Abstract
Uropathogenc Escherichia coli (UPEC) CFT073 has a pathogenicity-associated island (PAICFT073), which causes pyelonephritis and cystitis. Using PCR method, we found the prrA gene of PAICFT073 in E. coli O157:H7 EDL933. Further detailed PCR screening of 38 open reading frames, the right and left junction sequences of PAICFT073, revealed that it is the prrA-modD-yc73-fepC gene cluster but not the PAICFT073 present in E. coli O157:H7 EDL933. A rapid preliminary analysis suggested that the prrA-modD-yc73-fepC gene cluster of the PAICFT073, is present in 43 strains of E. coli O157:H7 containing Shiga toxin (Stx) gene but absent in 19 strains of E. coli O157:H7 without Stx gene. A strict co-occurrence of the prrA-modD-yc73-fepC gene cluster and Stx genes was observed, regardless of their origin. The prrA-modD-yc73-fepC gene cluster encode proteins probably involved in iron uptake system, which strongly suggests the importance of iron metabolism in the Stx-mediated virulence. In addition, the prrA-modD-yc73-fepC gene cluster may be used as a diagnostic marker to distinguish E. coli O157:H7 strains containing Stx gene from that without Stx gene, and possibly to quickly detect other pathogenic gram-negative bacteria containing the Stx gene.
Uropathogenic Escherichia coli (UPEC) strains produce hemolysin, P fimbriae, and aerobactin; exhibit serum resistance, and are encapsulated (10, 14). These features, usually absent from the typical fecal strains, imply the presence of a unique set of virulence determinants in UPEC strains, which are different from the virulence determinants of diarrheagenic E. coli. Many of these virulence determinants are encoded by gene clusters located on the pathogenicity-associated islands (8). Recently, Guyer et al. described the pathogenicity-associated island (PAICFT073), which contains 44 open reading frames (ORFs). Among them, 4 encode the hemolysin, 11 encode P fimbriae, and 19 show no homology to UPEC J96 or E. coli K-12 entries. Four genes (prrA, modD, fepC, and yc73), located on the PAICFT073 near the left junction, encode proteins homologous to the TonB-dependent outer membrane receptor, the ATP-binding subunits of the molybdate transporter (ModD), the ferric enterobactin transport ATP-binding protein (FepC), and a similar Haemophilus influenzae yc73 protein (4). Near the right junction, R1 and R2 genes encode an apparent antiterminator with homology to a gene in the sac operon of Bacillus subtilis, and a homolog of the maltose- and glucose-specific component II (5).
Enterohemorrhagic E. coli (EHEC) O157:H7 is a novel and increasingly important class of enteric pathogens causing intestinal and renal disease, such as hemorrhagic colitis and hemolytic-uremic syndrome. During the last 10 years, it has caused numerous sporadic causes and several massive outbreaks (9, 15, H. Watanabe, A. Wada, Y. Inagaki, K. Itoh, and K. Tamura, Letter, Lancet 348:831–832, 1996). The major virulence factors are Shiga toxins (Stx), which are responsible for death and many other symptoms in patients (12). We demonstrate here that the prrA-modD-yc73-fepC gene cluster of the PAICFT073 is present exclusively in E. coli O157:H7 containing Stx gene but is absent from E. coli O157:H7 without the Stx gene.
At the initial study to screen the published pathogenicity islands of gram-negative bacteria in diarrheagenic E. coli, we happen to identify prrA gene of PAICFT073 in E. coli O157:H7 EDL933. In order to prove whether prrA homologous gene only or PAICFT073 is present in E. coli O157:H7, screening for PAICFT073 genes by PCR method was conducted. To set up the PCR conditions for a specific detection of the PAICFT073 genes and its boundary sequences, various primers were designed according to the published sequences (5) and are presented in Table 1, including the hlyABCD for hemolysin, papABCDFHIJK for P pilus, and the genes related to insertion sequences, transposons, and hypothetical proteins, including Hp1-4 and R1-16 (Table 1). E. coli CFT073, used as a positive control, was isolated from the blood and urine of a woman with acute pyelonephritis (5). Laboratory strain E. coli HB101 was used as a negative control. E. coli O157:H7 EDL933 was used as reference strain to study in detail the presence of PAICFT073 genes of E. coli O157:H7 strains containing the Stx gene. PCR was performed with 30 cycles of reaction composed each of a denaturing step at 94°C for 1 min, an annealing step at the temperature as indicated in Table 1, and an elongation step at 72°C for various times (see Table 1), as well as one final extension step at 72°C for 10 min.
TABLE 1.
