Abstract
Breast cancers are a diverse group of diseases and are often characterized by their expression of receptors for hormones such as estrogen and progesterone. Recently another steroid hormone receptor, the glucocorticoid receptor (GR) has been shown to be a key player in breast cancer progression, metastasis, and treatment. These receptors bind to chromatin to elicit transcriptional changes within cells, which are often inhibited by the structure of chromatin itself. Chromatin remodeling proteins, such as Brahma-related gene 1 (BRG1), function to overcome this physical inhibition of transcription factor function and have been linked to many cancers including breast cancer. Recent efforts to understand the interactions of BRG1 and GR, including genomic and single cell analyses, within breast cancers may give insight into personalized medicine and other potential treatments.
Introduction:
Genetic information is packaged in cells as chromatin, a hierarchically condensed structure of DNA and histone proteins in the nucleus. The condensation of DNA into chromatin creates a structural barrier that may inhibit DNA binding proteins from accomplishing their functions. ATP-dependent chromatin remodeling complexes such as the SWItch/Sucrose Non-Fermentable (SWI/SNF) complex alter the contacts between DNA and histones to allow for critical processes such as DNA repair, recombination, and gene expression [1,2]. This alteration in chromatin structure permits transcription factors including nuclear receptors (NRs) to bind and mediate a variety of biological processes [1,3]. The SWI/SNF complex regulates NR mediated transcription, often by direct interaction with NRs. The MMTV promoter model in breast cancer cells was utilized to characterize the requirement for SWI/SNF chromatin remodeling in glucocorticoid (GR), progesterone (PR), and androgen receptor (AR) mediated activation [4,5]. While the mechanisms of NR and SWI/SNF interactions have been extensively reviewed [6–9], recent work utilizing next-generation sequencing technology has characterized the combinatorial roles that SWI/SNF and NRs play in mediating gene expression changes. As the SWI/SNF complex may function as a shared protein complex for nuclear receptors, studies directed towards this platform may be informative for the function of multiple nuclear receptors. This review will highlight the recent advances of work relevant to understanding the roles of the SWI/SNF complex and GR in breast cancer and provide insight into the promising future of studying these proteins within the context of this disease.
BRG1, BAF complex, and Cancer:
SWI/SNF is a large, multiprotein complex with diverse and variable subunit composition [10,11]. Initially, two distinct SWI/SNF complexes were identified in mammalian cells, BAF and PBAF. These complexes both contain a central ATPase (BRG1 or BRM) and several common BRG1-BRM associated factor (BAF) subunits, and the particular complex compositions can be seen in Table 1. While the BAF and PBAF complexes perform similar biochemical functions, different biological roles have been described for each. PBAF-specific subunits are often associated with cell differentiation and cell-type identity regulation, while BAF complex specific roles have been characterized in development, self-renewal, and pluripotency [12]. The recent description of two additional complexes (embryonic stem cell-specific BAF and non-canonical BAF) have emphasized the importance for correct assembly of unique complexes for specific biological processes [13–15].
Table 1. SWI/SNF proteins that compose mammalian BAF, PBAF, or ncBAF complexes.
The complexes that each protein has been categorized into is marked by color and column.
| Subunit | BAF | PBAF | ncBAF | Complex Identity |
|---|---|---|---|---|
| SMARCA4/2 | X | X | X | Common |
| SMARCC1/2 | X | X | X | BAF and PBAF |
| SMARCD1/2/3 | X | X | X | BAF specific |
| BCL7A/B/C | X | X | X | PBAF specific |
| ACTB | X | X | X | BAF and ncBAF |
| ACTL6A | X | X | X | ncBAF specfic |
| SMARCB1 | X | X | ||
| SMARCE1 | X | X | ||
| ARID1A/B | X | |||
| DPF1/2/3 | X | |||
| SS18/1L | X | X | ||
| PBRM1 | X | |||
| PHF10 | X | |||
| BRD7 | X | |||
| ARID2 | X | |||
| GLTSCR1/1L | X | |||
| BRD9 | X |
Advances in cryo-electron microscopy have allowed for determination of the structure of the SWI/SNF complex. A 3.7 angstrom structure of an in vitro assembled BAF complex revealed that the ATPase domain of BRG1 is engaged with nucleosomal DNA, with some subunits modeled into the structure rather than de novo determined. This interaction is guided in place by the ARP (actin-related protein) module and the HSA (helicase/SANT associated) domain of BRG1. This HSA-ARP module bridges the ATPase to the base module, which contains the N-terminal region of BRG1 and the other auxiliary subunits. This base module serves several functions, including nucleosome binding by BAF47, stabilization of the core by ARID1A, scaffolding by BAF155 and BAF170, and organizational support by BAF60 and BAF57 [16]. Recent studies on the structure of yeast remodelers including SWI/SNF and RSC have revealed similar structures [17–19]. These efforts have allowed preliminary hypotheses for the mechanisms of chromatin remodeling, which require further testing to confirm.
