agreed with a recent study of HLA Class I and II effects on early-onset and late-onset myasthenia gravis risk in the Norwegian, Swedish, and Italian populations (23). This served as an additional replication cohort and underscored the veracity of our results.
Identifying pathogenic autoantibodies is a critical step in defining any autoimmune disease (24). In addition to providing insight into the antigenic target, the ability to screen patients for the relevant autoantibody allows for a more precise diagnosis of specific syndromes and disease subtypes (25). The field’s central issue is that the current methods used to discover novel autoantibodies are time-consuming, involving immunoblotting or precipitation of the antigen by the patient’s serum or binding of antibodies in cell-based assays (26). Our analyses identified association signals in the CHRNA1 and CHRNB1 loci that encode the proteins that are the antigenic targets of some acetylcholine receptor antibodies. We propose that the corollary approach is also valid in some cases: GWAS can be used as a screening tool to narrow the search scope for autoantibodies. Thus, genes located within associated loci and encoding proteins expressed within the tissue damaged in patients with a particular autoimmune disease are candidates for more directed autoantibody discovery efforts using enzyme-linked immunosorbent assays.
We found genetic evidence linking myasthenia gravis with other autoimmune diseases, including rheumatoid arthritis, multiple sclerosis, type 1 diabetes, and ulcerative colitis. Epidemiological data have extensively reported that patients with myasthenia gravis and their family members are at increased risk of other autoimmune diseases (27). Nonetheless, using genetic data to map these commonalities represents a powerful approach in neurological disease, and our results point to the molecular basis for this overlap. An intriguing possibility is that medications used to treat one of the linked autoimmune diseases may also help patients with myasthenia gravis. In contrast, we found only weak evidence of overlap with neurological disorders.
Over 90% of drugs fail during clinical development, driving up drug development costs and, ultimately, healthcare expenditures (28). An emerging theme in the pharmaceutical industry is that drugs targeting proteins and pathways with genetic evidence are more likely to succeed (5). Based on this, we used our genetic data to prioritize gene targets and pathways that may be amenable to therapeutic intervention (14). Additional experimental evidence in preclinical disease models is required to assess their relevance for treating myasthenia gravis. However, the present scope of research in myasthenia gravis is limited to studies involving the destruction of acetylcholine receptors by autoantibodies. Our results serve as an alternative starting point for generating data-driven hypotheses that can be tested for their ability to modify disease risk.
Our study has limitations. Several loci identified in the discovery cohorts failed to replicate in our follow-up cohort, likely reflecting our replication cohort's small size and limited power to detect an association. Furthermore, the CTLA4 locus identified in our previous GWAS (7) was subsignificant in our new analysis (rs231770, P = 2.46 × 10−3, OR = 1.12, 95% CI = 1.04–1.21). The acetylcholine receptor antibody status of the UK Biobank samples used in the replication cohort was not available, further adding to the heterogeneity of this group and perhaps accounting for this failure of replication. Our initial nomination of CTLA4 may have been a false-positive finding, or it may reflect variability in the alleles influencing myasthenia gravis risk across different populations. Although these loci remain biologically plausible, more extensive studies are required to confirm their association.
Another constraint is the difficulty in identifying a single gene responsible for disease within the HLA locus, leading us to rely instead on the association between haplotypes and disease susceptibility (29). By extension, TWAS analysis also underestimates the complexity of the linkage disequilibrium landscape in the HLA region. A full exploration of this locus will likely require long-range sequencing to resolve this repetitive and GC-rich genomic region and determine which variants and genes within HLA are driving the disease association (30).
Finally, our study focused on European ancestry individuals, as this is the only population in which extensive collections of myasthenia gravis patients were available. As myasthenia gravis is found worldwide, future research should focus on recruiting patients and healthy controls from diverse, non-European individuals and patients with anti-MuSK and other autoantibodies.
In conclusion, we identified loci relevant to the pathogenesis of myasthenia gravis and confirmed the different genetic architectures among early-onset and late-onset cases. Among these, the discovery of risk variants within the CHRNA1 and CHRNB1 genes that encode subunits of the acetylcholine receptor is remarkable, and it provides insights into the mechanisms that may trigger the disease.
Materials and Methods
Participants and Study Design.
