Abstract
The arrangement of transcription factor (TF) binding motifs (syntax) is an important part of the cis-regulatory code, yet remains elusive. We introduce a deep learning model, BPNet, that uses DNA sequence to predict base-resolution ChIP-nexus binding profiles of pluripotency TFs. We develop interpretation tools to learn predictive motif representations and identify soft syntax rules for cooperative TF binding interactions. Strikingly, Nanog preferentially binds with helical periodicity, and TFs often cooperate in a directional manner, which we validate using CRISPR-induced point mutations. Our model represents a powerful general approach to uncover the motifs and syntax of cis-regulatory sequences in genomics data.
Introduction
Understanding the cis-regulatory code of the genome is vital for understanding when and where genes are expressed and how genetic variation and somatic mutations affect disease. Despite extensive efforts to map millions of putative enhancers in a wide variety of cell types and tissues1-3, identifying the critical bases that alter their regulatory information remains a major challenge. It is known that short sequence motifs are critical for the binding of sequence-specific transcription factors (TFs), but how motif combinations and their syntactic arrangements influence TF binding in vivo is not well understood. For example, two or more strictly spaced motifs may form composite motifs that provide a platform for DNA-mediated cooperativity between the corresponding TFs4. However, whether less strict (“soft”) motif spacing preferences exist within enhancers and influence TF cooperativity is not clear. The precise rules of the cis-regulatory code remain to be elucidated.
Experimental manipulations of enhancer sequences, such as mutations or synthetic designs, strongly support the existence of motif syntax5-12. However, genome-wide analyses have rarely identified statistically overrepresented motif syntax rules, questioning whether they exist and impose evolutionarily constraints on enhancer function13-17. One limitation is that motif instances are typically identified as overrepresented sequences matching position weight matrix (PWM) models18-21. When patterns are discovered computationally22-28, they are difficult to validate experimentally and the mechanism by which they might affect TF cooperativity is not clear. For example, overrepresented instances of strict motif spacings are sometimes associated with retrotransposons that contain multiple TF binding motifs23,24. On the other hand, when experimental TF binding data are available, i.e. from chromatin immunoprecipitation experiments coupled to sequencing (ChIP-seq)29-34, inference of motif syntax is still limited by the low resolution of putative binding events identified using peak-callers29-34.
There is hence a critical need for a general method that can identify cis-regulatory motif syntax based on genome-wide experimental data. Recently, convolutional neural networks (CNNs) have been applied towards accurately predicting diverse molecular phenotypes including TF binding from DNA sequence35-38. The advantage of CNNs is that they can learn flexible predictive models composed of hierarchical layers of arbitrarily complex, non-linear pattern detectors, allowing them to capture de novo sequence motifs and their higher-order organizational context without making strong prior biological assumptions. However, the complexity of these models makes them particularly challenging to interpret. While several methods have been developed to visualize TF binding motifs from trained CNNs35,36,38-42, methods for extracting the rules by which motif syntax informs TF binding are lacking43.
Another critical limitation is the resolution of current CNN models. State-of-the-art models of TF binding predict binary binding events35-37 or low-resolution continuous binding signal averaged across 100-200 bp windows44. This can limit the ability to learn motif syntax that promotes TF cooperativity43, which likely exists in ChIP-seq experiments. For example, TFs sometimes bind indirectly to motifs of other TFs16,24,45-47. TF cooperativity is even more apparent when the resolution of ChIP-seq is improved by adding an exonuclease digestion step (ChIP-exo)48. ChIP-exo methods such as ChIP-nexus generate base-resolution footprints precisely over the motif instances bound by the TF in vivo49,50 and these footprints differ between directly and indirectly bound motifs50,51. ChIP-nexus profiles have also provided evidence that TFs may help the binding of another TF nearby52. Although the full extent of TF cooperativity at the level of binding is not known, these results indicate that ChIP-seq data, and especially ChIP-nexus data, are a useful readout for cis-regulatory motif syntax, if the data are modeled at sufficiently high resolution.
To discover motif syntax, we developed a novel CNN called BPNet that models the relationship between cis-regulatory sequence and TF binding profiles at base resolution. We studied the pluripotency TFs Oct4, Sox2, Nanog and Klf4 in the well-characterized mouse embryonic stem cell (ESC) model, generating ChIP-nexus data for maximum resolution. We trained base-resolution BPNet models on these ChIP-nexus profiles with high predictive performance, on par with concordance between replicate experiments. We extended model interpretation methods to extract new motif representations that are not based on statistical over-representation but directly summarize the predictive influence on TF binding. We then developed methods that use the trained BPNet model as an in-silico oracle to measure how the distance between motif pairs affects TF cooperativity. We find that strict motif spacings in the genome are mainly due to retrotransposons, but that TF cooperativity depends on preferential soft motif syntax that is in agreement with experimentally characterized protein-protein or nucleosome interactions in ESCs. We also observe unexpected rules of TF binding cooperativity, including a broad preference for Nanog to bind DNA with helical periodicity, and perform experimental validations.
These results suggest that end-to-end neural network models trained on high-resolution genomics data, coupled with a dedicated suite of interpretation tools, can serve as a powerful tool for discovering the critical bases within cis-regulatory sequences and identifying the underlying motif syntax associated with TF cooperativity.
Results
BPNet predicts TF binding profiles from sequence
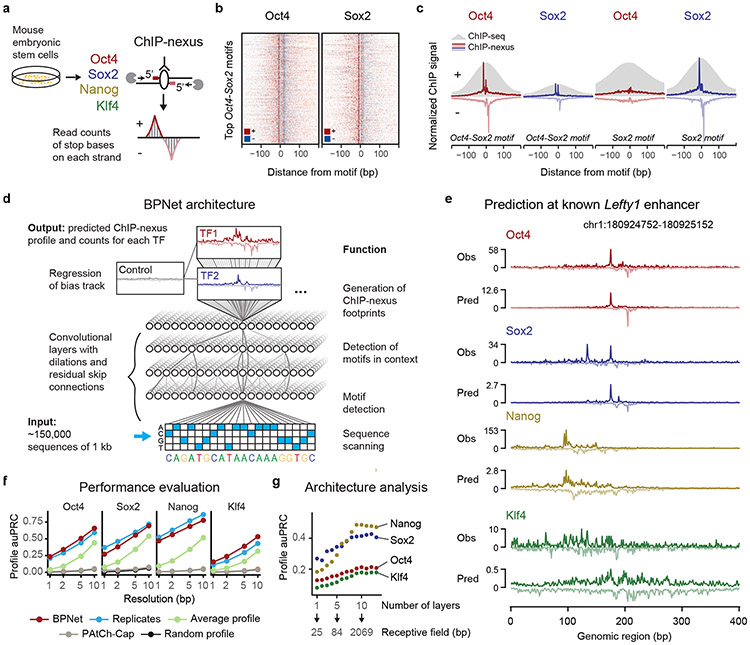
We performed ChIP-nexus experiments for Oct4, Sox2, Nanog and Klf4 in mouse ESCs and obtained genome-wide strand-specific base-resolution profiles for each TF (Fig. 1a). As shown for previous TF ChIP-nexus data49, the profiles at known TF binding motifs showed consistent stereotypical footprints across various genomic regions, as illustrated by the binding of Oct4 and Sox2 to the composite Oct4-Sox2 motif53 (Fig. 1b). These footprints not only had higher resolution compared to ChIP-seq data, but also displayed increased motif specificity. For example, the Sox2 motif showed a sharp ChIP-nexus footprint for Sox2 but not for Oct4, while ChIP-seq data showed binding signal for both (Fig. 1c). We identified 147,974 genomic regions of 1 kb length exhibiting statistically significant and reproducible enrichment of ChIP-nexus signal for Oct4, Sox2, Nanog or Klf4.
Fig. 1: BPNet predicts ChIP-nexus signal at base resolution.
a) ChIP-nexus experiments were performed on Oct4, Sox2, Nanog and Klf4 in mouse ESCs. After digestion of the 5’ DNA ends with lambda exonuclease, strand-specific stop sites were mapped to the genome at base resolution. Bound sites exhibit a distinct footprint of aligned reads, where the positive (+) strand peak occurs many bases before the negative (−) strand peak. b) Profile heatmaps of Oct4 and Sox2 ChIP-nexus data at the 500 Oct4-Sox2 motifs with the most ChIP-nexus reads (color depth for each strand represents normalized signal intensity). c) The average Oct4 and Sox2 ChIP-nexus footprints and ChIP-seq profiles at the 500 Oct4-Sox2 or Sox2 motifs with the most reads. The ChIP-nexus data have higher resolution and show less unspecific binding of Oct4 to the Sox2 motif. d) Architecture of the convolutional neural network (BPNet) that was trained to simultaneously predict the ChIP-nexus read counts at each strand for all TFs from 1 kb DNA sequences, while being prevented from learning information already explained by a bias track (PAtCh-Cap control). e) Observed and predicted ChIP-nexus read counts for the Lefty enhancer located on the held-out test chromosome 8. f) BPNet predicts the positions of high ChIP-nexus signal within the profiles at replicate-level accuracy as measured by the area under precision-recall curve (auPRC) at resolutions from 1 to 10 bp in held-out test chromosomes 1, 8 and 9. Results for the average ChIP-nexus profile, the PAtCh-Cap control profile and a randomized profile are shown as control. g) More convolutional layers (x-axis) increase the number of input bases considered for profile prediction at each position (receptive field) and this yields increasingly more accurate profile shape predictions on the tuning chromosomes 2-4 (measured in auPRC as above), showing that larger sequence context is important
In contrast to all current deep learning models for TF binding, we designed BPNet to directly predict the raw base-resolution binding profiles from DNA sequence. Binding profiles can be decomposed into the total signal (read counts) and the profile shape (base-resolution distribution of reads). We reasoned that the profile shape should be predictable from 1-kb genomic sequences since minimal enhancer activity can typically be reproduced outside its genomic context with sequences of <500 bases54,55. The total signal however could be influenced by factors that are not modeled, including chromatin state and higher-order chromatin organization.
To achieve high prediction accuracy, BPNet was designed with the following properties (Fig. 1d). (1) BPNet is a CNN that uses 25 bp wide filters in the first convolutional layer to scan the 1-kb region for relevant sequence motifs, followed by nine dilated convolutional layers with residual skip connections56,57 and exponential dilation in every layer44,58 to learn increasingly complex predictive sequence patterns with a 1-kb receptive field. To preserve base resolution, pooling is not used. (2) BPNet uses multi-task learning to jointly train on the strand-specific ChIP-nexus profiles of all four TFs. (3) Experimental control data are used as an auxiliary input (PAtCh-CAP for ChIP-nexus data59). The signal from this track is regressed out during training, which prevents BPNet from learning these experimental biases. (4) BPNet uses a multi-scale loss function to separately evaluate the predictions of profile shape (using a multinomial negative log-likelihood loss) and total read counts (using a mean squared error loss). Model training, hyperparameter tuning and performance evaluation were performed on different sets of genomic regions in distinct chromosomes.
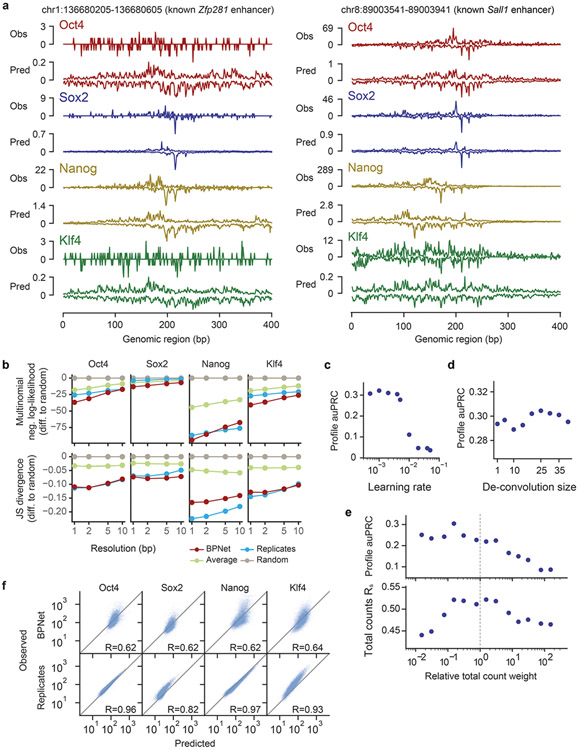
To evaluate predictive performance, we inspected individual enhancers located on held-out test chromosomes such as those associated with the genes Lefty160, Zfp28161 and Sall162,63 and found that the predicted and observed ChIP-nexus profiles were noticeably similar, with highly concordant summits of footprints (Fig. 1e, Extended Data Fig. 1a). We then systematically compared the positions of high ChIP-nexus counts between predicted versus observed profiles in all regions of the held-out test set. Strikingly, the positional concordance at resolutions ranging from 1-10 bp was on par with replicate experiments and substantially better than randomized profiles, average profiles and the control track (PAtCh-Cap) (Fig. 1f). Other measures of profile concordance confirmed the high prediction performance (Extended Data Fig. 1b). We also confirmed that mappability of regions did not bias the predictions (Supplementary Fig. 1). These results show that BPNet accurately learned to predict the ChIP-nexus binding profiles of all four TFs from DNA sequence.
To identify key components for the high prediction performance, we systematically varied the network architecture (Fig. 1g, Extended Data Fig. 1c-e). We found that the large number of convolutional layers was critical for predicting all four ChIP-nexus data sets and was particularly important for Nanog (Fig. 1g). This indicates that the learned sequence patterns required to predict ChIP-nexus profiles span over larger sequence regions beyond individual motifs64, especially in the case of Nanog. We also found that the relative priority of the profile versus total count prediction tasks during training affected prediction performance. Up-weighting the profile prediction task improved the performance of the profile predictions. However, irrespective of the relative task weights, the model’s performance for total count prediction (Rs = 0.62) did not match replicate-concordance (Rs = 0.94, Extended Data Fig. 1f). These results are consistent with our assumption that longer sequences or other measurements such as local chromatin state may be required for optimal prediction of total TF occupancy64, but that local sequence context (1 kb) is sufficient to accurately predict the shape of ChIP-nexus profiles.
A suite of model interpretation tools for TF binding motifs
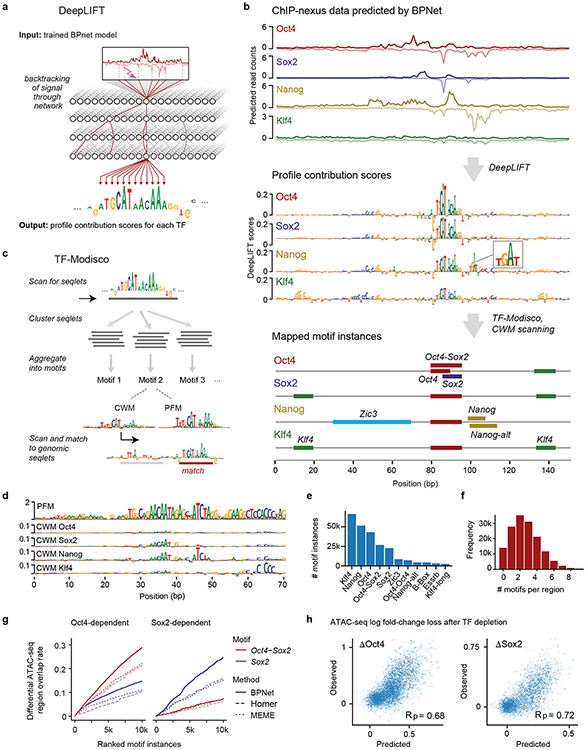
We next set out to extract the sequence features that were predictive of TF binding from the trained BPNet model. We extended our previously developed tool DeepLIFT65 to quantify the contribution of each base within an input sequence to the entire predicted ChIP-nexus profile of each TF (Fig. 2a, Methods). These TF-specific contribution scores are illustrated at the distal Oct4 enhancer where all four TFs show strong predicted footprints matching the observed ChIP-nexus footprints (Fig. 2b top, Supplementary Fig. 2a).
