Abstract
Background
Epidemiological studies have reported conflicting findings on the potential adverse effects of long-term antihypertensive medication use on cancer risk. Naturally occurring variation in genes encoding antihypertensive drug targets can be used as proxies for these targets to examine the effect of their long-term therapeutic inhibition on disease outcomes.
Methods and findings
We performed a mendelian randomization analysis to examine the association between genetically proxied inhibition of 3 antihypertensive drug targets and risk of 4 common cancers (breast, colorectal, lung, and prostate). Single-nucleotide polymorphisms (SNPs) in ACE, ADRB1, and SLC12A3 associated (P < 5.0 × 10−8) with systolic blood pressure (SBP) in genome-wide association studies (GWAS) were used to proxy inhibition of angiotensin-converting enzyme (ACE), β-1 adrenergic receptor (ADRB1), and sodium-chloride symporter (NCC), respectively. Summary genetic association estimates for these SNPs were obtained from GWAS consortia for the following cancers: breast (122,977 cases, 105,974 controls), colorectal (58,221 cases, 67,694 controls), lung (29,266 cases, 56,450 controls), and prostate (79,148 cases, 61,106 controls). Replication analyses were performed in the FinnGen consortium (1,573 colorectal cancer cases, 120,006 controls). Cancer GWAS and FinnGen consortia data were restricted to individuals of European ancestry. Inverse-variance weighted random-effects models were used to examine associations between genetically proxied inhibition of these drug targets and risk of cancer. Multivariable mendelian randomization and colocalization analyses were employed to examine robustness of findings to violations of mendelian randomization assumptions. Genetically proxied ACE inhibition equivalent to a 1-mm Hg reduction in SBP was associated with increased odds of colorectal cancer (odds ratio (OR) 1.13, 95% CI 1.06 to 1.22; P = 3.6 × 10−4). This finding was replicated in the FinnGen consortium (OR 1.40, 95% CI 1.02 to 1.92; P = 0.035). There was little evidence of association of genetically proxied ACE inhibition with risk of breast cancer (OR 0.98, 95% CI 0.94 to 1.02, P = 0.35), lung cancer (OR 1.01, 95% CI 0.92 to 1.10; P = 0.93), or prostate cancer (OR 1.06, 95% CI 0.99 to 1.13; P = 0.08). Genetically proxied inhibition of ADRB1 and NCC were not associated with risk of these cancers. The primary limitations of this analysis include the modest statistical power for analyses of drug targets in relation to some less common histological subtypes of cancers examined and the restriction of the majority of analyses to participants of European ancestry.
Conclusions
In this study, we observed that genetically proxied long-term ACE inhibition was associated with an increased risk of colorectal cancer, warranting comprehensive evaluation of the safety profiles of ACE inhibitors in clinical trials with adequate follow-up. There was little evidence to support associations across other drug target–cancer risk analyses, consistent with findings from short-term randomized controlled trials for these medications.
In a Mendelian randomization analysis, James Yarmolinsky and colleagues investigate associations between genetically-proxied inhibition of antihypertensive drug targets and breast, colorectal, lung, and prostate cancer risk.
Author summary
Why was this study done?
Angiotensin-converting enzyme (ACE) inhibitors, beta blockers, and thiazide diuretics are commonly prescribed antihypertensive medications.
Some epidemiological studies have suggested that long-term use of these medications can increase cancer risk, though findings have been conflicting.
Germline genetic variation in genes encoding drug targets can be used to proxy the effect of long-term modulation of these targets on disease endpoints (“drug-target mendelian randomization”).
What did the researchers do and find?
We used drug-target mendelian randomization to examine the association of genetically proxied inhibition of the drug targets of ACE inhibitors, beta blockers, and thiazide diuretics with risk of 4 of the most common adult cancers (breast, colorectal, lung, and prostate) in up to 289,612 cancer cases and 291,224 controls.
We found evidence that genetically proxied inhibition of the drug target for ACE inhibitors was associated with an increased risk of colorectal cancer.
There was little evidence that genetically proxied inhibition of the drug target for ACE inhibitors was associated with risk of the other cancers examined or evidence for an association of genetically proxied inhibition of drug targets for beta blockers and thiazide diuretics with risk of all 4 cancers examined.
What do these findings mean?
These findings provide support for a link between long-term inhibition of the drug target for ACE inhibitors and colorectal cancer risk, highlighting the need to evaluate the safety profiles of these medications in clinical trials with adequate follow-up length.
Prior to confirmation of an effect of ACE inhibitor use on colorectal cancer risk in clinical trials, these findings should not alter clinical practice for ACE inhibitor prescribing.
Introduction
Angiotensin-converting enzyme (ACE) inhibitors are commonly prescribed antihypertensive medications [1]. These medications lower blood pressure by inhibiting the conversion of angiotensin I to angiotensin II, a vasoconstrictor and the primary effector molecule of the renin–angiotensin system (RAS). Though clinical trials have supported the relative safety of these medications in the short term (median follow-up of 3.5 years), concerns have been raised that long-term use of these medications could increase risk of cancer [2,3]. These safety concerns relate to the multifaceted role of ACE, which cleaves various other substrates beyond angiotensin I, including several peptides that have proliferative effects. For example, ACE inhibition leads to the accumulation of bradykinin, an inflammatory mediator involved in tumor growth and metastasis [4]. In addition, substance P is elevated in ACE inhibitor users, which can promote tumor proliferation, migration, and angiogenesis [5,6].
Some observational epidemiological studies have suggested potential adverse effects of long-term use of these drugs on risk of common cancers (i.e., breast, colorectal, lung, and prostate) [7–10], though findings have been largely inconsistent (i.e., null and protective associations have also been reported for the relationship between ACE inhibitor use and cancer risk) [11–15]. Interpretation of the epidemiological literature is challenging for several reasons. First, pharmaco-epidemiological studies are susceptible to residual confounding due to unmeasured or imprecisely measured confounders, including those related to indication [16]. Second, several studies examining ACE inhibitor use and cancer risk have included prevalent drug users, which can introduce bias because prevalent users are “survivors” of the early period of pharmacotherapy and because covariates at study entry can be influenced by prior medication use [12,17–20]. Third, some prior studies may have suffered from time-related biases, including immortal time bias, which can arise because of misalignment of the start of follow-up, eligibility, and treatment assignment of participants [17,18,20,21]. These biases can produce illusory results in favor of the treatment group, while other biases often pervasive in the pharmaco-epidemiological literature (e.g., detection bias due to more intensive clinical monitoring and testing of individuals receiving treatment) can alternatively generate upward-biased effect estimates among those receiving treatment.
Along with ACE inhibitors, β blockers and thiazide diuretics are commonly prescribed antihypertensive medications that lower blood pressure through pathways independent to that of ACE (i.e., β blockers bind to β-adrenergic receptors, inhibiting the binding of norepinephrine and epinephrine to these receptors; thiazide diuretics promote sodium and water excretion by inhibiting sodium reabsorption in renal tubules) [4]. Some in vitro and epidemiological studies have suggested potential chemopreventive effects of these medications on cancer risk, though findings have been inconclusive [22–30].
Naturally occurring variation in genes encoding antihypertensive drug targets can be used as proxies for these targets to examine the effect of their therapeutic inhibition on disease outcomes (“mendelian randomization”) [31,32]. Such an approach should be less prone to conventional issues of confounding as germline genetic variants are randomly assorted at meiosis. In addition, mendelian randomization analysis permits the effect of long-term modulation of drug targets on cancer risk to be examined. Drug-target mendelian randomization can therefore be used to mimic the effect of pharmacologically modulating a drug target in clinical trials and has been used previously to anticipate clinical benefits and adverse effects of therapeutic interventions [33–36].
We used a mendelian randomization approach to examine the effect of long-term inhibition of the drug targets for ACE inhibitors (ACE; angiotensin-converting enzyme), β blockers (ADRB1; beta-1 adrenergic receptor), and thiazide diuretic agents (NCC; sodium-chloride symporter) on risk of overall and subtype-specific breast, colorectal, lung, and prostate cancer.
Methods
Study populations
For primary analyses, summary genetic association data were obtained from 4 cancer genome-wide association study (GWAS) consortia. Summary genetic association estimates for overall and estrogen receptor (ER)–stratified breast cancer risk in up to 122,977 cases and 105,974 controls were obtained from the Breast Cancer Association Consortium (BCAC) [37]. Summary genetic association estimates for overall and site-specific colorectal cancer risk in up to 58,221 cases and 67,694 controls were obtained from an analysis of the Genetics and Epidemiology of Colorectal Cancer Consortium (GECCO), ColoRectal Transdisciplinary Study (CORECT), and Colon Cancer Family Registry (CCFR) [38]. Summary genetic association estimates for overall and histological subtype-stratified lung cancer risk in up to 29,266 cases and 56,450 controls were obtained from an analysis of the Integrative Analysis of Lung Cancer Risk and Etiology (INTEGRAL) team of the International Lung Cancer Consortium (ILCCO) [39]. Summary genetic association estimates for overall and advanced prostate cancer risk in up to 79,148 cases and 61,106 controls were obtained from the Prostate Cancer Association Group to Investigate Cancer Associated Alterations in the Genome (PRACTICAL) consortium [40]. These analyses were restricted to participants of European ancestry.
For replication analyses, summary genetic association data were obtained on 1,573 colorectal cancer cases and 120,006 controls of European ancestry from the Finngen consortium. We also examined whether findings could be extended to individuals of East Asian ancestry by obtaining summary genetic association data on 23,572 colorectal cancer cases and 48,700 controls of East Asian ancestry from a GWAS meta-analysis of the Asia Colorectal Cancer Consortium and the Korean National Cancer Center CRC Study 2 [41].
Further information on statistical analysis, imputation, and quality control measures for these studies is available in the original publications. All studies contributing data to these analyses had the relevant institutional review board approval from each country, in accordance with the Declaration of Helsinki, and all participants provided informed consent.
Instrument construction
To generate instruments to proxy ACE, ADRB1, and NCC inhibition, we pooled summary genetic association data from 2 previously published GWAS of systolic blood pressure (SBP) using inverse-variance weighted fixed-effects models in METAL [42]. The first GWAS was a meta-analysis of ≤757,601 individuals of European descent in the UK Biobank and International Consortium of Blood Pressure-Genome Wide Association Studies (ICBP) [43]. The second GWAS was performed in 99,785 individuals in the Genetic Epidemiology Research on Adult Health and Aging (GERA) cohort, of whom the majority (81.0%) were of European ancestry [44]. Both GWAS were adjusted for age, sex, body mass index (BMI), and antihypertensive medication use. Estimates that were genome-wide significant (P < 5.0 × 10−8) in pooled analyses (N ≤ 857,386) and that showed concordant direction of effect across both GWAS were then used to generate instruments.
To proxy ADRB1 inhibition, 8 single-nucleotide polymorphisms (SNPs) associated with SBP at genome-wide significance and within ±100 kb windows from ADRB1 were obtained. To proxy NCC inhibition, 1 SNP associated with SBP at genome-wide significance and within a ±100-kb window from SLC12A3 (alias for NCC) was obtained. For both of these drug targets, SNPs used as proxies were permitted to be in weak linkage disequilibrium (r2 < 0.10) with each other to increase the proportion of variance in each respective drug target explained by the instrument, maximizing instrument strength.
Since pooled GWAS estimates were obtained from analyses adjusted for BMI, which could induce collider bias, we also examined constructing instruments using summary genetic association data from a previous GWAS of SBP in 340,159 individuals in UK Biobank without adjustment for BMI or antihypertensive medication use (S1 Table) [45].
We explored construction of genetic instruments to proxy ACE inhibition using 2 approaches: (i) by obtaining genome-wide significant variants in weak linkage disequilibrium (r2 < 0.10) in or within ±100 kb from ACE that were associated with SBP in previously described pooled GWAS analyses (resulting in 2 SNPs); and (ii) by obtaining genome-wide significant variants in weak linkage disequilibrium (r2 < 0.10) in or within ±100 kb from ACE that were associated with serum ACE concentrations in a GWAS of 4,174 participants in the Outcome Reduction with Initial Glargine INtervention (ORIGIN) study (resulting in 14 SNPs) [46]. Approximately 46.6% of participants in the ORIGIN study were of European ancestry, and 53.4% were of Latin American ancestry. Effect allele frequencies for these 14 SNPs were broadly similar across both ancestries (S2 Table). We then compared the proportion of variance in either SBP or serum ACE concentrations explained (r2) across each respective instrument to prioritize the primary instrument to proxy ACE inhibition. The “serum ACE concentrations instrument” (r2 = 0.34 to 0.39, F = 2,156.5 to 2,594.9) was prioritized as our primary instrument to examine genetically proxied ACE inhibition because of stronger instrument strength as compared to the “SBP instrument” (r2 = 0.02, F = 128.5). In sensitivity analyses, we also examined the association between genetically proxied ACE inhibition and cancer endpoints using the “SBP instrument.”
As an additional instrument construction step, we also performed a post hoc comparison of the proportion of variance in serum ACE concentrations explained by both instruments and found that the serum ACE concentrations instrument explained a larger proportion of the variance in this trait than the SBP instrument (r2 = 0.28, F = 759.9).
To validate the serum ACE concentrations instrument, we examined the association between genetically proxied ACE inhibition and (i) SBP; (ii) risk of stroke in the MEGASTROKE consortium (40,585 cases; 406,111 controls of European ancestry); (iii) risk of coronary artery disease in the CARDIoGRAMplusC4D consortium (60,801 cases; 123,504 controls, 77% of whom were of European ancestry); and (iv) risk of type 2 diabetes in the DIAGRAM consortium (N = 74,124 cases; 824,006 controls of European ancestry) and compared the direction of effect estimates obtained with those reported for ACE inhibitor use in meta-analyses of randomized controlled trials [47–49]. Likewise, we validated ADRB1 and NCC instruments by examining the association between inhibition of these targets and risk of stroke and coronary artery disease, as reported in meta-analyses of clinical trials [49].
For analyses in individuals of East Asian ancestry, 1 cis-acting variant (rs4343) associated with ACE activity (P = 3.0 × 10−25) in a GWAS of 623 individuals with young onset hypertension of Han Chinese descent was obtained [50]. In the Japanese Biobank (N = 136,597), the A allele of rs4343 has previously been shown to associate with lower SBP (−0.26 mm Hg SBP, 95% CI −0.11 to −0.42; P = 6.7 × 10−4) [51]. This variant explained 0.008% of the variance of SBP (F = 11.6).
Mendelian randomization primary and sensitivity analyses
Inverse-variance weighted random-effects models (permitting heterogeneity in causal estimates) were employed to estimate causal effects of genetically proxied drug target inhibition on cancer risk [52]. These models were adjusted for weak linkage disequilibrium between SNPs (r2 < 0.10) with reference to the 1,000 Genomes Phase 3 reference panel [53,54]. If underdispersion in causal estimates generated from individual genetic variants was present, the residual standard error was set to 1.
Mendelian randomization analysis assumes that the genetic instrument used to proxy a drug target (i) is associated with the drug target (“relevance”); (ii) does not share a common cause with the outcome (“exchangeability”); and (iii) affects the outcome only through the drug target (“exclusion restriction”).
We tested the “relevance” assumption by generating estimates of the proportion of variance of each drug target explained by the instrument (r2) and F-statistics. F-statistics can be used to examine whether results are likely to be influenced by weak instrument bias, i.e., reduced statistical power when an instrument explains a limited proportion of the variance in a drug target. As a convention, an F-statistic of at least 10 is indicative of minimal weak instrument bias [55].
We evaluated the “exclusion restriction” assumption by performing various sensitivity analyses. First, we performed colocalization to examine whether drug targets and cancer endpoints showing nominal evidence of an association in MR analyses (P < 0.05) share the same causal variant at a given locus. Such an analysis can permit exploration of whether drug targets and cancer outcomes are influenced by distinct causal variants that are in linkage disequilibrium with each other, indicative of horizontal pleiotropy (an instrument influencing an outcome through pathways independent to that of the exposure), a violation of the exclusion restriction criterion [56]. Colocalization analysis was performed by generating ±300 kb windows from the top SNP used to proxy each respective drug target. As a convention, a posterior probability of ≥0.80 was used to indicate support for a configuration tested. An extended description of colocalization analysis including assumptions of this method is presented in S1 Methods.
For analyses showing evidence of colocalization across drug target and cancer endpoint signals, we then examined whether there was evidence of an association of genetically proxied inhibition of that target with previously reported risk factors for the relevant cancer endpoint (i.e., BMI, low-density lipoprotein cholesterol, total cholesterol, iron, insulin-like growth factor 1, alcohol intake, standing height, and physical activity for colorectal cancer risk) [57–65]. If there was evidence for an association between a genetically proxied drug target and previously reported risk factor (P < 0.05), this could reflect vertical pleiotropy (i.e. “mediated pleiotropy” where an instrument has an effect on 2 or more traits that influence an outcome via the same biological pathway) or horizontal pleiotropy. In the presence of an association with a previously reported risk factor, multivariable mendelian randomization can then be used to examine the association of drug target inhibition in relation to cancer risk, accounting for this risk factor [45].
As an additional post hoc sensitivity analysis, we also evaluated whether SNPs used to instrument ADRB1 and NCC inhibition were also expression quantitative trait loci (eQTLs) for the genes encoding these proteins. Instrument validation and cancer endpoint mendelian randomization analyses were then repeated by restricting instruments to SNPs showing evidence of being eQTLs for these targets (Additional information on these sensitivity analyses is provided in S2 Methods).
Finally, iterative leave-one-out analysis was performed iteratively removing 1 SNP at a time from instruments to examine whether findings were driven by a single influential SNP.
To account for multiple testing across primary drug target analyses, a Bonferroni correction was used to establish a P value threshold of <0.0014 (false positive rate = 0.05/36 statistical tests [3 drug targets tested against 12 cancer endpoints]), which we used as a heuristic to define “strong evidence,” with findings between P ≥ 0.0014 and P < 0.20 defined as “weak evidence.”