Myasthenia gravis samples were collected from collaborators in the United States and Italy. The US samples were collected from January 2010 to January 2011 from patients attending myasthenia gravis clinics at 14 centers throughout North America (7). Our group previously published a GWAS analysis based on 972 of the US cases and 1,977 of the US control subjects (7). These genotype and phenotype data are available on the database of Genotype and Phenotype (dbGaP) web portal (accession number phs000726). Blood samples were collected from Italian patients at the Catholic University Rome and Cisanello Hospital, Pisa. The diagnosis of myasthenia gravis was based on standard clinical criteria of characteristic fatigable weakness and electrophysiological and/or pharmacological abnormalities and confirmed by the presence of anti-acetylcholine receptor antibodies (7). Patients with positive test results for antibodies to muscle-specific kinase (anti-MuSK) were excluded from enrollment. We used publicly available genotype data from the dbGaP web portal for the US and Italian neurologically normal individuals for the control cohort. Written consent was obtained from all subjects enrolled in this study. This study was approved by the institutional review boards of all participating institutions, including Johns Hopkins University, the National Institute on Aging (protocol 03-AG-N329), the University of Pisa, and the Catholic University of Rome.
For replication, we used myasthenia gravis cases identified within the UK Biobank based on the ICD10 code G70.0 (31). Only samples identified as European and White British were included in the analysis. The control subjects were selected from a cohort of over 400,000 in a ratio of 20 controls for each case, using the MatchIt (version 3.0.2) R library. The control subjects were matched to cases using the “nearest” method based on age, gender, and uniform manifold approximation and projection (UMAP) components 1 and 2 to account for population structure.
Procedures.
The US and Italian myasthenia gravis cases were genotyped on HumanOmniExpress arrays (Illumina Inc.) at the National Institute on Aging. The UK Biobank samples were genotyped on UK BiLEVE Axiom or UK Biobank Axiom arrays (Affymetrix, ThermoFisher Scientific Inc.). The Illumina and Affymetrix arrays assayed over 730,000 and 850,000 SNPs.
Genome-Wide Association Analysis.
The current study analyzed data from 1,873 myasthenia gravis cases and 36,370 controls. This cohort included 972 cases and 1,977 controls used in a previous GWAS of myasthenia gravis (7). Genotype data from the US and Italian samples were processed using a standard quality control pipeline (32). The pipeline avoids P value inflation due to imbalances in the case and control cohort numbers. Samples were excluded for the following reasons: 1) excessive heterozygosity rate (exceeding +/− 0.15 F-statistic), a measure of DNA contamination; 2) low call rate (≤95%); 3) discordance between reported sex and genotypic sex; 4) duplicate samples (determined by pi-hat statistics > 0.8); and 5) non-European ancestry based on principal components analysis compared to the HapMap 3 Genome Reference Panel. Related samples (defined as having a pi-hat > 0.125) were included for imputation, but one pair member was removed before the association testing. Plotting of the principal components demonstrated that the study samples were centered around the European cohort of HapMap3, indicating the absence of population stratification (SI Appendix, Fig. S10).
Variants were excluded from the analysis for the following reasons: 1) monomorphic SNPs; 2) palindromic SNPs; 3) variants that showed nonrandom missingness between cases and controls (P value ≤ 1.0 × 10−4); 4) variants with haplotype-based nonrandom missingness (P value ≤ 1.0 × 10−4); 5) variants with an overall missingness rate of ≥5.0%; 6) nonautosomal variants (X, Y, and mitochondrial chromosomes); and 7) variants that significantly departed from Hardy-Weinberg equilibrium in the control cohort (P value ≤ 1.0 × 10−10).
Imputation was performed for the US and Italian cohorts against the Trans-Omics for Precision Medicine (TopMed) imputation reference panel (hg38) via minimac4 using data phased by Eagle v2.4 provided by TopMed Imputation Server (33). The US cohort was divided equally into two subsets before imputation. After imputation, variants were included in the analysis if they met the following criteria: 1) imputation quality score (R2) > 0.3 and 2) minor allele frequency > 0.001. The UK Biobank cohort was previously imputed using the Human Reference Consortium, UK10K, and 1000 Genomes reference panels (hg19) (34). After applying quality control filters, there were ∼32 to 42 million SNPs in 964 US myasthenia gravis cases and 35,580 US controls and 919 Italian cases and 2,478 Italian controls. Index variants from the discovery phase analysis were extracted from the UK Biobank dataset. Genomic positions were lifted over to hg38 before the association analysis using the 354 myasthenia cases and 7,080 controls. All samples were of European descent, and only unrelated samples were included in the analysis (SI Appendix, Table S6).