Fig. 2: TF motifs and their genomic instances can be accurately derived from BPNet using interpretation tools.
a) DeepLIFT recursively decomposes the predicted TF-specific binding output of the model and quantifies the contribution of each base of the input DNA sequence by backtracking the prediction through the network. b) Procedure for inferring and mapping predictive motif instances using the known distal Oct4 enhancer (chr17:35504453-35504603) as an example. From the predicted ChIP-nexus profile for each TF (top), DeepLIFT derives TF-specific profile contribution scores (middle). Regions with high contribution scores (called seqlets) resemble TF binding motifs. Seqlets are annotated by scanning the contribution scores with motifs discovered by TF-MoDISco (bottom). c) To discover motifs, TF-MoDISco scans for seqlets, extends the seqlets to 70 bp, performs pairwise alignments and clusters the seqlets. For each cluster, a motif is derived as contribution weight matrix (CWM), obtained by averaging the contribution scores of each of the 4 bases at each position across all aligned seqlets. The corresponding position frequency matrix (PFM) is the frequency of bases at each position. Motif instances are identified by scanning the CWM for each motif for high scoring matches across the profile contribution scores in the genomic regions. d) Example of a motif (N6) where the PFM differs from the CWMs. The PFM indicates that it is a repeat sequence (RLTR9E), while the CWM for each TF highlights the sequences that contribute to binding. e) Number of motif instances in thousands (k) found in the ~150,000 genomic regions for the 11 representative motifs. f) Histogram of the number mapped motif instances in thousands (k) found per region. g) Evaluation of the mapped motifs using previously identified regions that lose ATAC-seq signal in response to either Oct4 or Sox2 depletion (but not both)77. BPNet motif instances of Oct4-Sox2 and Sox2 (ranked by contribution scores) outperformed those obtained by HOMER and MEME (ranked by PWM match scores). h) A linear model based on the bottleneck layer of the trained BPNet model makes accurate quantitative predictions of the log fold-change loss in ATAC-seq signal upon depletion of Oct4 (ΔOct4) or Sox2 (ΔSox2). Results are shown with Pearson correlation coefficient (Rp) for the test chromosomes 1, 8 and 9 that were held out during training. See Supplementary Fig. 7b for a similar model based on motif instance features.
Subsequences with high contribution scores, which we call seqlets, often resemble TF binding motifs (Fig. 2b middle). One prominent seqlet matches the composite Oct4-Sox2 motif, which has previously been mapped to this exact position in the Oct4 enhancer66. This motif has high contribution scores for Oct4 and Sox2, which are directly bound to the motif, and slightly lower scores for Nanog and Klf4 (Fig. 2b middle), indicating that the Oct4-Sox2 motif could be indirectly important for the binding of other TFs.
Other seqlets did not readily match known motifs. For example, we found a TGAT sequence in the middle of the Nanog footprint (highlighted in Fig. 2a middle), but it was unclear whether it is a Nanog motif since previous reports on its consensus have been conflicting47,67-72. These results demonstrate the ability of contribution scores to highlight TF binding motifs, but also indicate the need to identify and characterize the motifs more systematically.
Next, we used TF-MoDISco41 to systematically discover and summarize recurring predictive sequence patterns into consolidated motifs from the sequences of all bound regions and their associated base-resolution contribution scores. For each TF, TF-MoDISCo uses contribution scores to identify, align and cluster seqlets across all bound sequences into consolidated motifs (Fig. 2c). For each cluster, a novel motif representation called contribution weight matrix (CWM) is derived by averaging the contribution scores of each of the four possible bases at every position across the seqlets. A more traditional position frequency matrix (PFM) representation, which contains the normalized base frequencies instead of the average contribution scores, is also calculated (see Supplementary Note on CWMs and PFMs/PWMs).
TF-MoDISco discovered 51 motifs, but 18 of them had unusually long PFMs (>40 bp) with high information content (30-100 bits) (Fig. 2d, Extended Data Fig. 2a). This implies that the genomic instances of these motifs share near identical base composition across the entire length of the pattern (despite being discovered by uniquely mappable ChIP-nexus reads). Indeed, we found that the majority of them (>80%) overlapped with annotated repeat elements (Extended Data Fig. 2b). The most common were long-terminal repeats (LTRs) of endogenous retrotransposon viruses (ERVs), including those of the ERVK, ERVL and the ERVL-MaLR family (Extended Data Fig. 2c). Remarkably, the corresponding CWM representations of these long PFMs were quite different. Instead of long stretches of uniformly overrepresented bases, the CWMs highlighted the shorter subsequences predictive of TF binding (Fig. 2d, Extended Data Fig. 2c). This difference between CWM and PFM representations provides a means to discover and pinpoint bound motifs within retrotransposons.
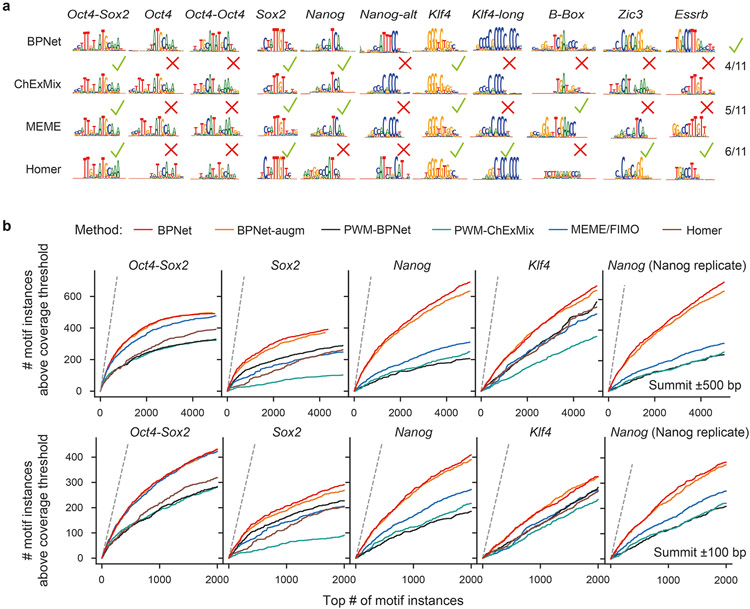
The remaining 33 motifs were all interpretable TF binding motifs, but contained subsets with subtle differences, leading us to select 11 representative motifs for further analysis (Extended Data Fig. 2d, Supplementary Fig. 3). These motifs include the well-known Oct4-Sox2, Sox2, and Klf4 motifs, as well as the Zic3 and Esrrb motifs, which bind pluripotency TFs that we did not profile. All motifs were overall robustly discovered by TF-MoDISCo from five different BPNet models trained on different subsets of ChIP-nexus peak regions (Supplementary Fig. 4).
Using the 11 representative motifs, we then comprehensively mapped and labeled all predictive motif instances in the bound genomic regions. We scanned the base-resolution contribution scores of all regions and annotated predictive motif instances that had high contribution scores and high match scores to the CWM (Fig. 2c). In total, we obtained 241,005 unique motif instances in the 147,974 genomic regions, with Klf4 motifs occurring most frequently (Fig. 2e). Altogether, 72,696 regions (48.1%) have at least three motif instances, and 20,352 regions (13.5%) have at least 5 motif instances (Fig. 2f). These genome-wide motif annotations are in agreement with motif instances supported by previous independent validation experiments73-75 (Supplementary Fig. 2b-d) and provide a strong foundation for analyzing genome-wide motif syntax and characterizing known functional enhancers in mouse ESCs (Fig. 2b bottom, Supplementary Fig. 5).
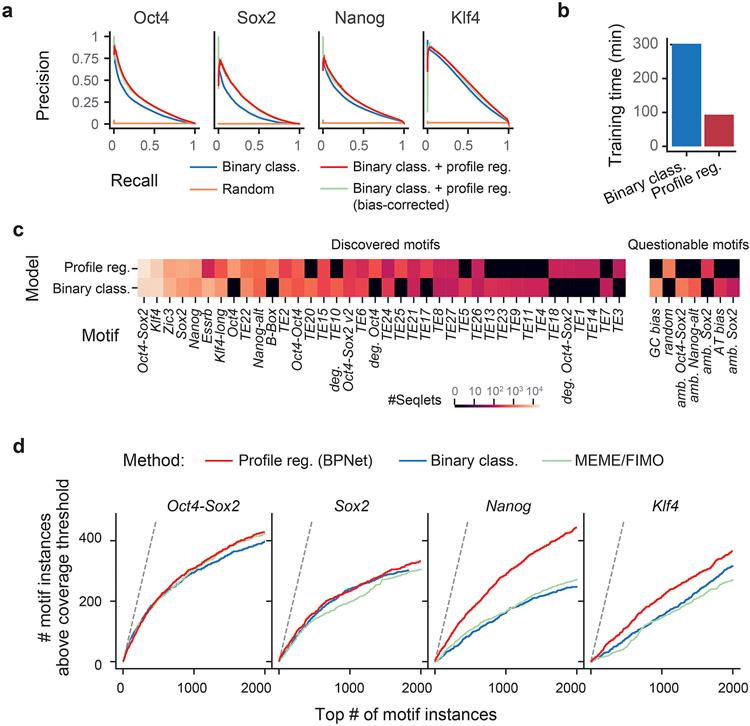
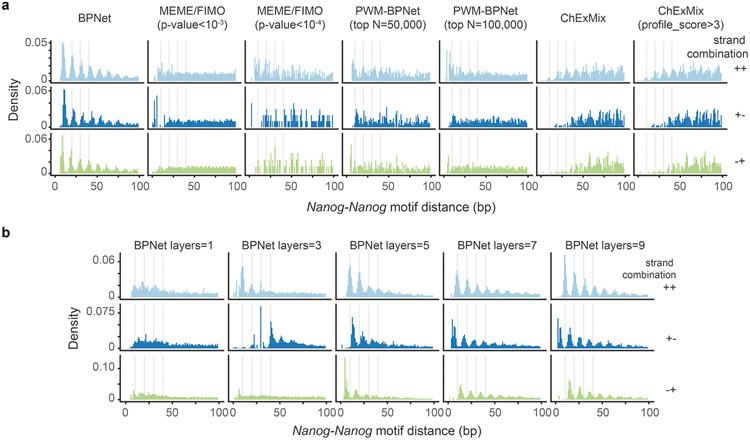
The motif maps derived from BPNet outperformed those obtained by traditional approaches such as PWM scanning, assessed by ChIP-nexus footprint height (Extended Data Fig. 3, Supplementary Note). BPNet correctly identified more motif instances supported by footprints in sequences from held-out test chromosomes than MEME18-21 or HOMER76, especially for the short Nanog motif. The improved performance is achieved because PWM-based motif scanning methods compute match scores using only sequence similarity, while BPNet’s method also incorporates the predictive contribution scores derived from the entire 1 kb sequence (Supplementary Fig. 6). The higher motif accuracy requires BPNet to be trained on base-resolution profiles, rather than coarse-resolution binary (bound vs. unbound) labels (Supplementary Note, Extended Data Fig. 4). This suggests that BPNet leverages the profiles to learn the importance of motif instances in their larger sequence context, thereby reducing the false discovery rate.
Our method also outperformed traditional methods when using an independent, previously published ATAC-seq data set77 for evaluation. After induced depletion of Oct4 or Sox2, regions with differential chromatin accessibility (as defined by the authors) overlapped more Oct4-Sox2 and Sox2 motif instances ranked by BPNet contribution scores than those ranked by motif scores from MEME or HOMER (Fig. 2g, Supplementary Fig. 7a). These results support the high accuracy of the BPNet mapped motif instances relative to those obtained from traditional motif discovery and scanning methods. They also confirm the link between the in vivo binding of Oct4 and Sox2 and their effect on chromatin accessibility.
Finally, we found that the quantitative changes in ATAC-seq signal after Oct4 and Sox2 depletion can also be accurately predicted from BPNet TF binding models. Specifically, linear models trained using the sequence features encoded in the final convolutional layer of the BPNet model were able to accurately predict differential accessibility (Fig. 2h). These models outperformed linear models trained using only the inferred motif instances (Supplementary Fig. 7b). These results indicate that the complete sequence representation learned by BPNet encodes predictive features beyond linear, additive effects of the motif instances. Hence, we set out to identify higher-order sequence features such as motif syntax.
Composite motifs and indirect binding footprints
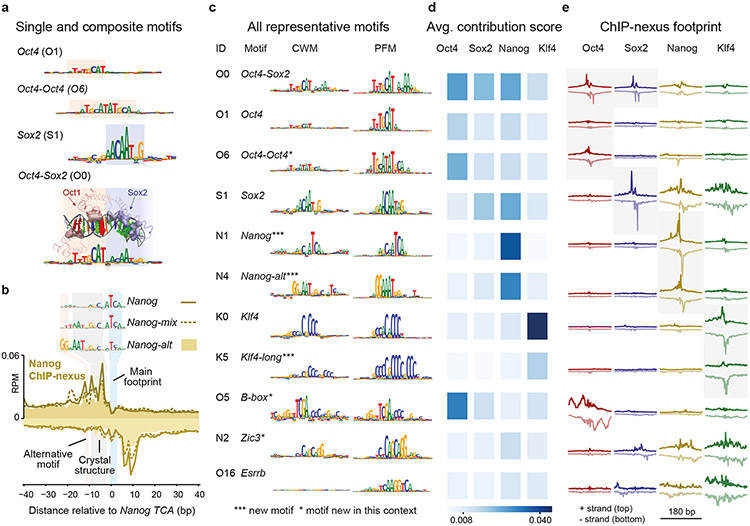
As a first step towards identifying motif syntax, we inspected the motifs identified by TF-MoDISCo for composite motifs, the simplest form of motif syntax. Indeed, we not only discovered the Sox2 motif and the monomeric Oct4 motif78, but also the composite Oct4-Oct4 motif (Fig. 3a), a near-palindromic motif that resembles the MORE and PORE motifs bound by Oct4 homodimers79,80. This motif has not previously been shown to be bound in ESCs in vivo, but is known to be important during neuronal differentiation81. Finally, we rediscovered the Oct4-Sox2 motif, in which the bases with high contribution scores correspond to the specific DNA contacts made by the heterodimer (based on the Oct1-Sox2 crystal structure)53,82,83 (Fig. 3a). Thus, we discovered composite motifs that are consistent with known structural data.
Fig. 3: Discovery of composite motifs and indirect binding footprints.
a) The CWMs of Oct4, Oct4-Oct4, Sox2 and Oct4-Sox2 were identified by TF-MoDISco as separate motifs (motif IDs = first letter of the TF + number, e.g. O1 discovered for Oct4), highlighting its ability to identify composite motifs. The CWM of the Oct4-Sox2 composite motif correlates with the structure of Oct1 and Sox2 bound to the Oct4-Sox2 motif. For visualization, the amino acids of Oct1 and Sox2 that contact DNA are shown as solid, and the atoms in the DNA bases, shown as colored spheres, are sized according to the contribution scores shown in the CWM below. b) Nanog ChIP-nexus binding footprints were associated with three Nanog motif variants (shown as CWM). For all motifs, the main footprint was found at the TCA sequence. The CWM of Nanog-mix (N5) and Nanog-alt (N4) contain a sequence that matches the sequence AATGGGC bound by Nanog in a crystal structure84. The CWM of Nanog-alt contains an alternative GG. c) The discovered representative short motifs contain known motifs, new motifs and known motifs new in this context. All sequence logos share the same y-axis. The B-box mediates RNA polymerase III transcription117,118 and is associated with high levels of Oct4 binding upstream and downstream of tRNA (Extended Data Fig. 5e,f). d) The average contribution score of the motif is shown for each TF. The highest score may indicate the TF that binds directly. e) The TF’s average ChIP-nexus footprint better indicates whether the motif is directly bound (sharp profile, marked with gray background), indirectly bound (fuzzy profile) or not bound at all. The footprints for each TF share the same y-axis.
