Abstract
Infections caused by Escherichia coli isolates expressing adhesins of the Dr family are associated with diarrhea and urinary tract infections, and these E. coli strains recognize the complement regulatory protein decay-accelerating factor (DAF) as their receptor. Clustering of the DAF receptor at the sites of bacterial adherence to epithelial cells is proposed as an alternative to PCR assay for rapid detection of Dr-positive E. coli.
Escherichia coli isolates that bear mannose-resistant hemagglutinins, such as Dr fimbriae and related adhesins, have attracted attention due to their association with two common diseases: urinary tract infection and diarrhea. Epidemiological studies suggest that children and pregnant women are prone to recurrent or persistent infections caused by these organisms. As many as 50% of isolates from children with cystitis, 50% of isolates from patients with protracted diarrhea, and 30% of isolates from pregnant women with pyelonephritis express Dr adhesins (2, 9). Hallmarks of infection associated with E. coli expressing Dr adhesins, such as persistence of infection, tendency to recur, and positive association with ampicillin resistance, justify laboratory efforts aimed at rapid detection of Dr-positive E. coli (1, 5). The Dr family of adhesins consists of the afimbrial and fimbrial adhesins Dr, Dr-II, F1845, AFA-1, and AFA-III. The Dr adhesins recognize the Dra antigen of the Cromer blood group system (10). The subsequent mapping of the Dra antigen to the exposed domains of decay-accelerating factor (DAF) led to the discovery that DAF was the cellular receptor for Dr-positive E. coli (13). DAF is a complement regulatory protein found on erythrocytes and on most nucleated mammalian cells. It protects the cells from autologous complement-mediated damage by preventing the formation and accelerating the decay of C3 convertases (7). DAF consists of four short consensus repeat (SCR) domains of 60 amino acids each followed by a serine/threonine-rich domain, and it is attached to the cell membrane by a glycosylphosphatidylinositol anchor. The Dr operons involved in the biogenesis of various Dr adhesins share a conserved genetic organization with a high degree of homology in the accessory subunits and a great range of diversity among their adhesin subunits (11). Despite a low level of similarity in the adhesin genes, all Dr adhesins recognize distinct but overlapping regions on the extracellular domains of DAF (8). One characteristic feature of DAF and other glycosylphosphatidylinositol-anchored proteins is lateral mobility on the cell surface (14). Binding of recombinant E. coli strains bearing Dr, Dr-II, F1845, AFA-I, or AFA-III adhesins to HeLa cells or of a clinical Dr-positive strain to the human intestinal cell line Caco-2 results in the clustering of receptors around the adhering bacteria (4). Current detection of Dr-positive E. coli is based on agglutination assays with human erythrocytes in the presence of mannose analogues and inhibition of agglutination with anti-DAF SCR3 antibodies. Dr-positive isolates also can be detected by PCR with primers designed to amplify a 750-bp fragment of the afaB gene, which encodes a periplasmic chaperone protein involved in the biogenesis of Dr adhesins. This primer set has been demonstrated to identify all of the Dr adhesins known to date (15). The aim of the present study was to propose a rapid assay to identify E. coli strains expressing Dr adhesins based on their novel ability to cluster DAF receptors at the sites of bacterial adherence. We provide evidence that the clustering of DAF on infected epithelial cells is a reliable and consistent phenomenon that might be used in clinical laboratories for the rapid detection of E. coli strains bearing adhesins of the Dr family.
A collection of 80 uropathogenic E. coli strains isolated from pregnant patients with pyelonephritis and 78 strains of E. coli from patients with traveler's diarrhea were used to verify the DAF clustering approach. Owing to the nature of the study and limitations in data representation, we did not examine uropathogenic isolates from other patient populations or any pediatric isolates. However, our collection reflects two major population sources of Dr-positive E. coli isolates: pregnant women with pyelonephritis and adult patients with diarrhea.
