Abstract
An evaluation of fluorescent amplified fragment length polymorphism (FAFLP) for typing Neisseria gonorrhoeae using 52 isolates revealed that its discriminatory power approached that of Opa-typing in identifying epidemiologically linked isolates. Automated, accurate sizing of FAFLP amplified fragments permits objective data analysis and storage, making it an attractive method for large surveillance projects.
Elucidating relationships between isolates of Neisseria gonorrhoeae can contribute to the identification of outbreaks and sexual networks, the recognition of temporally or geographically predominant strains, and the monitoring of antibiotic resistance. Auxotyping and serotyping are commonly applied to N. gonorrhoeae and can serve as a preliminary screen of heterogeneity. Combined auxotype/serovar classification improves resolution but does not always distinguish between epidemiologically related and unrelated isolates (16). Molecular typing methods meet the need for higher resolution (3, 5, 10, 13, 14, 16, 18, 19, 22). Of these, Opa-typing (16), pulsed-field gel electrophoresis (18–20), and sequencing of the entire por gene (3) currently give the best discrimination between strains of N. gonorrhoeae (23). Sequencing of the por gene produces data that are easy to analyze and store in a database but involves at least four sequencing reactions and the associated cost. Opa-typing and pulsed-field gel electrophoresis are both less costly but generate data as restriction fragment patterns that are difficult to analyze objectively and are not easily stored in a database. Multilocus sequence typing is a useful typing method for several species of bacteria including Neisseria meningitidis. However, when applied to N. gonorrhoeae, sequencing of housekeeping genes was not informative because the species was found to be too uniform (21).
Amplified fragment length polymorphism (AFLP) analysis is a PCR-based genome sampling technique that reproducibly generates a fragment profile for each bacterial clone (11, 24). In its fluorescent format (FAFLP) it generates patterns of fragments that are accurately sized using internal lane standards. Using each fragment size as an individual character, data can be reduced to binary form and stored in a database in a spreadsheet format. FAFLP has been used successfully to investigate outbreaks of Streptococcus pyogenes, Staphylococcus aureus, and N. meningitidis (6–8). Here we have applied FAFLP to a collection of N. gonorrhoeae isolates and investigated its congruence with auxotyping, serotyping, and Opa-typing.
Fifty-two isolates (auxotyped and serotyped previously by standard methods [4, 12]) were used in this study. These represented six serovars (IA1, IA2, IA6, IB2, IB3, and IB16) and five auxotypes (A, arginine requiring; AHU, arginine, hypoxanthine, and uracil requiring; NR, nonrequiring; P, proline requiring; and PA°U, proline requiring, arginine requiring [not satisfied by ornithine], and uracil requiring), a total of 17 auxotype/serovar classes. Eleven isolates were from sexual contacts: three linked cases from Brighton and eight cases (one linked pair and two triplets from a single outbreak) from Bristol. The 41 other isolates, with no known epidemiological links, were from patients in three United Kingdom cities (Bristol, Leeds, and London) collected over a 6-year period.
N. gonorrhoeae strains were grown on heated blood agar plates overnight at 37°C in 5% CO2. Genomic DNA was extracted by standard methods (1). Plasmid DNA was purified as described previously (2) from three isolates (14890, 6346, and 7777). These collectively contained examples of gonococcal plasmids present in some of the isolates included in the present study (the cryptic, conjugative, tetracycline resistance, and African and Asian forms of the β-lactamase-producing plasmids).
Opa-typing was performed as described previously (17) on duplicate DNA preparations of isolates that were indistinguishable by FAFLP. FAFLP was carried out on duplicate DNA preparations as previously described for N. meningitidis (7), except that the primers Eco+A (5′GACTGCGTACCAATTCA) and Mse+0 (5′GATGAGTCCTGAGTAA) were used. These primers were used for N. gonorrhoeae in place of those previously published for N. meningitidis as they produced more discriminatory fragments in the size range analyzed (data not shown). Also, 4 μl (instead of 2 μl) of ligated DNA was used in the PCR in order to increase signal strength. Thermal cycling conditions were as follows: predenaturation at 94°C for 2 min, followed by 30 cycles of denaturation at 94°C for 20 s, a 30-s annealing step, and extension at 72°C for 2 min. The annealing temperature for the first cycle, 66°C, was subsequently reduced by 1°C for the next 9 cycles, with the remaining 20 cycles at an annealing temperature of 56°C. This was followed by a 30-min incubation at 60°C. FAFLP was also carried out on subcultures of two isolates (8005 and 14890) before and after 11 passages and on plasmid preparations of three strains (6346, 7777, and 14890). For all isolates, each amplified-fragment size within the range 60 to 500 bp was assigned as an individual character, and the presence/absence was scored for each isolate using Genotyper software (Applied Biosystems). These binary data, stored in an Excel spreadsheet and then exported as text, were then analyzed by using an in-house software package described previously to calculate pairwise distances between isolates (7). These data were exported into Neighbor, a part of the Phylip package, and a tree was generated using the neighbor-joining method.