Determination of the composition, assembly and structure of SWI/SNF complexes is critical for human health as human SWI/SNF subunits and associated proteins are mutated in nearly 20% of human cancers [20]. Recent efforts have characterized the organization of human SWI/SNF complexes, which are assembled in modular fashion [21]. Cancer-relevant deleterious mutations of SWI/SNF proteins have provided additional insight into complex assembly and chromatin interaction [22]. Cancer-relevant truncations or mutations of ARID1A also disrupted complex formation [21] and loss of BAF47 altered the genomic distribution of the BAF complex [23]. Catalytically inactivating mutations of BRG1 and BRM are also found in human cancers [24]. BAF complexes containing catalytically inactive BRG1 protein did not bind to all chromatin sites bound by complexes containing WT BRG1. Rather, the binding of complexes with catalytically inactive BRG1 was biased toward sites bound by PBAF complexes. Furthermore, sites of SWI/SNF chromatin interaction were linked to gene expression profiles associated with tumor-suppressor pathways [22], providing insight into how SWI/SNF mutations result in cancer and suggesting that SWI/SNF chromatin interactions could be targeted for cancer treatment. More systematic investigations of individual subunit losses have also demonstrated how losses of these subunits can lead to cancers and highlights novel synthetic lethalities [25]. Extensive efforts have also demonstrated that elevated levels of complex members such as BRG1 are found in cancers of the breast, brain, colon, and pancreas. TGCA analysis demonstrates very low BRG1 mutation frequency in invasive breast cancers, but elevated expression occurred much more frequently [24,26–30]. BRG1 has been demonstrated to bind at promoters of genes that are overexpressed in primary breast tumors and invasive breast cancer cell lines and activate their transcription directly. The BRG1 complexes formed at these locations work in concert with EP300 and often PARP1 to direct the transcription of proliferation and DNA repair genes in breast cancer cells [31,32]. Direct studies have shown that knockdown of BRG1 decreased breast cancer cell proliferation. The additional observation that BRG1 knockdown also sensitizes triple negative breast cancer cells to chemotherapeutics suggests BRG1 may be a valuable target in breast cancers where the normal frontline treatment has been focused on nuclear receptors. These observations highlight the fact that BRG1 plays critical roles in breast cancer biology independent nuclear receptors, and more focus should be placed on understanding these functions in future studies [33].
Glucocorticoid Receptor Function and Breast Cancer:
Nuclear hormone receptors elicit the transcriptional response to hormones and have well-established roles in many hormone-dependent and hormone-regulated cancer types. Breast cancers are commonly stratified by the expression status of Estrogen Receptor (ER) and PR, and the expression of AR and GR have also been demonstrated to play a role in the prognosis of breast cancers. Furthermore, ER, GR, and PR are heterogeneously expressed within individual tumors indicating that treatments targeting hormone receptors will have non-uniform effects on breast cancer cells [34–36]. As such, crosstalk between receptors has become the focus of many recent studies [37]. In breast cancer cells, antagonists of ER and PR are commonly used treatments, and activation of GR through glucocorticoid treatment is widely utilized as a co-treatment to alleviate side effects of chemotherapy. GR was also recently described to negatively modulate PR in breast cancer and this crosstalk may also be a key focus for therapeutics [38]. However, recent work has suggested that activation of GR promotes breast cancer metastasis, underscoring the need for more research into the function of GR in breast cancer progression [39].
GR expression is nearly ubiquitous in humans, however the transcriptional responses to glucocorticoids in different cells types are diverse and distinct. GR elicits a robust transcriptional response in both human and mouse breast cancer cell lines which has been broadly studied to elucidate mechanisms of GR activity. Mapping GR chromatin interactions in breast cancer cells has revealed that GR largely interacts with regions of chromatin that are at least partially accessible prior to hormone treatment [40–42]. This suggested that GR chromatin interactions are prefigured by other factors and/or epigenetic features of chromatin. Indeed, many GR binding sites are occupied by the BAF complex, pioneer factors such as FOXA1, and the enhancer-associated histone variant H2A.Z prior to hormone treatment [42–44]. These pre-patterning events were not independent, as GR preferentially bound to pioneer factor binding sites or to H2A.Z containing nucleosomes that were also enriched for the presence of BRG1 [42,44]. GR also interacted with other transcription factors including ER and PR, suggesting the transcriptional response to glucocorticoids in breast cancer is directed, in part, via the pre-patterning of GR chromatin interactions by many factors [45,46].
