Significance
Genetic mutations fuel organismal evolution but can also cause disease. As proteins are the cell’s workhorses, the ways in which mutations can disrupt their structure, stability, function, and interactions have been studied extensively. However, proteins evolve and function in a cellular context, and our ability to relate changes in protein sequence to cell-level phenotypes remains limited. In particular, the molecular mechanism underlying most disease-associated mutations is unknown. Here, we show that mutations changing a protein’s surface chemistry can dramatically impact its supramolecular self-assembly and localization in the cell. These results highlight the complex nature of genotype–phenotype relationships with a simple system.
Keywords: protein evolution, protein interactions, genotype–phenotype map
Abstract
Understanding the molecular consequences of mutations in proteins is essential to map genotypes to phenotypes and interpret the increasing wealth of genomic data. While mutations are known to disrupt protein structure and function, their potential to create new structures and localization phenotypes has not yet been mapped to a sequence space. To map this relationship, we employed two homo-oligomeric protein complexes in which the internal symmetry exacerbates the impact of mutations. We mutagenized three surface residues of each complex and monitored the mutations’ effect on localization and assembly phenotypes in yeast cells. While surface mutations are classically viewed as benign, our analysis of several hundred mutants revealed they often trigger three main phenotypes in these proteins: nuclear localization, the formation of puncta, and fibers. Strikingly, more than 50% of random mutants induced one of these phenotypes in both complexes. Analyzing the mutant’s sequences showed that surface stickiness and net charge are two key physicochemical properties associated with these changes. In one complex, more than 60% of mutants self-assembled into fibers. Such a high frequency is explained by negative design: charged residues shield the complex from self-interacting with copies of itself, and the sole removal of the charges induces its supramolecular self-assembly. A subsequent analysis of several other complexes targeted with alanine mutations suggested that such negative design is common. These results highlight that minimal perturbations in protein surfaces’ physicochemical properties can frequently drive assembly and localization changes in a cellular context.
Understanding genotype to phenotype relationships is crucial to predict the molecular consequences of mutations (1). At the protein level, alanine scans have revealed how individual residues contribute to protein function, stability, and binding affinity (2–4). More recently, systematic mappings have been widely used to connect sequence variability to changes in protein structure (5, 6), stability (7–9), solubility (10), and functionality (2, 11–14). Similar efforts have been made to map the impact of mutations in protein–ligand (15, 16) and protein–protein interactions (17–21).
However, mutations can impact proteins beyond their stability, function, or existing interactions with specific partners or ligands. Sequences can also encode how proteins distribute spatially in cells, either by addressing them to membrane-bound compartments (22) or by inducing their self-assembly into large polymeric structures (23–27) and membraneless compartments (28, 29). While changes in protein self-assembly and localization can serve a functional purpose in adaptation (30–36), they can also lead to disease (37). For example, the supramolecular self-assembly of hemoglobin and γD-crystallin cause sickle-cell disease and cataracts, respectively (38, 39). The mislocalization of nuclear proteins TDP-43 and FUS in the cytosol is associated with amyotrophic lateral sclerosis disease (40, 41), and the mislocalization of Ataxin-3 to the nucleus has been implicated in spinocerebellar ataxia type 3 disease (42). It is therefore critical to characterize principles by which mutations can trigger such supramolecular self-assembly and mislocalization.
Symmetry is frequent in proteins (37, 43) and is a crucial property promoting their self-assembly into high-order structures (44–50). Indeed, a strong enrichment in symmetric homo-oligomers among natural filament-forming proteins has been reported (37). Previous work has also shown that point mutations to two hydrophobic amino acids—leucine and tyrosine—frequently led symmetric homo-oligomers to assemble into high-order assemblies. However, whether other types of amino acids would display a similar potential, whether they would do so often, and whether additional phenotypes of assembly and localization could emerge upon mutation remains unknown.
Here, we assess the potential of mutations to trigger such changes in protein assembly and localization in vivo. We targeted two homo-oligomeric protein complexes and randomly mutated three neighboring residues at the surface of each complex. We expressed the mutants fused to a fluorescent protein to track their spatial distribution in yeast cells. We found that a vast sequence space led to changes in protein assembly and localization in both proteins with three predominant phenotypes: nuclear localization, the formation of filaments, and the formation of puncta. Sequencing of the mutants revealed that increasing surface stickiness frequently promoted nuclear localization in one of the two proteins. Surprisingly, in the other protein, a loss of negatively charged residues was sufficient to trigger protein self-assembly, with fibers frequently forming regardless of the type of mutation, including to alanine and glycine. We also observed that four out of eight additional complexes analyzed underwent supramolecular self-assembly or a change in cellular localization when surface charges were mutated to alanine, implying that negative design against supramolecular self-assembly and mislocalization is common among symmetric homo-oligomers.
Results
A Plasmid Library of Surface Mutants Shows Frequent Self-Assembly and Nuclear Localization Phenotypes.
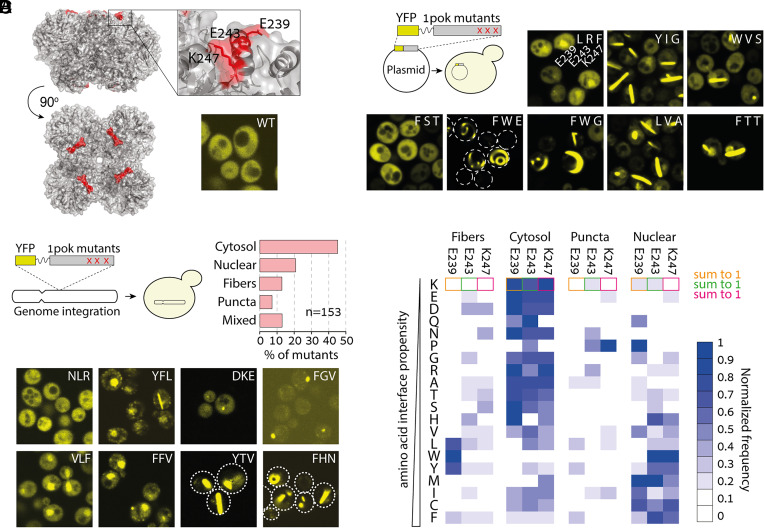
We sought to characterize how frequently mutations can trigger new protein assemblies, change protein localization, and identify which types of mutations are most likely to do so. We initially focused on a homo-octameric dipeptidase from Escherichia coli, hereafter referred to by its Protein Data Bank (PDB) code 1pok (Fig. 1A). To track the self-assembly and localization of the protein in cells, we fused the subunit forming the octamer to a yellow fluorescent protein (YFP; further details of the constructs are provided in SI Appendix, Table S1). We then created a plasmid library by site-saturation mutagenesis of three solvent-exposed residues (E239/E243/K247) located in an alpha helix (Fig. 1A). We specifically targeted those residues because a triple leucine mutant at these positions was previously observed to form fibers. We transformed the plasmid library in yeast cells and imaged them by fluorescence microscopy (Fig. 1B). While the wild-type protein exhibited a homogeneous and cytosolic localization, the mutants frequently formed micrometer-long filaments or puncta or localized to the nucleus (Fig. 1 A and B).
