Abstract
Following the outbreak and subsequent pandemic of coronavirus disease 2019 (COVID-19), clinical diagnostic laboratories worldwide sought accurate and reliable testing methodologies. However, many laboratories were and still are hindered by a number of factors, including an unprecedented demand for testing, reagent and laboratory supply shortages and availability of qualified staff. To respond to these concerns, two separate laboratory-developed tests were validated for detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) using two different specimen types. In addition, these assays target different genomic regions of SARS-CoV-2, allowing for viral detection and mitigating genetic variation. Lower limit of detection and clinical evaluation studies showed detection of SARS-CoV-2 at 500 cp/mL with nasopharyngeal and saliva samples. These multiplexed RT-qPCR assays, although based on modified CDC, New York State Department of Health, and World Health Organization Emergency Use Authorization tests, allow for higher throughput and rapid turnaround time, benefiting patients, clinicians, and communities as a whole. These cost-effective tests also use readily obtainable reagents, circumventing commercial assay supply chain issues. The laboratory-developed tests described here have improved patient care and are highly adaptable should the need arise at other clinical diagnostic laboratories. Furthermore, the foundation and design of these assays may be modified in the future for detection of COVID-19 variants or other RNA-based viral detection tests.
Following the first reports of a novel coronavirus disease from the city of Wuhan in the Hubei province of China on December 31, 2019, and subsequent World Health Organization designation as a global pandemic, clinical diagnostic laboratories around the world had to rapidly adapt and modify protocols required to detect the causative agent.1 , 2 Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is an approximately 30 kb positive-sense, single-stranded, enveloped, pneumotropic RNA beta-coronavirus. The SARS-CoV-2 genome encodes 31 proteins, including 16 nonstructural proteins (resulting from the proteolytic cleavage of the two large polyproteins ORF1a and ORF1b), 11 accessory proteins, and four structural proteins, including two glycoproteins (spike and membrane), an envelope protein, and a nucleocapsid protein.3, 4, 5 The clinical presentation of SARS-CoV-2 infection ranges from mild or moderate “flu-like” symptoms to severe respiratory and multiorgan failure.6 Although other beta-coronaviruses such as lineage A OC43 and HKU1 often result in only mild to moderate symptoms, lineage B SARS-CoV and SARS-CoV-2, in addition to lineage C MERS-CoV, can cause exacerbated symptoms and poor prognostic, and often fatal, outcomes in vulnerable populations.7 , 8
The global toll of the COVID-19 pandemic has resulted in >246 million diagnoses and >5.7 million deaths (WHO Situation Report, https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19---8-february-2022, last accessed Feburary 14, 2022). Although substantial advancements have been made in vaccination programs throughout much of the world, vaccine delivery in many countries continues to be hindered. In addition, therapeutic treatments are still in the early stages of development and delivery. Testing has now become a worldwide requirement and, as a result, accurate and timely molecular diagnostic tools are a necessity, with >100 nucleic acid–based US Food and Drug Administration (FDA) Emergency Use Authorization (EUA) tests now in use.9 , 10 This requirement for timely identification of individuals infected with SARS-CoV-2 is now a necessity, not only for triaging the care of infected individuals but also for isolation and contact tracing efforts aimed at slowing the spread of the pandemic. Although this magnitude and variety of testing have been a boon for identifying infected individuals and preventing SARS-CoV-2 transmission, many clinical laboratories throughout the world found themselves hindered by shortages in manufactured tests, supply chain issues, and a lack of qualified staff necessary to perform the assays.11 Furthermore, most clinical laboratories rely on commercially manufactured tests performed on companion instrumentation for routine viral detection in contrast to the laboratory method known as RT-qPCR, widely recognized as the most sensitive and specific means of detecting RNA viruses.
We sought to validate laboratory-developed RT-qPCR tests (LDTs) for detection of SARS-CoV-2 based on protocols from the New York State Department of Health and the National Reference Center for Respiratory Viruses at the Institut Pasteur in Paris, France. The goals were to validate modifications to these tests that increased the testing capacity and throughput, expanded validated sample types, and decreased labor and costs while still providing analytically sensitive and specific diagnostic tools to enhance patient care. Two independent nucleic acid–based LDTs were validated, both of which target separate genomic regions of SARS-CoV-2, and one of which can use two different patient specimen types.
The first assay, SARS-CoV-2 nucleocapsid LDT, uses RNA from nasopharyngeal specimens extracted with either the EZ1 Advanced XL instrument and the EZ1 Virus Mini Kit 2.0 (Qiagen, Germantown, MD) or, alternatively, the KingFisher Flex instrument (Thermo Fisher Scientific, Waltham, MA) and either the MagMAX Viral/Pathogen II KIT (Thermo Fisher Scientific) or the NucleoMag Virus kit (Macherey-Nagel, Düren, Germany). RT-qPCR for the SARS-CoV-2 nucleocapsid LDT is performed on the Applied Biosystems 7500 Fast Dx Real-Time PCR Instrument (Thermo Fisher Scientific) and analyzed by using Sequence Detection Software version 1.4.1 (Thermo Fisher Scientific). The second assay, SARS-CoV-2 RNA-dependent RNA polymerase (RdRp) LDT, allows for the use of nasopharyngeal or saliva specimens. RNA for this assay is extracted via the KingFisher Flex instrument using either of the aforementioned kits, and RT-qPCR is performed by using the CFX96 Real-Time PCR Detection System (Bio-Rad, Hercules, CA). Analysis is performed by using CFX Maestro Software version 1.1 (Bio-Rad).
Manual resulting for non-interfaced assays is a significant workload issue; an Excel 2016 (Microsoft Corporation, Redmond, WA) spreadsheet with Visual Basic for Applications–based macro-embedded scripts was therefore engineered to import and review all data after analyses from either platform (Supplemental Codes S1–S9). After a manual review of growth curves, use of these scripts automated the interpretation and reporting process. This allowed for patient results to be immediately reported to the laboratory information system.
Herein, we describe the laboratory studies used to evaluate the performance, analytical sensitivity/lower limit of detection (LoD), and specificity of these assays. Validation studies were performed and compared with three other EUAs authorized by the FDA (Table 1 ). The New York SARS-CoV-2 Real-time Reverse Transcriptase (RT)-PCR Diagnostic Panel (Wadsworth EUA) is a variation of the CDC 2019-Novel Coronavirus Real-Time RT-PCR Diagnostic Panel and was authorized by the FDA under an EUA on February 29, 2020 (http://www.fda.gov/media/135661/download, last accessed November 3, 2021). The Clarifi COVID-19 Test Kit (Quadrant Biosciences, Syracuse, NY) was authorized by the FDA under an EUA on September 22, 2020 (http://www.fda.gov/media/142376/download, last accessed November 3, 2021). A minority of specimens were compared with the TaqPath COVID-19 Combo Kit (TaqPath EUA; Thermo Fisher Scientific) and was authorized by the FDA under an EUA on March 13, 2020 (http://www.fda.gov/media/136113/download, last accessed November 3, 2021). In addition, two specimens described in this study were reflexed to the Simplexa COVID-19 Direct real-time RT-PCR kit (DiaSorin Molecular, Cypress, CA), which was authorized by the FDA under an EUA on March 20, 2020 (http://www.fda.gov/media/136286/download, last accessed November 3, 2021). Following the conclusion of these studies and submission for evaluation by the New York State Department of Health, we were able to implement both molecular tests, substantially improving our workflow, throughput, turnaround time, and overall patient care. Furthermore, the foundation of these nucleic acid–based LDTs may be used in the future should other genomic regions of SARS-CoV-2, or perhaps other RNA viruses, require targeting for accurate molecular diagnoses.
Table 1.
Details of LDTs and EUAs Describing Specimen Type, Extraction Methods, SARS-CoV-2 Genomic Targets, and RT-qPCR Method
| Assay | LDT/EUA | Specimen type | RNA extraction | SARS-CoV-2 genomic targets | RT-qPCR |
|---|---|---|---|---|---|
| SARS-CoV-2 nucleocapsid LDT | LDT | Nasopharyngeal | KingFisher Flex or EZ1 Advanced XL | N | Applied Biosystems 7500 Fast Dx Real-Time PCR Instrument |
| SARS-CoV-2 RdRp LDT | LDT | Nasopharyngeal and saliva | KingFisher Flex | RdRp | CFX96 |
| New York SARS-CoV-2 Real-time Reverse Transcriptase (RT)-PCR Diagnostic Panel | EUA | Nasopharyngeal | EZ1 Advanced XL | N | Applied Biosystems 7500 Fast Dx Real-Time PCR Instrument |
| Clarifi COVID-19 Test Kit EUA | EUA | Saliva | Zymo Research RNA Isolation Kit | RdRp | CFX96 |
| Thermo Fisher TaqPath COVID-19 Combo Kit EUA | EUA | Nasopharyngeal | KingFisher Flex | ORF1ab, S, N | Applied Biosystems 7500 Fast Dx Real-Time PCR Instrument |
| DiaSorin Molecular Simplexa COVID-19 | EUA | Nasopharyngeal | Liaison MDX | ORF1ab, S | Liaison MDX (DiaSorin Molecular) |
EUA, Emergency Use Authorization; LDT, laboratory-developed test; RdRp, RNA-dependent RNA polymerase; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Materials and Methods
SARS-CoV-2 Nucleocapsid LDT Primer and Probe Design
Sequences for both forward and reverse primers targeting two regions (N1 and N2) of the SARS-CoV-2 nucleocapsid (N) gene (http://www.ncbi.nlm.nih.gov/nuccore; accession number NC_045512.2) were obtained from the CDC 2019-Novel Coronavirus Real-time RT-PCR Panel (http://www.cdc.gov/coronavirus/2019-ncov/lab/virus-requests.html, last accessed November 3, 2021); the N1 probe sequence, 5′-FAM reporter dye, and the 3′-BHQ1 quencher were also procured from this panel. In the CDC protocol, all probes are labeled with a 5′-FAM reporter dye and a 3′-BHQ1 quencher, preventing multiplexing of all three reactions within the same well and limiting assay throughput. To permit multiplexing, the N1 probe was allowed to retain the 5′-FAM reporter dye and 3′-BHQ1 quencher, whereas the N2 probe and HRP probes were labeled with different reporter dyes and quenchers. The N2 probe includes a 5′-ABY reporter dye and a 3′-QSY quencher, and the sequence remains equivalent to that of the CDC design. Both the forward and reverse primer sequences for the human RNAse P (HRP) internal control were derived from the CDC panel (http://www.ncbi.nlm.nih.gov/nuccore; accession number NM_006413.5). However, the reverse primer (HRP-R-S3) was modified to restrict the amplification of any RNAse P genomic DNA by designing a primer that spans exons one and two.12 The HRP probe sequence design was also derived from this panel, but the 5′-FAM reporter dye was replaced with JUN, and the 3′-BHQ-1 quencher was replaced with QSY. All forward and reverse primers, as well as the N1 probe, were synthesized by Integrated DNA Technologies, Inc. (Coralville, IA). The N2-ABY and HRP-JUN probes were synthesized by Thermo Fisher Scientific (Table 2 ).
