Abstract
Amplified fragment length polymorphism analysis seems well suited for studying the epidemiology of isolates of Neisseria gonorrhoeae obtained from patients attending the Sexually Transmitted Disease Outpatient Clinic in Amsterdam, The Netherlands. It shows potential to identify the core group of transmitters.
Neisseria gonorrhoeae can maintain itself at low endemicity as a result of transmission within a so-called high-risk core group (1, 8). These high-frequency transmitters serve as a reservoir of infection and should therefore be the focus for targeted measures to control this and other sexually transmitted diseases (STDs). Whether the recent increase in Amsterdam, The Netherlands (3), reflects the introduction of one or more new strains into the core group population is not known, and accurate typing is needed to better understand the epidemiology of N. gonorrhoeae. Molecular methods investigated as an alternative for the rather complicated, laborious, and time-consuming auxotyping and serological characterization include pulsed-field gel electrophoresis (PFGE) (9, 10), arbitrarily primed PCR (9), amplified ribosomal DNA restriction analysis (9), and opa typing (2, 6, 9). According to the Simpson discriminatory index of diversity (i.e., the ability of a typing method to distinguish between unrelated strains), the combination of PFGE and opa typing has shown the highest discriminatory index (9). The present study sought to assess the applicability of the amplified fragment length polymorphism (AFLP) technique (7) to the typing of N. gonorrhoeae at the strain level and its feasibility for molecular epidemiological studies.
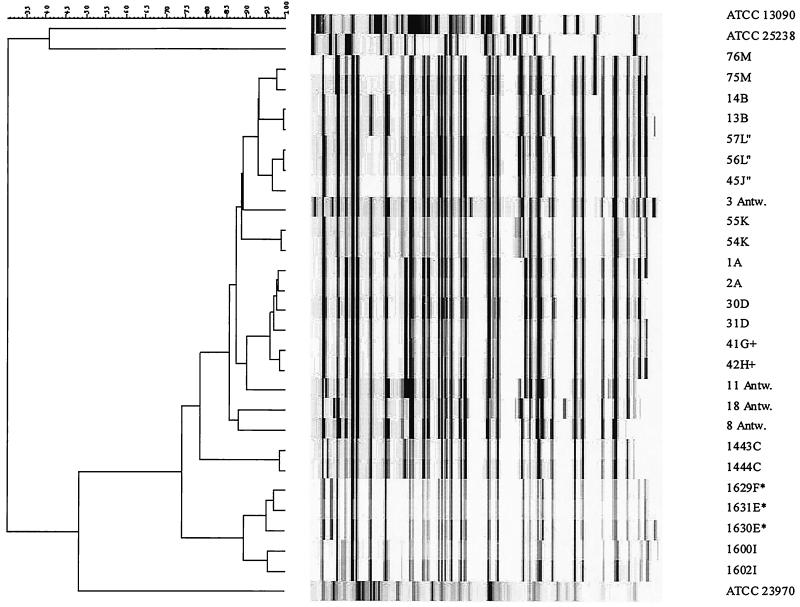
Based on the availability of samples, 13 patients with culture-positive pharyngeal, urogenital, and/or anorectal gonococcal infections were selected for study in the first quarter of 1999. Of these, 3 were women (all heterosexual) and 10 were men (6 homosexual, 1 bisexual, and 3 heterosexual). The age distribution was 20 to 59 years; nine patients reported having had previous STDs. At the STD clinic, samples for gonococcal cultures were collected, directly inoculated onto GC-Lectagar plates (Becton Dickinson Biosciences, Sparks, Md.), incubated at 37°C in candle jars with moist wads of cotton, and transported to the laboratory on the same day. After incubation at 37°C for 48 h, suspected colonies were then subjected to Gram staining and identified biochemically (4). Isolates were subcultured and stored until use at −70°C in 20% (wt/vol) glycerol in microbank vials (Nunc A/S, Roskilde, Denmark). Antibiotic sensitivity testing of penicillin, tetracycline, azithromycin, and ciprofloxacin was performed by the disk diffusion method according to the recommendations of the National Committee for Clinical Laboratory Standards. Reference strains used were Moraxella catarrhalis ATCC 25238, Neisseria lactamica ATCC 23970, and Neisseria meningitidis ATCC 13090. In addition, four strains of N. gonorrhoeae known to be temporally and geographically diverse (3 Antw., 1979-Thailand; 8 Antw., 1989-United Kingdom; 18 Antw., 1989-Gambia; and 11 Antw., 1993-United Kingdom) and characterized by other typing methods were used to evaluate the AFLP genotyping analysis for N. gonorrhoeae (9). To assess the genomic stability