Fig. 1.
A mutant library reveals a wide diversity of localization phenotypes. (A) Structure of an octameric protein complex from E. coli (PDB code 1POK). Expression of the wild-type homomer fused to a YFP exhibits cytosolic localization in yeast cells. (B) We created a plasmid library with random mutations at three solvent-exposed positions located at the apex of 1pok and transformed the library into yeast cells. Fluorescence microscopy images of cells showing various phenotypes. The identity of the mutations for the residues 239/243/247 is overlaid onto each image. (C) A different mutant library was created using a genome integration cassette. Subsequent transformation of the library into yeast cells gave genomically integrated mutants. Cells expressing the mutants showed four main localization phenotypes with frequencies given in the bar plot. (Bottom) We show fluorescence microscopy images of yeast cells displaying eight representative phenotypes. The mutations at residues 239/243/247 are overlaid onto each image. (D) Frequency of amino acids at each position (indicated on Top) normalized across phenotypes. Amino acids observed in mutants (y-axis, Left) are ordered by increasing interaction propensity or “stickiness” (51). The sum of frequencies for a given mutation equals 1.
We subsequently PCR amplified the regions harboring the mutations to sequence them and relate the various phenotypes to specific amino acid identities. However, sequencing showed that a majority of clones were cotransformants of up to four different plasmids. Consequently, isolation of cells harboring a unique plasmid required multiple streaking steps, making it impossible to evaluate the different phenotypes’ frequencies and relate them to genotypes. Nevertheless, sequencing 20 isolated mutants suggested that filaments and nuclear localization were frequent outcomes of increasing the protein’s surface hydrophobicity (Fig. 1B). Notably, one mutant (F/W/E) showed a phenotype with curved filaments growing along the plasma membrane and the nuclear envelope.
Genome-Integrated Mutant Libraries Allow Quantifying Phenotype Frequencies.
To overcome the problem of cotransformation with multiple plasmids, we created a second library based on a cassette suitable for genome integration. With this library, each transformant carried a unique set of mutations. In agreement with our initial results, the fluorescence microscopy of 220 isolated mutants revealed the frequent formation of fibers and puncta as well as nuclear localization (Dataset S1). Sequencing enabled us to relate the phenotype of each mutant to the underlying mutations. After excluding strains containing deletions, insertions, and stop codons, we obtained 153 unique variants exhibiting four main phenotypes (Fig. 1C): 20.9% displayed nuclear localization, 13.1 and 7.2% showed fibers and puncta, respectively, and 45.7% of the mutants remained cytosolic like the wild-type sequence. In addition, 13.1% of mutants showed a combination of phenotypes such as cytosolic localization with rare puncta or a combination of both fibers and puncta.
We next examined whether specific mutations were associated with particular phenotypes. Given a mutated position (e.g., E239), we calculated the frequency of a target amino acid (e.g., mutation to L) for each of the four main phenotypes, excluding mutants showing mixed phenotypes. For example, out of 14 mutants harboring a leucine at position E239, nine formed fibers, four formed puncta, one was cytosolic, and none exhibited nuclear localization. Thus, the normalized frequency of leucine-associated fibers at position E239 is 9/14 = 0.64. These normalized frequencies for the four phenotypes reveal that fiber formation is predominantly associated with a mutation to L, W, or Y at position 239 (Fig. 1D). The cytosolic phenotype was enriched in mutations to polar amino acids and was favored by mutations to amino acids with low interaction propensity [i.e., nonsticky amino acids as defined previously (51)]. Interestingly, nuclear localization appears driven by interaction propensity in agreement with a recent report (52). Lastly, puncta formation did not show a strong association with a specific set of amino acids except for a large proportion of mutations to proline, suggesting that the local unfolding of the alpha helix might partially explain this phenotype (Fig. 1D). Thus, considering the protein 1pok, mutations readily induced changes in its localization and self-assembly phenotypes, and the physicochemical properties of the mutated residues were associated with these changes. Notably, sticky amino acids promoted both new self-assemblies and nuclear localization.
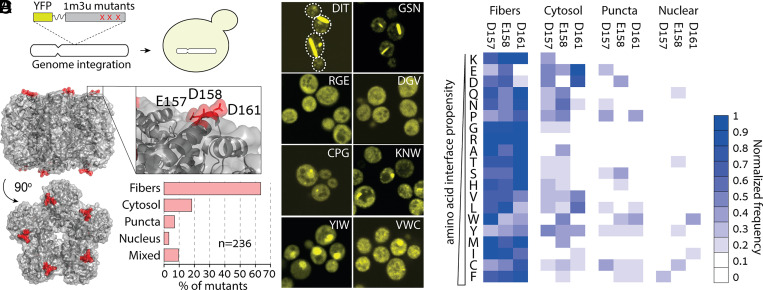
To assess whether mutations could reproducibly induce new assembly and localization phenotypes, we targeted a different protein (Fig. 2A): a homo-decameric ketopantoate methyltransferase from E. coli, hereinafter referred to by its PDB identifier 1m3u (Fig. 2B). Likewise, we created a library of mutants at three solvent-exposed residues located in an alpha helix. We picked 400 strains, of which 237 carried a unique sequence without stop codons, insertions, or deletions. The phenotypes of the mutants fell into the same four main groups (Dataset S2), with a staggering 63.6% of mutants forming fibers, 2.5% localizing to the nucleus, 6.4% forming puncta, 9.7% showing mixed localizations, and only 17.8% remaining cytosolic (Fig. 2 B and C). The heatmap of a mutation’s normalized frequency for these four phenotypes revealed a different picture than that of 1pok: all types of amino acids promoted fiber formation (Fig. 2D). Similarly, the cytosol, puncta, and nuclear localization groups did not exhibit a clear association with a specific class of amino acids (Fig. 2D).
Fig. 2.
E. coli ketopantoate transferase shows a striking tendency for fiber formation upon mutation. (A) We created a YFP-fused 1m3u mutant library in a cassette for genome integration and transformed it into yeast cells. (B) Structure of the decameric ketopantoate methyltransferase from E. coli (PDB code 1M3U). The mutant library targeted three solvent-exposed residues in an alpha helix located at the apex of the complex. Upon expression in yeast cells, mutants displayed four main phenotypes: cytosolic, nuclear, puncta, and fiber, with the latter found in >60% of the mutants. (C) Fluorescence microscopy images of yeast cells expressing eight representative mutants. The identity of mutated residues at positions 157/158/161 is indicated within each image. (D) Heatmap showing the normalized frequency of mutations of each amino acid type across the four phenotypes.
