ABSTRACT
Gastric cancer (GC) is one of the leading types of fatal cancer worldwide. Epigenetic manipulation of cancer cells is a useful tool to better understand gene expression regulatory mechanisms and contributes to the discovery of novel biomarkers. Our research group recently reported a list of 83 genes that are potentially modulated by DNA methylation in GC cell lines. Herein, we further explored the regulation of one of these genes, LRRC37A2, in clinical samples. LRRC37A2 expression was evaluated by RT-qPCR, and DNA methylation was studied using next-generation bisulphite sequencing in 36 GC and paired adjacent nonneoplastic tissue samples. We showed that both reduced LRRC37A2 mRNA levels and increased LRRC37A2 exon methylation were associated with undifferentiated and poorly differentiated tumours. Moreover, LRRC37A2 gene expression and methylation levels were inversely correlated at the +45 exon CpG site. We suggest that DNA hypermethylation may contribute to reducing LRRC37A2 expression in undifferentiated and poorly differentiated GC. Therefore, our results show how some genes may be useful to stratify patients who are more likely to benefit from epigenetic therapy.Abbreviations: AR: androgen receptor; 5-AZAdC: 5-aza-2'-deoxycytidine; B2M: beta-2-microglobulin; GAPDH: glyceraldehyde-3-phosphate dehydrogenase; GC: gastric cancer; GLM: general linear model; LRRC37A2: leucine-rich repeat containing 37 member A2; SD: standard deviation; TFII-I: general transcription factor II-I; TSS: transcription start site; XBP1: X-box binding protein 1
KEYWORDS: DNA methylation, gene expression, cancer therapy, LRRC37A2
Introduction
Gastric cancer (GC) is the third leading cause of cancer-related death worldwide [1]. The major contributors to the high mortality rates of GC are late diagnosis and lack of effective therapies to combat disease heterogeneity [2]. The understanding of epigenomic regulation has provided further evidence for the application of epigenetic drugs for solid tumour treatment [3], with several agents progressing to clinical trials with gastrointestinal cancer patients [3,4]. In addition, the elucidation of gene expression regulatory mechanisms may help to identify patients who are more likely to benefit from the use of epigenetic drugs. In this context, our research group previously assessed the gene expression profile of GC cells [5] treated with the DNA demethylating agent 5-aza-2'-deoxycytidine (5-AZAdC) and identified 83 genes potentially modulated by DNA methylation [6]. Leucine-rich repeat containing 37 member A2 (LRRC37A2) was one of the upregulated genes, with fold change>1.5-fold that presented CpG island near the TSS.
Although the LRR superfamily is composed of a heterogeneous group of proteins involved in the differentiation and development of normal nervous tissues [7] and the immune response [8], studies have reported that deregulation of LRR genes is critical in tumorigenesis [7,9]. In particular, the role of LRRC37A2 and its underlying mechanism of regulation remain unknown in GC.
Here, we report a detailed analysis of the LRRC37A2 DNA methylation profile in a panel of GC and matched control tissue samples using next-generation sequencing of bisulphite-converted DNA. Moreover, we provide further evidence that DNA methylation impacts the transcript level of LRRC37A2 in GC, especially in undifferentiated and poorly differentiated tumours.
Materials and methods
Subjects and sample preparation
Thirty-six matched pairs of GC and adjacent nonneoplastic tissues (control group) were obtained from patients with gastric adenocarcinoma who underwent gastric resection in João de Barros Barreto University Hospital (HUJBB) and São Paulo Hospital (HSP), Brazil. None of the patients had a history of exposure to either chemotherapy or radiotherapy prior to surgery or the cooccurrence of diagnosed cancers. Written informed consent with the approval of the ethics committees of HUJBB and HSP was obtained from all patients before sample collection (Ethics Committee number 0511/09).
The genomic DNA and RNA from fresh snap-frozen tissues in liquid nitrogen were isolated and quantified as previously reported, and RNA conversion to cDNA was performed [6].