We did not identify the composite Sox2-Nanog motif71 and found no evidence that this motif was bound in our ChIP-nexus data (Supplementary Fig. 8a). Instead, we identified three Nanog motifs: Nanog, Nanog-alt and Nanog-mix, the latter of which is partially similar to the first two. All have a main footprint around a TCA core sequence (Fig. 3b). Our primary Nanog motif resembles a previously identified Nanog motif from a thermodynamic model of ChIP-seq data72. Consistent with direct binding, a closely matching sequence (GCCATCA) is bound by Nanog in an EMSA gel shift assay72. Nanog-alt and Nanog-mix contain the sequence to which monomeric Nanog is bound in a crystal structure (AATGGGC)84. Given these two separate direct DNA contacts, the observed Nanog binding footprint likely represents Nanog binding as a homodimer85. But since Nanog-alt contains an additional GG to the left (Fig. 3b), we cannot rule out the existence of an unknown Nanog binding partner (but it is not Sox2 or Pbx, see Supplementary Fig. 8b,c).
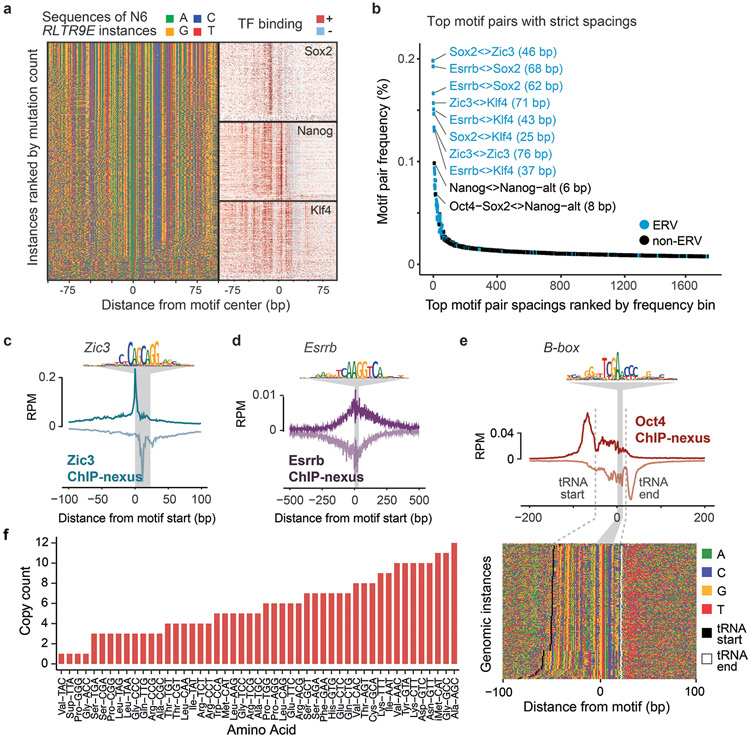
The majority of composite motifs, however, came from retrotransposons. This is consistent with previous observations that retrotransposons may contain multiple ancestral TF binding sites86-90 (Extended Data Fig. 5a). Among all motif pairs, the top 1% most frequent distances mapped in 83% to ERVs and were often larger than 20 bp (Extended Data Fig. 5b, Supplementary Fig. 9), which exceeds the typical distance between motifs found in composite motifs that promote TF cooperativity91,92. This suggests that overrepresented strict motif spacings alone are not a reliable indicator of functional motif syntax.
We next analyzed whether the 11 motifs showed evidence beyond strict motif spacings for mediating cooperative TF interactions (Fig. 3c). By inspecting the contribution scores (Fig. 3d), we found that many motifs were predicted to contribute to the binding of other TFs. Moreover, we discovered motifs of pluripotency TFs that we did not profile, including the Zic3 and Esrrb motifs, which we validated with additional ChIP-nexus experiments (Extended Data Fig. 5c-f). Thus, BPNet predicts that Oct4, Sox2, Nanog, and Klf4 frequently bind with the help of motifs from other TFs.
One explanation for this observation is that TFs may be indirectly recruited to motifs of other TFs50,51. We therefore inspected the average ChIP-nexus binding footprints of all TFs at all motifs (Fig. 3e). We found that TFs directly bound to their motifs showed sharp average ChIP-nexus footprints (marked in gray in Fig. 3e), but that TFs also showed broader, more fuzzy footprints at other motifs, which we attribute to indirect binding. The level of indirect TF occupancy correlated with the contribution score for the TF (Fig. 3d,e), suggesting that the indirect footprints are predicted by BPNet.
Notably, the indirect footprints tended to occur in an asymmetric or directional manner (Fig. 3d,e). For example, Nanog was bound indirectly to the Sox2 motif, but Sox2 was not detected at the Nanog motif. Since Sox2 and Nanog have been shown to physically interact with each other71,93, this suggests that these TFs indeed cooperate in some way, but not through a composite motif. We therefore set out to systematically analyze how motif pairs influence cooperative binding, as a means to identify functional motif syntax.
Interpreting BPNet reveals cooperative TF interactions
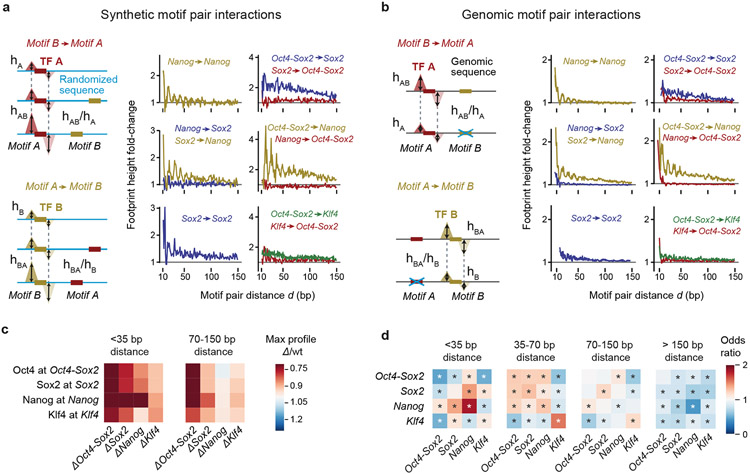
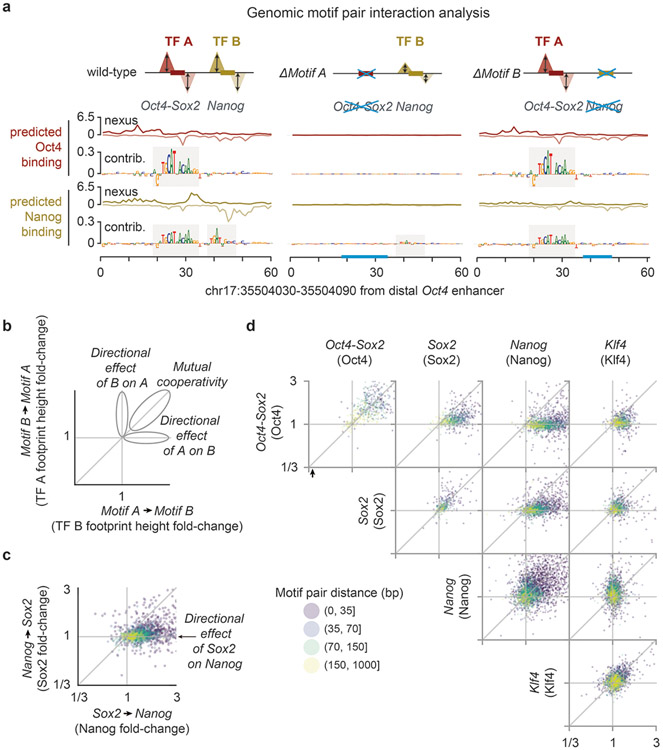
By training on base-resolution profiles, BPNet learned rules of TF cooperativity that we could extract by interrogating the trained model in silico like an oracle. We developed two complementary in-silico motif interaction analysis approaches that measure how the binding of a TF to its motif is affected by a second motif as a function of their relative distance (Fig. 4, Extended Data Fig. 6, Supplementary Fig. 10). We focused on the motifs most strongly bound by each of the four TFs: Oct4-Sox2 (bound by Oct4), Sox2, Nanog, and Klf4. The first approach uses synthetically designed sequences (Fig. 4a), while the second mutates naturally occurring non-overlapping motifs in genomic sequences (Fig. 4b).
Fig. 4: In-silico motif interaction analysis reveals TF cooperativity and motif syntax.
a) In the synthetic motif interaction analysis, Motif A is inserted into random sequences and the average profile for TF A is predicted by BPNet. The footprint’s summits are recorded (dotted lines) and the height (hA) is measured at this position. Motif B is then inserted at a specific distance (d) from Motif A into a new set of random sequences and the average predicted footprint height is measured at the reference summit position (hAB at dotted lines). The interaction of Motif B->Motif A as a function of d is quantified as the footprint height fold-change (hAB /hA) after correcting hAB for shoulder effects or indirect binding footprints from the nearby motif (Supplementary Fig. 10a). The interaction of Motif A->Motif B is obtained in an analogous way. The results show functions consistent with protein-range interactions between Nanog and Sox2 or nucleosome-range interactions exerted by the Oct4-Sox2 motif (bound by Oct4) on the binding of Sox2, Nanog or Klf4 on their respective motifs. Results are shown for the +/+ orientation of the two motifs (see Supplementary Fig. 10c for all motif pair orientations and Supplementary Fig. 10b for the frequency of motif pairs). b) In the genomic motif interaction analysis, naturally occurring instances of Motif A and Motif B as determined by CWM scanning are used. The average predicted footprint height and position of TF A is measured in the presence of Motif B (hAB) and after replacing Motif B with random bases (hA at dotted lines). The same corrected footprint height fold-change hAB /hA or hBA /hB as a function of d is used to quantify the interaction. The results from the average of all motif orientations is similar to those in the synthetic motif interaction analysis. c) Quantification of the results shown in (b) as heat map. The distances <35 bp is shown as representative for protein-range interactions, while 70-150 bp is shown as representative for nucleosome-range interactions. d) Odds by which two motifs are found within a specified distance from each other divided by the odds the two motifs would be found in the proximity by chance (observed by permuting the region index). * denotes p-value <10−5 using Pearson's Chi-squared test (Supplementary Methods).
In the synthetic approach, Motif A is embedded in random DNA sequences and the BPNet model is used to predict the fold-change in binding of TF A due to the addition of Motif B at a range of distances from Motif A (Fig. 4a, Supplementary Videos 1-6). The procedure is then repeated by anchoring Motif B and predicting the fold-change in binding of TF B as a function of distance to Motif A. The robustness of the results was confirmed by the reproducibility of the patterns across five models trained independently on different subsets of regions (Supplementary Fig. 11).
Using the synthetic approach on all motif pair combinations, we observed distance-dependent cooperative TF interactions (Fig. 4a). They were distinct for each motif pair but independent of strand orientation (Supplementary Fig. 10b,c). For example, predicted Nanog binding at the Nanog motif was strongly enhanced when another Nanog motif was nearby, but interestingly, the distance-dependent enhancement exhibited a periodic pattern (Fig. 4a). A similar periodic binding dependency was observed for Nanog when a Sox2 motif was nearby. The magnitude of this interaction was strongest at close distances (<35 bp), thus it could be mediated by protein-protein interactions between Sox2 and Nanog71,93 or DNA-mediated allostery4,94. For larger inter-motif distances, the impact on Nanog binding rapidly diminished, but was still elevated further away in the presence of a Sox2 motif (but not a Nanog motif). This was not true the other way around since Sox2 binding to its motif was not enhanced by a nearby Nanog motif (Fig. 4a). Thus, BPNet predicts that Sox2 and Nanog interact and that this cooperative interaction is directional, consistent with the indirect footprints we observed.
The motif interaction functions also suggested that the Oct4-Sox2 motif mediates its effect through increased DNA accessibility in chromatin, consistent with Oct4 and Sox2 being pioneer TFs73,77,95,96. First, Oct4-Sox2 strongly enhanced the predicted binding of Sox2, Nanog, and to a lesser extent Klf4, at nucleosome-range distances of 150 bp (Fig. 4a). Second, these interactions were directional since the motifs of the other TFs did not substantially impact the predicted binding of Oct4 to the Oct4-Sox2 motif, consistent with a hierarchical requirement for pioneer TFs to come first and make the region accessible for other TFs. Our results therefore suggest that motifs can be classified in a given context by their strength as pioneer motifs, i.e. the Oct4-Sox2 is a stronger pioneer motif than Sox2.
We observed very similar distance-dependent cooperative interactions for all motif pairs using a complementary motif mutagenesis approach for genomic sequences (Fig. 4b, Extended Data Fig. 6). Here, we used the original genomic sequences and predicted the binding profile of TF A to Motif A before and after replacing Motif B with a random sequence (Motif B −> Motif A) and vice versa. The effect sizes were smaller than in the synthetic approach, likely because the genomic motif instances were often of lower affinity than the ideal motifs used in the synthetic approach. It is also possible that motif mutations can be buffered by the additional motifs present in genomic sequences. However, the distance relationship and the directionality of the cooperative interactions were again very similar (Extended Data Fig. 6). These relationships can also be summarized as a heat map using the distance intervals of <35 bp and 70-150 bp, which highlight the interactions in protein-range and nucleosome-range respectively (Fig. 4c).
These results suggest the existence of soft motif syntax: rather than requiring strict inter-motif distances for cooperative binding, interactions between two motifs occur in a flexible but distance-dependent fashion that is specific for each motif pair. To obtain further evidence, we asked whether the preferred inter-motif distances are observed in naturally occurring genomic regions. We removed retrotransposons containing strictly spaced motifs and analyzed whether motif pairs co-occur more frequently than expected by chance at certain distances (Fig. 4d, Supplementary Fig. 10b). The Nanog motifs were most strongly overrepresented at short distances to Sox2 and other Nanog motifs (<35 bp), consistent with their protein-range interactions. At nucleosome-distance (70-150 bp), the Oct4-Sox2 motif still co-occurred with Nanog, consistent with its pioneering role. Although BPNet is designed to capture potential motif interactions up to 1 kb apart, we did not identify significantly overrepresented motif pairs beyond 150 bp (Fig. 4d). Altogether, we detected genome-wide soft preferences for motif spacings that correspond to some extent with detected cooperative binding interactions and thus are likely functionally relevant soft motif syntax.
