Abstract
Recently the most infectious disease is the novel Coronavirus disease (COVID 19) creates a devastating effect on public health in more than 200 countries in the world. Since the detection of COVID19 using reverse transcription-polymerase chain reaction (RT-PCR) is time-consuming and error-prone, the alternative solution of detection is Computed Tomography (CT) images. In this paper, Contrast Limited Histogram Equalization (CLAHE) was applied to CT images as a preprocessing step for enhancing the quality of the images. After that, we developed a novel Convolutional Neural Network (CNN) model that extracted 100 prominent features from a total of 2482 CT scan images. These extracted features were then deployed to various machine learning algorithms — Gaussian Naive Bayes (GNB), Support Vector Machine (SVM), Decision Tree (DT), Logistic Regression (LR), and Random Forest (RF). Finally, we proposed an ensemble model for the COVID19 CT image classification. We also showed various performance comparisons with the state-of-art methods. Our proposed model outperforms the state-of-art models and achieved an accuracy, precision, and recall score of 99.73%, 99.46%, and 100%, respectively.
Keywords: CLAHE, Convolutional Neural Network, COVID19, Ensemble learning, Feature scaling, Guassian Naive Bayes, Support Vector Machine, Soft voting
1. Introduction
At the end of December 2019, the world’s catastrophic Coronavirus Disease (COVID19) was first observed in Wuhan, China which is known as a respiratory disease caused by a severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (Ai, et al., 2020, Jaiswal et al., 2020). Now 220 million people are infected by COVID19 worldwide, and among them, 97,757 people are in critical condition. Almost 2 million people have died in the last two years around the world (Worldometer, 2021). The initial symptoms of COVID19 infected patients are dry cough, loss of taste sensation, fever, headache, diarrhoea, short breathing, sore throat, tiredness, and mild to moderate respiratory illness [31]. In the initial steps, the medical experts first guess a patient, whereas a patient has COVID19 infected or not using these symptoms. When a person have some of these symptoms, they are examined by medical experts and performed tests such as CT scan, chest X-ray, etc. Finally, doctors or medical experts inspect COVID19 from these tests (Mohamud, Mohamed, Ali, & Adam, 2020). This disease can spread when a COVID 19 infected patient sneezes or coughs travelling through the air and transmitted to the regular person through the nose or mouth. When this virus infects a person, it may take 5 to 6 days to show the symptoms of this disease (Tang, Schmitz, Persing, & Stratton, 2020). It may easy to recover when the disease is early detected. Still, the people who have chronic respiratory disease, heart diseases, diabetes, etc., may face difficulty recovering from this disease (Ahuja, Panigrahi, Dey, Rajinikanth, & Gandhi, 2021). This disease may be more life-threatening for older people than the younger generation. Since the virus is transmitted from an infected patient to a normal person, the only way to stop it is to quarantine the infected person. COVID19 can be detected from respiratory samplings by using Real-Time RT-PCR (Wang, et al., 2020). But the detection of this disease using RT-PCR is very time-consuming i.e. its take around 4 to 6 h for processing the samples (Pathak, Shukla, Tiwari, Stalin, & Singh, 2020) It also gives error-prone results i.e. high false-negative rates (Shah, et al., 2020, Zu, et al., 2020). For these drawbacks of RT-PCR COVID19 detection creates challenges in preventing the expansion of infection.
An alternative solution is to detect SARS-CoV-2 from different types of radiological imaging methods such as CT scans or chest X-ray images (Singh et al., 2020, Xie, et al., 2020). Using these techniques, the COVID19 patients can be detected quickly and quarantine infected patients timely and overcome this critical situation. But there is a problem with chest X-ray images that cannot be detected in the soft tissues (Tingting, Junqian, Lintai, & Yong, 2019). We can handle this problem by using chest CT scans which can be discriminated the soft tissues accurately (Jaiswal et al., 2020). A radiology expert is required to detect COVID19 infected patients from these chest CT scan images but it requires a lot of time and maybe defective. Hence it is necessary to design a decision support tool based on deep learning (DL) for the automatic prediction of COVID19 patients quickly and efficiently. DL plays a vital role in solving different types of problem for instances image classification (Islam, Siddique, Rahman, & Jabid, 2018), speech recognition, object detection (Szegedy, Toshev, & Erhan, 2013), disease detection (Nahiduzzaman, Islam, et al., 2021, Nahiduzzaman et al., 2019), etc. In a particular field like in image classification convolutional neural network gives astonishing results (Krizhevsky, Sutskever, & Hinton, 2017). For this reason, the main focus of our task is to automatic detection of COVID19 patients from chest CT scan images using CNN.
In overall, the paper shows the following contributions:
-
•
Enhanced the quality of the CT scan images using CLAHE.
-
•
Built a novel CNN for extracting the most relevant features from the CT scan images.
-
•
Proposed a soft voting ensemble learning model for improving the classification performance than previous works in terms of accuracy, precision, recall, AUC.
In the next sections, we describe the previous works in this field. Section 3 represents the proposed architecture of our model. The performance analysis of our task is presented in Section 4. Finally, Section 5 draws the main conclusion of this paper.