Quantitative polymerase chain reaction
TaqMan gene expression assays (Thermo Fisher Scientific, Waltham, MA, USA) were used to evaluate the expression of LRRC37A2 (Hs03805446_mH) in triplicate. Expression data were normalized to GAPDH (Hs99999905_m1) and B2M (Hs00984230_m1), as described previously by our research group [10]. The mRNA levels were analysed using the ∆Ct method.
Next-generation sequencing
Primers were designed by using MethPrimer software [11] for the LRRC37A2 CpG island, which includes 8 CpG sites surrounding the transcription start site (TSS):5'-AAGAGTGTTTTGGTGATATGGAGTAG-3', and 3'-CCACAATAATTACCACATAAAAAAAA-5' (Figure 1a).
Figure 1.
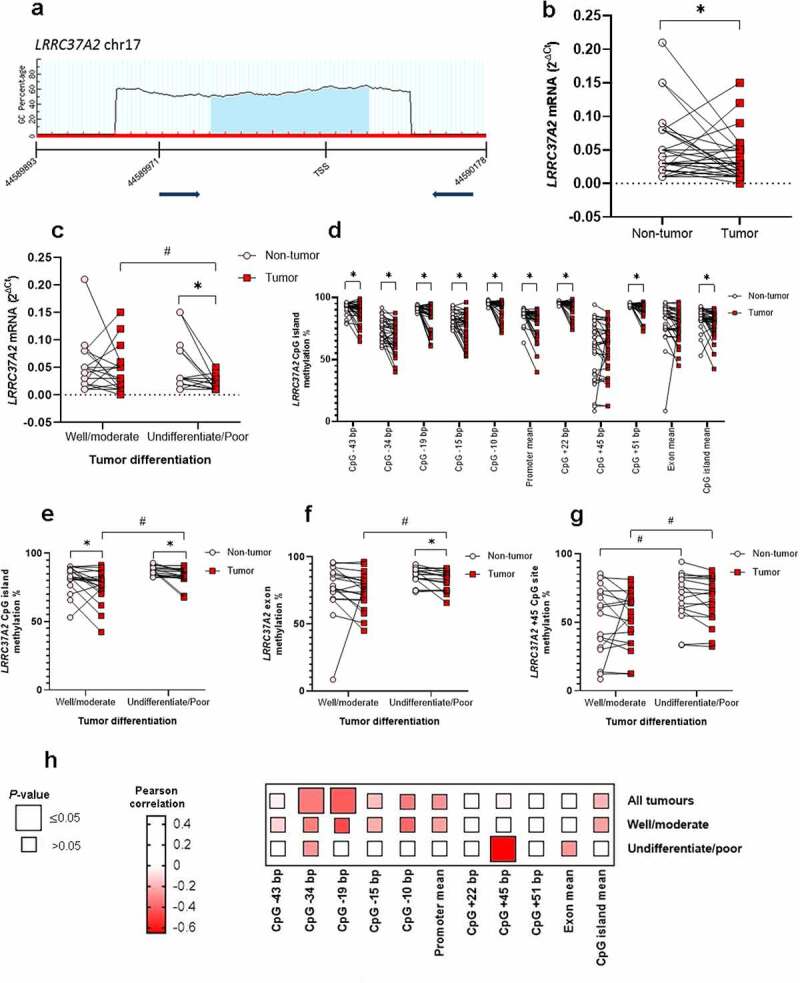
LRRC37A2 mRNA and methylation levels in GC. a: schematic diagram of CpG dinucleotide density across the LRRC37A2 locus and the location of the investigated CpG dinucleotides. b: reduced LRRC37A2 mRNA in GC compared with paired adjacent nonneoplastic tissue samples. c: reduced LRRC37A2 mRNA levels associated with undifferentiated and poorly differentiated GC. In this subset of tumours, significantly reduced LRRC37A2 mRNA levels were also observed compared with paired adjacent nonneoplastic tissue samples. d: significant differences in LRRC37A2 methylation levels across CpG sites between GC and the corresponding adjacent nonneoplastic tissue samples. e: increased LRRC37A2 CpG island methylation associated with undifferentiated and poorly differentiated tumours. f: increased LRRC37A2 exon methylation associated with undifferentiated and poorly differentiated tumours. In this subset of tumours, significantly increased LRRC37A2 mRNA levels were also observed compared with paired adjacent nonneoplastic tissue samples. g: increased methylation at the +45 CpG exon site in undifferentiated and poorly differentiated tumours and the corresponding adjacent nonneoplastic samples compared with well and moderately differentiated tumours and adjacent nonneoplastic samples. h: correlation analysis between LRRC37A2 gene expression and methylation levels.