Intriguingly, GR binding also resulted in the recruitment of transcription factors and chromatin remodelers upon hormone treatment. Sites bound by GR exhibited increased enrichment of BRG1, pioneer factors, H2A.Z, and chromatin accessibility after hormone treatment. Thus, while GR bound to pre-patterned regions of chromatin, it also induced further chromatin remodeling and transcription factor recruitment [42–44]. GR also appeared to be capable of acting as a pioneer factor at some binding sites, as hormone treatment and GR binding resulted in new FOXA1 and ER binding sites, an indication of pioneer factor activity [40,43,45]. Furthermore, GR was also able to bind to some regions of the genome that did not appear to be pre-patterned by chromatin accessibility or the binding of other factors [47]. As such, there are multiple mechanisms of GR interaction with chromatin that can be classified by the chromatin and transcription factor environment in both untreated and hormone-treated breast cancer cells [42,44]. Understanding the mechanisms by which GR regulates transcription and its subsequent influences on ER and PR activity, is critical for enhancing the efficient clinical targeting of hormone signaling for breast cancer treatment.
Technological Advances toward Precision Medicine
Single cell genomic experiments such as single cell RNAseq (scRNA-seq) represent a new frontier in cancer and molecular genetics. During the last decade, single cell gene expression analysis has been widely used to interrogate gene expression signatures, tumor and cellular heterogeneity, and cell type compositions in many disease models [48]. In breast cancer models, these analyses have provided insights that may impact prognosis and treatment. For instance, scRNA-seq studies have demonstrated that heterogeneity in immune cell populations within tumors have expanded the diversity of breast cancer, demonstrating the contribution of immune cells to breast cancer phenotypes [49,50]. Indeed, scRNA-seq in primary triple negative breast cancer cells and single cell QRT-PCR in the luminal MCF7 cell line revealed that each population contained a subpopulation of stem-like cells with signatures associated with treatment/drug resistance and metastasis [51,52]. The transcriptional response to drug treatments in breast cancer cells was also demonstrated to be heterogeneous, as scRNA-seq in luminal T47D breast cancer cells treated with the synthetic glucocorticoid Dexamethasone revealed striking levels of heterogeneity [53]. Taken together, these single cell transcriptomic experiments demonstrated the importance in understanding how the heterogeneity of breast cancer cells contributes to disease progression and prognosis (Figure 1).
Figure 1: Levels of Heterogeneity in Human Breast Cancer.
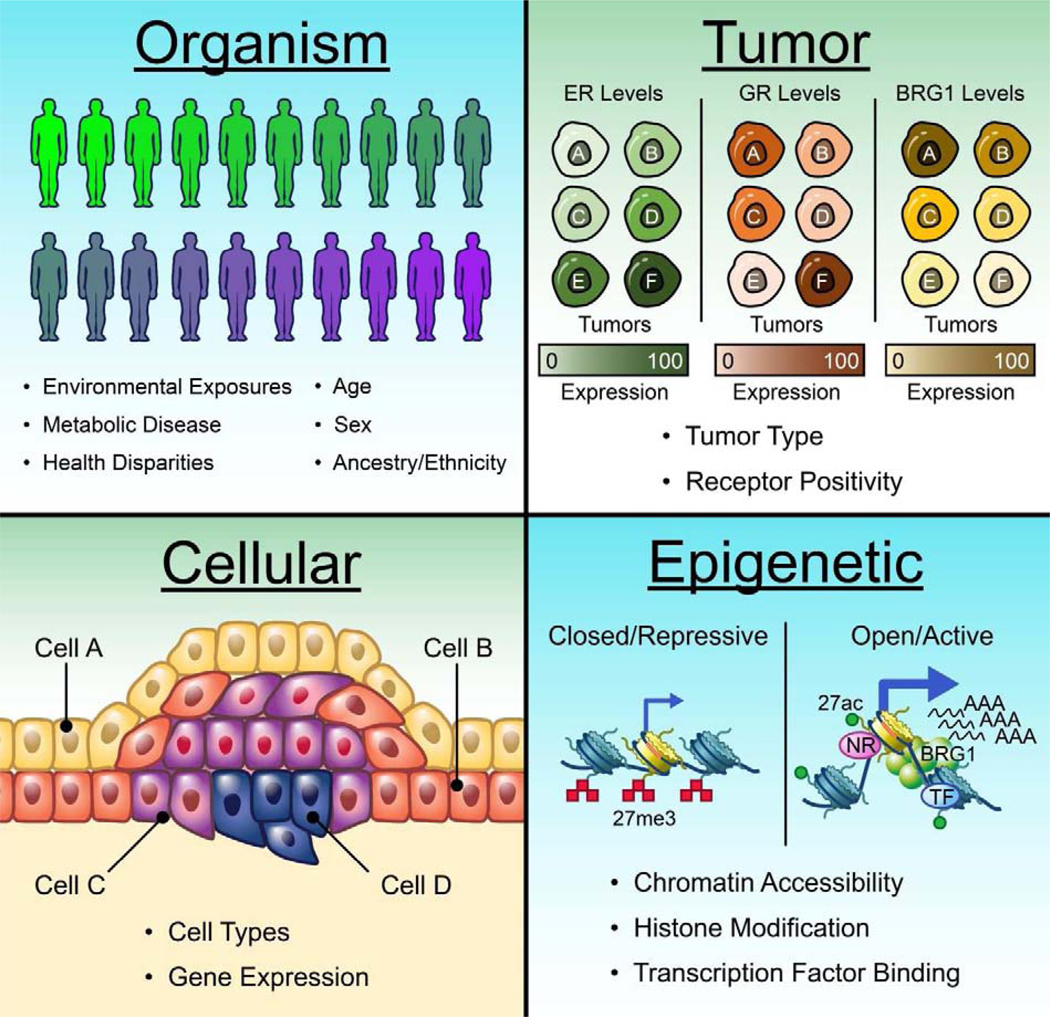