Colon transcriptome-wide GRS analysis
To explore potential mechanisms governing associations and to further evaluate potential violations of mendelian randomization assumptions, we examined associations between a genetic risk score for serum ACE concentrations and gene expression profiles in normal (i.e., nonneoplastic) colon tissue samples. Gene expression analysis was performed using data from the University of Barcelona and the University of Virginia Genotyping and RNA Sequencing Project (BarcUVa-Seq) [66]. This analysis was restricted to 445 individuals (mean age 60 years, 64% female, 95% of European ancestry) who participated in a Spanish colorectal cancer risk screening program that obtained a normal colonoscopy result (i.e., macroscopically normal colon tissue, with no malignant lesions). Further information on RNA-Seq data processing and quality control is presented in S3 Methods.
To perform transcriptome-wide analyses, weighted genetic risk scores (wGRS) to proxy serum ACE concentrations were constructed using 14 ACE SNPs in Plink v1.9 [67]. Expression levels for 21,482 genes (expressed as inverse normal transformed trimmed mean of M-values) were regressed on the standardized wGRS and adjusted for sex, the top 2 principal components of genetic ancestry, sequencing batch, probabilistic estimation of expression residuals (PEER) factors, and colon anatomical location. To account for multiple testing, a Bonferroni correction was used to establish a P value threshold of <2.33 × 10−6 (false positive rate = 0.05/21,482 statistical tests).
Bioinformatic follow-up of findings from transcriptome-wide analysis was performed to further interrogate downstream perturbations of the ACE wGRS on gene expression profiles using gene set enrichment analysis and coexpression network analysis. In brief, these methods can either evaluate whether expression levels of genes associated with the ACE wGRS are enriched in relation to an a priori defined set of genes based on curated functional annotation (gene set enrichment analysis) or permit the identification of clusters of genes (termed “modules” and assigned arbitrary color codes), which show a coordinated expression pattern associated with the wGRS (coexpression network analysis). Further information on gene set enrichment and coexpression network analysis is presented in S4 Methods.
There was no formal prespecified protocol for this study. All analyses described above were decided a priori except those designated as “post hoc” where additional sensitivity analyses were performed in response to peer review comments. This study is reported as per the Guidelines for strengthening the reporting of mendelian randomization studies (STROBE-MR) checklist (S1 STROBE Checklist) [68]. All statistical analyses were performed using R version 3.3.1.
Results
Across the 3 drug targets that we examined, conservative estimates of F-statistics for their respective genetic instruments ranged from 269.1 to 2,156.5, suggesting that our analyses were unlikely to suffer from weak instrument bias. Characteristics of genetic variants in ACE, ADRB1, and SLC12A3 used to proxy each pharmacological target are presented in Table 1. Estimates of r2 and F-statistics for each target are presented in S3 Table.
Table 1. Characteristics of SBP lowering genetic variants in ACE, ADRB1, and SLC12A3.
| Target | Effect allele/Noneffect Allele | Effect Allele Frequency | Effect (SE) | P value |
|---|---|---|---|---|
| ACE | ||||
| rs4343 | A/G | 0.45 | −0.63 (0.02) | 1.53 × 10−213 |
| rs12452187 | A/G | 0.60 | −0.23 (0.02) | 2.53 × 10−27 |
| rs79480822 | C/T | 0.93 | −0.55 (0.05) | 6.37 × 10−24 |
| rs3730025 | G/A | 0.01 | −0.80 (0.09) | 4.32 × 10−19 |
| rs11655956 | C/G | 0.08 | −0.35 (0.04) | 1.06 × 10−15 |
| rs118121655 | G/A | 0.96 | −0.54 (0.07) | 3.10 × 10−15 |
| rs4365 | G/A | 0.97 | −0.58 (0.08) | 7.06 × 10−12 |
| rs4968771 | G/A | 0.08 | −0.22 (0.03) | 1.78 × 10−11 |
| rs12150648 | G/A | 0.96 | −0.39 (0.06) | 1.88 × 10−10 |
| rs80311894 | T/G | 0.97 | −0.46 (0.07) | 2.60 × 10−10 |
| rs118138685 | C/G | 0.04 | −0.40 (0.07) | 2.44 × 10−9 |
| rs13342595 | C/T | 0.23 | −0.14 (0.02) | 2.48 × 10−9 |
| rs28656895 | T/C | 0.23 | −0.14 (0.02) | 3.77 × 10−9 |
| rs4968780 | C/A | 0.05 | −0.28 (0.05) | 1.86 × 10−8 |
| ADRB1 | ||||
| rs1801253 | G/C | 0.23 | −0.41 (0.03) | 8.07 × 10−43 |
| rs11196549 | G/A | 0.96 | −0.62 (0.07) | 2.53 × 10−19 |
| rs4918889 | G/C | 0.17 | −0.30 (0.04) | 7.53 × 10−18 |
| rs460718 | A/G | 0.33 | −0.24 (0.03) | 2.21 × 10−17 |
| rs11196597 | G/A | 0.86 | −0.27 (0.04) | 3.07 × 10−12 |
| rs143854972 | G/A | 0.94 | −0.39 (0.06) | 4.35 × 10−11 |
| rs17875473 | C/T | 0.91 | −0.28 (0.05) | 9.04 × 10−9 |
| rs10787510 | A/G | 0.48 | −0.15 (0.03) | 2.01 × 10−8 |
| NCC | ||||
| rs35797045 | A/C | 0.05 | −0.35 (0.06) | 4.85 × 10−8 |
Effect (SE) represents change in serum ACE concentrations per additional copy of the effect allele for ACE analysis and change in SBP per additional copy of the effect allele for ADRB1 and NCC analyses. In analyses of genetically proxied ACE inhibition and colorectal cancer risk, 1 SNP (rs8064760) was not available in the colorectal cancer dataset. Two SNPs associated with SBP used to proxy ACE inhibition in sensitivity analyses were as follows: rs8077276 (effect allele/noneffect allele: A/G, effect (se): −0.27 (0.03), effect allele frequency: 0.62; P value: 4.47 × 10−22) and rs28656895 (effect allele/noneffect allele: T/C, effect (se): −0.19 (0.03), effect allele frequency: 0.23; P value: 3.37 × 10−9).
ACE, angiotensin-converting enzyme; ADRB1, β-1 adrenergic receptor; NCC, sodium-chloride symporter; SBP, systolic blood pressure; SNP, single-nucleotide polymorphism.
Instrument validation
Findings from genetic instrument validation analyses for drug targets were broadly concordant (i.e., in direction of effect) with findings from meta-analyses of randomized trials for these medications. Genetically proxied ACE inhibition was associated with lower SBP (mm Hg per SD lower serum ACE concentration: −0.40, 95% CI −0.21 to −0.59, P = 4.2 × 10−5) and a lower risk of type 2 diabetes (odds ratio (OR) equivalent to 1 mm Hg lower SBP: 0.90, 95% CI 0.85 to 0.95, P = 1.3 × 10−4). There was weak evidence for an association of genetically proxied ACE inhibition with lower risk of stroke (OR 0.94, 95% CI 0.88 to 1.01; P = 0.06) and coronary artery disease (OR 0.95, 95% CI 0.89 to 1.02; P = 0.16).
Genetically proxied ADRB1 inhibition was associated with lower risk of coronary artery disease (per 1 mm Hg lower SBP: OR 0.95, 95% CI 0.92 to 0.98; P = 1.5 × 10−3) and weakly associated with risk of stroke (OR 1.03, 95% CI 0.99 to 1.07; P = 0.18).
Genetically proxied NCC inhibition was associated with lower risk of coronary artery disease (per 1 mm Hg lower SBP: OR 0.81, 95% CI 0.81, 95% CI 0.71 to 0.93, P = 3.2 × 10−3) and was weakly associated with lower risk of stroke (OR 0.89, 95% CI 0.78 to 1.02; P = 0.10).
Genetically proxied ACE inhibition and cancer risk
Genetically proxied ACE inhibition was associated with an increased odds of colorectal cancer (OR equivalent to 1 mm Hg lower SBP: 1.13, 95% CI 1.06 to 1.22; P = 3.6 × 10−4). Likewise, in analyses using SBP SNPs in ACE, genetically proxied SBP lowering via ACE inhibition was associated with an increased odds of colorectal cancer (OR equivalent to 1 mm Hg lower SBP: 1.11, 95% CI 1.04 to 1.18; P = 1.3 × 10−3). When scaled to represent SBP lowering achieved in clinical trials of ACE inhibitors for primary hypertension (equivalent to 8 mm Hg lower SBP), this represents an OR of 2.74 (95% CI 1.58 to 4.76) [69]. In site-specific analyses, this association was stronger for colon cancer risk (OR 1.18, 95% CI 1.07 to 1.31; P = 9.7 × 10−4) than rectal cancer risk (OR 1.07, 95% CI 0.97 to 1.18; P = 0.16). Similar associations were found across risk of proximal colon cancer (OR 1.23, 95% CI 1.10 to 1.37; P = 1.9 × 10−4) and distal colon cancer (OR 1.15, 95% CI 1.03 to 1.27; P = 0.01).
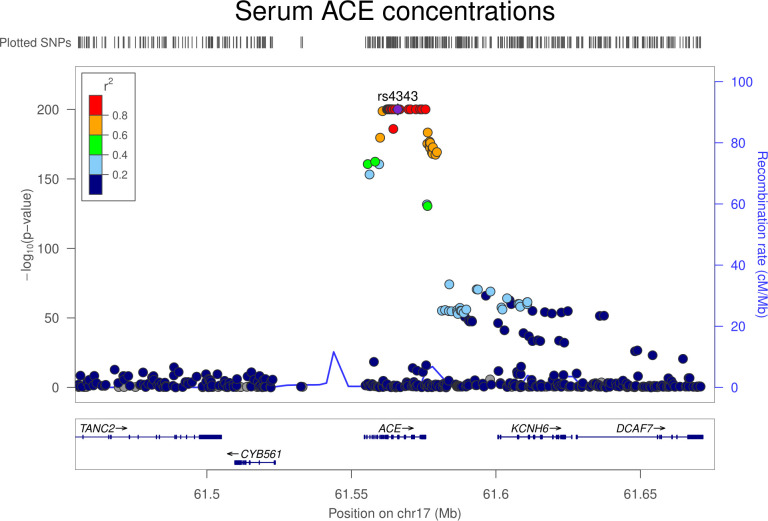
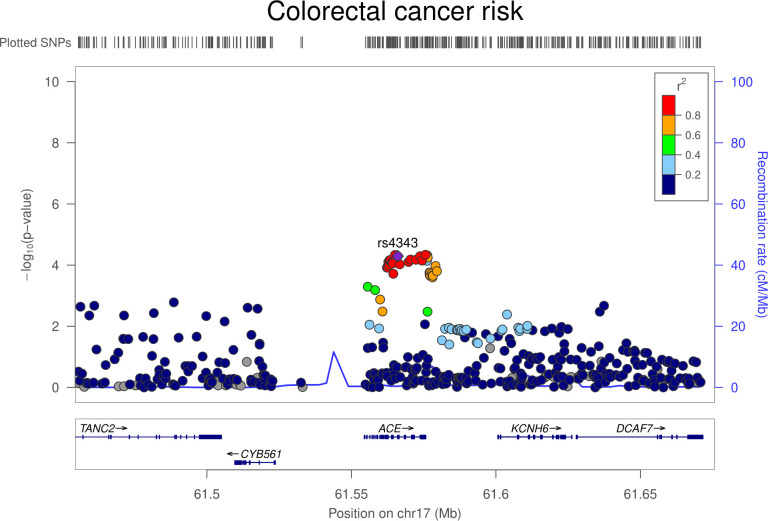
Colocalization analysis suggested that serum ACE and colorectal cancer associations had a 91.4% posterior probability of sharing a causal variant within the ACE locus (S4 Table). Regional Manhattan plots examining the association of all SNPs ±300 kb from the top SNP for serum ACE concentrations (rs4343) for their association with serum ACE concentrations (Fig 1) and with colorectal cancer risk (Fig 2) did not appear to support the presence of 2 or more independent causal variants driving associations across either trait.
Fig 1. Regional Manhattan plot of associations of SNPs with serum ACE concentrations ±300 kb from the SNP used to proxy serum ACE concentrations (rs4343) in the ACE region.
ACE, angiotensin-converting enzyme; Mb, Megabase; SNP, single-nucleotide polymorphism.
Fig 2. Regional Manhattan plot of associations of SNPs with colorectal cancer risk ±300 kb from the SNP used to proxy serum ACE concentrations (rs4343) in the ACE region.
ACE, angiotensin-converting enzyme; Mb, Megabase; SNP, single-nucleotide polymorphism.
In mendelian randomization analyses examining the association of genetically proxied ACE inhibition with 8 previously reported colorectal cancer risk factors, there was little evidence to support associations (S5 Table). There was also little evidence to support an association of genetically proxied SBP with colorectal cancer risk (OR per 1 mmHg lower SBP: 1.00, 95% CI 0.99 to 1.01; P = 0.50), suggesting a potential mechanism-specific effect of this drug target on colorectal cancer risk.
Additionally, results of analyses that iteratively removed one SNP at a time from the instrument and recalculated the overall mendelian randomization estimate were consistent, suggesting that associations were not being driven through individual influential SNPs (S6 Table).
There was little evidence that genetically proxied ACE inhibition was associated with risk of breast cancer (OR 0.98, 95% CI 0.94 to 1.02; P = 0.35) or lung cancer (OR 1.01, 95% CI 0.92 to 1.10; P = 0.93) and weak evidence for an association with prostate cancer risk (OR 1.06, 95% CI 0.99 to 1.13; P = 0.08). Likewise, there was little evidence of association of genetically proxied ACE inhibition with these cancers in histological subtype-stratified analyses (Table 2).
Table 2. Association between genetically proxied ACE inhibition and risk of overall and subtype-specific breast, colorectal, prostate, and lung cancer risk.
| Outcome | N (cases, controls) | OR (95% CI) | P value |
|---|---|---|---|
| Breast cancer | 122,977; 105,974 | 0.98 (0.94–1.02) | 0.35 |
| ER+ Breast cancer | 69,501; 105,974 | 0.99 (0.94–1.04) | 0.76 |
| ER− Breast cancer | 21,468; 105,974 | 0.97 (0.90–1.05) | 0.47 |
| Colorectal cancer | 58,221; 67,694 | 1.13 (1.06–1.22) | 3.6 × 10−4 |
| Colon cancer | 32,002; 64,159 | 1.18 (1.07–1.31) | 9.7 × 10−4 |
| Rectal cancer | 16,212; 64,159 | 1.07 (0.97–1.18) | 0.16 |
| Lung cancer | 29,863; 55,586 | 1.01 (0.92–1.10) | 0.93 |
| Lung adenocarcinoma | 11,245; 54,619 | 1.02 (0.91–1.15) | 0.70 |
| Small cell lung carcinoma | 2,791; 20,580 | 0.96 (0.76–1.20) | 0.71 |
| Squamous cell lung cancer | 7,704; 54,763 | 0.97 (0.81–1.16) | 0.73 |
| Prostate cancer | 79,148; 61,106 | 1.06 (0.99–1.13) | 0.08 |
| Advanced prostate cancer | 15,167; 58,308 | 1.05 (0.94–1.17) | 0.37 |
ACE, angiotensin-converting enzyme; CI, confidence interval; ER, estrogen receptor; OR, odds ratio, SBP, systolic blood pressure.
OR represents the exponential change in odds of cancer per genetically proxied inhibition of ACE equivalent to a 1-mm Hg decrease in SBP.
Genetically proxied ADRB1 inhibition and cancer risk
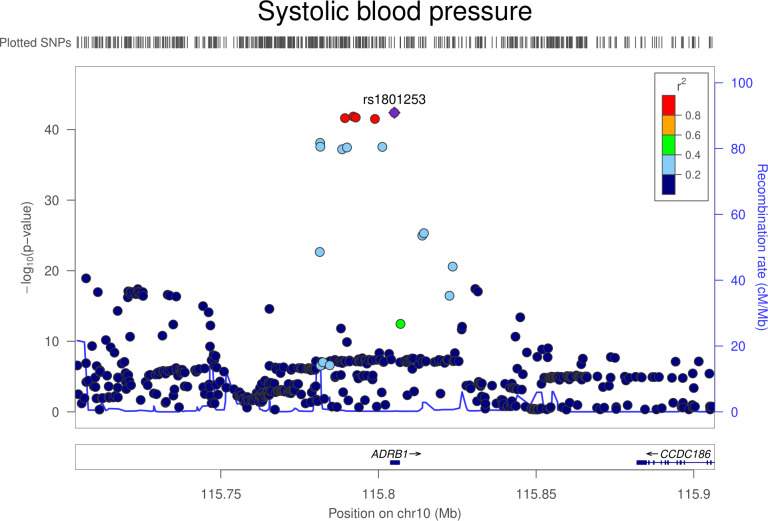
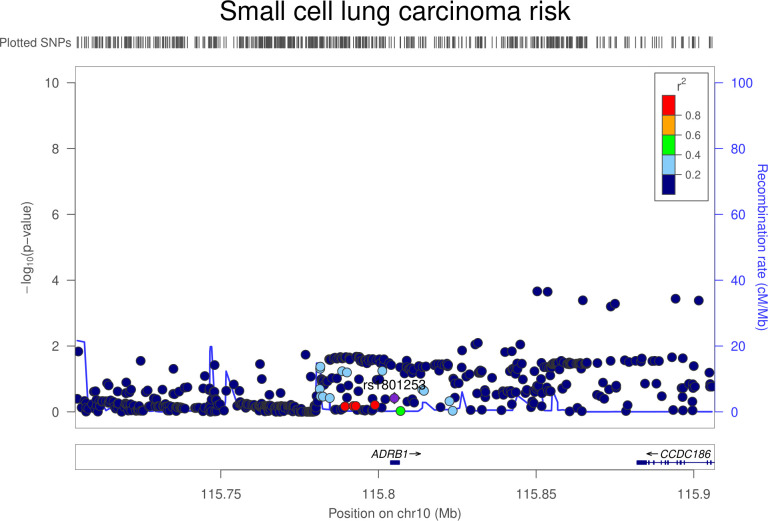
There was little evidence that genetically proxied ADRB1 inhibition was associated with overall risk of breast, colorectal, lung, or prostate cancer (Table 3). In lung cancer subtype-stratified analyses, there was weak evidence to suggest an association of genetically proxied ADRB1 inhibition with lower risk of small cell lung carcinoma (OR equivalent to 1 mm Hg lower SBP: 0.87, 95% CI 0.79 to 0.96; P = 0.008). Colocalization analysis suggested that ADRB1 and small cell lung carcinoma were unlikely to share a causal variant within the ADRB1 locus (1.5% posterior probability of a shared causal variant) (S7 Table, Figs 3 and 4). Findings for overall and subtype-specific cancer risk did not differ markedly when using an instrument for ADRB1 inhibition constructed from a GWAS unadjusted for BMI (S8 Table). In sensitivity analyses restricting the ADRB1 instrument to SNPs that are eQTLs for ADRB1, findings were consistent with those obtained when using the primary instrument for this target (S10 Table).