Data are expressed as the mean ± SD* Significant difference between groups by repeated-measures GLM# Significant difference between groups by univariate GLMTSS: transcription start site.
DNA samples were bisulphite-converted using EpiTect Fast DNA Bisulphite kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. A 201 bp fragment corresponding to the LRRC37A2 CpG island was PCR amplified at an annealing temperature of 58°C.
Sequencing was performed using the IonTorrent PGMTM platform (Thermo Fisher Scientific) and data were processed as described previously [6]. In detail, Bismark version 0.16.3 [12] was used to align the reads to the human reference genome (hg19) restricted to the target region. A minimum of 100 CpG measurements across samples was required for the region set. Non-CpG methylation was used as an internal upper-bound estimate of the inefficiency of bisulphite conversion. All samples presented less than 1% methylation in the CHG and CHH contexts.
Statistical analysis
The Shapiro-Wilk normality test was used to evaluate the distribution of all data. Data were not normally distributed and were transformed (z-score). Repeated-measures or univariate general linear models (GLMs), and Pearson correlation tests were employed to analyse the data. Two-sided tests were performed, and 95% confidence intervals were reported.
Results
LRRC37A2 is downregulated in gastric cancer
To first confirm that LRRC37A2 may have a role in gastric carcinogenesis, we evaluated its mRNA expression in a panel of clinical samples (Supplementary Table 1). The expression of LRRC37A2 was reduced in GC tissue samples compared with the controls (P< 0.01; Figure 1b). Undifferentiated and poorly differentiated GC showed reduced LRRC37A2 expression in comparison with well and moderately differentiated tumours (P= 0.05; Figure 1c and Supplementary Table 1). In addition, low LRRC37A2 mRNA levels were detected in undifferentiated and poorly differentiated tumours compared with the respective controls (P< 0.01; Figure 1c).
LRRC37A2 is methylated in gastric cancer
We next evaluated the LRRC37A2 methylation pattern in CpG sites in the promoter and exon portions. Overall, high LRRC37A2 methylation (~80%) was observed in the studied specimens; however, GC samples presented significantly decreased methylation in the CpG island (P< 0.01) and promoter regions (P< 0.01) compared with controls (Figure 1). A detailed analysis of LRRC37A2 methylation revealed significantly decreased methylation at all the specific CpG sites (P< 0.01) in the GC compared with controls, except at the +45 location (Figure 1d).
By grouping the patients according to clinicopathological features, we observed that LRRC37A2 CpG island and exon methylation percentages were increased in patients with undifferentiated and poorly differentiated tumours (P= 0.03 and P= 0.04, respectively; Supplementary Table 1, Figure 1e,Figure 1f). LRRC37A2 CpG island, promoter, and exon methylation percentages were also increased in patients with lymph node metastasis (P= 0.04, P= 0.05, and P= 0.03, respectively; Supplementary Table 1). Furthermore, LRRC37A2 CpG and promoter methylation percentages were increased in patients with TNM stage III and IV tumours (P= 0.01 and P= 0.02, respectively; Supplementary Table 1). Last, LRRC37A2 exon methylation was increased in patients with early onset (P= 0.03; Supplementary Table 1).