Organisms: many factors contribute to the heterogeneity observed between individuals. Differential environmental exposures, metabolic diseases, health disparities, age, sex, and ancestry/ethnicity all have impacts on breast cancer progression, prognosis, and treatment. Tumors: distinct tumor types and grades exhibit heterogeneous characteristics including the status of hormone receptor expression/activity as well as the expression of other factors and mutation burden. Cellular: the diverse cell types of mammary tissue give rise to different types of tumors. Furthermore, heterogeneity between and within different cell types in individual tumors confound treatment strategies targeting the activity of specific genes/proteins. Epigenetics: heterogeneity in the chromatin landscape and transcription factor interactions result in further cellular heterogeneity within tumors. These levels of heterogeneity are likely inter-related and further understanding of each level will be critical for improving breast cancer treatment strategies.
Beyond scRNA-seq, other single cell assays have provided critical insight into the interplay between tumor and immune cell populations. Single cell proteomics using mass cytometry has generated an atlas of the relationship between tumor and immune cells in breast cancer [54]. This experiment revealed that the phenotypes of tumor and immune cells within breast cancers and the relationships between these cell types stratified patients and underscored the importance of characterization of the tumor ecosystem. Such advances in the understanding of the patient-specific heterogeneity in the tumor ecosystem are critical to inform breast cancer treatments and facilitate the use of precision medicine.
Recent technological advances have also paved the way for examination of the epigenetic landscape in single cells. Single cell ChIP-seq has been used to examine the profiles of active and repressive histone modifications in breast cancer cells [55]. As with scRNA-seq, these experiments revealed a subpopulation of cells that shared a common chromatin signature with drug-resistant cells. Single cell analysis of chromatin accessibility has been performed across a variety of adult mouse tissues, in developing mouse mammary glands, and in mouse breast cancer models [56–58]. These studies demonstrate the potential for the analysis of epigenetic characteristics in single cells as a tool for interrogating the regulatory mechanisms underlying the heterogeneity observed in the transcriptomic profile of breast cancer cells. Furthermore, various “multi-omic” single cell experiments have been developed to simultaneously perform combinations of transcriptomic, epigenomic, and proteomic assays, providing great promise for future experiments [59,60]. Such interrogation of the epigenetic and transcriptomic environment in single cells has the potential to provide novel insight into the role of GR (as well as ER and PR) and the SWI/SNF complex in breast cancer. Recognizing and understanding cellular heterogeneity in breast cancer will in turn inform clinical treatments based on inter-tumor heterogeneity and provide new tools and targets for patient-specific precision medicine.