Overall, both mutant libraries imply that self-assembly and nuclear localization can frequently emerge by surface mutations in homo-oligomers. Indeed, less than 50% of random mutants remained cytosolic in both libraries.
Stickiness and Charge Are the Main Determinants Modulating Protein Assembly and Nuclear Localization at the Level of Single Cells.
We have seen so far that mutations in both homo-oligomers frequently triggered their self-assembly and nuclear localization. However, the relationship between physicochemical changes introduced by the mutations and changes in assembly and localization appeared different for the two proteins studied, which motivates a more quantitative analysis of this genotype–phenotype relationship. Notably, a significant number of strains showed incomplete penetrance of a particular phenotype or even mixed phenotypes. For example, while some strains displayed fibers in most of the cells, in other strains, only a small percentage of cells contained fibers (SI Appendix, Fig. S1).
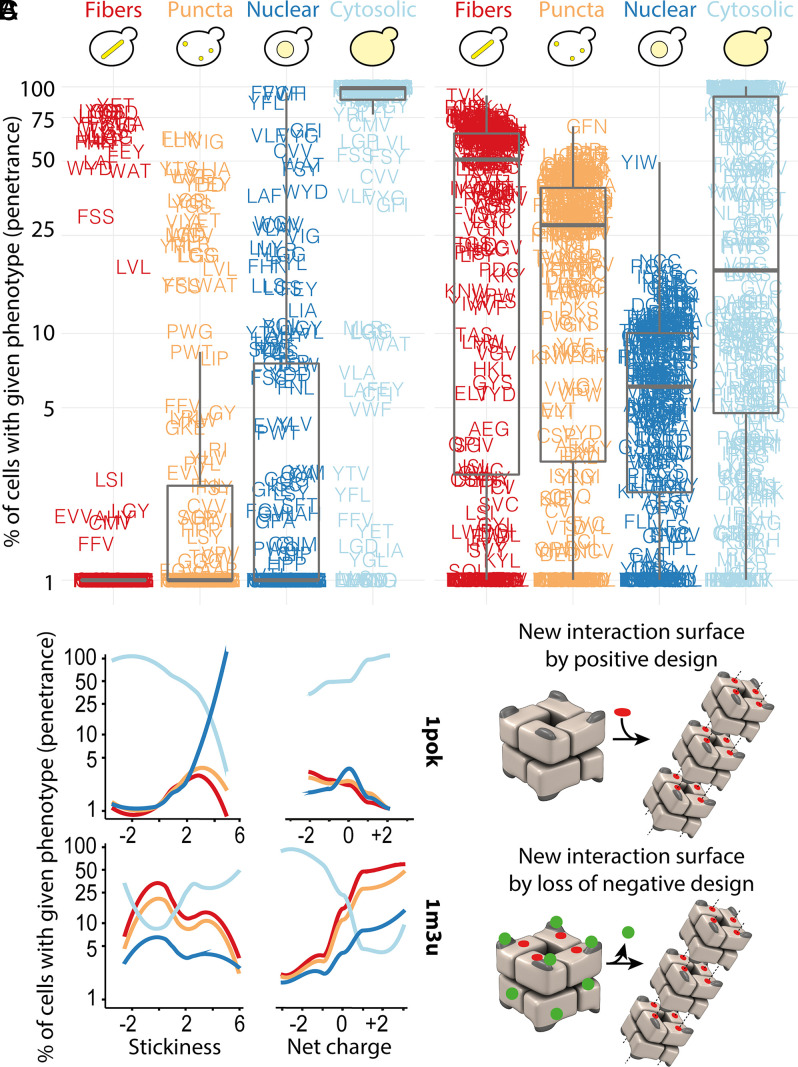
To capture these phenotypic differences in a quantitative manner, we automatically analyzed the fluorescence properties within single cells and assigned each cell to one or more of the four possible phenotypic categories (Materials and Methods and SI Appendix, Fig. S2). This approach enabled us to calculate the fraction of cells exhibiting a particular phenotype (i.e., a phenotype’s penetrance) for each mutant strain. The penetrance distribution for each phenotype is shown in Fig. 3A. We observe, for example, that fibers are identified in 1.9, 18.6, and 79.6% of cells for the 1pok mutants L/G/Y, L/V/L, and Y/G/L, respectively (with each triplet corresponding to the positions E239/E243/K247).
Fig. 3.
Physicochemical properties of amino acids trigger different responses in 1pok and 1m3u. (A) Percentage of cells of a given mutant of 1pok and 1m3u displaying fibers, puncta, cytosolic, and nuclear localization (phenotype penetrance). (B) Phenotype penetrance as a function of the summed interaction propensity and net charge of the mutated amino acids for 1pok and 1m3u. (C) Phenotypic changes require an increased stickiness in the case of 1pok, whereas the sole removal of charge appears sufficient to induce phenotypic changes in 1m3u, suggesting that charges represent a feature of negative design in this complex.
We analyzed the association between the penetrance of each phenotype and two physicochemical features of the mutated amino acids: their summed interaction propensity and their net charge (Fig. 3B and SI Appendix, Fig. S3). Confirming our initial observation, both proteins exhibited strikingly different patterns. On the one hand, the summed interaction propensity of the triplet was a strong predictor of cytosolic localization in the case of 1pok: 27 and 22 mutants showed an interaction propensity score <0 or >3, and among them, 99 and 46% of cells showed a cytosolic localization, respectively (P = 7e-6, Welch’s t test). In contrast, considering 1m3u, 38 and 37 mutants have an interaction propensity score below 0 or above 3, and these exhibit a similar fraction of cells with cytosolic localization (33.2 and 46.6%, respectively, P = 0.15). On the other hand, the cytosolic localization of 1m3u was well explained by the net charge of the mutations: among 27 and 61 mutants with a net charge ≤−1 or ≥+1, the fraction of cells showing a cytosolic localization was significantly different (77.2 and 14.2%, respectively, P = 8.2e-11). In contrast, the net charge failed to predict cytosolic localization for 1pok. Using the same charge criteria, we find 24 and 26 mutants among which cytosolic localization occurs in 79 and 94% of cells, respectively (P = 0.1).
We extended this correlation analysis by considering 20 additional amino acid features, including secondary structure formation propensity, size, hydrophobicity, etc. (SI Appendix, Fig. S4 and Dataset S3). This analysis confirmed that 1pok and 1m3u respond differently to specific physicochemical changes. While the interaction propensity, hydrophobicity, and aromaticity of the residues were the main properties triggering changes in 1pok localization and assembly, 1m3u was primarily impacted by the net charge as we saw before. We carried out a partial correlation analysis to identify these properties’ independent contributions (SI Appendix, Fig. S5). This analysis also confirmed the same picture, whereby stickiness is the primary determinant associated with the penetrance of all four phenotypes in 1pok. In contrast, the net charge of the mutations was the main determinant in the case of 1m3u.