The impact of DNA methylation on LRRC37A2 gene expression
To provide further evidence that LRRC37A2 is regulated by epigenetic processes, we performed correlation analysis between mRNA and methylation levels. Overall, LRRC37A2 gene expression and DNA methylation at the specific −34 (r = −0.34, P= 0.05) and −19 (r = −0.42, P= 0.02) promoter CpG sites were inversely correlated (Figure 1h and Supplementary Table 2).
Considering the consistently reduced LRRC37A2 gene expression and increased DNA methylation in undifferentiated and poorly differentiated tumours, we analysed gene expression and methylation data correlation in this subset of tumours. We observed that LRRC37A2 expression and DNA methylation at the specific +45 exon CpG site were inversely correlated (r = −0.65, P< 0.01; Figure 1h and Supplementary Table 2). In well and moderately differentiated tumours, no significant correlation between gene expression and DNA methylation data was observed (Figure 1h).
We also observed that undifferentiated and poorly differentiated tumours and their paired controls presented increased levels of methylation specifically at the +45 CpG site in relation to well and moderately differentiated tumours (P= 0.03 and P= 0.01, respectively; Figure 1g).
Discussion
In this study, we assessed in clinical samples the expression and methylation of LRRC37A2, a candidate target modulated by DNA methylation originally identified by our research group in GC cell lines [6]. Our results showed reduced LRRC37A2 mRNA levels in GC, especially in undifferentiated and poorly differentiated GC, a subset of GC that tend to grow quickly and show a more invasive phenotype than well and moderately differentiated GC [13]. Therefore, we demonstrated that LRRC37A2 is downregulated in GC and may be involved in tumour cell differentiation.
To evaluate whether DNA methylation is the mechanism implied in the reduction of LRRC37A2 mRNA levels in GC, we analysed CpG sites surrounding the TSS individually and grouped by promoter and exon portions. We first observed decreased methylation in the CpG island and promoter regions, as well as at all the specific CpG sites (except at the +45 location) in the GC compared with controls. Although promoter CpG islands are typically devoid of methylation, studies have demonstrated hypermethylation of CpG islands in nonneoplastic gastric mucosae in relation to GC [14–16]. Chronic inflammation and ageing seem to be closely associated with increased methylation in these samples [17].
Our results also showed high levels of methylation in the entire CpG island and in the specific exon region associated with undifferentiated and poorly differentiated tumours. Gene expression and methylation correlation analysis reinforced these results and revealed the +45 site as a possible methylated exon CpG site involved in LRRC37A2 downregulation in undifferentiated and poorly differentiated GC. These findings corroborate a previous study in which DNA methylation downstream of the TSS (first exon) was shown to be the most critical for transcriptional silencing [18].
To predict transcription factors binding the +45 exon CpG site, we used the PROMO server with TRANSFAC version 8.3 [19,20], and we found that this CpG site falls within the recognition sequence of general transcription factor II–I (TFII-I), X-box binding protein 1 (XBP1), and androgen receptor (AR). These findings suggest that methylation at the +45 exon CpG site may inhibit the binding of transcription factors and repress LRRC37A2 expression.
Taking the gene expression and methylation results together, our study shows that increased methylation of LRRC37A2, more specifically at the +45 exon CpG site, may be a mechanism of gene silencing in undifferentiated and poorly differentiated GC. Thus, our results show how specific genes may be useful to stratify patients who are more likely to benefit from epigenetic therapy.