Primers used for amplifying PAICFT073 genes PCR products obtained in E. coli O157:H7 EDL933 and UPEC CFT073a
| Gene(s) | Orientation | Primer sequence | Size (bp) | Annealing temp (°C) | Extension time (min) | Presence of gene inb:
|
|
|---|---|---|---|---|---|---|---|
| EHEC EDL933 | UPEC CFT073 | ||||||
| O241 plus prrA (left joint) | F | CTGGTGGTGTCATACGCTAA | 1,654 | 55.6 | 1 | − | + |
| R | GTAACGCTGTCGGAAGAGGC | ||||||
| prrA | F | ATGGTGTTGATGGGCTGGC | 479 | 55 | 1 | + | + |
| R | CCCTGAAAAGTCGGCTGTATC | ||||||
| modD | F | TGCCTGTTGCGGACTAAAT | 378 | 55 | 1 | + | + |
| R | GTGCTGGAGTGGAGTTTGC | ||||||
| yc73 | F | GCGAAGCCGTGCCTGATTAT | 437 | 56 | 1 | + | + |
| R | CAGACCTTTACCTTCAGCAA | ||||||
| fepC | F | TACCTGGATAATGCTGTCGG | 347 | 55 | 1 | + | + |
| R | ATGGTGTTGATGGGGCTG GC | ||||||
| HP1* | F | AGTGGCTCAGGCTCTCATTT | 506 | 56.3 | 1 | + | + |
| R | TGGCATCATCGTTGGTCGTG | ||||||
| HP2* | F | CGTATTTTTCAGCGACTCCT | 181 | 51.3 | 1 | + | + |
| R | CACTTTAGGACAATGGGTTA | ||||||
| HP3* | F | TCGTTGCTGCCTCCTGTGAA | 216 | 54.9 | 1 | + | + |
| R | GCTGATACTGAGGGTTTCCG | ||||||
| HP4* | F | CGGCCATGTTTTCATTTTCC | 222 | 58.7 | 1 | + | + |
| R | ATGGTCAGGGAGGTCAGCAG | ||||||
| hlyC | F | CCAGTTCCCCATTACACAGA | 311 | 51.4 | 1 | − | + |
| R | CACCCTGATGGCTCTGAATA | ||||||
| hlyA | F | ATTAGTCCCCTCTCATTCCT | 1,100 | 52.1 | 2 | − | + |
| R | ATTAGTCCCCTCTCATTCCT | ||||||
| hlyB | F | GTTGTGGTGGTGTTTGAGAT | 111 | 52.3 | 1 | + | + |
| R | TGCCAGGATTAGCCAGTGAA | ||||||
| hlyD | F | AGGCTGGAACAAACTCGGTA | 755 | 52.7 | 1 | − | + |
| R | TATCGGGTGTAAGGAAAGGC | ||||||
| papG | F | TTTGCGAGTGGAGTGTATTT | 622 | 50.7 | 1 | − | + |
| R | TACCTAACCCAACCGAAAAT | ||||||
| papF | F | ATCGTTGCTTCTGACATCGG | 311 | 52.3 | 1 | − | + |
| R | TTGACATTCCTTTTCCCTGA | ||||||
| papE | F | GAGTCAAAATGGAAATCACG | 204 | 48.7 | 1 | − | + |
| R | AAAGTTATCGCAGTCCCAAT | ||||||
| papK | F | CGCTCTTTTACTGTTTGCCG | 466 | 57.8 | 1 | − | + |
| R | ACTCTTCGTCCGCAGGGCTT | ||||||
| papJ | F | TCCATACTTTCCTGCGGGCT | 370 | 56.1 | 1 | − | + |
| R | ATAACAAGATGGTCACAGCC | ||||||
| papD | F | ACAAACAACTGCCCTATCTT | 424 | 52.5 | 1 | − | + |
| R | TCACCTTCCTCTGCCTGCTT | ||||||
| papC | F | TTATCTGTTCCGTGCCATTC | 769 | 57.2 | 1 | − | + |
| R | TTCCCGACTGCTGTAATCAT | ||||||
| papH | F | TTGGCTGTGTGTTTGTTCAT | 461 | 54.9 | 1 | − | + |
| R | CGCTTCTTCATTACCCGTCA | ||||||
| papA | F | ATGCTGCTCCAACTATTCCA | 393 | 53 | 11 | + | + |
| R | CGTTTTCACCATCTTTCAGG | ||||||
| papB | F | GAAGTCATCAGTCGGTCAGG | 254 | 52.6 | 1 | − | + |
| R | GCAAGAGCATTCAGCCGTAT | ||||||
| pap1 | F | TTCAAAAACCAGTATGTCGC | 140 | 52.2 | 1 | − | + |
| R | GGAGGGAAAACCGCAGAAAT | ||||||
| R15* | F | CCAGCCTTCCCAGCAATCGT | 256 | 57.0 | 1 | + | + |
| R | GGCACCATCCATCACAGCGA | ||||||
| R14* | F | ACCCTGATTCCTTCCCGTAA | 164 | 52.1 | 1 | + | + |
| R | TATTACCATTGTCAGCAGCA | ||||||
| R13* | F | AACTCCGCCTTCGCAAAATA | 266 | 57.1 | 1 | + | + |
| R | CGGGCAGTTCGTATGGTTCT | ||||||
| R12* | F | CATCTCTCCCAGTCATTACG | 207 | 54.0 | 1 | − | + |
| R | CCCTGTTGAAAGTTGGCGTC | ||||||
| R11 | F | GAGGCGTATTGTTATTGTTG | 253 | 50.7 | 1 | − | + |
| R | CTTCTGATTGGTAGGCTTGC | ||||||