Nanog binding has a strong ~10.5-bp periodic pattern
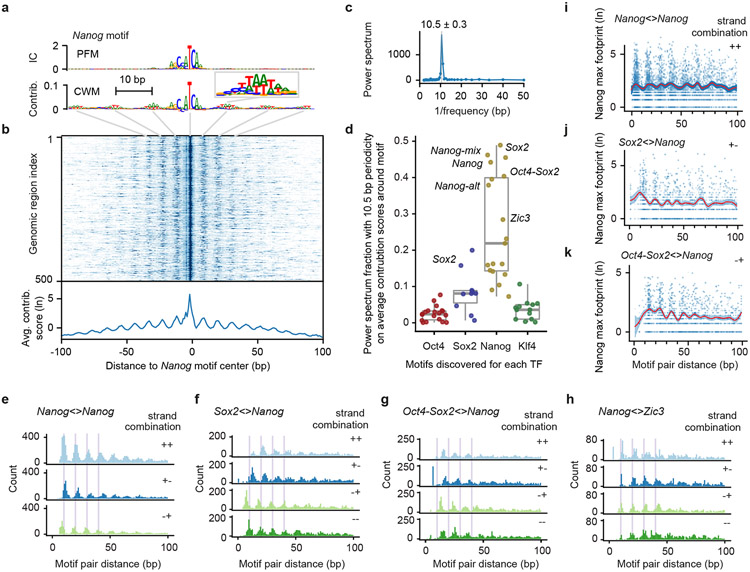
The most remarkable soft motif syntax we observed was a ~10.5 bp periodicity associated with Nanog. We first observed periodicity in the full-length CWM of the Nanog motif, which showed flanking A/T bases in a periodic pattern (Fig. 5a). This pattern is not seen in the corresponding PFM representation, suggesting that the A/T bases are not statistically overrepresented, but when present, contribute strongly to the Nanog binding predictions. The strong periodic pattern is confirmed in the individual contribution scores of Nanog motif instances, shown as heat map and average contribution scores (Fig. 5b). A Fourier power spectrum analysis of the contribution scores around the Nanog motif revealed strong periodicity averaging around 10.5 bp (+/− 0.3 bp) (Fig. 5c), which falls within the observed 10-11 bp periodicity of the DNA helix observed in vitro and in vivo97-100. This helical periodicity was also found for other motifs important for predicting Nanog binding, including Nanog-mix, Nanog-alt, Sox2, Oct4-Sox2 and Zic3. But the same motifs did not predict periodic binding for other TFs, suggesting that the helical periodicity is specific for Nanog binding (Fig. 5d), consistent with its behavior in the in-silico motif interaction analysis.
Fig. 5: Pervasive helical periodicity between Nanog and partner motifs.
a) The CWM, but not the PFM, of the main Nanog motif has periodically occurring contributing bases in the flanks (example in enlarged window). b) A heat map of the contribution scores of the individual Nanog instances also show this periodic pattern, the average of which is shown below. c) A Fourier power spectrum of the average contribution score around Nanog motif instances (after subtracting the smoothed signal) reveals an average periodicity of 10.5 +/− 0.3 bp. d) Fraction of the power spectrum with 10.5 bp periodicity of the average contribution scores around the motifs discovered for each TF (19 for Oct4, 10 for Sox2, 19 for Nanog, and 13 for Klf4) shows that the helical periodicity is specific for Nanog binding. Important motifs are labelled; unlabeled high-scoring motifs are from retrotransposons. The box-plots mark the median, the upper and lower quartiles, and the 1.5x interquartile range (whiskers). e) The pairwise spacing of Nanog motif instances in all possible orientations also show a periodic pattern (++ includes the −− orientation). f-h) Heterologous motif combinations of Nanog with Sox2, Oct4-Sox2 and Zic3 also show a preferred spacing with the same periodicity. The distance between two motifs is always kept positive by placing the second motif in the pair downstream of the first motif. All 4 motif orientations are considered: + denotes the motif lies on the forward strand and − denotes the motifs on the reverse strand. i-k) Nanog ChIP-nexus signal at the reference summit position for each motif instance across every motif pair (blue dots), with the smooth curve fit (B-splines) depicted as a red line and the 95% confidence intervals depicted as blue ribbon. Number of data-points used to estimate 50 smoothing parameters for each plot: 8930 for Nanog<>Nanog, 4011 for Sox2<>Nanog, and 4947 for Oct4-Sox2<>Nanog. Nanog on average binds higher when Nanog motifs have the preferred inter-motif distance.
To obtain further evidence of this periodicity, we tested whether Nanog’s soft syntax was naturally found in genomic DNA sequence. Indeed, the pairwise distance between our mapped Nanog motif instances showed a strong helical spacing preference for multiples of ~10.5 bp, independent of motif orientation (Fig. 5e). This periodicity was reproducibly inferred from five independent models on different subsets of the binding data (Supplementary Fig. 12a). Despite being present in genomic DNA, this pattern had not been discovered previously47,67-72, presumably because it is difficult to find with traditional methods and requires BPNet’s large receptive field to learn motifs in a larger sequence context (Extended Data Fig. 7).
The in-silico motif interaction analysis also predicted enhanced periodic binding cooperativity of Nanog in the presence of other motifs. In support of this, the mapped genomic distances between Nanog and either Sox2 or Oct4-Sox2 motif instances also showed strong preferred distances of helical periodicity regardless of motif orientation (Fig. 5f-g). This was also true for the distances between Nanog and Zic3, indicating that Zic3 is an additional interaction partner (Fig. 5h). Furthermore, the Nanog ChIP-nexus profiles themselves also showed this periodic pattern (Fig. 5i-k, Supplementary Fig. 12b, Supplementary Fig. 13). The signal in the original data likely explains how BPNet was able to learn the preferred binding pattern of Nanog during training.
The helical periodicity suggests that Nanog binding is enhanced when the relevant partner motifs are found on the same side of the DNA. Since Nanog physically interacts with Sox271,93 and preferentially interacts at protein-protein distance in our in-silico motif interaction analysis, it is possible that Nanog engages in cooperative protein-protein interactions similar to those observed for the lambda and lac repressors101,102. Alternatively, the helical periodicity could be due to preferred binding of Nanog to nucleosomal DNA from the solvent surface, which has been observed for some homeodomain TFs103,104.
Altogether, we identified helical periodicity as a strong cis-regulatory motif syntax for Nanog, a biophysical parameter that BPNet was not explicitly trained on. This result demonstrates the power of neural networks to discover novel patterns de novo without making explicit prior assumptions about the nature of the sequence features.
CRISPR validates the motif syntax between Nanog and Sox2
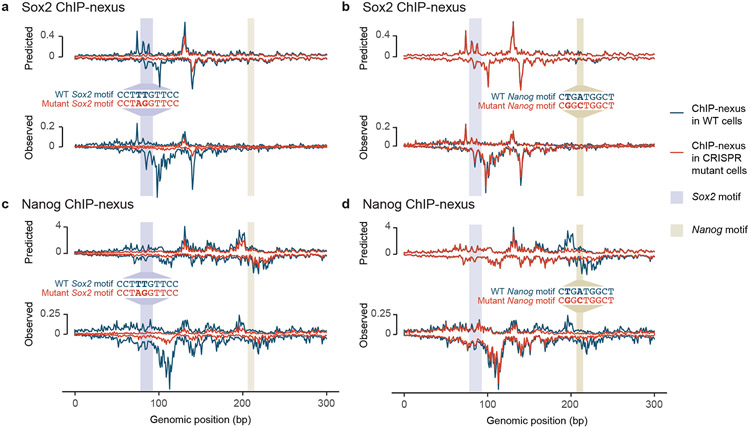
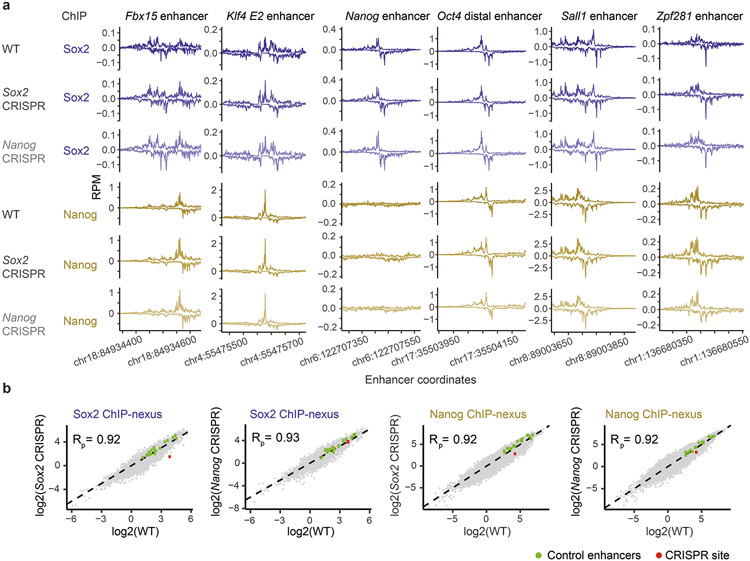
To experimentally validate the motif syntax identified by BPNet, we performed targeted point mutations in mapped motifs and compared the observed changes in the ChIP-nexus profiles to those predicted by BPNet (Fig. 6). Since the most striking motif syntax was the helical periodicity of Nanog and the directional cooperativity with Sox2, and since the Nanog motif had been uncertain before47,67-72, we selected a genomic region that has a Nanog and Sox2 motif, as well as periodic Nanog binding. Using CRISPR/Cas9 and homologous recombination, we performed two-base substitutions in either the Sox2 motif (TTG to AGG) or the Nanog motif (TGA to GGC). We then performed Sox2 and Nanog ChIP-nexus experiments on wild-type and mutant ESCs, using three independently derived clones per motif mutation. All replicate experiments were highly correlated and possessed indistinguishable normalized binding profiles and counts across known enhancers (Extended Data Fig. 8, Supplementary Fig. 14).
Fig. 6: CRISPR mutations in a Sox2 and Nanog motif validate BPNet’s predictions.
(a-d) A Sox2 motif and Nanog motif in a selected genomic region were mutated through CRISPR/Cas9 and homologous recombination in mouse ESCs. Predicted and observed ChIP-nexus profiles (+ strand above zero, − strand below zero) in reads per million (RPM) are shown for wild-type cells and mutant cells across 300 bp (chr10:85,539,550-85,539,850). a) Upon mutating the Sox2 motif, the Sox2 footprint is lost as predicted. b) In contrast, mutating the Nanog motif does not noticeably affect Sox2 binding. c) Consistent with directional cooperativity, the Sox2 mutation does however affect Nanog binding, which is reduced throughout the region as predicted. d) Similarly, mutating the Nanog motif not only abrogates the Nanog footprint, but also results in reduced binding nearby as predicted. See Extended Data Fig. 8, Supplementary Fig. 14 for reproducibility validations.
We then examined how the binding profiles were affected by the mutations. As expected, mutating the Sox2 motif specifically abolished the corresponding Sox2 binding footprint (Fig. 6a). However, mutating the Nanog motif did not affect Sox2 binding (Fig. 6b), while mutating the Sox2 motif strongly affected Nanog binding (Extended Data Fig. 8b). Nanog binding was almost completely lost near the Sox2 mutation and still reduced at the nearby Nanog motif (Fig. 6c).
This directional cooperativity is strikingly consistent with the results from the in-silico motif interaction analysis performed across all genomic sequences (Fig. 4b) and with the asymmetry observed in the indirect binding footprints of Nanog and Sox2 (Fig. 3c). In addition, the short-range cooperativity of Nanog was confirmed. Namely, when the Nanog motif was mutated, not only was the corresponding footprint of Nanog abrogated as expected, but the surrounding periodic Nanog binding was also reduced as predicted (Fig. 6d).
Altogether, these results confirm that the derived syntax rules are predictive and applicable to individual examples. This demonstrate that BPNet can be used to derive novel, testable biological hypotheses on how the cis-regulatory motif syntax influences TF binding.
Discussion
Here we introduced BPNet, a versatile and interpretable deep learning tool to learn TF motifs and the rules of syntax that best predict experimental data at base resolution. To leverage the unprecedented resolution of BPNet and showcase its ability to reveal novel biological insights, we applied it to ChIP-nexus data in ESCs. The results were not only consistent with previous findings, but revealed new details and principles of cis-regulatory motif syntax. We found that TF binding is guided by soft syntax rules, which follow clear inter-motif distance-dependent relationships consistent with protein-protein interactions16,105, or nucleosome-mediated cooperativity106. Such soft syntax rules represent an intermediate between the strict motif syntax associated with the original enhanceosome model107,108 and the very flexible syntax suggested by the billboard model14. The TF cooperativity associated with specific motif pairs was often directional and consistent with motifs mediating the role of pioneer TFs with different strengths. Finally, we observed a strong preference for Nanog to bind with ~10.5 bp helical periodicity. Helical periodicity has long been thought to be a possible element of the cis-regulatory code25,27,101,102,107,109-112. Our finding that the helical periodicity is motif-encoded and TF-specific provides a guidance for identifying this feature for other TFs in the future.
As we will outline below, BPNet represents a new paradigm for discovering relevant motifs and syntax rules underlying the cis-regulatory code. Through several important design innovations (Supplementary Note), as well as extensive quality control and rigorous evaluations to ensure that the method works as intended, BPNet outperforms both traditional methods and previous deep learning models (Supplementary Note). BPNet outperforms traditional methods because it infers predictive patterns in a larger sequence context and does not rely on overrepresented sequence patterns. BPNet outperforms previous neural networks by modeling TF binding profiles at base resolution, which enables it to learn subtle cooperative interactions between motifs (Extended Data Fig. 4). The result is a powerful and general computational framework for deciphering the cis-regulatory code from a variety of genomics assays.
An important innovation was the development of tools that make the trained BPNet model interpretable. Computational models in regulatory genomics have long grappled with an inherent tradeoff between prediction accuracy and interpretability, but the BPNet framework enables both. The key to enhancing interpretability was the distillation of predictive motif representations and context-aware motif instances from the entire neural network, rather than direct interpretation of millions of cryptic, partially redundant parameters of the trained model. Importantly, by using BPNet as an in-silico oracle, we systematically predicted the effect of mutated sequences or synthetic sequence designs, which enables us to extract the influence of pairwise motif spacing on TF cooperativity. The precise oracle predictions, which are not possible with classical models, allow less scalable in vivo experiments such as the CRISPR editing experiments to be performed on the most interesting and promising observations.
The advantage of BPNet over classical methods is that it detects motifs and their syntax in a fundamentally different way. Classical methods for motif discovery rely on motifs being overrepresented over background sequences18-21. Similarly, existing approaches to infer syntax rules use summary statistics of overrepresented co-occurrence patterns1,23,113. These methods have limited statistical power to test individual features present in complex cis-regulatory sequences (Supplementary Note). By contrast, BPNet’s vast network capacity allows it to learn complex predictive rules agnostically based on their ability to accurately predict relevant experimental profiles, without explicitly defining features a priori. This allows the discovery of relatively rare but nonetheless predictive motifs (e.g. Oct4-Oct4), as well as predictive syntax features, such as helical periodicity or the direction of TF cooperativity, that were not known to be relevant for these TFs.
BPNet’s approach of modeling the entire cis-regulatory sequence is better suited for deciphering the combinatorial requirements for TF binding in vivo. Traditionally, a TF binding site is defined by its strong affinity in in-vitro experiments or by statistically significant sequence matches to PWM models. In both cases, a selection is typically made by arbitrary thresholds before the role of motif combinations, syntax and sequence context is considered113,114. However, our results suggest that in vivo, TF binding to a motif instance is by itself a highly cooperative process that depends on neighboring motifs and syntax. Indeed, this explains how enhancer function can critically depend on low-affinity binding sites10,52,115. The fact that BPNet discovered subtle predictive patterns that are not strong matches to PWM motif models (e.g. the predictive bases in the flanks of Nanog motifs) and outperformed classical methods for identifying motif instances relevant in vivo (Fig. 2g-h, Supplementary Note) suggests that modeling putative motif instances within their cis-regulatory context is an important advantage.