Table 3. Association between genetically proxied ADRB1 inhibition and risk of overall and subtype-specific breast, colorectal, prostate, and lung cancer risk.
| Outcome | N (cases, controls) | OR (95% CI) | P value |
|---|---|---|---|
| Breast cancer | 122,977; 105,974 | 1.01 (0.99–1.04) | 0.38 |
| ER+ Breast cancer | 69,501; 105,974 | 1.01 (0.98–1.04) | 0.44 |
| ER− Breast cancer | 21,468; 105,974 | 0.98 (0.94–1.02) | 0.38 |
| Colorectal cancer | 58,221; 67,694 | 0.98 (0.96–1.01) | 0.31 |
| Colon cancer | 32,002; 64,159 | 0.99 (0.95–1.03) | 0.63 |
| Rectal cancer | 16,212; 64,159 | 1.00 (0.95–1.04) | 0.84 |
| Lung cancer | 29,863; 55,586 | 1.01 (0.96–1.07) | 0.64 |
| Lung adenocarcinoma | 11,245; 54,619 | 0.98 (0.91–1.04) | 0.48 |
| Small cell lung carcinoma | 2,791; 20,580 | 0.87 (0.79–0.96) | 0.008 |
| Squamous cell lung cancer | 7,704; 54,763 | 0.98 (0.91–1.06) | 0.67 |
| Prostate cancer | 79,148; 61,106 | 1.00 (0.96–1.03) | 0.73 |
| Advanced prostate cancer | 15,167; 58,308 | 1.00 (0.94–1.06) | 0.97 |
ADRB1, β-1 adrenergic receptor; CI, confidence interval; ER, estrogen receptor; OR, odds ratio, SBP, systolic blood pressure.
OR represents the exponential change in odds of cancer per genetically proxied inhibition of ADRB1 equivalent to a 1-mm Hg decrease in SBP.
Fig 3. Regional Manhattan plot of associations of SNPs with SBP ±300 kb from the SNP used to proxy SBP (rs1801253) in the ADRB1 region.
Mb, Megabase; SBP, systolic blood pressure; SNP, single-nucleotide polymorphism.
Fig 4. Regional Manhattan plot of associations of SNPs with small cell lung carcinoma risk ±300 kb from the SNP used to proxy SBP (rs1801253) in the ADRB1 region.
Mb, Megabase; SBP, systolic blood pressure; SNP, single-nucleotide polymorphism.
Genetically proxied NCC inhibition and cancer risk
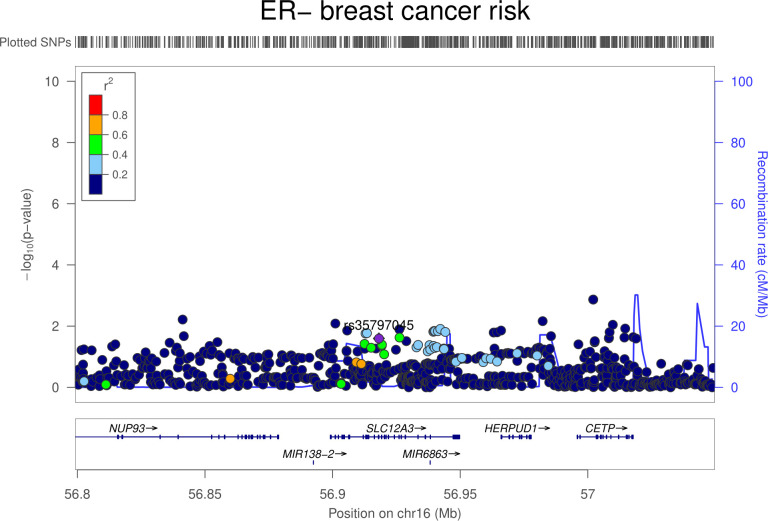
There was little evidence that genetically proxied NCC inhibition was associated with overall risk of breast, colorectal, lung, or prostate cancer (Table 4). In ER–stratified breast cancer analyses, there was weak evidence that NCC inhibition was associated with an increased risk of ER− breast cancer (OR equivalent to 1 mm Hg lower SBP: 1.20, 95% CI 1.02 to 1.40; P = 0.03). Colocalization analysis provided little support for NCC and ER− breast cancer association sharing a causal variant within the SLC12A3 locus (11.5% posterior probability of a shared causal variant) (S11 Table, Figs 5 and 6).
Table 4. Association between genetically proxied NCC inhibition and risk of overall and subtype-specific breast, colorectal, prostate, and lung cancer risk.
| Outcome | N (cases, controls) | OR (95% CI) | P value |
|---|---|---|---|
| Breast cancer | 122,977; 105,974 | 1.08 (0.99–1.18) | 0.08 |
| ER+ Breast cancer | 69,501; 105,974 | 1.06 (0.95–1.18) | 0.28 |
| ER− Breast cancer | 21,468; 105,974 | 1.20 (1.02–1.40) | 0.03 |
| Colorectal cancer | 58,221; 67,694 | 1.09 (0.96–1.23) | 0.19 |
| Colon cancer | 32,002; 64,159 | 1.03 (0.89–1.19) | 0.69 |
| Rectal cancer | 16,212; 64,159 | 1.13 (0.94–1.36) | 0.20 |
| Lung cancer | 29,863; 55,586 | 1.09 (0.89–1.33) | 0.38 |
| Lung adenocarcinoma | 11,245; 54,619 | 1.01 (0.81–1.26) | 0.95 |
| Small cell lung carcinoma | 2,791; 20,580 | 1.12 (0.76–1.53) | 0.57 |
| Squamous cell lung cancer | 7,704; 54,763 | 1.00 (0.78–1.29) | 0.99 |
| Prostate cancer | 79,148; 61,106 | 1.08 (0.96–1.19) | 0.18 |
| Advanced prostate cancer | 15,167; 58,308 | 1.05 (0.86–1.28) | 0.63 |
CI, confidence interval; ER, estrogen receptor; NCC, sodium-chloride symporter; OR, odds ratio, SBP, systolic blood pressure.
OR represents the exponential change in odds of cancer per genetically proxied inhibition of NCC equivalent to a 1-mm Hg decrease in SBP.
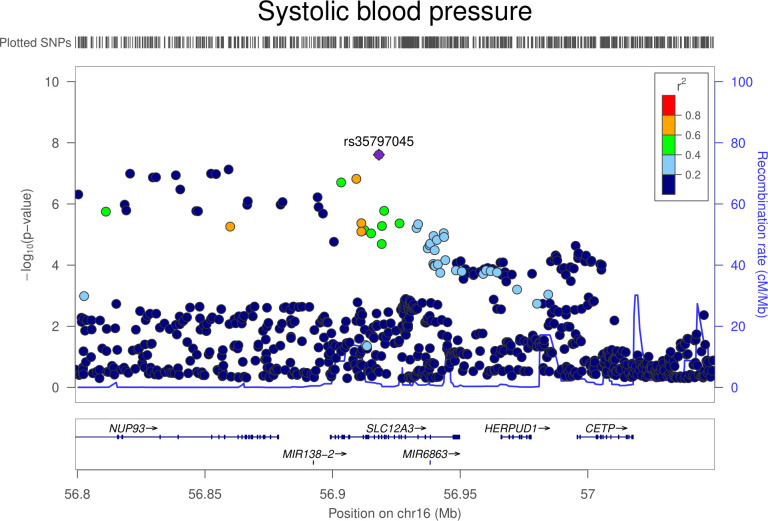
Fig 5. Regional Manhattan plot of associations of SNPs with SBP ±300 kb from the SNP used to proxy SBP (rs35797045) in the SLC12A3 region.
Mb, Megabase; SBP, systolic blood pressure; SNP, single-nucleotide polymorphism.
Fig 6. Regional Manhattan plot of associations of SNPs with ER− breast cancer risk ±300 kb from the SNP used to proxy SBP (rs35797045) in the SLC12A3 region.
ER, estrogen receptor; Mb, Megabase; SBP, systolic blood pressure; SNP, single-nucleotide polymorphism.
Replication analysis in Europeans and exploratory analysis in East Asians
Findings for genetically proxied ACE inhibition and colorectal cancer risk were replicated in an independent sample of 1,571 colorectal cancer cases and 120,006 controls of European ancestry in the Finngen consortium (1.40, 95% CI 1.02 to 1.92; P = 0.035). In analyses of 23,572 colorectal cancer cases and 48,700 controls of East Asian descent, there was little evidence of association of genetically proxied ACE inhibition and colorectal cancer risk (OR 0.97, 95% CI 0.88 to 1.07; P = 0.59).
Colon gene expression analysis
In transcriptome-wide analyses, the serum ACE wGRS was most strongly associated with ACE expression levels in the colon (trimmed mean of M-values [TMMs] per SD increase in wGRS: −0.42, 95% CI −0.49 to −0.36; P = 2.29 × 10−31). Genetically proxied ACE expression in the colon was associated with increased odds of colorectal cancer (OR per SD increase in expression: 1.02, 95% CI 1.00 to 1.04; P = 0.01). However, colocalization analysis suggested that colon ACE expression and colorectal cancer risk were unlikely to share a causal variant within the ACE locus (29.1% posterior probability of a shared causal variant) (S12 Table, Figs 7 and 8). The serum ACE wGRS was also associated with expression levels of CYB561 (TMMs per SD increase in wGRS: −0.17, 95% CI −0.21 to −0.12; P = 8.28 × 10−11) and FTSJ3 (TMMs per SD increase in wGRS: −0.19, 95% CI −0.24 to −0.13; P = 2.95 × 10−10) in the colon after correction for multiple testing. ACE, CYB561, and FTSJ3 are neighboring genes on chromosome 17, suggesting that associations between the ACE wGRS and CYB561 and FTSJ3 could be driven through their coexpression. Genetically proxied CYB561 expression in the colon was associated with increased odds of colorectal cancer (OR per SD increase in expression: 1.06, 95% CI 1.02 to 1.10; P = 0.005). However, multivariable mendelian randomization analysis examining the association of genetically proxied ACE inhibition with colorectal cancer risk adjusting for CYB561 expression in the colon was consistent with univariable analyses (OR 1.13, 95% CI: 0.96 to 1.32; P = 0.14). Genetically proxied FTSJ3 expression in the colon was not associated with odds of colorectal cancer (OR per SD increase in expression: 1.00, 95% CI 0.98 to 1.03; P = 0.77).
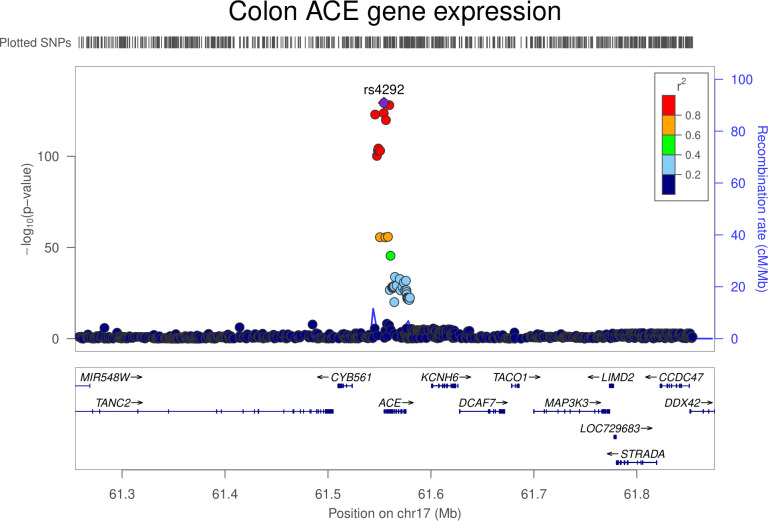
Fig 7. Regional Manhattan plot of associations of SNPs with colon ACE expression ±300 kb from the SNP used to proxy colon ACE expression (rs4292) in the ACE region.
ACE, angiotensin-converting enzyme; Mb, Megabase; SNP, single-nucleotide polymorphism.
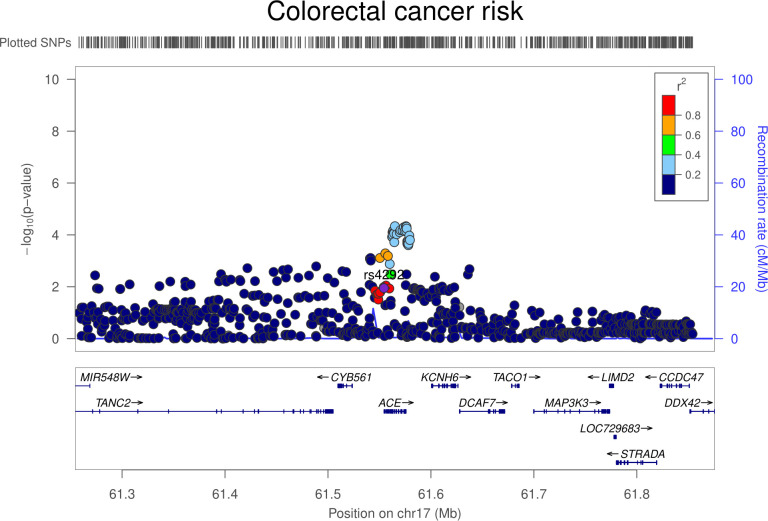
Fig 8. Regional Manhattan plot of associations of SNPs with colorectal cancer risk ±300 kb from the SNP used to proxy colon ACE expression (rs4292) in the ACE region.
ACE, angiotensin-converting enzyme; Mb, Megabase; SNP, single-nucleotide polymorphism.
In gene set enrichment analysis of genes whose expression was associated with the serum ACE wGRS (P < 5.0 × 10−3), there was evidence for enrichment of expression of genes relating to memory CD8 T cells (as compared to effector CD8 T cells) in the immunologic signatures database (GSE10239) (P = 1.35 × 10−6) but little evidence for expression of other gene sets or pathways after correction for multiple testing.
In coexpression network analysis, 30 distinct modules were defined. ACE was in the black module along with another 659 genes. This module was correlated with the ACE wGRS (r = −0.11; P = 0.03). Gene set enrichment analysis of genes located in the black module showed evidence of enrichment in susceptibility genes for colorectal cancer (P = 1.00 × 10−3).
Complete findings from transcriptome-wide GRS and gene set enrichment analyses, along with genes from the black module from coexpression network analysis are presented in S11–S14 Tables.
Discussion
In this mendelian randomization analysis of up to 289,612 cancer cases and 291,224 controls, genetically proxied long-term ACE inhibition was associated with an increased risk of colorectal cancer. This association was restricted to cancer of the colon, with similar associations across the proximal and distal colon. There was little evidence to support associations of genetically proxied ACE inhibition with risk of other cancers. Genetically proxied ADRB1 and NCC inhibition were not associated with risk of breast, colorectal, lung or prostate cancer.
Our findings for genetically proxied ACE inhibition and colorectal cancer risk are not consistent with some previous conventional observational analyses. A meta-analysis of 7 observational studies reported a protective association of ACE inhibitor use with colorectal cancer risk (OR 0.81 95% CI 0.70 to 0.92), though with substantial heterogeneity across studies (I2 = 71.1%) [14]. Interpretation of these findings is complicated by variable use of prevalent drug users, heterogenous comparator groups (both active controls and nondrug users), and the potential for immortal time bias across most included studies. Further, this meta-analysis did not include data from an earlier large Danish population-based case–control analysis with 15,560 colorectal cancer cases and 62,525 controls, which reported an increased risk of colorectal cancer (OR 1.30, 95% CI 1.22 to 1.39) among long-term users of ACE inhibitors (≥1,000 daily doses within 5 years of study entry), as compared to never-users [7].
The potential mechanisms underpinning an association between genetically proxied ACE inhibition and colorectal cancer risk are unclear. ACE is a multifaceted enzyme, capable of cleaving several different peptide substrates with potential roles in carcinogenesis [70]. Along with ACE inhibition leading to an accumulation of bradykinin and substance P, both potential inducers of tumor proliferation, ACE inhibition can also lead to an increase in Ac-SDKP, an endogenous antifibrotic peptide that is capable of inducing angiogenesis [71]. The observed restriction of an association of genetically proxied ACE inhibition with risk of colon, but not rectal, cancer is consistent with evidence that mRNA and protein levels of ACE are enriched in the colon but not in rectal tissue [72]. There was limited evidence of association of a serum ACE genetic risk score with distinct gene expression profiles in transcriptome-wide analyses. However, gene set enrichment analysis of these findings suggested enriched expression of genes involved in immunological pathways relating to memory CD8 T cells and coexpression network analysis identified ACE expression in a cluster of coexpressed genes enriched for colorectal cancer risk susceptibility genes (e.g., LAMA5, PNKD, TOX2, PLEKHG6) [73]. These findings suggest potential future avenues of exploration to uncover mechanistic pathways linking ACE with colorectal cancer risk.
Meta-analyses of randomized trials have not reported increased rates of cancer among ACE inhibitor users, though these analyses have not reported findings separately for colorectal cancer [2,3]. Potential discrepancies in findings for colorectal cancer between this mendelian randomization analysis and previous clinical trials could reflect the relatively short duration of these trials (median follow-up of 3.5 years) given long induction periods of colorectal cancer. For example, the “adenocarcinoma sequence” proposes that transformation of normal colorectal epithelium to an adenoma and ultimately to invasive and metastatic cancer may occur over the course of several decades [74,75]. Consistent with this long induction period, in randomized controlled trials examining the chemopreventive effect of aspirin on colorectal cancer risk, protective effects of aspirin are not seen until 7 years after initiation of treatment, with clear risk reductions becoming apparent only after 10 years of follow-up [76]. It is therefore possible that adverse effects of ACE inhibition on colorectal cancer risk may likewise not emerge until many years after treatment initiation. Alternatively, it may be possible that an effect of ACE inhibition on cancer is restricted solely to the earliest stages of the adenoma-carcinoma sequence and therefore may not influence cancer risk among largely middle-aged participants of clinical trials if dysplasia is already present. Finally, it is possible that lower levels of circulating ACE concentrations may influence colorectal cancer risk only during a particular critical or sensitive period of the life course (e.g., in childhood or adolescence), given some evidence to suggest a potential role of early-life factors in colorectal carcinogenesis [77].