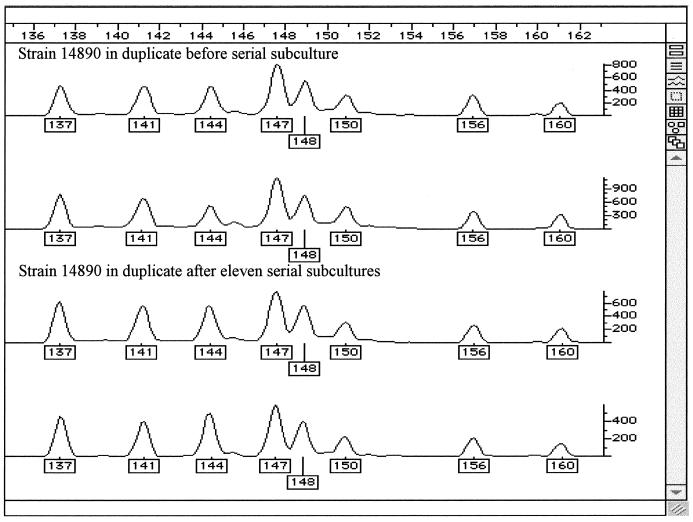
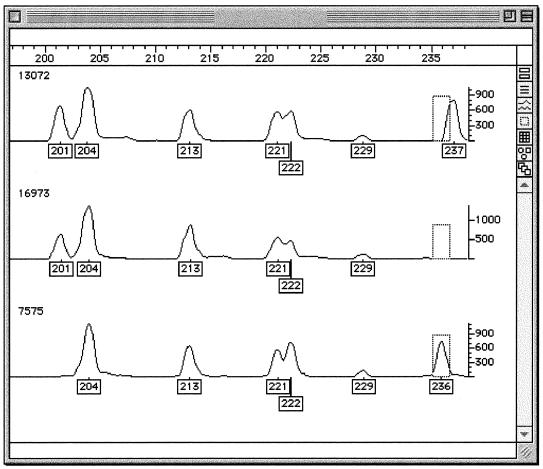
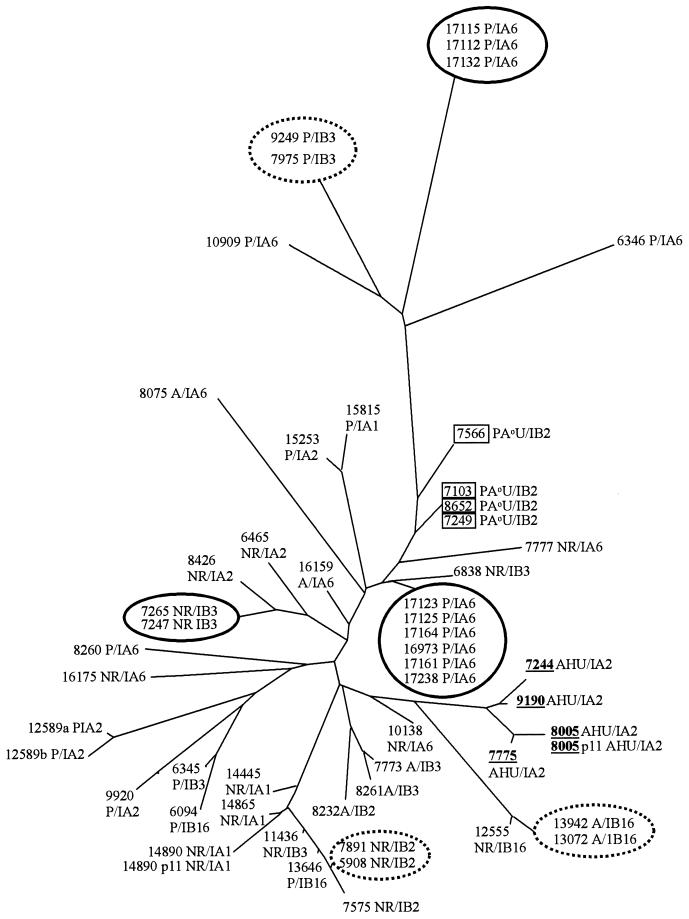
Duplicate analyses of all isolates were identical with the exception of one strain (12589), which differed by a single amplified fragment from the original result (possibly poor quality DNA may have partially inhibited the restriction digestion). Two strains (14890 and 8005) examined before and after serial subculture gave identical fragment amplification patterns. Figure 1 shows trace data from strain 14890 in duplicate before and after serial subculture. A total of 83 DNA fragments were identified, 40 of which were common to all isolates. Figure 2 shows trace data demonstrating the presence/absence of fragments of differing size generated from the genomes of three unrelated strains. Analysis of the FAFLP profiles using the neighbor-joining method yielded the tree shown in Fig. 3. Trees were also generated using different algorithms in the Phylip package, including UPGMA (unweighted pair-group method with arithmetic mean), which generated trees with exactly the same topology (data not shown). FAFLP profiles of the plasmid preparations gave four amplified fragments which, since these coincided with chromosomally derived amplified fragments, were excluded from the analysis.
