Abstract
The human gut microbiome plays a central role in health and disease. Environmental factors, such as lifestyle and diet, are known to shape the gut microbiome as well as the reservoir of resistance genes that these microbes harbour; the resistome. In this study we assessed whether long-term dietary habits within a single geographical region (the Netherlands) impact the human gut resistome. Faecal samples from Dutch omnivores, pescatarians, vegetarians and vegans were analysed by metagenomic shotgun sequencing (MSS) (n = 149) and resistome capture sequencing approach (ResCap) (n = 64). Among all diet groups, 119 and 145 unique antibiotic resistance genes (ARGs) were detected by MSS or ResCap, respectively. Five or fifteen ARGs were shared between all diet groups, based on MSS and ResCap, respectively. The total number of detected ARGs by MSS or ResCap was not significantly different between the groups. MSS also revealed that vegans have a distinct microbiome composition, compared to other diet groups. Vegans had a lower abundance of Streptococcus thermophilus and Lactococcus lactis compared to pescatarians and a lower abundance of S. thermophilus when compared to omnivores. In summary, our study showed that long-term dietary habits are not associated with a specific resistome signature.
Subject terms: Molecular biology, Antimicrobials, Antibiotics, Microbiology, Microbial communities, Metagenomics, Microbiome
Introduction
The human gut microbiome is a complex ecosystem composed of bacteria, fungi, viruses and phages. It not only plays a central role in nutrient acquisition, but it also affects our state of health and disease1–3. Many factors are known to influence its composition, which can be either host-derived, such as age or immunological and pathological disorders4,5, or exposure to environmental factors (the exposome), including diet6–12. Correlations have been observed between diets rich in protein and animal-fats and the high relative abundance of Bacteroides, as opposed to carbohydrate-rich diets and the high abundance of Prevotella13,14. In addition, increased abundance of Prevotella and Lachnospira was correlated with fiber- and vegetable-rich diets15,16. The majority of studies that have observed these diet-induced effects on the microbiome either compare participants from different geographic areas or involve short term dietary intervention studies14,17–19. When studying the effect of long-term dietary habits within a single community on the gut microbiome of vegans, vegetarians and omnivores, Losasso et al. only observed differences in bacteria that are present in low abundance, and part of the families Bacteroides, Lachnospiraceae, and Ruminococcaceae20. This study used 16S rRNA gene sequencing to determine the microbiome composition, which has insufficient resolution to allow for comparisons at the species level.
The human gut microbiome is also an important reservoir of antibiotic resistance genes (ARGs)21–24. It is therefore important to understand how long-term dietary habits not only impact the microbiome composition, but also the composition of the total of ARGs, the resistome, in the human gut. Advances in high-throughput sequencing have allowed in depth studies of the human gut resistome. The gut resistome of healthy humans can typically contain over 100 unique ARGs, with the most abundant ARGs being those that encode for tetracycline resistance, followed by macrolide and beta lactam resistance genes25,26. Just as with the microbiome, several factors are known to alter the resistome composition. Orally administered antibiotics are known to select for bacteria that are resistant to these antibiotics, therefore increasing the abundance of these ARGs in the gut27,28. This antibiotic-driven enrichment can take place in either the general population or more specific, in clinical settings, where effects on the resistome were observed in a matter of days29–31. Additionally, the human gut resistome can be affected by environmental factors such as international travel and living conditions. This has been shown in studies comparing urbanized with agricultural populations, in which the use of antibiotics plays an important role32,33. Finally, meat contaminated with bacteria carrying ARGs as a result of antibiotic usage in livestock, has been highlighted as a possible transmission route for resistant bacteria and could therefore influence the gut resistome as well34–38. More specifically, zoonotic pathogens such as species of Salmonella and Campylobacter and certain types of Escherichia coli (e.g. Shiga-toxin-producing E. coli (STEC)) are known for causing foodborne infections and are frequent carriers of ARGs39–42.
In this study we assessed whether long-term dietary habits within a single geographical region impact the human gut resistome in the general Dutch population. Using metagenomic shotgun sequencing (MSS), we were able to detect 877 unique bacterial species and an extensive resistome composed of 119 unique ARGs, in the gut microbiome of healthy Dutch residents. resistome capture sequencing approach (ResCap) was applied for a subset of samples and revealed 145 unique ARGs, thereby surpassing the detection limits of MSS. Despite the high resolution of the sequencing data, the total number of ARGs detected by MSS or ResCap per diet group was not significantly different in the general Dutch population.
Results
Diet-associated differences in the gut microbiome
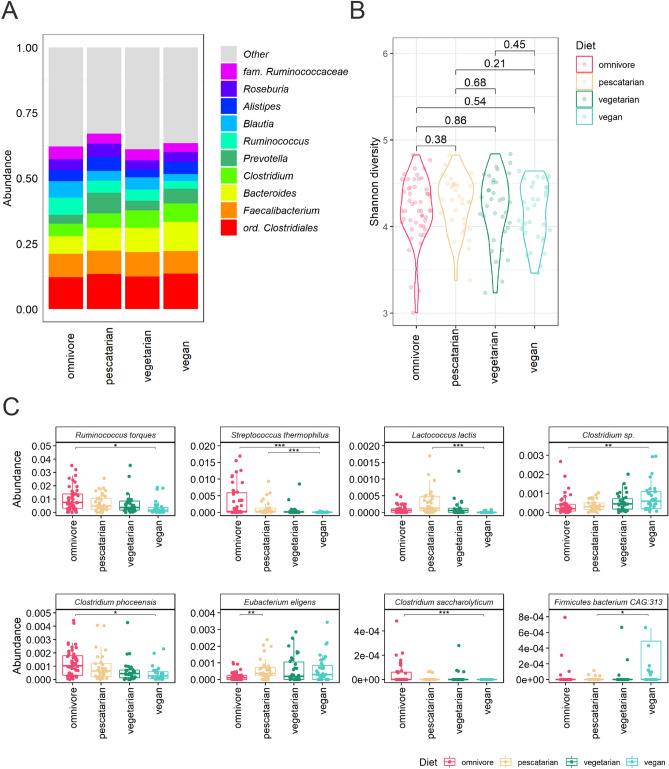
Before determining diet-associated resistome differences, we first assessed the effect of long-term dietary habits on the gut microbiome, as this ecological niche is an important reservoir of antibiotic resistance genes. Faecal samples from 149 Dutch individuals were selected based on their dietary habits and categorized in four matched diet groups: (1) omnivores, (2) pescatarians, (3) vegetarians and (4) vegans (Table 1). Faecal samples from these four groups were used for metagenomic shotgun sequencing (MSS) in order to study the effect of dietary habits on the gut microbiome. The mOTUs2-based taxonomic binning method revealed that the top 10 most abundant genera did not differ between the diet groups (Fig. 1a). In all our study groups, the most abundant taxa belong to the order of Clostridiales and genera Faecalibacterium, Bacteroides, Clostridium and Prevotella, which matches with the gut microbiome composition observed in previous studies43,44. In addition, the gut microbiome diversity, expressed as Shannon index, was not significantly different between diet groups, indicating that the total species diversity is highly similar among diet groups (Fig. 1b). Statistical analysis of the inter-diet group beta diversity based principal component analysis (PCA) using Aitchison distance, Bray–Curtis distance, or Jaccard distance further revealed that diet was not a main driver of the observed variance in microbiome composition between the samples (figure s1).