Our largely null findings for genetically proxied ACE inhibition and risk of breast, lung, and prostate cancer risk are not consistent with some previous observational reports that compared ACE inhibitor users to nonusers or to users of β blockers or thiazide diuretics [7,9,17]. However, our findings for genetically proxied ACE inhibition are in agreement with those from short-term randomized controlled trials for these site-specific cancers and suggest that long-term use of these drugs may not influence cancer risk, though we cannot rule out small effects from their long-term use [3]. Likewise, our findings for ADRB1 and NCC are in agreement with short-term trial data reporting no association of β blockers and thiazide diuretics use with overall cancer risk [2].
Strengths of this analysis include the use of cis-acting variants in genes encoding antihypertensive drug targets to proxy inhibition of these targets, which should minimize confounding, the employment of various sensitivity analyses to rigorously assess for violations of mendelian randomization assumptions, and the use of a summary-data mendelian randomization approach, which permitted us to leverage large-scale genetic data from several cancer GWAS consortia, enhancing statistical power and precision of causal estimates. As with prior mendelian randomization analyses of antihypertensive drug targets that used similar approaches to instrument construction to our analysis, the general concordance of estimates of the effect of these instruments on cardiometabolic endpoints with those reported in prior clinical trials for these medications supports the plausibility of these instruments [78,79]. Finally, the use of germline genetic variants as proxies for antihypertensive drug targets facilitated evaluation of the effect of the long-term inhibition of these targets, which may be more representative of the typically decades-long use of antihypertensive therapy as compared to periods of medication use typically examined in conventional observational studies and randomized trials. Despite the evidence suggesting a link between genetically proxied ACE inhibition and colorectal cancer, however, these findings cannot demonstrate a causal relationship between ACE inhibitor use and colorectal cancer risk; only a randomized controlled trial could establish this relationship.
There are several limitations to these analyses. First, mendelian randomization analyses are restricted to examining on-target (i.e., target-mediated) effects of therapeutic interventions. Second, statistical power was likely limited in some analyses of less common cancer subtypes. Limited statistical power in analyses of genetically proxied ACE inhibition and colorectal cancer risk in East Asians (instrumented by rs4343) may also have accounted for the lack of association between these traits within this population. Identification of stronger genetic instruments for ACE inhibition in East Asian populations can help to uncover whether the lack of transportability of ACE and colorectal cancer findings from Europeans to East Asians reflects lower statistical power in the latter or differences in local LD structure across ancestries. In addition, statistical power can be limited in colocalization analysis, which can reduce the probability of shared causal variants across traits examined being detected (i.e., leading to “false negative” findings). We therefore cannot rule out the possibility that some colocalization findings suggesting low posterior probabilities of shared causal variants across traits (e.g., colocalization analyses for genetically proxied NCC inhibition and ER− breast cancer risk) reflected the limited power of this approach. Third, while we were able to perform sensitivity analyses for ADRB1 inhibition by restricting instruments to SNPs that were also eQTLs for ADRB1, we were unable to find evidence that the SNP used to instrument NCC (rs35797045) was also an eQTL for the gene encoding this target. Fourth, while these analyses did not account for previously reported associations of genetically proxied elevated SBP with antihypertensive medication use within the colorectal cancer datasets analyzed, such correction would be expected to strengthen, rather than attenuate, findings presented in this study [80]. Likewise, in “instrument validation” analyses for ADRB1, our inability to recapitulate known effects of ADRB1 inhibition (via beta blockers) on stroke risk could reflect the aforementioned inability to account for blood pressure medication users in the stroke datasets analyzed. It is also possible that these findings reflect the presence of horizontal pleiotropy in the instrument biasing the effect estimate toward the null and/or a nontarget-mediated effect of these medications on stroke risk. Fifth, though mixed-ancestry GWAS were used to construct instruments for serum ACE concentrations, effect allele frequencies for variants used in this instrument were similar across European and Latin American ancestry participants, suggesting that mendelian randomization findings were unlikely to be influenced by confounding through residual population stratification. Sixth, effect estimates presented make the additional assumptions of linearity and the absence of gene–environment and gene–gene interactions. Seventh, our genetically proxied ADRB1 findings are of greater relevance to second generation β blockers (e.g., atenolol and metoprolol), which selectively inhibit ADRB1 as compared to first generation β blockers (e.g., propranolol and nadolol), which equally inhibit ADRB1 and ADRB2 [81]. Future mendelian randomization analyses examining the potential effects of long-term first generation β blocker use incorporating both ADRB1 and ADRB2 variants is warranted. Eighth, we cannot rule out findings presented being influenced by canalization (i.e., compensatory processes being generated during development that counter the phenotypic impact of genetic variants being used as instruments). Finally, while various sensitivity analyses were performed to examine exchangeability and exclusion restriction violations, these assumptions are unverifiable.
Colorectal cancer is the third most common cause of cancer globally [82]. Given the prevalence of ACE inhibitor use in high- and middle-income countries and growing use in low-income countries, and the often long-term nature of antihypertensive therapy, these findings, if replicated in subsequent clinical trials, may have important implications for choice of antihypertensive therapy [4]. Importantly, given that hypertension is more prevalent among those who are overweight or obese (risk factors for colorectal cancer), these findings suggest that long-term use of this medication could increase colorectal cancer risk among populations who are already at elevated risk of this disease. There are different types and classes of ACE inhibitors, which vary in their pharmacodynamic and pharmacokinetic properties [83]. These differing pharmacological properties (e.g., differential absorption rate, affinity for tissue-bound ACE, and plasma half-life) can influence therapeutic benefit (or experience of adverse effects) of ACE inhibitors [84,85]. Future evaluation of the potential effects of long-term ACE inhibitor use on cancer risk should therefore include assessment of whether findings are specific to individual agents or classes of ACE inhibitors. As data on circulating levels of ADRB1 and NCC become available in future GWAS, there would be merit in evaluating whether findings presented in this analysis can be replicated when using alternate instruments developed from protein quantitative loci for these targets. Further work is warranted to unravel molecular mechanisms underpinning the association of ACE with colorectal cancer risk. In addition, extension of the analyses presented in this study to a survival framework could inform on whether concurrent use of ACE inhibitors may have an adverse effect on prognosis among colorectal cancer patients. Finally, findings from this analysis should be “triangulated” by employing other epidemiological designs with orthogonal (i.e., nonoverlapping) sources of bias to each other to further evaluate the association of ACE inhibition and colorectal cancer risk [86].
Conclusions
Our mendelian randomization analyses suggest that genetically proxied long-term inhibition of ACE is associated with increased risk of colorectal cancer. Evaluation of ACE inhibitor use in randomized controlled trials with sufficient follow-up data can inform on the long-term safety of these medications. Our findings provide human genetic support to results from short-term randomized trials suggesting that long-term use of β blockers and thiazide diuretics may not influence risk of common cancers.
Supporting information
(DOCX)
Footnote: No SNPs at genome-wide significance (P < 5 × 10−8) were available to instrument NCC. Effect (SE) represents change in SBP per additional copy of the effect allele. ADRB1, β-1 adrenergic receptor; BMI, body mass index; NCC, sodium-chloride symporter; SBP, systolic blood pressure; SNP, single-nucleotide polymorphism.
(DOCX)
Footnote: ACE, angiotensin-converting enzyme; EUR, European participants; GWAS, genome-wide association study; LA, Latin American participants; SNP, single-nucleotide polymorphism.
(DOCX)
Footnote: Range represents r2 and F-stats across instruments applying a linkage disequilibrium threshold of <0.01 to <0.10.
(DOCX)
Footnote: H0 = neither serum ACE concentrations nor colorectal cancer risk has a genetic association in the region, H1 = only serum ACE concentrations has a genetic association in the region, H2 = only colorectal cancer risk has a genetic association in the region, H3 = both serum ACE concentrations and colorectal cancer risk are associated but have different causal variants, H4 = both serum ACE concentrations and colorectal cancer risk are associated and share a single causal variant. ACE, angiotensin-converting enzyme.
(DOCX)
Footnote: Effect represents the unit change in colorectal cancer risk factor per genetically proxied inhibition of ACE equivalent to a 1-mm Hg decrease in SBP. For analyses of genetically proxied ACE inhibition and low-density lipoprotein cholesterol, 2 SNPs (rs12452187 and rs11655956) were not available, and 1 SNP (rs118138685) was removed because of palindromic alleles with ambiguous effect allele frequencies not permitting strands to be matched. For analyses of iron, 1 SNP (rs11655956) was not included because of palindromic alleles with ambiguous effect allele frequencies. For analyses of insulin-like growth factor 1, 2 SNPs (rs11655956 and rs118138685) were not included because of palindromic alleles with ambiguous effect allele frequencies. For analyses of alcohol intake, 2 SNPs (rs12452187 and rs12150648) were not available in the outcome dataset. ACE, angiotensin-converting enzyme; SBP, systolic blood pressure; SNP, single-nucleotide polymorphism.
(DOCX)
Footnote: OR represents the exponential change in odds of cancer per genetically proxied inhibition of ACE equivalent to a 1 mmHg decrease in SBP. ACE, angiotensin-converting enzyme; OR, odds ratio; SBP, systolic blood pressure; SNP, single-nucleotide polymorphism.
(DOCX)
Footnote: Marginal = SNP associations that are unconditioned (on either the sentinel SNP or additional conditionally independent genome-wide significant SNP for each respective trait), * = Sentinel SNP, † = Conditionally independent and significant (P < 5 × 10−8) SNP, H0 = neither SBP (in ADRB1) nor small cell lung carcinoma risk has a genetic association in the region, H1 = only SBP (in ADRB1) has a genetic association in the region, H2 = only small cell lung carcinoma risk has a genetic association in the region, H3 = both SBP (in ADRB1) and small cell lung carcinoma risk are associated but have different causal variants, H4 = both SBP (in ADRB1) and small cell lung carcinoma risk are associated and share a single causal variant. ADRB1, β-1 adrenergic receptor; SBP, systolic blood pressure; SNP, single-nucleotide polymorphism.
(DOCX)
Footnote: OR represents the exponential change in odds of cancer per genetically proxied inhibition of ADRB1 equivalent to a 1-mm Hg decrease in SBP. ADRB1, β-1 adrenergic receptor; BMI, body mass index; GWAS, genome-wide association study; OR, odds ratio; SBP, systolic blood pressure.
(DOCX)
Footnote: ADRB1, β-1 adrenergic receptor; EA, effect allele; eQTL, expression quantitative trait locus; NCC, sodium-chloride symporter; NEA, noneffect allele; SE, standard error; NES, normalized effect size (obtained from GTex V8); SNP, single-nucleotide polymorphism; TMMs, trimmed mean of M-values (obtained from Barc-UVa-Seq); Z, Z-statistic (obtained from blood EQTL gen).
(DOCX)
Footnote: Scaled to represent ADRB1 inhibition equivalent of a 1-mm Hg SBP reduction. * Neither rs1801253 (nor a high LD proxy) was available in PRACTICAL for prostate cancer risk. ADRB1, β-1 adrenergic receptor; eQTLs, expression quantitative trait loci; SBP, systolic blood pressure.
(DOCX)
Footnote: Marginal = SNP associations that are unconditioned (on either the sentinel SNP or additional conditionally independent genome-wide significant SNP for each respective trait), * = Sentinel SNP, † = Conditionally independent and significant (P < 5 × 10−8) SNP, H0 = neither SBP (in SLC12A3) nor ER− breast cancer risk has a genetic association in the region, H1 = only SBP (in SLC12A3) has a genetic association in the region, H2 = only ER− breast cancer risk has a genetic association in the region, H3 = both SBP (in SLC12A3) and ER− breast cancer risk are associated but have different causal variants, H4 = both SBP (in SLC12A3) and ER− breast cancer risk are associated and share a single causal variant. ER, estrogen receptor; SBP, systolic blood pressure; SNP, single-nucleotide polymorphism.
(DOCX)
Footnote: Marginal = SNP associations that are unconditioned (on either the sentinel SNP or additional conditionally independent genome-wide significant SNP for each respective trait), * = Sentinel SNP, † = Conditionally independent and significant (P < 5 × 10−8) SNP, H0 = neither colon ACE concentrations nor colorectal cancer risk has a genetic association in the region, H1 = only colon ACE concentrations has a genetic association in the region, H2 = only colorectal cancer risk has a genetic association in the region, H3 = both colon ACE concentrations and colorectal cancer risk are associated but have different causal variants, H4 = both colon ACE concentrations and colorectal cancer risk are associated and share a single causal variant. ACE, angiotensin-converting enzyme; SNP, single-nucleotide polymorphism.
(DOCX)
Caption: gene_id = ENSEMBL gene id, beta = effect estimate, se = standard error, pval (not adjusted for multiple testing), R2 = proportion of variance explained by serum ACE wGRS, z-score = test-statistic for association between serum ACE wGRS and gene expression, p.adjust = p-value adjusted for multiple testing using a Bonferroni correction, chr = chromosome of gene, start = location of start of gene, end = location of end of gene, strand = coding strand, gene_type = type of gene examined. ACE, angiotensin-converting enzyme; GRS, genetic risk score; wGRS, weighted genetic risk score.
(XLSX)
Caption: Category = one of 9 “major collections” included in the MSigDB, GeneSet = name of gene set as provided by MSigDB, N_genes = number of genes in gene set, N_overlap = number of overlapping genes from wGRS analysis in gene set, p = p-value (unadjusted for multiple testing), adjP = p-value (adjusted for multiple testing), genes = genes from wGRS analysis that overlap with gene set, link = link to further information on gene set. ACE, angiotensin-converting enzyme; MSigDB, Molecular Signatures Database; wGRS, weighted genetic risk score.
(XLSX)
Caption: N/A.
(XLSX)
Caption: Category = one of 9 “major collections” included in the MSigDB, GeneSet = name of gene set as provided by MSigDB, N_genes = number of genes in gene set, N_overlap = number of genes located in the black module of the coexpression network in gene set, p = p-value (unadjusted for multiple testing), adjP = p-value (adjusted for multiple testing), genes = genes from black module of the coexpression network that overlap with gene set, link = link to further information on gene set. MSigDB, Molecular Signatures Database.
(XLSX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
The authors would like to thank the participants of the individual studies contributing to the BCAC, GECCO, CORECT, CCFR, INTEGRAL-ILCCO, PRACTICAL, Finngen, BioBank Japan, Asia Colorectal Cancer Consortium, ICBP, DIAGRAM, CARDIoGRAMplusC4D, and MEGASTROKE consortia and the Genetic Epidemiology Research on Adult Health, UK Biobank, and the Korean National Cancer Center CRC Study 2. The authors would also like to acknowledge the investigators of these consortia and studies for generating the data used for this analysis. The authors would like to acknowledge the following investigators of the OncoArray and GAME-ON1KG INTEGRAL-ILCCO analyses: Maria Teresa Landi, Victoria Stevens, Ying Wang, Demetrios Albanes, Neil Caporaso, Paul Brennan, Christopher I Amos, Sanjay Shete, Rayjean J Hung, Heike Bickeböller, Angela Risch, Richard Houlston, Stephen Lam, Adonina Tardon, Chu Chen, Stig E Bojesen, Mattias Johansson, H-Erich Wichmann, David Christiani, Gadi Rennert, Susanne Arnold, John K. Field, Loic Le Marchand, Olle Melander, Hans Brunnström, Geoffrey Liu, Angeline Andrew, Lambertus A Kiemeney, Hongbing Shen, Shan Zienolddiny, Kjell Grankvist, Mikael Johansson, M Dawn Teare, Yun-Chul Hong, Jian-Min Yuan, Philip Lazarus, Matthew B Schabath, Melinda C Aldrich. Cancer consortia-specific funding and acknowledgments is presented in S1 Text.