2. Literature review
Several research works and studies have been performed to detect COVID19 patients from chest CT scan images in the last two years. Zhang, Satapathy, Zhang, and Wang (2021) proposed a model where DenseNet and the optimization of transfer learning setting (OTLS) strategy were combined to create a revolutionary method. They achieved an accuracy of 96.30 ±0.56 and specificity of 96.25 ±1. Wang, Nayak, Guttery, Zhang, and Zhang (2021) proposed a structure that achieved a more outstanding performance. Firstly, pre-trained models (PTMs) were utilized to learn features, and a unique (L, 2) transfer feature learning approach was suggested to extract them. Secondly, they introduced a pre-trained network selection approach for fusion to choose the best two models defined by PTM and NLR. Thirdly, discriminant correlation analysis was developed to help fuse the two features from the two models via deep chest CT (CCT) fusion. They achieved the best sensitivity, precision, and F1-score of 98.30%, 97.38%, and 97.62%, respectively. Mishra, Das, Roy, and Bandyopadhyay (2020) detected the COVID19 patients using different transfer learning (TL) algorithms. They have also merged the results of several deep CNN algorithms using a decision fusion-based approach to produce a final result. They used 744 CT scan images (347 COVID19 vs 397 Non-COVID19) and achieved an average accuracy of 88.34%, the area under the curve (AUC) of 88.32%, and an F1 score of 86.7%. Jaiswal et al. (2020) proposed pre-trained TL algorithms known as VGG16, Inception ResNet, ResNet 152V2, and DenseNet201 to classify patients as infected with COVID19 or not. They used 2492 CT scan images (1262 COVID19 vs 1230 Non-COVID19) and achieved the highest accuracy, specificity, precision, recall, F1 score, and AUC of 96.25%, 96.21%, 96.29%, 96.29%, 96.29%, and 97% respectively using DenseNet201. Wang, Liu, and Dou (2020) identified the COVID19 accurately using a joint learning framework known as redesigned COVID-Net, which performed separate feature normalization. They used 2492 CT scan images as Site A and 568 CT scan images as Site B for their model. They achieved an accuracy, precision, recall, F1 score, and AUC of 90.83±0.93%, 90.87±1.29%, 85.89±1.05%, 95.75±0.43%, and 96.24±0.35%, respectively for Site A. They achieved an accuracy, precision, recall, F1 score, and AUC of 78.69±1.54%, 78.83±1.43%, 79.71±1.42%, 78.02±1.34%, and 85.32±0.32%, respectively for Site B.
Yang, et al. (2020) developed a model for improving the accuracy by using contrastive self-supervised and multitask learning. They achieved an accuracy of 89%, a F1 score of 90%, and an AUC of 98%. Loey, Manogaran, and Khalifa (2020) proposed a deep transfer learning model for the detection of COVID-19 from chest CT scan images. They used classical data augmentation techniques and Conditional Generative Adversarial Nets (CGAN) for data processing. Five TL algorithms, including AlexNet, VGGNet16, VGGNet19, GoogleNet, and ResNet50, were used to detect COVID19. The best performance was achieved by ResNet50 with an accuracy of 82.91%, a sensitivity of 77.66%, and a specificity of 82.62%. Sarker, Islam, Hannan, and Ahmed (2021) proposed DenseNet121 for prediction of COVID19. They achieved an accuracy of 87% and also built a website that marks the infected area from the radiology images. Panwar, et al. (2020) proposed a transfer learning algorithm which is VGG19, for the detection of COVID19 patients from both chest X-ray and CT scan images. They also used Gradient Weighted Class Activation Mapping (Grad-CAM) to visualize the infected area. They achieved an overall accuracy of 95.61%. Amyar, Modzelewski, Li, and Ruan (2020) proposed a new multitask deep learning model to identify COVID19 lesions from chest CT scan images. They were tried to improve segmentation and classification performance and achieved an accuracy of 94.67%, a sensitivity of 96%, specificity of 92%, and Receiver Operating Characteristics (ROC) curve of 97%.
Narin, Kaya, and Pamuk (2020) proposed a deep convolutional neural network that automatically detects COVID19 from chest X-ray images. They further used transfer learning algorithms such as ResNet50, InceptionV3, and Inception-ResNetV2 that trained on 100 images (50 COVID19 vs 50 Non-COVID19) and achieved accuracy 97% and 87% using InceptionV3 and Inception-ResNetV2 respectively. Hemdan, Shouman, and Karar (2020) proposed a deep CNN that automatically detects COVID19 from chest X-ray images. They used VGG16 that trained on a limited dataset with 50 images (25 COVID19 vs 25 Non-COVID19) and achieved an accuracy of 90%. Kumar, Mishra, and Singh (2020) has developed a deep transfer learning algorithm called DeQueezeNet for the detection of COVID19 from chest X-ray images. They achieved an accuracy of 94.52% with a precision of 90.48%.
3. Proposed model
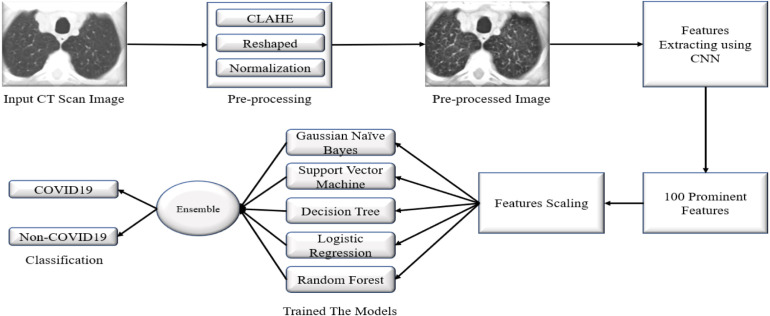
Deep learning has dramatically been used in medical imaging in the last few decades. We have collected the CT scan images of COVID19 patients in this work. The images were not cleared; for this reason, we need to preprocess our data using various methods. Then we developed a novel deep CNN for extracting the most discriminant features from the images. After extracting the features, we preprocessed these features then applied several well-known machine learning algorithms — GNB, SVM, DT, LR, and RF. In addition to this algorithm, a voting ensemble-based approach has been considered to make a final prediction. The main idea of this voting approach is that errors in the particular algorithm can be reduced by merging the particular decisions through a majority voting scheme (Maclin, 2016, Polikar and Polikar, 2006). Finally, we enhanced the overall performance of these algorithms using this ensemble method. Fig. 1 shows our proposed model to detect the COVID19 from CT scan images.