of N. gonorrhoeae during laboratory passage, AFLP patterns were studied both before and after laboratory passage. Five strains (2 of the 22 studied and 3 single nonrelated isolates) of N. gonorrhoeae were subcultured every other day for 15 days. On days 1, 2, 4, 6, 8, and 15, chromosomal DNA was isolated by standard methods and subjected to AFLP analysis (5). There were no striking changes in the AFLP pattern of any isolate after six passages during 15 days. Thus, the AFLP patterns were conserved following in vitro passage. The 22 isolates studied were obtained from various sites: urethra (n = 10), tonsil (n = 6), rectal swabs (n = 4), and cervix (n = 2). For nine patients, two isolates from different sites could be typed. In these cases, the paired isolates were identical with regard to sensitivity to penicillin, tetracycline, azithromycin, and ciprofloxacin. This was also the case for the three pairs representing known sexual contacts (E-F, G-H, and J-L). Figure 1 shows the AFLP patterns of N. gonorrhoeae isolates obtained from 13 different patients. Comparison of the strains obtained from two different sites of individual patients revealed that these pairs had indistinguishable AFLP patterns. Figure 1 also shows that AFLP patterns of N. gonorrhoeae isolates from persons known to be sexual partners were the same within each pair. Cluster analysis showed correlation levels of >90% between the partners, irrespective of the anatomical source of the isolates. Unrelated strains from different geographic origins and different years of collection did not cluster with the presented N. gonorrhoeae isolates, nor did the ATCC reference strains. The AFLP technique appears to be reliable and reproducible, as indicated by the identical AFLP patterns of N. gonorrhoeae isolates from different sites in index patients and those of index patients and their sexual partners. Cluster analysis of isolates from the same patient but from different locations revealed AFLP patterns with 90 to 95% homology. Using standardization by defining windows of discrimination as described earlier (7), these percentages are considered to represent identical strains. The same is true for isolates from known sexual contacts. Temporally as well as geographically diverse strains showed 80 to 90% homology, indicating different strains from the same species. AFLP patterns of other members of the family Neisseriaceae showed far less homology, ranging from 40 to 50% and indicating isolates of the same genus but of different species.
FIG. 1.
Fluorescently labeled AFLP patterns and dendrogram of 29 strains, including 22 clinical N. gonorrhoeae isolates, obtained after amplification of EcoA and MseO templates. The patterns were analyzed by Pearson correlation, and a dendrogram was constructed with the UPGMA (unweighted pair group method with averages) BioNumerics software package (Applied Maths, Kortrijk, Belgium). Percentages of similarity are shown above the dendrogram. The strain numbers and patient codes are shown on the right. Known sexual contacts are indicated by asterisks, plus signs, and quotation marks. ATCC 23970, N. lactamica; ATCC 13090, N. meningitidis; ATCC 25238, Moraxella catarrhalis. 3 Antw., 1979-Thailand; 8 Antw., 1989-United Kingdom; 11 Antw., 1993-United Kingdom; 18 Antw., 1989-Gambia.