The PRACTICAL Consortium
Rosalind A. Eeles1,2, Christopher A. Haiman3, Zsofia Kote-Jarai1, Fredrick R. Schumacher4,5, Sara Benlloch6,1, Ali Amin Al Olama6,7, Kenneth Muir8,9, Sonja I. Berndt10, David V. Conti3, Fredrik Wiklund11, Stephen Chanock10, Ying Wang12, Victoria L. Stevens12, Catherine M. Tangen13, Jyotsna Batra14,15, Judith A. Clements14,15, APCB BioResource (Australian Prostate Cancer BioResource)14,15, Henrik Grönberg11, Nora Pashayan16,17, Johanna Schleutker18,19, Demetrius Albanes10, Stephanie Weinstein10, Alicja Wolk20, Catharine M. L. West21, Lorelei A. Mucci22, Géraldine Cancel-Tassin23,24, Stella Koutros10, Karina Dalsgaard Sørensen25,26, Eli Marie Grindedal27, David E. Neal28,29,30, Freddie C. Hamdy31,32, Jenny L. Donovan33, Ruth C. Travis34, Robert J. Hamilton35,36, Sue Ann Ingles37, Barry S. Rosenstein38,39, Yong-Jie Lu40, Graham G. Giles41,42,43, Adam S. Kibel44, Ana Vega45,46,47, Manolis Kogevinas48,49,50,51, Kathryn L. Penney52, Jong Y. Park53, Janet L. Stanford54,55, Cezary Cybulski56, Børge G. Nordestgaard57,58, Sune F. Nielsen57,58, Hermann Brenner59,60,61, Christiane Maier62, Jeri Kim63, Esther M. John64, Manuel R. Teixeira65,66,67, Susan L. Neuhausen68, Kim De Ruyck69, Azad Razack70, Lisa F. Newcomb54,71, Davor Lessel72, Radka Kaneva73, Nawaid Usmani74,75, Frank Claessens76, Paul A. Townsend77,78, Jose Esteban Castelao79, Monique J. Roobol80, Florence Menegaux81, Kay-Tee Khaw82, Lisa Cannon-Albright83,84, Hardev Pandha78, Stephen N. Thibodeau85, David J. Hunter86, Peter Kraft87, William J. Blot88,89, Elio Riboli90
1 The Institute of Cancer Research, London, SM2 5NG, UK
2 Royal Marsden NHS Foundation Trust, London, SW3 6JJ, UK
3 Center for Genetic Epidemiology, Department of Preventive Medicine, Keck School of Medicine, University of Southern California/Norris Comprehensive Cancer Center, Los Angeles, CA 90015, USA
4 Department of Population and Quantitative Health Sciences, Case Western Reserve University, Cleveland, OH 44106–7219, USA
5 Seidman Cancer Center, University Hospitals, Cleveland, OH 44106, USA
6 Centre for Cancer Genetic Epidemiology, Department of Public Health and Primary Care, University of Cambridge, Strangeways Research Laboratory, Cambridge CB1 8RN, UK
7 University of Cambridge, Department of Clinical Neurosciences, Stroke Research Group, R3, Box 83, Cambridge Biomedical Campus, Cambridge CB2 0QQ, UK
8 Division of Population Health, Health Services Research and Primary Care, University of Manchester, Oxford Road, Manchester, M13 9PL, UK
9 Warwick Medical School, University of Warwick, Coventry, CV4 7AL, UK
10 Division of Cancer Epidemiology and Genetics, National Cancer Institute, NIH, Bethesda, Maryland, 20892, USA
11 Department of Medical Epidemiology and Biostatistics, Karolinska Institute, SE-171 77 Stockholm, Sweden
12 Department of Population Science, American Cancer Society, 250 Williams Street, Atlanta, GA 30303, USA
13 SWOG Statistical Center, Fred Hutchinson Cancer Research Center, Seattle, WA 98109, USA
14 Australian Prostate Cancer Research Centre-Qld, Institute of Health and Biomedical Innovation and School of Biomedical Sciences, Queensland University of Technology, Brisbane QLD 4059, Australia
15 Translational Research Institute, Brisbane, Queensland 4102, Australia
16 Department of Applied Health Research, University College London, London, WC1E 7HB, UK
17 Centre for Cancer Genetic Epidemiology, Department of Oncology, University of Cambridge, Strangeways Laboratory, Worts Causeway, Cambridge, CB1 8RN, UK
18Institute of Biomedicine, University of Turku, Finland
19 Department of Medical Genetics, Genomics, Laboratory Division, Turku University Hospital, PO Box 52, 20521 Turku, Finland
20 Department of Surgical Sciences, Uppsala University, 75185 Uppsala, Sweden
21 Division of Cancer Sciences, University of Manchester, Manchester Academic Health Science Centre, Radiotherapy Related Research, The Christie Hospital NHS Foundation Trust, Manchester, M13 9PL UK
22 Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, MA 02115, USA
23 CeRePP, Tenon Hospital, F-75020 Paris, France
24 Sorbonne Universite, GRC n°5, AP-HP, Tenon Hospital, 4 rue de la Chine, F-75020 Paris, France
25 Department of Molecular Medicine, Aarhus University Hospital, Palle Juul-Jensen Boulevard 99, 8200 Aarhus N, Denmark
26 Department of Clinical Medicine, Aarhus University, DK-8200 Aarhus N
27 Department of Medical Genetics, Oslo University Hospital, 0424 Oslo, Norway
28 Nuffield Department of Surgical Sciences, University of Oxford, Room 6603, Level 6, John Radcliffe Hospital, Headley Way, Headington, Oxford, OX3 9DU, UK
29 University of Cambridge, Department of Oncology, Box 279, Addenbrooke’s Hospital, Hills Road, Cambridge CB2 0QQ, UK
30 Cancer Research UK, Cambridge Research Institute, Li Ka Shing Centre, Cambridge, CB2 0RE, UK
31 Nuffield Department of Surgical Sciences, University of Oxford, Oxford, OX1 2JD, UK
32 Faculty of Medical Science, University of Oxford, John Radcliffe Hospital, Oxford, UK
33 Population Health Sciences, Bristol Medical School, University of Bristol, BS8 2PS, UK
34 Cancer Epidemiology Unit, Nuffield Department of Population Health, University of Oxford, Oxford, OX3 7LF, UK
35 Dept. of Surgical Oncology, Princess Margaret Cancer Centre, Toronto ON M5G 2M9, Canada
36 Dept. of Surgery (Urology), University of Toronto, Canada
37 Department of Preventive Medicine, Keck School of Medicine, University of Southern California/Norris Comprehensive Cancer Center, Los Angeles, CA 90015, USA
38 Department of Radiation Oncology and Department of Genetics and Genomic Sciences, Box 1236, Icahn School of Medicine at Mount Sinai, One Gustave L. Levy Place, New York, NY 10029, USA
39 Department of Genetics and Genomic Sciences, Icahn School of Medicine at Mount Sinai, New York, NY 10029–5674, USA
40 Centre for Cancer Biomarker and Biotherapeutics, Barts Cancer Institute, Queen Mary University of London, John Vane Science Centre, Charterhouse Square, London, EC1M 6BQ, UK
41 Cancer Epidemiology Division, Cancer Council Victoria, 615 St Kilda Road, Melbourne, VIC 3004, Australia
42 Centre for Epidemiology and Biostatistics, Melbourne School of Population and Global Health, The University of Melbourne, Grattan Street, Parkville, VIC 3010, Australia
43 Precision Medicine, School of Clinical Sciences at Monash Health, Monash University, Clayton, Victoria 3168, Australia
44 Division of Urologic Surgery, Brigham and Womens Hospital, 75 Francis Street, Boston, MA 02115, USA
45 Fundación Pública Galega Medicina Xenómica, Santiago de Compostela, 15706, Spain
46 Instituto de Investigación Sanitaria de Santiago de Compostela, Santiago De Compostela, 15706, Spain
47 Centro de Investigación en Red de Enfermedades Raras (CIBERER), Spain
48 ISGlobal, Barcelona, Spain
49 IMIM (Hospital del Mar Medical Research Institute), Barcelona, Spain
50 CIBER Epidemiología y Salud Pública (CIBERESP), 28029 Madrid, Spain
51 Universitat Pompeu Fabra (UPF), Barcelona, Spain
52 Channing Division of Network Medicine, Department of Medicine, Brigham and Women’s Hospital/Harvard Medical School, Boston, MA 02115, USA
53 Department of Cancer Epidemiology, Moffitt Cancer Center, 12902 Magnolia Drive, Tampa, FL 33612, USA
54 Division of Public Health Sciences, Fred Hutchinson Cancer Research Center, Seattle, Washington, 98109–1024, USA
55 Department of Epidemiology, School of Public Health, University of Washington, Seattle, Washington 98195, USA
56 International Hereditary Cancer Center, Department of Genetics and Pathology, Pomeranian Medical University, 70–115 Szczecin, Poland
57 Faculty of Health and Medical Sciences, University of Copenhagen, 2200 Copenhagen, Denmark
58 Department of Clinical Biochemistry, Herlev and Gentofte Hospital, Copenhagen University Hospital, Herlev, 2200 Copenhagen, Denmark
59 Division of Clinical Epidemiology and Aging Research, German Cancer Research Center (DKFZ), D-69120, Heidelberg, Germany
60 German Cancer Consortium (DKTK), German Cancer Research Center (DKFZ), D-69120 Heidelberg, Germany
61 Division of Preventive Oncology, German Cancer Research Center (DKFZ) and National Center for Tumor Diseases (NCT), Im Neuenheimer Feld 460, 69120 Heidelberg, Germany
62 Humangenetik Tuebingen, Paul-Ehrlich-Str 23, D-72076 Tuebingen, Germany
63 The University of Texas M. D. Anderson Cancer Center, Department of Genitourinary Medical Oncology, 1515 Holcombe Blvd., Houston, TX 77030, USA
64 Departments of Epidemiology & Population Health and of Medicine, Division of Oncology, Stanford Cancer Institute, Stanford University School of Medicine, Stanford, CA 94304 USA
65 Department of Genetics, Portuguese Oncology Institute of Porto (IPO-Porto), 4200–072 Porto, Portugal
66 Biomedical Sciences Institute (ICBAS), University of Porto, 4050–313 Porto, Portugal
67 Cancer Genetics Group, IPO-Porto Research Center (CI-IPOP), Portuguese Oncology Institute of Porto (IPO-Porto), 4200–072 Porto, Portugal
68 Department of Population Sciences, Beckman Research Institute of the City of Hope, 1500 East Duarte Road, Duarte, CA 91010, 626-256-HOPE (4673)
69 Ghent University, Faculty of Medicine and Health Sciences, Basic Medical Sciences, Proeftuinstraat 86, B-9000 Gent
70 Department of Surgery, Faculty of Medicine, University of Malaya, 50603 Kuala Lumpur, Malaysia
71 Department of Urology, University of Washington, 1959 NE Pacific Street, Box 356510, Seattle, WA 98195, USA
72 Institute of Human Genetics, University Medical Center Hamburg-Eppendorf, D-20246 Hamburg, Germany
73 Molecular Medicine Center, Department of Medical Chemistry and Biochemistry, Medical University of Sofia, Sofia, 2 Zdrave Str., 1431 Sofia, Bulgaria
74 Department of Oncology, Cross Cancer Institute, University of Alberta, 11560 University Avenue, Edmonton, Alberta, Canada T6G 1Z2
75 Division of Radiation Oncology, Cross Cancer Institute, 11560 University Avenue, Edmonton, Alberta, Canada T6G 1Z2
76 Molecular Endocrinology Laboratory, Department of Cellular and Molecular Medicine, KU Leuven, BE-3000, Belgium
77 Division of Cancer Sciences, Manchester Cancer Research Centre, Faculty of Biology, Medicine and Health, Manchester Academic Health Science Centre, NIHR Manchester Biomedical Research Centre, Health Innovation Manchester, Univeristy of Manchester, M13 9WL
78 The University of Surrey, Guildford, Surrey, GU2 7XH, UK
79 Genetic Oncology Unit, CHUVI Hospital, Complexo Hospitalario Universitario de Vigo, Instituto de Investigación Biomédica Galicia Sur (IISGS), 36204, Vigo (Pontevedra), Spain
80 Department of Urology, Erasmus University Medical Center, 3015 CE Rotterdam, The Netherlands
81 "Exposome and Heredity", CESP (UMR 1018), Faculté de Médecine, Université Paris-Saclay, Inserm, Gustave Roussy, Villejuif
82 Clinical Gerontology Unit, University of Cambridge, Cambridge, CB2 2QQ, UK
83 Division of Epidemiology, Department of Internal Medicine, University of Utah School of Medicine, Salt Lake City, Utah 84132, USA
84 George E. Wahlen Department of Veterans Affairs Medical Center, Salt Lake City, Utah 84148, USA
85 Department of Laboratory Medicine and Pathology, Mayo Clinic, Rochester, MN 55905, USA
86 Nuffield Department of Population Health, University of Oxford, United Kingdom
87 Program in Genetic Epidemiology and Statistical Genetics, Department of Epidemiology, Harvard School of Public Health, Boston, MA, USA
88 Division of Epidemiology, Department of Medicine, Vanderbilt University Medical Center, 2525 West End Avenue, Suite 800, Nashville, TN 37232 USA
89 International Epidemiology Institute, Rockville, MD 20850, USA
90 Department of Epidemiology and Biostatistics, School of Public Health, Imperial College London, SW7 2AZ, UK
MEGASTROKE consortium
Rainer Malik 1, Ganesh Chauhan 2, Matthew Traylor 3, Muralidharan Sargurupremraj 4,5, Yukinori Okada 6,7,8, Aniket Mishra 4,5, Loes Rutten-Jacobs 3, Anne-Katrin Giese 9, Sander W van der Laan 10, Solveig Gretarsdottir 11, Christopher D Anderson 12,13,14,14, Michael Chong 15, Hieab HH Adams 16,17, Tetsuro Ago 18, Peter Almgren 19, Philippe Amouyel 20,21, Hakan Ay 22,13, Traci M Bartz 23, Oscar R Benavente 24, Steve Bevan 25, Giorgio B Boncoraglio 26, Robert D Brown, Jr. 27, Adam S Butterworth 28,29, Caty Carrera 30,31, Cara L Carty 32,33, Daniel I Chasman 34,35, Wei-Min Chen 36, John W Cole 37, Adolfo Correa 38, Ioana Cotlarciuc 39, Carlos Cruchaga 40,41, John Danesh 28,42,43,44, Paul IW de Bakker 45,46, Anita L DeStefano 47,48, Marcel den Hoed 49, Qing Duan 50, Stefan T Engelter 51,52, Guido J Falcone 53,54, Rebecca F Gottesman 55, Raji P Grewal 56, Vilmundur Gudnason 57,58, Stefan Gustafsson 59, Jeffrey Haessler 60, Tamara B Harris 61, Ahamad Hassan 62, Aki S Havulinna 63,64, Susan R Heckbert 65, Elizabeth G Holliday 66,67, George Howard 68, Fang-Chi Hsu 69, Hyacinth I Hyacinth 70, M Arfan Ikram 16, Erik Ingelsson 71,72, Marguerite R Irvin 73, Xueqiu Jian 74, Jordi Jiménez-Conde 75, Julie A Johnson 76,77, J Wouter Jukema 78, Masahiro Kanai 6,7,79, Keith L Keene 80,81, Brett M Kissela 82, Dawn O Kleindorfer 82, Charles Kooperberg 60, Michiaki Kubo 83, Leslie A Lange 84, Carl D Langefeld 85, Claudia Langenberg 86, Lenore J Launer 87, Jin-Moo Lee 88, Robin Lemmens 89,90, Didier Leys 91, Cathryn M Lewis 92,93, Wei-Yu Lin 28,94, Arne G Lindgren 95,96, Erik Lorentzen 97, Patrik K Magnusson 98, Jane Maguire 99, Ani Manichaikul 36, Patrick F McArdle 100, James F Meschia 101, Braxton D Mitchell 100,102, Thomas H Mosley 103,104, Michael A Nalls 105,106, Toshiharu Ninomiya 107, Martin J O’Donnell 15,108, Bruce M Psaty 109,110,111,112, Sara L Pulit 113,45, Kristiina Rannikmäe 114,115, Alexander P Reiner 65,116, Kathryn M Rexrode 117, Kenneth Rice 118, Stephen S Rich 36, Paul M Ridker 34,35, Natalia S Rost 9,13, Peter M Rothwell 119, Jerome I Rotter 120,121, Tatjana Rundek 122, Ralph L Sacco 122, Saori Sakaue 7,123, Michele M Sale 124, Veikko Salomaa 63, Bishwa R Sapkota 125, Reinhold Schmidt 126, Carsten O Schmidt 127, Ulf Schminke 128, Pankaj Sharma 39, Agnieszka Slowik 129, Cathie LM Sudlow 114,115, Christian Tanislav 130, Turgut Tatlisumak 131,132, Kent D Taylor 120,121, Vincent NS Thijs 133,134, Gudmar Thorleifsson 11, Unnur Thorsteinsdottir 11, Steffen Tiedt 1, Stella Trompet 135, Christophe Tzourio 5,136,137, Cornelia M van Duijn 138,139, Matthew Walters 140, Nicholas J Wareham 86, Sylvia Wassertheil-Smoller 141, James G Wilson 142, Kerri L Wiggins 109, Qiong Yang 47, Salim Yusuf 15, Najaf Amin 16, Hugo S Aparicio 185,48, Donna K Arnett 186, John Attia 187, Alexa S Beiser 47,48, Claudine Berr 188, Julie E Buring 34,35, Mariana Bustamante 189, Valeria Caso 190, Yu-Ching Cheng 191, Seung Hoan Choi 192,48, Ayesha Chowhan 185,48, Natalia Cullell 31, Jean-François Dartigues 193,194, Hossein Delavaran 95,96, Pilar Delgado 195, Marcus Dörr 196,197, Gunnar Engström 19, Ian Ford 198, Wander S Gurpreet 199, Anders Hamsten 200,201, Laura Heitsch 202, Atsushi Hozawa 203, Laura Ibanez 204, Andreea Ilinca 95,96, Martin Ingelsson 205, Motoki Iwasaki 206, Rebecca D Jackson 207, Katarina Jood 208, Pekka Jousilahti 63, Sara Kaffashian 4,5, Lalit Kalra 209, Masahiro Kamouchi 210, Takanari Kitazono 211, Olafur Kjartansson 212, Manja Kloss 213, Peter J Koudstaal 214, Jerzy Krupinski 215, Daniel L Labovitz 216, Cathy C Laurie 118, Christopher R Levi 217, Linxin Li 218, Lars Lind 219, Cecilia M Lindgren 220,221, Vasileios Lioutas 222,48, Yong Mei Liu 223, Oscar L Lopez 224, Hirata Makoto 225, Nicolas Martinez-Majander 172, Koichi Matsuda 225, Naoko Minegishi 203, Joan Montaner 226, Andrew P Morris 227,228, Elena Muiño 31, Martina Müller-Nurasyid 229,230,231, Bo Norrving 95,96, Soichi Ogishima 203, Eugenio A Parati 232, Leema Reddy Peddareddygari 56, Nancy L Pedersen 98,233, Joanna Pera 129, Markus Perola 63,234, Alessandro Pezzini 235, Silvana Pileggi 236, Raquel Rabionet 237, Iolanda Riba-Llena 30, Marta Ribasés 238, Jose R Romero 185,48, Jaume Roquer 239,240, Anthony G Rudd 241,242, Antti-Pekka Sarin 243,244, Ralhan Sarju 199, Chloe Sarnowski 47,48, Makoto Sasaki 245, Claudia L Satizabal 185,48, Mamoru Satoh 245, Naveed Sattar 246, Norie Sawada 206, Gerli Sibolt 172, Ásgeir Sigurdsson 247, Albert Smith 248, Kenji Sobue 245, Carolina Soriano-Tárraga 240, Tara Stanne 249, O Colin Stine 250, David J Stott 251, Konstantin Strauch 229,252, Takako Takai 203, Hideo Tanaka 253,254, Kozo Tanno 245, Alexander Teumer 255, Liisa Tomppo 172, Nuria P Torres-Aguila 31, Emmanuel Touze 256,257, Shoichiro Tsugane 206, Andre G Uitterlinden 258, Einar M Valdimarsson 259, Sven J van der Lee 16, Henry Völzke 255, Kenji Wakai 253, David Weir 260, Stephen R Williams 261, Charles DA Wolfe 241,242, Quenna Wong 118, Huichun Xu 191, Taiki Yamaji 206, Dharambir K Sanghera 125,169,170, Olle Melander 19, Christina Jern 171, Daniel Strbian 172,173, Israel Fernandez-Cadenas 31,30, W T Longstreth, Jr 174,65, Arndt Rolfs 175, Jun Hata 107, Daniel Woo 82, Jonathan Rosand 12,13,14, Guillaume Pare 15, Jemma C Hopewell 176, Danish Saleheen 177, Kari Stefansson 11,178, Bradford B Worrall 179, Steven J Kittner 37, Sudha Seshadri 180,48, Myriam Fornage 74,181, Hugh S Markus 3, Joanna MM Howson 28, Yoichiro Kamatani 6,182, Stephanie Debette 4,5, Martin Dichgans 1,183,184.
