Abstract
Here, we present the complete mitochondrial genome of Pachliopta aristolochiae, a Common Rose butterfly from Malaysia. The sequence was generated using Illumina NovaSeq 6000 sequencing platform. The mitogenome is 15,235bp long, consisting of 13 protein-coding genes, 22 transfer RNAs, two ribosomal RNAs, and two D-loop regions. The total base composition was (81.6%), with A (39.3%), T (42.3%), C (11.0%) and G (7.3%). The gene order of the three tRNAs was trnM-trnI-trnQ, which differs from the ancestral insect gene order trnI-trnQ-trnM. Phylogenetic tree analysis revealed that the sequenced Pachliopta aristolochiae in this data is closely related to Losaria neptunus (NC 037868), with highly supported ML and BI analysis. The data presented in this work can provide useful resources for other researchers to study deeper into the phylogenetic relationships of Lepidoptera and the diversification of the Pachliopta species. Also, as one of the bioindicator species, this data can be used to assess environmental changes in the terrestrial and aquatic ecosystem via enviromental DNA approahes. The mitogenome of Pachliopta aristolochiae is available in GenBank under the accession number MZ781228.
Keywords: Pachliopta aristolochiae, Mitogenome, Lepidoptera, Papilionidae, Malaysia
Specifications Table
| Subject | Genomics |
| Specific subject area | Lepidoptera, Papilionidae, Mitogenomics |
| Type of data |
|
| How the data were acquired | Whole genome shotgun sequencing using Illumina NovaSeq 6000 platform with 150 paired-end mode (PE150) |
| Data format | Raw and analyzed |
| Parameters for data collection | Genomic DNA was extracted from fresh tissue sample of Pachliopta aristolochiae using the Qiagen Blood and Tissue Kit (Qiagen, Valencia, CA) and fragmented using a Bioruptor® system. The library was prepared using NEBNext® Ultra™ II DNA Library Prep Kit for Illumina®. The sample was then sent for sequencing using the Illumina NovaSeq 6000 platform with 150 paired-end mode (PE150). |
| Description of data collection | The assembly was done using NOVOPlasty v.4.2 and run through a PALEOMIX BAM pipeline to assess the mitogenome mapping. Annotation was done using the MITOS v2 web server and the predicted protein-coding genes were further verified using the Open Reading Frame (ORF) Finder. The circular mitogenome map was generated using OGDRAW. PhyloSuite v1.2.2 was used to extract, align and concatenate 13 protein-coding genes from 22 Lepidoptera mitogenomes prior to phylogenetic analysis. IQ-Tree and MrBayes v3.2.7 programs were used to build the phylogenetic trees using Maximum-Likelihood (ML) and Bayesian Inference (BI) probability method. PartitionFinder v2.2.1 was used to set the best partitioning schemes for the dataset. The resulting phylogenetic trees were visualized using Figtree v1.4.4. |
| Data source location | The sample Pachliopta aristolochiae (voucher no: DIB022) was collected from Sungai Semawak Taman Negara Endau-Rompin Johor, Malaysia (5.62 N, 100.46 E) in March 2019. |
| Data accessibility | Repository name: NCBI BioProject Data identification number: PRJNA753627 Direct URL to data: http://www.ncbi.nlm.nih.gov/bioproject/753627 Repository name: NCBI GenBank Data identification number: MZ781228 Direct URL to data: https://www.ncbi.nlm.nih.gov/nuccore/mz781228 Repository name: Mendeley Data Data identification number: 10.17632/n52pmth7cc.2 Direct URL to data: https://data.mendeley.com/datasets/n52pmth7cc/2 |
Value of the Data
-
•
The sequenced mitochondrial genome of the Common Rose butterfly, Pachliopta aristolochiae in this data represents the Pachliopta species originating from Malaysia.
-
•
As one of the bioindicator species, this mitogenome data can be used to assess environmental changes in the terrestrial and aquatic ecosystem via environmental DNA approaches.
-
•
The additional mitogenome data of Pachliopta aristolochiae generated can also provide the relevant information needed for other researchers to study deeper into the phylogenetic relationships of Lepidoptera and the diversification of the Pachliopta species.