Finally, BPNet is designed to be a general and versatile end-to-end approach adaptable to a number of genomic assays. It is ideally suited to learn from high-resolution genomic data, but its base-resolution output is still beneficial for lower resolution data since it does not discard any information present in the training data profiles. For example, we successfully trained BPNet models on ChIP-seq profiles for the same TFs and obtained motifs that were highly similar, including a periodic Nanog motif (Extended Data Fig. 9 and 10, Supplementary Note). The number and accuracy of motif instances was lower than those from ChIP-nexus profiles models, but better than those from models trained on coarse-resolution binary binding labels (Extended Data Fig. 10c,d). Similarly, we found that BPNet can accurately model base-resolution DNase-seq profiles116. This suggests that applying BPNet to existing compendia of ChIP-seq, DNase-seq and ATAC-seq data, such as those generated by ENCODE will improve the systematic mapping of cis-regulatory motifs and their rules of syntax in a variety of cellular contexts. To foster the broad application of BPNet, we have made the entire software framework available with documentation and tutorials.
Learning motifs and syntax-dependent regulatory influence for a variety of genomic assays in many cell types will build a more complete understanding of the cis-regulatory code and reveal how specific bases influence the various molecular steps associated with enhancer function. At the same time, these models will provide opportunities to pinpoint causal quantitative trait and disease-associated genetic variants and understand the molecular mechanisms by which they alter gene regulation. Ultimately, the ability to decipher the cis-regulatory code will unlock an enormous amount of information underlying organismal development, its maintenance and pinpoint therapeutic intervention opportunities for diseases.
Online method
Cell culture
R1 ESCs were cultured on 0.1% gelatin-coated plates without feeder cells in N2B27 medium (DMEM/F12 with 1:1 mix of GlutaMax/N2 and Neurobasal medium/B27, Invitrogen) supplemented with 2 mM L-Glutamine (Stemcell Technologies), 1x 2-Mercaptoethanol (Millipore), 1x NEAA (Stemcell Technologies), 3 μM CHIR99021 (Stemcell Technologies), 1 μM PD0325901 (Stemcell Technologies), 0.033% BSA solution (Invitrogen) and 107 U/ml LIF (Millipore).
ChIP-nexus, PAtCh-Cap and ChIP-seq experiments
For each ChIP-nexus experiment, 10 million ESCs were used. Cells were washed with PBS and cross-linked with 1% formaldehyde (Fisher Scientific) in PBS for 10 min at room temperature. The reaction was quenched with 125 mM glycine. Fixed cells were washed twice with cold PBS, resuspended in cold lysis buffer (15 mM HEPES (pH 7.5), 140 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 1% Triton X-100, 0.5% N-lauroylsarcosine, 0.1% sodium deoxycholate, 0.1% SDS), incubated for 10 min on ice and sonicated with a Bioruptor Pico (Diagenode) for five cycles of 30 s on and 30 s off. The ChIP-nexus procedure and data processing were performed as previously described49 except that the ChIP-nexus adaptor mix contained four fixed barcodes (ACTG, CTGA, GACT, TGAC) and that the PCR library amplification was performed directly after the circularization of the purified DNA fragments (without addition of the oligo and BamHI digestion). PAtCh-Cap was performed as described59with 10% of sheared chromatin from 10 million ESCs. ChIP-seq experiments were performed as described119 with 10 million ESCs per ChIP.
For each ChIP, 5 μg antibody was coupled to 50 μl Protein A or Protein G Dynabeads (Invitrogen). The following antibodies were used: α-Oct3/4 (Santa Cruz, sc-8628), α-Sox2 (Santa Cruz, sc-17320), α-Sox2 (Active Motif, 39843), α-Nanog (Santa Cruz, sc-30328), α-Klf4 (R&D Systems, AF3158), α-Klf4 (Abcam, ab106629), α-Esrrb (Abcam, ab19331), α-Pbx 1/2/3 (Santa Cruz, sc-888), and α-Zic3 (Abcam, ab222124). For all experiments, at least two biological replicates were prepared, i.e. the experiments were performed on different days starting with cells from a different passage number. Single-end sequencing was performed on either an Illumina HiSeq instrument (50 cycles) or NextSeq 500 instrument (75 cycles).
Mutation of binding motifs using CRISPR/Cas9 technology
Using mouse R1 ESCs, the predicted Nanog motif on chr10: 85,539,756-85,539,765 (mm10) was mutated from CTGATGGCT (wildtype) to CGGCTGGCT (mutant). The predicted Sox2 motif on chr10: 85,539,634-85,539,643 (mm10) was mutated from CCTTTGTTCC (wildtype) to CCTAGGTTCC (mutant). Guide RNA (gRNA) target sites were designed using CCTop target predictor tool120 by evaluating the predicted on-target efficiency score and the off-target potential121. The single-stranded donor oligonucleotides (ssODN) were designed containing ~40 bases of homology from the targeted cut site (gRNA and ssODN sequences are shown in Supplementary Table 3). A ribonucleoprotein (RNP) complex was formed by combining 90 pmol of gRNA (ordered as Alt-R sgRNA; IDT, USA) and 10 pmol of Cas9 HiFi protein (IDT) and hybridizing for 10 min at room temperature. The RNP was combined with 100 pmol of ssODN donor and delivered to cells by Neon electroporation (1500 V, 10 ms, 3 pulses; Neon Transfection System, Model MPK5000, Life Technologies). Single cells were screened for the expected mutations through paired-end sequencing on an Illumina MiSeq instrument (250 cycles). On-target indel frequency and expected mutations were analyzed using CRIS.py122. Only clones with the intentional mutation and sequence alignments above 90% were chosen for future experiments.
Per target site, three monoclonal cell lines were selected and used as replicate experiments: clones B07, B09 and F10 for the mutant Nanog motif, and clones B07, B11 and C10 for the mutant Sox2 motif. For the wild-type R1 ESCs control samples, at least two biological replicates were prepared as above. ChIP-nexus was performed as described above with 20 million ESCs and 5 μg α-Nanog (Abcam, ab214549) or α-Sox2 (Active Motif, 39843) per replicate. The following fixed fixed barcodes were used: AGTC, CAGT, GTCA, TCAG. Single-end sequencing was performed on an Illumina NovaSeq instrument (100 cycles) to obtain a coverage of ~400 million reads per experiment.
ChIP-nexus data processing pipeline
Random barcodes and fixed barcodes were trimmed off the reads and reassigned to FASTQ labels using nimnexus (v0.1.1). The adapters were then trimmed using cutadapt (v1.8.1)123. Next, the reads were aligned with Bowtie (v1.1.12)124,125 using the command bowtie --chunkmbs 512 -k 1 -m 1 -v 2 --best --strata to the mouse genome assembly mm10. Mutant samples were aligned to a modified mm10 genome that accommodated the CRISPR changes. Mapping stats were computed using SAMtools flagstat (v1.2)126. Reads were filtered using SAMtools view to remove unmapped reads and mates, non-primary alignments, PCR or optical duplicates (-F 1804) and reads that failed platform or vendor quality checks or had poor mapping quality (MAPQ <30). Reads aligned to the same position with the same barcode, CIGAR string and the SAM flag were de-duplicated using nimnexus dedup (v0.1.1). The total number of final (filtered) aligned reads was 243M for Oct4, 140M for Sox2, 214M for Nanog and 176M for Klf4. The final filtered BAM file was converted to tagAlign format (BED 3+3) using bedtools `bamtobed` (v2.26)127. Cross-correlation scores were obtained for each file using phantompeakqualtools (v1.2)128. BigWig tracks containing the strand-specific number of aligned 5' read ends (pooled across all replicates) were generated using bedtools genomecov −5 -bg -strand <+/−>, followed by bedGraph to BigWig conversion using UCSC bedGraphToBigWig version 4129.
Peaks were called using MACS2 (v2.1.1.20160309) by extending 5’-ends of reads on each strand using a 150 bp window (±75 bp) and then computing coverage of extended reads across both strands (shift=−75, extsize=150). For each TF, peak calling was performed on filtered, aligned reads from each replicate using a relaxed p-value threshold of 0.1 and retaining the top 300,000 peaks as described128. Relaxed peak calls were similarly performed on pseudo-replicates, which were obtained by pooling filtered, aligned reads from all replicates for a TF and randomly splitting the pooled reads into two balanced pseudo-replicates. Peaks overlaping the blacklisted regions listed in https://www.encodeproject.org/files/ENCFF547MET/ were excluded. The Irreproducible Discovery Rate (IDR) framework was used to obtain reproducible peaks across the true replicates and pseudo-replicates130. The set with the larger number of peaks was defined as the IDR optimal peaks for each TF: 25,849 for Oct4, 10,999 for Sox2, 56,459 for Nanog, and 57,601 for Klf4. Regions of 1 kb centered on the peak summits were used as inputs to BPNet. All samples passed quality control metrics used in the ENCODE TF ChIP-seq pipeline128 (Supplementary Table 1).
The nim-nexus code is available at https://github.com/Avsecz/nimnexus/. The ChIP-nexus pipeline performing the described steps (e.g. turning the raw reads in the FASTQ format to BigWig coverage tracks and the called peaks) is available at https://github.com/kundajelab/chip-nexus-pipeline. A detailed pipeline specification is available at https://docs.google.com/document/d/1h9lZ0GyVWd02RCmtaFWSaSFzrcNHoH_OgyPHMpU7b04. ChIP-seq datasets were processed using the ENCODE ChIP-seq pipeline https://github.com/ENCODE-DCC/chip-seq-pipeline2/releases/tag/v1.2.2. It is identical to the ChIP-nexus pipeline except that it uses the SPP peak caller29 and does not use barcodes for read de-duplication.
BPNet architecture
BPNet is a sequence-to-profile convolutional neural network that uses one-hot-encoded DNA sequence (A=[1,0,0,0], C=[0,1,0,0], G=[0,0,1,0], T=[0,0,0,1]) with adjustable length as input to predict base-resolution read count profiles as output. For flexibility, the architecture of BPNet can be compartmentalized into the body and multiple task-specific output heads. The body of BPNet consists of a sequence of convolutional layers with residual skip connections and ReLU activations57. The first convolutional layer uses 64 filters of width 25 bp, followed by 9 dilated convolutional layers (64 filters of width 3 in each layer) where the dilation rate (number of skipped positions in the convolutional filter) doubles at every layer. This results in a receptive field of +/−1034 bp for any position in the genome. The output of the final convolutional layer within the BPNet body (also referred to as the bottleneck activation map) serves as input for two output heads per TF: i) a deconvolutional layer (filter width=25, typical ChIP-nexus footprint width) predicting the strand-specific probabilities of observing a particular read at a particular position in the input sequence (shape or profile prediction) and ii) a global average pooling layer followed by the fully connected layer predicting the total number of read counts aligned to the input sequence for each strand (total read count prediction). The training occurs for all TF ChIP-nexus experiments together in a multi-task fashion. BPNet architecture (without bias correction) implementation in Keras 2.2.4 is provided in Supplementary Methods.
BPNet loss function
Let kobs be the vector of length L of observed read counts for a particular strand and a particular task (i.e., TF) along the sequence of length L. Let ppred be the vector of length L of predicted probabilities along the sequence, such that and let be the total number of observed counts and npred the total number of predicted counts for the sequence. The following loss function is used for each sequence, strand and task:
The first term evaluates the error in the shape of the predicted profile. It is the multinomial negative log-likelihood of observed base read counts given the predicted probabilities and the total number of observed counts. The second term evaluates the squared error of the log total number of reads in the region. During BPNet training, the total loss function is the sum of individual loss functions across both strands, all input sequences and all tasks.
The key hyper-parameter is λ. In Supplementary methods (Relationship between the Poisson log-likelihood, mean-squared error and multinomial log likelihood), we show that if , where is the average number of total counts in our training set, the profile loss and the total count loss will be roughly given equal weight. To upweight the profile predictions relative to the total count predictions, with α < 1 can be used.
BPNet’s control for biases
Experimental assays often have small biases that can be measured by control experiments (input for ChIP-seq and PAtCh-CAP for ChIP-nexus59). To prevent the sequence-to-profile model from learning these non-informative bias signals, the model tries to explain the target experimental track (e.g., the Oct4 profile) using both the sequence-based model predictions for specific head h and the control experiment track ctl:
where is a neural network based transformation of the control track trying to explain data for head h. The integration with the control data therefore occurs after the task-specific model head . We require that if the control track is 0 (i.e. bias not present) so that the model represents the bias-free part of the signal. Each head/track will have a different bias transformation either by having different parameters or even a different architecture for . For the total count prediction head, is simply , where nctl is the total number of reads from the control experiment in the modeled local region. For the profile prediction head, is a weighted sum of i) the raw counts and ii) a smoothed version of the raw counts using a sliding window sum of 50 bp (since control data are often sparse). During model training, the parameters of are also trained to best explain the output using the control track. This framework easily integrates multiple control tracks, or control tracks predicted from sequence using a bias model learned on other data such as deproteinized genomic DNA for DNase-seq131.
BPNet training and hyper-parameter tuning
ChIP-nexus profiles of Oct4, Sox2, Nanog and Klf4 were used to train and evaluate BPNet. Regions from mouse chromosomes 2, 3 and 4 (20%) were used as the tuning set for hyper-parameter tuning. Regions from chromosomes 1, 8 and 9 (20%) were used as the test set for the performance evaluation (Supplementary Methods). The remaining regions were used for model training. Hyper-parameters were manually adjusted to yield best performance on the tuning set. All neural network models were implemented and trained in Keras (v2.2.4)132 (TensorFlow backend v1.6) using the Adam optimizer133 (learning rate = 0.004) and early stopping with patience of 5 epochs.
DeepLIFT contribution scores for sequence-to-profile models
DeepLIFT is a feature attribution method for computing the contribution of each base (feature) in an input sequence to a specific scalar output prediction from a neural network model65. DeepLIFT decomposes the difference between the output prediction from an input sequence versus that of a neutral reference input sequence as an additive combination of contribution scores of all bases (D features) in the input sequence:
where ci is the contribution of feature i in input x to the model output prediction f(x) compared to model prediction f(r) based on the reference input r.
The output of BPNet for each head is however not a scalar, but a tensor of 2D L x S, where L is the sequence length and S is the number of output channels or strands for ChIP-nexus. We therefore needed to adapt DeepLIFT and defined the profile contribution score of a base with respect to the entire output profile as follows:
where pis is the predicted probability values for position i and strand s, obtained by normalizing the profile predictions on the logit scale using the softmax function along the sequence axis: p = Softmax(f(x)). cis is the contribution score of the base with respect to the (scalar) profile prediction on the logit scale at position i and strand s. A weighted sum is used to ensure that positions with high predicted profile output values are given more weight, but has the disadvantage that it would normally require the contribution scores to be computed L x S (=2,000) times for each 1 kb input sequence per TF. To drastically speed up this computation, we exploit the backpropagation algorithm used in DeepLIFT and the additive decomposition of DeepLIFT scores. We define a new TensorFlow operation as follows:
where Const denotes the tf.stop_gradients operation which treats the wrapped expression pi(x) as a constant. By applying DeepLIFT to , we obtain the desired result in a single DeepLIFT backpropagation step:
Pseudo-code of the described operation in TensorFlow code is:
wn = tf.reduce_mean(tf.reduce_sum(tf.stop_gradient(tf.nn.softmax(f, dim=−2)) * f, axis=−2), axis=−1).
For the reference input r, all zeroes were used since it showed the highest correlation with in-silico mutagenesis contribution scores, defined as the weighted sum of the profile prediction changes at all profile locations after introducing a mutation at a particular position. The DeepLIFT contribution scores were computed with TensorFlow v1.6 using the DeepExplain implementation of DeepLIFT (repository fork available at https://github.com/kundajelab/DeepExplain/, with commit hash: 738c7145e915a7a48f3a4248d088bcc2e1a94614).