Acknowledgements
We thank the members of the Chromatin and Gene Expression Group and the Epigenetics and Stem Cell Biology Laboratory for their ongoing support. This research was supported by the Intramural Research Program of the National Institute of Environmental Health Sciences Z01 ES071006–20.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Aoyagi S, Trotter KW, Archer TK: ATP-dependent chromatin remodeling complexes and their role in nuclear receptor-dependent transcription in vivo. Vitam Horm 2005, 70:281–307. [DOI] [PubMed] [Google Scholar]
- 2.Wu Q, Lian JB, Stein JL, Stein GS, Nickerson JA, Imbalzano AN: The BRG1 ATPase of human SWI/SNF chromatin remodeling enzymes as a driver of cancer. Epigenomics 2017, 9:919–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kinyamu HK, Archer TK: Modifying chromatin to permit steroid hormone receptor-dependent transcription. Biochim Biophys Acta 2004, 1677:30–45. [DOI] [PubMed] [Google Scholar]
- 4.Fryer CJ, Archer TK: Chromatin remodelling by the glucocorticoid receptor requires the BRG1 complex. Nature 1998, 393:88–91. [DOI] [PubMed] [Google Scholar]
- 5.Huang ZQ, Li J, Sachs LM, Cole PA, Wong J: A role for cofactor-cofactor and cofactor-histone interactions in targeting p300, SWI/SNF and Mediator for transcription. Embo j 2003, 22:2146–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.King HA, Trotter KW, Archer TK: Chromatin remodeling during glucocorticoid receptor regulated transactivation. Biochim Biophys Acta 2012, 1819:716–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gadaleta RM, Magnani L: Nuclear receptors and chromatin: an inducible couple. J Mol Endocrinol 2014, 52:R137–149. [DOI] [PubMed] [Google Scholar]
- 8.Green CD, Han JD: Epigenetic regulation by nuclear receptors. Epigenomics 2011, 3:59–72. [DOI] [PubMed] [Google Scholar]
- 9.Kishimoto M, Fujiki R, Takezawa S, Sasaki Y, Nakamura T, Yamaoka K, Kitagawa H, Kato S: Nuclear receptor mediated gene regulation through chromatin remodeling and histone modifications. Endocr J 2006, 53:157–172. [DOI] [PubMed] [Google Scholar]
- 10.Wang W, Cote J, Xue Y, Zhou S, Khavari PA, Biggar SR, Muchardt C, Kalpana GV, Goff SP, Yaniv M, et al. : Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. Embo j 1996, 15:5370–5382. [PMC free article] [PubMed] [Google Scholar]
- 11.Wang W, Xue Y, Zhou S, Kuo A, Cairns BR, Crabtree GR: Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev 1996, 10:2117–2130. [DOI] [PubMed] [Google Scholar]
- 12.Hodges C, Kirkland JG, Crabtree GR: The Many Roles of BAF (mSWI/SNF) and PBAF Complexes in Cancer. Cold Spring Harb Perspect Med 2016, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ho L, Ronan JL, Wu J, Staahl BT, Chen L, Kuo A, Lessard J, Nesvizhskii AI, Ranish J,Crabtree GR: An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc Natl Acad Sci U S A 2009, 106:5181–5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang X, Li B, Li W, Ma L, Zheng D, Li L, Yang W, Chu M, Chen W, Mailman RB, et al. : Transcriptional repression by the BRG1-SWI/SNF complex affects the pluripotency of human embryonic stem cells. Stem Cell Reports 2014, 3:460–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alpsoy A, Dykhuizen EC: Glioma tumor suppressor candidate region gene 1 (GLTSCR1) and its paralog GLTSCR1-like form SWI/SNF chromatin remodeling subcomplexes. J Biol Chem 2018, 293:3892–3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. He S, Wu Z, Tian Y, Yu Z, Yu J, Wang X, Li J, Liu B, Xu Y: Structure of nucleosome-bound human BAF complex. Science 2020, 367:875–881.32001526 This structural study of the human BAF complex provides insight into how the complex is organized for binding to chromatin to allow for remodeling activity. The study provides a model for future studies of BAF complexing binding and function across mammalian cells.