Fig. 1.
Proposed framework for detection of COVID19.
3.1. Preprocessing
Image preprocessing is an important task for getting a better result. Various methods had developed so far for enhancing medical images. We utilized CLAHE for image enhancement. Primarily, CLAHE was developed for the image enhancement of medical images of low-contrast (Pisano, et al., 1998). Clipping the histogram at a user-defined value called clip limit restricts the amplification in CLAHE. The clipping level controls how much noise in the histogram should be smoothed and, as a result, how much contrast should be increased. We used a colour version of CLAHE. For this, we kept the clipping limit of 2.0 and the tile grid size of ().
-
•
First, we converted our RGB image into a LAB image
-
•
After that, we utilized the CLAHE method to L channel
-
•
Then merged the enhanced L channel with A and B to get enhanced LAB image
-
•
Finally, that enhanced LAB image was converted back into the enhanced RGB image
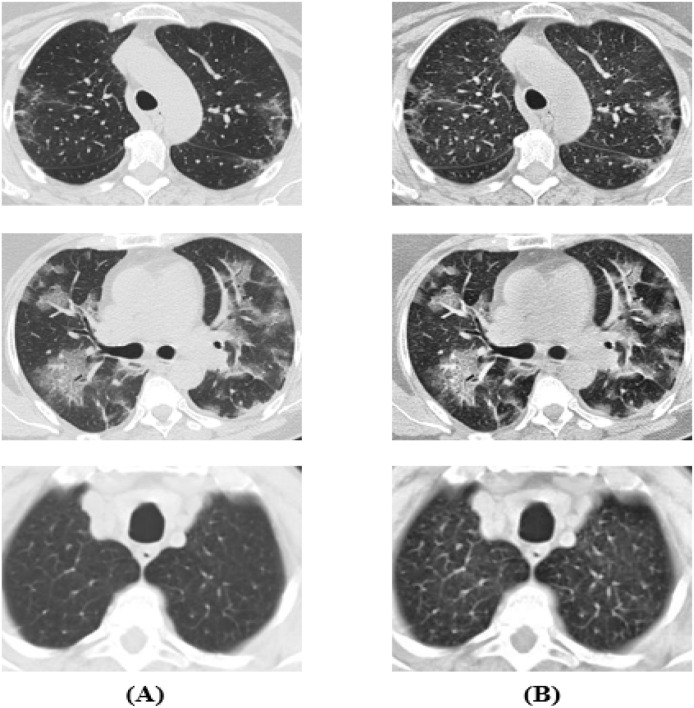
After that, all the images were resized into () as the images in the dataset come with various resolutions. Finally, we performed normalization on each image. In Fig. 2, we can see some original CT scan images and their corresponding enhanced CT scan images with the CLAHE method.
Fig. 2.
(A) Original image, (B) Image after applying CLAHE.
3.2. Feature extraction
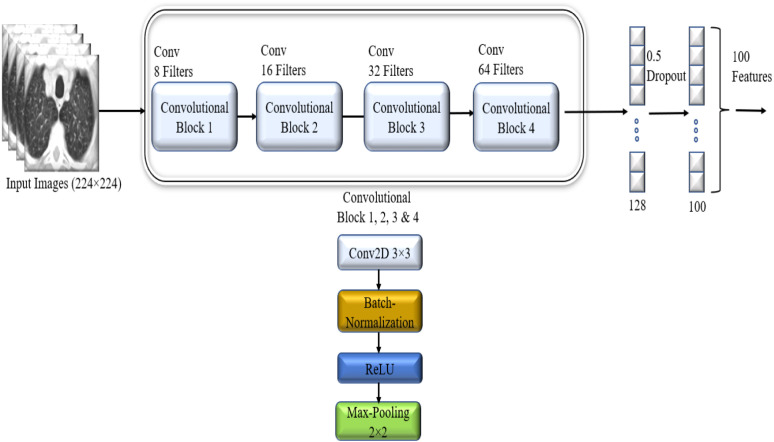
Feature Engineering is the most critical part of the classification. Feature extraction using image processing techniques are erroneous and tedious. As the features for COVID19 from CT scan images are complex, we used a deep convolutional neural network for extracting 100 prominent features for COVID19 identification. Fig. 3 shows our deep CNN model for extracting the features. We used four convolution layers, followed by batch normalization and max-pooling layers. After about 100 epochs, our model learned the parameters for COVID19 classification. During training time, the learning rate was 0.001, and ‘Adam’ was used as optimizer and a dropout with a 0.50 probability for getting more generalized results. After completing the learning process, we further deployed our dataset to the trained CNN model. We extracted the 100 prominent features of every image from the second last dense layer composed of 100 neurons. Table 1 shows the summary of our CNN model.
Fig. 3.
Feature extracted using CNN model.
Table 1.