Table 1.
Study participants characteristics included in metagenomic shotgun sequencing.
| Characteristics | Omnivore | Pescatarian | Vegetarian | Vegan |
|---|---|---|---|---|
| Participants, n = 149 | 50 | 33 | 34 | 32 |
| Age in years, median (10th–90th percentile) | 47 (29–59) | 51 (29–62) | 45 (28–62) | 37 (29–56) |
| Male participants,(percentage) | 16 (32%) | 12 (36%) | 11 (32%) | 11 (34%) |
| Participants with pets, (percentage) | 28 (56%) | 16 (48%) | 18 (53%) | 18 (56%) |
| Participants using medication*, (percentage) | 15 (30%) | 15 (45%) | 7 (21%) | 8 (25%) |
*Medication other than antibiotics, proton pump inhibitors, insulin and cancer treatment.
Figure 1.
Association of dietary habits and the gut microbiome composition. (A) Relative abundance of the 10 most abundant bacterial genera per diet group. (B) Alpha diversity per diet group expressed by Shannon index. Differences in alpha diversity between diet groups were compared using Wilcoxon rank-sum tests. (C) ANCOM-BC analysis for differential relative abundance of bacterial species between diet groups. Abundance was plotted on the relative abundance scale from 0.00 to 1.00. Adjusted p values are indicated by *< 0.05, **< 0.01 and ***< 0.001.
We further explored potential differences in the gut microbiome composition between diet groups by using supervised analysis to compare the abundance of bacterial species using ANCOM-BC45. Compared to the omnivores, vegans had lower abundance of Ruminococcus torques (padj = 5.0E−02), Streptococcus thermophilus (padj = 4.7E−06), Clostridium sp. (padj = 6.5E−03), Clostridium phoceensis (padj = 3.7E−02) and Clostridium saccharolyticum (padj = 1.6E−03) (Fig. 1c). Similarly, Streptococcus thermophilus (padj = 1.5E−04), Lactococcus lactis (padj = 7.0E−07) and Firmicutes bacterium CAG:313 (padj = 2.7E−02) were less abundant in vegans compared to pescatarians. Finally, Eubacterium eligens (padj = 2.9E−03) was more abundant in the microbiome of pescatarians when compared to omnivores (Fig. 1c).
In addition to mOTUs2 we also applied MetaPhlAn3 to profile the microbiome composition. In concordance with the results of the mOTUs2, using MetaPhlAn3 we did not observe differences in the top 10 most abundant genera between the diet groups (figure s2). Using MetaPhlAn3 for differential abundance analysis, we also found a lower relative abundance of R. torques, S. thermophilus and C. saccharolyticum in vegans compared to omnivores and higher abundance of E. eligens in pescatarians compared to omnivores (figure s3). Furthermore, the MetaPhlAn3 approach detected a higher relative abundance of Lactobacillus delbrueckii, Coprococcus comes, Dorea formicigenerans, Dorea longicatena, Lawsonibacter asaccharolyticus and Phascolarctobacterium CAG:266 in omnivores compared to vegans (figure s3).
Composition of the gut resistome across diet groups based on metagenomic shotgun sequencing
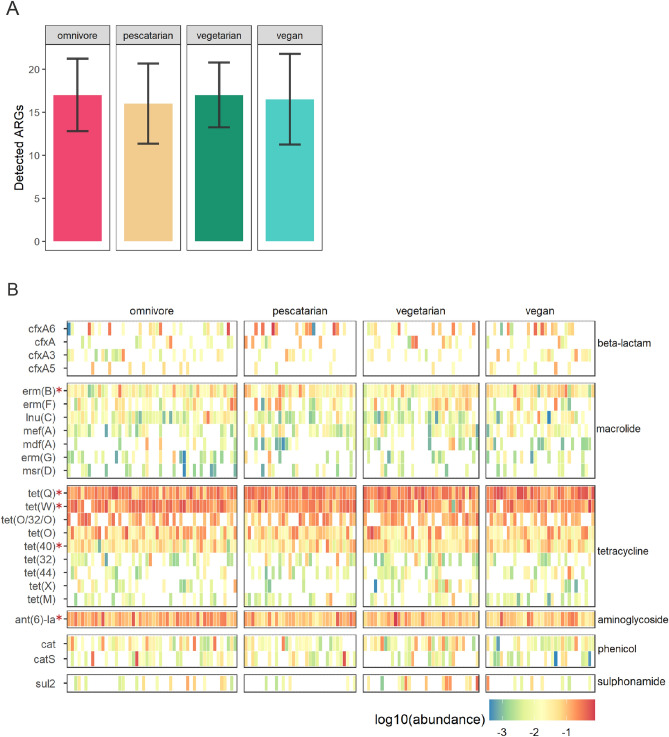
We next set out to investigate whether long-term dietary habits impacted the gut resistome composition. Using MSS we were able to identify 119 unique antibiotic resistance genes (ARGs) among all diet groups. The total number of detected ARGs was not significantly different between the groups, with an average of 17 ± 4 genes found in omnivores, 16 ± 5 in pescatarians, 17 ± 4 in vegetarians and 17 ± 5 in vegans (Fig. 2a). Among these, five were consistently detected in all diet groups (detected in 95% of the samples), namely the aminoglycoside resistance gene ant(6)-Ia, the macrolide-lincosamide-streptogramin B resistance gene erm(B), and the tetracycline resistance genes tet(40), tet(Q) and tet(W) (Fig. 2b).
Figure 2.
Antibiotic resistance gene distribution in the gut resistome through metagenomic shotgun sequencing. (A) Average number and standard deviation of antibiotic resistance genes (ARGs) detected in each diet group. (B) Heatmap depicting ARGs abundance per participant. Each column denotes a study participant, clustered per diet group. Rows represent the relative abundance of ARG classes. ARGs present at least in 10% of the participants are shown. ARGs present in the resistome of at least 95% of the participants are indicated by the asterisks.