| R10 | F | ATTGTCGCCCTTGGTCTCAT | 222 | 51.3 | 1 | − | + |
| R | GGCAGTTCCATCAAGTTTAT | ||||||
| R9 | F | TAGTTATTCTTCGCCTGTCT | 352 | 50.8 | 1 | − | + |
| R | TATTTCAGCAGGACACTACC | ||||||
| R8 | F | GTTGGGGTCTCAGGCACACT | 200 | 54.8 | 1 | − | + |
| R | GGCAGCACAGGAAGCGGAAT | ||||||
| R7 | F | GTGGCGGTTGTGTTGTTATC | 173 | 55.1 | 1 | − | + |
| R | TGTCAGCCTCTACGAAACGC | ||||||
| R6* | F | GTTGTGCTGGGTGGTGAGAG | 369 | 56.6 | 1 | + | + |
| R | CTGATTGTTACGGTTGTGCC | ||||||
| R5 | F | CGGCAACTCTGTGAAACGAC | 482 | 56.4 | 1 | − | + |
| R | CTTGTTACTGCCTTCGCTGT | ||||||
| R4* | F | TCCTCAGCAAATACCGACCA | 683 | 50.7 | 1 | + | + |
| R | TGGCTCTCTTCCGTCAATGC | ||||||
| R3 | F | GGATAACCAATAGCAGAACA | 716 | 48.7 | 1 | − | + |
| R | CCCAGTGTGATGTATTCTAT | ||||||
| R2(malX) | F | GCCGATAATGACTTGTAGGG | 504 | 53.5 | 1 | − | + |
| R | CCACTGCTGTTTGTCTTCCA | ||||||
| R1 | F | GCGACAACTCAATAATCCGT | 482 | 50.0 | 1 | − | + |
| R | TGGACAGGAGGTTATCATTT | ||||||
| R1 plus R2 | F | AGCCTTTCTGTTTTGAGCAT | 1,525 | 51.0 | 2 | − | + |
| R | TCGCTACTATTGATTCTTGC | ||||||
| R1 plus f447 (right joint) | F | CCGCAAGAATCAATAGTAGC | 564 | 50.7 | 1 | − | + |
| R | CTGGCGAGAAGGGGATAATG | ||||||
The genes are listed in the order of their position on PAICFT071. The PCR was performed by using the forward primer (F) or the reverse primer (R), with the annealing temperature and the extension time as indicated.
The presence (+) or absence (−) of the genes listed in column 1 is indicated for each strain.
Under the conditions used, 41 PAICFT073 genes were successfully amplified with the expected sizes for UPEC CFT071. Interestingly, 25 of 41 primer pairs, including primers for junction regions, did not yield any fragment for EDL933, including the primes for hlyACD and papBCDFHIJK. However, 15 genes were amplified in the reference Stx-positive strain E. coli O157:H7 EDL933, including HP1-4, R13-15, R6, R4, hlyB, and papA (Table 1). It is reasonable that most of the homologous sequences are present in E. coli K-12. The R4 homologous gene, iha, has been identified in E. coli O157:H7 (18). The HP1 and HP2 represent the IS600 hypothetical 31- and 11-kDa proteins HP3 and HP4 are hypothetical proteins identified in E. coli K-12. R15, R14, R12, and R6 are related to insertion sequences and transposons. R13 is homologous to the 12.7-kDa protein of E. coli K-12 (5). We cannot explain why the papA and hlyB fragments were synthesized in E. coli O157:H7 EDL933 at present time.
The boundary sequences of PAICFT073 were also analyzed. The prrA gene is inside the left junction, next to L4. Just inside the right junction, the R1 gene is linked to f447 (5). Both gene L4 and gene f447 are sequences of E. coli K-12. The primers were designed for detection of the left junction (L4-prrA) and right junction (R1-f447) (Table 1). The left and right junctions of PAICFT071 could not be amplified from strain E. coli O157:H7 EDL933. Furthermore, the PCR experiment with R1-R2 primers did not yield any product in 62 strains of E. coli O157:H7, which are linked at the right junction of PAICFT073 (Table 2).
TABLE 2.