Features extraction model summary.
| Layer name | Output shape |
|---|---|
| Input Layer | (224, 224, 3) |
| Conv2D | (222, 222, 8) |
| Batch_Normalization, Activation(ReLU) | (222, 222, 8) |
| MaxPooling2D | (111, 111, 8) |
| Conv2D_1 | (109, 109, 16) |
| Batch_Normalization_1, Activation_1(ReLU) | (109, 109, 16) |
| MaxPooling2D_1 | (54, 54, 16) |
| Conv2D_2 | (52, 52, 32) |
| Batch_Normalization_2, Activation_2(ReLU) | (52, 52, 32) |
| MaxPooling2D_2 | (26, 26, 16) |
| Conv2D_3 | (26, 26, 64) |
| Batch_Normalization_3, Activation_3(ReLU) | (26, 26, 64) |
| MaxPooling2D_3 | (13, 13, 64) |
| Flatten | (10816) |
| Dense1 | (128) |
| Batch_Normalization_4, Activation_4(ReLU) | (128) |
| Dropout | (128) |
| Dense2 | (100) |
3.3. Feature scaling
Feature scaling keeps the data’s independent features into a normalized range. It is done during data pre-processing to deal with highly varying magnitudes, values, or units. If feature scaling is not accomplished, a machine learning algorithm would consider larger values to be higher and smaller values to be lower, regardless of the unit of measurement. Having featured on a similar scale can help the gradient descent converge more quickly towards the minima. Various techniques have been developed for normalizing features. We used the standard scaler technique for standardizing the features, which improves the performance of the models (Nahiduzzaman, Goni, et al., 2021).
3.4. Gaussian Naive Bayes
Naive Bayes classifier is a probabilistic machine learning model that is used for binary (two-class) and multi-class classification problems. The classifier is based on the Bayes theorem:
| (1) |
The variable is the class variable and variable X represent the parameters/features. Where . Naive Bayes classifier assumed that attributes are independent of each other. So,
| (2) |
In the case of gaussian naive bayes, the conditional probability comes from a normal distribution like
| (3) |
3.5. Support vector machine
SVM is a supervised machine learning algorithm used for classification and regression problems. It performs classification by finding the hyper-plane that differentiates the classes very well. It finds the hyper-plane by maximizing the margin. In the kernel trick method, kernel function transforms low dimensional input space to a higher-dimensional space, i.e. it converts not separable problem to separable problem. It is primarily helpful in non-linear separable problems. We used sigmoid as a kernel function.
3.6. Decision tree classifier
A decision tree is a flowchart-like tree structure with an internal node representing a function (or attribute), a branch representing a decision law, and each leaf node representing the result. The root node is at the very top of a decision tree. It learns to partition based on the value of an attribute. Recursive partitioning is a method of partitioning the tree recursively. This flowchart-like form assists in making decisions. It is a flowchart diagram-style visualization that highly reflects human thought. As a result, decision trees are simple to comprehend and perceive.
3.7. Logistic Regression
Logistic Regression is a widely used mathematical method for predicting binary outcomes ( or 1). Linear regression is helpful for forecasting continuous-valued outcomes, whereas logistic regression is suitable for categorical outcomes (binomial/multinomial values of y). The standard logistic function, which is an S-shaped curve given by the equation:
| (4) |
3.8. Random forest
Random forest is a supervised learning algorithm. It creates a “forest” out of a set of decision trees, which are typically trained using the “bagging” technique. The bagging method’s general premise is that combining several learning models improves the final outcome. Moreover, It can handle by forming multiple numbers decisions tress during training and output is provided by class mode or averaging the individual tree’s prediction (Ho, 1995). Random forests can handle the overfitting problem of training data for decision trees (Hastie, Tibshirani, & Friedman, 2001).
3.9. Ensemble learning
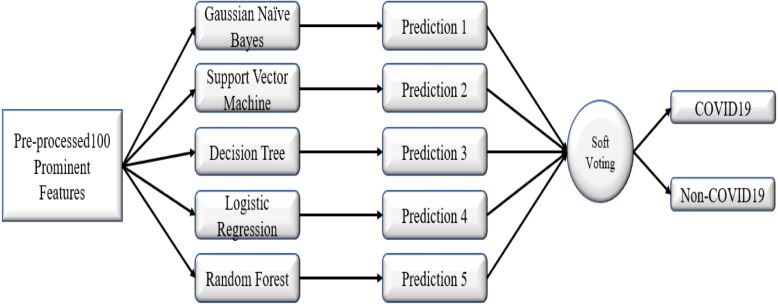
The ensemble model is created by strategically combining base models to create a robust model. The ensemble model employs a mixture of learning algorithms to solve a classification/regression problem that cannot be solved easily by either of the individual models. One can achieve more outstanding performance than a particular model using ensemble learning (Wolpert, 1992). Here we used soft voting ensemble learning. First, we trained base models — GNB, SVM, DT, LR, RF using training data. After training, we tested our models’ performance using test data, where each model gave an individual prediction. These models’ predictions act as an additional input to our ensemble learning that acts as a combined model trained to make the final prediction. Fig. 4 shows our proposed ensemble learning model.
Fig. 4.
Proposed ensemble learning model.
4. Performance analysis & experimental results
4.1. Dataset
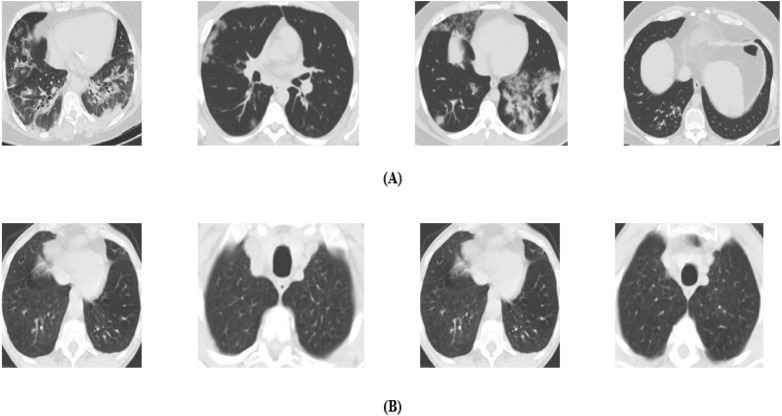
We collected the SARS-CoV-2 CT scan dataset from Kaggle (PlamenEduardo, 2020), but the original dataset was collected by Angelov and Almeida Soares (2020) from hospitals of Sao Paulo, Brazil. The dataset contains a total of 2482 CT scan images in which the total number of COVID19 infected patients are 1252, and Non-COVID19 patients are 1230. Although the patients who are not infected by SARS-CoV-2 but are infected by other pulmonary diseases. The CT scan images of COVID19 infected patients were collected from a total of 60 patients in which the total number of male patients is 32 and females are 28. At the same time, the CT scan images of Non-COVID19 patients were collected from 60 patients, including 30 male and 30 female patients. Fig. 5 shows some samples of our collected dataset.