1 Institute for Stroke and Dementia Research (ISD), University Hospital, LMU Munich, Munich, Germany
2 Centre for Brain Research, Indian Institute of Science, Bangalore, India
3 Stroke Research Group, Division of Clinical Neurosciences, University of Cambridge, UK
4 INSERM U1219 Bordeaux Population Health Research Center, Bordeaux, France
5 University of Bordeaux, Bordeaux, France
6 Laboratory for Statistical Analysis, RIKEN Center for Integrative Medical Sciences, Yokohama, Japan
7 Department of Statistical Genetics, Osaka University Graduate School of Medicine, Osaka, Japan
8 Laboratory of Statistical Immunology, Immunology Frontier Research Center (WPI-IFReC), Osaka University, Suita, Japan
9 Department of Neurology, Massachusetts General Hospital, Harvard Medical School, Boston, MA, USA
10 Laboratory of Experimental Cardiology, Division of Heart and Lungs, University Medical Center Utrecht, University of Utrecht, Utrecht, Netherlands
11 deCODE genetics/AMGEN inc, Reykjavik, Iceland
12 Center for Genomic Medicine, Massachusetts General Hospital (MGH), Boston, MA, USA
13 J. Philip Kistler Stroke Research Center, Department of Neurology, MGH, Boston, MA, USA
14 Program in Medical and Population Genetics, Broad Institute, Cambridge, MA, USA
15 Population Health Research Institute, McMaster University, Hamilton, Canada
16 Department of Epidemiology, Erasmus University Medical Center, Rotterdam, Netherlands
17 Department of Radiology and Nuclear Medicine, Erasmus University Medical Center, Rotterdam, Netherlands
18 Department of Medicine and Clinical Science, Graduate School of Medical Sciences, Kyushu University, Fukuoka, Japan
19 Department of Clinical Sciences, Lund University, Malmö, Sweden
20 Univ. Lille, Inserm, Institut Pasteur de Lille, LabEx DISTALZ-UMR1167, Risk factors and molecular determinants of aging-related diseases, F-59000 Lille, France
21 Centre Hosp. Univ Lille, Epidemiology and Public Health Department, F-59000 Lille, France
22 AA Martinos Center for Biomedical Imaging, Department of Radiology, Massachusetts General Hospital, Harvard Medical School, Boston, MA, USA
23 Cardiovascular Health Research Unit, Departments of Biostatistics and Medicine, University of Washington, Seattle, WA, USA
24 Division of Neurology, Faculty of Medicine, Brain Research Center, University of British Columbia, Vancouver, Canada
25 School of Life Science, University of Lincoln, Lincoln, UK
26 Department of Cerebrovascular Diseases, Fondazione IRCCS Istituto Neurologico "Carlo Besta", Milano, Italy
27 Department of Neurology, Mayo Clinic Rochester, Rochester, MN, USA
28 MRC/BHF Cardiovascular Epidemiology Unit, Department of Public Health and Primary Care, University of Cambridge, Cambridge, UK
29 The National Institute for Health Research Blood and Transplant Research Unit in Donor Health and Genomics, University of Cambridge, UK
30 Neurovascular Research Laboratory, Vall d’Hebron Institut of Research, Neurology and Medicine Departments-Universitat Autònoma de Barcelona, Vall d’Hebrón Hospital, Barcelona, Spain
31 Stroke Pharmacogenomics and Genetics, Fundacio Docència i Recerca MutuaTerrassa, Terrassa, Spain
32 Children’s Research Institute, Children’s National Medical Center, Washington, DC, USA
33 Center for Translational Science, George Washington University, Washington, DC, USA
34 Division of Preventive Medicine, Brigham and Women’s Hospital, Boston, MA, USA
35 Harvard Medical School, Boston, MA, USA
36 Center for Public Health Genomics, Department of Public Health Sciences, University of Virginia, Charlottesville, VA, USA
37 Department of Neurology, University of Maryland School of Medicine and Baltimore VAMC, Baltimore, MD, USA
38 Departments of Medicine, Pediatrics and Population Health Science, University of Mississippi Medical Center, Jackson, MS, USA
39 Institute of Cardiovascular Research, Royal Holloway University of London, UK & Ashford and St Peters Hospital, Surrey UK
40 Department of Psychiatry, The Hope Center Program on Protein Aggregation and Neurodegeneration (HPAN), Washington University, School of Medicine, St. Louis, MO, USA
41 Department of Developmental Biology, Washington University School of Medicine, St. Louis, MO, USA
42 NIHR Blood and Transplant Research Unit in Donor Health and Genomics, Department of Public Health and Primary Care, University of Cambridge, Cambridge, UK
43 Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK
44 British Heart Foundation, Cambridge Centre of Excellence, Department of Medicine, University of Cambridge, Cambridge, UK
45 Department of Medical Genetics, University Medical Center Utrecht, Utrecht, Netherlands
46 Department of Epidemiology, Julius Center for Health Sciences and Primary Care, University Medical Center Utrecht, Utrecht, Netherlands
47 Boston University School of Public Health, Boston, MA, USA
48 Framingham Heart Study, Framingham, MA, USA
49 Department of Immunology, Genetics and Pathology and Science for Life Laboratory, Uppsala University, Uppsala, Sweden
50 Department of Genetics, University of North Carolina, Chapel Hill, NC, USA
51 Department of Neurology and Stroke Center, Basel University Hospital, Switzerland
52 Neurorehabilitation Unit, University and University Center for Medicine of Aging and Rehabilitation Basel, Felix Platter Hospital, Basel, Switzerland
53 Department of Neurology, Yale University School of Medicine, New Haven, CT, USA
54 Program in Medical and Population Genetics, The Broad Institute of Harvard and MIT, Cambridge, MA, USA
55 Department of Neurology, Johns Hopkins University School of Medicine, Baltimore, MD, USA
56 Neuroscience Institute, SF Medical Center, Trenton, NJ, USA
57 Icelandic Heart Association Research Institute, Kopavogur, Iceland
58 University of Iceland, Faculty of Medicine, Reykjavik, Iceland
59 Department of Medical Sciences, Molecular Epidemiology and Science for Life Laboratory, Uppsala University, Uppsala, Sweden
60 Division of Public Health Sciences, Fred Hutchinson Cancer Research Center, Seattle, WA, USA
61 Laboratory of Epidemiology and Population Science, National Institute on Aging, National Institutes of Health, Bethesda, MD, USA
62 Department of Neurology, Leeds General Infirmary, Leeds Teaching Hospitals NHS Trust, Leeds, UK
63 National Institute for Health and Welfare, Helsinki, Finland
64 FIMM—Institute for Molecular Medicine Finland, Helsinki, Finland
65 Department of Epidemiology, University of Washington, Seattle, WA, USA
66 Public Health Stream, Hunter Medical Research Institute, New Lambton, Australia
67 Faculty of Health and Medicine, University of Newcastle, Newcastle, Australia
68 School of Public Health, University of Alabama at Birmingham, Birmingham, AL, USA
69 Department of Biostatistical Sciences, Wake Forest School of Medicine, Winston-Salem, NC, USA
70 Aflac Cancer and Blood Disorder Center, Department of Pediatrics, Emory University School of Medicine, Atlanta, GA, USA
71 Department of Medicine, Division of Cardiovascular Medicine, Stanford University School of Medicine, CA, USA
72 Department of Medical Sciences, Molecular Epidemiology and Science for Life Laboratory, Uppsala University, Uppsala, Sweden
73 Epidemiology, School of Public Health, University of Alabama at Birmingham, USA
74 Brown Foundation Institute of Molecular Medicine, University of Texas Health Science Center at Houston, Houston, TX, USA
75 Neurovascular Research Group (NEUVAS), Neurology Department, Institut Hospital del Mar d’Investigació Mèdica, Universitat Autònoma de Barcelona, Barcelona, Spain
76 Department of Pharmacotherapy and Translational Research and Center for Pharmacogenomics, University of Florida, College of Pharmacy, Gainesville, FL, USA
77 Division of Cardiovascular Medicine, College of Medicine, University of Florida, Gainesville, FL, USA
78 Department of Cardiology, Leiden University Medical Center, Leiden, the Netherlands
79 Program in Bioinformatics and Integrative Genomics, Harvard Medical School, Boston, MA, USA
80 Department of Biology, East Carolina University, Greenville, NC, USA
81 Center for Health Disparities, East Carolina University, Greenville, NC, USA
82 University of Cincinnati College of Medicine, Cincinnati, OH, USA
83 RIKEN Center for Integrative Medical Sciences, Yokohama, Japan
84 Department of Medicine, University of Colorado Denver, Anschutz Medical Campus, Aurora, CO, USA
85 Center for Public Health Genomics and Department of Biostatistical Sciences, Wake Forest School of Medicine, Winston-Salem, NC, USA
86 MRC Epidemiology Unit, University of Cambridge School of Clinical Medicine, Institute of Metabolic Science, Cambridge Biomedical Campus, Cambridge, UK
87 Intramural Research Program, National Institute on Aging, National Institutes of Health, Bethesda, MD, USA
88 Department of Neurology, Radiology, and Biomedical Engineering, Washington University School of Medicine, St. Louis, MO, USA
89 KU Leuven–University of Leuven, Department of Neurosciences, Experimental Neurology, Leuven, Belgium
90 VIB Center for Brain & Disease Research, University Hospitals Leuven, Department of Neurology, Leuven, Belgium
91 Univ.-Lille, INSERM U 1171. CHU Lille. Lille, France
92 Department of Medical and Molecular Genetics, King’s College London, London, UK
93 SGDP Centre, Institute of Psychiatry, Psychology & Neuroscience, King’s College London, London, UK
94 Northern Institute for Cancer Research, Paul O’Gorman Building, Newcastle University, Newcastle, UK
95 Department of Clinical Sciences Lund, Neurology, Lund University, Lund, Sweden
96 Department of Neurology and Rehabilitation Medicine, Skåne University Hospital, Lund, Sweden
97 Bioinformatics Core Facility, University of Gothenburg, Gothenburg, Sweden
98 Department of Medical Epidemiology and Biostatistics, Karolinska Institutet, Stockholm, Sweden
99 University of Technology Sydney, Faculty of Health, Ultimo, Australia
100 Department of Medicine, University of Maryland School of Medicine, MD, USA
101 Department of Neurology, Mayo Clinic, Jacksonville, FL, USA
102 Geriatrics Research and Education Clinical Center, Baltimore Veterans Administration Medical Center, Baltimore, MD, USA
103 Division of Geriatrics, School of Medicine, University of Mississippi Medical Center, Jackson, MS, USA
104 Memory Impairment and Neurodegenerative Dementia Center, University of Mississippi Medical Center, Jackson, MS, USA
105 Laboratory of Neurogenetics, National Institute on Aging, National institutes of Health, Bethesda, MD, USA
106 Data Tecnica International, Glen Echo MD, USA
107 Department of Epidemiology and Public Health, Graduate School of Medical Sciences, Kyushu University, Fukuoka, Japan
108 Clinical Research Facility, Department of Medicine, NUI Galway, Galway, Ireland
109 Cardiovascular Health Research Unit, Department of Medicine, University of Washington, Seattle, WA, USA
110 Department of Epidemiology, University of Washington, Seattle, WA
111 Department of Health Services, University of Washington, Seattle, WA, USA
112 Kaiser Permanente Washington Health Research Institute, Seattle, WA, USA
113 Brain Center Rudolf Magnus, Department of Neurology, University Medical Center Utrecht, Utrecht, The Netherlands
114 Usher Institute of Population Health Sciences and Informatics, University of Edinburgh, Edinburgh, UK
115 Centre for Clinical Brain Sciences, University of Edinburgh, Edinburgh, UK
116 Fred Hutchinson Cancer Research Center, University of Washington, Seattle, WA, USA
117 Department of Medicine, Brigham and Women’s Hospital, Boston, MA, USA
118 Department of Biostatistics, University of Washington, Seattle, WA, USA
119 Nuffield Department of Clinical Neurosciences, University of Oxford, UK
120 Institute for Translational Genomics and Population Sciences, Los Angeles Biomedical Research Institute at Harbor-UCLA Medical Center, Torrance, CA, USA
121 Division of Genomic Outcomes, Department of Pediatrics, Harbor-UCLA Medical Center, Torrance, CA, USA
122 Department of Neurology, Miller School of Medicine, University of Miami, Miami, FL, USA
123 Department of Allergy and Rheumatology, Graduate School of Medicine, the University of Tokyo, Tokyo, Japan
124 Center for Public Health Genomics, University of Virginia, Charlottesville, VA, USA
125 Department of Pediatrics, College of Medicine, University of Oklahoma Health Sciences Center, Oklahoma City, OK, USA
126 Department of Neurology, Medical University of Graz, Graz, Austria
127 University Medicine Greifswald, Institute for Community Medicine, SHIP-KEF, Greifswald, Germany
128 University Medicine Greifswald, Department of Neurology, Greifswald, Germany
129 Department of Neurology, Jagiellonian University, Krakow, Poland
130 Department of Neurology, Justus Liebig University, Giessen, Germany
131 Department of Clinical Neurosciences/Neurology, Institute of Neuroscience and Physiology, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden
132 Sahlgrenska University Hospital, Gothenburg, Sweden
133 Stroke Division, Florey Institute of Neuroscience and Mental Health, University of Melbourne, Heidelberg, Australia
134 Austin Health, Department of Neurology, Heidelberg, Australia
135 Department of Internal Medicine, Section Gerontology and Geriatrics, Leiden University Medical Center, Leiden, the Netherlands
136 INSERM U1219, Bordeaux, France
137 Department of Public Health, Bordeaux University Hospital, Bordeaux, France
138 Genetic Epidemiology Unit, Department of Epidemiology, Erasmus University Medical Center Rotterdam, Netherlands
139 Center for Medical Systems Biology, Leiden, Netherlands
140 School of Medicine, Dentistry and Nursing at the University of Glasgow, Glasgow, UK
141 Department of Epidemiology and Population Health, Albert Einstein College of Medicine, NY, USA
142 Department of Physiology and Biophysics, University of Mississippi Medical Center, Jackson, MS, USA
143 A full list of members and affiliations appears in the Supplementary Note
144 Department of Human Genetics, McGill University, Montreal, Canada
145 Department of Pathophysiology, Institute of Biomedicine and Translation Medicine, University of Tartu, Tartu, Estonia
146 Department of Cardiac Surgery, Tartu University Hospital, Tartu, Estonia
147 Clinical Gene Networks AB, Stockholm, Sweden
148 Department of Genetics and Genomic Sciences, The Icahn Institute for Genomics and Multiscale Biology Icahn School of Medicine at Mount Sinai, New York, NY, USA
149 Department of Pathophysiology, Institute of Biomedicine and Translation Medicine, University of Tartu, Biomeedikum, Tartu, Estonia
150 Integrated Cardio Metabolic Centre, Department of Medicine, Karolinska Institutet, Karolinska Universitetssjukhuset, Huddinge, Sweden
151 Clinical Gene Networks AB, Stockholm, Sweden
152 Sorbonne Universités, UPMC Univ. Paris 06, INSERM, UMR_S 1166, Team Genomics & Pathophysiology of Cardiovascular Diseases, Paris, France
153 ICAN Institute for Cardiometabolism and Nutrition, Paris, France
154 Department of Biomedical Engineering, University of Virginia, Charlottesville, VA, USA
155 Group Health Research Institute, Group Health Cooperative, Seattle, WA, USA
156 Seattle Epidemiologic Research and Information Center, VA Office of Research and Development, Seattle, WA, USA
157 Cardiovascular Research Center, Massachusetts General Hospital, Boston, MA, USA
158 Department of Medical Research, Bærum Hospital, Vestre Viken Hospital Trust, Gjettum, Norway
159 Saw Swee Hock School of Public Health, National University of Singapore and National University Health System, Singapore
160 National Heart and Lung Institute, Imperial College London, London, UK
161 Department of Gene Diagnostics and Therapeutics, Research Institute, National Center for Global Health and Medicine, Tokyo, Japan
162 Department of Epidemiology, Tulane University School of Public Health and Tropical Medicine, New Orleans, LA, USA
163 Department of Cardiology, University Medical Center Groningen, University of Groningen, Netherlands
164 MRC-PHE Centre for Environment and Health, School of Public Health, Department of Epidemiology and Biostatistics, Imperial College London, London, UK
165 Department of Epidemiology and Biostatistics, Imperial College London, London, UK
166 Department of Cardiology, Ealing Hospital NHS Trust, Southall, UK
167 National Heart, Lung and Blood Research Institute, Division of Intramural Research, Population Sciences Branch, Framingham, MA, USA
168 A full list of members and affiliations appears at the end of the manuscript
169 Department of Phamaceutical Sciences, Collge of Pharmacy, University of Oklahoma Health Sciences Center, Oklahoma City, OK, USA
170 Oklahoma Center for Neuroscience, Oklahoma City, OK, USA
171 Department of Pathology and Genetics, Institute of Biomedicine, The Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden
172 Department of Neurology, Helsinki University Hospital, Helsinki, Finland
173 Clinical Neurosciences, Neurology, University of Helsinki, Helsinki, Finland
174 Department of Neurology, University of Washington, Seattle, WA, USA