Table 2.
Primer and Probe Sets for Genomic Regions Targeted by the SARS-CoV-2 Nucleocapsid LDT and SARS-CoV-2 RdRp LDT
| Target | Name | Sequence | Nucleotide position |
|---|---|---|---|
| N | SARS-CoV-2 N1-F | 5′-GACCCCAAAATCAGCGAAAT-3′ | 28,287–28,306 |
| SARS-CoV-2 N1-R | 5′-TCTGGTTACTGCCAGTTGAATCTG-3′ | 28,335–28,358 | |
| SARS-CoV-2 N1-Probe | 5′-FAM-ACCCCGCATTACGTTTGGTGGACC-BHQ1-3′ | 28,309–28,332 | |
| SARS-CoV-2 N2-F | 5′-TTACAAACATTGGCCGCAAA-3′ | 29,164–29,183 | |
| SARS-CoV-2 N2-R | 5′-GCGCGACATTCCGAAGAA-3′ | 29,213–29,230 | |
| SARS-CoV-2 N2-Probe | 5′-ABY-ACAATTTGCCCCCAGCGCTTCAG-QSY-3′ | 29,188–29,210 | |
| RdRp | nCoV_IP2-F | 5′-ATGAGCTTAGTCCTGTTG-3′ | 12,690–12,707 |
| nCoV_IP2-R | 5′-CTCCCTTTGTTGTGTTGT-3′ | 12,780–12,797 | |
| nCoV_IP2-Probe | 5′-HEX-AGATGTCTT/ZEN/GTGCTGCCGGTA-IABkFQ-3′ | 12,717–12,737 | |
| nCoV_IP4-F | 5′-GGTAACTGGTATGATTTCG-3′ | 14,080–14,098 | |
| nCoV_IP4-R | 5′-CTGGTCAAGGTTAATATAGG-3′ | 14,167–14,186 | |
| nCoV_IP4-Probe | 5′-HEX-TCATACAAA/ZEN/CCACGCCAGG-IABkFQ-3′ | 14,105–14,123 | |
| RNase P | Human RP-F | 5′-AGATTTGGACCTGCGAGCG-3′ | g.90,872,001_90,872,019 |
| Human RP-R-S3 | 5′-ATAGCAACAACTGAATAGCCAAGG-3′ | c.94_117 | |
| Human RP-Probe | 5′-JUN-TTCTGACCTGAAGGCTCTGCGCG-QSY-3′ | g.90,872,022_90,872,044 | |
| PPIA | PPIA-F | 5′-CATCCTAAAGCATACGGGTCC-3′ | g.44,799,776_44,799,796 |
| PPIA-R | 5′-TCTTTCACTTTGCCAAACACC-3′ | g.44,801,305_44,801,325 | |
| PPIA-Probe | 5′-Cy5-TGCTTGCCATCCAACCACTCAGTC-IAbRqSp-3′ | c.398_421 |
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) targets accession number NC_045512.2; RNaseP accession number NM_006413.5; peptidylprolyl isomerase A (PPIA) accession number NM_021130 (all available from https://www.ncbi.nlm.nih.gov/nuccore). Reference Genome Assembly: GRCh38/hg38 (available from https://www.ncbi.nlm.nih.gov/assembly).
LDT, laboratory-developed test; RdRp, RNA-dependent RNA polymerase.
SARS-CoV-2 RdRp LDT Primer and Probe Design
Primers and probes targeting two regions of the SARS-CoV-2 RNA-dependent RNA polymerase (RdRp) gene (accession number NC_045512.2) (IP2 and IP4) were designed by the National Reference Center for Respiratory Viruses at the Institut Pasteur (https://www.who.int/docs/default-source/coronaviruse/real-time-rt-pcr-assays-for-the-detection-of-sars-cov-2-institut-pasteur-paris.pdf?sfvrsn=3662fcb6_2, last accessed November 3, 2021).13 Both probes targeting these regions use a 5′-HEX probe and a 3′-IABkFQ quencher. Peptidylprolyl isomerase A (PPIA) serves as an internal control, with primers and probes pre-designed by Integrated DNA Technologies, Inc., to detect all splice variants across exons 4 to 5 (http://www.ncbi.nlm.nih.gov/nuccore; accession number NM_021130). The peptidylprolyl isomerase A probe contains a 5′-Cy5 probe and a 3′-IAbRqSp quencher. All primers and probes were synthesized by Integrated DNA Technologies, Inc. (Table 2).
Specimen Collection
Nasopharyngeal specimens were collected by using flocked swabs [BD (Franklin Lakes, NJ) and Hardy Diagnostics [Santa Maria, CA] via standard methods and transferred to tubes containing 1 to 3 mL of universal transport media or viral transport media. All nasopharyngeal specimens were transported at room temperature and stored at 4°C for 72 hours or −80°C for >72 hours. Saliva specimens were collected by using the ORAcollect RNA collection device (DNA Genotek, Ottawa, ON, Canada). For the saliva samples, different regions of the mouth were self-swabbed under the supervision of a health care professional. Swabs were then transferred to the collection tube and mixed with stabilizing reagent. Upon specimen receipt, saliva swabs were incubated at 60°C for 2 hours to heat-inactivate SARS-CoV-2 and enhance protease activity, allowing for specimens to be handled outside of a biological safety cabinet. Saliva specimens were transported and stored at room temperature, where they are stable for up to 1 week as indicated by the manufacturer.
Using a program designed with a .NET framework, specimen label barcodes were scanned, and their accession number was linked to corresponding patient identifiers and information from the SUNY Upstate LIS system (Sunquest CoPathPlus version 6.3.0077; Sunquest Information Systems, Tucson, AZ). Patient batches were downloaded as a flat file (CSV), which was subsequently imported to a macro-based Excel 2016 worksheet (Microsoft Corporation). This macro-based Excel 2016 worksheet allowed for direct exportation of all thermal cycler setup information (Supplemental Tables S1 and S2).
RNA Extraction
RNA extracted from nasopharyngeal specimens on the EZ1 Advanced XL was performed with 120 μL of patient sample input and a 120 μL elution using the EZ1 Virus Mini Kit 2.0. Regardless of specimen type, extraction of RNA on the KingFisher Flex instrument used a 200 μL sample input and a 50 μL elution. KingFisher Flex RNA extractions were performed by using the MagMAX Viral/Pathogen II Kit or the NucleoMag Viral Kit according to manufacturers’ instructions. RNA extraction using the Zymo Research Quick-RNA Viral Kit (Irvine, CA) was performed according to the manufacturer’s instructions using a sample input of 100 μL and a 30 μL elution.
SARS-CoV-2 Nucleocapsid LDT RT-qPCR
Reverse transcription of RNA eluates, cDNA synthesis, and target amplification were performed by using an Applied Biosystems 7500 Fast Dx Real-Time PCR Instrument. For each reaction, 5 μL of RNA eluate was combined with 5 μL of 4× TaqPath 1-Step RT-qPCR Master Mix (Thermo Fisher Scientific), 1 μL 20× primer/probe mix, and 9 μL nuclease-free water. The 20× primer/probe mix was generated to allow for 0.9 μmol/L working concentrations of N1 and N2 forward and reverse primers, 0.25 μmol/L working concentrations of N1-FAM and N2-ABY probes, 0.1 μmol/L working concentrations of HRP forward and reverse primers, and a 0.1 μmol/L working concentration of the HRP-JUN probe. Cycling conditions were based on the New York State Department of Health Wadsworth EUA from March 7, 2020 [New York SARS-CoV-2 Real-time Reverse Transcriptase (RT)-PCR Diagnostic Panel], with some modifications: uracil N-glycosylase incubation at 25°C for 1 minute, reverse transcription incubation at 50°C for 15 minutes, enzyme activation at 95°C for 2 minutes, and 42 cycles of amplification at 95°C for 3 seconds and 60°C for 30 seconds. RNA extraction controls were prepared from human embryonic lung cells (ZeptoMetrix Corporation, Buffalo, NY) using the EZ1 Advanced XL instrument. Positive control was prepared from quantified, purified genomic 2019-nCoV/USA-WA1/2020 RNA (American Type Culture Collection, Manassas, VA) combined with human embryonic lung cell RNA eluate, allowing for a final working concentration of 5 cp/μL and detection of the N1, N2, and HRP targets simultaneously. No template control was composed of nuclease-free water used to generate the master mix.
SARS-CoV-2 RdRp LDT RT-qPCR
Using a CFX96 thermal cycler, RNA eluates were reverse transcribed to cDNA, and RdRp gene targets were amplified. For each reaction, 5 μL of RNA eluate was combined with 5 μL 4× Reliance Supermix (Bio-Rad), 1 μL 20× primer/probe mix, and 9 μL nuclease-free water. Then, 20× primer/probe mix was generated at stock concentrations resulting in working concentrations of 0.9 μmol/L IP2 and IP4 forward and reverse primers, 0.25 μmol/L IP2-HEX and IP4-HEX probes, 0.3 μmol/L PPIA forward and reverse primers, and 0.083 μmol/L of peptidylprolyl isomerase A–Cy5 probe. The cycling protocol included a reverse transcription cycle at 50°C for 20 minutes, an enzyme inactivation and polymerase activation cycle at 95°C for 10 minutes, and 45 cycles of amplification at 95°C for 10 seconds followed by 59°C for 20 seconds. The RNA extraction control, positive control, and no template control were prepared in the same manner as for the SARS-CoV-2 nucleocapsid assay.