Motif discovery using TF-MoDISco
TF-MoDISco (v0.5.1.1) was run on DeepLIFT profile contribution scores for each TF separately (using all 1 kb peak regions bound by the TF on autosomes). Significant seqlets were selected by computing contribution scores over a width of 21 bp and using the FDR threshold of 0.01 (target_seqlet_fdr). The null distribution was estimated from 4,800 randomly selected peaks with contribution scores computed on reshuffled sequences while preserving dinucleotide counts. A total of 145,748 non-overlapping significant seqlets were identified. Due to memory constraints (250 GB), 50,000 seqlets were used for each TF during the clustering/motif-discovery phase of TF-MoDISco. For all discovered motifs, the PFM and CWM are computed from the aligned seqlets by averaging the base frequencies and the contribution scores, respectively. See Supplementary Methods for more details.
Clustering of discovered motifs
Motifs were aligned to each other in all possible offsets and strand combinations, and a pairwise distance metric was generated using the smallest continuous Jaccard distance metric41 on the PFM information content between each motif pair. Hierarchical clustering was performed in scipy (v1.2.1) using the Ward variance minimization algorithm134 (method='ward') and optimal leaf ordering135 (Extended Data Fig. 2d). From these clusters, 11 representative TF motifs were manually selected.
CWM scanning to identify motif instances
Once BPNet is trained, it is possible but not necessary to use the experimentally measured ChIP-nexus profiles during model interpretation. For the mapping of motifs with TF-MoDISco and CWM scanning, no experimental information was used. CWM scanning was developed because TF-MoDISco only analyzes 50,000 seqlets per run. Trimmed CWMs were used to scan the contribution scores of all 147,974 peak regions (as done by TF-MoDISco) and computing the following similarity metric. Let denote the CWM of length LW and denote the contribution scores for one-hot-encoded sequence s of length LS≥ LW. The contribution score Ci,b for base b at position i is 0 if base b was not observed in the actual sequence (i.e. if si,b = 0). We decompose the similarity metric between the CWM scanning position i of the contribution scores into the 'contrib' score, computed as the L1 norm of the contribution scores at positions between i and i+LW in the scanned sequence:
and the 'match' score, which represents its similarity to the CWM computed using the continuous Jaccard distance metric41 between the CWM and L1 normalized contribution scores:
At each position i, the maximum 'match' score (Scorematch) between wCWM and its reverse-complement version is chosen. To call motif instances from the CWM scanning scores, three criteria were defined based on thresholds identified from the TF-MoDISco’s corresponding seqlets: (i) The 'match' score >20th percentile of those of the seqlets. This stringent threshold more effectively discriminates between similar motifs. (ii) The 'contrib' score is higher than the seqlets lowest 'contrib' score. (iii) The log odds score with respect to the PWM derived from the PFM is larger than 0.
In-silico motif interaction analysis
In the synthetic approach, two consensus motifs were inserted into 128 random background sequences of 1 kb: Motif A at the center and Motif B downstream at distance d between the motif centers (max at 160 bp). The average strand-specific ChIP-nexus profile predictions PAB for the TF that binds Motif A was then obtained using the trained BPNet model as oracle. Additional profiles were predicted by i) inserting only the Motif A in the center (PA), ii) inserting only the Motif B d-bases downstream of the center (PB), and iii) not inserting any motif (PØ). The strand-specific summit (maximum) location of the footprint was then determined for each strand from profile PA within 35 bp of the Motif A center. These summit locations were used to determine the footprint height h within all four profiles to obtain hA, hB, hAB, and hØ. The influence of Motif B on Motif A was then defined by the corrected binding fold-change (hAB - (hB - hØ)) / hA as a function of d. The procedure was repeated to quantify the influence of Motif A on the binding of TF B to Motif B. In the genomic motif interaction approach, the motif pair interactions were calculated in the same way using motif instances that were mapped by CWM scanning in genomic sequences underlying ChIP-nexus peaks, excluding motif instances overlapping retrotransposons. Instead of inserting motifs into the random sequence, motifs were removed from the genomic sequence by replacing them with random sequences (see also Supplementary Methods, Supplementary Fig. 10).
Reproducibility
All ChIP-nexus and ChIP-seq replicate experiments passed quality control metrics used by ENCODE128 (Supplementary Table 1). For Sox2 and Nanog, we used two different antibodies for each with reproducible results: the initial wild-type Sox2 ChIP-nexus experiments used two different antibodies (sc-17320 and Active Motif 39843) with IDR rescue ratio of <2; the wild-type and CRISPR Nanog ChIP-nexus experiments also used two different antibodies (sc-30328 and ab-214549) with consistent Nanog footprints on Nanog motifs (Extended Data Figure 3). The entire pipeline, including the training of BPNet, computing the contribution scores, obtaining motif representations, and analysing motif interactions, was performed in 5-fold cross-validations, which support our claims (Supplementary Information, Supplementary Fig. 4, Supplementary Fig. 11, Supplementary Fig. 12). The CRISPR mutant and wild-type experiments were consistent in profile and counts at control enhancers (Extended Data Fig. 8), and replicate experiments were highly reproducible (Supplementary Fig. 14).
Data Availability Statement
The raw sequencing data are available from GEO under the accession number GSE137193. Data used to train, evaluate and interpret the BPNet models are found on ZENODO at https://doi.org/10.5281/zenodo.3371215. Trained BPNet models and all the model interpretation results are on ZENODO at https://doi.org/10.5281/zenodo.3371163. The BPNet model trained on ChIP-nexus data is available on Kipoi under the name "BPNet-OSKN" (http://kipoi.org/models/BPNet-OSKN/). Genome browser tracks showing observed/predicted ChIP-nexus signal and the contribution scores for all factors are available at https://genome.ucsc.edu/s/mlweilert/mesc_OSKN_tracks. ATAC-seq data in mouse ESCs used in Fig. 2 and Supplementary Fig. 7 have been obtained from GSE134680. Blacklisted regions used to filter genomic coordinates throughout the analysis are available at https://www.encodeproject.org/files/ENCFF547MET. RepeatMasker mm10 annotations are from http://www.repeatmasker.org/genomes/mm10/RepeatMasker-rm405-db20140131/mm10.fa.out.gz. The NMR structure 1O4X used to render Sox2 and Oct1 in Fig. 3 is available at https://www.rcsb.org/structure/1o4x. TRANSFAC (v7.0) was used to identify the TFIIIC B-box discussed in Fig. 3. The PH0134.1 Pbx PWM used for motif validation in Supplementary Fig. 8 and Extended Data Fig. 5 was obtained from JASPAR at http://jaspar.genereg.net/api/v1/matrix/PH0134.1.jaspar. The MA0141.1 Esrrb PWM used in Extended Data Fig. 5 was obtained from JASPAR at http://jaspar.genereg.net/api/v1/matrix/MA0141.1.jaspar. The tRNA database GtRNAdb (v2.0, release 17.1) annotations and associated tRNAscan-SE scores used in Extended Data Fig. 5 are from http://gtrnadb.ucsc.edu/GtRNAdb_archives/release17/genomes/eukaryota/Mmusc10/mm10-tRNAs.tar.gz.
Code Availability Statement
The BPNet software package is available at https://github.com/kundajelab/bpnet/. Code to reproduce the results is available at https://github.com/kundajelab/bpnet-manuscript (https://doi.org/10.5281/zenodo.4294813). The ChIP-nexus data processing pipeline is available at https://github.com/kundajelab/chip-nexus-pipeline. Software to trim and de-duplicate ChIP-nexus reads is available at https://github.com/Avsecz/nimnexus/.
Extended Data
Extended Data Fig. 1. Additional performance evaluation of BPNet’s predictions of ChIP-nexus data.
a) Observed and predicted ChIP-nexus read counts mapping to the forward strand (dark) and the reverse strand (light) for the Zfp281 and Sall1 enhancers located on the held-out (test) chromosome 1. b) Alternative profile shape evaluation metrics showing the difference to random predictions: multinomial negative log-likelihood and Jensen-Shannon (JS) divergence. Both metrics were computed at different resolutions (from 1 bp to 10 bp windows) in held-out test chromosomes 1, 8 and 9. c) auPRC of profile predictions is high across various learning rates on the tuning set chromosomes 2, 3 and 4, demonstrating the robustness of the model. d) The deconvolutional layer slightly improves the profile predictive performance compared to a point-wise convolutional layer (deconvolution size=1). e) auPRC of profile predictions (top) and the Spearman correlation of total count predictions (bottom) for a range of different relative total count weight α in the BPNet loss function parameterized as λ = α/2 n_obs. Relative weight of 1 (center) denotes equal weighting of the counts and profile loss functions. The best performance is obtained for α < 1, showing that putting more weight to profile predictions aids both profile and count predictions. f) Observed and predicted total read counts for BPNet (top) and replicate experiments (bottom) across the four studied TFs along with the Spearman correlation coefficient.
Extended Data Fig. 2. Removal of long motifs in retrotransposons and clustering of motifs by similarity.
a) Among all motifs discovered by TF-MoDISco, 18 motifs display unusually high information content (IC) of >30 bits (green). The expected short motifs are shown in gray. b) Histogram of the overlap of short motifs (gray) and long motifs (green) with repeat elements shows that long motifs overlap >80% with annotated retrotransposons. c) Long motifs with their PFM, ID, fraction of motif instances overlapping with a repeat and the most frequent (top class) RepeatMasker annotation. Highlighted within the repeat elements are potential motif instances of Oct4-Sox2, Sox2, Nanog and Klf4 as indicated by the CWMs. d) To identify a set of representative motifs from the 33 short motifs discovered for different TFs (information content <30 bit, shown in Supplementary Fig. 3) and remove redundant short motifs, motifs were clustered by similarity using hierarchical clustering. The results were then manually inspected to select clusters that separate known motifs that are distinct (e.g. Oct4-Oct4 resembles the known MORE and PORE motifs that bind Oct4 homodimers, which is different from the monomerically bound Oct4 motif). Among very similar motifs within a cluster, we then selected the most abundant motif that was discovered for the most relevant TF if known). The 11 representative motifs that we selected are shown on the left. Non-canonical motifs were given a name (Nanog-alt for Nanog alternative, Klf4-long for longer Klf4).
Extended Data Fig. 3. BPNet and TF-MoDISco outperform traditional methods in motif discovery and the mapping of motif instances.
a) Motifs discovered by ChExMix, HOMER and MEME for Oct4, Sox2, Nanog and Klf4 ChIP-nexus peaks that are closest to the 11 primary representative BPNet motifs (top row). Green checkmark denotes whether the discovered motif is similar to the BPNet motif. b) Number of motif instances located up to 500 bp (top) or 100 bp (bottom) away from the ChIP-nexus peak summits showing a strong ChIP-nexus footprint. Only motif instances in peaks from held-out test chromosomes (1, 8 and 9) were used for the evaluation. (x-axis) top N motif instances from each of the methods were sorted in descending order of scores (PWM log odds score or CWM contrib score). For BPNet-augm, the center of the genomic region for which the contribution scores were computed was randomly jittered up to 200 bp away from the peak summit. This augmentation prevents BPNet from using the positional information of the peak summit. In the final column (Nanog replicate), the Nanog ChIP-nexus footprint was measured by a separate biological replicate using a different antibody (α-Nanog from Abcam, ab214549), which was not used during training or evaluation.
Extended Data Fig. 4. BPNet training on ChIP-nexus profiles is faster and yields more accurate motif instances than a binary classification model.
a) Predictive performance as measured by the precision-recall curve of the binary classification models predicting the presence or absence of ChIP-nexus peaks from 1 kb DNA sequences evaluated across the held-out (tuning/validation) chromosomes 2, 3 and 4. The model trained to classify the sequences is outperformed when the model is trained to also predict the ChIP-nexus profiles from DNA sequence (without or without profile bias-correction) in addition to classifying them is shown in blue (without or without profile bias-correction) in light blue and with bias-correction in dark blue). b) Training time of the binary classification model trained genome-wide and the sequence-to-profile model (BPNet) trained in ChIP-nexus peaks. c) Detected motifs by TF-MoDISco using the contribution scores in ChIP-nexus peaks of the sequence-to-profile BPNet (profile reg.) or the binary classification model (binary class). A light color denotes a high number of seqlets for each motif. Motifs not discovered or motifs supported by less than 100 seqlets are shown in black. Questionable motifs are displayed separately on the right. d) The number of motif instances (500 bp within ChIP-nexus peak summit) showing a ChIP-nexus footprint (y-axis) within the top N motif instances with highest contribution scores (x-axis) from the held-out (test) chromosomes 1, 8 and 9. A site was considered to show a ChIP-nexus footprint if the number of reads at the position of the aggregate footprint summit (averaged across both strands) is higher than the 90th percentile value of all motif instances detected by the profile regression model for the corresponding TF (i.e. same as in Extended Data Fig. 3b).
Extended Data Fig. 5. Strict motif spacings are found on retrotransposons and indirectly bound motifs can be validated.
a) To show that TF binding occurs with strict spacings in retrotransposons and that this is likely ancestral, the RLTR9E N6 motif is shown as an example. Sequences of the individual instances in the genome were sorted by the Kimura distance from the consensus motif, with the most similar sequences on top (which are likely more ancestral). Nanog, Sox2 and Klf4 ChIP-nexus binding footprints are shown in the same order on the right (+ strand reads in red, − strand reads in blue), revealing that the binding site spacing is largely constant across all sequences. b) Analysis of the most frequent distances between motif pairs (with >500 co-occurrences, distance measured at the trimmed motifs’ centers). The top 1% most frequent distances mapped in 83% to ERVs and were often longer than 20 bp. c) To validate the identified Zic3 motif instances, Zic3 ChIP-nexus experiments were performed. The average signal across the Zic3 instances reveals a strong Zic3 binding footprint. d) A similar validation was performed for the Esrrb motif instances, revealing that the Esrrb ChIP-nexus signal is present but more diffuse at the discovered Esrrb motif instances. e) To better understand the binding of Oct4 to the B-box, which is frequently found in tRNA, tRNA-overlapping B-box motif instances were reoriented to match the transcriptional direction and sorted by tRNA gene start proximity. This reveals Oct4 binding at tRNA gene start/stop sites. f) Amino acid anti-codons and their copy count of the tRNAs that overlapped with the B-box motif instances.
Extended Data Fig. 6. Additional genomic in-silico interaction analyses confirm the directional effects.
a) Example genomic in-silico mutagenesis analysis at the distal Oct4 enhancer. Predicted ChIP-nexus profiles and the contribution scores greatly decrease at both motifs (Oct4-Sox2 and Nanog) when erasing the Oct4-Sox2 motif (through random sequence insertion). By contrast, when the Nanog motif is erased (right), the predicted profile and the contribution scores of Oct4-Sox2 motif remain intact. b) Such directional effect of motifs can be quantified by the corrected binding fold change (Supplementary Fig. 10a) for all motif pairs in the genome and visualized as a scatterplot. c) Example scatterplot for the interaction between Sox2 and Nanog. Sox2 shows a positive directional effect on Nanog most profound for short motif distances (<35 bp). d) Predicted binding fold changes for all motif pairs in genomic sequences.
Extended Data Fig. 7. Helical periodicity of Nanog motifs is not discovered with traditional methods and requires BPNet’s large receptive field.
a) The pairwise spacing of Nanog motif instances located up to 100 bp away from the ChIP-nexus peak summits in all possible strand orientations (rows) for different methods and/or thresholds (columns). Results for all chromosomes are shown. b) The pairwise spacing of Nanog motif instances when BPNet is trained with different numbers of convolutional layers (Fig. 1g). BPNet with only a single convolutional layer (first column) is unable to capture the 10 bp periodicity due to the limited receptive field similar to PWMs.