1. Data Description
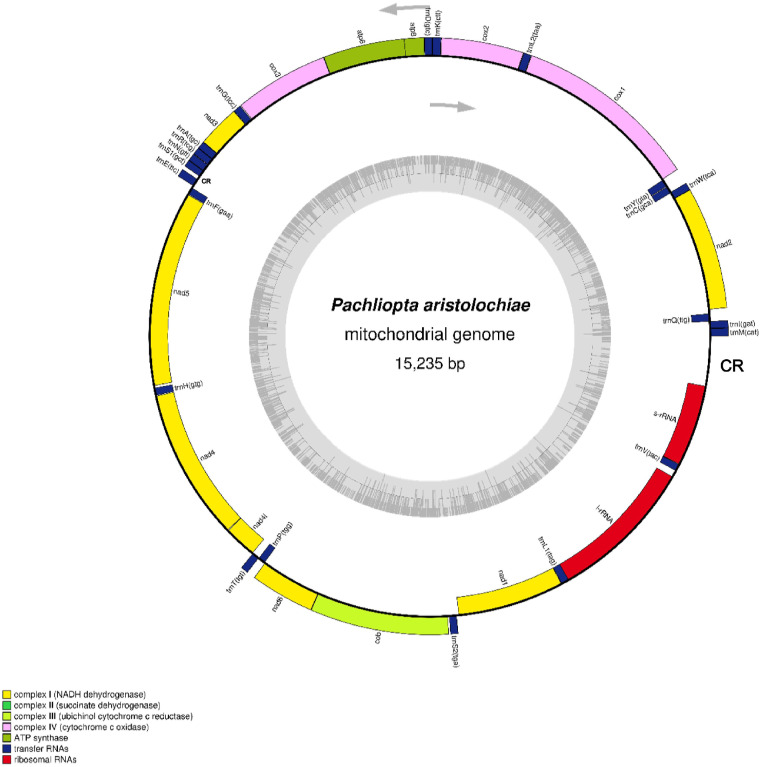
The Common Rose butterfly, Pachliopta aristolochiae mitogenome is a circular DNA with a total of 15,235bp in length (Fig. 1). Table 1 shows the statistical data information for the sequence reads. The mitogenome encodes 13 protein-coding genes (PCGs), 22 transfer RNAs, 2 ribosomal RNAs, and two D-loop regions (Table 2). The gene order of P.aristolochiae located between the D-loop and NAD2 was trnM-trnI-trnQ, which had been observed in most Lepidoptera mitogenomes, however, it differs from that of the ancestral insect gene order, trnI-trnQ-trnM [1]. The total size of the PCGs was 11,178bp in length and the tRNAs were 1,452bp long, ranging from 60bp to 71bp. Meanwhile, the sizes for the 12S and 16S RNAs are 719bp and 1280bp respectively. The majority of the PCGs (NAD2, COX1, COX2, ATP8, ATP6, COX3, NAD3, NAD6, CYTB) are scattered on the heavy strand, and NAD5, NAD4, NAD4l, NAD1 are on the light strand. Out of 13 PCGs, 12 were initiated by the typical ATN codon except for COX1 which uses the CGA start codon. Contrary to the start codon, two PCGs (COX2 and NAD4) were terminated with the incomplete stop codon T and the others were terminated by either TAA or TAG stop codon. The phenomena of incomplete termination codon had been observed in most Lepidoptera mitogenomes, and are associated with the polyadenylation process [2]. The mitogenome of P. aristolochiae showed an AT content of 81.64% with the base composition of A (39.3%), T (42.3%), C (11.0%), and G (7.3%) as shown in Table 3. The nucleotide skew statistics of the whole mitogenome indicates a high occurrence of T over A, and C over G with an AT-skew of -0.037 and GC-skew of -0.202.
Fig. 1.
Mitogenome map of Pachliopta aristolochiae generated using OGDRAW [3]. The genes scattered on the heavy strand are shown on the outer side of the circle, while the inner side shows those that are scattered on the light strand. The arrows indicate the direction of gene transcription. CR represents the control region (D-loop).
Table 1.