- 17.Patel AB, Moore CM, Greber BJ, Luo J, Zukin SA, Ranish J, Nogales E: Architecture of the chromatin remodeler RSC and insights into its nucleosome engagement. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ye Y, Wu H, Chen K, Clapier CR, Verma N, Zhang W, Deng H, Cairns BR, Gao N, Chen Z: Structure of the RSC complex bound to the nucleosome. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wagner FR, Dienemann C, Wang H, Stützer A, Tegunov D, Urlaub H, Cramer P: Structure of SWI/SNF chromatin remodeller RSC bound to a nucleosome. Nature 2020, 579:448–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kadoch C, Hargreaves DC, Hodges C, Elias L, Ho L, Ranish J, Crabtree GR: Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat Genet 2013, 45:592–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mashtalir N, D’Avino AR, Michel BC, Luo J, Pan J, Otto JE, Zullow HJ, McKenzie ZM, Kubiak RL, St Pierre R, et al. : Modular Organization and Assembly of SWI/SNF Family Chromatin Remodeling Complexes. Cell 2018, 175:1272–1288.e1220.30343899 This study provided critical insights into the assembly of the mammalian SWI/SNF complex, one of the major barriers in the evolving understanding of SWI/SNF function. The authors also characterized disease-associated mutations and how these mutations alter complex organization, providing a model for how these mutations may contribute to disease phenotypes.
- 22.Pan J, McKenzie ZM, D’Avino AR, Mashtalir N, Lareau CA, St Pierre R, Wang L, Shilatifard A, Kadoch C: The ATPase module of mammalian SWI/SNF family complexes mediates subcomplex identity and catalytic activity-independent genomic targeting. Nat Genet 2019, 51:618–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakayama RT, Pulice JL, Valencia AM, McBride MJ, McKenzie ZM, Gillespie MA, Ku WL,Teng M, Cui K, Williams RT, et al. : SMARCB1 is required for widespread BAF complex-mediated activation of enhancers and bivalent promoters. Nat Genet 2017, 49:1613–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guerrero-Martínez JA, Reyes JC: High expression of SMARCA4 or SMARCA2 is frequently associated with an opposite prognosis in cancer. Scientific Reports 2018, 8:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schick S, Rendeiro AF, Runggatscher K, Ringler A, Boidol B, Hinkel M, Májek P, Vulliard L,Penz T, Parapatics K, et al. : Systematic characterization of BAF mutations provides insights into intracomplex synthetic lethalities in human cancers. Nature Genetics 2019, 51:1399–1410.31427792 The authors systematically characterized how the loss of individual SWI/SNF subunits alter chromatin structure and gene expression. These studies give insight into how different subcomplexes function and offers approaches for therapeutics focused on cancers driven by mutations in SWI/SNF proteins.
- 26.Bai J, Mei P, Zhang C, Chen F, Li C, Pan Z, Liu H, Zheng J: BRG1 is a prognostic marker and potential therapeutic target in human breast cancer. PLoS One 2013, 8:e59772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jubierre L, Soriano A, Planells-Ferrer L, Paris-Coderch L, Tenbaum SP, Romero OA,Moubarak RS, Almazan-Moga A, Molist C, Roma J, et al. : BRG1/SMARCA4 is essential for neuroblastoma cell viability through modulation of cell death and survival pathways. Oncogene 2016, 35:5179–5190. [DOI] [PubMed] [Google Scholar]
- 28.Watanabe T, Semba S, Yokozaki H: Regulation of PTEN expression by the SWI/SNF chromatin-remodelling protein BRG1 in human colorectal carcinoma cells. British Journal of Cancer 2010, 104:146–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin S, Jiang T, Ye L, Han Z, Liu Y, Liu C, Yuan C, Zhao S, Chen J, Wang J, et al. : The chromatin-remodeling enzyme BRG1 promotes colon cancer progression via positive regulation of WNT3A. Oncotarget 2016, 7:86051–86063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu X, Tian X, Wang F, Ma Y, Kornmann M, Yang Y: BRG1 promotes chemoresistance of pancreatic cancer cells through crosstalking with Akt signalling. Eur J Cancer 2014, 50:2251–2262. [DOI] [PubMed] [Google Scholar]
- 31.Sobczak M, Pitt AR, Spickett CM, Robaszkiewicz A: PARP1 Co-Regulates EP300-BRG1-Dependent Transcription of Genes Involved in Breast Cancer Cell Proliferation and DNA Repair. Cancers (Basel) 2019, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sobczak M, Pietrzak J, Ploszaj T, Robaszkiewicz A: BRG1 Activates Proliferation and Transcription of Cell Cycle-Dependent Genes in Breast Cancer Cells. Cancers (Basel) 2020, 12. Using breast cancer lines, the authors demonstrated that BRG1 binds to the promoters of cell cycle related genes and regulates their expression. These efforts also show BRG1 prevents assembly of repressive complexes at the promoters of these genes.