Extended Data Fig. 8. The ChIP-nexus data on CRISPR-mutated ESCs are highly reproducible.
a) Nanog and Sox2 ChIP-nexus profiles normalized to reads per million (RPM) show highly similar profiles and read counts across known enhancer regions for wild-type (Wt) and CRISPR ESCs with either a mutated Sox2 motif (Sox2 CRISPR) or mutated Nanog motif (Nanog CRISPR) at a selected genomic region (chr10: 85,539,626-85,539,777). b) Pairwise comparisons of ChIP-nexus RPM counts between Wt and CRISPR ESCs at bound genomic regions (151 bp centered on the respective motif) with Sox2 ChIP-nexus counts on Sox2 motifs and Nanog ChIP-nexus counts on Nanog motifs (motifs based on the original model). The bulk data (gray) are highly correlated and known enhancer regions as shown in Supplementary Fig. 5 (green) are highly reproducible between ESC lines. Note the specific loss of counts in the selected mutated genomic region (red) over wild-type. Pearson correlations (Rp) between groups are shown in the top left of each scatter plot.
Extended Data Fig. 9. The base-resolution BPNet model can be trained on ChIP-seq profiles.
a) Observed read counts (Obs) and Predicted read counts (Pred) for BPNet trained on ChIP-seq data for the Zfp281 and Lefty1 enhancers located on the held-out (test) chromosome 1, with forward strand reads (dark) and reverse strand reads (light). For Obs, a sliding window of 50 bp was used to smooth the raw 5' end read counts (line); raw counts are shown as points on the bottom at y=0. b) BPNet predicts the ChIP-seq profile shape better than replicates. Multinomial log-likelihood difference compared to the constant model was used to evaluate the profile shape quality at different resolutions (from 1 bp to 10 bp windows) in held-out chromosomes 1, 8 and 9. A log-likelihood of 0 corresponds to the constant model. Multinomial log-likelihood was conditioned on the observed number of total counts as in the training loss. c) Total counts in 1 kb regions can be predicted by BPNet (red) at decent accuracy (measured by Pearson correlation with log(1+observed values)). They do not surpass replicate performance (blue), but are well above the Input control (grey). d) Obs and Pred as in panel a, as well as contribution scores for the known Oct4 enhancer. Motif instances derived by CWM scanning are highlighted with a green box.
Extended Data Fig. 10. BPNet trained on ChIP-seq discovers similar motifs and recovers the Nanog motif periodicity.
a) BPNet applied to ChIP-seq discovers the majority of the motifs identified by BPNet applied to ChIP-nexus data. The models 'ChIP-nexus profile cr' and 'ChIP-seq profile cr' were trained on the union of the ChIP-nexus/seq peaks predicting Oct4, Sox2, and Nanog binding and were interpreted on the intersection of the ChIP-nexus/seq peaks. b) The pairwise spacing of Nanog motif instances derived from the ChIP-seq profile model in all possible strand orientations shows helical periodicity (similar to Extended Data Fig. 7a). c) Motif instance calling with CWM scanning has higher accuracy for BPNet trained on ChIP-nexus data than for BPNet trained on ChIP-seq data (evaluated on the union of the ChIP-nexus/seq peaks, 500 bp around the peak summit using ChIP-nexus footprints as ground truth). d) Training a sequence-to-profile model on ChIP-seq data yields more accurate motif instances (500 bp around the ChIP-seq peak summits using ChIP-nexus footprints as ground truth) than training a binary classification model or using a PWM scanning approach using FIMO for motifs derived directly from ChIP-nexus data. See Extended Data Fig. 3b, 4d and Supplementary Note for more details.
Supplementary Material
Supplementary Data 1 Clustered motifs and their labels. Motifs were obtained by TF-MoDISco ran on BPNet models trained on 6 different datasets: i) seq/profile.peaks-union (ChIP-seq profile model trained on a union of ChIP-nexus/ChIP-seq peaks), ii) seq/binary (binary classification model trained on genome-wide ChIP-seq peaks), iii) seq/profile (ChIP-seq profile model trained in ChIP-nexus peaks), iv) nexus/profile.peaks-union (ChIP-nexus profile model trained on a union of ChIP-nexus/ChIP-seq peaks), v) nexus/binary (binary classification model trained on genome-wide ChIP-nexus peaks), vi) nexus/profile (ChIP-nexus profile model trained in ChIP-nexus peaks). Each motif logo shows the sequence information content of a PFM. The logo title consists of the manually assigned motif label (e.g. TE1, Oct4-Sox2) and the motif ID composed from the model name, the task name and TF-MoDISco motif ID (e.g. seq/profile/Nanog/m0_p13).
Supplementary Table 1 1) List of all ChIP-nexus and ChIP-seq replicate experiments and the associated quality-control metrics. 2) Further binary classification metrics corresponding to Extended Data Fig. 4a. 3) Sequences of guide RNA and single-stranded oligo donors for CRISPR mutations.
Supplementary Video 1 BPNet profile predictions averaged across 128 random sequences with two motifs inserted at different positions. Centers of the motifs are marked by the vertical gray line. Motif distance is shown on the right. For each motif, the predicted profile of the corresponding TF is shown on the y-axis.
Supplementary Video 2 See Supplementary Video 1
Supplementary Video 3 See Supplementary Video 1
Supplementary Video 4 See Supplementary Video 1
Supplementary Video 5 See Supplementary Video 1
Supplementary Video 6 See Supplementary Video 1
Source Data Fig. 5 Motif 10 bp periodicity for all motifs visualized in Fig. 5d.
Acknowledgements
We thank Mike Levine and Robb Krumlauf for comments and Johnny Israeli for initial technical help. This work was funded by the Stowers Institute for Medical Research (SIMR), the NIH grant 1R01HG010211 to J.Z., and the NIH grants 1DP2GM123485, 1U01HG009431 and 1R01HG009674 to A.K.. Ž.A. was supported by the German Bundesministerium für Bildung und Forschung (BMBF) through the project MechML (01IS18053F). A.S. was supported by the Stanford BioX Fellowship and HHMI International Student Research Fellowship. Illumina sequencing was performed at SIMR (Anoja Perera, Michael Peterson) and the University of Kansas Medical Center Genomics Core supported by the NIH grants U54HD090216, S10OD021743 and COBRE P30GM122731-03. Generation of the CRISPR/Cas9 mouse ESC lines was performed by the following Cores at SIMR: Genome Engineering (Kym Delventhal, Brandon Miller, Kyle Weaver), Tissue Culture (Chongbei Zhao, Alexis Murray, Yan Wang, Olga Kenzior, Qian Jiang, Skyler Hime, Sonia Gosh) and Cytometry (Jeff Haug, Dustin DeGraffenreid).
Footnotes
Conflict of Interests statement
J.Z. owns a patent on ChIP-nexus (Patent No. 10287628). All other authors declare no competing interests.
References
- 1.Gerstein MB et al. Architecture of the human regulatory network derived from ENCODE data. Nature 489, 91–100 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roadmap Epigenomics Consortium et al. Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morgunova E & Taipale J Structural perspective of cooperative transcription factor binding. Curr. Opin. Struct. Biol 47, 1–8 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Zinzen RP, Senger K, Levine M & Papatsenko D Computational models for neurogenic gene expression in the Drosophila embryo. Curr. Biol 16, 1358–1365 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Fiore C & Cohen BA Interactions between pluripotency factors specify cis-regulation in embryonic stem cells. Genome Res. 26, 778–786 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sayal R, Dresch JM, Pushel I, Taylor BR & Arnosti DN Quantitative perturbation-based analysis of gene expression predicts enhancer activity in early Drosophila embryo. elife 5, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erceg J et al. Subtle changes in motif positioning cause tissue-specific effects on robustness of an enhancer’s activity. PLoS Genet. 10, e1004060 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crocker J & Ilsley GR Using synthetic biology to study gene regulatory evolution. Curr. Opin. Genet. Dev 47, 91–101 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Farley EK et al. Suboptimization of developmental enhancers. Science 350, 325–328 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Swanson CI, Evans NC & Barolo S Structural rules and complex regulatory circuitry constrain expression of a Notch- and EGFR-regulated eye enhancer. Dev. Cell 18, 359–370 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu F & Posakony JW Role of architecture in the function and specificity of two Notch-regulated transcriptional enhancer modules. PLoS Genet. 8, e1002796 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lusk RW & Eisen MB Evolutionary mirages: selection on binding site composition creates the illusion of conserved grammars in Drosophila enhancers. PLoS Genet. 6, e1000829 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kulkarni MM & Arnosti DN Information display by transcriptional enhancers. Development 130, 6569–6575 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Liberman LM & Stathopoulos A Design flexibility in cis-regulatory control of gene expression: synthetic and comparative evidence. Dev. Biol 327, 578–589 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Junion G et al. A transcription factor collective defines cardiac cell fate and reflects lineage history. Cell 148, 473–486 (2012). [DOI] [PubMed] [Google Scholar]
- 17.King DM, Maricque BB & Cohen BA Synthetic and genomic regulatory elements reveal aspects of cis-regulatory grammar in Mouse Embryonic Stem Cells. BioRxiv (2018). doi: 10.1101/398107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bailey TL et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–8 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hughes JD, Estep PW, Tavazoie S & Church GM Computational identification of cis-regulatory elements associated with groups of functionally related genes in Saccharomyces cerevisiae. J. Mol. Biol 296, 1205–1214 (2000). [DOI] [PubMed] [Google Scholar]
- 20.Pavesi G, Mereghetti P, Mauri G & Pesole G Weeder Web: discovery of transcription factor binding sites in a set of sequences from co-regulated genes. Nucleic Acids Res. 32, W199–203 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thijs G et al. A higher-order background model improves the detection of promoter regulatory elements by Gibbs sampling. Bioinformatics 17, 1113–1122 (2001). [DOI] [PubMed] [Google Scholar]
- 22.Cheng Q et al. Computational identification of diverse mechanisms underlying transcription factor-DNA occupancy. PLoS Genet. 9, e1003571 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo Y, Mahony S & Gifford DK High resolution genome wide binding event finding and motif discovery reveals transcription factor spatial binding constraints. PLoS Comput. Biol 8, e1002638 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang J et al. Sequence features and chromatin structure around the genomic regions bound by 119 human transcription factors. Genome Res. 22, 1798–1812 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee D, Karchin R & Beer MA Discriminative prediction of mammalian enhancers from DNA sequence. Genome Res. 21, 2167–2180 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Erives A & Levine M Coordinate enhancers share common organizational features in the Drosophila genome. Proc Natl Acad Sci USA 101, 3851–3856 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Papatsenko D, Goltsev Y & Levine M Organization of developmental enhancers in the Drosophila embryo. Nucleic Acids Res. 37, 5665–5677 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ng FSL et al. Constrained transcription factor spacing is prevalent and important for transcriptional control of mouse blood cells. Nucleic Acids Res. 42, 13513–13524 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kharchenko PV, Tolstorukov MY & Park PJ Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat. Biotechnol 26, 1351–1359 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rozowsky J et al. PeakSeq enables systematic scoring of ChIP-seq experiments relative to controls. Nat. Biotechnol 27, 66–75 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo Y et al. Discovering homotypic binding events at high spatial resolution. Bioinformatics 26, 3028–3034 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuan PF et al. A Statistical Framework for the Analysis of ChIP-Seq Data. J. Am. Stat. Assoc 106, 891–903 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hartonen T, Sahu B, Dave K, Kivioja T & Taipale J PeakXus: comprehensive transcription factor binding site discovery from ChIP-Nexus and ChIP-Exo experiments. Bioinformatics 32, i629–i638 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Alipanahi B, Delong A, Weirauch MT & Frey BJ Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning. Nat. Biotechnol 33, 831–838 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Zhou J & Troyanskaya OG Predicting effects of noncoding variants with deep learning-based sequence model. Nat. Methods 12, 931–934 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quang D & Xie X FactorNet: A deep learning framework for predicting cell type specific transcription factor binding from nucleotide-resolution sequential data. Methods 166, 40–47 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bogard N, Linder J, Rosenberg AB & Seelig G A deep neural network for predicting and engineering alternative polyadenylation. Cell 178, 91–106.e23 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kelley DR, Snoek J & Rinn JL Basset: learning the regulatory code of the accessible genome with deep convolutional neural networks. Genome Res. 26, 990–999 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lanchantin J, Singh R, Wang B & Qi Y Deep motif dashboard: visualizing and understanding genomic sequences using deep neural networks. Pac. Symp. Biocomput 22, 254–265 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shrikumar A et al. TF-MoDISco v0.4.2.2-alpha: Technical Note. arXiv (2018). [Google Scholar]
- 42.Jha A, Aicher JK, Singh D & Barash Y Improving interpretability of deep learning models: splicing codes as a case study. BioRxiv (2019). doi: 10.1101/700096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Greenside P, Shimko T, Fordyce P & Kundaje A Discovering epistatic feature interactions from neural network models of regulatory DNA sequences. Bioinformatics 34, i629–i637 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kelley DR et al. Sequential regulatory activity prediction across chromosomes with convolutional neural networks. Genome Res. 28, 739–750 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gordân R, Hartemink AJ & Bulyk ML Distinguishing direct versus indirect transcription factor-DNA interactions. Genome Res. 19, 2090–2100 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mariani L, Weinand K, Vedenko A, Barrera LA & Bulyk ML Identification of Human Lineage-Specific Transcriptional Coregulators Enabled by a Glossary of Binding Modules and Tunable Genomic Backgrounds. Cell Syst. 5, 187–201.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bailey TL & Machanick P Inferring direct DNA binding from ChIP-seq. Nucleic Acids Res. 40, e128 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rhee HS & Pugh BF Comprehensive genome-wide protein-DNA interactions detected at single-nucleotide resolution. Cell 147, 1408–1419 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.He Q, Johnston J & Zeitlinger J ChIP-nexus enables improved detection of in vivo transcription factor binding footprints. Nat. Biotechnol 33, 395–401 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamada N, Lai WKM, Farrell N, Pugh BF & Mahony S Characterizing protein-DNA binding event subtypes in ChIP-exo data. Bioinformatics 35, 903–913 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Starick SR et al. ChIP-exo signal associated with DNA-binding motifs provides insight into the genomic binding of the glucocorticoid receptor and cooperating transcription factors. Genome Res. 25, 825–835 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Papagianni A et al. Capicua controls Toll/IL-1 signaling targets independently of RTK regulation. Proc Natl Acad Sci USA 115, 1807–1812 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reményi A et al. Crystal structure of a POU/HMG/DNA ternary complex suggests differential assembly of Oct4 and Sox2 on two enhancers. Genes Dev. 17, 2048–2059 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Banerji J, Rusconi S & Schaffner W Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell 27, 299–308 (1981). [DOI] [PubMed] [Google Scholar]
- 55.Spitz F & Furlong EEM Transcription factors: from enhancer binding to developmental control. Nat. Rev. Genet 13, 613–626 (2012). [DOI] [PubMed] [Google Scholar]
- 56.Jaganathan K et al. Predicting Splicing from Primary Sequence with Deep Learning. Cell 176, 535–548.e24 (2019). [DOI] [PubMed] [Google Scholar]
- 57.He K, Zhang X, Ren S & Sun J Deep residual learning for image recognition. in IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 770–778 (IEEE, 2016). doi: 10.1109/CVPR.2016.90 [DOI] [Google Scholar]
- 58.Van Den Oord A et al. WaveNet: A generative model for raw audio. SSW 125, (2016). [Google Scholar]