FIG. 1.
Genotyper outputs of strain 14890 after serial subculture for the size range 135 to 163 bp. The boxed numbers under the peaks of the traces are the fragment sizes in base pairs assigned by comparison with the standard curve generated with the internal size standard. The panels show fragment sizes in base pairs before subculture (top two panels) and after 11 subcultures (bottom two panels).
FIG. 2.
Genotyper output of FAFLP analysis of unrelated strains 13072 (top panel), 16973 (middle panel), and 7575 (bottom panel) in the size range of 197- to 237-bp fragments. It is clear that the fragment at 201 bp (boxed) is present for strains 13072 and 16973 but not for strain 7575. Also, the fragment present in the dashed box at 236 bp is present for strain 7575 but absent for the other two strains and the fragment present at 237 bp for strain 13072 is absent for the other two strains.
FIG. 3.
FAFLP-generated distance tree for 52 strains of N. gonorrhoeae. FAFLP was carried out in duplicate for all strains. These gave identical profiles except for strain 12589, which differed by one fragment (12589a and 12589b). Epidemiologically related strains (solid circle) were indistinguishable by FAFLP. Epidemiologically unrelated strains of the auxotype AHU (underlined), and to some extent PA°U (boxed), clustered together by FAFLP. Strains of other auxotypes did not group together. Three strains of the auxotype PA°U (7103, 7249, and 8652) were indistinguishable by FAFLP but differed by two to three fragments when analyzed by Opa-typing. Three pairs of strains not known to be epidemiologically related (dashed circle) were indistinguishable by FAFLP or Opa-typing. The phenotype of strains is given as auxotype/serovar. Auxotypes are as follows: A, arginine requiring; AHU, arginine, hypoxanthine, and uracil requiring; NR, nonrequiring; P, proline requiring; and PA°U, proline requiring, arginine requiring (not satisfied by ornithine), and uracil requiring. Serovars are as follows: IA, Porin IA, IB, and Porin IB, with numbers designated according to the established scheme (12).
FAFLP subdivided the 17 auxotype/serovar classes of N. gonorrhoeae into a total of 39 distinguishable groups. Epidemiologically unrelated strains of the auxotype AHU (underlined in Fig. 3) and to some extent of auxotype PA°U (boxed in Fig. 3) formed clusters by FAFLP. No clustering of strains from other auxotypes or individual serovars was observed. Seven groups of isolates that were indistinguishable by FAFLP were Opa-typed. Sets of isolates from sexual contacts and from an outbreak (solid circles in Fig. 3) were indistinguishable by both FAFLP and Opa-typing. Three pairs of isolates not known to be epidemiologically linked were indistinguishable by auxotype, serotype, and both genotypic methods (dashed circles in Fig. 3). These were 7975 and 9249, isolated in Bristol and Leeds, respectively, within 6 months of each other; 13072 and 13942, isolated in Bristol within 8 months of each other; and 5908 and 7891, isolated in London within 17 months of each other. Three strains of the PA°U auxotype (7103, 7249, and 8652) were indistinguishable by FAFLP but gave two or three band differences between each other when analyzed by Opa-typing.
FAFLP is more discriminatory than a combined auxotyping and serotyping classification; and using the enzyme and primer combination described, it approaches the discrimination achieved with Opa-typing. As FAFLP analysis generates accurately sized fragment patterns that can easily be reduced to binary data and stored in a database in a spreadsheet format, it is an excellent tool for genotyping large numbers of isolates.