Prevalence of prrA-modD-yc73-fepC genes in E. coli O157:H7 strains with or without the Stx genea
| Strain group and source | No. of strains tested | No. of strains
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| hlyAb | stx1 | stx2 | exe | prrA | modD | yc73 | fepC | R1 plus R2 | ||
| Control | ||||||||||
| Positive control, UPEC CFT073 | 1 | 1− | 1− | 1− | 1− | 1+ | 1+ | 1+ | 1+ | 1+ |
| Negative control, E. coli HB101 | 1 | 1− | 1− | 1− | 1− | 1− | 1− | 1− | 1− | 1− |
| E. coli O157:H7 strains containing Stx gene | ||||||||||
| Patients (United States) | 4 | 4+ | 3+1− | 3+1− | 4+ | 4+ | 3+1− | 2+2− | 4+ | 4− |
| Patients (China) | 5 | 5+ | 5+ | 5+ | 5+ | 5+ | 5+ | 5+ | 5+ | 5− |
| Patients (Japan) | 6 | 6+ | 6+ | 4−2+ | 6+ | 6+ | 5+1− | 5+1− | 6+ | 6− |
| Pigs (Jiangsu Province, China) | 7 | 7+ | 7− | 7+ | 7+ | 7+ | 4+3− | 7+ | 7+ | 7− |
| Chicks (Jiangsu Province, China) | 5 | 5+ | 1+ | 5+ | 5+ | 5+ | 4+1− | 5+ | 5+ | 5− |
| Goats (Jiangsu Province, China) | 16 | 16+ | 16− | 16+ | 15+1− | 16+ | 15+1− | 16+ | 16+ | 16− |
| E. coli O157:H7 strains without Stx gene | ||||||||||
| Pigs (Jiangsu Province, China) | 6 | 6− | 6− | 6− | 6− | 6− | 6− | 6− | 6− | 6− |
| Pigs (Fijian Province, China) | 4 | 4− | 4− | 4− | 4− | 4− | 4− | 4− | 4− | 4− |
| Chicks (Jiangsu Province, China) | 4 | 4− | 4− | 4− | 4− | 4− | 4− | 4− | 4− | 4− |
| Goats (Jiangsu Province, China) | 5 | 5− | 5− | 5− | 5− | 5− | 4−1+ | 5− | 5− | 5− |
| Totals | ||||||||||
| E. coli O157:H7 strains containing Stx gene | 43 | 43+ | 15+ | 38+ | 43+ | 43+ | 37+6− | 40+3− | 43+ | 43− |
| E. coli O157:H7 strains without Stx gene | 19 | 19− | 19− | 19− | 19− | 19− | 18−1+ | 19− | 19− | 19− |
The number positive (+) and the number negative (−) indicate the numbers of strains showing the presence and absence of the gene analyzed, respectively.
That is, EHEC-hlyA.
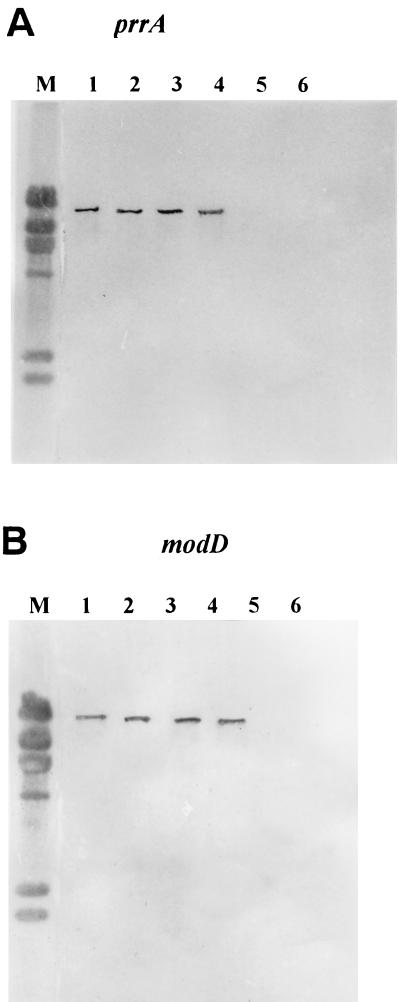
EDL933 and several other E. coli O157:H7 isolates were selected for Southern hybridization, including two clinical strains containing Stx gene (strain 223 and 143) and one animal isolate without Stx gene (PC02). The chromosomal DNA of tested strains was extracted by lysozyme-sodium dodecyl sulfate-proteinase K method, which were further purified by the phenol and chloroform extraction method. It was digested with restriction enzyme EcoRI and separated on a 0.9% agarose gel. The digested genomic DNA fragments were transferred from the gel to Zeta-Probe BT blotting membranes (Bio-Rad Laboratories, Richmond, Calif.). The PCR products (prrA, modD, yc73, and fepC) of E. coli CFT073, obtained from agarose gels by QIAquick Gel Extraction kit (Gene Company Limited, Beijing, China), were used as probes in the hybridization assays. Digoxigenin labeling of the probes and hybridization were performed with a DNA labeling and detection kit (Promega, Beijing, China). After prehybridization at 68°C for 2 h and the addition of a heat-denatured probe, the blots were incubated overnight (for ca. 16 h) at 68°C in the absence of formamide. The detection was performed according to the manufacturer's instructions. Digoxigenin labeling of PCR fragments of the prrA and modD gene were used as the probes. Southern hybridization was performed with a DNA labeling and detection kit (Boehringer, Mannheim, Germany). One DNA fragment of E. coli O157:H7 EDL933 and other isolates containing the Stx gene hybridized with the probes, of which the molecular size was identical to that of UPEC CFT073 in our condition (Fig. 1). No positive signal was observed for the animal isolate without the Stx gene, as well as for E. coli HB101. Hybridization with probes of yc73 and fepC gave the same results (data not shown). These results thus revealed that not all of the pathogenicity-associated island PAICFT073 is present in E. coli O157:H7 containing Stx gene, an idea supported by the recently published genome sequence data of E. coli O157:H7 EDL933 (16).