Sequencing data of Pachliopta aristolochiae mitogenome.
| Pachliopta aristolochiae | |
|---|---|
| Raw reads | 10,102,746 |
| Trimmed reads | 10,102,675 |
| Ave. read length | 149.5 |
| Mapped reads | 17,890 |
| % mapped reads | 0.002 |
| Depth of coverage (X) | 175.72 |
Table 2.
Gene features of Pachliopta aristolochiae mitogenome.
| Position |
|||||
|---|---|---|---|---|---|
| Gene (anticodon) | Start | Stop | Direction | Size | Start/Stop codon |
| trnM(cat) | 1 | 67 | F | 67 | |
| trnI(gat) | 67 | 130 | F | 64 | |
| trnQ(ttg) | 128 | 196 | R | 69 | |
| NAD2 | 231 | 1244 | F | 1014 | ATT/TAA |
| trnW(tca) | 1243 | 1307 | F | 65 | |
| trnC(gca) | 1300 | 1365 | R | 66 | |
| trnY(gta) | 1368 | 1434 | R | 67 | |
| COX1 | 1437 | 2967 | F | 1531 | CGA/TAA |
| trnL2(taa) | 2968 | 3034 | F | 67 | |
| COX2 | 3035 | 3716 | F | 682 | ATG/T |
| trnK(ctt) | 3717 | 3787 | F | 71 | |
| trnD(gtc) | 3787 | 3853 | F | 67 | |
| ATP8 | 3854 | 4021 | F | 168 | ATT/TAA |
| ATP6 | 4015 | 4692 | F | 678 | ATG/TAA |
| COX3 | 4692 | 5477 | F | 786 | ATG/TAA |
| trnG(tcc) | 5481 | 5546 | F | 66 | |
| NAD3 | 5547 | 5900 | F | 354 | ATA/TAG |
| trnA(tgc) | 5899 | 5963 | F | 65 | |
| trnR(tcg) | 5963 | 6024 | F | 62 | |
| trnN(gtt) | 6025 | 6089 | F | 65 | |
| trnS1(gct) | 6089 | 6148 | F | 60 | |
| D-loop | 6148 | 6192 | F | 45 | |
| trnE(ttc) | 6178 | 6246 | F | 69 | |
| trnF(gaa) | 6265 | 6330 | R | 66 | |
| NAD5 | 6333 | 8048 | R | 1716 | ATT/TAA |
| trnH(gtg) | 8067 | 8133 | R | 67 | |
| NAD4 | 8137 | 9472 | R | 1336 | ATG/T |
| NAD4l | 9474 | 9764 | R | 291 | ATG/TAA |
| trnT(tgt) | 9767 | 9831 | F | 65 | |
| trnP(tgg) | 9832 | 9896 | R | 65 | |
| NAD6 | 9899 | 10432 | F | 534 | ATT/TAA |
| CYTB | 10432 | 11580 | F | 1149 | ATG/TAA |
| trnS2(tga) | 11593 | 11657 | F | 65 | |
| NAD1 | 11674 | 12612 | R | 939 | ATG/TAA |
| trnL1(tag) | 12613 | 12683 | R | 71 | |
| 16S rRNA | 12659 | 13963 | R | 1280 | |
| trnV(tac) | 14021 | 14083 | R | 63 | |
| 12S rRNA | 14084 | 14802 | R | 719 | |
| D - loop | 14816 | 15235 | F | 420 | |
Table 3.
Base composition and AT/GC skewness for each gene region of Pachliopta aristolochiae mitogenome.