Altogether, these results reveal that interaction propensity and charges are central modulators for self-assembly and nuclear localization in these two proteins. These results also highlight that changes in such properties on protein surfaces can trigger different responses in symmetric proteins. For some surface patches like that targeted in 1pok, self-assembly phenotypes may arise as a response to increments in surface hydrophobicity or stickiness, a change we can regard as positive design in the context of protein interactions. For other surface patches, like that targeted in 1m3u, neutralizing surface charges triggers new phenotypes. Remarkably, in this context, the target amino acid’s identity has little influence on the outcome, indicating that the sole removal of charges drives new phenotypes, particularly self-assembly into fibers in this case. This observation implies that charged residues at the targeted surface in 1m3u act as “negative design” elements (Fig. 3C), which we examine next.
Charged Gatekeeper Residues Protect against Uncontrolled Assembly and Relocalization In Vivo.
The desolvation of hydrophobic residues that accompanies their burial at protein interfaces is energetically favorable (53). For this reason, it is expected that mutations to hydrophobic amino acids are more likely to trigger new interaction sites than mutations to hydrophilic ones. For instance, in sickle-cell disease, a charged and polar glutamic acid is mutated to a hydrophobic valine in hemoglobin’s beta chain, inducing its assembly into filaments (38). This scenario is consistent with the mutant library of 1pok, whereby self-assembly is associated with positive design in the form of a hydrophobic amino acid required at position 239. In contrast, the self-assembly of 1m3u into fibers and puncta occurred not only in response to mutations toward hydrophobic residues but also with polar and charged residues (SI Appendix, Fig. S6A). For example, mutants N/S/S, T/D/R, and R/Q/T (at positions D157/D158/D161) consisted exclusively of polar residues and were among the fiber-forming variants (Dataset S2). These mutants suggest that the sole elimination of the negatively charged residues suffices to create a new self-interaction and induce the self-assembly of 1m3u into fibers. This idea is reminiscent of negative design (54–59) in which gatekeeper residues prevent proteins from folding into nonnative conformations (60, 61) and from engaging in nonnative interactions (54). Indeed, removing elements of negative design can promote intermolecular contacts and protein crystallization (62).
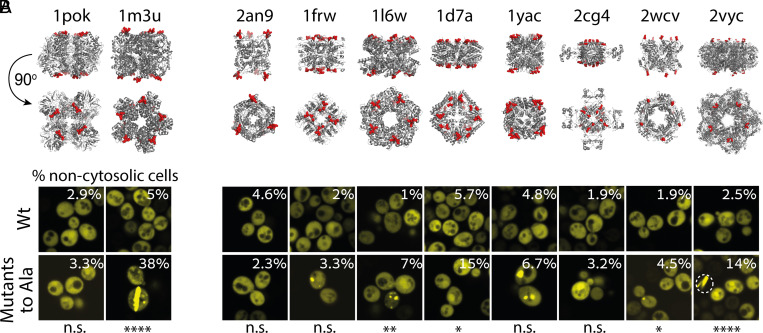
To test whether disabling the negatively charged residues sufficed to trigger new self-interactions, we created specific triple mutants to alanine. Interestingly, the mutations drove the formation of fibers and puncta in 1m3u, whereas 1pok remained soluble (Fig. 4A). We observed a similar outcome when examining the effect of triple glycine mutants of 1pok and 1m3u that existed in the mutant libraries (Datasets S1 and S2 and SI Appendix, Fig. S7). Taken together, these results suggest that the self-assembly of 1pok into fibers requires positive design, whereas removal of charges at the surface of 1m3u is sufficient to trigger its assembly into fibers.
Fig. 4.
Negative design against self-assembly is common in symmetric proteins. (A) Fluorescence microscopy images of yeast cells expressing the wild-type and the alanine mutants in 1pok and 1m3u. (B) Fluorescence microscopy images of yeast cells expressing eight additional symmetric complexes and their alanine mutant variants. The percentage of cells with a noncytosolic phenotype for each strain is overlaid onto the image. Statistical significance: ****P < 0.0001,***P < 0.001, **P < 0.01, *P < 0.05, and n.s. not significant.
To investigate this idea further, we created alanine mutants for eight additional homomers, which we refer to by their PDB code (Fig. 4B and SI Appendix, Table S2). We introduced two to four mutations at positions previously shown to trigger the formation of fibers when mutated to leucine or tyrosine (44). The mutations to alanine increased the penetrance of noncytosolic phenotypes in seven out of the eight complexes, of which four show statistical significance. These alanine mutations resulted in more frequent puncta (1l6w, 1frw, 1yac, and 2wcv), fibers (2vyc), and nuclear localization (1d7a and 2cg4, Fig. 4B and SI Appendix, Fig. S7). Thus, the sole neutralization of surface charges can, by itself, often trigger new protein assemblies and change a protein’s subcellular localization.
Importantly, these observations describe the properties of the surface patches targeted by mutations. To examine whether these two proteins exhibit different self-assembly potentials, we overexpressed the wild-type sequences. We reasoned that the higher apparent potential of 1m3u to self-assemble might drive a concentration-dependent assembly. Indeed, expression from a multicopy plasmid led 1m3u to assemble into fibers in a majority of cells, whereas it had comparatively little effect on the assembly of 1pok (SI Appendix, Fig. S6B).
Interestingly, even though local changes in stickiness and charges were identified as the main physicochemical properties modulating the assembly of these two proteins, both protein surfaces display remarkably similar stickiness values and charge distribution (SI Appendix, Fig. S6C). Thus, the nature of the different mutational routes required for self-assembly cannot be fully explained by physicochemical properties of the surface. Additional structural factors such as surface topology must be at play, and future work will be needed to understand and model these factors.
Discussion and Conclusions
Here, we show that mutations at the surface of two symmetric complexes frequently trigger their self-assembly into puncta and fibers as well as changes in their subcellular localization. Furthermore, we report two distinct mutational pathways driving these changes: one involves mutations increasing surface stickiness, another the sole removal of surface charges. This result prompted us to analyze eight additional symmetric complexes and ask whether mutating surface charges to alanine could suffice to trigger new assembly and localization phenotypes. We detected such changes in seven out of eight protein complexes investigated, and for four, the changes were statistically significant. These mutations were located at the apex of the quaternary structure (Fig. 4). In previous work, we predicted computationally that these regions are subject to negative design (44, 49), and here, we validated this prediction experimentally. Future work will be needed to identify whether mutations in other parts of the structure (i.e., farther from the “apex”) also frequently trigger changes in protein assembly and subcellular localization phenotypes.