Prototype clinical strains of E. coli that express adhesins of the Dr family and produce DAF clustering on HeLa cells were included as positive controls (strains IH11128 [Dr], 7372 [Dr-II], C1845 [F1845], KSS2 [AFA-I], and A30 [AFA-III]). The Dr-negative E. coli strain DR14, which was developed by insertional mutagenesis of the Dr operon in the clinical pyelonephritis strain IH11128, was included as a negative control (3). The Dr-negative mutant did not display any binding to HeLa cells, and therefore no DAF clustering was observed. Shigella flexneri SA100 and Salmonella enterica serovar Typhimurium STYP2000, strains which bind to HeLa cells but do not exploit DAF as a receptor, were also included as negative controls. An agglutination assay with a 3% suspension of human erythrocytes in phosphate-buffered saline (PBS) was performed with and without 1.1 mM methyl-d-mannoside (Sigma Chemical Co., St. Louis, Mo.) to select isolates that display the mannose-resistant hemagglutination (MRHA) phenotype. All MRHA-positive isolates were tested in the hemagglutination inhibition assay with human erythrocytes and with human erythrocytes preincubated with anti-DAF monoclonal antibody IH4. The assay allows for the detection of E. coli isolates that recognize DAF as their receptor. All isolates were further subjected to PCR and tested for adherence and the ability to produce DAF clustering on HeLa cells. Primers that amplify a 750-bp fragment of the afaB gene, conserved within Afa-I and related operons, were used to identify Dr-positive strains as previously described (12). HeLa cells for the DAF clustering assay (DCA) were grown in minimal essential medium (Gibco BRL, Grand Island, N.Y.) on glass coverslips placed in 12-well tissue culture plates. Confluent monolayers were washed with PBS and infected for 1 h with 1 ml of bacterial suspension (optical density at 600 nm, 2.0) diluted 1:10 in PBS containing 1.1 mM methyl-d-mannoside (Sigma Chemical Co.). The 1-h incubation time was selected based on experiments with recombinant Dr-positive E. coli strains and selected Dr-positive clinical isolates. Bacterial suspensions have been incubated with HeLa cells for time periods ranging from 15 min to 3 h. DAF clustering has been known to occur as early as 15 min postinfection (3). We found that 1 h of incubation was optimal for evaluating DAF clustering on HeLa cells infected with clinical isolates. Incubation times extended to 2 or 3 h were not advantageous and sometimes resulted in the overloading of HeLa cells with adherent bacteria. After incubation at 37°C in 5% CO2, bacterial suspensions were discarded and monolayers were washed twice with PBS, fixed for 10 min at room temperature with 3.7% formaldehyde, and washed again with PBS. Fixed HeLa cells were subjected to 30 min of immunofluorescence staining with phycoerythrin (PE)-conjugated anti-human DAF antibody (PE-labeled anti-human CD55, clone IA10, catalog no. 33575X; PharMingen, San Diego, Calif.) diluted 1:50 in PBS. Cells then were washed twice with PBS and incubated for 20 min at 37°C with RNase, DNase-free (Roche, Indianapolis, Ind.), diluted 1:5 in 2× SSC solution containing 300 mM NaCl and 30 mM sodium citrate (pH 7.0) to prevent cytoplasmic RNA-associated background staining. Finally, cells were incubated for 1 min with Sytox Green (Molecular Probes, Inc., Eugene, Oreg.) diluted 1:2,000 in deionized water, air dried, and placed on microscopic slides in mounting medium (Cytoseal 60; Stephens Scientific, Riverdale, N.J.). Slides were examined under a fluorescence microscope (Nikon Eclipse 600; Nikon, Melville, N.Y.) equipped with appropriate filters and the U-III photomicrographic system. Two investigators independently screened 20 randomly selected microscopic fields for the presence of DAF clusters.