| Gene | Size (bp) | A% | G% | T% | C% | A+T% | AT skew | GC skew |
|---|---|---|---|---|---|---|---|---|
| Whole mitogenome | 15,235 | 39.3 | 7.3 | 42.3 | 11.0 | 81.6 | −0.037 | −0.202 |
| Protein coding | 11,178 | 33.5 | 10.1 | 46.8 | 9.6 | 80.3 | −0.166 | 0.025 |
| tRNA | 1,452 | 43.0 | 10.5 | 39.1 | 7.5 | 82.1 | 0.048 | 0.167 |
| rRNA | 2,024 | 43.6 | 10.4 | 40.8 | 5.2 | 84.4 | 0.033 | 0.333 |
| D-loop (major) | 365 | 46.3 | 1.6 | 49.6 | 2.5 | 95.9 | −0.034 | −0.220 |
| D-loop (minor) | 45 | 46.7 | 2.2 | 51.1 | 0.0 | 97.8 | −0.045 | 1.000 |
Two D-loop regions were found in the sequenced mitogenome of P.aristolochiae for this data. The first region was found at the position 6148bp to 6192bp, located between trnS1 and trnE. This region is 45bp long, which contained a string of microsatellite-like element (AT). Meanwhile, the second D-loop region was 420bp long, located between 12S rRNA and trnM, spanning a conserved ATAGA motif, followed by a poly-T stretch, and a microsatellite-like element (AT)₉ and (TA)₆ after the motif ATTTA, as commonly found in all Lepidoptera mitogenomes [4]. Fig. 2 describe the features of the two D-loop regions.
Fig. 2.
Features of the two D-loop regions of Pachliopta aristolochiae mitogenome located between trnS1 and trnE, as well as 12S rRNA and trnM. Conserved motifs ‘ATAGA’ and ‘ATTTA’ are indicated in red and blue respectively. Poly-T stretch is indicated in green while microsatellite-like elements (TA)n and (AT)n are shown in yellow.
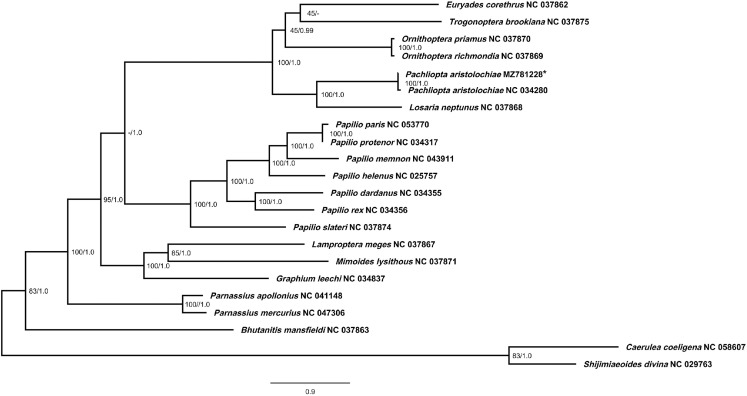
Maximum-Likelihood (ML) and Bayesian Inference (BI) probability tree were generated using 13 PCGs of 22 Lepidoptera mitogenomes from the family Papilionidae and Lycaenidae obtained from GenBank, including the sequenced P. aristolochiae in this data (Table 4). The resulting trees yielded identical topology under the ML and BI analysis (Fig. 3). Most of the nodes are highly supported with bootstrap value of more than 70% in ML analysis, and a Bayesian posterior probability of more than 0.95 in BI analysis. The sequence P. aristolochiae (MZ781228) in this study is clustered with the previously sequenced P. aristolochiae (NC 034280) and are closely related to Losaria neptunus (NC 037868), supported with a bootstrap value of 100% in ML and 1.0 posterior probability value in BI. A BLASTn analysis was also conducted to compare between the two mitogenomes of P.aristolochiae, where P.aristolochiae (MZ781228) in this data is 99.42% similar to P. aristolochiae (NC 034280) deposited in GenBank.
Table 4.
Lepidoptera mitogenomes used to build the phylogenetic tree analysis. The sequenced P.aristolochiae in this data is indicated by (*), with GenBank Accession No. MZ781228.