Analysis
After completion of the SARS-CoV-2 nucleocapsid assay, the threshold was initially set at 1.0 × 104 relative fluorescence units. For most runs, this threshold was manually adjusted based on a fluorescence background. After analysis, cycle threshold (CT) values were exported as a flat file (CSV) and imported into the Excel 2016 worksheet (Supplemental Table S1). An automatic threshold of 500 relative fluorescence units for the HEX signal was set for the SARS-CoV-2 RdRp assay. CT values were also exported as a flat file (CSV) and imported into the worksheet (Supplemental Table S2). A CT value of 40 was used as a cutoff for positive samples in both LDTs. Based on the imported CT values for the internal control and the SARS-CoV-2 target gene, patient interpretations were generated. After manual review of the CT values and interpretations, results were exported to the laboratory information system via a macro-enabled Excel 2016 spreadsheet (Supplemental Tables S1 and S2).
Results
CDC-designed primers and probes targeting two regions of the SARS-CoV-2 nucleocapsid gene allow for qualitative detection of viral nucleic acid from nasopharyngeal specimens. Amplification of the 71 bp N1 target occurs 943 bp upstream from the 66 bp N2 amplification target. N1 detection occurs at an emission of 517 nm via a 5′-FAM reporter dye and a 3′-BHQ1 quencher, whereas N2 is detected at an emission of 580 nm using a 5′-ABY reporter dye and a 3′-QSY quencher.
Although CDC-designed primers and probes were used for qualitative detection of virus in nasopharyngeal specimens, primers and probes designed by the National Reference Center for Respiratory Viruses at the Institut Pasteur allowed for qualitative viral detection from both nasopharyngeal and saliva specimens. These primers and probes target two regions of the SARS-CoV-2 RdRp gene. The IP2 primers amplify a 108 bp target, whereas the IP4 primers amplify a 107 bp target located 1282 bp upstream from the IP2 target site. Probes designed with a 5′-HEX reporter dye and a 3′-IABkFQ quencher for both IP2 and IP4 allow for target detection at an emission of 559 nm.
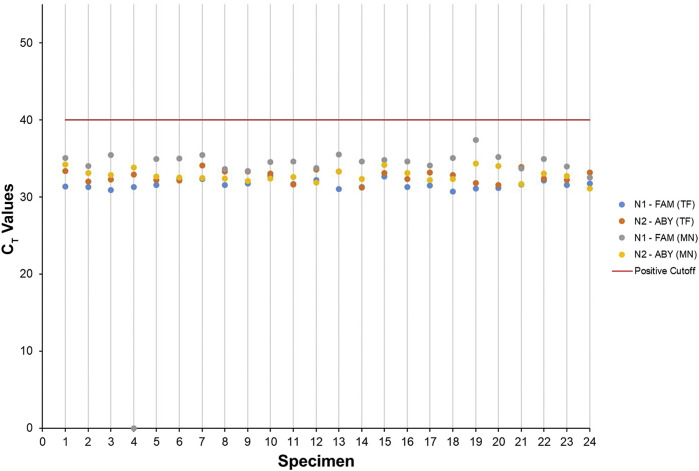
The analytical sensitivity of the SARS-CoV-2 nucleocapsid LDT was assessed by performing an LoD study using a CT value of 40 as a cutoff for positive specimens. This resulted in an LoD of 500 cp/mL for both the N1 and N2 gene targets using the MagMAX Viral/Pathogen II Kit and the KingFisher Flex in 100% (25 of 25) of nasopharyngeal specimens tested (Table 3 and Figure 1 ). In addition, in the same assay but using the NucleoMag Viral Kit, an LoD of 500 cp/mL was achieved in 96% (24 of 25) of nasopharyngeal specimens tested (Table 4 and Figure 1). Further testing was performed by using the EZ1 Advanced XL in conjunction with the EZ1 Virus Mini Kit 2.0. An LoD of 1000 cp/mL was achieved in 95% (19 of 20) of nasopharyngeal specimens tested (Table 5 ).
Table 3.
An LoD of 500 cp/mL Was Detected in 100% of Samples Extracted Using the KingFisher Flex Extraction Instrument and MagMAX Viral/Pathogen II Kit
| Specimen | CT values |
Interpretation | ||
|---|---|---|---|---|
| N1-FAM | N2-ABY | HRP-JUN | ||
| 1 | 31.37 | 33.39 | 25.17 | Positive |
| 2 | 31.30 | 32.00 | 28.53 | Positive |
| 3 | 30.92 | 32.26 | 28.79 | Positive |
| 4 | 31.28 | 32.95 | 26.89 | Positive |
| 5 | 31.55 | 32.23 | 27.76 | Positive |
| 6 | 32.32 | 32.14 | 30.22 | Positive |
| 7 | 32.35 | 34.06 | 26.24 | Positive |
| 8 | 31.57 | 33.32 | 32.76 | Positive |
| 9 | 31.77 | 33.32 | 27.26 | Positive |
| 10 | 32.70 | 33.03 | 30.85 | Positive |
| 11 | 31.67 | 31.61 | 28.10 | Positive |
| 12 | 32.22 | 33.60 | 29.46 | Positive |
| 13 | 31.05 | 33.29 | 26.91 | Positive |
| 14 | 31.28 | 31.26 | 32.10 | Positive |
| 15 | 32.64 | 33.10 | 30.32 | Positive |
| 16 | 31.32 | 32.34 | 28.41 | Positive |
| 17 | 31.47 | 33.18 | 25.42 | Positive |
| 18 | 30.71 | 32.86 | 26.21 | Positive |
| 19 | 31.08 | 31.79 | 28.09 | Positive |
| 20 | 31.19 | 31.58 | 28.92 | Positive |
| 21 | 31.61 | 33.89 | 29.09 | Positive |
| 22 | 32.12 | 32.39 | 38.00 | Positive |
| 23 | 31.53 | 32.23 | 28.69 | Positive |
| 24 | 31.72 | 33.17 | 28.16 | Positive |
| 25 | 32.01 | 33.17 | 29.60 | Positive |
| Mean CT Values | 31.63 | 32.73 | 28.88 | |
NATrol SARS-related Coronoavirus 2 (SARS-CoV-2) External Run Control (ZeptoMetrix) from SARS-CoV-2 isolate: USA-WA1/2020 was supplied at a concentration of 50,000 cp/mL and transferred to previously tested, SARS-CoV-2 negative nasopharyngeal specimen aliquots resulting in a final concentration of 500 cp/mL. Nasopharyngeal specimen aliquots were extracted using the KingFisher Flex extraction instrument and MagMAX Viral/Pathogen II kit and subsequently tested for SARS-CoV-2 using the SARS-CoV-2 Nucleocapsid LDT. SARS-CoV-2 was detected in 100% of extracted specimens at a limit of detection (LoD) of 500 cp/mL. All 25 samples are listed with their CT values and interpretation.
Figure 1.
Severe acute respiratory syndrome coronavirus 2 limit of detection studies performed on nasopharyngeal specimens confirmed detection of both N1 and N2 targets using the KingFisher Flex and two different extraction methods. CT values for each specimen from the limit of detection study are represented as a scatter plot comparing the MagMAX Viral/Pathogen II kit (TF) with the NucleoMag Viral kit (MN).
Table 4.
An LoD of 500 cp/mL Was Detected in 96% of Samples Extracted using the KingFisher Flex Extraction Instrument and NucleoMag Viral Kit
| Specimen | CT values |
Interpretation | ||
|---|---|---|---|---|
| N1-FAM | N2-ABY | HRP-JUN | ||
| 1 | 35.10 | 34.24 | 27.75 | Positive |
| 2 | 34.05 | 33.15 | 30.44 | Positive |
| 3 | 35.45 | 32.84 | 30.68 | Positive |
| 4 | 0.00 | 33.86 | 30.66 | Inconclusive |
| 5 | 34.93 | 32.64 | 29.54 | Positive |
| 6 | 35.03 | 32.53 | 32.40 | Positive |
| 7 | 35.48 | 32.47 | 30.91 | Positive |
| 8 | 33.63 | 32.40 | 35.07 | Positive |
| 9 | 33.35 | 32.08 | 29.02 | Positive |
| 10 | 34.53 | 32.43 | 29.67 | Positive |
| 11 | 34.64 | 32.61 | 29.59 | Positive |
| 12 | 33.79 | 31.91 | 31.69 | Positive |
| 13 | 35.53 | 33.29 | 29.28 | Positive |
| 14 | 34.59 | 32.37 | 32.53 | Positive |
| 15 | 34.78 | 34.18 | 31.68 | Positive |
| 16 | 34.64 | 33.10 | 30.11 | Positive |
| 17 | 34.11 | 32.21 | 28.32 | Positive |
| 18 | 35.04 | 32.32 | 28.31 | Positive |
| 19 | 37.38 | 34.33 | 29.77 | Positive |
| 20 | 35.21 | 34.06 | 31.42 | Positive |
| 21 | 33.71 | 31.70 | 30.51 | Positive |
| 22 | 34.91 | 33.08 | 35.31 | Positive |
| 23 | 33.94 | 32.71 | 29.57 | Positive |
| 24 | 32.56 | 31.09 | 28.70 | Positive |
| Mean CT Values | 34.63 | 32.82 | 30.54 | |
NATrol SARS-related Coronoavirus 2 (SARS-CoV-2) External Run Control (ZeptoMetrix) from SARS-CoV-2 isolate: USA-WA1/2020 was supplied at a concentration of 50,000 cp/mL and transferred to previously tested, SARS-CoV-2 negative nasopharyngeal specimen aliquots resulting in a concentration of 500 cp/mL. Nasopharyngeal aliquots were extracted using the KingFisher Flex Extraction instrument and NucleoMag Viral kit and subsequently tested for SARS-CoV-2 using the SARS-CoV-2 Nucleocapsid LDT. A limit of detection (LoD) of 500 cp/mL was detected in 96% of extracted specimens. All 24 samples are listed with their CT values and interpretation.
Table 5.