Statistical Analysis.
Logistic regression with dosage data was performed for the following cohorts: (i) all samples; (ii) early-onset myasthenia gravis, defined as an age of onset <40 y; and (iii) late-onset myasthenia gravis with onset age ≥40 y. Results were additionally stratified by sex in the early-onset and late-onset cohorts. The covariates were adjusted in each analysis with the optimum combination (age, sex, and principal components to account for population structure) as implemented by the step function (version 3.5.2) in R (SI Appendix, Table S6). GWAS was performed separately in the US, Italian, and UK cohorts. Systematic biases were assessed using quantile-quantile plots and genomic inflation factors (SI Appendix, Fig. S11). Meta-analysis was performed under the fixed-effects model, and variants with high heterogeneity (HetISq > 80) or minor allele frequency <1% were removed. The genome-wide significance level threshold was P value < 5.0 × 10−8, and the suggestive significance level was defined as P value < 5.0 × 10−7. The threshold for significance in the replication cohort was set as P value < 0.1 based on Bayesian inference of having the same direction effect as observed in the discovery cohort and correcting according to the number of variants selected for replication (P value threshold = 0.1/7 loci = 0.0143). Our previous GWAS analyzing 972 cases and 1,977 controls (7) had 5.2% power to detect significant risk variants under an additive model assuming a minor allele frequency of 0.05, an OR risk of 1.6, a disease prevalence of 20 per 100,000, and a significance threshold of 5.0 × 10−8. In contrast, our current effort involving 1,873 cases and 36,370 controls had 92.9% power to detect loci under the same assumptions (SI Appendix, Fig. S12).
Transcriptome-Wide Association Analysis.
Transcriptome-wide imputation was performed using the FUSION pipeline to predict the tissue-specific gene expression based on our GWAS summary statistics (9). Precomputed gene expression weights on GTEx (version 7) data from skeletal muscle, whole blood, and tibial nerve were used to estimate a gene’s association to disease risk (10). Gene association was considered significant if the following criteria were met: 1) TWAS P value < 0.05/number of genes per tissue type (skeletal muscle, n = 7,409 genes; whole blood, n = 6,007 genes; and tibial nerve, n = 9,657 genes) and 2) permutation P values < 0.05. Colocalization PPH4 was computed for genes with P value < 1.0 × 10−4 using the built-in COLOC function in FUSION. A PPH4 of ≥75% was considered strong evidence for the expression quantitative trait loci (eQTL)-GWAS pair influencing both the expression and the GWAS trait in a particular region.
HLA Fine-Mapping.
The HLA region was fine-mapped using the CookHLA (35) and HATK tools (36). HLA alleles were imputed against the SNP2HLA preformatted 1000 Genomes European reference panel mapped against the IMGT-HLA database.
Genetic Correlation Analysis.
The shared genetic risks between myasthenia gravis and other diseases were estimated using the Linkage Disequilibrium Score Regression method (13) using the LDSC tool (available on https://github.com/bulik/ldsc). Default parameters were used for analysis and using the precomputed linkage disequilibrium (LD) scores derived from the 1000 Genomes European dataset. We selected GWASes for each disease based on the availability of the corresponding summary statistics in the GWAS catalog (https://www.ebi.ac.uk/gwas/) and MRC IEU Open GWAS Project (https://gwas.mrcieu.ac.uk/) websites. Study selection was based on the availability within these repositories. The autoimmune summary statistics downloaded from the GWAS catalog were Crohn’s disease: GCST004132, rheumatoid arthritis: GCST002318, systemic lupus erythematosus: GCST005831, type 1 diabetes: GCST005536, and ulcerative colitis: GCST004133. Summary statistics downloaded from MRC IEU Open GWAS Project were multiple sclerosis: ieu-b-18, celiac disease: ebi-a-GCST000612, psoriasis: ukb-b-10537, and hypothyroidism: ukb-b-19732. Summary statistics from neurological disorders were obtained from Jansen et al. (37), Nalls et al. (38), Van Rheenen et al. (39), and Chia et al. (40).
Prioritization of Therapies for Myasthenia Gravis.
Priority Index analysis (R package Pi [version 2.2.1]) was applied to identify targets that may be amenable to therapy (i.e., druggable) for myasthenia gravis (14). Summary statistics from the overall GWAS were filtered to retain variants with minor allele frequency > 0.05 and P values > 0.05. Enrichment analysis of the prioritized genes was then performed using the REACTOME database (https://www.reactome.org/) to identify the immune and signal transduction pathways overrepresented by these genes (14).