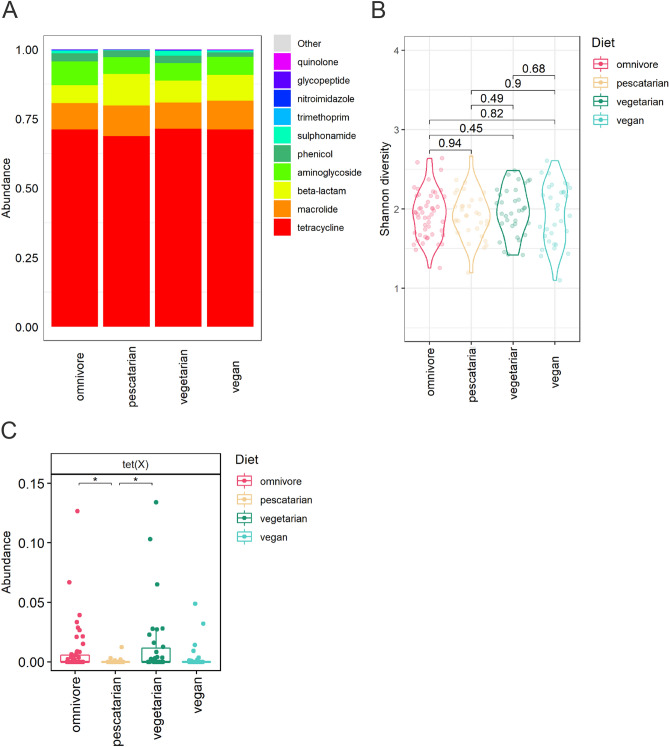
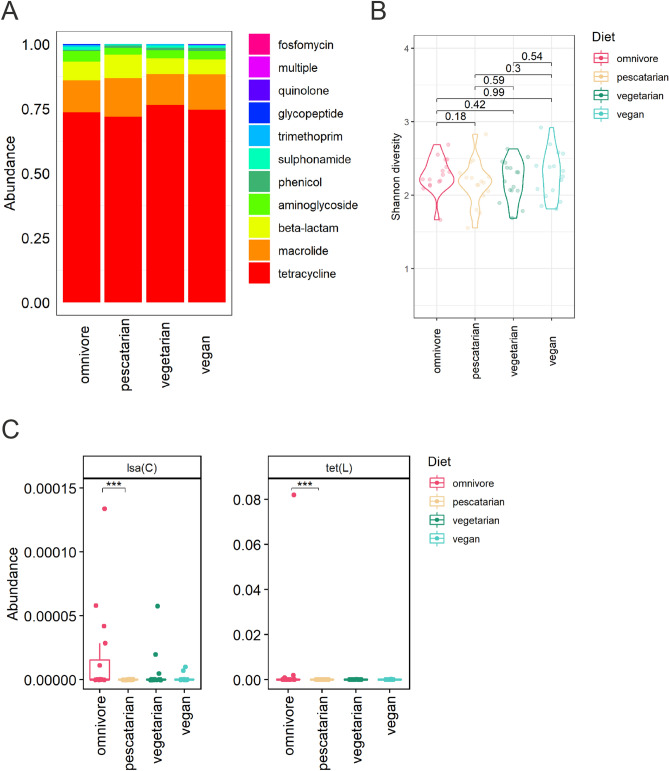
The most abundant genes detected encode for resistance to the classes of tetracyclines, macrolides, beta-lactams, aminoglycosides and phenicols. There were no differences when comparing the overall top 10 most abundant ARGs between the diet groups (Fig. 3a). Also, the resistome diversity, expressed by Shannon index, was not significantly different between the diet groups (Fig. 3b). In addition, diet does not seem to be the main driver of the observed variance in resistome composition between the samples when analysed by beta diversity based principal component analysis (PCA, figure s4). Supervised analysis using ANCOM-BC only revealed a significant difference of the abundance of tet(X), which is present in low abundance in pescatarians in comparison to omnivores (padj = 4.4E−02) and vegetarians (padj = 1.0E−02) (Fig. 3c). Overall, based on MSS, the resistome was highly similar among the studied diet groups.
Figure 3.
Association of dietary habits and the gut resistome composition through metagenomic shotgun sequencing. (A) Relative abundance of the 10 most abundant gene classes encoding antibiotic resistance, summarized per diet group. (B) Alpha diversity per diet group expressed by Shannon index. Differences in alpha diversity between diet groups were compared using Wilcoxon rank-sum tests. (C) Differential abundance analysis using ANCOM-BC plotted on relative abundance scale from 0.00 to 1.00. Adjusted p values below 0.05 are indicated by *.
Higher antibiotic resistance gene detection sensitivity of ResCap compared to metagenomic shotgun sequencing
Although MSS was able to detect a large variety of ARGs, these only represented 0.06 ± 0.03% of the total number of reads. This indicates that the proportion of ARGs is relatively low when compared to the total gene pool present in the samples. In order to improve the sensitivity to detect ARGs, we applied the probe based resistome capture sequencing approach (ResCap), on a subset (64/149; 43%) of samples (Table 2)46. ResCap was able to greatly enrich the number ARGs specific reads, with 40.5 ± 15.2% of the total number of reads sequenced mapping to ARGs.
Table 2.
Participant characteristics of the subset selected for ResCap.
| Characteristics | Omnivore | Pescatarian | Vegetarian | Vegan |
|---|---|---|---|---|
| Participants, n = 64 | 16 | 16 | 16 | 16 |
| Age in years, median (10th–90th percentile) | 46 (27–57) | 42 (31–59) | 44 (30–59) | 43 (33–56) |
| Male participants, (percentage) | 8 (50%) | 8 (50%) | 8 (50%) | 9 (56%) |
| Participants with pets, (percentage) | 9 (56%) | 11 (69%) | 8 (50%) | 8 (50%) |
| Participants using medication*, (percentage) | 4 (25%) | 5 (31%) | 6 (38%) | 2 (13%) |
*Medication other than antibiotics, proton pump inhibitors, insulin and cancer treatment.
To evaluate the sensitivity of ResCap compared to MSS, we compared the observed number of ARGs identified per sequencing depth, by using rarefaction curves in the same 16 samples per diet group, subjected to both MSS and ResCap. Overall, ResCap was able to detect a higher number of ARGs than MSS. Even at 70 M reads per sample, MSS did not reach the level of sensitivity that ResCap was able to achieve (figure s5a). Where MSS detected 16 ± 5 to 18 ± 4 ARGs per diet group with a sequencing depth of 70 M reads, ResCap resulted in the detection of 33 ± 8 to 39 ± 7 ARGs per diet group with a sequencing depth of 20 M reads (figure s5b).