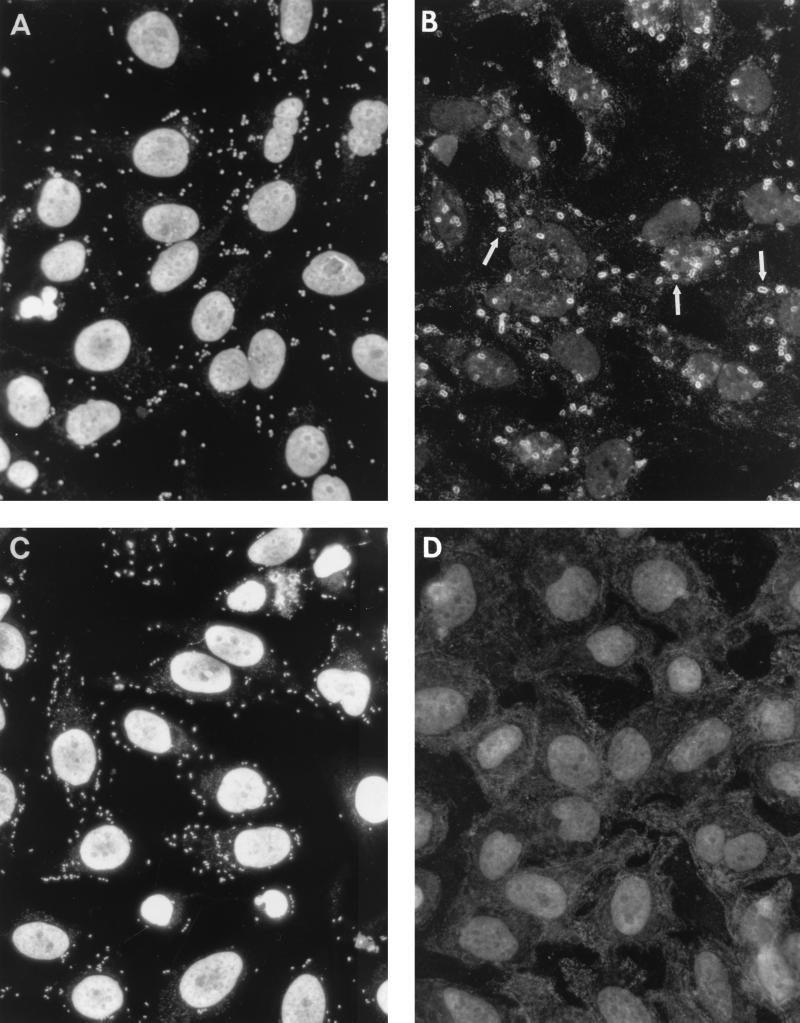
Hemagglutination assays revealed that 62 (77%) of 80 gestational pyelonephritis isolates and 24 (31%) of 78 diarrheal isolates displayed the MRHA phenotype. Blocking the SCR3 domain of DAF with IH4 antibody inhibited agglutination of 15 (19%) gestational pyelonephritis isolates and 8 (10%) diarrheal isolates. PCR studies demonstrated the presence of afaB amplicons in 15 gestational pyelonephritis isolates and 8 diarrheal isolates that displayed MRHA inhibited by IH4 antibodies (Table 1). In addition, afaB sequences were detected in 9 gestational pyelonephritis isolates with MRHA not inhibited by IH4 antibodies. Adherence assays showed that both afaB-positive and afaB-negative E. coli strains were able to bind to HeLa cells when incubated in PBS in the presence of mannose analogue. DAF staining demonstrated that the ability to produce clustering is a unique feature of E. coli strains positive in the PCR assay. The clustering of DAF manifested itself as intense staining that outlined individual bacterial cells (Fig. 1B). Double staining with anti-CD55 (DAF) antibody conjugated with PE and Sytox Green allowed colocalization of DAF clustering at the sites of bacterial adherence. The bacterial cell was stained green and was outlined by a red stain denoting DAF clustering. afaB-positive E. coli cells cultured on Luria-Bertani agar or on McConkey agar were positive by DCA. The afaB-negative gestational pyelonephritis and diarrheal E. coli isolates that bound to HeLa cells (Fig. 1C) were not able to produce the characteristic appearance of clustered receptors (Fig. 1D). DCA was also negative with clinical Shigella and Salmonella strains. The replacement of anti-DAF PE-conjugated antibody with mouse immunoglobulin G conjugated with PE resulted in a lack of DAF staining.
TABLE 1.
Correlation between amplification of the afaB region and DCA in gestational pyelonephritis and diarrheal E. coli isolates
| Source of E. coli isolates | No. (%) of isolatesa:
|
||||||
|---|---|---|---|---|---|---|---|
| With MRHA phenotype | Showing indicated result with anti-DAF monoclonal antibody IH4
|
Positive by PCR with afaB primers
|
Positive by DCA
|
||||
| Inhibited | Not inhibited | Inhibited | Not inhibited | Inhibited | Not inhibited | ||
| Gestational pyelonephritis (n = 80) | 62 (77) | 15 (19) | 47 (59) | 15 (19) | 9 (11) | 15 (19) | 9 (11) |
| Diarrhea (n = 78) | 24 (31) | 8 (10) | 15 (19) | 8 (10) | 0 | 8 (10) | 0 |
“Inhibited” and “not inhibited” refer to the status of inhibition of the MRHA phenotype by IH4.
FIG. 1.