Fig. 5.
CT scan images of: (A) COVID19 infected, (B) Non-COVID19 infected patients.
4.2. Data splitting
The main idea behind the machine learning algorithm is that we first need to learn our algorithm using some CT scan images called training data. After that, to calculate the performance of our model, we need to use some new CT scan images called test data that have not been used for training. So using this testing data, we can evaluate the efficiency of our model. We divided 2482 CT scan images into training and testing sets. We used 15% images for testing and 85% images for training. Table 2 shows our data splitting.
Table 2.
Datasets splitting into training & testing sets.
| Disease type | Training | Testing |
|---|---|---|
| COVID19 (0) | 1053 | 185 |
| Non-COVID19 (1) | 1056 | 188 |
| Total | 2109 | 373 |
4.3. Quantitative analysis
To perform a quantitative analysis of the machine learning algorithms, we considered various evaluation metrics, for instance, accuracy (Acc), precision (P), recall (R), f1-score, and the area under the curve (AUC). For classification report, true positive (TP) means that COVID19 infected patient detected as COVID19, true negative (TN) means that non-COVID19 infected patient detected as non-COVID19, false positive (FP) means that non-COVID19 infected patient wrongly detected as COVID19, and false negative (FN) means that COVID19 infected patient wrongly detected as a non-COVID19. The equation of difference performance measure matrices (Menditto et al., 2007, Powers, 2020) are given below.
| (5) |
| (6) |
| (7) |
| (8) |
4.4. Results analysis
The experiments were done at the Pycharm Community Edition19 (2020.2.3x64) software. All the machine learning models had been implemented using Keras with TensorFlow as a backend. The training and testing phases were performed on a 64-bit Windows 10 Pro operating system with 32 GB RAM, NVIDIA GeForce GTX 1650 SUPER 4 GB GPU, and Intel(R) Core(TM) i7-6700 CPU @3.40 GHz. The code is available in the GitHub repository:
https://github.com/robiulRUET/COVID19Detection2.
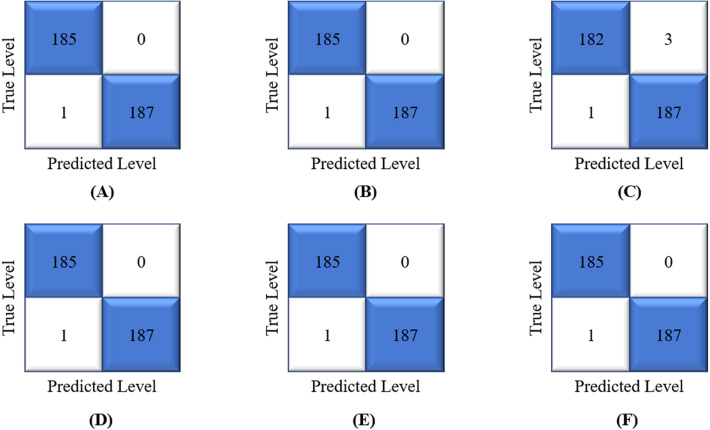
We trained our machine learning algorithms — GNB, SVM, DT, LR, RF using 2109 CT scan images of COVID19 and non-COVID19 infected patients. Then we tested all these models using 373 CT scan images where COVID19 and non-COVID19 infected patients were 185 and 188, respectively. Finally, we performed a soft voting ensemble-based approach considered to make a final detection. We used a confusion matrix for each model to evaluate the robustness of each model by determining the accuracy, precision, recall, f1-score, and AUC. In the case of the medical sector, the recall should be maximized because the patient who has been infected by COVID19 must be detected as COVID19 accurately. Fig. 6 shows the confusion matrix of each model.
Fig. 6.
Confusion matrices for (A) GNB, (B) SVM, (C) DT, (D) LR, (E) RF, and (F) Ensemble model.
The average accuracy of GNB, SVM, DT, RF, LR, and ensemble models are 99.73%, 99.73%, 98.43%, 99.73%, 99.73% and 99.73%, respectively. Table 3 shows the classification performance measures of all models.
Table 3.