The two mutational pathways we observed also occur among natural protein assemblies. Indeed, numerous protein-forming filaments are stabilized by the interaction of specific amino acids that are typically hydrophobic or aromatic. For example, glutamic acid to valine mutation is causing hemoglobin to form filaments and leads to sickle-cell disease (38). In another example, the polymerization of yeast hexokinase into filaments occurs when a tyrosine inserts in a hydrophobic pocket because of a ligand-triggered conformational change (30). Similar interactions are also driving the filament formation of human inosine-5′-monophosphate dehydrogenase (IMPDH) (63), the E. coli and human cytidine triphosphate synthetase (CTPS) (64), and the drug-induced polymerization of the oncogenic transcription factor BCL6 (65).
Interestingly, the increase in self-assembly propensity seen in quiescent cells is associated with changes in pH (34, 66) and might be reminiscent of the negative design principle observed in this work. Indeed, a decrease in pH may neutralize surface charges and unlock cryptic interaction sites, promoting self-assembly in those conditions. Similarly, oxidative damage associated with aging cells promotes aggregation through charge neutralization (67) and also could trigger protein assembly. Future experiments aimed to map the sequence–assembly relationship in different growth conditions or at different pH will help singling out sequence determinants for such condition-dependent protein assembly.
Certain mutations triggered self-assembly in a subpopulation of cells only. We ascertained that such incomplete penetrance was not just a consequence of a technical bias of our imaging mode (SI Appendix, Fig. S8 and Materials and Methods). Instead, it could be linked to differences in expression levels across individual cells. Alternatively, factors such as partial misfolding and conformational heterogeneity associated to, for example, prolines mutations (68, 69) or rare nucleation events (70), might explain the low penetrance of some puncta and fiber phenotypes. More work will be needed to examine these hypotheses.
A striking number of mutants of the 1pok and 1m3u libraries and the alanine mutants of two additional complexes (1d7a and 2cg4) exhibited nuclear localization. The nuclear pore complex has a diffusion barrier composed of intrinsically disordered proteins rich in phenylalanine and glycine. It acts as a molecular sieve with a passive-diffusion size limit of about 40 kDa. Larger macromolecules do not efficiently cross the diffusion barrier and must be ferried through by nuclear transporters (71). All the protein complexes used in this work considerably exceed the limit of passive diffusion (SI Appendix, Tables S1 and S2). Specifically, 1m3u and 1pok have molecular weights in the range of 280 and 320 kDa, respectively. Since they lack a nuclear localization signal, they should not be recognized by nuclear transporters, and, therefore, they should be excluded from the nucleus. However, in agreement with previous reports (52, 72), we found that surface hydrophobicity promotes the nuclear localization of 1pok despite its large size. The dysregulation of nucleocytoplasmic localization is implicated in neurodegenerative disorders as well as cancer (73). Our results imply that such dysregulation of localization can occur through noncanonical pathways that will need to be characterized in future work.
This work paves the way to understand how changes in a protein’s surface chemistry can impact its spatial distribution in the cell by modulating its self-assembly state and nucleocytoplasmic localization. A surprisingly wide sequence space drove assembly and localization changes in the two proteins that we studied. The fact that proteins are marginally soluble (74) means we could expect such assembly and localization changes to be common in proteomes. The observations we made also bear implications in terms of proteome function and evolution. While protein localization is regarded as being regulated by linear motifs, changes in protein surface chemistry may drive such changes during evolution both in health and disease.
Materials and Methods
Selection of the Proteins.
The homo-oligomers used in this work were selected based on specific criteria described previously (44). Details of their structure, gene names, PDB accession numbers, number of subunits, symmetry, theoretical isoelectric point, and the number of positive and negative charges are given in SI Appendix, Tables S1 and S2.
Cloning Procedures, Mutagenesis, and Gene Libraries Construction.
Genes coding for the 10 dihedral homomers used in this work were amplified from the E. coli strain K12 and cloned as previously described (44). Additionally, the genes encoding the structures 1POK and 1M3U were cloned downstream of the yeast glyceraldehyde-3-phosphate dehydrogenase (GPD) promoter into M3925 plasmids for genome integration (75). Molecular cloning was performed using the polymerase incomplete primer extension method (76). The 10 homomers were fused to a YFP (Venus) (77) spaced by a flexible linker (sequence: GGGGSGGGGS) to track their localization in vivo. To introduce alanine mutations, the 10 homomers were subjected to site-directed mutagenesis (QuikChange, Agilent). The 1pok episomal gene library was based on the plasmid p413 GPD (78). The genomically integrated mutant libraries were based on the plasmid M3925 GPD (trp1::KanMX3). Site-saturation mutagenesis of the targeted residues (SI Appendix, Table S1) was achieved using oligos with randomized bases in the codons targeted for mutagenesis. All residues targeted for mutagenesis were solvent exposed with >25% of relative accessible surface area. For the overexpression experiment, both 1POK and 1M3U wild-type genes were cloned on the high-copy (2-μm) plasmid, p423 GPD (78). The oligonucleotides used for cloning, mutagenesis, and mutant library construction were ordered from Integrated DNA Technologies (IDT).
Resolving Multiple Vector Transformants.
Sequencing of yeast colonies after the transformation of the p413 plasmid-based 1pok gene library revealed that most of them bore up to four different plasmids. To resolve individual sequences, single colonies were diluted in phosphate-buffered saline (PBS) and streaked on agar plates with synthetic complete medium lacking histidine (SC-His). This dilution-streaking procedure was iterated three times for every mutant before they were sequenced.
Yeast Strains and Media.
The parent yeast strain used in this work was BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0). Plasmids p413 and p423 encoding the wild-type proteins and the alanine mutants as well as the 1pok gene library were transformed into BY4741 cells using the LiAc method (79). Transformants were grown in SC-His agar plates and were inoculated into 384-well plates containing SC-His with 15% glycerol and stored −80 °C after growing to saturation. For the 1pok and 1m3u gene libraries based on the M3925 plasmid, the cassette for genomic integration was amplified as a linear fragment prior to its transformation into yeast cells. The transformants were grown in SC + G418 (300 μg/mL) agar plates and were also inoculated into 384-well plates, grown in SC + G418 (200 μg/mL) 15% glycerol, and stored at −80 °C.
Microscopy Screenings and Sample Preparation.