Our study is the first to investigate the possible involvement of DNA methylation in the regulation of LRRC37A2 transcription; however, there are three limitations that should be addressed. First, LRRC37A2 is contained in a core duplication region on chromosome 17 [8], and the commercial assay available for gene expression analysis also detects another LRR family member, the LRRC37A gene. Furthermore, the methylation results were only specific for LRRC37A2, once the alignment of reads was restricted to the region of interest. Second, a correction for multiple comparisons (multiple CpG sites being evaluated simultaneously) was not carried out in the analysis of DNA methylation data. Because no similar study has been published previously, we chose to prioritize the biological effect rather than reject the involvement of an epigenetic event in GC due to statistical rigour (type II error). Third, it is known that the traditional bisulphite sequencing technique does not distinguish between 5-methylcytosine (5mC) and 5-hydroxymethylcytosine (5hmC) [21,22]. Although the presence of 5hmC in the mammalian genome is less abundant than 5mC [23], and a previous study, which had performed genome-wide profiling of 5hmC in GC, did not identify our CpG island as a differentially hydroxymethylated region [24–27], the quantification of 5hmC at LRRC37A2 CpG island and the determination of its impact on gene expression in our samples would contribute to our findings.
Supplementary Material
Acknowledgments
We would like to thank Brunno dos Santos Pereira, Renata Sanches de Almeida, and Camila Albuquerque Pinto for help in sample collection and Brunno dos Santos Pereira for help in generating the final figures. We are particularly grateful to the study participants.
Funding Statement
This study was supported by the Fundação de Amparo à Pesquisa do Estado de São Paulo [FAPESP, Processo n° 2016/19953-6 to FW, Processo n° 2017/06227-8 to JCG, Processo n° 2007/02470-3 to MFL, Processo n° 2010/11174-1 to DQC, and Processo n° 2009/07145-9 to MACS], the Conselho Nacional de Desenvolvimento Científico e Tecnológico [CNPq; to FW, DQC, JCG, RRB, and MACS], and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior [CAPES; to COG and ACA].
Disclosure statement
The authors declare that they have no conflict of interest.
Author contribution
Conception and design: FW, MFL, RRB, and MACS. Sample collection: FW, LCS, DQC, JCG, COG, ACA, ESC, PPA, LGL, and CHA. Pathological analysis: RA and SD. Molecular experiments: FW, JCG, LCS, MFL, DQC, COG, ESC, and ACA. Data analysis: FW, JCG, LCS, SP, and JK. Writing, review, and/or revision of the manuscript: all authors. Administrative, technical, or material support: MACS and RRB.
Ethical Statements
Written informed consent with the approval of the ethics committees of João de Barros Barreto University Hospital and São Paulo Hospital was obtained from all patients before sample collection (Ethics Committee number 0511/09).
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. [DOI] [PubMed] [Google Scholar]
- [2].Padmanabhan N, Ushijima T, Tan P.. How to stomach an epigenetic insult: the gastric cancer epigenome. Nat Rev Gastroenterol Hepatol. 2017;14(8):467–478. [DOI] [PubMed] [Google Scholar]
- [3].Cheng Y, He C, Wang M, et al. Targeting epigenetic regulators for cancer therapy: mechanisms and advances in clinical trials. Signal Transduct Target Ther. 2019;4:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Abdelfatah E, Kerner Z, Nanda N, et al. Epigenetic therapy in gastrointestinal cancer: the right combination. Therap Adv Gastroenterol. 2016;9(4):560–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Leal MF, Martins Do Nascimento JL, da Silva CE, et al. Establishment and conventional cytogenetic characterization of three gastric cancer cell lines. Cancer Genet Cytogenet. 2009;195(1):85–91. [DOI] [PubMed] [Google Scholar]
- [6].Wisnieski F, Santos LC, Calcagno DQ, et al. The impact of DNA demethylation on the upregulation of the NRN1 and TNFAIP3 genes associated with advanced gastric cancer. J Mol Med. 2020;98:707–717. [DOI] [PubMed] [Google Scholar]