Dietary habits are not associated with differences in resistome composition assessed by ResCap
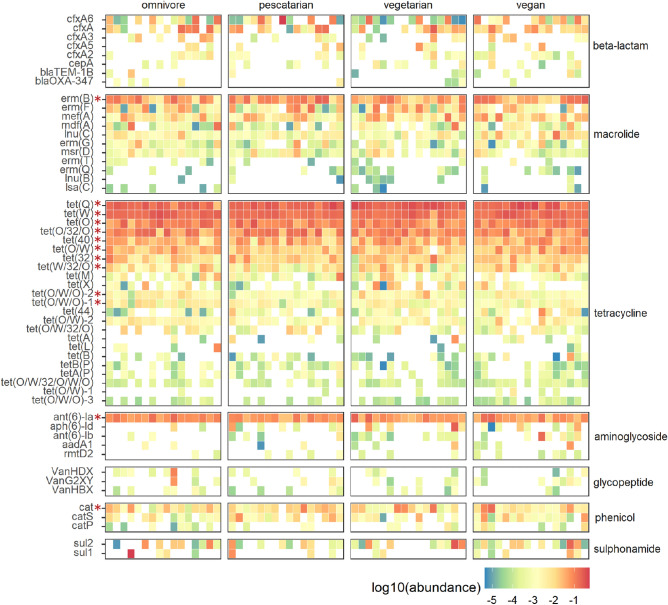
A total of 145 ARGs were detected using ResCap, from which 86 ARGs were also detected by MSS (table s1). The majority of genes detected by both methods included tetracycline resistance genes (19 genes, 22%), followed by beta-lactam resistance (19 genes, 22%) and macrolide resistance (15 genes, 17%). The ARGs detected by ResCap and not MSS include mainly beta-lactam (28/59; 47%), tetracycline (9/59; 15%) and aminoglycoside resistance genes (9/59; 15%). In contrast, MSS revealed 32 genes that went undetected by ResCap, including beta-lactam resistance (10 genes, 31%), nitroimidazole resistance (8 genes, 25%), and tetracycline resistance (4 genes, 13%). Of the 145 genes detected by ResCap, 15 ARGs were detected between all diet groups (detected in 95% of the samples) (Fig. 4). These 15 genes included the five genes (ant(6)-Ia, erm(B), tet(40), tet(Q) and tet(W)) that were also detected in all diet groups by MSS. In addition to these five, the chloramphenicol resistance gene cat, the lincomycin resistance gene lnu(C), the macrolide resistance gene mef(A), and the tetracycline resistance genes tet(32), tet(O), tet(O/32/O), tet(O/W), tet(O/W/O)-1, tet(O/W/O)-2 and tet(W/32/O) were detected in all diet groups using ResCap (Fig. 4).
Figure 4.
Antibiotic resistance gene distribution in the gut resistome as detected by ResCap. Each column denotes a participant, clustered in columns according to diet group. Rows are categorized by antibiotic resistance gene (ARG) classes. ARGs present at least in 10% of the participants are shown. ARGs that are present in the resistome of at least 95% of the participants are indicated by the asterisks.
The total number of ARGs detected by ResCap per diet group was not significantly different, with an average of 36 ± 5 genes found in omnivores, 32 ± 9 in pescatarians, 40 ± 10 in vegetarians and 40 ± 10 in vegans (figure s5b). The most abundant ARGs identified encode for tetracycline, macrolide, beta-lactam and aminoglycoside resistance (Fig. 5a). No differences were observed when comparing the overall top 10 most abundant ARG. No significant differences were observed when comparing the resistome diversity as calculated by Shannon index (Fig. 5b). Beta-diversity based PCA further confirmed that diet is not a main driver of the observed variance in resistome composition between the samples (figure s6). Supervised analysis using ANCOM-BC revealed that the abundance of two ARGs, lsa(C) and tet(L), were significantly different between the diet groups pescatarians and omnivores (Fig. 5c). However, these genes are present in very low abundance (median below 0.05%) and in low prevalence (< 15%). Based on these results we can conclude that differences in dietary habits did not result in significant differences in resistome composition.
Figure 5.
Association of dietary habits and the gut resistome composition through ResCap. (A) Relative abundance of the ten most abundant antibiotic resistance gene classes, summarized per diet group. (B) Gene diversity per diet group, shown by Shannon index. (C) Differential abundance analysis using ANCOM-BC plotted on the scale 0.00–1.00%. Adjusted p values below 0.001 are indicated by ***.
Discussion
By using the combined power of metagenomic shotgun sequencing (MSS) and ResCap we were able to detect over 850 different bacterial species and more than 100 unique antibiotic resistance genes (ARGs) in the gut microbiome of participants from the general Dutch population with distinct dietary habits. Our results show that long-term dietary habits are not associated with specific resistome signatures.
To investigate the gut resistome in humans, we used two techniques, MSS and ResCap. Using genomic DNA from human faecal samples, Lanza et al. found that ResCap results in a two-fold increase in gene diversity compared to MSS. In addition, they reported a 279-fold increase in the amount of reads that mapped to the ResCap targeted genes46. Results of our study are comparable to Lanza et al., as we observed a two-fold increase in gene diversity when comparing the number of ARGs detected by ResCap to MSS. Moreover, ResCap resulted in 40.5 ± 15.2% of the reads mapping to ARGs, compared to 0.06 ± 0.03% for MSS, thus indicating a 675-fold change. In a similar way, Macedo et al. also reported an increase of mapped reads of 200-fold, when comparing ResCap to MSS performed on genomic DNA from soil and manure samples47. Lastly, Guitor et al. applied a custom ARG probe-database and compared its efficiency to capture the resistome over MSS. They compared both systems using a DNA pool composed of four bacterial species and observed a 100-fold increase of target specific reads48. ResCap and MSS sequencing results differed in the number of ARGs that they were able to detect. The 59 ARGs that were detected by ResCap and not MSS suggests that the capture-based approach offers increased sensitivity. However, MSS revealed 32 genes that went undetected by ResCap. From these, 14 genes (44%) were not included in the probe library, including nitroimidazole resistance genes nimA, nimB, nimC, nimD, nimE, nimF, nimH and nimJ (table s1). We currently have no explanation why we were not able to detect the 14 genes for which probes were present in the ResCap library and that were detected by MSS but not by ResCap.
We observed limited differences in the resistome composition between the diet groups, independently of the detection approach. A previous MSS based comparison of the gut resistome in the Chinese, Danish and Spanish general population, revealed the overall high abundance of ARGs that confer resistance to tetracycline, followed by macrolide, β-lactam and aminoglycoside resistance26. This is also observed in our study which included samples from only Dutch population. When using ResCap as high-resolution method to profile the gut resistome, only two ARGs (lsa(C) and tet(L)) were differentially abundant between omnivores and pescatarians. Since these genes are observed in low abundance and prevalence, the biological relevance of this finding is unclear. MSS revealed that tet(X) was more abundant in the resistome of omnivores compared to pescatarians. It is most likely that the increased sample size (n = 149 vs n = 64 samples included for MSS and ResCap respectively) resulted in the detection of differentially abundant tet(X) by MSS, but not by ResCap. This suggests that the small sample size for ResCap is a potential limitation of the current study.
Two methods, mOTUs2 and MetaPhlAn3 were used to study the impact of dietary habits on the gut microbiome49,50. This showed that long-term dietary habits did not result in significant differences in the top 10 most abundant genera, nor in differences in gut microbiome diversity and that diet was not a main driver of the observed variance in microbiome composition between the samples. Both methods were largely in concordance regarding which genera were differentially abundant using a supervised analysis where only a few compositional differences between the microbiome of omnivores, pescatarians and vegetarians were detected. Variation between methods include the detection of differentially abundant Clostridium species by the mOTUs2, compared to differentially abundant species from the family of Lachnospiraceae, by the MetaPhlAn3. This variation could be attributed to the distinct ways in which the tools correct for genomic sequences from yet unknown species. While mOTUs2 makes use of marker genes, using its clusters of orthologues groups of proteins approach (COGs), MetaPhlAn3 instead estimates the ‘unknown’ portion by using the average gene length and genome length.