Diffuse-type adherence (A) and DAF clustering (arrows) (B) on HeLa cells infected with a urinary isolate of gestational afaB-positive E. coli. Receptors accumulate at the sites of adherence and outline the shape of bacterial cells. afaB-negative E. coli (C) binds to HeLa cells but is not able to produce DAF clustering (D).
DCA allows for rapid detection of uropathogenic and diarrheal E. coli isolates that express adhesins of the Dr family and recognize DAF as their receptor. The assay is based on the phenomenon of the accumulation of DAF at the sites of bacterial adherence. A similar diagnostic approach, based on cytoskeleton rearrangements, has been proposed as a tool for detecting enteropathogenic and enterohemorrhagic E. coli strains of diarrheal origin. Accumulation of actin on Hep-2 or CaCo cells at the sites of bacterial binding indicates the presence of attaching and effacing lesions, a hallmark of infection due to enteropathogenic or enterohemorrhagic E. coli strains (6).
DCA is a simple procedure that can be accomplished in less than 2 h in a common laboratory setting. We found that all DCA-positive E. coli strains grown on Luria-Bertani agar produced the same intensity of DAF clustering when bacterial suspensions were prepared from overnight cultures grown on MacConkey agar, a selective medium frequently used in clinical laboratories. All assays—MRHA, MRHA inhibition, PCR, and DCA—were done twice, and the results were entirely reproducible. The staining for DAF clustering was originally performed in two steps with initial anti-DAF monoclonal antibody IH4 (kindly provided by D. Lublin) followed by incubation with goat anti-mouse antibody conjugated with Texas red (Molecular Probes, Inc.). This assay has been adapted for clinical laboratory application by a one-step staining procedure using commercial anti-DAF (CD55) monoclonal antibody conjugated with PE. Counterstaining to visualize adherent bacterial cells is not required for the detection of DCA-positive cells. The fluorescent microscope equipped with the appropriate filters required for examining the results is often standard laboratory equipment. Reading of the results does not demand specific skills. Any number of clusters found in a few microscopic fields is interpreted as a DCA-positive strain and indicates the ability of E. coli to produce one of the adhesins of the Dr family.
All gestational pyelonephritis and diarrheal E. coli isolates that produced DAF clustering were found to be positive by PCR with primers amplifying the afaB 750-bp sequences. None of the afaB-negative isolates that displayed adherence to HeLa cells were able to produce DAF clustering. Our results of Dr-positive E. coli strains in the gestational population are comparable to a previous study, although the incidence of Dr-positive E. coli isolates in the diarrheal specimens was low. We believe that the rate of reported detection of Dr-positive E. coli in different clinical conditions may vary based on the population studied, geographical location, outbreak situations, and methods employed. DCA demonstrates a high sensitivity and specificity that is similar to PCR. In addition, performance of DCA in a clinical laboratory is cheaper and requires less personnel skill than the PCR technique.
Identification of E. coli that recognizes DAF as a receptor based on the inhibition of hemagglutination with anti-DAF antibodies cannot be recommended for routine use due to the method's lack of sensitivity. The DAF monoclonal antibody inhibition assay does not allow for the detection of Dr-positive E. coli that recognizes domains other than the SCR3 domain of DAF. Inhibition of MRHA also presents other inconveniences, such as the lack of availability of fresh human erythrocytes and the difficulties associated with the assessment of agglutination results. In conclusion, DCA is a rapid, specific, and cost-effective procedure that offers a reasonable alternative to DNA amplification-based assays.
Acknowledgments
We thank John Mathewson for providing the collection of diarrheal E. coli strains, David Niesel for the Shigella flexneri strain, and Johnny Peterson for the Salmonella enterica serovar Typhimurium strain.
This work was supported in part by grant 2RO1DK42029 from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDKD).