Supplementary Material
Acknowledgments
This work was supported in part by the Intramural Research Programs of the NIH, National Institute on Aging (Z01-AG000949-02). The work was also supported by the Myasthenia Gravis Foundation (D.B.D. and B.J.T.), a generous bequest by Geraldine Weinrib, and a gift from Philip Swift. Support was provided by Mr. and Mrs. Don Brandon and the Department of Neurology, University of Kansas Medical Center. The dataset(s) used for the analyses described in this manuscript were obtained from the Age-Related Eye Disease Study (AREDS) Database found at https://www.nei.nih.gov/research/clinical-trials/age-related-eye-disease-studies-aredsareds2 through dbGaP accession number phs000001.v3.p1. Funding support for AREDS was provided by the National Eye Institute (N01-EY-0-2127). We thank the AREDS participants and the AREDS Research Group for their valuable contribution to this research. The Framingham Heart Study is conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI) in collaboration with Boston University (Contract No. N01-HC-25195 and HHSN268201500001I). This manuscript was not prepared in collaboration with investigators of the Framingham Heart Study and does not necessarily reflect the opinions or views of the Framingham Heart Study, Boston University, or NHLBI. Funding to support the Omni cohort recruitment, retention, and examination was provided by NHLBI Contract N01-HC-25195 and HHSN268201500001I as well as NHLBI grants R01-HL070100, R01-HL076784, R01-HL-49869, and U01-HL-053941. Research support to collect data and develop an application to support this project was provided by 3P50CA093459, 5P50CA097007, 5R01ES011740, and 5R01CA133996. The Women’s Health Initiative (WHI) program is funded by NHLBI, NIH, US Department of Health and Human Services through contracts HHSN268201600018C, HHSN268201600001C, HHSN268201600002C, HHSN268201600003C, and HHSN268201600004C. This manuscript was not prepared in collaboration with investigators of the WHI, has not been reviewed and/or approved by WHI, and does not necessarily reflect the opinions of the WHI investigators or the NHLBI. Funding support for WHI GARNET was provided through the NHGRI Genomics and Randomized Trials Network (GARNET) (Grant Number U01 HG005152). Assistance with phenotype harmonization and genotype cleaning as well as with general study coordination was provided by the GARNET Coordinating Center (U01 HG005157). Assistance with data cleaning was provided by the National Center for Biotechnology Information. Funding support for genotyping, which was performed at the Broad Institute of MIT and Harvard, was provided by the NIH Genes, Environment and Health Initiative (GEI) (U01 HG004424). The datasets used for the analyses described in this manuscript were obtained from dbGaP at https://www.ncbi.nlm.nih.gov/sites/entrez?db=gap through dbGaP accession numbers phs000001, phs000007, phs000101, phs000187, phs000196, phs000248, phs000292, phs000304, phs000315, phs000368, phs000394, phs000397, phs000404, phs000421, phs000428, phs000454, phs000615, phs000675, phs000801, and phs000869. The authors acknowledge the contribution of data from Hepatitis C Pathogenesis and the Human Genome supported by 1X01HG005271-01 and R01DA013324 and accessed through dbGaP to the analysis presented in this publication. Funding support for the Genes and Blood Clotting Study was provided through the NIH/NHLBI (R37 HL039693). The Genes and Blood Clotting Study is one of the Phase 3 studies as part of the Gene Environment Association Studies (GENEVA) under GEI. Assistance with genotype cleaning was provided by the GENEVA Coordinating Center (U01 HG004446). Funding support for DNA extraction and genotyping, which was performed at the Broad Institute, was provided by NIH/NHLBI (R37 HL039693). Additional support was provided by the Howard Hughes Medical Institute. The dataset(s) used for the analyses described in this manuscript were obtained from the dbGaP found at https://www.ncbi.nlm.nih.gov/gap through dbGaP accession number phs000368. Samples and associated phenotype data for the Genome-Wide Association Study (GWAS) of