As part of a short-term diet intervention study, David et al. associated the consumption of animal derived products with an increased relative abundance of Alistipes, Bilophila and Bacteroides and a decrease in the relative abundance of Roseburia, Eubacterium rectale and Ruminococcus bromii14. Although our study included similar diet groups with omnivores consuming animal derived products, and vegans that do not, we did not observe similar findings. When compared to pescatarians, we detected a lower relative abundance of Eubacterium in omnivores, namely Eubacterium eligens. Chung et al. described E. eligens to be a pectin degrading specialist, which is a major component of plant cell walls and increased plant consumption might therefore explain the lower abundance of E. eligens in omnivores51. The non-concordance between the studies by David et al. and our study might be explained by differences in resolution of 16S rRNA gene sequencing used in David et al., as compared to MSS in our study, but maybe more importantly this illustrates the difference between an intervention-based study compared to our study design, where long-term diet habits were studied.
More recently, De Angelis et al. compared the gut microbiome, proteome and metabolome of Italian omnivores, vegetarians and vegans based on 16S rRNA gene profiling. Similar to our study, they showed only slight differences in the abundance of bacterial families between the diet groups. Ruminococcaceae were found to be most abundant in omnivores, while Lachnospira were associated with vegans and vegetarians52. Similarly, Losasso et al. observed differences in the relative abundance of Ruminococcaceae when comparing the microbiome of vegans, vegetarians, and omnivores20, using 16S rRNA profiling. Although the observations are limited to the family level, these studies align with our results, where omnivores were found to have a higher relative abundance of R. torques when compared to vegans. In contrast to these dietary studies, we observed differences in the relative abundance of lactic acid bacteria when comparing the microbiome between diet groups. This difference could be explained by the relatively high consumption of dairy products in the Netherlands53. Omnivores and pescatarians showed higher relative abundance of Streptococcus thermophilus when compared to vegans. Similarly, Lactococcus lactis was present in higher relative abundance in the microbiome of pescatarians compared to vegans. Both S. thermophilus as L. lactis have been associated with the consumption of dairy products54. Zhernakova et al. also detected a specific association between the consumption of buttermilk and the abundance of Leuconostoc mesenteroides and L. lactis, when assessing factors contributing to variation of the gut microbiome composition in the Dutch and Belgium population12,55. Since dairy products are excluded from the vegan diet, it could explain the low abundance of these lactic acid bacteria in this group.
To conclude, using ResCap we were able to increasingly detect resistance genes in complex samples like human faeces in our study, compared to MSS, and found that the gut microbiome of humans included in this study represented a large reservoir of 145 different antibiotic resistance genes in the studied population. However, differences in long-term dietary habits in the Dutch population did not result in significant differences in resistome composition between the four diet groups. When comparing the microbiome composition of diet groups in the Dutch population, mainly the vegan diet was associated with a distinct taxonomic composition. Since geographic location likely has an impact on the microbiome and resistome composition in the general population, future studies involving human populations from other geographic regions, are needed to determine the generalizability of our findings in the Dutch population.
Methods
Participant inclusion
In this study, faecal samples from 149 Dutch individuals from a previous study in the general population (the “NLD-VEGA-study”) were selected based on their dietary habits (Table 1) and categorized in four different diet groups: (1) omnivores, (2) pescatarians, (3) vegetarians and (4) vegans56. Groups were matched for sex, age, education level, medication usage and keeping animals (Table 1). Differences between groups were compared by the Kruskal–Wallis or X2 test where appropriate. All participants had a Dutch nationality, were born in the Netherlands and lived in urban areas. None of them had used antibiotics, insulin, proton pump inhibitors or drugs related to cancer treatment and chemotherapy in the past 3 months.
Ethical approval
The original “NLD-VEGA-study” protocol was approved by the medical ethics committee of the University Medical Center Utrecht, the Netherlands (no. 15–561/C). All experiments were performed in accordance with relevant guidelines and regulations.
Classification of diet groups
Diet groups were defined based on the following criteria: omnivores consumed meat at least three times per week, for the past 6 months or more. Pescatarians consumed fish and animal derived products but did not consume meat in the past 6 months or more. Vegetarians consumed animal derived products but did not consume meat or fish in the past 6 months or more. Vegans did not consume meat, fish, or animal derived products in the past 6 months or more. Furthermore, pescatarians, vegetarians and vegans did not prepare the restricted products described for house members or pets or have house members that consumed those.
Sample collection, storage and DNA extraction
Faecal samples were sent by regular mail and transported for a maximum of 24 h before storage at − 80 °C. Samples were divided into aliquots of 0.2 g, thereby introducing one freeze–thaw cycle. Samples were thawed a second time and used for DNA extraction, using a modified protocol of the QIAamp fast DNA stool mini kit (Qiagen, Venlo, the Netherlands) as described by Knudsen et al.57. In brief, 0.2 g faeces were added to ‘lysing matrix A, 2 ml tubes’ (MP biomedicals, Landsmeer, the Netherlands), containing 1 ml InhibitEx buffer (Qiagen). Beat beating was applied at 3.5 m/s for 30 s, followed by 30 s incubation on ice and one final beat beating step, using the FastPrep-24 (MP biomedicals). After 7 min of incubation at 95 °C, the fast DNA stool mini kit protocol (Qiagen) was resumed at the proteinase K treatment step. Total DNA was quantified by Picogreen assay (Thermo Fisher Scientific, Waltham, MA, USA).
Metagenomic shotgun sequencing and Microbiome data processing
Samples were sent to Baseclear B.V. (Leiden, the Netherlands) for metagenomic shotgun sequencing (MSS), together with a DNA extraction (negative) control and mock control (ZymoBIOMICS Microbial Community Standard). The NovaSeq 6000 (Illumina, San Diego, USA) was used, with the S1, 2 × 150 bp paired-end kit (Illumina) and the company protocol/standard settings. Raw reads were trimmed by Trimmomatic v0.39 (options: slidingwindow:4:15 minlen:70) and used for taxonomic profiling with either mOTUs2 version 2.5.0 or MetaPhlAn3 using default settings49,50,58. Samples contained on average 70.7 M ± 11.3 M reads, while the negative control contained less than 300 k reads. The MSS results of the mock control contained a total of 89 M reads, and matched with the expected mock composition (table s2).