FIG. 1.
Southern hybridization profile of the EcoRI-digested chromosomal DNA with prrA and modD probes. Lanes: M, molecular standard (DNA digested with HindIII); 1, E. coli CFT073 (positive control); 2, E. coli O157:H7 EDL933; 3, E. coli O157:H7 223; 4, E. coli O157:H7 143; 5, E. coli O157:H7 PC02 (animal isolate without Stx gene); 6, E. coli HB101 (negative control). (A) Hybridization with prrA probe. (B) Hybridization with modD probe.
A rapid preliminary analysis suggested a co-occurrence between the prrA-modD-yc73-fepC gene cluster of PAICFT073 and Stx gene in the E. coli O157:H7 strains. Therefore, we carried out a detailed screening to verify this correlation. We analyzed a total of 62 E. coli O157:H7 isolates, including 4 isolated from the diarrheal patients in the United States (11), 6 isolated from diarrheal patients in Japan (22), and 52 isolated in China (23–25). All of the isolates were reconfirmed in our laboratory by serological methods for O157 and H7 antigens, as well as by PCR methods for genes of Shiga toxin 1 (Stx1), Shiga toxin 2 (stx2), EHEC attachment and effacing (eae), and hemolysin (EHEC-hlyA) (3, 20). The results are shown in Table 2. In 43 E. coli O157:H7 strains containing the Stx gene, the frequencies of the detection of the prrA, modD, yc73, or fepC were 100, 86, 93, and 100%, respectively. In remarkable contrast, none of the prrA, yc73, and fepC genes was detected in the 19 E. coli O157:H7 strains without the Stx gene, and the modD gene was detected only in one of them. In 43 strains of E. coli O157:H7 containing Stx gene, 37 displayed prrA-modD-yc73-fepC-positive PCR pattern, 5 strains were prrA-yc73-fepC positive, 2 strains were prrA-modD-fepC positive, and one strain showed a prrA-fepC-positive PCR pattern.
In order to confirm the PCR results, Southern blots was conducted for nine strains containing Stx gene with various PCR patterns and three E. coli O157:H7 strains without the Stx gene. UPEC CFT073 and E. coli HB101 were used as positive and negative controls, respectively. The purified chromosome DNA was blotted on a nitrocellulose filter. The PCR products (prrA, modD, yc73, and fepC) of E. coli CFT073, obtained from agarose gels, were used as probes in the hybridization assays. All E. coli O157:H7 strains containing the Stx gene tested hybridized with probes of prrA, modD, yc73, and fepC, regardless of the PCR patterns. Among these, three were PCR negative for modD genes, and two were PCR negative for yc73 or for modD-yc73 genes, respectively. These probes did not hybridize with the chromosome of the negative control HB101 under the conditions used. It seems that all of the E. coli O157:H7 strains containing the Stx gene have a prrA-modD-yc73-fepC gene cluster, and the negative PCR results might be due to the variation in primer sequences of the targeted genes (Table 3). The E. coli O157:H7 strain without the Stx gene that yielded modD PCR product was failed to hybridize with modD DNA probe. It is reasonable to assume that the primers for modD gene may yield a false result on some occasions. Therefore, these results demonstrate for the first time a strict correlation between the presence of the prrA-modD-yc73-fepC gene cluster in E. coli O157:H7 strains containing Stx gene(s). It should be noted that one strain in 43 E. coli O157:H7 isolates containing Stx gene had no eae gene detected.
TABLE 3.