A liquid handling robot (Tecan Evo 200) was used to perform plate-to-plate transfers of cells by operating a pin tool (FP1 pins, V&P Scientific). The cells were inoculated from glycerol stocks into 384-well polypropylene plates containing SC-His (for strains containing p413 plasmids) or SC + G418 (200 μg/mL) (for strains of the genome-integrated libraries) and grown until they reached saturation. Then, cells from the saturated cultures were inoculated into Greiner 384-well glass-bottom optical imaging plates containing the appropriate selection media for each case. The cells were grown for at least 6 h until they reached logarithmic growth (an optical density of ∼0.5 to 1). The imaging was performed using an automated Olympus microscope ×83 coupled to a spinning-disk confocal scanner (Yokogawa W1) with a 60×, 1.35 numerical aperture, oil immersion objective (UPLSAPO 60XO, Olympus). The fluorescence excitation was achieved with a 488-nm laser (Toptica, 100 mW), and the emission used a 525/28 filter (Chroma). We recorded 16-bit images for brightfield and green fluorescent protein (GFP) channels on a Hamamatsu Flash 4 V2 camera. Hardware autofocus (Olympus Z-Drift Compensation System) was used to maintain the focus during the imaging experiments. A motorized XY stage with a piezo stage (Mad City Labs) was used to perform the automated imaging and for acquiring Z-stacks. For a set of 163 mutants of the 1m3u library, we acquired Z-stacks with seven slices for the GFP channel. We then used the max-intensity projection for the automatic assignment of cell phenotypes, which was compared to the assignments obtained based on the center slice (SI Appendix, Fig. S8A).
Automatic Assignment of Cell Phenotypes.
The cells were identified and segmented, and their fluorescent signal (median, average, minimum, and maximum) as well as additional cell properties were determined using custom algorithms (80, 81) in Fiji (82) and exported as tabulated files.
The fibers, puncta, and nuclear localization were identified in each cell independently in a multistep process: 1) We calculated the maximum and median fluorescence intensity of pixels in a given cell. 2) Cells in which the maximal intensity was below 200 or cells not containing a markedly bright region (maximum/median < 2.5) were assigned as cytosolic. 3) We identified the largest four regions composed of pixels with an intensity 2.5-fold above the cell median intensity. If such regions of interest (ROI) existed and showed a circularity above 0.4 and an area above 4 pixels, the cell was assigned one or more of the noncytosolic phenotypes (fibers, nuclear, or puncta) based on the criteria of the ROI fluorescence and shape following a decision tree described in SI Appendix, Fig. S2B. In those cells, for each ROI found (up to the fourth largest), a phenotype was assigned to the ROI. Finally, the phenotype of the cell was determined as the union of the phenotypes of all the ROIs. This procedure was optimized based on a dataset of 1,834 cells that we annotated manually.
To image the strains, we recorded a single focal plane corresponding to ∼1 μm in depth. Thus, some puncta or fibers in a cell could be missed if they were situated outside of the focal plane. Such underestimation would not impact our analysis if it is comparable across strains. To control whether this is the case, we imaged again 163 mutants from the 1m3u library using seven focal planes. We subsequently analyzed the Z-projection of those images, which captures the entire cellular volume (SI Appendix, Fig. S8). The phenotype penetrance values of fibers and puncta that we obtained on this repeat matched the original values obtained with a single plane, indicating that the center plane provides a reliable estimate for the fraction of cells with puncta and fibers. We did not compare the predictions of nuclear localization because only a few strains (n = 4, Fig. 2) were assigned such a phenotype by manual inspection of the data. The identities of the strains used in this analysis are given in Dataset S2.
Data Analysis.
The 20 numerical scales representing physicochemical and biochemical properties of AAs (Dataset S3) were selected from https://www.genome.jp/aaindex/, except for interaction propensities scales “Levy” taken from ref. 83 and “Villegas–Levy” from ref. 84. Briefly, while the Levy scale was derived from amino acid frequencies at protein surfaces and interfaces, the Villegas–Levy stickiness scale is derived from the fractional area amino acids occupy at interfaces and surfaces. Data from sequencing was tabulated and aggregated with phenotypic information (Datasets S1–S3). The physicochemical properties of the mutants and their relationship to phenotypic information was analyzed using R (85).
Supplementary Material
Acknowledgments
We thank Benjamin Dubreuil for helping with the data analysis, H. Greenblatt for helping with the computer infrastructure, and members of the laboratory for discussions throughout the realization of this work. This work was supported by the European Research Council under the European Union’s Horizon 2020 research and innovation program (Grant Agreement No. 819318), by the Israel Science Foundation (Grant No. 1452/18), by a research grant from A.-M. Boucher, and by research grants from the Estelle Funk Foundation, the Estate of Fannie Sherr, the Estate of Albert Delighter, the Merle S. Cahn Foundation, Mrs. Mildred S. Gosden, the Estate of Elizabeth Wachsman, and the Arnold Bortman Family Foundation. H.G.S. received support from the Koshland Foundation. T.L. acknowledges support from the Israeli Council for Higher Education via the Weizmann Data Science Research Center and from a research grant from the Estate of Tully and Michele Plesser.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2101117119/-/DCSupplemental.
Data Availability
All the study data and statistics are included in the article and supporting information. Raw image data and analysis scripts are available from the authors upon request.