| Family | Subfamily | Species | GenBank Accession No. |
|---|---|---|---|
| Papilionidae | Papilioninae | Papilio paris | NC 053770 |
| Papilionidae | Parnassiinae | Parnassius mercurius | NC 047306 |
| Papilionidae | Papilioninae | Papilio memnon | NC 043911 |
| Papilionidae | Parnassiinae | Parnassius apollonius | NC 041148 |
| Papilionidae | Papilioninae | Pachliopta aristolochiae | NC 034280 |
| Papilionidae | Papilioninae | Papilio protenor | NC 034317 |
| Papilionidae | Papilioninae | Papilio dardanus | NC 034355 |
| Papilionidae | Papilioninae | Papilio rex | NC 034356 |
| Papilionidae | Papilioninae | Graphium leechi | NC 034837 |
| Papilionidae | Papilioninae | Papilio helenus | NC 025757 |
| Papilionidae | Papilioninae | Euryades corethrus | NC 037862 |
| Papilionidae | Parnassiinae | Bhutanitis mansfieldi | NC 037863 |
| Papilionidae | Papilioninae | Lamproptera meges | NC 037867 |
| Papilionidae | Papilioninae | Losaria neptunus | NC 037868 |
| Papilionidae | Papilioninae | Ornithoptera richmondia | NC 037869 |
| Papilionidae | Papilioninae | Ornithoptera priamus | NC 037870 |
| Papilionidae | Papilioninae | Mimoides lysithous | NC 037871 |
| Papilionidae | Papilioninae | Papilio slateri | NC 037874 |
| Papilionidae | Papilioninae | Trogonoptera brookiana | NC 037875 |
| Papilionidae | Papilioninae | Pachliopta aristolochiae* | MZ781228 |
| Lycaenidae | Polyommatinae | Caerulea coeligena | NC 058607 |
| Lycaenidae | Polyommatinae | Shijimiaeoides divina | NC 029763 |
Fig. 3.
Phylogenetic tree of Pachliopta aristolochiae (MZ781228), indicated by asterisk (*) and 21 other Lepidoptera mitogenomes built using Maximum-Likelihood (ML) and Bayesian Inference (BI) approach. Bootstrap support values were indicated on each tree node, showing the results of ML and BI analysis. Caerulea coeligena (NC 058607) and Shijimiaeoides divina (NC 029763) from the family Lycaenidae were used as outgroups.
2. Experimental Design, Materials and Methods
2.1. Sample collection, DNA extraction and pre-processing
The sample Pachliopta aristolochiae (voucher no: DIB022) was collected from Sungai Semawak Taman Negara Endau-Rompin Johor, Malaysia (5.62 N, 100.46 E) in March 2019. The genomic DNA was extracted from a fresh tissue sample using Qiagen Blood and Tissue Kit (Qiagen, Valencia, CA) and was fragmented using a Bioruptor® system [5]. The library preparation was done using NEBNext® Ultra™ II DNA Library Prep Kit for Illumina®, following the manufacturer's instructions. Then, the library was sent for sequencing using the Illumina NovaSeq 6000 platform with 150 paired-end mode (PE150). A total of 10,102,764 raw reads were obtained and firstly verified using the FastQC program for quality assessment (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Next, the raw reads were trimmed for sequencing adapters, low-quality bases as well as Ns [6,7] using AdapterRemoval v2.3.2 [8]. Sequences with quality score of 20 and above were retained. Both the forward and reverse reads were interleaved into a single file before using PALEOMIX [9].
2.2. Mitogenome assembly, annotation and sequence analysis
The mitogenome was assembled using the NOVOPlasty v.4.2 [10] program with the default parameter. The reference sequence and seed input were taken from BOLD public data (http://barcodinglife.org/), with the sequence ID BKKP127-18.COI-5P. Next, the assembled mitogenome was run through PALEOMIX BAM pipeline [9] using default parameters to remove reads shorter than 15 bp after trimming. The mitogenome annotation was carried out using MITOS v2 web server [11], with reference set ‘RefSeq 81 Metazoa’ and genetic code ‘5’ for invertebrates. Then, the predicted proteins were verified using the Open Reading Frame (ORF) Finder (https://www.ncbi.nlm.nih.gov/orffinder/) server using BLASTP. To improve the genome annotation, the predicted proteins from MITOS v2 web server [11] and ORF Finder were aligned with the reference sequence of Pachliopta aristolochiae (NC 034280) in GenBank using Jalview 2 v11.1.4 [12]. Tablet software [13] was used to manually check for insertion and deletion of bases, as well as the sequence coverage. The total base compositions were calculated using BioEdit [14]. The AT/GC skewness was calculated as follows: AT skew= (A-T)/(A+T) and GC skew=(G-C)/(G+C), where each letter represents the total percentage of the respective base count. The annotated mitogenome sequence file was converted into GenBank format using GB2sequin web application [15]. The GenBank file format was then used to generate the circular mitogenome map using OGDRAW [3].