175 Albrecht Kossel Institute, University Clinic of Rostock, Rostock, Germany
176 Clinical Trial Service Unit and Epidemiological Studies Unit, Nuffield Department of Population Health, University of Oxford, Oxford, UK
177 Department of Genetics, Perelman School of Medicine, University of Pennsylvania, PA, USA
178 Faculty of Medicine, University of Iceland, Reykjavik, Iceland
179 Departments of Neurology and Public Health Sciences, University of Virginia School of Medicine, Charlottesville, VA, USA
180 Department of Neurology, Boston University School of Medicine, Boston, MA, USA
181 Human Genetics Center, University of Texas Health Science Center at Houston, Houston, TX, USA
182 Center for Genomic Medicine, Kyoto University Graduate School of Medicine, Kyoto, Japan
183 Munich Cluster for Systems Neurology (SyNergy), Munich, Germany
184 German Center for Neurodegenerative Diseases (DZNE), Munich, Germany
185 Boston University School of Medicine, Boston, MA, USA
186 University of Kentucky College of Public Health, Lexington, KY, USA
187 University of Newcastle and Hunter Medical Research Institute, New Lambton, Australia
188 Univ. Montpellier, Inserm, U1061, Montpellier, France
189 Centre for Research in Environmental Epidemiology, Barcelona, Spain
190 Department of Neurology, Università degli Studi di Perugia, Umbria, Italy
191 Department of Medicine, University of Maryland School of Medicine, Baltimore, MD, USA
192 Broad Institute, Cambridge, MA, USA
193 Univ. Bordeaux, Inserm, Bordeaux Population Health Research Center, UMR 1219, Bordeaux, France
194 Bordeaux University Hospital, Department of Neurology, Memory Clinic, Bordeaux, France
195 Neurovascular Research Laboratory. Vall d’Hebron Institut of Research, Neurology and Medicine Departments-Universitat Autònoma de Barcelona. Vall d’Hebrón Hospital, Barcelona, Spain
196 University Medicine Greifswald, Department of Internal Medicine B, Greifswald, Germany
197 DZHK, Greifswald, Germany
198 Robertson Center for Biostatistics, University of Glasgow, Glasgow, UK
199 Hero DMC Heart Institute, Dayanand Medical College & Hospital, Ludhiana, India
200 Atherosclerosis Research Unit, Department of Medicine Solna, Karolinska Institutet, Stockholm, Sweden
201 Karolinska Institutet, Stockholm, Sweden
202 Division of Emergency Medicine, and Department of Neurology, Washington University School of Medicine, St. Louis, MO, USA
203 Tohoku Medical Megabank Organization, Sendai, Japan
204 Department of Psychiatry, Washington University School of Medicine, St. Louis, MO, USA
205 Department of Public Health and Caring Sciences / Geriatrics, Uppsala University, Uppsala, Sweden
206 Epidemiology and Prevention Group, Center for Public Health Sciences, National Cancer Center, Tokyo, Japan
207 Department of Internal Medicine and the Center for Clinical and Translational Science, The Ohio State University, Columbus, OH, USA
208 Institute of Neuroscience and Physiology, the Sahlgrenska Academy at University of Gothenburg, Goteborg, Sweden
209 Department of Basic and Clinical Neurosciences, King’s College London, London, UK
210 Department of Health Care Administration and Management, Graduate School of Medical Sciences, Kyushu University, Japan
211 Department of Medicine and Clinical Science, Graduate School of Medical Sciences, Kyushu University, Japan
212 Landspitali National University Hospital, Departments of Neurology & Radiology, Reykjavik, Iceland
213 Department of Neurology, Heidelberg University Hospital, Germany
214 Department of Neurology, Erasmus University Medical Center
215 Hospital Universitari Mutua Terrassa, Terrassa (Barcelona), Spain
216 Albert Einstein College of Medicine, Montefiore Medical Center, New York, NY, USA
217 John Hunter Hospital, Hunter Medical Research Institute and University of Newcastle, Newcastle, NSW, Australia
218 Centre for Prevention of Stroke and Dementia, Nuffield Department of Clinical Neurosciences, University of Oxford, UK
219 Department of Medical Sciences, Uppsala University, Uppsala, Sweden
220 Genetic and Genomic Epidemiology Unit, Wellcome Trust Centre for Human Genetics, University of Oxford, Oxford, UK
221 The Wellcome Trust Centre for Human Genetics, Oxford, UK
222 Beth Israel Deaconess Medical Center, Boston, MA, USA
223 Wake Forest School of Medicine, Wake Forest, NC, USA
224 Department of Neurology, University of Pittsburgh, Pittsburgh, PA, USA
225 BioBank Japan, Laboratory of Clinical Sequencing, Department of Computational biology and medical Sciences, Graduate school of Frontier Sciences, The University of Tokyo, Tokyo, Japan
226 Neurovascular Research Laboratory, Vall d’Hebron Institut of Research, Neurology and Medicine Departments-Universitat Autònoma de Barcelona. Vall d’Hebrón Hospital, Barcelona, Spain
227 Department of Biostatistics, University of Liverpool, Liverpool, UK
228 Wellcome Trust Centre for Human Genetics, University of Oxford, Oxford, UK
229 Institute of Genetic Epidemiology, Helmholtz Zentrum München—German Research Center for Environmental Health, Neuherberg, Germany
230 Department of Medicine I, Ludwig-Maximilians-Universität, Munich, Germany
231 DZHK (German Centre for Cardiovascular Research), partner site Munich Heart Alliance, Munich, Germany
232 Department of Cerebrovascular Diseases, Fondazione IRCCS Istituto Neurologico “Carlo Besta”, Milano, Italy
233 Karolinska Institutet, MEB, Stockholm, Sweden
234 University of Tartu, Estonian Genome Center, Tartu, Estonia, Tartu, Estonia
235 Department of Clinical and Experimental Sciences, Neurology Clinic, University of Brescia, Italy
236 Translational Genomics Unit, Department of Oncology, IRCCS Istituto di Ricerche Farmacologiche Mario Negri, Milano, Italy
237 Department of Genetics, Microbiology and Statistics, University of Barcelona, Barcelona, Spain
238 Psychiatric Genetics Unit, Group of Psychiatry, Mental Health and Addictions, Vall d’Hebron Research Institute (VHIR), Universitat Autònoma de Barcelona, Biomedical Network Research Centre on Mental Health (CIBERSAM), Barcelona, Spain
239 Department of Neurology, IMIM-Hospital del Mar, and Universitat Autònoma de Barcelona, Spain
240 IMIM (Hospital del Mar Medical Research Institute), Barcelona, Spain
241 National Institute for Health Research Comprehensive Biomedical Research Centre, Guy’s & St. Thomas’ NHS Foundation Trust and King’s College London, London, UK
242 Division of Health and Social Care Research, King’s College London, London, UK
243 FIMM-Institute for Molecular Medicine Finland, Helsinki, Finland
244 THL-National Institute for Health and Welfare, Helsinki, Finland
245 Iwate Tohoku Medical Megabank Organization, Iwate Medical University, Iwate, Japan
246 BHF Glasgow Cardiovascular Research Centre, Faculty of Medicine, Glasgow, UK
247 deCODE Genetics/Amgen, Inc., Reykjavik, Iceland
248 Icelandic Heart Association, Reykjavik, Iceland
249 Institute of Biomedicine, the Sahlgrenska Academy at University of Gothenburg, Goteborg, Sweden
250 Department of Epidemiology, University of Maryland School of Medicine, Baltimore, MD, USA
251 Institute of Cardiovascular and Medical Sciences, Faculty of Medicine, University of Glasgow, Glasgow, UK
252 Chair of Genetic Epidemiology, IBE, Faculty of Medicine, LMU Munich, Germany
253 Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, Nagoya, Japan
254 Department of Epidemiology, Nagoya University Graduate School of Medicine, Nagoya, Japan
255 University Medicine Greifswald, Institute for Community Medicine, SHIP-KEF, Greifswald, Germany
256 Department of Neurology, Caen University Hospital, Caen, France
257 University of Caen Normandy, Caen, France
258 Department of Internal Medicine, Erasmus University Medical Center, Rotterdam, Netherlands
259 Landspitali University Hospital, Reykjavik, Iceland
260 Survey Research Center, University of Michigan, Ann Arbor, MI, USA
261 University of Virginia Department of Neurology, Charlottesville, VA, USA
The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health and Social Care.
Abbreviations
- ACE
angiotensin-converting enzyme
- ADRB1
β-1 adrenergic receptor
- BCAC
Breast Cancer Association Consortium
- BMI
body mass index
- CCFR
Colon Cancer Family Registry
- CORECT
ColoRectal Transdisciplinary Study
- eQTLs
expression quantitative trait loci
- ER
estrogen receptor
- GECCO
Genetics and Epidemiology of Colorectal Cancer Consortium
- GERA
Genetic Epidemiology Research on Adult Health and Aging
- GWAS
genome-wide association study
- ICBP
International Consortium of Blood Pressure
- ILCCO
International Lung Cancer Consortium
- INTEGRAL
Integrative Analysis of Lung Cancer Risk and Etiology
- NCC
sodium-chloride symporter
- OR
odds ratio
- ORIGIN
Outcome Reduction with Initial Glargine Intervention
- PEER
probabilistic estimation of expression residuals
- PRACTICAL
Prostate Cancer Association Group to Investigate Cancer Associated Alterations in the Genome consortium
- RAS
renin–angiotensin system
- SBP
systolic blood pressure
- SNP
single-nucleotide polymorphism
- TMM
trimmed mean of M-values
- wGRS
weighted genetic risk score
Data Availability
Approval was received to use restricted summary genetic association data from GECCO, INTEGRAL ILCCO, and PRACTICAL consortia after submitting a proposal to access this data. Summary genetic association data from these consortia can be accessed by contacting GECCO (kafdem@fredhutch.org, INTEGRAL ILCCO (rayjean.hung@lunenfeld.ca) (https://ilcco.iarc.fr/), and PRACTICAL (practical@icr.ac.uk). Summary genetic association data from the BarcUVa-Seq analyses included in this manuscript can be downloaded from https://doi.org/10.34810/data132 (“eqtls_annot_hg38.txt.xz”). Summary genetic association data from the Finngen consortium can be accessed by visiting https://www.finngen.fi/en/access_results. To obtain data from the Asia Colorectal Cancer Consortium and the Korean National Cancer Center CRC Study, please contact the Vanderbilt Epidemiology Center (epidemiology@vumc.org). All other relevant data are within the manuscript and its Supporting Information files.
Funding Statement
JY is supported by a Cancer Research UK Population Research Postdoctoral Fellowship (C68933/A28534) (https://www.cancerresearchuk.org/). JY, EEV, VT, GDS, and RMM are supported by Cancer Research UK (C18281/A29019) programme grant (the Integrative Cancer Epidemiology Programme) (https://www.cancerresearchuk.org/). JY, TGR, VMW, EEV, VT, GDS, and RMM are part of the Medical Research Council Integrative Epidemiology Unit at the University of Bristol which is supported by the Medical Research Council (MC_UU_00011/1, MC_UU_00011/3, MC_UU_00011/6, and MC_UU_00011/4) and the University of Bristol (https://mrc.ukri.org/; https://www.bristol.ac.uk/). RMM is also supported by the NIHR Bristol Biomedical Research Centre which is funded by the NIHR and is a partnership between University Hospitals Bristol NHS Foundation Trust and the University of Bristol (https://www.nihr.ac.uk/). EEV is supported by Diabetes UK (17/0005587) and the World Cancer Research Fund (WCRF UK), as part of the World Cancer Research Fund International grant programme (IIG_2019_2009) (https://www.diabetes.org.uk/; https://www.wcrf.org/). MOS received a post-doctoral fellowship from the Spanish Association Against Cancer (AECC) Scientific Foundation (https://www.aecc.es/es). VDO, MOS, and VM are part of the group 55 of CIBERESP and AGAUR 2017SGR723 (https://www.ciberesp.es/en). CIA is a Cancer Prevention Research Institute of Texas Research Scholar and is supported by RR170048 (https://www.cprit.state.tx.us/). INTEGRAL-ILCCO is supported by a National Institutes of Health grant (U19 CA203654) (https://www.nih.gov/). The International Lung Cancer Consortium is supported by a National Cancer Institute grant (U19CA203654) (https://www.cancer.gov/). GC is supported by R01 NIH/NCI CA143237 and CA204279. Cancer consortia funding is presented in the Supplementary Material. The funders of this study played no role in the design, data collection and analysis, decision to publish, or preparation of this manuscript.
References
- 1.Mahmoudpour SH, Asselbergs FW, Souverein PC, de Boer A. Maitland-van der Zee AH. Prescription patterns of angiotensin-converting enzyme inhibitors for various indications: A UK population-based study. Br J Clin Pharmacol. 2018;84(10):2365–72. doi: 10.1111/bcp.13692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bangalore S, Kumar S, Kjeldsen SE, Makani H, Grossman E, Wetterslev J, et al. Antihypertensive drugs and risk of cancer: network meta-analyses and trial sequential analyses of 324 168 participants from randomised trials. Lancet Oncol. 2011;12(1):65–82. doi: 10.1016/S1470-2045(10)70260-6 [DOI] [PubMed] [Google Scholar]
- 3.Effects of telmisartan, irbesartan, valsartan, candesartan, and losartan on cancers in 15 trials enrolling 138,769 individuals. J Hypertens. 2011;29(4):623–35. doi: 10.1097/HJH.0b013e328344a7de [DOI] [PubMed] [Google Scholar]
- 4.Sanidas E, Velliou M, Papadopoulos D, Fotsali A, Iliopoulos D, Mantzourani M, et al. Antihypertensive Drugs and Risk of Cancer. Between Scylla and Charybdis. Am J Hypertens. 2020. doi: 10.1093/ajh/hpaa098 [DOI] [PubMed] [Google Scholar]
- 5.Mayordomo C, García-Recio S, Ametller E, Fernández-Nogueira P, Pastor-Arroyo EM, Vinyals L, et al. Targeting of substance P induces cancer cell death and decreases the steady state of EGFR and Her2. J Cell Physiol. 2012;227(4):1358–66. doi: 10.1002/jcp.22848 [DOI] [PubMed] [Google Scholar]
- 6.Coveñas R, Muñoz M. Cancer progression and substance P. Histol Histopathol. 2014;29(7):881–90. doi: 10.14670/HH-29.881 [DOI] [PubMed] [Google Scholar]
- 7.Hallas J, Christensen R, Andersen M, Friis S, Bjerrum L. Long term use of drugs affecting the renin-angiotensin system and the risk of cancer: a population-based case-control study. Br J Clin Pharmacol. 2012;74(1):180–8. doi: 10.1111/j.1365-2125.2012.04170.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hicks BM, Filion KB, Yin H, Sakr L, Udell JA, Azoulay L. Angiotensin converting enzyme inhibitors and risk of lung cancer: population based cohort study. BMJ. 2018;363:k4209. doi: 10.1136/bmj.k4209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kemppainen KJ, Tammela TL, Auvinen A, Murtola TJ. The association between antihypertensive drug use and incidence of prostate cancer in Finland: a population-based case-control study. Cancer Causes Control. 2011;22(10):1445–52. doi: 10.1007/s10552-011-9819-3 [DOI] [PubMed] [Google Scholar]
- 10.Lin SY, Lin CL, Lin CC, Hsu WH, Lin CD, Wang IK, et al. Association between Angiotensin-Converting Enzyme Inhibitors and Lung Cancer-A Nationwide, Population-Based, Propensity Score-Matched Cohort Study. Cancers (Basel). 2020;12(3). doi: 10.3390/cancers12030747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Devore EE, Kim S, Ramin CA, Wegrzyn LR, Massa J, Holmes MD, et al. Antihypertensive medication use and incident breast cancer in women. Breast Cancer Res Treat. 2015;150(1):219–29. doi: 10.1007/s10549-015-3311-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ronquist G, Rodríguez LA, Ruigómez A, Johansson S, Wallander MA, Frithz G, et al. Association between captopril, other antihypertensive drugs and risk of prostate cancer. Prostate. 2004;58(1):50–6. doi: 10.1002/pros.10294 [DOI] [PubMed] [Google Scholar]
- 13.Boudreau DM, Koehler E, Rulyak SJ, Haneuse S, Harrison R, Mandelson MT. Cardiovascular medication use and risk for colorectal cancer. Cancer Epidemiol Biomark Prev. 2008;17(11):3076–80. doi: 10.1158/1055-9965.EPI-08-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen X, Yi CH, Ya KG. Renin-angiotensin system inhibitor use and colorectal cancer risk and mortality: A dose-response meta analysis. J Renin-Angiotensin-Aldosterone Syst. 2020;21(3):1470320319895646. doi: 10.1177/1470320319895646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheung KS, Chan EW, Seto WK, Wong ICK, Leung WK. ACE (Angiotensin-Converting Enzyme) Inhibitors/Angiotensin Receptor Blockers Are Associated With Lower Colorectal Cancer Risk: A Territory-Wide Study With Propensity Score Analysis. Hypertension. 2020;76(3):968–75. doi: 10.1161/HYPERTENSIONAHA.120.15317 [DOI] [PubMed] [Google Scholar]
- 16.McMahon AD. Approaches to combat with confounding by indication in observational studies of intended drug effects. Pharmacoepidemiol Drug Saf. 2003;12(7):551–8. doi: 10.1002/pds.883 [DOI] [PubMed] [Google Scholar]
- 17.Kedika R, Patel M, Pena Sahdala HN, Mahgoub A, Cipher D, Siddiqui AA. Long-term use of angiotensin converting enzyme inhibitors is associated with decreased incidence of advanced adenomatous colon polyps. J Clin Gastroenterol. 2011;45(2):e12–6. doi: 10.1097/MCG.0b013e3181ea1044 [DOI] [PubMed] [Google Scholar]