References
- 1.Ng P. C., Henikoff S., Predicting the effects of amino acid substitutions on protein function. Annu. Rev. Genomics Hum. Genet. 7, 61–80 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Fowler D. M., et al. , High-resolution mapping of protein sequence-function relationships. Nat. Methods 7, 741–746 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cunningham B. C., Wells J. A., High-resolution epitope mapping of hGH-receptor interactions by alanine-scanning mutagenesis. Science 244, 1081–1085 (1989). [DOI] [PubMed] [Google Scholar]
- 4.Weiss G. A., Watanabe C. K., Zhong A., Goddard A., Sidhu S. S., Rapid mapping of protein functional epitopes by combinatorial alanine scanning. Proc. Natl. Acad. Sci. U.S.A. 97, 8950–8954 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang W., Petrosino J., Hirsch M., Shenkin P. S., Palzkill T., Amino acid sequence determinants of beta-lactamase structure and activity. J. Mol. Biol. 258, 688–703 (1996). [DOI] [PubMed] [Google Scholar]
- 6.Suckow J., et al. , Genetic studies of the Lac repressor. XV: 4000 single amino acid substitutions and analysis of the resulting phenotypes on the basis of the protein structure. J. Mol. Biol. 261, 509–523 (1996). [DOI] [PubMed] [Google Scholar]
- 7.Rocklin G. J., et al. , Global analysis of protein folding using massively parallel design, synthesis, and testing. Science 357, 168–175 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schlinkmann K. M., et al. , Critical features for biosynthesis, stability, and functionality of a G protein-coupled receptor uncovered by all-versus-all mutations. Proc. Natl. Acad. Sci. U.S.A. 109, 9810–9815 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim I., Miller C. R., Young D. L., Fields S., High-throughput analysis of in vivo protein stability. Mol. Cell. Proteomics 12, 3370–3378 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bolognesi B., et al. , The mutational landscape of a prion-like domain. Nat. Commun. 10, 4162 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo H. H., Choe J., Loeb L. A., Protein tolerance to random amino acid change. Proc. Natl. Acad. Sci. U.S.A. 101, 9205–9210 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Firnberg E., Labonte J. W., Gray J. J., Ostermeier M., A comprehensive, high-resolution map of a gene’s fitness landscape. Mol. Biol. Evol. 31, 1581–1592 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rockah-Shmuel L., Tóth-Petróczy Á., Tawfik D. S., Systematic mapping of protein mutational space by prolonged drift reveals the deleterious effects of seemingly neutral mutations. PLoS Comput. Biol. 11, e1004421 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Després P. C., Dubé A. K., Seki M., Yachie N., Landry C. R., Perturbing proteomes at single residue resolution using base editing. Nat. Commun. 11, 1871 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ernst A., et al. , Coevolution of PDZ domain-ligand interactions analyzed by high-throughput phage display and deep sequencing. Mol. Biosyst. 6, 1782–1790 (2010). [DOI] [PubMed] [Google Scholar]
- 16.Taylor N. D., et al. , Engineering an allosteric transcription factor to respond to new ligands. Nat. Methods 13, 177–183 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sidhu S. S., Koide S., Phage display for engineering and analyzing protein interaction interfaces. Curr. Opin. Struct. Biol. 17, 481–487 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Whitehead T. A., et al. , Optimization of affinity, specificity and function of designed influenza inhibitors using deep sequencing. Nat. Biotechnol. 30, 543–548 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jardine J. G., et al. , HIV-1 broadly neutralizing antibody precursor B cells revealed by germline-targeting immunogen. Science 351, 1458–1463 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cohen-Khait R., Schreiber G., Low-stringency selection of TEM1 for BLIP shows interface plasticity and selection for faster binders. Proc. Natl. Acad. Sci. U.S.A. 113, 14982–14987 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Diss G., Lehner B., The genetic landscape of a physical interaction. eLife 7, e32472 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huh W.-K., et al. , Global analysis of protein localization in budding yeast. Nature 425, 686–691 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Alberti S., Halfmann R., King O., Kapila A., Lindquist S., A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell 137, 146–158 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cid-Samper F., et al. , An integrative study of protein-RNA condensates identifies scaffolding RNAs and reveals players in fragile X-associated tremor/ataxia syndrome. Cell Rep. 25, 3422–3434.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vernon R. M., et al. , Pi-Pi contacts are an overlooked protein feature relevant to phase separation. eLife 7, e31486 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J., et al. , A molecular grammar governing the driving forces for phase separation of prion-like RNA binding proteins. Cell 174, 688–699.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hughes M. P., et al. , Atomic structures of low-complexity protein segments reveal kinked β sheets that assemble networks. Science 359, 698–701 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hyman A. A., Weber C. A., Jülicher F., Liquid-liquid phase separation in biology. Annu. Rev. Cell Dev. Biol. 30, 39–58 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Gomes E., Shorter J., The molecular language of membraneless organelles. J. Biol. Chem. 294, 7115–7127 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stoddard P. R., et al. , Polymerization in the actin ATPase clan regulates hexokinase activity in yeast. Science 367, 1039–1042 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barry R. M., et al. , Large-scale filament formation inhibits the activity of CTP synthetase. eLife 3, e03638 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Duong-Ly K. C., et al. , T cell activation triggers reversible inosine-5′-monophosphate dehydrogenase assembly. J. Cell Sci. 131, jcs223289 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim C.-W., et al. , Induced polymerization of mammalian acetyl-CoA carboxylase by MIG12 provides a tertiary level of regulation of fatty acid synthesis. Proc. Natl. Acad. Sci. U.S.A. 107, 9626–9631 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petrovska I., et al. , Filament formation by metabolic enzymes is a specific adaptation to an advanced state of cellular starvation. eLife 10.7554/eLife.02409 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Narayanaswamy R., et al. , Widespread reorganization of metabolic enzymes into reversible assemblies upon nutrient starvation. Proc. Natl. Acad. Sci. U.S.A. 106, 10147–10152 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Noree C., et al. , A quantitative screen for metabolic enzyme structures reveals patterns of assembly across the yeast metabolic network. Mol. Biol. Cell 30, 2721–2736 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garcia-Seisdedos H., Villegas J. A., Levy E. D., Infinite assembly of folded proteins in evolution, disease, and engineering. Angew. Chem. Int. Ed. 58, 5514–5531 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eaton W. A., Hofrichter J., Sickle cell hemoglobin polymerization. Adv. Protein Chem. 40, 63–279 (1990). [DOI] [PubMed] [Google Scholar]
- 39.Boatz J. C., Whitley M. J., Li M., Gronenborn A. M., van der Wel P. C. A., Cataract-associated P23T γD-crystallin retains a native-like fold in amorphous-looking aggregates formed at physiological pH. Nat. Commun. 8, 15137 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barmada S. J., et al. , Cytoplasmic mislocalization of TDP-43 is toxic to neurons and enhanced by a mutation associated with familial amyotrophic lateral sclerosis. J. Neurosci. 30, 639–649 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kwiatkowski T. J. Jr., et al. , Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science 323, 1205–1208 (2009). [DOI] [PubMed] [Google Scholar]
- 42.Antony P. M. A., et al. , Identification and functional dissection of localization signals within ataxin-3. Neurobiol. Dis. 36, 280–292 (2009). [DOI] [PubMed] [Google Scholar]