ResCap sequencing
Sixteen samples per diet group were selected for in-depth resistome analysis using the ResCap targeted sequence capture panel consisting of probes targeting genes that confer resistance to antibiotics, metals, biocides and included probes that target relaxase genes46. The capture panel contains probes against 7963 resistance genes and was expanded, by the addition of probes against the following mcr genes (mcr1.1, mcr1.2, mcr1.3, mcr1.4, mcr1.5, mcr1.6, mcr1.7, mcr1.8, mcr1.9, mcr1.10, mcr2.1, mcr2.2, mcr2.3, mcr3.1, mcr3.2.1, mcr3.2.2, mcr3.3.1, mcr3.3.2, mcr3.4.1, mcr3.4.2, mcr3.5.1, mcr3.5.2, mcr3.6, mcr4, mcr5, mcr6, mcr7 and mcr8) (Roche ID: OID41815). The subset of samples selected per diet group were matched for sex, age, education level, medication usage and having pets (Table 2). Differences between groups were tested by the Kruskal–Wallis or X2 test where appropriate. ResCap was performed according to the supplied protocol. In brief, 0.8–1.0 µg DNA was used for fragmentation using the KAPA HyperPlus Kit v4.17 (Roche, Woerden, The Netherlands) to generate 400 bp fragments. End repair, A-tailing and adapter ligation were performed as described by the SeqCap EZ HyperCap User’s Guide v2.3. Pools of 12 samples were used for hybridization and capture using an extended version of the ResCap probe collection as described in the original publication46. Sample pools were sequenced on a NextSeq500 (Illumina), using high output and paired-ends of 2 × 150 bp.
Resistome data processing
ResCap and MSS data were trimmed using Trim Galore version 0.6.4 with standard settings59. KMA version 1.3.4 was used to align sequences to the Resfinder database version of 2020-06-0260. For KMA, paired-end reads were used as input by using -ipe, together with the options: -tmp, -1t1, -cge, -apm p, -ef. The resulting list of detected genes and their abundance was trimmed by applying a cut-off of 90% identity (called query Identity) and of 80% coverage (called template coverage). Finally, the output value depth was used for subsequent analysis, which represents the amount of aligned base pairs, while correcting for gene length. Analysis of the MSS negative control for antibiotic resistance genes, containing less than 300 k reads, revealed the low abundance of antibiotic resistance genes tet(L) and aadD.
Data analysis
Analysis of sequencing data was performed in R version 4.0 and the functions of the packages phyloseq and ggplot261–63. The top 10 abundances of the microbiome and resistome were plotted using aggregate top taxa and plotting functions of the microbiome package64. Shannon index was calculated using the alpha diversity functions of the microbiome package and plotting functions of the microbiomeutilities package65. Aitchison, Bray–Curtis and Jaccard distance PCA were generated using the transform function of the microbiome package and ordinate (RDA) and plot ordination functions of the phyloseq package. Correlations of sample dissimilarity and diets were tested using PERMANOVA with the adonis function at 999 permutations of the vegan package66. Basic accessor functions of phyloseq were used to generate heatmaps. Differential abundance analysis was performed using ANCOM-BC version 1.0.2, with Bonferroni correction for false discovery rate and an alpha of 0.05 as a threshold for significance45.
Consent to publish
In this study, faecal samples from 149 Dutch individuals from a previous study in the population at large (the “vegastudy”) were selected. All participants provided written informed consent for the initial vegastudy and had given permission to use their faecal samples for further research.
Supplementary Information
Acknowledgements
The authors thank Angela van Hoek for her insights on the analysis and for corrections of the manuscript. We would like to thank Dr. Philip Clausen (Technical University of Denmark) for the technical assistance on KMA (k-mer alignment).
Abbreviations
- ARGs
Antibiotic resistance genes
- MSS
Metagenomic shotgun sequencing
- PCA
Principal component analysis
- ResCap
Resistome capture sequencing approach
Author contributions
P.B.S., F.L.P., R.J.L.W., E.F., E.G., C.M.D., E.D. and S.F. all contributed to the study design and implementation. Lab work was performed by P.B.S., R.Q.J.P., M.V. and M.C.V. P.B.S., J.H., S.A.S., M.R.C.R., R.J.L.W. and F.L.P. performed data analysis, statistical analysis and interpretation. All authors read and approved the final manuscript.
Funding
This work was supported by the Netherlands Centre of One Health (NCOH metagenome call), the Dutch Ministry of Health, Welfare and Sport (Grant number V/050305/AA and the Strategic Program RIVM (SPR)) and Utrecht Exposome Hub of Utrecht Life Sciences (www.uu.nl/exposome), funded by the Executive Board of Utrecht University.
Data availability
The 149 MSS and 64 ResCap sequencing files have been deposited in the European Nucleotide Archive repository under the study accession no. PRJEB45944 and PRJEB46230, respectively. R scripts to reproduce the analysis reported in this study can be found at; https://gitlab.com/PB_Stege/diet_microbiome_resistome/.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Joost Hordijk and Sudarshan A. Shetty.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-05817-4.
References
- 1.Wampach L, Heintz-Buschart A, Hogan A, Muller EEL, Narayanasamy S, Laczny CC, et al. Colonization and succession within the human gut microbiome by archaea, bacteria, and microeukaryotes during the first year of life. Front. Microbiol. 2017;8:738. doi: 10.3389/fmicb.2017.00738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Manor O, Dai CL, Kornilov SA, Smith B, Price ND, Lovejoy JC, et al. Health and disease markers correlate with gut microbiome composition across thousands of people. Nat. Commun. 2020;11(1):5206. doi: 10.1038/s41467-020-18871-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fan Y, Pedersen O. Gut microbiota in human metabolic health and disease. Nat. Rev. Microbiol. 2021;19(1):55–71. doi: 10.1038/s41579-020-0433-9. [DOI] [PubMed] [Google Scholar]
- 4.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486(7402):222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weng YJ, Gan HY, Li X, Huang Y, Li ZC, Deng HM, et al. Correlation of diet, microbiota and metabolite networks in inflammatory bowel disease. J. Dig. Dis. 2019;20(9):447–459. doi: 10.1111/1751-2980.12795. [DOI] [PubMed] [Google Scholar]