2.3. Phylogenetic analysis
A total of 21 available Lepidoptera mitogenomes from the family Papilionidae and Lycaenidae were obtained from GenBank (Table 4). Caerulea coeligena (NC 058607) and Shijimiaeoides divina (NC 029763) from the family Lycaenidae were used as outgroups. The PCGs of each Lepidoptera mitogenomes were firstly extracted using the PhyloSuite v1.2.2 [16] platform. The 13 protein-coding genes were then aligned in batches using the MAFFT program integrated into PhyloSuite [16] and were concatenated. Phylogenetic analyses were performed using Maximum-Likelihood (ML) and Bayesian Inference (BI) approach using the IQ-Tree [17] program implemented in PhyloSuite v1.2.2 [16] and MrBayes v3.2.7 [18] respectively. PartitionFinder v2.1.1 [19] was used to determine the best partitioning schemes for the dataset. Maximum-Likelihood (ML) tree was built using 5000 ultrafast bootstrapping with 1000 iterations, and the best substitution model was determine by PartitionFinder v2.1.1 [19]. For Bayesian Inference (BI) analysis, each partition was set to the GTR substitution model (nst=6) with gamma distributed rate variation across sites (rates=invgamma) and a proportion of invariable sites (GTR + Γ + I). The analysis was carried out for 10,000,000 generations with 4 chains, sampled every 1000 generations with a burn-in of 25% until the average standard deviation of split frequencies are less than 0.01. Tracer v1.7.2 was used to ensure sufficient parameter sampling and that the Estimated Sample Size (ESS) is more than 200 [20]. Both resulting trees were visualized using Figtree v1.4.4 (http://tree.bio.ed.ac.uk/software/figtree/).
CRediT authorship contribution statement
Marylin Miga: Conceptualization, Methodology, Data curation, Software, Validation, Writing – original draft. Puteri Nur Syahzanani Jahari: Data curation, Conceptualization, Methodology, Software, Validation, Writing – review & editing. Chan Vei Siang: Methodology, Software. Kamarul Rahim Kamarudin: Methodology. Mohd Shahir Shamsir: Methodology, Formal analysis, Resources, Funding acquisition. Lili Tokiman: Methodology. Sivachandran Parimannan: Formal analysis, Resources, Funding acquisition. Heera Rajandas: Formal analysis, Resources, Funding acquisition. Farhan Mohamed: Methodology, Software. Faezah Mohd Salleh: Conceptualization, Methodology, Resources, Writing – review & editing, Supervision, Funding acquisition.
Declaration of competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors are deeply indebted to the Johor National Parks Corporation for providing the research permit TNJ700-2/1/1.
This work was supported by the Program Konsortium Kecemerlangan Penyelidikan (JPT(BKPI)1000/016/018/25 (60)) provided by the Ministry of Higher Education of Malaysia and UTM-Transdisciplinary Research Grant (Q.J130000.3514.06G57/Q.J130000.3554.05G69) by Universiti Teknologi Malaysia.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.dib.2021.107740.
Appendix. Supplementary materials
References
- 1.Cao Y.Q., Ma C., Chen J.Y., Yang D.R. The complete mitochondrial genomes of two ghost moths, Thitarodes renzhiensis and Thitarodes yunnanensis: the ancestral gene arrangement in Lepidoptera. BMC Genomics. 2012;13:1–13. doi: 10.1186/1471-2164-13-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen L., Wahlberg N., Liao C.Q., Bin Wang C., Ma F.Z., Huang G.H. Fourteen complete mitochondrial genomes of butterflies from the genus Lethe (Lepidoptera, Nymphalidae, Satyrinae) with mitogenome-based phylogenetic analysis. Genomics. 2020;112:4435–4441. doi: 10.1016/j.ygeno.2020.07.042. [DOI] [PubMed] [Google Scholar]
- 3.Lohse M., Drechsel O., Kahlau S., Bock R. OrganellarGenomeDRAW-a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 2013;41:575–581. doi: 10.1093/nar/gkt289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun Y., Huang H., Zhang X., Xia J., Geng J., Zhang K. The complete mitochondrial genome of the Papilio paris (Lepidoptera : Papilionidae) Mitochondrial DNA Part B. 2020;5:733–735. doi: 10.1080/23802359.2020.1715281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shearing technologies, Bioruptor (R), (n.d.). https://www.diagenode.com/en/categories/bioruptor-shearing-device. Accessed September 30, 2021.