Polycystic Ovary Syndrome Phenotypes were provided by Andrea Dunaif, MD. We acknowledge the contribution of data from Genetic Architecture of Smoking and Smoking Cessation accessed through dbGaP. Funding support for genotyping, which was performed at the Center for Inherited Disease Research (CIDR), was provided by 1 X01 HG005274-01. CIDR is fully funded through a federal contract from NIH to Johns Hopkins University, contract number HHSN268200782096C. Assistance with genotype cleaning as well as with general study coordination was provided by the GENEVA Coordinating Center (U01 HG004446). Funding support for collection of datasets and samples was provided by the Collaborative Genetic Study of Nicotine Dependence (P01 CA089392) and the University of Wisconsin Transdisciplinary Tobacco Use Research Center (P50 DA019706, P50 CA084724). The dataset(s) used for the analyses described in this manuscript were obtained from the Genetics of Fuchs’ Endothelial Corneal Dystrophy (FECD) Study through dbGaP accession number phs000421. The grants that have funded the enrollment of the cases and controls to be used in this GWAS are R01EY016514 (DUEC, Principal Investigator (PI): Gordon Klintworth), R01EY016482 (CWRU, PI: Sudha Iyengar), and 1X01HG006619-01 (PI: Sudha Iyengar, Natalie Afshari). We thank the FECD participants and the FECD Research Group for their valuable contribution to this research. The authors acknowledge the contribution of data from CIDR-NIDA Study of HIV Host Genetics accessed through dbGaP. Funding support for genotyping, which was performed at the CIDR, was provided by 1 X01 HG005275-01A1. Funding support for collection of datasets and samples was provided by National Institute on Drug Abuse (NIDA) grants R01DA026141(Johnson); R01DA004212 (Watters); U01DA006908 (Watters); and R01DA009532 (Bluthenthal) as well as the San Francisco Department of Public Health; SAMHSA; and HRSA. The GWAS of Non-Hodgkin Lymphoma project was supported by the intramural program of the Division of Cancer Epidemiology and Genetics, National Cancer Institute, NIH. The datasets have been accessed through the NIH dbGaP under accession no. phs000801. A full list of acknowledgments can be found in the supplementary note [S. I. Berndt et al., Genome-wide association study identifies multiple risk loci for chronic lymphocytic leukemia. Nat. Genet. 45, 868–876 (2013), PMID: 23770605]. This study made use of data generated by investigators in the BEACON consortium through a grant funded by NIH (RO1CA136725) to Thomas L. Vaughan and David C. Whiteman (multiple PIs). In support of this work, T.L.V. was also supported by NIH grant KO5CA124911 and D.C.W. by a Future Fellowship grant FT0990987 from the Australia Research Council. R.H.R. is supported through philanthropic support from Dr. Peter Buck. Additional collaborators, sources of support, and origin of the data and biospecimens are listed in the following publication: D. M. Levine et al. A genome-wide association study identifies new susceptibility loci for esophageal adenocarcinoma and Barrett’s esophagus. Nat Genet. 45, 1487–1493 (2013). This research has been conducted using the UK Biobank Resource under application number 33601. This study used the high-performance computational capabilities of the Biowulf Linux cluster at NIH, Bethesda, MD (https://hpc.nih.gov/). We thank the Laboratory of Neurogenetics (NIH) staff for their collegial support and technical assistance.
Footnotes
Competing interest statement: R.J.B. served as a consultant for NuFactor and Momenta Pharmaceutical and receives research support from PTC Therapeutics, Ra Pharma, Orphazyme, Sanofi Genzyme, FDA OOPD, NIH, and Patient-Centered Outcomes Research Institute (PCORI). M.B. reports grant support from Muscular Dystrophy Association, ALS Association, ALS Recovery Fund, Kimmelman Estate, Target ALS, Eli Lilly and Company, and NIH during the conduct of the study. He also reports grant support from FDA, Centers for Disease Control and Prevention (CDC), and DOD; research support from Alexion Pharmaceuticals, UCB, Cytokinetics, Neuraltus, Biogen, and Orphazyme A/S; and personal fees from NMD