- 33.Wu Q, Sharma S, Cui H, LeBlanc SE, Zhang H, Muthuswami R, Nickerson JA, ImbalzanoAN: Targeting the chromatin remodeling enzyme BRG1 increases the efficacy of chemotherapy drugs in breast cancer cells. Oncotarget 2016, 7:27158–27175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bardou VJ, Arpino G, Elledge RM, Osborne CK, Clark GM: Progesterone receptor status significantly improves outcome prediction over estrogen receptor status alone for adjuvant endocrine therapy in two large breast cancer databases. J Clin Oncol 2003, 21:1973–1979. [DOI] [PubMed] [Google Scholar]
- 35.Belova L, Delgado B, Kocherginsky M, Melhem A, Olopade OI, Conzen SD: Glucocorticoid receptor expression in breast cancer associates with older patient age. Breast Cancer Res Treat 2009, 116:441–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harvey JM, Clark GM, Osborne CK, Allred DC: Estrogen receptor status by immunohistochemistry is superior to the ligand-binding assay for predicting response to adjuvant endocrine therapy in breast cancer. J Clin Oncol 1999, 17:1474–1481. [DOI] [PubMed] [Google Scholar]
- 37.Truong TH, Lange CA: Deciphering Steroid Receptor Crosstalk in Hormone-Driven Cancers. Endocrinology 2018, 159:3897–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ogara MF, Rodriguez-Segui SA, Marini M, Nacht AS, Stortz M, Levi V, Presman DM, Vicent GP, Pecci A: The glucocorticoid receptor interferes with progesterone receptor-dependent genomic regulation in breast cancer cells. Nucleic Acids Res 2019, 47:10645–10661.31598691 The authors show GR and PR can form in the same complex and bind to several regions at the same time in breast cancer cells. These studies indicated that GR can inhibit the gene expression changes mediated by PR activation that are normally associated with cell proliferation. The results confirm the importance of understanding crosstalk between nuclear receptors in cancer cells.
- 39. Obradović MMS, Hamelin B, Manevski N, Couto JP, Sethi A, Coissieux M-M, Münst S, Okamoto R, Kohler H, Schmidt A, et al. : Glucocorticoids promote breast cancer metastasis. Nature 2019, 567:540–544.30867597 An multi-omics based approach reveals that activation of Glucocorticoid signaling may promote breast cancer metastatis. This study demonstrates the urgent need for further examination of the role of GR in breast cancer, as Glucocorticoids are commonly used as a co-treatment in breast cancer patients.
- 40.John S, Sabo PJ, Thurman RE, Sung MH, Biddie SC, Johnson TA, Hager GL, Stamatoyannopoulos JA: Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat Genet 2011, 43:264–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Burd CJ, Archer TK: Chromatin architecture defines the glucocorticoid response. Mol Cell Endocrinol 2013, 380:25–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hoffman JA, Trotter KW, Ward JM, Archer TK: BRG1 governs glucocorticoid receptor interactions with chromatin and pioneer factors across the genome. Elife 2018, 7. Genomic analysis of GR, BRG1, pioneer factors, and the chromatin landscape in human luminal breast cancer cell line. This work demonstrates that BRG1 is required for the proper transcriptional response to Dexamethasone and that BRG1 plays a role in establishing chromatin environments that specify the interactions of GR with chromatin and pioneer factors.
- 43.Swinstead EE, Miranda TB, Paakinaho V, Baek S, Goldstein I, Hawkins M, Karpova TS, Ball D, Mazza D, Lavis LD, et al. : Steroid Receptors Reprogram FoxA1 Occupancy through Dynamic Chromatin Transitions. Cell 2016, 165:593–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Johnson TA, Chereji RV, Stavreva DA, Morris SA, Hager GL, Clark DJ: Conventional and pioneer modes of glucocorticoid receptor interaction with enhancer chromatin in vivo. Nucleic Acids Res 2018, 46:203–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miranda TB, Voss TC, Sung MH, Baek S, John S, Hawkins M, Grontved L, Schiltz RL,Hager GL: Reprogramming the chromatin landscape: interplay of the estrogen and glucocorticoid receptors at the genomic level. Cancer Res 2013, 73:5130–5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scheschowitsch K, Leite JA, Assreuy J: New Insights in Glucocorticoid Receptor Signaling—More Than Just a Ligand-Binding Receptor. Front Endocrinol (Lausanne) 2017, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Klemm SL, Shipony Z, Greenleaf WJ: Chromatin accessibility and the regulatory epigenome. Nat Rev Genet 2019, 20:207–220. [DOI] [PubMed] [Google Scholar]