- 6.Jakobsson HE, Jernberg C, Andersson AF, Sjölund-Karlsson M, Jansson JK, Engstrand L. Short-term antibiotic treatment has differing long-term impacts on the human throat and gut microbiome. PLoS One. 2010;5(3):e9836. doi: 10.1371/journal.pone.0009836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Queipo-Ortuño MI, Boto-Ordóñez M, Murri M, Gomez-Zumaquero JM, Clemente-Postigo M, Estruch R, et al. Influence of red wine polyphenols and ethanol on the gut microbiota ecology and biochemical biomarkers. Am. J. Clin. Nutr. 2012;95(6):1323–1334. doi: 10.3945/ajcn.111.027847. [DOI] [PubMed] [Google Scholar]
- 8.Pasolli E, Asnicar F, Manara S, Zolfo M, Karcher N, Armanini F, et al. Extensive unexplored human microbiome diversity revealed by over 150,000 genomes from metagenomes spanning age, geography, and lifestyle. Cell. 2019;176(3):649–62.e20. doi: 10.1016/j.cell.2019.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bressa C, Bailén-Andrino M, Pérez-Santiago J, González-Soltero R, Pérez M, Montalvo-Lominchar MG, et al. Differences in gut microbiota profile between women with active lifestyle and sedentary women. PLoS One. 2017;12(2):e0171352. doi: 10.1371/journal.pone.0171352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rogers MA, Greene MT, Saint S, Chenoweth CE, Malani PN, Trivedi I, et al. Higher rates of Clostridium difficile infection among smokers. PLoS One. 2012;7(7):e42091. doi: 10.1371/journal.pone.0042091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keohane DM, Ghosh TS, Jeffery IB, Molloy MG, O'Toole PW, Shanahan F. Microbiome and health implications for ethnic minorities after enforced lifestyle changes. Nat. Med. 2020;26(7):1089–1095. doi: 10.1038/s41591-020-0963-8. [DOI] [PubMed] [Google Scholar]
- 12.Falony G, Joossens M, Vieira-Silva S, Wang J, Darzi Y, Faust K, et al. Population-level analysis of gut microbiome variation. Science. 2016;352(6285):560–564. doi: 10.1126/science.aad3503. [DOI] [PubMed] [Google Scholar]
- 13.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334(6052):105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505(7484):559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Filippis F, Pellegrini N, Vannini L, Jeffery IB, La Storia A, Laghi L, et al. High-level adherence to a Mediterranean diet beneficially impacts the gut microbiota and associated metabolome. Gut. 2016;65(11):1812–1821. doi: 10.1136/gutjnl-2015-309957. [DOI] [PubMed] [Google Scholar]
- 16.Kovatcheva-Datchary P, Nilsson A, Akrami R, Lee YS, De Vadder F, Arora T, et al. Dietary fiber-induced improvement in glucose metabolism is associated with increased abundance of prevotella. Cell Metab. 2015;22(6):971–982. doi: 10.1016/j.cmet.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 17.Schnorr SL, Candela M, Rampelli S, Centanni M, Consolandi C, Basaglia G, et al. Gut microbiome of the Hadza hunter-gatherers. Nat. Commun. 2014;5:3654. doi: 10.1038/ncomms4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meslier V, Laiola M, Roager HM, De Filippis F, Roume H, Quinquis B, et al. Mediterranean diet intervention in overweight and obese subjects lowers plasma cholesterol and causes changes in the gut microbiome and metabolome independently of energy intake. Gut. 2020;69(7):1258–1268. doi: 10.1136/gutjnl-2019-320438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bonder MJ, Tigchelaar EF, Cai X, Trynka G, Cenit MC, Hrdlickova B, et al. The influence of a short-term gluten-free diet on the human gut microbiome. Genome Med. 2016;8(1):45. doi: 10.1186/s13073-016-0295-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Losasso C, Eckert EM, Mastrorilli E, Villiger J, Mancin M, Patuzzi I, et al. Assessing the influence of vegan, vegetarian and omnivore oriented westernized dietary styles on human gut microbiota: A cross sectional study. Front. Microbiol. 2018;9:317. doi: 10.3389/fmicb.2018.00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Penders J, Stobberingh EE, Savelkoul PH, Wolffs PF. The human microbiome as a reservoir of antimicrobial resistance. Front. Microbiol. 2013;4:87. doi: 10.3389/fmicb.2013.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Araos R, Battaglia T, Ugalde JA, Rojas-Herrera M, Blaser MJ, D'Agata EMC. Fecal microbiome characteristics and the resistome associated with acquisition of multidrug-resistant organisms among elderly subjects. Front. Microbiol. 2019;10:2260. doi: 10.3389/fmicb.2019.02260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rose G, Shaw AG, Sim K, Wooldridge DJ, Li MS, Gharbia S, et al. Antibiotic resistance potential of the healthy preterm infant gut microbiome. PeerJ. 2017;5:e2928. doi: 10.7717/peerj.2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Milanović V, Osimani A, Aquilanti L, Tavoletti S, Garofalo C, Polverigiani S, et al. Occurrence of antibiotic resistance genes in the fecal DNA of healthy omnivores, ovo-lacto vegetarians and vegans. Mol. Nutr. Food Res. 2017;61:9. doi: 10.1002/mnfr.201601098. [DOI] [PubMed] [Google Scholar]
- 25.Moore AM, Ahmadi S, Patel S, Gibson MK, Wang B, Ndao MI, et al. Gut resistome development in healthy twin pairs in the first year of life. Microbiome. 2015;3:27. doi: 10.1186/s40168-015-0090-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu Y, Yang X, Qin J, Lu N, Cheng G, Wu N, et al. Metagenome-wide analysis of antibiotic resistance genes in a large cohort of human gut microbiota. Nat. Commun. 2013;4:2151. doi: 10.1038/ncomms3151. [DOI] [PubMed] [Google Scholar]
- 27.Li J, Rettedal EA, van der Helm E, Ellabaan M, Panagiotou G, Sommer MOA. Antibiotic treatment drives the diversification of the human gut resistome. Genom. Proteom. Bioinform. 2019;17(1):39–51. doi: 10.1016/j.gpb.2018.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oldenburg CE, Hinterwirth A, Sié A, Coulibaly B, Ouermi L, Dah C, et al. Gut resistome after oral antibiotics in preschool children in burkina faso: A randomized, controlled trial. Clin. Infect. Dis. 2020;70(3):525–527. doi: 10.1093/cid/ciz455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palleja A, Mikkelsen KH, Forslund SK, Kashani A, Allin KH, Nielsen T, et al. Recovery of gut microbiota of healthy adults following antibiotic exposure. Nat. Microbiol. 2018;3(11):1255–1265. doi: 10.1038/s41564-018-0257-9. [DOI] [PubMed] [Google Scholar]
- 30.Buelow E, Gonzalez TB, Versluis D, Oostdijk EA, Ogilvie LA, van Mourik MS, et al. Effects of selective digestive decontamination (SDD) on the gut resistome. J. Antimicrob. Chemother. 2014;69(8):2215–2223. doi: 10.1093/jac/dku092. [DOI] [PubMed] [Google Scholar]
- 31.Willmann M, Vehreschild M, Biehl LM, Vogel W, Dörfel D, Hamprecht A, et al. Distinct impact of antibiotics on the gut microbiome and resistome: A longitudinal multicenter cohort study. BMC Biol. 2019;17(1):76. doi: 10.1186/s12915-019-0692-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mancabelli L, Milani C, Lugli GA, Turroni F, Ferrario C, van Sinderen D, et al. Meta-analysis of the human gut microbiome from urbanized and pre-agricultural populations. Environ. Microbiol. 2017;19(4):1379–1390. doi: 10.1111/1462-2920.13692. [DOI] [PubMed] [Google Scholar]