An LoD of 1000 cp/mL Was Detected in 96% of Samples Extracted Using an EZ1 Advanced XL Extraction Instrument and EZ1 Virus Mini Kit 2.0
| Specimen | CT values |
Interpretation | ||
|---|---|---|---|---|
| N1-FAM | N2-ABY | HRP-JUN | ||
| EZ1-1 | 35.74 | 35.70 | 38.04 | Positive |
| EZ1-2 | 35.11 | 36.83 | 22.51 | Positive |
| EZ1-3 | 34.78 | 34.43 | 28.13 | Positive |
| EZ1-4 | 35.08 | 34.04 | 26.08 | Positive |
| EZ1-5 | 34.38 | 35.58 | 30.80 | Positive |
| EZ1-6 | 0.00 | 34.25 | 27.45 | Inconclusive |
| EZ1-7 | 33.34 | 34.22 | 25.19 | Positive |
| EZ1-8 | 35.03 | 36.97 | 24.72 | Positive |
| EZ1-9 | 35.86 | 35.33 | 31.90 | Positive |
| EZ1-10 | 36.09 | 35.21 | 29.97 | Positive |
| EZ1-11 | 34.50 | 38.35 | 24.53 | Positive |
| EZ1-12 | 35.84 | 35.05 | 31.22 | Positive |
| EZ1-13 | 33.52 | 33.59 | 28.96 | Positive |
| EZ1-14 | 33.07 | 32.89 | 25.59 | Positive |
| EZ1-15 | 33.85 | 34.69 | 28.53 | Positive |
| EZ1-16 | 35.01 | 35.39 | 37.46 | Positive |
| EZ1-17 | 34.01 | 35.20 | 30.70 | Positive |
| EZ1-18 | 36.51 | 35.80 | 31.09 | Positive |
| EZ1-19 | 34.33 | 34.54 | 29.02 | Positive |
| EZ1-20 | 34.53 | 34.51 | 38.02 | Positive |
| Mean CT Values | 33.03 | 35.13 | 29.50 | |
NATtrol SARS-Related Coronavirus 2 (SARS-CoV-2) External Run Control (ZeptoMetrix Corporation) from severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) isolate: USA-WA1/2020 was supplied at a concentration of 50,000 cp/mL and transferred to nasopharyngeal specimen aliquots resulting in a concentration of 1000 cp/mL. Nasopharyngeal specimen aliquots were extracted and subsequently tested for SARS-CoV-2 by using the SARS-CoV-2 nucleocapsid laboratory-developed test. All 20 samples are listed with their CT values and interpretation.
LoD, limit of detection.
A clinical evaluation comparing the SARS-CoV-2 nucleocapsid LDT versus the Wadsworth EUA was performed by using RNA extracted from nasopharyngeal specimens on the EZ1 Advanced XL and the EZ1 Virus Mini Kit 2.0. Direct concordance for positive and negative samples was 90.3% (28 of 31) and 100% (31 of 31), respectively. Taken together, a concordance level of 95.2% was reached (Table 6 ). The original reported CT values for all three discordant positive samples in the Wadsworth EUA assay were >34. Two of these three specimens were tested by using the Simplexa COVID-19 Direct assay and found to be in agreement with the SARS-CoV-2 nucleocapsid LDT.
Table 6.
An Overall Concordance of Level of 95.2% Was Reached When Comparing the SARS-CoV-2 Nucleocapsid LDT with the Wadsworth EUA
| Wadsworth EUA results | N1 nucleocapsid LDT | N1 Wadsworth EUA | N2 nucleocapsid LDT | N2 Wadsworth EUA | HRP nucleocapsid LDT | HRP Wadsworth EUA | Concordance between SARS-CoV-2 nucleocapsid LDT and Wadsworth EUA |
|---|---|---|---|---|---|---|---|
| Early Ct (<30) | 12.71 | 14.28 | 10.99 | 14.61 | 17.37 | 20.44 | Concordant |
| 18.18 | 17.50 | 16.52 | 18.13 | 23.83 | 22.35 | Concordant | |
| 19.61 | 18.84 | 17.93 | 18.17 | 23.11 | 21.02 | Concordant | |
| 23.00 | 21.45 | 21.35 | 21.43 | 28.16 | 27.01 | Concordant | |
| 25.26 | 21.96 | 22.49 | 23.30 | 24.78 | 22.61 | Concordant | |
| 25.72 | 23.52 | 23.02 | 25.16 | 24.10 | 22.46 | Concordant | |
| 25.43 | 23.60 | 22.68 | 24.82 | 25.49 | 25.34 | Concordant | |
| 27.68 | 25.37 | 26.12 | 27.72 | 23.40 | 22.14 | Concordant | |
| 27.69 | 25.41 | 24.84 | 26.85 | 23.99 | 22.99 | Concordant | |
| 32.38 | 29.73 | 30.94 | 31.86 | 24.23 | 21.29 | Concordant | |
| Late Ct (>30) | 33.55 | 31.00 | 30.75 | 33.72 | 28.04 | 25.34 | Concordant |
| 31.41 | 31.14 | 30.39 | 30.74 | 27.74 | 23.68 | Concordant | |
| 32.91 | 31.49 | 31.88 | 34.44 | 24.74 | 25.16 | Concordant | |
| 32.70 | 31.61 | 32.03 | 32.32 | 21.77 | 22.13 | Concordant | |
| 33.38 | 31.81 | 32.89 | 35.74 | 23.24 | 23.53 | Concordant | |
| 34.28 | 31.82 | 32.35 | 32.09 | 22.86 | 22.07 | Concordant | |
| 33.84 | 32.09 | 30.51 | 35.06 | 24.29 | 24.02 | Concordant | |
| 34.21 | 32.10 | 32.86 | 32.40 | 22.47 | 23.03 | Concordant | |
| 33.13 | 32.35 | 28.78 | 31.52 | 25.22 | 26.03 | Concordant | |
| 34.64 | 32.39 | 32.35 | 31.84 | 24.67 | 23.41 | Concordant | |
| 36.55 | 32.62 | 33.56 | 32.65 | 24.56 | 22.46 | Concordant | |
| 34.15 | 33.23 | 32.86 | 34.00 | 23.31 | 21.12 | Concordant | |
| 34.25 | 33.45 | 31.21 | 36.95 | 23.28 | 22.21 | Concordant | |
| 34.16 | 33.82 | 33.33 | 33.22 | 22.92 | 23.47 | Concordant | |
| 35.21 | 34.13 | 32.87 | 35.08 | 23.84 | 21.58 | Concordant | |
| 36.54 | 34.54 | 34.26 | 33.74 | 34.30 | 33.31 | Concordant | |
| 32.60 | 34.67 | 31.43 | 33.00 | 21.96 | 22.56 | Concordant | |
| 35.46 | 35.09 | 32.77 | 33.92 | 25.35 | 25.08 | Concordant | |
| 0.00 | 35.76 | 39.87 | 38.36 | 22.22 | 20.94 | Discordant: Inconclusive via SARS-CoV-2 Nucleocapsid LDT; positive with Wadsworth EUA | |
| 0.00 | 35.97 | 0.00 | 0.00 | 23.35 | 21.34 | Discordant: Negative via SARS-CoV-2 nucleocapsid LDT; inconclusive via Wadsworth EUA; reported as negative from reflex testing via Simplexa COVID-19 Direct assay | |
| 37.44 | Not detected | 36.47 | 34.42 | 27.94 | 25.77 | Discordant: Inconclusive via Wadsworth EUA; positive via SARS-CoV-2 Nucleocapsid LDT; reported as positive from reflex testing via Simplexa COVID-19 Direct assay |
Nasopharyngeal patient specimens were extracted by using an EZ1 Advanced XL Instrument and the EZ1 Virus Mini Kit 2.0. CT values and concordance results are listed for each patient sample. The three discordant samples are provided with notes describing Wadsworth Emergency Use Authorization (EUA) results and any subsequent reflex testing via the Simplexa COVID-19 Direct assay.
HRP, human RNAse P; LDT, laboratory-developed test; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
An additional clinical evaluation was performed by comparing the SARS-CoV-2 nucleocapsid LDT versus the Wadsworth EUA assay by using two different nucleic acid extraction instruments. RNA was extracted from nasopharyngeal specimens using the KingFisher Flex and the MagMAX Viral/Pathogen II Kit for the SARS-CoV-2 nucleocapsid LDT, whereas the Wadsworth EUA assay used RNA extracted with the EZ1 Advanced XL and the EZ1 Virus Mini Kit 2.0. A total of 69 nasopharyngeal specimens (37 positive samples and 32 negative samples) were compared, and an overall concordance rate of 95.6% was achieved. A 100% concordance (32 of 32) was reached with negative nasopharyngeal specimens, whereas a 91.89% concordance rate (34 of 37) was achieved with positive nasopharyngeal specimens (Table 7 ). The three discordant samples were subsequently retested by using the TaqPath EUA and found to be negative.
Table 7.