Pharma, Ra Pharmaceuticals, Mitsubishi Tanabe, Avexis, UCB, and Denali outside the submitted work. V.C. served as a consultant for review and expert testimony for the Department of Health and Human Services and the Department of Justice under the Vaccine Injury and Compensation Program. Dr. Chaudhry has received a royalty for total neuropathy score (TNS) patented (through Johns Hopkins University) for the license of TNS use from AstraZeneca, Genentech, Seattle Genetics, Calithera Biosciences, Merrimack Pharmaceuticals, Levicept, and Acetylon Pharmaceuticals. M.M.D. serves or recently served as a consultant for Argenx Pharmaceuticals, Catalyst, CSL Behring, Kezar, Momenta, NuFactor, RMS Medical, Sanofi Genzyme, Shire Takeda, and Spark Therapeutics. Dr. Dimachkie received grants from Alexion Pharmaceuticals, Alnylam Pharmaceuticals, Amicus, Biomarin, Bristol-Myers Squibb, Catalyst, CSL Behring, FDA/OOPD, GlaxoSmithKline, Genentech, Grifols, Mitsubishi Tanabe Pharma, Muscular Dystrophy Association (MDA), NIH, Novartis, Sanofi Genzyme, Octapharma, Orphazyme, Sarepta Therapeutics, Shire Takeda, Spark, UCB Biopharma, Viromed, and TMA. A.E. was a member of the advisory board for Alexion Pharmaceuticals, a scientific award jury member for Grifols, and safety data monitor for UCB. M.F. has received honoraria for serving on advisory boards for Argenx Pharmaceuticals and Alexion Pharmaceuticals. M.F. also has research support from Catalyst, Ra pharma, Amicus, Orphazyme, Alexion Pharmaceuticals, Momenta, and Alnylam. J.F.H. reports research support and grants from Alexion Pharmaceuticals and Argenx Pharmaceuticals, Centers for Disease Control and Prevention, MDA, NIH (including the National Institute of Neurologic Disorders and Stroke and the National Institute of Arthritis and Musculoskeletal and Skin Disease), PCORI, and Ra Pharmaceuticals (now UCB Biosciences); honoraria from Alexion Pharmaceuticals, Argenx Pharmaceuticals, Immunovant, Ra Pharmaceuticals (now UCB Biosciences), Regeneron Pharmaceuticals, and Viela Bio; and nonfinancial support from Alexion Pharmaceuticals, Argenx Pharmaceuticals, Ra Pharmaceuticals (now UCB Biosciences), and Toleranzia. P.J.T. holds patents on the clinical testing and therapeutic intervention for the hexanucleotide repeat expansion of C9orf72 and has received grant funding from the Helsinki University Hospital, Finnish Academy, Sigrid Juselius Foundation, EU Marie Curie action, Biogen Finland, Roche Finland, Merck Finland, Sanofi Genzyme Finland, and Novartis Finland and has made consultations to Alexion Pharmaceuticals, Biogen, Novartis, Orion, Roche, Sanofi Genzyme, Santen, and Teva. H.J.K. is funded by the MDA (508240) and NIH grant U54NS115054; is a consultant for Alnylam Pharmaceuticals, Ra Pharmaceuticals, and UCB Pharmaceuticals; and is CEO of ARC Biotechnology, LLC, which receives support from the NIH (R41NS110331). He serves on the Editorial Board of Experimental Neurology. M.M.M. has received honoraria as a speaker and/or moderator from Alnylam, Akcea, Pfizer, and CSL Behring. She has served on Advisory Boards for Pfizer, Alnylam, and Akcea. She serves as an investigator for clinical trials with Alnylam and Biogen. S.M. has served on advisory board meetings for Alexion Pharmaceuticals and Argenx Pharmaceuticals. M.P. served on the advisory boards for CSL Behring, Alexion Pharmaceuticals, and Argenx Pharmaceuticals and has been a consultant for Momenta Pharmaceuticals. D.P.R. receives research funding from a Sponsored Research Agreement from Cabaletta Bioscience. B.J.T. holds patents on the clinical testing and therapeutic intervention for the hexanucleotide repeat expansion of C9orf72 and has received research grants from the Myasthenia Gravis Foundation, the Robert Packard Center for ALS Research, the ALS Association, the Italian Football Federation, the CDC, the MDA, Merck, and Microsoft Research. B.J.T. receives funding through the Intramural Research Program at NIH.
A complete list of the International Myasthenia Gravis Genomics Consortium is available in the SI Appendix.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2108672119/-/DCSupplemental.