- 18.Mansouri D, McMillan DC, Roxburgh CS, Crighton EM, Horgan PG. The impact of aspirin, statins and ACE-inhibitors on the presentation of colorectal neoplasia in a colorectal cancer screening programme. Br J Cancer. 2013;109(1):249–56. doi: 10.1038/bjc.2013.292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ray WA. Evaluating medication effects outside of clinical trials: new-user designs. Am J Epidemiol. 2003;158(9):915–20. doi: 10.1093/aje/kwg231 [DOI] [PubMed] [Google Scholar]
- 20.Lever AF, Hole DJ, Gillis CR, McCallum IR, McInnes GT, MacKinnon PL, et al. Do inhibitors of angiotensin-I-converting enzyme protect against risk of cancer? Lancet. 1998;352(9123):179–84. doi: 10.1016/S0140-6736(98)03228-0 [DOI] [PubMed] [Google Scholar]
- 21.Suissa S, Azoulay L. Metformin and the risk of cancer: time-related biases in observational studies. Diabetes Care. 2012;35(12):2665–73. doi: 10.2337/dc12-0788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perron L, Bairati I, Harel F, Meyer F. Antihypertensive drug use and the risk of prostate cancer (Canada). Cancer Causes Control. 2004;15(6):535–41. doi: 10.1023/B:CACO.0000036152.58271.5e [DOI] [PubMed] [Google Scholar]
- 23.Largent JA, McEligot AJ, Ziogas A, Reid C, Hess J, Leighton N, et al. Hypertension, diuretics and breast cancer risk. J Hum Hypertens. 2006;20(10):727–32. doi: 10.1038/sj.jhh.1002075 [DOI] [PubMed] [Google Scholar]
- 24.Jansen L, Below J, Chang-Claude J, Brenner H, Hoffmeister M. Beta blocker use and colorectal cancer risk: population-based case-control study. Cancer. 2012;118(16):3911–9. doi: 10.1002/cncr.26727 [DOI] [PubMed] [Google Scholar]
- 25.Li CI, Daling JR, Tang MT, Haugen KL, Porter PL, Malone KE. Use of antihypertensive medications and breast cancer risk among women aged 55 to 74 years. JAMA Intern Med. 2013;173(17):1629–37. doi: 10.1001/jamainternmed.2013.9071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vezina RM, Lesko SM, Rosenberg L, Shapiro S. Calcium channel blocker use and the risk of prostate cancer. Am J Hypertens. 1998;11(12):1420–5. doi: 10.1016/s0895-7061(98)00176-9 [DOI] [PubMed] [Google Scholar]
- 27.Gómez-Acebo I, Dierssen-Sotos T, Palazuelos C, Pérez-Gómez B, Lope V, Tusquets I, et al. The Use of Antihypertensive Medication and the Risk of Breast Cancer in a Case-Control Study in a Spanish Population: The MCC-Spain Study. PLoS ONE. 2016;11(8):e0159672. doi: 10.1371/journal.pone.0159672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tenenbaum A, Motro M, Jonas M, Fisman EZ, Grossman E, Boyko V, et al. Is diuretic therapy associated with an increased risk of colon cancer? Am J Med. 2001;110(2):143–5. doi: 10.1016/s0002-9343(00)00674-4 [DOI] [PubMed] [Google Scholar]
- 29.Lin CS, Lin WS, Lin CL, Kao CH. Carvedilol use is associated with reduced cancer risk: A nationwide population-based cohort study. Int J Cardiol. 2015;184:9–13. doi: 10.1016/j.ijcard.2015.02.015 [DOI] [PubMed] [Google Scholar]
- 30.Monami M, Filippi L, Ungar A, Sgrilli F, Antenore A, Dicembrini I, et al. Further data on beta-blockers and cancer risk: observational study and meta-analysis of randomized clinical trials. Curr Med Res Opin. 2013;29(4):369–78. doi: 10.1185/03007995.2013.772505 [DOI] [PubMed] [Google Scholar]
- 31.Smith GD, Ebrahim S. ’Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol. 2003;32(1):1–22. doi: 10.1093/ije/dyg070 [DOI] [PubMed] [Google Scholar]
- 32.Yarmolinsky J, Wade KH, Richmond RC, Langdon RJ, Bull CJ, Tilling KM, et al. Causal Inference in Cancer Epidemiology: What Is the Role of Mendelian Randomization? Cancer Epidemiol Biomark Prev. 2018;27(9):995–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ference BA, Majeed F, Penumetcha R, Flack JM, Brook RD. Effect of naturally random allocation to lower low-density lipoprotein cholesterol on the risk of coronary heart disease mediated by polymorphisms in NPC1L1, HMGCR, or both: a 2 × 2 factorial Mendelian randomization study. J Am Coll Cardiol. 2015;65(15):1552–61. doi: 10.1016/j.jacc.2015.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ference BA, Robinson JG, Brook RD, Catapano AL, Chapman MJ, Neff DR, et al. Variation in PCSK9 and HMGCR and Risk of Cardiovascular Disease and Diabetes. N Engl J Med. 2016;375(22):2144–53. doi: 10.1056/NEJMoa1604304 [DOI] [PubMed] [Google Scholar]
- 35.Plenge RM, Scolnick EM, Altshuler D. Validating therapeutic targets through human genetics. Nat Rev Drug Discov. 2013;12(8):581–94. doi: 10.1038/nrd4051 [DOI] [PubMed] [Google Scholar]
- 36.Swerdlow DI, Preiss D, Kuchenbaecker KB, Holmes MV, Engmann JE, Shah T, et al. HMG-coenzyme A reductase inhibition, type 2 diabetes, and bodyweight: evidence from genetic analysis and randomised trials. Lancet. 2015;385(9965):351–61. doi: 10.1016/S0140-6736(14)61183-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Michailidou K, Lindström S, Dennis J, Beesley J, Hui S, Kar S, et al. Association analysis identifies 65 new breast cancer risk loci. Nature. 2017;551(7678):92–4. doi: 10.1038/nature24284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huyghe JR, Bien SA, Harrison TA, Kang HM, Chen S, Schmit SL, et al. Discovery of common and rare genetic risk variants for colorectal cancer. Nat Genet. 2019;51(1):76–87. doi: 10.1038/s41588-018-0286-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McKay JD, Hung RJ, Han Y, Zong X, Carreras-Torres R, Christiani DC, et al. Large-scale association analysis identifies new lung cancer susceptibility loci and heterogeneity in genetic susceptibility across histological subtypes. Nat Genet. 2017;49(7):1126–32. doi: 10.1038/ng.3892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schumacher FR, Al Olama AA, Berndt SI, Benlloch S, Ahmed M, Saunders EJ, et al. Association analyses of more than 140,000 men identify 63 new prostate cancer susceptibility loci. Nat Genet. 2018;50(7):928–36. doi: 10.1038/s41588-018-0142-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu Y, Kweon SS, Cai Q, Tanikawa C, Shu XO, Jia WH, et al. Identification of Novel Loci and New Risk Variant in Known Loci for Colorectal Cancer Risk in East Asians. Cancer Epidemiol Biomark Prev. 2020;29(2):477–86. doi: 10.1158/1055-9965.EPI-19-0755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190–1. doi: 10.1093/bioinformatics/btq340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Evangelou E, Warren HR, Mosen-Ansorena D, Mifsud B, Pazoki R, Gao H, et al. Genetic analysis of over 1 million people identifies 535 new loci associated with blood pressure traits. Nat Genet. 2018;50(10):1412–25. doi: 10.1038/s41588-018-0205-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoffmann TJ, Ehret GB, Nandakumar P, Ranatunga D, Schaefer C, Kwok PY, et al. Genome-wide association analyses using electronic health records identify new loci influencing blood pressure variation. Nat Genet. 2017;49(1):54–64. doi: 10.1038/ng.3715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Burgess S, Thompson SG. Multivariable Mendelian Randomization: The Use of Pleiotropic Genetic Variants to Estimate Causal Effects. Am J Epidemiol. 2015;181(4):251–60. doi: 10.1093/aje/kwu283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pigeyre M, Sjaarda J, Chong M, Hess S, Bosch J, Yusuf S, et al. ACE and Type 2 Diabetes Risk: A Mendelian Randomization Study. Diabetes Care. 2020;43(4):835–42. doi: 10.2337/dc19-1973 [DOI] [PubMed] [Google Scholar]
- 47.Malik R, Chauhan G, Traylor M, Sargurupremraj M, Okada Y, Mishra A, et al. Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat Genet. 2018;50(4):524–37. doi: 10.1038/s41588-018-0058-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mahajan A, Taliun D, Thurner M, Robertson NR, Torres JM, Rayner NW, et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat Genet. 2018;50(11):1505–13. doi: 10.1038/s41588-018-0241-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wright JM, Musini VM, Gill R. First-line drugs for hypertension. Cochrane Database Syst Rev. 2018;4(4):Cd001841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chung CM, Wang RY, Chen JW, Fann CS, Leu HB, Ho HY, et al. A genome-wide association study identifies new loci for ACE activity: potential implications for response to ACE inhibitor. Pharmacogenomics J. 2010;10(6):537–44. doi: 10.1038/tpj.2009.70 [DOI] [PubMed] [Google Scholar]
- 51.Kanai M, Akiyama M, Takahashi A, Matoba N, Momozawa Y, Ikeda M, et al. Genetic analysis of quantitative traits in the Japanese population links cell types to complex human diseases. Nat Genet. 2018;50(3):390–400. doi: 10.1038/s41588-018-0047-6 [DOI] [PubMed] [Google Scholar]
- 52.Bowden J, Del Greco MF, Minelli C, Davey Smith G, Sheehan N, Thompson J. A framework for the investigation of pleiotropy in two-sample summary data Mendelian randomization. Stat Med. 2017;36(11):1783–802. doi: 10.1002/sim.7221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, et al. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. doi: 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Burgess S, Zuber V, Valdes-Marquez E, Sun BB, Hopewell JC. Mendelian randomization with fine-mapped genetic data: Choosing from large numbers of correlated instrumental variables. Genet Epidemiol. 2017;41(8):714–25. doi: 10.1002/gepi.22077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Burgess S, Thompson SG. Avoiding bias from weak instruments in Mendelian randomization studies. Int J Epidemiol. 2011;40(3):755–64. doi: 10.1093/ije/dyr036 [DOI] [PubMed] [Google Scholar]
- 56.Wallace C. Statistical testing of shared genetic control for potentially related traits. Genet Epidemiol. 2013;37(8):802–13. doi: 10.1002/gepi.21765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Murphy N, Carreras-Torres R, Song M, Chan AT, Martin RM, Papadimitriou N, et al. Circulating Levels of Insulin-like Growth Factor 1 and Insulin-like Growth Factor Binding Protein 3 Associate With Risk of Colorectal Cancer Based on Serologic and Mendelian Randomization Analyses. Gastroenterology. 2020;158(5):1300–12.e20. doi: 10.1053/j.gastro.2019.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mariosa D, Carreras-Torres R, Martin RM, Johansson M, Brennan P. Commentary: What can Mendelian randomization tell us about causes of cancer? Int J Epidemiol. 2019;48(3):816–21. doi: 10.1093/ije/dyz151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kho PF, Amant F, Annibali D, Ashton K, Attia J, Auer PL, et al. Mendelian randomization analyses suggest a role for cholesterol in the development of endometrial cancer. Int J Cancer. 2020. doi: 10.1002/ijc.33206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cornish AJ, Law PJ, Timofeeva M, Palin K, Farrington SM, Palles C, et al. Modifiable pathways for colorectal cancer: a mendelian randomisation analysis. Lancet Gastroenterol Hepatol. 2020;5(1):55–62. doi: 10.1016/S2468-1253(19)30294-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Prins BP, Kuchenbaecker KB, Bao Y, Smart M, Zabaneh D, Fatemifar G, et al. Genome-wide analysis of health-related biomarkers in the UK Household Longitudinal Study reveals novel associations. Sci Rep. 2017;7(1):11008. doi: 10.1038/s41598-017-10812-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kettunen J, Demirkan A, Würtz P, Draisma HH, Haller T, Rawal R, et al. Genome-wide study for circulating metabolites identifies 62 loci and reveals novel systemic effects of LPA. Nat Commun. 2016;7:11122. doi: 10.1038/ncomms11122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Benyamin B, Esko T, Ried JS, Radhakrishnan A, Vermeulen SH, Traglia M, et al. Novel loci affecting iron homeostasis and their effects in individuals at risk for hemochromatosis. Nat Commun. 2014;5:4926. doi: 10.1038/ncomms5926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Doherty A, Smith-Byrne K, Ferreira T, Holmes MV, Holmes C, Pulit SL, et al. GWAS identifies 14 loci for device-measured physical activity and sleep duration. Nat Commun. 2018;9(1):5257. doi: 10.1038/s41467-018-07743-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu M, Jiang Y, Wedow R, Li Y, Brazel DM, Chen F, et al. Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat Genet. 2019;51(2):237–44. doi: 10.1038/s41588-018-0307-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Díez-Obrero V, Dampier CH, Moratalla-Navarro F, Devall M, Plummer SJ, Díez-Villanueva A, et al. Genetic effects on transcriptome profiles in colon epithelium provide functional insights for genetic risk loci. Cell Mol Gastroenterol Hepatol. 2021. doi: 10.1016/j.jcmgh.2021.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Skrivankova VW, Richmond RC, Woolf BAR, Yarmolinsky J, Davies NM, Swanson SA, et al. Strengthening the Reporting of Observational Studies in Epidemiology Using Mendelian Randomization: The STROBE-MR Statement. JAMA. 2021;326(16):1614–21. doi: 10.1001/jama.2021.18236 [DOI] [PubMed] [Google Scholar]
- 69.Heran BS, Wong MM, Heran IK, Wright JM. Blood pressure lowering efficacy of angiotensin converting enzyme (ACE) inhibitors for primary hypertension. Cochrane Database Syst Rev. 2008;2008(4):Cd003823. doi: 10.1002/14651858.CD003823.pub2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Okwan-Duodu D, Landry J, Shen XZ, Diaz R. Angiotensin-converting enzyme and the tumor microenvironment: mechanisms beyond angiogenesis. Am J Physiol Regul Integr Comp Physiol. 2013;305(3):R205–15. doi: 10.1152/ajpregu.00544.2012 [DOI] [PubMed] [Google Scholar]
- 71.Wang D, Carretero OA, Yang XY, Rhaleb NE, Liu YH, Liao TD, et al. N-acetyl-seryl-aspartyl-lysyl-proline stimulates angiogenesis in vitro and in vivo. Am J Physiol Heart Circ Physiol. 2004;287(5):H2099–105. doi: 10.1152/ajpheart.00592.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. doi: 10.1126/science.1260419 [DOI] [PubMed] [Google Scholar]
- 73.Law PJ, Timofeeva M, Fernandez-Rozadilla C, Broderick P, Studd J, Fernandez-Tajes J, et al. Association analyses identify 31 new risk loci for colorectal cancer susceptibility. Nat Commun. 2019;10(1):2154. doi: 10.1038/s41467-019-09775-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Armaghany T, Wilson JD, Chu Q, Mills G. Genetic alterations in colorectal cancer. Gastrointest Cancer Res. 2012;5(1):19–27. [PMC free article] [PubMed] [Google Scholar]
- 75.Sievers CK, Grady WM, Halberg RB, Pickhardt PJ. New insights into the earliest stages of colorectal tumorigenesis. Expert Rev Gastroenterol Hepatol. 2017;11(8):723–9. doi: 10.1080/17474124.2017.1330150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rothwell PM, Wilson M, Elwin CE, Norrving B, Algra A, Warlow CP, et al. Long-term effect of aspirin on colorectal cancer incidence and mortality: 20-year follow-up of five randomised trials. Lancet. 2010;376(9754):1741–50. doi: 10.1016/S0140-6736(10)61543-7 [DOI] [PubMed] [Google Scholar]
- 77.Clarke MA, Joshu CE. Early Life Exposures and Adult Cancer Risk. Epidemiol Rev. 2017;39(1):11–27. doi: 10.1093/epirev/mxx004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Georgakis MK, Gill D, Webb AJS, Evangelou E, Elliott P, Sudlow CLM, et al. Genetically determined blood pressure, antihypertensive drug classes, and risk of stroke subtypes. Neurology. 2020;95(4):e353–e61. doi: 10.1212/WNL.0000000000009814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gill D, Georgakis MK, Koskeridis F, Jiang L, Feng Q, Wei WQ, et al. Use of Genetic Variants Related to Antihypertensive Drugs to Inform on Efficacy and Side Effects. Circulation. 2019;140(4):270–9. doi: 10.1161/CIRCULATIONAHA.118.038814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wu Y, Byrne EM, Zheng Z, Kemper KE, Yengo L, Mallett AJ, et al. Genome-wide association study of medication-use and associated disease in the UK Biobank. Nat Commun. 2019;10(1):1891. doi: 10.1038/s41467-019-09572-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chung JF, Lee SJ, Sood AK. Immunological and pleiotropic effects of individual β-blockers and their relevance in cancer therapies. Expert Opin Investig Drugs. 2016;25(5):501–5. doi: 10.1517/13543784.2016.1164141 [DOI] [PubMed] [Google Scholar]
- 82.Fitzmaurice C, Abate D, Abbasi N, Abbastabar H, Abd-Allah F, Abdel-Rahman O, et al. Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived With Disability, and Disability-Adjusted Life-Years for 29 Cancer Groups, 1990 to 2017: A Systematic Analysis for the Global Burden of Disease Study. JAMA Oncol. 2019;5(12):1749–68. doi: 10.1001/jamaoncol.2019.2996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Reid JL. From kinetics to dynamics: Are there differences between ACE inhibitors? Eur Heart J. 1997;18(suppl_E):14–8. doi: 10.1016/s0195-668x(97)90004-x [DOI] [PubMed] [Google Scholar]
- 84.Brown NJ, Vaughan DE. Angiotensin-converting enzyme inhibitors. Circulation. 1998;97(14):1411–20. doi: 10.1161/01.cir.97.14.1411 [DOI] [PubMed] [Google Scholar]
- 85.Dinicolantonio JJ, Lavie CJ, O’Keefe JH. Not all angiotensin-converting enzyme inhibitors are equal: focus on ramipril and perindopril. Postgrad Med. 2013;125(4):154–68. doi: 10.3810/pgm.2013.07.2687 [DOI] [PubMed] [Google Scholar]
- 86.Lawlor DA, Tilling K, Davey SG. Triangulation in aetiological epidemiology. Int J Epidemiol. 2016;45(6):1866–86. doi: 10.1093/ije/dyw314 [DOI] [PMC free article] [PubMed] [Google Scholar]