An Overall Concordance Level of 91.89% Was Achieved When Comparing the SARS-CoV-2 Nucleocapsid LDT with the Wadsworth EUA
| Wadsworth EUA results | N1 nucleocapsid LDT | N1 Wadsworth EUA | N2 nucleocapsid LDT | N2 Wadsworth EUA | HRP nucleocapsid LDT | HRP Wadsworth EUA | Concordance between SARS-CoV-2 nucleocapsid LDT and Wadsworth EUA | Notes |
|---|---|---|---|---|---|---|---|---|
| Early Ct (<30) | 13.61 | 13.20 | 10.32 | 12.23 | 12.30 | 22.71 | Concordant | |
| Positives | 14.22 | 13.77 | 12.20 | 13.14 | 14.11 | 20.93 | Concordant | |
| 14.51 | 14.03 | 13.08 | 13.31 | 15.08 | 21.67 | Concordant | ||
| 16.16 | 16.53 | 14.85 | 15.02 | 19.86 | 25.95 | Concordant | ||
| 16.40 | 16.86 | 15.23 | 16.59 | 20.37 | 25.32 | Concordant | ||
| 17.93 | 18.14 | 17.58 | 17.57 | 19.60 | 22.22 | Concordant | ||
| 18.62 | 19.56 | 16.75 | 17.38 | 22.78 | 24.15 | Concordant | ||
| 20.71 | 22.52 | 20.79 | 21.06 | 20.21 | 22.12 | Concordant | ||
| 23.09 | 24.32 | 21.03 | 23.77 | 24.71 | 25.33 | Concordant | ||
| 24.01 | 24.61 | 21.97 | 23.00 | 25.66 | 26.02 | Concordant | ||
| 23.47 | 24.56 | 21.50 | 23.38 | 23.44 | 24.47 | Concordant | ||
| 26.63 | 25.63 | 24.82 | 24.16 | 27.46 | 25.28 | Concordant | ||
| 24.62 | 27.28 | 22.80 | 25.64 | 26.79 | 20.19 | Concordant | ||
| 25.01 | 29.63 | 23.26 | 28.24 | 27.52 | 25.86 | Concordant | ||
| 27.21 | 29.87 | 27.25 | 31.19 | 23.89 | 24.84 | Concordant | ||
| Late Ct (>30) | 31.10 | 30.76 | 28.52 | 32.09 | 28.45 | 26.93 | Concordant | |
| Positives | 31.20 | 30.81 | 32.82 | 30.67 | 20.43 | 19.69 | Concordant | |
| 27.37 | 31.35 | 27.60 | 30.01 | 27.02 | 26.07 | Concordant | ||
| 0.00 | 31.38 | 0.00 | 30.70 | 21.72 | 20.68 | Discordant | Negative in TaqPath EUA with KingFisher Flex Extraction | |
| 29.84 | 31.54 | 30.55 | 32.27 | 22.59 | 22.83 | Concordant | ||
| 29.45 | 31.82 | 29.22 | 31.14 | 28.22 | 27.37 | Concordant | ||
| 28.83 | 31.82 | 29.95 | 31.43 | 22.09 | 23.57 | Concordant | ||
| 32.17 | 32.03 | 31.41 | 30.25 | 20.42 | 22.13 | Concordant | ||
| 34.08 | 32.06 | 32.49 | 32.51 | 33.85 | 28.47 | Concordant | ||
| 30.43 | 32.44 | 31.42 | 34.39 | 22.82 | 22.30 | Concordant | ||
| 29.36 | 32.87 | 27.27 | 31.59 | 22.24 | 23.14 | Concordant | ||
| 34.50 | 32.89 | 34.94 | 35.17 | 24.30 | 26.15 | Concordant | ||
| 30.04 | 33.35 | 29.96 | 32.29 | 25.09 | 26.14 | Concordant | ||
| 0.00/0.00 | 34.00 | 0.00/0.00 | 35.89 | 22.24/21.62 | 21.77 | Discordant X2 | Negative in TaqPath EUA with KingFisher Flex Extraction | |
| 31.09 | 34.09 | 31.21 | 35.82 | 28.51 | 28.00 | Concordant | ||
| 33.87 | 34.58 | 0.00 | 33.62 | 22.92 | 24.19 | Discordant | Negative in TaqPath EUA with KingFisher Flex Extraction | |
| 32.95 | 35.05 | 36.34 | 35.12 | 25.81 | 27.29 | Concordant | ||
| 31.74 | 35.07 | 31.46 | 35.69 | 24.55 | 27.44 | Concordant | ||
| 33.84 | 35.11 | 31.65 | 34.78 | 28.27 | 30.11 | Concordant | ||
| 34.07 | 36.17 | 31.81 | 31.33 | 32.17 | 27.11 | Concordant | ||
| 32.87 | 35.52 | 36.40 | 34.41 | 22.23 | 24.22 | Concordant | ||
| 33.42 | 36.63 | 32.07 | 34.21 | 29.01 | 27.57 | Concordant |
A clinical evaluation was performed comparing the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) nucleocapsid laboratory-developed test (LDT) with the Wadsworth Emergency Use Authorization (EUA). RNA was extracted by using the KingFisher Flex Extraction instrument with the MagMAX Viral/Pathogen II kit for the SARS-CoV-2 nucleocapsid LDT. RNA extraction for the Wadsworth EUA was performed with the EZ1 Advanced XL instrument and the EZ1 Virus Mini Kit 2.0. CT values and concordance results are listed for each patient sample. Notes for discordant samples are provided in the last column.
HRP, human RNAse P.
To determine analytical sensitivity when comparing the SARS-CoV-2 nucleocapsid LDT versus the Wadsworth EUA, two Wadsworth EUA runs and three SARS-CoV-2 nucleocapsid LDT runs were performed (Table 8 ). Each run used a series of SARS-CoV-2 dilutions in negative patient RNA eluates, ranging from 0.5 cp/μL to 50,000 cp/μL. Upon comparison, acceptable correlation levels were achieved for both the N1 and N2 reactions, showing successful assay efficiency. Moreover, this supports our multiplexing design because no competitive inhibition between primer pairs and/or probes was observed. This also led us to confidently hypothesize that an LoD of 500 cp/mL would be most feasible as CT values >30 were observed in reactions with lower SARS-CoV-2 concentrations.
Table 8.
An Analytical Sensitivity Assay Shows Acceptable Results When Comparing the SARS-CoV-2 Nucleocapsid LDT with the Wadsworth EUA
| SARS-CoV-2 (cp/μL) | N1 reaction |
N2 reaction |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Wadsworth EUA |
Nucleocapsid LDT |
Wadsworth EUA |
Nucleocapsid LDT |
|||||||
| Run 1 | Run 2 | Run 1 | Run 2 | Run 3 | Run 1 | Run 2 | Run 1 | Run 2 | Run 3 | |
| 50,000 | 21.30 | 21.16 | 21.07 | 20.83 | 21.08 | 21.71 | 22.00 | 19.70 | 19.41 | 19.30 |
| 50,000 | 21.35 | 21.03 | 21.14 | 20.83 | 21.04 | 21.63 | 21.94 | 19.68 | 19.47 | 19.25 |
| 50,000 | 21.41 | 20.99 | 21.18 | 20.99 | 21.11 | 21.68 | 21.84 | 19.69 | 19.35 | 19.21 |
| Mean | 21.35 | 21.06 | 21.13 | 20.88 | 21.08 | 21.67 | 21.93 | 19.69 | 19.41 | 19.25 |
| SD | 0.06 | 0.09 | 0.06 | 0.09 | 0.04 | 0.04 | 0.08 | 0.01 | 0.06 | 0.05 |
| % CV | 0.26 | 0.42 | 0.26 | 0.44 | 0.17 | 0.19 | 0.37 | 0.05 | 0.31 | 0.23 |
| 5000 | 24.47 | 24.10 | 24.42 | 24.41 | 24.64 | 25.10 | 25.27 | 23.09 | 23.10 | 22.73 |
| 5000 | 24.52 | 24.37 | 24.93 | 24.09 | 24.53 | 25.10 | 25.31 | 23.26 | 22.83 | 22.80 |
| 5000 | 24.44 | 24.28 | 24.13 | 24.16 | 24.48 | 25.03 | 25.14 | 22.85 | 22.86 | 22.63 |
| Mean | 24.48 | 24.25 | 24.49 | 24.22 | 24.55 | 25.08 | 25.24 | 23.07 | 22.93 | 22.72 |
| SD | 0.04 | 0.14 | 0.41 | 0.17 | 0.08 | 0.04 | 0.09 | 0.21 | 0.15 | 0.09 |
| % CV | 0.17 | 0.57 | 1.65 | 0.69 | 0.33 | 0.16 | 0.35 | 0.89 | 0.65 | 0.38 |
| 500 | 28.04 | 27.74 | 27.66 | 27.21 | 27.30 | 28.53 | 29.08 | 26.37 | 26.11 | 25.91 |
| 500 | 28.04 | 27.63 | 27.77 | 27.72 | 27.55 | 28.60 | 29.09 | 26.47 | 26.40 | 26.04 |
| 500 | 28.07 | 27.57 | 27.74 | 26.99 | 27.37 | 28.42 | 29.21 | 26.36 | 26.07 | 25.87 |
| Mean | 28.05 | 27.65 | 27.72 | 27.31 | 27.41 | 28.52 | 29.13 | 26.40 | 26.19 | 25.94 |
| SD | 0.02 | 0.09 | 0.06 | 0.37 | 0.13 | 0.09 | 0.07 | 0.06 | 0.18 | 0.09 |
| % CV | 0.06 | 0.31 | 0.21 | 1.37 | 0.47 | 0.32 | 0.25 | 0.23 | 0.69 | 0.34 |
| 50 | 31.88 | 30.94 | 30.99 | 29.60 | 32.06 | 32.20 | 33.24 | 30.01 | 29.57 | 30.18 |
| 50 | 31.16 | 31.25 | 30.90 | 30.80 | 31.90 | 32.35 | 33.25 | 30.04 | 29.62 | 30.26 |
| 50 | 31.94 | 30.96 | 31.34 | 31.05 | 30.61 | 32.05 | 32.11 | 30.15 | 30.33 | 29.31 |
| Mean | 31.66 | 31.05 | 31.08 | 30.48 | 31.52 | 32.20 | 32.87 | 30.07 | 29.84 | 29.92 |
| SD | 0.43 | 0.17 | 0.23 | 0.78 | 0.80 | 0.15 | 0.66 | 0.07 | 0.43 | 0.53 |
| % CV | 1.37 | 0.56 | 0.75 | 2.54 | 2.52 | 0.47 | 1.99 | 0.25 | 1.42 | 1.76 |
| 5 | 34.63 | 33.34 | 34.10 | 33.49 | 36.57 | 34.60 | 31.53 | 33.59 | 32.56 | 34.23 |
| 5 | 36.11 | 33.31 | 34.05 | 35.83 | 34.27 | 35.70 | 36.25 | 33.16 | 36.41 | 33.16 |
| 5 | 35.10 | 35.62 | 34.09 | 33.48 | 33.45 | 34.69 | 34.94 | 33.32 | 33.88 | 32.83 |
| Mean | 35.28 | 34.09 | 34.08 | 34.27 | 34.76 | 35.00 | 35.60 | 33.36 | 34.28 | 33.41 |
| SD | 0.76 | 1.33 | 0.03 | 1.35 | 1.62 | 0.61 | 0.93 | 0.22 | 1.96 | 0.73 |
| % CV | 2.14 | 3.89 | 0.08 | 3.95 | 4.65 | 1.75 | 2.60 | 0.65 | 5.71 | 2.19 |
| 0.5 | – | 35.20 | – | 34.65 | – | – | – | 35.82 | 35.15 | – |
| 0.5 | – | 35.16 | – | – | – | – | – | – | – | – |
| 0.5 | – | – | – | – | 36.81 | – | – | – | – | 35.23 |
| Mean | NA | 35.18 | NA | NA | NA | NA | NA | NA | NA | NA |
| SD | NA | 0.03 | NA | NA | NA | NA | NA | NA | NA | NA |
| % CV | NA | 0.08 | NA | NA | NA | NA | NA | NA | NA | NA |
Two individual Wadsworth Emergency Use Authorization (EUA) runs and three individual severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) nucleocapsid laboratory-developed test (LDT) runs were performed. The Wadsworth EUA positive control, a synthetic RNA transcript containing N1 and N2 RT-PCR amplicon sequences (Bio-Synthesis, Lewisville, TX), was used for generating dilutions ranging from 0.5 cp/μL to 50,000 cp/μL. Concentrations of SARS-CoV-2 diluted in negative patient RNA eluates are displayed in the left column. Wadsworth EUA runs 1 to 2 and SARS-CoV-2 nucleocapsid LDT runs 1 to 3 for the N1 reaction are displayed on the left, and the N2 reactions are displayed on the right. CT values for each reaction are listed in addition to the mean value, SD, and % CV.