- 43.Marsh J. A., Teichmann S. A., Structure, dynamics, assembly, and evolution of protein complexes. Annu. Rev. Biochem. 84, 551–575 (2015). [DOI] [PubMed] [Google Scholar]
- 44.Garcia-Seisdedos H., Empereur-Mot C., Elad N., Levy E. D., Proteins evolve on the edge of supramolecular self-assembly. Nature 548, 244–247 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Padilla J. E., Colovos C., Yeates T. O., Nanohedra: Using symmetry to design self assembling protein cages, layers, crystals, and filaments. Proc. Natl. Acad. Sci. U.S.A. 98, 2217–2221 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.King N. P., et al. , Computational design of self-assembling protein nanomaterials with atomic level accuracy. Science 336, 1171–1174 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Suzuki Y., et al. , Self-assembly of coherently dynamic, auxetic, two-dimensional protein crystals. Nature 533, 369–373 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grueninger D., et al. , Designed protein-protein association. Science 319, 206–209 (2008). [DOI] [PubMed] [Google Scholar]
- 49.Empereur-Mot C., Garcia-Seisdedos H., Elad N., Dey S., Levy E. D., Geometric description of self-interaction potential in symmetric protein complexes. Sci. Data 6, 64 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ben-Sasson A. J., et al. , Design of biologically active binary protein 2D materials. Nature 589, 468–473 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Levy E. D., De S., Teichmann S. A., Cellular crowding imposes global constraints on the chemistry and evolution of proteomes. Proc. Natl. Acad. Sci. U.S.A. 109, 20461–20466 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Frey S., et al. , Surface properties determining passage rates of proteins through nuclear pores. Cell 174, 202–217.e9 (2018). [DOI] [PubMed] [Google Scholar]
- 53.Chothia C., Janin J., Principles of protein-protein recognition. Nature 256, 705–708 (1975). [DOI] [PubMed] [Google Scholar]
- 54.Doye J. P. K., Louis A. A., Vendruscolo M., Inhibition of protein crystallization by evolutionary negative design. Phys. Biol. 1, 9–13 (2004). [DOI] [PubMed] [Google Scholar]
- 55.Pechmann S., Levy E. D., Tartaglia G. G., Vendruscolo M., Physicochemical principles that regulate the competition between functional and dysfunctional association of proteins. Proc. Natl. Acad. Sci. U.S.A. 106, 10159–10164 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bolon D. N., Grant R. A., Baker T. A., Sauer R. T., Specificity versus stability in computational protein design. Proc. Natl. Acad. Sci. U.S.A. 102, 12724–12729 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Berezovsky I. N., Zeldovich K. B., Shakhnovich E. I., Positive and negative design in stability and thermal adaptation of natural proteins. PLoS Comput. Biol. 3, e52 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Noivirt-Brik O., Horovitz A., Unger R., Trade-off between positive and negative design of protein stability: From lattice models to real proteins. PLoS Comput. Biol. 5, e1000592 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Richardson J. S., Richardson D. C., Natural β-sheet proteins use negative design to avoid edge-to-edge aggregation. Proc. Natl. Acad. Sci. U.S.A. 99, 2754–2759 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.DeGrado W. F., Summa C. M., Pavone V., Nastri F., Lombardi A., De novo design and structural characterization of proteins and metalloproteins. Annu. Rev. Biochem. 68, 779–819 (1999). [DOI] [PubMed] [Google Scholar]
- 61.Hecht M. H., Richardson J. S., Richardson D. C., Ogden R. C., De novo design, expression, and characterization of Felix: A four-helix bundle protein of native-like sequence. Science 249, 884–891 (1990). [DOI] [PubMed] [Google Scholar]
- 62.Derewenda Z. S., Rational protein crystallization by mutational surface engineering. Structure 12, 529–535 (2004). [DOI] [PubMed] [Google Scholar]
- 63.Anthony S. A., et al. , Reconstituted IMPDH polymers accommodate both catalytically active and inactive conformations. Mol. Biol. Cell 28, 2600–2608 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lynch E. M., et al. , Human CTP synthase filament structure reveals the active enzyme conformation. Nat. Struct. Mol. Biol. 24, 507–514 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Słabicki M., et al. , Small-molecule-induced polymerization triggers degradation of BCL6. Nature 588, 164–168 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Munder M. C., et al. , A pH-driven transition of the cytoplasm from a fluid- to a solid-like state promotes entry into dormancy. eLife 5, e09347 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.de Graff A. M. R., Hazoglou M. J., Dill K. A., Highly charged proteins: The Achilles’ heel of aging proteomes. Structure 24, 329–336 (2016). [DOI] [PubMed] [Google Scholar]
- 68.Torbeev V. Y., Hilvert D., Both the cis-trans equilibrium and isomerization dynamics of a single proline amide modulate β2-microglobulin amyloid assembly. Proc. Natl. Acad. Sci. U.S.A. 110, 20051–20056 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zosel F., Mercadante D., Nettels D., Schuler B., A proline switch explains kinetic heterogeneity in a coupled folding and binding reaction. Nat. Commun. 9, 3332 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Krishnan R., Lindquist S. L., Structural insights into a yeast prion illuminate nucleation and strain diversity. Nature 435, 765–772 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lin D. H., Hoelz A., The structure of the nuclear pore complex: An update. Annu. Rev. Biochem. 88, 725–783 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Naim B., Zbaida D., Dagan S., Kapon R., Reich Z., Cargo surface hydrophobicity is sufficient to overcome the nuclear pore complex selectivity barrier. EMBO J. 28, 2697–2705 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hung M.-C., Link W., Protein localization in disease and therapy. J. Cell Sci. 124, 3381–3392 (2011). [DOI] [PubMed] [Google Scholar]
- 74.Vecchi G., et al. , Proteome-wide observation of the phenomenon of life on the edge of solubility. Proc. Natl. Acad. Sci. U.S.A. 117, 1015–1020 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Voth W. P., Jiang Y. W., Stillman D. J., New ‘marker swap’ plasmids for converting selectable markers on budding yeast gene disruptions and plasmids. Yeast 20, 985–993 (2003). [DOI] [PubMed] [Google Scholar]
- 76.Klock H. E., Koesema E. J., Knuth M. W., Lesley S. A., Combining the polymerase incomplete primer extension method for cloning and mutagenesis with microscreening to accelerate structural genomics efforts. Proteins 71, 982–994 (2008). [DOI] [PubMed] [Google Scholar]
- 77.Nagai T., et al. , A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 20, 87–90 (2002). [DOI] [PubMed] [Google Scholar]
- 78.Mumberg D., Müller R., Funk M., Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156, 119–122 (1995). [DOI] [PubMed] [Google Scholar]
- 79.Knop M., et al. , Epitope tagging of yeast genes using a PCR-based strategy: More tags and improved practical routines. Yeast 15, 963–972 (1999). [DOI] [PubMed] [Google Scholar]
- 80.Matalon O., Steinberg A., Sass E., Hausser J., Levy E. D., Reprogramming protein abundance fluctuations in single cells by degradation. bioRxiv [Preprint] (2018) 10.1101/260695. Accessed 5 February 2018. [DOI]
- 81.Heidenreich M., et al. , Designer protein assemblies with tunable phase diagrams in living cells. Nat. Chem. Biol. 16, 939–945 (2020). [DOI] [PubMed] [Google Scholar]
- 82.Schindelin J., et al. , Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Levy E. D., A simple definition of structural regions in proteins and its use in analyzing interface evolution. J. Mol. Biol. 403, 660–670 (2010). [DOI] [PubMed] [Google Scholar]
- 84.Villegas J. A., Levy E. D., Desolvation energy explains partitioning of client proteins into condensates. bioRxiv [Preprint] (2021) 10.1101/2021.08.16.456554. Accessed 17 August 2021. [DOI]
- 85.R Core Team, R: A language and environment for statistical computing. http://www.R-project.org/. Accessed 12 December 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the study data and statistics are included in the article and supporting information. Raw image data and analysis scripts are available from the authors upon request.