- 48.Gonzalez-Silva L, Quevedo L, Varela I: Tumor Functional Heterogeneity Unraveled by scRNA-seq Technologies. Trends Cancer 2020, 6:13–19. [DOI] [PubMed] [Google Scholar]
- 49.Azizi E, Carr AJ, Plitas G, Cornish AE, Konopacki C, Prabhakaran S, Nainys J, Wu K,Kiseliovas V, Setty M, et al. : Single-Cell Map of Diverse Immune Phenotypes in the Breast Tumor Microenvironment. Cell 2018, 174:1293–1308.e1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chung W, Eum HH, Lee HO, Lee KM, Lee HB, Kim KT, Ryu HS, Kim S, Lee JE, Park YH, et al. : Single-cell RNA-seq enables comprehensive tumour and immune cell profiling in primary breast cancer. Nat Commun 2017, 8:15081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Karaayvaz M, Cristea S, Gillespie SM, Patel AP, Mylvaganam R, Luo CC, Specht MC,Bernstein BE, Michor F, Ellisen LW: Unravelling subclonal heterogeneity and aggressive disease states in TNBC through single-cell RNA-seq. Nat Commun 2018, 9:3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Prieto-Vila M, Usuba W, Takahashi RU, Shimomura I, Sasaki H, Ochiya T, Yamamoto Y: Single-Cell Analysis Reveals a Preexisting Drug-Resistant Subpopulation in the Luminal Breast Cancer Subtype. Cancer Res 2019, 79:4412–4425. [DOI] [PubMed] [Google Scholar]
- 53. Hoffman JA, Papas BN, Trotter KW, Archer TK: Single-cell RNA sequencing reveals a heterogeneous response to Glucocorticoids in breast cancer cells. Commun Biol 2020, 3:126.32170217 Single-cell RNAseq analysis of a human luminal breast cancer cell line over a time course of Dexamethasone treatments. This work reveals that there is a striking degree of heterogeneity in the cellular response to hormone that varies over time.
- 54. Wagner J, Rapsomaniki MA, Chevrier S, Anzeneder T, Langwieder C, Dykgers A, Rees M,Ramaswamy A, Muenst S, Soysal SD, et al. : A Single-Cell Atlas of the Tumor and Immune Ecosystem of Human Breast Cancer. Cell 2019, 177:1330–1345.e1318.30982598 Single-cell proteomic analysis of tumor and immune cells in human breast cancer. This work examines the phenotypic diversity of individual tumors and demonstrates the importance of characterizing the tumor and immune ecosystem for precision medicine.
- 55. Grosselin K, Durand A, Marsolier J, Poitou A, Marangoni E, Nemati F, Dahmani A, Lameiras S, Reyal F, Frenoy O, et al. : High-throughput single-cell ChIP-seq identifies heterogeneity of chromatin states in breast cancer. Nat Genet 2019, 51:1060–1066.31152164 Single cell ChIP-seq for the active and repressive histone marks in patient-derived xenograft models of human breast cancer. This work shows that a subset of cells within treatment-sensitive tumors exhibit chromatin signatures similar to treatment-resistant tumor cells, revealing a new layer of intra-tumoral heterogeneity.
- 56.Cusanovich DA, Hill AJ, Aghamirzaie D, Daza RM, Pliner HA, Berletch JB, Filippova GN,Huang X, Christiansen L, DeWitt WS, et al. : A Single-Cell Atlas of In Vivo Mammalian Chromatin Accessibility. Cell 2018, 174:1309–1324.e1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chung CY, Ma Z, Dravis C, Preissl S, Poirion O, Luna G, Hou X, Giraddi RR, Ren B, Wahl GM: Single-Cell Chromatin Analysis of Mammary Gland Development Reveals Cell-State Transcriptional Regulators and Lineage Relationships. Cell Rep 2019, 29:495–510.e496.31597106 Examination of the chromatin landscape in fetal mouse mammary cells by single-cell ATACseq. This work demonstrates the utility of scATAC-seq in clustering mammary cells into basal and luminal clusters, and suggests that scATAC-seq could provide a novel approach for the identification of targets for breast cancer treatments.
- 58.Chen X, Miragaia RJ, Natarajan KN, Teichmann SA: A rapid and robust method for single cell chromatin accessibility profiling. Nat Commun 2018, 9:5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reyes M, Billman K, Hacohen N, Blainey PC: Simultaneous Profiling of Gene Expression and Chromatin Accessibility in Single Cells. Advanced Biosystems 2019, 3:1900065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hu Y, An Q, Sheu K, Trejo B, Fan S, Guo Y: Single Cell Multi-Omics Technology: Methodology and Application. Front Cell Dev Biol 2018, 6:28. [DOI] [PMC free article] [PubMed] [Google Scholar]