–, inconclusive result; NA, not applicable.
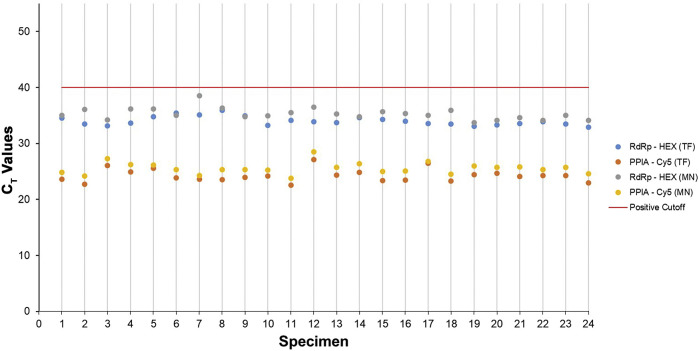
An LoD study for the SARS-CoV-2 RdRp LDT yielded similar results. Regardless of which extraction kit was used with the KingFisher Flex (the MagMAX Viral/Pathogen II Kit or the NucleoMag Viral Kit), virus was detected at 500 cp/mL in 100% (24 of 24) of saliva specimens (Table 9 and Figure 2 ). Moreover, the same study was performed by using nasopharyngeal specimens and detected SARS-CoV-2 at a level of 500 cp/mL in 96% of all specimens for both extraction kits (Table 10 ).
Table 9.
SARS-CoV-2 Was Detected at an LoD of 500 cp/mL in 100% of Saliva Specimens Tested
| Specimen | CT values |
Interpretation | CT values |
Interpretation | ||
|---|---|---|---|---|---|---|
| IP2/IP4 (HEX) | PPIA (Cy5) | IP2/IP4 (HEX) | PPIA (Cy5) | |||
| 1 | 34.55 | 23.64 | Positive | 35.04 | 24.84 | Positive |
| 2 | 33.50 | 22.73 | Positive | 36.07 | 24.18 | Positive |
| 3 | 33.13 | 26.03 | Positive | 34.23 | 27.29 | Positive |
| 4 | 33.66 | 24.96 | Positive | 36.14 | 26.21 | Positive |
| 5 | 34.79 | 25.60 | Positive | 36.20 | 26.12 | Positive |
| 6 | 35.46 | 23.85 | Positive | 35.01 | 25.34 | Positive |
| 7 | 35.09 | 23.58 | Positive | 38.51 | 24.27 | Positive |
| 8 | 35.94 | 23.55 | Positive | 36.38 | 25.35 | Positive |
| 9 | 34.93 | 23.93 | Positive | 34.77 | 25.32 | Positive |
| 10 | 33.24 | 24.17 | Positive | 34.94 | 25.26 | Positive |
| 11 | 34.13 | 22.53 | Positive | 35.55 | 23.78 | Positive |
| 12 | 33.93 | 27.15 | Positive | 36.53 | 28.53 | Positive |
| 13 | 33.74 | 24.33 | Positive | 35.27 | 25.78 | Positive |
| 14 | 34.61 | 24.82 | Positive | 34.79 | 26.40 | Positive |
| 15 | 34.28 | 23.41 | Positive | 35.67 | 24.97 | Positive |
| 16 | 34.00 | 23.42 | Positive | 35.33 | 25.12 | Positive |
| 17 | 33.55 | 26.50 | Positive | 35.00 | 26.81 | Positive |
| 18 | 33.50 | 23.27 | Positive | 35.91 | 24.55 | Positive |
| 19 | 33.07 | 24.47 | Positive | 33.76 | 25.97 | Positive |
| 20 | 33.33 | 24.72 | Positive | 34.16 | 25.73 | Positive |
| 21 | 33.54 | 24.10 | Positive | 34.59 | 25.82 | Positive |
| 22 | 33.86 | 24.27 | Positive | 34.16 | 25.30 | Positive |
| 23 | 33.46 | 24.26 | Positive | 35.04 | 25.74 | Positive |
| 24 | 32.94 | 23.00 | Positive | 34.17 | 24.60 | Positive |
| Mean CT values | 34.01 | 24.26 | 35.30 | 25.55 | ||
RNA was extracted from saliva specimens by using the KingFisher Flex extraction instrument and either the MagMAX Viral/Pathogen II kit (left columns) or the NucleoMag Viral kit (right columns). Specimens were subsequently tested for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) by using the SARS-CoV-2 RNA-dependent RNA polymerase laboratory-developed test. All CT values and interpretations are listed for each specimen along with the mean for each sample set.
LoD, limit of detection; PPIA, peptidylprolyl isomerase A.
Figure 2.
Severe acute respiratory syndrome coronavirus 2 was detected at a limit of detection of 500 cp/mL in 100% of saliva specimens tested using the KingFisher Flex and two different extraction methods. CT values for each saliva specimen from the limit of detection study are represented as a scatter plot comparing the MagMAX Viral/Pathogen II Kit (TF) with the NucleoMag Viral Kit (MN). PPIA, peptidylprolyl isomerase A; RdRp, RNA-dependent RNA polymerase.
Table 10.
SARS-CoV-2 Was Detected at an LoD of 500 cp/mL in 96% of Nasopharyngeal Specimens Tested
| MagMAX Viral Pathogen II kit |
NucleoMag Viral Kit |
|||||
|---|---|---|---|---|---|---|
| Specimen | CT values |
Interpretation | CT values |
Interpretation | ||
| IP2/IP4 (HEX) | PPIA (Cy5) | IP2/IP4 (HEX) | PPIA (Cy5) | |||
| 1 | 33.91 | 26.31 | Positive | 34.47 | 26.71 | Positive |
| 2 | 34.05 | 29.67 | Positive | 34.68 | 30.09 | Positive |
| 3 | 34.78 | 29.45 | Positive | 38.01 | 31.23 | Positive |
| 4 | 33.50 | 27.91 | Positive | 35.76 | 29.14 | Positive |
| 5 | 34.28 | 28.20 | Positive | 34.81 | 28.70 | Positive |
| 6 | 34.24 | 29.95 | Positive | 35.41 | 30.17 | Positive |
| 7 | 34.04 | 27.68 | Positive | 33.88 | 30.48 | Positive |
| 8 | 35.23 | 31.88 | Positive | 34.83 | 31.78 | Positive |
| 9 | 33.97 | 28.39 | Positive | 34.78 | 28.92 | Positive |
| 10 | 33.88 | 29.87 | Positive | 33.70 | 29.74 | Positive |
| 11 | 33.80 | 29.18 | Positive | 35.16 | 30.09 | Positive |
| 12 | 33.73 | 28.44 | Positive | 34.79 | 29.23 | Positive |
| 13 | 33.76 | 28.33 | Positive | 36.61 | 29.36 | Positive |
| 14 | 34.35 | 32.00 | Positive | 35.05 | 31.30 | Positive |
| 15 | 34.91 | 31.82 | Positive | 36.86 | 32.49 | Positive |
| 16 | 33.59 | 28.50 | Positive | 34.96 | 29.45 | Positive |
| 17 | 34.36 | 26.96 | Positive | 34.01 | 28.52 | Positive |
| 18 | 0.00 | 0.00 | Invalid | 33.79 | 27.88 | Positive |
| 19 | 34.30 | 28.57 | Positive | 35.94 | 28.60 | Positive |
| 20 | 33.02 | 29.49 | Positive | 34.76 | 30.75 | Positive |
| 21 | 34.24 | 27.21 | Positive | 33.84 | 29.82 | Positive |
| 22 | 34.32 | 35.30 | Positive | 35.15 | 0.00 | Presumptive Positive |
| 23 | 33.97 | 29.66 | Positive | 35.78 | 28.91 | Positive |
| 24 | 34.20 | 29.02 | Positive | 34.03 | 28.38 | Positive |
| 25 | 34.31 | 27.91 | Positive | |||
| Mean CT Values | 32.75 | 28.07 | 35.04 | 28.41 | ||
RNA was extracted from nasopharyngeal specimens by using the KingFisher Flex Extraction instrument and either the MagMAX Viral/Pathogen II kit or the NucleoMag Viral kit. Specimens were subsequently tested for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) by using the SARS-CoV-2 RNA-dependent RNA polymerase laboratory-developed test. All CT values and interpretations are listed for each specimen along with the mean for each sample set. One sample resulted as invalid due to no detection of PPIA. The presumptive positive sample was the result of no PPIA detection in conjunction with positive RNA-dependent RNA polymerase detection.
LoD, limit of detection; PPIA, peptidylprolyl isomerase A.
A clinical evaluation for both nasopharyngeal and saliva specimens using the SARS-CoV-2 RdRp LDT was also performed. RNA from each specimen type was extracted by using the KingFisher Flex and both the MagMAX Viral/Pathogen II Kit and the NucleoMag Viral Kit. Saliva specimen concordance was evaluated based on results from the Clarifi COVID-19 Test Kit EUA (Table 11 ). The Clarifi COVID-19 Test Kit EUA uses RNA from samples extracted with the Zymo Research RNA Isolation Kit and primers and probes targeting the same genomic regions of RdRp, albeit at different concentrations. Nasopharyngeal specimen concordance was compared with results from the Wadsworth EUA using RNA from specimens previously extracted by using the EZ1 Advanced XL and the EZ1 Virus Mini Kit 2.0. All negative (27 of 27) and positive (44 of 44) saliva specimens were 100% concordant, whereas 92.5% (37 of 40) concordance was observed for positive nasopharyngeal specimens and 100% (30 of 30) concordance in negative nasopharyngeal specimens (Table 12 ). Furthermore, the three discordant positive samples were found to be negative using the TaqPath EUA.
Table 11.