- [7].Li P, Xu G, Li G, et al. Function and mechanism of tumor suppressor gene LRRC4/NGL-2. Mol Cancer. 2014;13:266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Giannuzzi G, Siswara P, Malig M, et al. Evolutionary dynamism of the primate LRRC37 gene family. Genome Res. 2013;23(1):46–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Zhang H, Deng Z, Zhang D, et al. High expression of leucine rich repeat containing 8A is indicative of a worse outcome of colon cancer patients by enhancing cancer cell growth and metastasis. Oncol Rep. 2018;40(3):1275–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Wisnieski F, Calcagno DQ, Leal MF, et al. Reference genes for quantitative RT-PCR data in gastric tissues and cell lines. World J Gastroenterol. 2013;19(41):7121–7128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Li LC, Dahiya R.. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18(11):1427–1431. [DOI] [PubMed] [Google Scholar]
- [12].Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27(11):1571–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Dicken BJ, Bigam DL, Cass C, et al. Gastric adenocarcinoma: review and considerations for future directions. Ann Surg. 2005;241(1):27–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kang GH, Shim YH, Jung HY, et al. CpG island methylation in premalignant stages of gastric carcinoma. Cancer Res. 2001;61(7):2847–2851. [PubMed] [Google Scholar]
- [15].To KF, Leung WK, Lee TL, et al. Promoter hypermethylation of tumor-related genes in gastric intestinal metaplasia of patients with and without gastric cancer. Int J Cancer. 2002;102(6):623–628. [DOI] [PubMed] [Google Scholar]
- [16].Waki T, Tamura G, Tsuchiya T, et al. Promoter methylation status of E-cadherin, hMLH1, and p16 genes in nonneoplastic gastric epithelia. Am J Pathol. 2002;161(2):399–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kang GH, Lee HJ, Hwang KS, et al. Aberrant CpG island hypermethylation of chronic gastritis, in relation to aging, gender, intestinal metaplasia, and chronic inflammation. Am J Pathol. 2003;163(4):1551–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Brenet F, Moh M, Funk P, et al. DNA methylation of the first exon is tightly linked to transcriptional silencing. PLoS One. 2011;6(1):e14524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Messeguer X, Escudero R, Farre D, et al. PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics. 2002;18(2):333–334. [DOI] [PubMed] [Google Scholar]
- [20].Farre D, Roset R, Huerta M, et al. Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res. 2003;31(13):3651–3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Huang Y, Pastor WA, Shen Y, et al. The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS One. 2010;5(1):e8888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Nestor C, Ruzov A, Meehan R, et al. Enzymatic approaches and bisulfite sequencing cannot distinguish between 5-methylcytosine and 5-hydroxymethylcytosine in DNA. Biotechniques. 2010;48(4):317–319. [DOI] [PubMed] [Google Scholar]
- [23].Ponnaluri VK, Ehrlich KC, Zhang G, et al. Association of 5-hydroxymethylation and 5-methylation of DNA cytosine with tissue-specific gene expression. Epigenetics. 2017;12(2):123–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Liu H, Xu T, Cheng Y, et al. 5-hydroxymethylcytosine landscape in primary gastric adenocarcinoma. DNA Cell Biol. 2019;38(12):1460–1469. [DOI] [PubMed] [Google Scholar]
- [25].Lauren P. The two histological main types of gastric carcinoma: diffuse and so-called intestinal-type carcinoma. an attempt at a histo-clinical classification. Acta Pathol Microbiol Scand. 1965;64:31–49. [DOI] [PubMed] [Google Scholar]
- [26].Washington K. 7th edition of the AJCC cancer staging manual: stomach. Ann Surg Oncol. 2010;17(12):3077–3079. [DOI] [PubMed] [Google Scholar]
- [27].Wisnieski F, Calcagno DQ, Leal MF, et al. Differential expression of histone deacetylase and acetyltransferase genes in gastric cancer and their modulation by trichostatin A. Tumour Biol. 2014;35(7):6373–6381. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