Results of performance measures for machine learning models.
| Model name | Type | Precision | Recall | F1-score | Accuracy |
|---|---|---|---|---|---|
| GNB | COVID19 | 99% | 100% | 100% | – |
| Non-COVID19 | 99% | 100% | 99% | – | |
| Average | 99.46% | 100% | 99.73% | 99.73% | |
| SVM | COVID19 | 99% | 100% | 100% | – |
| Non-COVID19 | 99% | 100% | 99% | – | |
| Average | 99.46% | 100% | 99.73% | 99.73% | |
| DT | COVID19 | 99% | 98% | 99% | – |
| Non-COVID19 | 99% | 98% | 99% | – | |
| Average | 99.46% | 98.40% | 98.93% | 98.94% | |
| LR | COVID19 | 99% | 100% | 100% | – |
| Non-COVID19 | 99% | 100% | 99% | – | |
| Average | 99.46% | 100% | 99.73% | 99.73% | |
| RF | COVID19 | 99% | 100% | 100% | – |
| Non-COVID19 | 99% | 100% | 99% | – | |
| Average | 99.46% | 100% | 99.73% | 99.73% | |
| Ensemble Learning | COVID19 | 99% | 100% | 100% | – |
| Non-COVID19 | 99% | 100% | 99% | – | |
| Average | 99.46% | 100% | 99.73% | 99.73% |
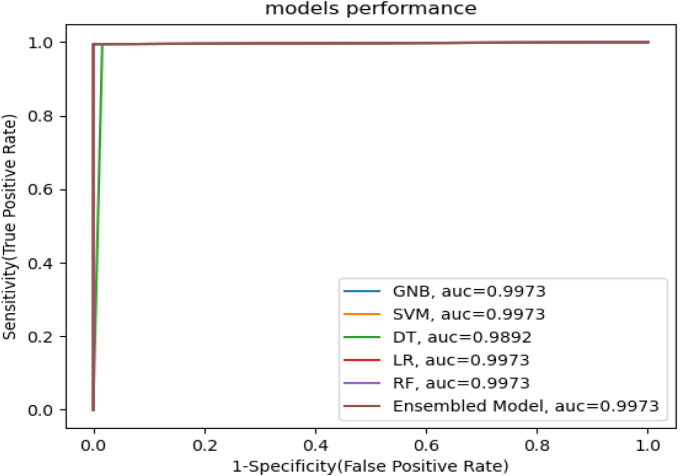
Fig. 7 demonstrates the AUC of our machine learning models, which serves as an evaluation metric to fine-tune the effectiveness of a machine learning model. The AUC of GNB, SVM, DT, LR, RF, and ensemble model are 99.73%, 99.73%, 98.92%, 99.73%, and 99.73% respectively.
Fig. 7.
Receiver Operating Characteristics (ROC) curve for machine learning models.
4.5. Performance comparison with previous works
This section shows how well our ensemble model works than the previous works in this field. We have already described the details of the previous works in Section 2. Jaiswal et al. (2020) used densenet201 for classification of COVID19 from SARS-CoV-2 chest CT scan images. They used 15% data for testing and achieved accuracy, precision, recall, f-measure, and AUC of 96.25%, 96.29%, 96.29%, 96.29%, and 97% respectively. We used the same data for testing and achieved an accuracy, precision, recall, f-measure, and AUC of 99.73%, 99.46%, 100%, 99.73%, and 99.73% respectively. Alshazly, Linse, Barth, and Martinetz (2021) used several deep transfer learning models for instance SqueezeNet, Inception, ResNet, ResNeXt, Xception, ShuffleNet, and DenseNet for training. They used 497 CT scan images (COVID19: 251, Non-COVID19: 246) for testing their model. They achieved an accuracy of 99.4%, a precision of 99.6%, recall of 99.8%, f1-score of 99.4%, using ResNet101. Using logistic regression, we used the same data for testing and achieved the highest accuracy, precision, recall, and f1-score of 99.79%, 100%, 99.59%, and 99.80% respectively. Table 4 shows the performance comparison with SOTA methods on the Kaggle SARS-CoV-2 CT scan dataset.
Table 4.
Performance comparison with previous works.
| Name | Precision | Recall | F1-score | AUC | Accuracy |
|---|---|---|---|---|---|
| Jaiswal et al. (2020) | 96.29% | 99.29% | 97% | 99.29% | 96.25% |
| Proposed Model | 99.46% | 100% | 99.73% | 99.73% | 99.73% |
| Alshazly et al. (2021) | 99.6% | 99.8% | 99.4% | – | 96.4% |
| Proposed Model | 100% | 99.59% | 99.80% | 99.79% | 99.79% |
| Panwar, et al. (2020) | 99% | 95% | 97% | – | 95.61% |
| Proposed Model | 98.76% | 100% | 99.38% | 99.38% | 99.38% |
| Yazdani, Minaee, Kafieh, Saeedizadeh, and Sonka (2020) | 91.6% | 90.3% | 91.6% | 97% | 92% |
| Proposed Model | 99% | 99.3% | 99.3% | 99.3% | 99.3% |
Panwar, et al. (2020) used VGG19 for detection of COVID19, and they also used grad-CAM for visualization. They used 320 CT scan images for testing where the total number of COVID19 and non-COVID19 infected patients are 151 and 169, respectively. They achieved an average accuracy, precision, recall, and f1-score of 95.61%, 0.99%, 0.95%, and 0.97%, respectively. We achieved the higher performance than their works and achieved an average accuracy, precision, recall, and f1-score of 99.38%, 98.76%, 100%, and 99.38%, respectively, using ensemble learning. Yazdani et al. (2020) proposed an attentional convolutional network (residual attention) that identifies the affected area of the chest from CT scan images. They used 1000 (COVID19: 488, Non-COVID19: 512) ct scan images for testing their model. They achieved an accuracy, precision, recall, f1-score, and AUC of 90.7%, 91.6%, 90.3%, 91.6%, and 97%, respectively. We used the same number of images for testing and achieved an optimistic accuracy, precision, recall, f1-score, and AUC of 99.30%, 99.00%, 99.30%, 99.30%, and 99.30% respectively.