All epidemiologically related isolates and three pairs not known to be related were indistinguishable by FAFLP and Opa-typing. Other studies have indicated that isolates sharing the same Opa-type are probably members of a single sexual network. Clones of N. gonorrhoeae sharing the same Opa-type have been isolated over temporal and geographical distances similar to those seen in the present study (17; C. A. Ison, I. Martin, S. Day, G. Bell, A. Ghani, G. Garnett, G. Kinghorn, J. Weber, and H. Ward, Abstr. 12th Meet. Int. Soc. Sex. Transm. Dis. Res., p. 80, 1997). The three pairs of isolates identified by both genotyping methods may therefore originate from related members of unrecognized sexual networks. Though there was a clear correlation between FAFLP clustering and the AHU auxotype, and possibly between FAFLP clustering and the PA°U auxotype, further isolates of these auxotypes should be analyzed to verify this. Other typing methods have shown the AHU and PA°U auxotype to be clonal (9, 14).
FAFLP is a rapid, highly discriminatory typing tool that is able to distinguish epidemiologically related from unrelated isolates of N. gonorrhoeae. It generates reproducible, accurately sized fragment patterns that can be stored in a database, and it is of particular value for objectively genotyping large numbers of isolates. The start-up costs of sequence-based and FAFLP-based typing are comparable, and the sample processing cost by FAFLP is US$10 per isolate (15) but US$40 for four sequencing reactions (needed for por sequence typing). By comparison, the start-up and sample costs of nonradioactive Opa-typing are minimal but the lack of accurate data analysis and storage is a serious constraint for large-scale typing projects.
Acknowledgments
We are grateful to C. A. Ison for serotyping five isolates, A. Turner for advice on strain selection, and P. Mortimer and A. Herring for critical review of the manuscript.
REFERENCES
- 1.Ausubel F M, Brent R, Kingstown R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology I. New York, N.Y: John Wiley & Sons; 1989. Preparation of genomic DNA from bacteria. [Google Scholar]
- 2.Bennett P M, Heritage J, Hawkey P M. An ultra-rapid method for the study of antibiotic resistance plasmids. J Antimicrob Chemother. 1986;18:421–424. doi: 10.1093/jac/18.3.421. [DOI] [PubMed] [Google Scholar]
- 3.Cooke S J, de la Paz H, Poh C L, Ison C A, Heckels J E. Variation within serovars of Neisseria gonorrhoeae detected by structural analysis of outer-membrane protein PIB and by pulsed-field gel electrophoresis. Microbiology. 1997;143:1415–1422. doi: 10.1099/00221287-143-4-1415. [DOI] [PubMed] [Google Scholar]
- 4.Copley C G, Egglestone S I. Auxotyping of N. gonorrhoeae isolated in the United Kingdom. J Med Microbiol. 1983;16:295–302. doi: 10.1099/00222615-16-3-295. [DOI] [PubMed] [Google Scholar]
- 5.Dasi M A, Camarena J J, Ledesma E, Garcia R, Moreno F, Nogueira J M. Random amplification of polymorphic DNA of penicillinase-producing Neisseria gonorrhoeae strains. Genitourin Med. 1993;69:404–405. doi: 10.1136/sti.69.5.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Desai M, Tanna A, Wall R, Efstratiou A, George R, Stanley J. Fluorescent amplified-fragment length polymorphism analysis of an outbreak of group A streptococcal invasive disease. J Clin Microbiol. 1998;36:3133–3137. doi: 10.1128/jcm.36.11.3133-3137.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goulding J, Hookey J V, Stanley J, Olver W, Neal K R, Ala'Aldeen D A A, Arnold C. Fluorescent amplified fragment length polymorphism genotyping of Neisseria meningitidis identifies clones associated with invasive disease. J Clin Microbiol. 2000;38:4580–4585. doi: 10.1128/jcm.38.12.4580-4585.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grady R, O'Neill G, Cookson B, Stanley J. Fluorescent amplified fragment length polymorphism analysis of the MRSA epidemic. FEMS Lett. 2000;187:27–30. doi: 10.1111/j.1574-6968.2000.tb09131.x. [DOI] [PubMed] [Google Scholar]