REFERENCES
- 1.Foxman B, Zhang L, Tallman P, Palin K, Rode C, Bloch C, Gillespie B, Marrs C F. Virulence characteristics of Escherichia coli causing first urinary tract infection predict risk of second infection. J Infect Dis. 1995;172:1536–1541. doi: 10.1093/infdis/172.6.1536. [DOI] [PubMed] [Google Scholar]
- 2.Giron J A, Jones T, Millan-Velasco F, Castro-Munoz E, Zarate L, Fry J, Frankel G, Moseley S L, Baudry B, Kaper J B. Diffuse-adhering Escherichia coli (DAEC) as a putative cause of diarrhea in Mayan children in Mexico. J Infect Dis. 1991;163:507–513. doi: 10.1093/infdis/163.3.507. [DOI] [PubMed] [Google Scholar]
- 3.Goluszko P, Moseley S L, Truong L D, Kaul A, Williford J R, Selvarangan R, Nowicki S, Nowicki B. Development of experimental model of chronic pyelonephritis with Escherichia coli 075:K5:H-bearing Dr fimbriae: mutation in the dra region prevented tubulointerstitial nephritis. J Clin Investig. 1997;99:1662–1672. doi: 10.1172/JCI119329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goluszko P, Selvarangan R, Popov V, Pham T, Wen J W, Singhal J. Decay-accelerating factor and cytoskeleton redistribution pattern in HeLa cells infected with recombinant Escherichia coli strains expressing Dr family of adhesins. Infect Immun. 1999;67:3989–3997. doi: 10.1128/iai.67.8.3989-3997.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hart, A., B. Nowicki, E. Pawelczyk, P. Goluszko, P. Urvil, and S. Nowicki. Ampicillin-resistant Escherichia coli in gestational pyelonephritis: increased occurrence and association with the colonization factor Dr adhesin. J. Infect. Dis., in press [DOI] [PubMed]
- 6.Knutton S, Baldwin T, Williams P H, McNeish A S. Actin accumulation at sites of bacterial adhesion to tissue culture cells: basis of a new diagnostic test for enteropathogenic and enterohemorrhagic Escherichia coli. Infect Immun. 1989;57:1290–1298. doi: 10.1128/iai.57.4.1290-1298.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lublin D M, Atkinson J P. Decay-accelerating factor: biochemistry, molecular biology, and function. Annu Rev Immunol. 1989;7:35–58. doi: 10.1146/annurev.iy.07.040189.000343. [DOI] [PubMed] [Google Scholar]
- 8.Nowicki B, Hart A, Coyne K E, Lublin D M, Nowicki S. Short consensus repeat-3 domain of recombinant decay-accelerating factor is recognized by Escherichia coli recombinant Dr adhesin in a model of a cell-cell interaction. J Exp Med. 1993;178:2115–2121. doi: 10.1084/jem.178.6.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nowicki B, Martens M, Hart A, Nowicki S. Gestational age-dependent distribution of Escherichia coli fimbriae in pregnant patients with pyelonephritis. Ann NY Acad Sci. 1994;730:290–291. doi: 10.1111/j.1749-6632.1994.tb44268.x. [DOI] [PubMed] [Google Scholar]
- 10.Nowicki B, Moulds J, Hull R, Hull S. A hemagglutinin of uropathogenic Escherichia coli recognizes the Dr blood group antigen. Infect Immun. 1988;56:1057–1060. doi: 10.1128/iai.56.5.1057-1060.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nowicki B, Selvarangan R, Nowicki S. Family of Escherichia coli Dr adhesins: decay-accelerating factor receptor recognition and invasiveness. J Infect Dis. 2001;183(Suppl 1):S24–S27. doi: 10.1086/318846. [DOI] [PubMed] [Google Scholar]
- 12.Pham T, Kaul A, Hart A, Goluszko P, Moulds J, Nowicki S, Lublin D M, Nowicki B J. dra-related X adhesins of gestational pyelonephritis-associated Escherichia coli recognize SCR-3 and SCR-4 domains of recombinant decay-accelerating factor. Infect Immun. 1995;63:1663–1668. doi: 10.1128/iai.63.5.1663-1668.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Telen M J, Hall E E, Green A M. Identification of human erythrocyte blood group antigens on decay-accelerating factor (DAF) and an erythrocyte phenotype negative for DAF. J Exp Med. 1988;167:1993–1998. doi: 10.1084/jem.167.6.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thomas J, Webb W, Davitz M A, Nussenzweig V. Decay accelerating factor diffuses rapidly on HeLa cell surfaces. Biophys J. 1987;51:522a. . (Abstract.) [Google Scholar]
- 15.Zhang L, Foxman B, Tallman P, Cladera E, Le Bouguenec C, Marrs C F. Distribution of drb genes coding for Dr binding adhesins among uropathogenic and fecal Escherichia coli isolates and identification of new subtypes. Infect Immun. 1997;65:2011–2018. doi: 10.1128/iai.65.6.2011-2018.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]