Confirmation of PCR results by DNA hybridizationa
| Source | Total no. of strains | No. of strains
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| stx |
prrA
|
modD
|
yc73
|
fepC
|
R1-R2
|
|||||||
| PCR | H | PCR | H | PCR | H | PCR | H | PCR | H | |||
| Patient | 3 | 3+ | 3+ | 3+ | 3+ | 3+ | 3+ | 3+ | 3+ | 3+ | 3− | 3− |
| Chick | 1 | 1+ | 1+ | 1+ | 1+ | 1+ | 1+ | 1+ | 1+ | 1+ | 1− | 1− |
| Patient | 1 | 1+ | 1+ | 1+ | 1− | 1+ | 1+ | 1+ | 1+ | 1+ | 1− | 1− |
| Pig | 1 | 1+ | 1+ | 1+ | 1− | 1+ | 1+ | 1+ | 1+ | 1+ | 1− | 1− |
| Goat | 1 | 1+ | 1+ | 1+ | 1− | 1+ | 1+ | 1+ | 1+ | 1+ | 1− | |
| Patient | 1 | 1+ | 1+ | 1+ | 1+ | 1+ | 1− | 1+ | 1+ | 1+ | 1− | 1− |
| Patient | 1 | 1+ | 1+ | 1+ | 1− | 1+ | 1− | 1+ | 1+ | 1+ | 1− | 1− |
| Pig | 2 | 2− | 2− | 2− | 2− | 2− | 2− | 2− | 2− | 2− | 2− | 2− |
| Goat | 2 | 2− | 2− | 2− | 1−1+ | 2− | 2− | 2− | 2− | 2− | 2− | 2− |
| E. coli HB101 | 1 | 1− | 1− | 1− | 1− | 1− | 1− | 1− | 1− | 1− | 1− | 1− |
| UPEC CFT073 | 1 | 1− | 1+ | 1+ | 1+ | 1+ | 1+ | 1+ | 1+ | 1+ | 1+ | 1+ |
PCR and Southern hybridization (H) results are compared. The number positive (+) and the number negative (−) indicate the numbers of strains showing the presence and absence of the gene analyzed, respectively.
In order to assess the similarity in the DNA sequence of the prrA-modD-yc73-fepC genes between UPEC CFT073 and E. coli O157:H7 EDL933, DNA fragments of the prrA, modD, yc73, and fepC were amplified by PCR using the chromosomal DNA UPEC CFT073 as a template and extracted from agarose gels by using the QIAquick Gel Extraction Kit and sequenced in both directions by using the Taq Dye-Deoxy-Cycle-Sequencing Kit and 373A DNA sequencer (Applied Biosystems, Foster City, Calif.). The sequences obtained were aligned with PAICFT073 sequences published by Guyer et al. with BLAST software (5). The DNA sequences of 479 bp (prrA) and 437 bp (yc73) of E. coli O157:H7 EDL933 were identical to that of UPEC CFT071. In 347 and 378 bp of the fepC and modD sequences, only two nucleotide mismatches were observed.
The PAICFT073 seems to be atypical. Guyer et al. stated that at the approximately 7 kb downstream of the left junction of PAICFT073, following the prrA-modD-yc73-fepC gene cluster, there are 8-kb sequences carrying six ORFs, which are identical to that found in the E. coli K-12 genome (5). The virulent hemolysin gene cluster hlyCABD follows this block. The segmentation is obvious with respect to unique PAICFT073 sequences and sequences of E. coli K-12 origin. It is reasonable to believe that the prrA-modD-yc73-fepC gene cluster, rather than PAICFT073, is present in E. coli O157:H7. Moreover, the prrA-modD-yc73-fepC gene cluster might not be a necessary part of PAICFT073 (5). Recently, Tarr et al. identified a tellurite resistance- and adherence-conferring island in E. coli O157:H7, in which the 99% DNA sequences of the gene iha are identical to R4 of PAICFT073, a putative exogenous ferric siderophore receptor (21). In regard to iron uptake, the tellurite resistance- and adherence-conferring island might have some relationship with the prrA-modD-yc73-fepC gene cluster in E. coli O157:H7 (21).
It seems that the prrA-modD-yc73-fepC gene cluster is linked with Shiga toxin gene, so that, it is probably crucial for virulence of E. coli O157:H7. Gyer et al. suggested that the prrA-modD-yc73-fepC gene cluster represent a TonB-dependent iron uptake system (5). It was suspected that the TonB system is necessary for all gram-negative organisms that dwell in the presence of oxygen. Indeed, the genes encoding homolog to E. coli TonB have been cloned and sequenced from Salmonella enterica serovar Typhimurium, Klebsiella pneumoniae, Enterobacter aerogenes, Serratia marcescens, Yersinia enterocolitica, H. influenzae, Pseudomonas putida, and others. TonB does play a role in addition to heme- and siderophore-mediated iron acquisition in vivo, and this function is related to the bacterial virulence, such as the intercellular spread of Shigella dysenteriae, abilities to produce invasive disease in an animal model of H. influenzae, and the requirement for mouse virulence of Y. enterocolitica (6, 7, 17). It has been demonstrated that the Shiga toxins of E. coli O157:H7 are also iron regulated (1, 2, 19). Calderwood and Mekalanos reported that the Shiga toxin operon was negatively regulated by fur gene product. In the DNA region between the −35 and −10 boxes of Shiga toxin, the 21-bp dyad repeat may represent an operator binding site for Fur protein in the presence of iron (1). The fepC gene has been known to encode ferric enterobactin transport ATP-binding protein (5). Payne and his colleagues have identified an iron transport system in E. coli O157:H7 strain EDL933. It has been known that E. coli O157:H7 can synthesize and transport enterobactin and had a ferric citrate transport system but lack the ability to produce or use aerobactin. It can use heme and hemoglobin, but not transferrin or lactoferrin, as iron sources. The heme utilization gene (chuA) encodes a 69-kDa outer membrane protein, for which the homologous one is also present in S. dysenteriae I, a Shiga toxin-producing species (13). However, the role played by the prrA-modD-yc73-fepC gene cluster in the virulence of E. coli O157:H7 should be clarified experimentally.