Whether AFLP analysis for N. gonorrhoeae has equal or superior discriminatory power compared to other fingerprinting techniques, like PFGE or opa typing, needs to be investigated. PFGE and opa typing have demonstrated their potential utility for gonococcal typing (10), but PFGE is not sufficiently discriminatory to distinguish the large array of genotypes expected in a recombining nonclonal population (10) and opa typing may not always define clones. The latter is a serious drawback because gonococci are naturally competent and may undergo intragenomic recombination as a result of the intrinsic hypervariability of opa genes and/or intergenomic recombination as a result of mixed infections (M. J. Gill and J. D. Ross, Editorial, Sex. Transm. Infect. 75:211–213, 1999). One of the important advantages of AFLP analysis is that only 10 to 50 ng of DNA is needed instead of the 200 ng of N. gonorrhoeae DNA needed for opa typing. This significantly reduces the labor necessary to propagate the gonococcal strains. Although the number of strains that we studied is relatively small, our data show that AFLP fingerprinting analysis is a genotyping method applicable to N. gonorrhoeae isolates with a high degree of reproducibility, which may allow compilation of genotype databases and the exchange of data among laboratories, as has been confirmed by the two collaborating laboratories (data not shown). International surveillance, a prerequisite for public health intervention, could therefore be within our reach.
Acknowledgments
We thank P. G. H. Peerbooms for critically reading the manuscript, M. van Looveren for providing the temporally and geographically diverse N. gonorrhoeae isolates, and L. D. Philips for editorial review.
REFERENCES
- 1.Brunham R C. The concept of core and its relevance to the epidemiology and control of sexually transmitted diseases. Sex Transm Dis. 2000;18:67–68. doi: 10.1097/00007435-199118020-00001. [DOI] [PubMed] [Google Scholar]
- 2.Camarena J J, Nogueira J M, Dasi M A, Moreno F, Garcia R, Ledesma E, Llorca J, Hernandez J. DNA amplification fingerprinting for subtyping Neisseria gonorrhoeae strains. Sex Transm Dis. 1995;22:128–136. doi: 10.1097/00007435-199503000-00009. [DOI] [PubMed] [Google Scholar]
- 3.Fennema J S, Cairo I, Coutinho R A. Substantial increase in gonorrhea and syphilis among clients of Amsterdam Sexually Transmitted Diseases Clinic. Ned Tijdschr Geneeskd. 2000;144:602–603. [PubMed] [Google Scholar]
- 4.Isenberg H D, editor. Clinical microbiology procedures handbook. Washington, D.C.: American Society for Microbiology; 1992. [Google Scholar]
- 5.Koeleman J G, Stoof J, Biesmans D J, Savelkoul P H, Vandenbroucke-Grauls C M. Comparison of amplified ribosomal DNA restriction analysis, random amplified polymorphic DNA analysis, and amplified fragment length polymorphism fingerprinting for identification of Acinetobacter genomic species and typing of Acinetobacter baumannii. J Clin Microbiol. 1998;36:2522–2529. doi: 10.1128/jcm.36.9.2522-2529.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O'Rourke M, Ison C A, Renton A M, Spratt B G. Opa-typing: a high-resolution tool for studying the epidemiology of gonorrhoea. Mol Microbiol. 1995;17:865–875. doi: 10.1111/j.1365-2958.1995.mmi_17050865.x. [DOI] [PubMed] [Google Scholar]
- 7.Savelkoul P H, Aarts H J, de Haas J, Dijkshoorn L, Duim B, Otsen M, Rademaker J L, Schouls L, Lenstra J A. Amplified-fragment length polymorphism analysis: the state of an art. J Clin Microbiol. 1999;37:3083–3091. doi: 10.1128/jcm.37.10.3083-3091.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Duynhoven Y T, van Klingeren B, van Santen-Verheuvel M G, van der Meijden W I, van de Laar M J. Molecular epidemiology of infections with Neisseria gonorrhoeae among visitors to a sexually transmitted diseases clinic. Sex Transm Dis. 1997;24:409–417. doi: 10.1097/00007435-199708000-00005. [DOI] [PubMed] [Google Scholar]
- 9.Van Looveren M, Ison C A, Ieven M, Vandamme P, Martin I M, Vermeulen K, Renton A, Goossens Evaluation of the discriminatory power of typing methods for Neisseria gonorrhoeae. J Clin Microbiol. 1999;37:2183–2188. doi: 10.1128/jcm.37.7.2183-2188.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xia M, Whittington W L, Holmes K K, Plummer F A, Roberts M C. Pulsed-field gel electrophoresis for genomic analysis of Neisseria gonorrhoeae. J Infect Dis. 1995;171:455–458. doi: 10.1093/infdis/171.2.455. [DOI] [PubMed] [Google Scholar]