Contributor Information
Collaborators: Yevgeniya Abramzon, Richard J. Barohn, Michael Benatar, Derrick Blackmore, Vinay Chaudhry, Adriano Chiò, Manisha Chopra, Andrea Corse, Anna De Rosa, Mazen M. Dimachkie, Amelia Evoli, Julaine Florence, Miriam Freimer, Melania Guida, James F. Howard, Jr, Theresa Jiwa, Henry J. Kaminski, John T. Kissel, Wilma J. Koopman, Bernadette Lipscomb, Michelangelo Maestri, Mariapaola Marino, Janice M. Massey, April McVey, Michelle M. Mezei, Srikanth Muppidi, Michael W. Nicolle, Joel Oger, Robert M. Pascuzzi, Mamatha Pasnoor, Alan Pestronk, Loredana Petrucci, Hannah A. Pliner, Carlo Provenzano, Alan E. Renton, Roberta Ricciardi, David P. Richman, Julie Rowin, Donald B. Sanders, Mario Sabatelli, Zaeem Siddiqi, Aimee Soloway, Laura Todi, Gil Wolfe, Charlie Wulf, Daniel B. Drachman, and Bryan J. Traynor
Data Availability
Summary GWAS statistics and the programming code used for analysis are deposited as Jupyter notebooks on https://github.com/ruthchia/MyastheniaGravis_AnalysisCode. Genotype data have been deposited in dbGAP (phs000726). Anonymized summary statistics data have been deposited in GWAS catalog (GCST90093061). Previously published data were used for this work [A. E. Renton et al., A genome-wide association study of myasthenia gravis. JAMA Neurol. 72, 396–404 (2015)].
References
- 1.Alshekhlee A., Miles J. D., Katirji B., Preston D. C., Kaminski H. J., Incidence and mortality rates of myasthenia gravis and myasthenic crisis in US hospitals. Neurology 72, 1548–1554 (2009). [DOI] [PubMed] [Google Scholar]
- 2.Carr A. S., Cardwell C. R., McCarron P. O., McConville J., A systematic review of population based epidemiological studies in Myasthenia Gravis. BMC Neurol. 10, 46 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vincent A., Palace J., Hilton-Jones D., Myasthenia gravis. Lancet 357, 2122–2128 (2001). [DOI] [PubMed] [Google Scholar]
- 4.Vincent A., Newsom-Davis J., Acetylcholine receptor antibody as a diagnostic test for myasthenia gravis: Results in 153 validated cases and 2967 diagnostic assays. J. Neurol. Neurosurg. Psychiatry 48, 1246–1252 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gilhus N. E., Myasthenia Gravis. N. Engl. J. Med. 375, 2570–2581 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Vincent A., Drachman D. B., Myasthenia gravis. Adv. Neurol. 88, 159–188 (2002). [PubMed] [Google Scholar]
- 7.Renton A. E., et al. , A genome-wide association study of myasthenia gravis. JAMA Neurol. 72, 396–404 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seldin M. F., et al. , Genome-wide association study of late-onset myasthenia gravis: Confirmation of TNFRSF11A and identification of ZBTB10 and three distinct HLA associations. Mol. Med. 21, 769–781 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gusev A., et al. , Integrative approaches for large-scale transcriptome-wide association studies. Nat. Genet. 48, 245–252 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.GTEx Consortium, The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 369, 1318–1330 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mao Z. F., et al. , Frequency of autoimmune diseases in myasthenia gravis: A systematic review. Int. J. Neurosci. 121, 121–129 (2011). [DOI] [PubMed] [Google Scholar]
- 12.Gotaas H. T., Skeie G. O., Gilhus N. E., Myasthenia gravis and amyotrophic lateral sclerosis: A pathogenic overlap. Neuromuscul. Disord. 26, 337–341 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Bulik-Sullivan B. K., et al. , Schizophrenia Working Group of the Psychiatric Genomics Consortium, LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 47, 291–295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fang H., et al. , ULTRA-DD Consortium, A genetics-led approach defines the drug target landscape of 30 immune-related traits. Nat. Genet. 51, 1082–1091 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ching K. H., et al. , Recombinant expression of the AChR-alpha1 subunit for the detection of conformation-dependent epitopes in Myasthenia Gravis. Neuromuscul. Disord. 21, 204–213 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lazaridis K., Tzartos S. J., Autoantibody specificities in myasthenia gravis; Implications for improved diagnostics and therapeutics. Front. Immunol. 11, 212 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Finsterer J., Congenital myasthenic syndromes. Orphanet J. Rare Dis. 14, 57 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pugliese A., et al. , The insulin gene is transcribed in the human thymus and transcription levels correlated with allelic variation at the INS VNTR-IDDM2 susceptibility locus for type 1 diabetes. Nat. Genet. 15, 293–297 (1997). [DOI] [PubMed] [Google Scholar]
- 19.Romi F., Hong Y., Gilhus N. E., Pathophysiology and immunological profile of myasthenia gravis and its subgroups. Curr. Opin. Immunol. 49, 9–13 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Giraud M., et al. , An IRF8-binding promoter variant and AIRE control CHRNA1 promiscuous expression in thymus. Nature 448, 934–937 (2007). [DOI] [PubMed] [Google Scholar]