5. Conclusion
In this work, we used binary classification to identify COVID 19 and non-COVID 19 infected patients from CT scan images. Firstly, we enhanced the image quality using CLAHE. Then, we designed an effective CNN for extracting the most discriminant features from images. The extracted features have been preprocessed, and then we made identification using several well-known machine learning algorithms. Furthermore, we enhanced the performance of the models by using an ensemble learning model that merged the identifications of each particular ML model. We achieved the highest accuracy than the previous works that are 99.73% while reducing the false positive rate to zero that is a recall of 100%. The detection of the COVID19 using our model is much faster and accurate than the traditional RT-PCR testing method. From the result, it can be revealed that our model has achieved an expected performance that can help the medical physicians correctly detect COVID19 infected patients and give the treatment to the patients timely. The main limitation of our work is that we trained our model on a tiny dataset because the dataset for COVID19 patients is limited. We will strive to collect a large dataset in the future. Another limitation is that we only employed existing machine learning algorithms to classify the COVID19 patient. We will attempt to develop a hybrid model for detecting COVID patients from vast datasets in the future.
CRediT authorship contribution statement
Md. Robiul Islam: Conception and design of study, Acquisition of data, Analysis and/or interpretation of data, Writing – original draft, Writing – review & editing. Md. Nahiduzzaman: Conception and design of study, Acquisition of data, Analysis and/or interpretation of data, Writing – original draft, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Md Robiul Islam and Md Nahiduzzaman contributed equally to this work.
References
- Ahuja S., Panigrahi B.K., Dey N., Rajinikanth V., Gandhi T.K. Deep transfer learning-based automated detection of COVID-19 from lung CT scan slices. Applied Intelligence: The International Journal of Artificial Intelligence, Neural Networks, and Complex Problem-Solving Technologies. 2021;51(1):571–585. doi: 10.1007/s10489-020-01826-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai T., Yang Z., Hou H., Zhan C., Chen C., Lv W., et al. Correlation of chest CT and RT-PCR testing for coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020;296(2):E32–E40. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alshazly H., Linse C., Barth E., Martinetz T. Explainable COVID-19 detection using chest CT scans and deep learning. Sensors. 2021;21(2):455. doi: 10.3390/s21020455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amyar A., Modzelewski R., Li H., Ruan S. Multi-task deep learning based CT imaging analysis for COVID-19 pneumonia: Classification and segmentation. Computers In Biology And Medicine. 2020;126 doi: 10.1016/j.compbiomed.2020.104037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelov P., Almeida Soares E. SARS-CoV-2 CT-scan dataset: A large dataset of real patients CT scans for SARS-CoV-2 identification. MedRxiv. 2020 [Google Scholar]
- Hastie T., Tibshirani R., Friedman J. The elements of statistical learning. Springer; 2001. (Springer series in statistics). [Google Scholar]
- Hemdan E.E.-D., Shouman M.A., Karar M.E. 2020. Covidx-net: A framework of deep learning classifiers to diagnose covid-19 in x-ray images. arXiv preprint arXiv:2003.11055. [Google Scholar]
- Ho T.K. Proceedings of 3rd international conference on document analysis and recognition (vol. 1) IEEE; 1995. Random decision forests; pp. 278–282. [Google Scholar]
- Islam M.T., Siddique B.N.K., Rahman S., Jabid T. 2018 International conference on intelligent informatics and biomedical sciences (vol. 3) IEEE; 2018. Image recognition with deep learning; pp. 106–110. [Google Scholar]
- Jaiswal A., Gianchandani N., Singh D., Kumar V., Kaur M. Classification of the COVID-19 infected patients using DenseNet201 based deep transfer learning. Journal Of Biomolecular Structure And Dynamics. 2020:1–8. doi: 10.1080/07391102.2020.1788642. [DOI] [PubMed] [Google Scholar]
- Krizhevsky A., Sutskever I., Hinton G.E. ImageNet classification with deep convolutional neural networks. Communications Of The ACM. 2017;60(6):84–90. [Google Scholar]
- Kumar S., Mishra S., Singh S.K. Deep transfer learning-based COVID-19 prediction using chest X-rays. MedRxiv. 2020 [Google Scholar]
- Loey M., Manogaran G., Khalifa N.E.M. A deep transfer learning model with classical data augmentation and cgan to detect covid-19 from chest ct radiography digital images. Neural Computing And Applications. 2020:1–13. doi: 10.1007/s00521-020-05437-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maclin R. 2016. Popular ensemble methods: An empirical study popular ensemble methods: An empirical study. [Google Scholar]
- Menditto A., Patriarca M., Magnusson B. Understanding the meaning of accuracy, trueness and precision. Accreditation and Quality Assurance. 2007;12(1):45–47. [Google Scholar]
- Mishra A.K., Das S.K., Roy P., Bandyopadhyay S. Identifying COVID19 from chest CT images: a deep convolutional neural networks based approach. Journal Of Healthcare Engineering. 2020;2020 doi: 10.1155/2020/8843664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamud M.F.Y., Mohamed Y.G., Ali A.M., Adam B.A. Loss of taste and smell are common clinical characteristics of patients with COVID-19 in Somalia: A retrospective double centre study. Infection And Drug Resistance. 2020;13:2631. doi: 10.2147/IDR.S263632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahiduzzaman M., Goni M.O.F., Anower M.S., Islam M.R., Ahsan M., Haider J., et al. A novel method for multivariant pneumonia classification based on hybrid CNN-PCA based feature extraction using extreme learning machine with CXR images. IEEE Access. 2021;9:147512–147526. [Google Scholar]