- 6.Jahari P.N.S., Abdul Malik N.F., Shamsir M.S., Gilbert M.T.P., Mohd Salleh F. The first complete mitochondrial genome data of Hippocampus kuda originating from Malaysia. Data Brief. 2020;31 doi: 10.1016/J.DIB.2020.105721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jahari P.N.S., Mohd Azman S., Munian K., Ahmad Ruzman N.H., Shamsir M.S., Richter S.R., Mohd Salleh F. Characterization of the mitogenomes of long-tailed giant rat, Leopoldamys sabanus and a comparative analysis with other Leopoldamys species. Mitochondrial DNA Part B Resour. 2021;6:502–504. doi: 10.1080/23802359.2021.1872433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schubert M., Lindgreen S., Orlando L. AdapterRemoval v2: Rapid adapter trimming, identification, and read merging. BMC Res. Notes. 2016;9:1–7. doi: 10.1186/s13104-016-1900-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schubert M., Ermini L., Der Sarkissian C., Jónsson H., Ginolhac A., Schaefer R., Martin M.D., Fernández R., Kircher M., McCue M., Willerslev E., Orlando L. Characterization of ancient and modern genomes by SNP detection and phylogenomic and metagenomic analysis using PALEOMIX. Nat. Protoc. 2014;9:1056–1082. doi: 10.1038/nprot.2014.063. [DOI] [PubMed] [Google Scholar]
- 10.Dierckxsens N., Mardulyn P., Smits G. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2017:45. doi: 10.1093/nar/gkw955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bernt M., Donath A., Jühling F., Externbrink F., Florentz C., Fritzsch G., Pütz J., Middendorf M., Stadler P.F. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013;69:313–319. doi: 10.1016/j.ympev.2012.08.023. [DOI] [PubMed] [Google Scholar]
- 12.Waterhouse A.M., Procter J.B., Martin D.M.A., Clamp M., Barton G.J. Jalview Version 2-A multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Milne I., Bayer M., Cardle L., Shaw P., Stephen G., Wright F., Marshall D. Tablet-next generation sequence assembly visualization. Bioinformatics. 2009;26:401–402. doi: 10.1093/bioinformatics/btp666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hall T., Biosciences I., Carlsbad C. BioEdit: an important software for molecular biology. GERF Bull. Biosci. 2011;2:60–61. [Google Scholar]
- 15.Lehwark P., Greiner S. GB2sequin - a file converter preparing custom GenBank files for database submission. Genomics. 2019;111:759–761. doi: 10.1016/j.ygeno.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 16.Zhang D., Gao F., Jakovlić I., Zou H., Zhang J., Li W.X., Wang G.T. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020;20:348–355. doi: 10.1111/1755-0998.13096. [DOI] [PubMed] [Google Scholar]
- 17.Trifinopoulos J., Nguyen L.T., von Haeseler A., Minh B.Q. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016;44:W232–W235. doi: 10.1093/nar/gkw256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ronquist F., Teslenko M., Van Der Mark P., Ayres D.L., Darling A., Höhna S., Larget B., Liu L., Suchard M.A., Huelsenbeck J.P. Mrbayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lanfear R., Frandsen P.B., Wright A.M., Senfeld T., Calcott B. Partitionfinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2017;34:772–773. doi: 10.1093/molbev/msw260. [DOI] [PubMed] [Google Scholar]
- 20.Rambaut A., Drummond A.J., Xie D., Baele G., Suchard M.A. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018;67:901–904. doi: 10.1093/sysbio/syy032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.