- 21.Altiok N., Bessereau J. L., Changeux J. P., ErbB3 and ErbB2/neu mediate the effect of heregulin on acetylcholine receptor gene expression in muscle: Differential expression at the endplate. EMBO J. 14, 4258–4266 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Si J., Luo Z., Mei L., Induction of acetylcholine receptor gene expression by ARIA requires activation of mitogen-activated protein kinase. J. Biol. Chem. 271, 19752–19759 (1996). [DOI] [PubMed] [Google Scholar]
- 23.Creary L. E., et al. , Next-generation sequencing identifies extended HLA class I and II haplotypes associated with early-onset and late-onset myasthenia gravis in Italian, Norwegian, and Swedish populations. Front. Immunol. 12, 667336 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Drachman D. B., How to recognize an antibody-mediated autoimmune disease: Criteria. Res. Publ. Assoc. Res. Nerv. Ment. Dis. 68, 183–186 (1990). [PubMed] [Google Scholar]
- 25.Dalmau J., Graus F., Antibody-mediated encephalitis. N. Engl. J. Med. 378, 840–851 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Ricken G., et al. , Detection methods for autoantibodies in suspected autoimmune encephalitis. Front. Neurol. 9, 841 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Green J. D., et al. , Epidemiological evidence for a hereditary contribution to myasthenia gravis: A retrospective cohort study of patients from North America. BMJ Open 10, e037909 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hay M., Thomas D. W., Craighead J. L., Economides C., Rosenthal J., Clinical development success rates for investigational drugs. Nat. Biotechnol. 32, 40–51 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Shiina T., Hosomichi K., Inoko H., Kulski J. K., The HLA genomic loci map: Expression, interaction, diversity and disease. J. Hum. Genet. 54, 15–39 (2009). [DOI] [PubMed] [Google Scholar]
- 30.Pollard M. O., Gurdasani D., Mentzer A. J., Porter T., Sandhu M. S., Long reads: Their purpose and place. Hum. Mol. Genet. 27 (R2), R234–R241 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sudlow C., et al. , UK biobank: An open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 12, e1001779 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nicolas A., et al. , ITALSGEN Consortium; Genomic Translation for ALS Care (GTAC) Consortium; ALS Sequencing Consortium; NYGC ALS Consortium; Answer ALS Foundation; Clinical Research in ALS and Related Disorders for Therapeutic Development (CReATe) Consortium; SLAGEN Consortium; French ALS Consortium; Project MinE ALS Sequencing Consortium, Genome-wide analyses identify KIF5A as a novel ALS gene. Neuron 97, 1268–1283.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taliun D., et al. , Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature 590, 290–299 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bycroft C., et al. , The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cook S., et al. , Accurate imputation of human leukocyte antigens with CookHLA. Nat. Commun. 12, 1264 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choi W., Luo Y., Raychaudhuri S., Han B., HATK: HLA analysis toolkit. Bioinformatics 37, 416–418 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jansen I. E., et al. , Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat. Genet. 51, 404–413 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nalls M. A., et al. , 23andMe Research Team; System Genomics of Parkinson’s Disease Consortium; International Parkinson’s Disease Genomics Consortium, Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: A meta-analysis of genome-wide association studies. Lancet Neurol. 18, 1091–1102 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Rheenen W., et al. , PARALS Registry; SLALOM Group; SLAP Registry; FALS Sequencing Consortium; SLAGEN Consortium; NNIPPS Study Group, Genome-wide association analyses identify new risk variants and the genetic architecture of amyotrophic lateral sclerosis. Nat. Genet. 48, 1043–1048 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chia R., et al. , American Genome Center, Genome sequencing analysis identifies new loci associated with Lewy body dementia and provides insights into its genetic architecture. Nat. Genet. 53, 294–303 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Summary GWAS statistics and the programming code used for analysis are deposited as Jupyter notebooks on https://github.com/ruthchia/MyastheniaGravis_AnalysisCode. Genotype data have been deposited in dbGAP (phs000726). Anonymized summary statistics data have been deposited in GWAS catalog (GCST90093061). Previously published data were used for this work [A. E. Renton et al., A genome-wide association study of myasthenia gravis. JAMA Neurol. 72, 396–404 (2015)].