- 59.Terooatea TW, Pozner A & Buck-Koehntop BA PAtCh-Cap: input strategy for improving analysis of ChIP-exo data sets and beyond. Nucleic Acids Res. 44, e159 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Whyte WA et al. Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature 482, 221–225 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Novo CL et al. Long-Range Enhancer Interactions Are Prevalent in Mouse Embryonic Stem Cells and Are Reorganized upon Pluripotent State Transition. Cell Rep. 22, 2615–2627 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Festuccia N et al. Esrrb extinction triggers dismantling of naïve pluripotency and marks commitment to differentiation. EMBO J. 37, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Moorthy SD et al. Enhancers and super-enhancers have an equivalent regulatory role in embryonic stem cells through regulation of single or multiple genes. Genome Res. 27, 246–258 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Avsec Ž et al. The Kipoi repository accelerates community exchange and reuse of predictive models for genomics. Nat. Biotechnol 37, 592–600 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shrikumar A, Greenside P & Kundaje A Learning Important Features Through Propagating Activation Differences. in 70, 3145–3153 (Proceedings of Machine Learning Research, 2017). [Google Scholar]
- 66.Chew J-L et al. Reciprocal transcriptional regulation of Pou5f1 and Sox2 via the Oct4/Sox2 complex in embryonic stem cells. Mol. Cell. Biol 25, 6031–6046 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen X et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133, 1106–1117 (2008). [DOI] [PubMed] [Google Scholar]
- 68.Mitsui K et al. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 113, 631–642 (2003). [DOI] [PubMed] [Google Scholar]
- 69.Loh Y-H et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet 38, 431–440 (2006). [DOI] [PubMed] [Google Scholar]
- 70.Salmon-Divon M, Dvinge H, Tammoja K & Bertone P PeakAnalyzer: genome-wide annotation of chromatin binding and modification loci. BMC Bioinformatics 11, 415 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gagliardi A et al. A direct physical interaction between Nanog and Sox2 regulates embryonic stem cell self-renewal. EMBO J. 32, 2231–2247 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.He X et al. A biophysical model for analysis of transcription factor interaction and binding site arrangement from genome-wide binding data. PLoS ONE 4, e8155 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xie L et al. A dynamic interplay of enhancer elements regulates Klf4 expression in naïve pluripotency. Genes Dev. 31, 1795–1808 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mistri TK et al. Dynamic changes in Sox2 spatio-temporal expression promote the second cell fate decision through Fgf4/Fgfr2 signaling in preimplantation mouse embryos. Biochem. J 475, 1075–1089 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tokuzawa Y et al. Fbx15 is a novel target of Oct3/4 but is dispensable for embryonic stem cell self-renewal and mouse development. Mol. Cell. Biol 23, 2699–2708 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Heinz S et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Friman ET et al. Dynamic regulation of chromatin accessibility by pluripotency transcription factors across the cell cycle. elife 8, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jolma A et al. DNA-binding specificities of human transcription factors. Cell 152, 327–339 (2013). [DOI] [PubMed] [Google Scholar]
- 79.Tomilin A et al. Synergism with the coactivator OBF-1 (OCA-B, BOB-1) is mediated by a specific POU dimer configuration. Cell 103, 853–864 (2000). [DOI] [PubMed] [Google Scholar]
- 80.Botquin V et al. New POU dimer configuration mediates antagonistic control of an osteopontin preimplantation enhancer by Oct-4 and Sox-2. Genes Dev. 12, 2073–2090 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mistri TK et al. Selective influence of Sox2 on POU transcription factor binding in embryonic and neural stem cells. EMBO Rep. 16, 1177–1191 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ambrosetti DC, Basilico C & Dailey L Synergistic activation of the fibroblast growth factor 4 enhancer by Sox2 and Oct-3 depends on protein-protein interactions facilitated by a specific spatial arrangement of factor binding sites. Mol. Cell. Biol 17, 6321–6329 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Merino F, Bouvier B & Cojocaru V Cooperative DNA Recognition Modulated by an Interplay between Protein-Protein Interactions and DNA-Mediated Allostery. PLoS Comput. Biol 11, e1004287 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hayashi Y et al. Structure-based discovery of NANOG variant with enhanced properties to promote self-renewal and reprogramming of pluripotent stem cells. Proc Natl Acad Sci USA 112, 4666–4671 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang J, Levasseur DN & Orkin SH Requirement of Nanog dimerization for stem cell self-renewal and pluripotency. Proc Natl Acad Sci USA 105, 6326–6331 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Todd CD, Deniz Ö, Taylor D & Branco MR Functional evaluation of transposable elements as enhancers in mouse embryonic and trophoblast stem cells. elife 8, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bourque G et al. Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genome Res. 18, 1752–1762 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kunarso G et al. Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat. Genet 42, 631–634 (2010). [DOI] [PubMed] [Google Scholar]
- 89.Sundaram V et al. Functional cis-regulatory modules encoded by mouse-specific endogenous retrovirus. Nat. Commun 8, 14550 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xie D et al. Rewirable gene regulatory networks in the preimplantation embryonic development of three mammalian species. Genome Res. 20, 804–815 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jankowski A, Szczurek E, Jauch R, Tiuryn J & Prabhakar S Comprehensive prediction in 78 human cell lines reveals rigidity and compactness of transcription factor dimers. Genome Res. 23, 1307–1318 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jolma A et al. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 527, 384–388 (2015). [DOI] [PubMed] [Google Scholar]
- 93.Mullin NP et al. Distinct Contributions of Tryptophan Residues within the Dimerization Domain to Nanog Function. J. Mol. Biol 429, 1544–1553 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kim S et al. Probing allostery through DNA. Science 339, 816–819 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Soufi A et al. Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell 161, 555–568 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Soufi A, Donahue G & Zaret KS Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell 151, 994–1004 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Winter DR, Song L, Mukherjee S, Furey TS & Crawford GE DNase-seq predicts regions of rotational nucleosome stability across diverse human cell types. Genome Res. 23, 1118–1129 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhong J et al. Mapping nucleosome positions using DNase-seq. Genome Res. 26, 351–364 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jin H, Rube HT & Song JS Categorical spectral analysis of periodicity in nucleosomal DNA. Nucleic Acids Res. 44, 2047–2057 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Drew HR et al. Structure of a B-DNA dodecamer: conformation and dynamics. Proc Natl Acad Sci USA 78, 2179–2183 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Müller J, Oehler S & Müller-Hill B Repression of lac promoter as a function of distance, phase and quality of an auxiliary lac operator. J. Mol. Biol 257, 21–29 (1996). [DOI] [PubMed] [Google Scholar]
- 102.Hochschild A & Ptashne M Cooperative binding of lambda repressors to sites separated by integral turns of the DNA helix. Cell 44, 681–687 (1986). [DOI] [PubMed] [Google Scholar]
- 103.Ghosh RP et al. Satb1 integrates DNA binding site geometry and torsional stress to differentially target nucleosome-dense regions. Nat. Commun 10, 3221 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhu F et al. The interaction landscape between transcription factors and the nucleosome. Nature 562, 76–81 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ptashne M Regulation of transcription: from lambda to eukaryotes. Trends Biochem. Sci 30, 275–279 (2005). [DOI] [PubMed] [Google Scholar]
- 106.Sun Y et al. Zelda overcomes the high intrinsic nucleosome barrier at enhancers during Drosophila zygotic genome activation. Genome Res. 25, 1703–1714 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Thanos D & Maniatis T Virus induction of human IFNβ gene expression requires the assembly of an enhanceosome. Cell 83, 1091–1100 (1995). [DOI] [PubMed] [Google Scholar]
- 108.Merika M & Thanos D Enhanceosomes. Curr. Opin. Genet. Dev 11, 205–208 (2001). [DOI] [PubMed] [Google Scholar]
- 109.Li Q & Wrange O Accessibility of a glucocorticoid response element in a nucleosome depends on its rotational positioning. Mol. Cell. Biol 15, 4375–4384 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sharon E et al. Inferring gene regulatory logic from high-throughput measurements of thousands of systematically designed promoters. Nat. Biotechnol 30, 521–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cai HN, Arnosti DN & Levine M Long-range repression in the Drosophila embryo. Proc Natl Acad Sci USA 93, 9309–9314 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cui F & Zhurkin VB Rotational positioning of nucleosomes facilitates selective binding of p53 to response elements associated with cell cycle arrest. Nucleic Acids Res. 42, 836–847 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Suryamohan K & Halfon MS Identifying transcriptional cis-regulatory modules in animal genomes. Wiley Interdiscip. Rev. Dev. Biol 4, 59–84 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Istrail S Eric Davidson’s Regulatory Genome for Computer Science: Causality, Logic, and Proof Principles of the Genomic cis-Regulatory Code. J. Comput. Biol 26, 653–684 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Slattery M et al. Absence of a simple code: how transcription factors read the genome. Trends Biochem. Sci 39, 381–399 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Tseng AM, Shrikumar A & Kundaje A Fourier-transform-based attribution priors improve the interpretability and stability of deep learning models for genomics. BioRxiv (2020). doi: 10.1101/2020.06.11.147272 [DOI] [Google Scholar]
- 117.Klemenz R, Stillman DJ & Geiduschek EP Specific interactions of Saccharomyces cerevisiae proteins with a promoter region of eukaryotic tRNA genes. Proc Natl Acad Sci USA 79, 6191–6195 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Oler AJ et al. Human RNA polymerase III transcriptomes and relationships to Pol II promoter chromatin and enhancer-binding factors. Nat. Struct. Mol. Biol 17, 620–628 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Online methods references
- 119.Koenecke N, Johnston J, He Q, Meier S & Zeitlinger J Drosophila poised enhancers are generated during tissue patterning with the help of repression. Genome Res. 27, 64–74 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Stemmer M, Thumberger T, Del Sol Keyer M, Wittbrodt J & Mateo JL Cctop: an intuitive, flexible and reliable crispr/cas9 target prediction tool. PLoS ONE 10, e0124633 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Labuhn M et al. Refined sgRNA efficacy prediction improves large- and small-scale CRISPR-Cas9 applications. Nucleic Acids Res. 46, 1375–1385 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Connelly JP & Pruett-Miller SM CRIS.py: A Versatile and High-throughput Analysis Program for CRISPR-based Genome Editing. Sci. Rep 9, 4194 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Martin M Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet j. 17, 10 (2011). [Google Scholar]
- 124.Li H & Durbin R Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Langmead B, Trapnell C, Pop M & Salzberg SL Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Li H et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Quinlan AR & Hall IM BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Landt SG et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 22, 1813–1831 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kent WJ, Zweig AS, Barber G, Hinrichs AS & Karolchik D BigWig and BigBed: enabling browsing of large distributed datasets. Bioinformatics 26, 2204–2207 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Li Q, Brown JB, Huang H & Bickel PJ Measuring reproducibility of high-throughput experiments. Ann. Appl. Stat 5, 1752–1779 (2011). [Google Scholar]
- 131.Yardımcı GG, Frank CL, Crawford GE & Ohler U Explicit DNase sequence bias modeling enables high-resolution transcription factor footprint detection. Nucleic Acids Res. 42, 11865–11878 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Chollet Francois and others. Keras. (2015). at <https://keras.io> [Google Scholar]
- 133.Kingma DP & Ba J Adam: A Method for Stochastic Optimization. (2014). [Google Scholar]
- 134.Ward JH Hierarchical Grouping to Optimize an Objective Function. J. Am. Stat. Assoc 58, 236–244 (1963). [Google Scholar]
- 135.Bar-Joseph Z, Gifford DK & Jaakkola TS Fast optimal leaf ordering for hierarchical clustering. Bioinformatics 17 Suppl 1, S22–9 (2001). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data 1 Clustered motifs and their labels. Motifs were obtained by TF-MoDISco ran on BPNet models trained on 6 different datasets: i) seq/profile.peaks-union (ChIP-seq profile model trained on a union of ChIP-nexus/ChIP-seq peaks), ii) seq/binary (binary classification model trained on genome-wide ChIP-seq peaks), iii) seq/profile (ChIP-seq profile model trained in ChIP-nexus peaks), iv) nexus/profile.peaks-union (ChIP-nexus profile model trained on a union of ChIP-nexus/ChIP-seq peaks), v) nexus/binary (binary classification model trained on genome-wide ChIP-nexus peaks), vi) nexus/profile (ChIP-nexus profile model trained in ChIP-nexus peaks). Each motif logo shows the sequence information content of a PFM. The logo title consists of the manually assigned motif label (e.g. TE1, Oct4-Sox2) and the motif ID composed from the model name, the task name and TF-MoDISco motif ID (e.g. seq/profile/Nanog/m0_p13).
Supplementary Table 1 1) List of all ChIP-nexus and ChIP-seq replicate experiments and the associated quality-control metrics. 2) Further binary classification metrics corresponding to Extended Data Fig. 4a. 3) Sequences of guide RNA and single-stranded oligo donors for CRISPR mutations.
Supplementary Video 1 BPNet profile predictions averaged across 128 random sequences with two motifs inserted at different positions. Centers of the motifs are marked by the vertical gray line. Motif distance is shown on the right. For each motif, the predicted profile of the corresponding TF is shown on the y-axis.
Supplementary Video 2 See Supplementary Video 1
Supplementary Video 3 See Supplementary Video 1
Supplementary Video 4 See Supplementary Video 1
Supplementary Video 5 See Supplementary Video 1
Supplementary Video 6 See Supplementary Video 1
Source Data Fig. 5 Motif 10 bp periodicity for all motifs visualized in Fig. 5d.
Data Availability Statement
The raw sequencing data are available from GEO under the accession number GSE137193. Data used to train, evaluate and interpret the BPNet models are found on ZENODO at https://doi.org/10.5281/zenodo.3371215. Trained BPNet models and all the model interpretation results are on ZENODO at https://doi.org/10.5281/zenodo.3371163. The BPNet model trained on ChIP-nexus data is available on Kipoi under the name "BPNet-OSKN" (http://kipoi.org/models/BPNet-OSKN/). Genome browser tracks showing observed/predicted ChIP-nexus signal and the contribution scores for all factors are available at https://genome.ucsc.edu/s/mlweilert/mesc_OSKN_tracks. ATAC-seq data in mouse ESCs used in Fig. 2 and Supplementary Fig. 7 have been obtained from GSE134680. Blacklisted regions used to filter genomic coordinates throughout the analysis are available at https://www.encodeproject.org/files/ENCFF547MET. RepeatMasker mm10 annotations are from http://www.repeatmasker.org/genomes/mm10/RepeatMasker-rm405-db20140131/mm10.fa.out.gz. The NMR structure 1O4X used to render Sox2 and Oct1 in Fig. 3 is available at https://www.rcsb.org/structure/1o4x. TRANSFAC (v7.0) was used to identify the TFIIIC B-box discussed in Fig. 3. The PH0134.1 Pbx PWM used for motif validation in Supplementary Fig. 8 and Extended Data Fig. 5 was obtained from JASPAR at http://jaspar.genereg.net/api/v1/matrix/PH0134.1.jaspar. The MA0141.1 Esrrb PWM used in Extended Data Fig. 5 was obtained from JASPAR at http://jaspar.genereg.net/api/v1/matrix/MA0141.1.jaspar. The tRNA database GtRNAdb (v2.0, release 17.1) annotations and associated tRNAscan-SE scores used in Extended Data Fig. 5 are from http://gtrnadb.ucsc.edu/GtRNAdb_archives/release17/genomes/eukaryota/Mmusc10/mm10-tRNAs.tar.gz.