Acknowledgments
We are grateful to Longfei Wu for critical reading of the manuscript. We thank James B. Kaper, Center for Vaccine Development, University of Maryland School of Medicine, for kindly providing the strain of E. coli O157:H7. We thank H. Watanabe, Department of Bacteriology, National Institute of Infectious Disease, Tokyo, Japan, for providing strains of E. coli O157:H7 isolated in Japan.
This work was supported by the Basic Research Program (G1999054101, to J.X.) from the Ministry of Science and Technology of the People's Republic of China and by a grant from the National Natural Science Foundation (no. 30070042, to J.X.).
REFERENCES
- 1.Calderwood S B, Mekalanos J J. Iron regulation of Shiga-like toxin expression in Escherichia coli is mediated by the fur locus. J Bacteriol. 1987;169:4759–4764. doi: 10.1128/jb.169.10.4759-4764.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chart H, Scotland S M, Rowe B. Production of vero cytotoxin by Escherichia coli and Shiga toxin by Shigella dysenteriae 1 as related to the growth medium and availability of iron. Zentbl Bakteriol. 1989;272:1–10. doi: 10.1016/s0934-8840(89)80086-6. [DOI] [PubMed] [Google Scholar]
- 3.Gannon V P, Rashed M, King R K, Thomas E J. Detection and characterization of the eae gene of Shiga-like toxin- producing Escherichia coli using polymerase chain reaction. J Clin Microbiol. 1993;31:1268–1274. doi: 10.1128/jcm.31.5.1268-1274.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guyer D M, Kao J S, Mobley H L. Genomic analysis of a pathogenicity island in uropathogenic Escherichia coli CFT073: distribution of homologous sequences among isolates from patients with pyelonephritis, cystitis, and catheter-associated bacteriuria and from fecal samples. Infect Immun. 1998;66:4411–4417. doi: 10.1128/iai.66.9.4411-4417.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guyer D M, Kao J S, Mobley H L. Genomic analysis of a pathogenicity island in uropathogenic Escherichia coli CFT073: distribution of homologous sequences among isolates from patients with pyelonephritis, cystitis, and catheter-associated bacteriuria and from fecal samples. Infect Immun. 1998;66:4411–4417. doi: 10.1128/iai.66.9.4411-4417.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hong M, Gleason Y, Wyckoff E E, Payne S M. Identification of two Shigella flexneri chromosomal loci involved in intercellular spreading. Infect Immun. 1998;66:4700–4710. doi: 10.1128/iai.66.10.4700-4710.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jarosik G P, Sanders J D, Cope L D, Muller-Eberhard U, Hansen E J. A functional tonB gene is required for both utilization of heme and virulence expression by Haemophilus influenzae type b. Infect Immun. 1994;62:2470–2477. doi: 10.1128/iai.62.6.2470-2477.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kao J S, Stucker D M, Warren J W, Mobley H L. Pathogenicity island sequences of pyelonephritogenic Escherichia coli CFT073 are associated with virulent uropathogenic strains. Infect Immun. 1997;65:2812–2820. doi: 10.1128/iai.65.7.2812-2820.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaper J B. Enterohemorrhagic Escherichia coli. Curr Opin Microbiol. 1998;1:103–108. doi: 10.1016/s1369-5274(98)80149-5. [DOI] [PubMed] [Google Scholar]
- 10.Kuhar I, Grabnar M, Zgur-Bertok D. Virulence determinants of uropathogenic Escherichia coli in fecal strains from intestinal infections and healthy individuals. FEMS Microbiol Lett. 1998;164:243–248. doi: 10.1111/j.1574-6968.1998.tb13093.x. [DOI] [PubMed] [Google Scholar]
- 11.Levine M M, Xu J G, Kaper J B, Lior H, Prado V, Tall B, Nataro J, Karch H, Wachsmuth K. A DNA probe to identify enterohemorrhagic Escherichia coli of O157:H7 and other serotypes that cause hemorrhagic colitis and hemolytic uremic syndrome. J Infect Dis. 1987;156:175–182. doi: 10.1093/infdis/156.1.175. [DOI] [PubMed] [Google Scholar]