Clinical Evaluation Was Performed Comparing the Clarifi COVID-19 Test Kit EUA with the Zymo Research RNA Isolation Kit and the SARS-CoV-2 RdRp LDT Using the KingFisher Flex Extraction Instrument with the MagMAX Viral/Pathogen II Kit
| Clarifi COVID-19 test kit EUA results |
Specimen | Clarifi COVID-19 test kit EUA |
SARS-CoV-2 RdRp LDT |
Concordance between Clarifi COVID-19 test kit EUA and SARS-CoV-2 RdRp LDT | |
|---|---|---|---|---|---|
| Positives | RdRp | RdRp | PPIA | ||
| Early Ct (<30) | 1 | 19.80 | 18.87 | 23.14 | Concordant |
| 2 | 20.30 | 18.03 | 23.20 | Concordant | |
| 3 | 24.80 | 23.42 | 23.39 | Concordant | |
| 4 | 25.40 | 24.85 | 24.47 | Concordant | |
| 5 | 26.50 | 24.14 | 23.15 | Concordant | |
| 6 | 27.10 | 25.23 | 22.45 | Concordant | |
| 7 | 27.70 | 23.19 | 24.80 | Concordant | |
| 8 | 27.80 | 26.28 | 23.86 | Concordant | |
| 9 | 28.20 | 26.98 | 22.23 | Concordant | |
| 10 | 28.00 | 31.93 | 23.36 | Concordant | |
| 11 | 28.10 | 25.08 | 23.72 | Concordant | |
| 12 | 29.10 | 26.32 | 22.27 | Concordant | |
| 13 | 29.60 | 26.82 | 23.53 | Concordant | |
| 14 | 29.80 | 27.26 | 24.28 | Concordant | |
| 15 | 29.99 | 27.65 | 22.73 | Concordant | |
| Late Ct (>30) | 16 | 30.00 | 28.09 | 24.28 | Concordant |
| 17 | 30.00 | 27.68 | 23.46 | Concordant | |
| 18 | 30.20 | 27.69 | 23.98 | Concordant | |
| 19 | 30.20 | 27.48 | 23.50 | Concordant | |
| 20 | 30.30 | 28.53 | 22.98 | Concordant | |
| 21 | 30.43 | 28.55 | 23.39 | Concordant | |
| 22 | 32.56 | 30.97 | 25.45 | Concordant | |
| 23 | 32.80 | 29.40 | 23.60 | Concordant | |
| 24 | 32.90 | 29.54 | 23.47 | Concordant | |
| 25 | 33.20 | 32.19 | 22.59 | Concordant | |
| 26 | 33.30 | 32.06 | 23.07 | Concordant | |
| 27 | 33.40 | 29.79 | 24.03 | Concordant | |
| 28 | 34.77 | 33.34 | 23.59 | Concordant | |
| 29 | 34.80 | 34.76 | 24.40 | Concordant | |
| 30 | 35.10 | 33.48 | 27.34 | Concordant | |
| 31 | 35.19 | 32.95 | 24.97 | Concordant | |
| 32 | 35.30 | 33.12 | 24.35 | Concordant | |
| 33 | 35.35 | 31.93 | 24.64 | Concordant | |
| 34 | 35.59 | 34.34 | 25.11 | Concordant | |
| 35 | 35.90 | 32.60 | 23.06 | Concordant | |
| 36 | 36.00 | 32.69 | 23.10 | Concordant | |
| 37 | 36.40 | 32.43 | 25.54 | Concordant | |
| 38 | 37.10 | 31.38 | 24.75 | Concordant | |
| 39 | 37.43 | 35.39 | 25.04 | Concordant | |
| 40 | 38.00 | 34.27 | 23.42 | Concordant | |
| 41 | 38.50 | 38.44 | 26.03 | Concordant | |
| 42 | 38.55 | 36.01 | 23.22 | Concordant | |
| 43 | 39.00 | 35.07 | 25.20 | Concordant | |
| 44 | 39.60 | 36.01 | 23.25 | Concordant | |
CT values and concordance results are listed for each patient specimen.
EUA, Emergency Use Authorization; LDT, laboratory-developed test; PPIA, peptidylprolyl isomerase A; RdRp, RNA-dependent RNA polymerase; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Table 12.
An Overall Concordance Level of 92.5% Was Achieved Comparing the SARS-CoV-2 RdRp LDT with the Wadsworth EUA
| Wadsworth EUA results | N1 Wadsworth EUA | N2 Wadsworth EUA | IP2/IP4 SARS-CoV-2 RdRp LDT | PPIA SARS-CoV-2 RdRp LDT | Concordance between SARS-CoV-2 RdRp LDT and Wadsworth EUA | Notes |
|---|---|---|---|---|---|---|
| Early Ct (<30) | 13.20 | 12.23 | 13.63 | 24.54 | Concordant | |
| Positives | 13.77 | 13.14 | 14.93 | 23.40 | Concordant | |
| 14.03 | 13.31 | 16.97 | 23.51 | Concordant | ||
| 16.53 | 15.02 | 16.59 | 28.98 | Concordant | ||
| 16.86 | 16.59 | 18.43 | 29.30 | Concordant | ||
| 18.14 | 17.57 | 19.73 | 26.06 | Concordant | ||
| 19.56 | 17.38 | 19.21 | 28.92 | Concordant | ||
| 22.52 | 21.06 | 22.67 | 23.91 | Concordant | ||
| 24.32 | 23.77 | 23.42 | 27.29 | Concordant | ||
| 24.61 | 23.00 | 24.59 | 28.09 | Concordant | ||
| 24.56 | 23.38 | 25.24 | 27.05 | Concordant | ||
| 25.63 | 24.16 | 27.21 | 29.72 | Concordant | ||
| 27.28 | 25.64 | 26.10 | 27.99 | Concordant | ||
| 29.63 | 28.24 | 26.34 | 30.41 | Concordant | ||
| 29.87 | 31.19 | 30.29 | 25.70 | Concordant | ||
| Late Ct (>30) | 30.76 | 32.09 | 31.39 | 29.51 | Concordant | |
| Positives | 30.81 | 30.67 | 32.90 | 23.41 | Concordant | |
| 31.35 | 30.01 | 30.22 | 28.80 | Concordant | ||
| 31.38 | 30.7 | 0.00 | 25.21 | Discordant | Negative in TaqPath EUA with KingFisher Flex Extraction | |
| 31.54 | 32.27 | 31.96 | 25.60 | Concordant | ||
| 31.82 | 31.14 | 31.69 | 29.14 | Concordant | ||
| 31.82 | 31.43 | 33.37 | 25.42 | Concordant | ||
| 32.03 | 30.25 | 34.13 | 23.41 | Concordant | ||
| 32.06 | 32.51 | 35.78 | 32.38 | Concordant | ||
| 32.44 | 34.39 | 32.38 | 26.28 | Concordant | ||
| 32.87 | 31.59 | 30.18 | 24.70 | Concordant | ||
| 32.89 | 35.17 | 36.50 | 27.64 | Concordant | ||
| 33.35 | 32.29 | 31.72 | 26.79 | Concordant | ||
| 33.53 | 33.2 | 36.64 | 23.68 | Concordant | ||
| 33.63 | 31.49 | 0.00 | 27.84 | Discordant | Negative in TaqPath EUA with KingFisher Flex Extraction | |
| 34 | 35.89 | 37.76 | 23.72 | Concordant | ||
| 34.09 | 35.82 | 34.57 | 29.02 | Concordant | ||
| 34.26 | 36.62 | 0.00 | 25.01 | Discordant | Negative in TaqPath EUA with KingFisher Flex Extraction | |
| 34.58 | 33.62 | 36.89 | 25.16 | Concordant | ||
| 35.05 | 35.12 | 35.49 | 29.19 | Concordant | ||
| 35.07 | 35.69 | 32.70 | 27.23 | Concordant | ||
| 35.11 | 34.78 | 33.59 | 28.73 | Concordant | ||
| 36.17 | 31.33 | 34.17 | 32.21 | Concordant | ||
| 35.52 | 34.41 | 37.54 | 26.08 | Concordant | ||
| 36.63 | 34.21 | 34.06 | 30.13 | Concordant |
A clinical evaluation was performed comparing the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA-dependent RNA polymerase (RdRp) laboratory-developed test (LDT) with the KingFisher Flex Extraction Instrument and the MagMAX Viral/Pathogen II Kit and the Wadsworth Emergency Use Authorization (EUA) using the EZ-1 Advanced XL instrument with the EZ1 Virus Mini Kit 2.0. CT values and concordance results are listed for each patient sample. Discordant samples are listed with an explanation under the Notes column.
PPIA, peptidylprolyl isomerase A.
Discussion
Accurate detection and diagnosis of SARS-CoV-2 infection are essential to minimizing transmission of the virus.14 , 15 Both the SARS-CoV-2 nucleocapsid LDT and the SARS-CoV-2 RdRp LDT allow for detection of the virus at an acceptable LoD regardless of specimen origin (nasopharyngeal or saliva). The SARS-CoV-2 nucleocapsid LDT was initially developed because our laboratory was specifically testing nasopharyngeal specimens, and a multiplexed diagnostic assay was needed. The singleplex assay (Wadsworth EUA) initially required more time, supplies, and technical work. The SARS-CoV-2 RdRp LDT was developed and validated following implementation of the SARS-CoV-2 nucleocapsid LDT. Saliva specimens are more readily obtainable and more often preferred than nasopharyngeal specimens. This also allows for circumvention of supply chain issues should complications arise from different vendors. The versatility and cost-effective nature of these assays permit clinical laboratories to perform them with tools and instruments often used in a variety of other molecular diagnostic tests. Furthermore, primers, probes and other reagents, although difficult to acquire during the early stages of the COVID-19 pandemic, can be readily obtained. Moreover, turnaround time compared with the singleplex, nucleic acid–based RT-qPCR assay (Wadsworth EUA) is greatly enhanced.
Patient-reporting criteria for the SARS-CoV-2 nucleocapsid LDT were based on the original Wadsworth EUA. Positive patient results require CT values <40 for the N1, N2, and HRP reaction. Should only one of the “N” reactions result as positive, the result is inconclusive, and the patient sample is reflexed for re-extraction and RT-qPCR. If the “N” reaction CT values are 0 or >40, in conjunction with an acceptable HRP CT value, a result of “SARS-CoV-2 Not Detected” is reported. In the SARS-CoV-2 RdRp LDT, a positive result for SARS-CoV-2 can only be reported according to the following criteria: HEX CT values ≤35 regardless of Cy5 CT values or HEX CT values between 35 and 40 paired with Cy5 CT values ≤35. A presumptive positive is reported when the HEX CT value is between 35 and 40 and paired with a Cy5 CT value >35 or no Cy5 CT value. Reporting a patient as “SARS-CoV-2 Not Detected” requires no HEX CT value or a HEX CT value >40 paired with a Cy5 CT value ≤35. This algorithm allows for confidence in reporting, as detection of the virus regardless of the Cy5 CT value leads to an interpretation of either positive or presumptive positive, aiding in the elimination of reporting false-negative results.