- Nahiduzzaman M., Islam M.R., Islam S.R., Goni M.O.F., Anower M.S., Kwak K.-S. Hybrid CNN-SVD based prominent feature extraction and selection for grading diabetic retinopathy using extreme learning machine algorithm. IEEE Access. 2021;9:152261–152274. [Google Scholar]
- Nahiduzzaman M., Nayeem M.J., Ahmed M.T., Zaman M.S.U. 2019 4th International Conference On Electrical Information And Communication Technology. IEEE; 2019. Prediction of heart disease using multi-layer perceptron neural network and support vector machine; pp. 1–6. [Google Scholar]
- Narin A., Kaya C., Pamuk Z. 2020. Automatic detection of coronavirus disease (covid-19) using x-ray images and deep convolutional neural networks. arXiv preprint arXiv:2003.10849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panwar H., Gupta P., Siddiqui M.K., Morales-Menendez R., Bhardwaj P., Singh V. A deep learning and grad-CAM based color visualization approach for fast detection of COVID-19 cases using chest X-ray and CT-scan images. Chaos, Solitons & Fractals. 2020;140 doi: 10.1016/j.chaos.2020.110190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak Y., Shukla P.K., Tiwari A., Stalin S., Singh S. Deep transfer learning based classification model for COVID-19 disease. Irbm. 2020 doi: 10.1016/j.irbm.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisano E.D., Zong S., Hemminger B.M., DeLuca M., Johnston R.E., Muller K., et al. Contrast limited adaptive histogram equalization image processing to improve the detection of simulated spiculations in dense mammograms. Journal Of Digital Imaging. 1998;11(4):193. doi: 10.1007/BF03178082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PlamenEduardo E.D. 2020. SARS-COV-2 ct-scan dataset. [Online]. https://www.kaggle.com/plameneduardo/sarscov2-ctscan-dataset. (Accessed: 21-March-2021) [Google Scholar]
- Polikar R., Polikar R. Ensemble based systems in decision making. IEEE Circuits and Systems Magazine. 2006;6:21–45. [Google Scholar]
- Powers D.M. 2020. Evaluation: from precision, recall and F-measure to ROC, informedness, markedness and correlation. arXiv preprint arXiv:2010.16061. [Google Scholar]
- Sarker L., Islam M.M., Hannan T., Ahmed Z. 2021. Covid-densenet: A deep learning architecture to detect covid-19 from chest radiology images. [Google Scholar]
- Shah V., Keniya R., Shridharani A., Punjabi M., Shah J., Mehendale N. Diagnosis of covid-19 using ct scan images and deep learning techniques. MedRxiv. 2020 doi: 10.1007/s10140-020-01886-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh D., Kumar V., Kaur M., et al. Classification of COVID-19 patients from chest CT images using multi-objective differential evolution–based convolutional neural networks. European Journal Of Clinical Microbiology & Infectious Diseases. 2020;39(7):1379–1389. doi: 10.1007/s10096-020-03901-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhal T. A review of coronavirus disease-2019 (COVID-19) The Indian Journal Of Pediatrics. 2020;87(4):281–286. doi: 10.1007/s12098-020-03263-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szegedy C., Toshev A., Erhan D. 2013. Deep neural networks for object detection. [Google Scholar]
- Tang Y.-W., Schmitz J.E., Persing D.H., Stratton C.W. Laboratory diagnosis of COVID-19: current issues and challenges. Journal Of Clinical Microbiology. 2020;58(6) doi: 10.1128/JCM.00512-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tingting Y., Junqian W., Lintai W., Yong X. Three-stage network for age estimation. CAAI Transactions On Intelligence Technology. 2019;4(2):122–126. [Google Scholar]
- Wang Z., Liu Q., Dou Q. Contrastive cross-site learning with redesigned net for COVID-19 CT classification. IEEE Journal Of Biomedical And Health Informatics. 2020;24(10):2806–2813. doi: 10.1109/JBHI.2020.3023246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.-H., Nayak D.R., Guttery D.S., Zhang X., Zhang Y.-D. COVID-19 classification by CCSHNet with deep fusion using transfer learning and discriminant correlation analysis. Information Fusion. 2021;68:131–148. doi: 10.1016/j.inffus.2020.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Xu Y., Gao R., Lu R., Han K., Wu G., et al. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020;323(18):1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolpert D.H. Stacked generalization. Neural Networks. 1992;5(2):241–259. [Google Scholar]
- Worldometer D.H. 2021. COVID-19 CORONAVIRUS PANDEMIC. [Online]. https://www.worldometers.info/coronavirus/?utm_campaign=homeAdvegas1?. (Accessed 22-February-2021) [Google Scholar]
- Xie X., Zhong Z., Zhao W., Zheng C., Wang F., Liu J. Chest CT for typical coronavirus disease 2019 (COVID-19) pneumonia: relationship to negative RT-PCR testing. Radiology. 2020;296(2):E41–E45. doi: 10.1148/radiol.2020200343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., He X., Zhao J., Zhang Y., Zhang S., Xie P. 2020. COVID-CT-dataset: a CT image dataset about COVID-19. arXiv preprint arXiv:2003.13865. [Google Scholar]
- Yazdani S., Minaee S., Kafieh R., Saeedizadeh N., Sonka M. 2020. Covid ct-net: Predicting covid-19 from chest ct images using attentional convolutional network. arXiv preprint arXiv:2009.05096. [Google Scholar]
- Zhang Y.-D., Satapathy S.C., Zhang X., Wang S.-H. Covid-19 diagnosis via DenseNet and optimization of transfer learning setting. Cognitive Computation. 2021:1–17. doi: 10.1007/s12559-020-09776-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu Z.Y., Jiang M.D., Xu P.P., Chen W., Ni Q.Q., Lu G.M., et al. Coronavirus disease 2019 (COVID-19): a perspective from China. Radiology. 2020;296(2):E15–E25. doi: 10.1148/radiol.2020200490. [DOI] [PMC free article] [PubMed] [Google Scholar]