- 12.Mainil J G, Duchesnes C J, Whipp S C, Marques L R, O'Brien A D, Casey T A, Moon H W. Shiga-like toxin production and attaching effacing activity of Escherichia coli associated with calf diarrhea. Am J Vet Res. 1987;48:743–748. [PubMed] [Google Scholar]
- 13.Mills M, Payne S M. Genetics and regulation of heme iron transport in Shigella dysenteriae and detection of an analogous system in Escherichia coli O157:H7. J Bacteriol. 1995;177:3004–3009. doi: 10.1128/jb.177.11.3004-3009.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morschhauser J, Vetter V, Emody L, Hacker J. Adhesin regulatory genes within large, unstable DNA regions of pathogenic Escherichia coli: cross-talk between different adhesin gene clusters. Mol Microbiol. 1994;11:555–566. doi: 10.1111/j.1365-2958.1994.tb00336.x. [DOI] [PubMed] [Google Scholar]
- 15.Park S, Worobo R W, Durst R A. Escherichia coli O157:H7 as an emerging foodborne pathogen: a literature review. Crit Rev Food Sci Nutr. 1999;39:481–502. doi: 10.1080/10408699991279259. [DOI] [PubMed] [Google Scholar]
- 16.Perna N, Plunkett III G, Burland V, Mau B, Glasner J D, Rose D J, Mayhew G F, Evans P S, Gregor J, Kirkpatrick H A, Pósfai G, Hackett J, Klink S, Boutin A, Shao Y, Miller L, Grotbeck E J, Davis N W, Lim A, Dimalanta E T, Potamousis K D, Apodaca J, Anantharaman T S, Lin J, Yen G, Schwartz D C, Welch R A, Blattner F R. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature. 2001;409:529–533. doi: 10.1038/35054089. [DOI] [PubMed] [Google Scholar]
- 17.Rakin A, Saken E, Harmsen D, Heesemann J. The pesticin receptor of Yersinia enterocolitica: a novel virulence factor with dual function. Mol Microbiol. 1994;13:253–263. doi: 10.1111/j.1365-2958.1994.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 18.Schmidt H, Geitz C, Tarr P I, Frosch M, Karch H. Non-O157:H7 pathogenic Shiga toxin-producing Escherichia coli: phenotypic and genetic profiling of virulence traits and evidence for clonality. J Infect Dis. 1999;179:115–123. doi: 10.1086/314537. [DOI] [PubMed] [Google Scholar]
- 19.Svinarich D M, Palchaudhuri S. Regulation of the SLT-1A toxin operon by a ferric uptake regulatory protein in toxinogenic strains of Shigella dysenteriae type 1. J Diarrhoeal Dis Res. 1992;10:139–145. [PubMed] [Google Scholar]
- 20.Takeshi K, Ikeda T, Kubo A, Fujinaga Y, Makino S, Oguma K, Isogai E, Yoshida S, Sunagawa H, Ohyama T, Kimura H. Direct detection by PCR of Escherichia coli O157 and enteropathogens in patients with bloody diarrhea. Microbiol Immunol. 1997;41:819–822. doi: 10.1111/j.1348-0421.1997.tb01934.x. [DOI] [PubMed] [Google Scholar]
- 21.Tarr P I, Bilge S S, Vary J C, Jr, Jelacic S, Habeeb R L, Ward T R, Baylor M R, Besser T E. Iha: a novel Escherichia coli O157:H7 adherence-conferring molecule encoded on a recently acquired chromosomal island of conserved structure. Infect Immun. 2000;68:1400–1407. doi: 10.1128/iai.68.3.1400-1407.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Watanabe H, Terajima J, Izumiya H, Wada A, Tamura K. Molecular analysis of enterohemorrhagic Escherichia coli isolates in Japan and its application to epidemiological investigation. Pediatr Int. 1999;41:202–208. doi: 10.1046/j.1442-200x.1999.4121043.x. [DOI] [PubMed] [Google Scholar]
- 23.Xu J-G, Jing H Q. Escherichia coli O157:H7 and Shiga-like-toxin-producing E. coli in China. World J Gastroenterol. 1999;5:191–194. doi: 10.3748/wjg.v5.i3.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu J G, Quan T S, Xiao D L, Fan T R, Li L M, Wang C A, Li W, Liu H M. Isolation and characterization of Escherichia coli O157:H7 strains in China. Curr Microbiol. 1990;20:299–303. [Google Scholar]
- 25.Xu J G, Qi G M. Clinical and epidemiological characters and identification of enterohemorrhagic Escherichia coli. Chung Hua Liu Hsing Ping Hsueh Tsa Chih. 1996;17:367–369. [PubMed] [Google Scholar]