As new variants of SARS-CoV-2 have emerged, it was confirmed that both the SARS-CoV-2 nucleocapsid LDT and SARS-CoV-2 RdRp LDT targets were unaffected by these mutations.16 Sequence analyses were performed on all major SARS-CoV-2 variants and it was found that C.37, which holds a P13L mutation in the N gene, may affect the SARS-CoV-2 nucleocapsid LDT, whereas the SARS-CoV-2 RdRp LDT remains unaffected. In addition, this effect may be negligible as it only involves one nucleotide change in the probe-binding region for the N1 reaction, whereas the N2 reaction is not affected. Several other assays, including many rapid tests, target regions that include mutations found in these variants, possibly impeding accurate clinical diagnosis of SARS-CoV-2 infection.17 Nonetheless, variants of SARS-CoV-2 could result in potential limitations for these assays. It should be noted that as new variants are identified over time, the targets used in these assays may require modification.
The SARS-CoV-2 nucleocapsid LDT and SARS-CoV-2 RdRp LDT have also shown higher analytical sensitivity compared with EUA PCR-based rapid tests. In several separate instances, we were able to detect the presence of SARS-CoV-2 in both nasopharyngeal and saliva specimens, whereas other FDA EUA rapid tests did not detect SARS-CoV-2 in these same samples. However, it should be noted that low-level positive findings are indeed subject to random sampling errors and are not always indicative of a difference in analytical or clinical sensitivity.
Limitations for both assays include test turnaround time and ample training. Upon specimen receipt and preparation, the extraction process requires approximately 40 minutes on the EZ1 Advanced XL and 25 minutes on the KingFisher Flex. Furthermore, both RT-qPCR programs are about 1.5 hours in length, followed by the time required for proper analysis, interpretation, and review. These assays also require training on multiple instruments using different software for analysis. Nonetheless, robust flexibility is found in the SARS-CoV-2 RdRp LDT, as it allows for two different specimen types to be tested simultaneously. In addition, the use of automated liquid-handlers enhances the process in a more rapid and efficient manner with less room for human error. However, it should be noted that these systems rely on several consumable materials, and supply chain issues could affect their utilization. Furthermore, the macro-enabled Excel 2016 worksheets that were developed for the SARS-CoV-2 RdRp LDT and the SARS-CoV-2 nucleocapsid LDT allowed for semi-automated resulting, thus accelerating test turnaround time.
Although SARS-CoV-2 rapid tests based on detection of SARS-CoV-2 antigens rather than viral RNA sequences are beneficial for timely diagnostic testing of clearly symptomatic individuals, the SARS-CoV-2 nucleocapsid and SARS-CoV-2 RdRp LDTs have great value in their superior sensitivity for detecting asymptomatic or mildly symptomatic individuals and flexibility with regard to specimen type. Diagnostic laboratories could introduce these assays into their workflows in an efficient manner with instruments and reagents that are often found in many molecular laboratories. This also aids in supply acquisition as many EUA rapid tests require the use of reagents specific to the manufacturer, and supply chain issues can hamper turnaround time. Moreover, these assays could be modified in the future to aid in the diagnosis of other viral targets that require nucleic acid–based testing. Overall, the SARS-CoV-2 nucleocapsid LDT and the SARS-CoV-2 RdRp LDT are cost-effective, analytically specific, and analytically sensitive assays that serve as accurate molecular diagnostic tests for COVID-19.
Footnotes
Supported by SUNY Upstate Medical University.
E.W.M. and C.M.L. contributed equally to this work.
Disclosures: None declared.
Supplemental material for this article can be found at http://doi.org/10.1016/j.jmoldx.2021.12.010.
Supplemental Data
Visual Basic for Applications commands and scripts for clearing of existing data and importing current patient data, provided as a TXT file (converted originally from a BAS file).
Visual Basic for Applications commands and scripts for exporting the run setup file provided as a TXT file (converted from a BAS file).
Visual Basic for Applications commands and scripts for importing the results file, followed by sorting and parsing of data, all provided as a TXT file (converted from a BAS file).
Visual Basic for Applications commands and scripts for QC, checking import errors, sending results to laboratory information system and providing e-mail notifications, all provided as a TXT file (converted from a BAS file).
Visual Basic for Applications commands and scripts for saving results as a PDF file with a timestamp indicating when results were sent to the laboratory information system, provided here as a TXT file (converted from a BAS file).
Visual Basic for Applications commands and scripts for the e-mail notification process, provided here as a TXT file (converted from a BAS file).
Visual Basic for Applications commands and scripts for sending results as a CSV file to the laboratory information system interface, provided as a TXT file (converted from a BAS file).
Functions to remove special characters not allowed by the laboratory information system provided as a TXT file (converted from a BAS file).
A macro allowing for protection or the removal of protection in the Excel 2016 worksheets, provided as a TXT file (converted from a BAS file).
References
- 1.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W., China Novel Coronavirus Investigating and Research Team A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L. A pneumonia outbreak associated with a new coronavirus of probably bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Finkel Y., Mizrahi O., Nachshon A., Weingarten-Gabbay S., Morgenstern D., Yahalom-Ronen Y., Tamir H., Achdout H., Stein D., Israeli O., Beth-Din A., Melamed S., Weiss S., Israely T., Paran N., Schwartz M., Stern-Ginossar N. The coding capacity of SARS-CoV-2. Nature. 2021;589:125–130. doi: 10.1038/s41586-020-2739-1. [DOI] [PubMed] [Google Scholar]
- 4.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Redondo N., Zaldívar-López S., Garrido J.J., Montoya M. SARS-CoV-2 accessory proteins in viral pathogenesis: knowns and unknowns. Front Immunol. 2021;12:708264. doi: 10.3389/fimmu.2021.708264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guan W.-J., Ni Z.-Y., Hu Y., Liang W.-H., Ou C.-Q., He J.-X., et al. China Medical Treatment Expert Group for Covid-19 Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yin Y., Wunderink R.G. MERS, SARS and other coronaviruses as causes of pneumonia. Respirology. 2018;23:130–137. doi: 10.1111/resp.13196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petrosillo N., Viceconte G., Ergonul O., Ippolito G., Petersen E. COVID-19, SARS and MERS: are they closely related? Clin Microbiol Infect. 2020;26:729–734. doi: 10.1016/j.cmi.2020.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Price T.K., Bowland B.C., Chandrasekaran S., Garner O.B., Yang S. Performance characteristics of severe acute respiratory syndrome coronavirus 2 RT-PCR tests in a single health system: analysis of >10,000 results from three different assays. J Mol Diagn. 2021;23:159–163. doi: 10.1016/j.jmoldx.2020.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu C.Y., Chan K.G., Yean C.Y., Ang G.Y. Nucleic acid-based diagnostic tests for the detection SARS-CoV-2: an update. Diagnostics (Basel) 2021;11:53. doi: 10.3390/diagnostics11010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang Y.-W., Schmitz J.E., Persing D.H., Stratton C.W. Laboratory diagnosis of COVID-19: current issues and challenges. J Clin Microbiol. 2020;58:e00512–e00520. doi: 10.1128/JCM.00512-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gadkar V.J., Goldfarb D.M., Young V., Watson N., Al-Rawahi G.N., Srigley J.A., Tilley P. Development and validation of a new triplex real-time quantitative reverse transcriptase-PCR assay for the clinical detection of SARS-CoV-2. Mol Cell Probes. 2021;58:101744. doi: 10.1016/j.mcp.2021.101744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Etievant S., Bal A., Escuret V., Brengel-Pesce K., Bouscambert M., Cheynet V., Generenaz L., Oriol G., Destras G., Billaud G., Josset L., Frobert E., Morfin F., Gaymard A. Performance assessment of SARS-CoV-2 PCR assays developed by WHO referral laboratories. J Clin Med. 2020;9:1871. doi: 10.3390/jcm9061871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Su Z., Wen J., McDonnell D., Goh E., Li X., Šegalo S., Ahmad J., Cheshmehzangi A., Xiang Y.-T. Vaccines are not yet a silver bullet: the imperative of continued communication about the importance of COVID-19 safety measures. Brain Behav Immun Health. 2021;12:100204. doi: 10.1016/j.bbih.2021.100204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Triggle C.R., Bansal D., Ding H., Islam M.M., Farag E.A.B.A., Hadi H.A., Sultan A.A. A comprehensive review of viral characteristics, transmission, pathophysiology, immune response, and management of SARS-CoV-2 and COVID-19 as a basis for controlling the pandemic. Front Immunol. 2021;12:631139. doi: 10.3389/fimmu.2021.631139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lauring A.S., Hodcroft E.B. Genetic variants of SARS-CoV-2—what do they mean? JAMA. 2021;325:529–531. doi: 10.1001/jama.2020.27124. [DOI] [PubMed] [Google Scholar]
- 17.Grubaugh N.D., Hodcroft E.B., Fauver J.R., Phelan A.L., Cevik M. Public health actions to control new SARS-CoV-2 variants. Cell. 2021;184:1127–1132. doi: 10.1016/j.cell.2021.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Visual Basic for Applications commands and scripts for clearing of existing data and importing current patient data, provided as a TXT file (converted originally from a BAS file).
Visual Basic for Applications commands and scripts for exporting the run setup file provided as a TXT file (converted from a BAS file).
Visual Basic for Applications commands and scripts for importing the results file, followed by sorting and parsing of data, all provided as a TXT file (converted from a BAS file).
Visual Basic for Applications commands and scripts for QC, checking import errors, sending results to laboratory information system and providing e-mail notifications, all provided as a TXT file (converted from a BAS file).
Visual Basic for Applications commands and scripts for saving results as a PDF file with a timestamp indicating when results were sent to the laboratory information system, provided here as a TXT file (converted from a BAS file).
Visual Basic for Applications commands and scripts for the e-mail notification process, provided here as a TXT file (converted from a BAS file).
Visual Basic for Applications commands and scripts for sending results as a CSV file to the laboratory information system interface, provided as a TXT file (converted from a BAS file).
Functions to remove special characters not allowed by the laboratory information system provided as a TXT file (converted from a BAS file).
A macro allowing for protection or the removal of protection in the Excel 2016 worksheets, provided as a TXT file (converted from a BAS file).