- 9.Gutjahr T S, O'Rourke M, Ison C A, Spratt B G. Arginine-, hypoxanthine-, uracil-requiring isolates of N. gonorrhoeae are a clonal lineage within a non-clonal population. Microbiology. 1997;143:633–640. doi: 10.1099/00221287-143-2-633. [DOI] [PubMed] [Google Scholar]
- 10.Hobbs M M, Alcorn T M, Davis R H, Fischer W, Thomas J C, Martin I, Ison C A, Sparling P F, Cohen M S. Molecular typing of Neisseria gonorrhoeae causing repeated infections: evolution of porin during passage within a community. J Infect Dis. 1999;179:371–381. doi: 10.1086/314608. [DOI] [PubMed] [Google Scholar]
- 11.Janssen P, Coopman R, Huys G, Swings J, Bleeker M, Vos P, Zabeau M, Kersters K. Evaluation of the DNA fingerprinting method AFLP as a new tool in bacterial taxonomy. Microbiology. 1996;142:1881–1893. doi: 10.1099/13500872-142-7-1881. [DOI] [PubMed] [Google Scholar]
- 12.Knapp J S, Tam M R, Nowinski R C, Holmes K K, Sandstrom E G. Serological classification of N. gonorrhoeae with use of monoclonal antibodies to gonococcal outer membrane protein I. J Infect Dis. 1984;150:44–48. doi: 10.1093/infdis/150.1.44. [DOI] [PubMed] [Google Scholar]
- 13.Li H, Dillon J A R. Utility of ribotyping, restriction endonuclease analysis and pulsed-field gel electrophoresis to discriminate between isolates of Neisseria gonorrhoeae of serovar IA-2 which require arginine, hypoxanthine or uracil for growth. J Med Microbiol. 1995;43:208–215. doi: 10.1099/00222615-43-3-208. [DOI] [PubMed] [Google Scholar]
- 14.Ng L K, Dillon J A. Typing by serovar, antibiogram, plasmid content, ribo-typing, and isoenzyme typing to determine whether Neisseria gonorrhoeae isolates requiring proline, citrulline, and uracil for growth are clonal. J Clin Microbiol. 1993;3:1555–1561. doi: 10.1128/jcm.31.6.1555-1561.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Olive D M, Bean P. Principles and applications of methods for DNA-based typing of microbial organisms. J Clin Microbiol. 1999;37:1661–1669. doi: 10.1128/jcm.37.6.1661-1669.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O'Rourke M, Ison C A, Renton A M, Spratt B G. Opa-typing: a high-resolution tool for studying the epidemiology of gonorrhoea. Mol Microbiol. 1995;17:865–875. doi: 10.1111/j.1365-2958.1995.mmi_17050865.x. [DOI] [PubMed] [Google Scholar]
- 17.Palmer, H. M., J. P. Leeming, and A. Turner. Investigation of an outbreak of ciprofloxacin resistant Neisseria gonorrhoeae using a simplified opa-typing method. Epidemiol. Infect., in press. [DOI] [PMC free article] [PubMed]
- 18.Poh C L, Lau Q C. Subtyping of N. gonorrhoeae auxotype-serovar groups by pulsed-field gel electrophoresis. J Med Microbiol. 1993;38:366–370. doi: 10.1099/00222615-38-5-366. [DOI] [PubMed] [Google Scholar]
- 19.Poh C L, Lau Q C, Chow V T K. Differentiation of Neisseria gonorrhoeae IB-3 and IB-7 serovars by direct sequencing of protein IB gene and pulsed-field gel electrophoresis. J Med Microbiol. 1995;43:201–207. doi: 10.1099/00222615-43-3-201. [DOI] [PubMed] [Google Scholar]
- 20.Poh C L, Loh G K, Tapsall J W. Resolution of clonal subgroups among Neisseria gonorrhoeae IB-2 and IB-6 serovars by pulsed-field gel electrophoresis. Genitourin Med. 1995;71:145–149. doi: 10.1136/sti.71.3.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spratt B G, Maiden M C J. Bacterial population genetics, evolution and epidemiology. Philos Trans R Soc Lond B Biol Sci. 1999;354:701–710. doi: 10.1098/rstb.1999.0423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thompson D K, Deal C D, Ison C A, Zenilman J M, Bash M C. A typing system for Neisseria gonorrhoeae based on biotinylated oligonucleotide probes to PIB gene variable regions. J Infect Dis. 2000;181:1652–1660. doi: 10.1086/315464. [DOI] [PubMed] [Google Scholar]
- 23.Van Looveren M, Ison C A, Ieven M, Vandamme P, Martin I M, Vermeulen K, Renton A, Goossens H. Evaluation of the discriminatory power of typing methods for N. gonorrhoeae. J Clin Microbiol. 1999;37:2183–2188. doi: 10.1128/jcm.37.7.2183-2188.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kulper M, Zabeau M. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]