- 33.von Wintersdorff CJ, Penders J, Stobberingh EE, Oude Lashof AM, Hoebe CJ, Savelkoul PH, et al. High rates of antimicrobial drug resistance gene acquisition after international travel, The Netherlands. Emerg. Infect. Dis. 2014;20(4):649–657. doi: 10.3201/eid2004.131718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pan H, Paudyal N, Li X, Fang W, Yue M. Multiple food-animal-borne route in transmission of antibiotic-resistant salmonella newport to humans. Front. Microbiol. 2018;9:23. doi: 10.3389/fmicb.2018.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaesbohrer A, Bakran-Lebl K, Irrgang A, Fischer J, Kämpf P, Schiffmann A, et al. Diversity in prevalence and characteristics of ESBL/pAmpC producing E. coli in food in Germany. Vet. Microbiol. 2019;233:52–60. doi: 10.1016/j.vetmic.2019.03.025. [DOI] [PubMed] [Google Scholar]
- 36.Clemente L, Manageiro V, Ferreira E, Jones-Dias D, Correia I, Themudo P, et al. Occurrence of extended-spectrum β-lactamases among isolates of Salmonella enterica subsp. enterica from food-producing animals and food products, in Portugal. Int. J. Food Microbiol. 2013;167(2):221–228. doi: 10.1016/j.ijfoodmicro.2013.08.009. [DOI] [PubMed] [Google Scholar]
- 37.Mughini-Gras L, Dorado-García A, van Duijkeren E, van den Bunt G, Dierikx CM, Bonten MJM, et al. Attributable sources of community-acquired carriage of Escherichia coli containing β-lactam antibiotic resistance genes: A population-based modelling study. Lancet Planet Health. 2019;3(8):e357–e369. doi: 10.1016/S2542-5196(19)30130-5. [DOI] [PubMed] [Google Scholar]
- 38.Evers EG, Pielaat A, Smid JH, van Duijkeren E, Vennemann FB, Wijnands LM, et al. Comparative exposure assessment of ESBL-producing Escherichia coli through meat consumption. PLoS One. 2017;12(1):e0169589. doi: 10.1371/journal.pone.0169589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramos S, Silva N, Dias D, Sousa M, Capelo-Martinez JL, Brito F, et al. Clonal diversity of ESBL-producing Escherichia coli in pigs at slaughter level in Portugal. Foodborne Pathog. Dis. 2013;10(1):74–79. doi: 10.1089/fpd.2012.1173. [DOI] [PubMed] [Google Scholar]
- 40.Meldrum RJ, Wilson IG. Salmonella and Campylobacter in United Kingdom retail raw chicken in 2005. J. Food Prot. 2007;70(8):1937–1939. doi: 10.4315/0362-028x-70.8.1937. [DOI] [PubMed] [Google Scholar]
- 41.Zhang S, Li S, Gu W, den Bakker H, Boxrud D, Taylor A, et al. Zoonotic source attribution of Salmonella enterica serotype typhimurium using genomic surveillance data, United States. Emerg. Infect. Dis. 2019;25(1):82–91. doi: 10.3201/eid2501.180835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Whitehouse CA, Young S, Li C, Hsu CH, Martin G, Zhao S. Use of whole-genome sequencing for Campylobacter surveillance from NARMS retail poultry in the United States in 2015. Food Microbiol. 2018;73:122–128. doi: 10.1016/j.fm.2018.01.018. [DOI] [PubMed] [Google Scholar]
- 43.King CH, Desai H, Sylvetsky AC, LoTempio J, Ayanyan S, Carrie J, et al. Baseline human gut microbiota profile in healthy people and standard reporting template. PLoS One. 2019;14(9):e0206484. doi: 10.1371/journal.pone.0206484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nishijima S, Suda W, Oshima K, Kim SW, Hirose Y, Morita H, et al. The gut microbiome of healthy Japanese and its microbial and functional uniqueness. DNA Res. 2016;23(2):125–133. doi: 10.1093/dnares/dsw002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin H, Peddada SD. Analysis of compositions of microbiomes with bias correction. Nat. Commun. 2020;11(1):3514. doi: 10.1038/s41467-020-17041-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lanza VF, Baquero F, Martínez JL, Ramos-Ruíz R, González-Zorn B, Andremont A, et al. In-depth resistome analysis by targeted metagenomics. Microbiome. 2018;6(1):11. doi: 10.1186/s40168-017-0387-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Macedo G, van Veelen HPJ, Hernandez-Leal L, van der Maas P, Heederik D, Mevius D, et al. Targeted metagenomics reveals inferior resilience of farm soil resistome compared to soil microbiome after manure application. Sci. Total Environ. 2021;770:145399. doi: 10.1016/j.scitotenv.2021.145399. [DOI] [PubMed] [Google Scholar]
- 48.Guitor AK, Raphenya AR, Klunk J, Kuch M, Alcock B, Surette MG, et al. Capturing the resistome: A targeted capture method to reveal antibiotic resistance determinants in metagenomes. Antimicrob. Agents Chemother. 2019;64:1. doi: 10.1128/AAC.01324-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Milanese A, Mende DR, Paoli L, Salazar G, Ruscheweyh HJ, Cuenca M, et al. Microbial abundance, activity and population genomic profiling with mOTUs2. Nat. Commun. 2019;10(1):1014. doi: 10.1038/s41467-019-08844-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Beghini F, McIver LJ, Blanco-Míguez A, Dubois L, Asnicar F, Maharjan S, et al. Integrating taxonomic, functional, and strain-level profiling of diverse microbial communities with bioBakery 3. Elife. 2021;10:25. doi: 10.7554/eLife.65088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chung WSF, Meijerink M, Zeuner B, Holck J, Louis P, Meyer AS, et al. Prebiotic potential of pectin and pectic oligosaccharides to promote anti-inflammatory commensal bacteria in the human colon. FEMS Microbiol. Ecol. 2017;93:11. doi: 10.1093/femsec/fix127. [DOI] [PubMed] [Google Scholar]
- 52.De Angelis M, Ferrocino I, Calabrese FM, De Filippis F, Cavallo N, Siragusa S, et al. Diet influences the functions of the human intestinal microbiome. Sci. Rep. 2020;10(1):4247. doi: 10.1038/s41598-020-61192-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Our world in data per capita milk consumption. https://ourworldindata.org/grapher/per-capita-milk-consumption. Accessed 17 July 2021.
- 54.Pasolli E, De Filippis F, Mauriello IE, Cumbo F, Walsh AM, Leech J, et al. Large-scale genome-wide analysis links lactic acid bacteria from food with the gut microbiome. Nat. Commun. 2020;11(1):2610. doi: 10.1038/s41467-020-16438-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhernakova A, Kurilshikov A, Bonder MJ, Tigchelaar EF, Schirmer M, Vatanen T, et al. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science. 2016;352(6285):565–569. doi: 10.1126/science.aad3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Meijs AP, Gijsbers EF, Hengeveld PD, Veenman C, van Roon AM, van Hoek A, et al. Do vegetarians less frequently carry ESBL/pAmpC-producing Escherichia coli/Klebsiella pneumoniae compared with non-vegetarians? J. Antimicrob. Chemother. 2020;75(3):550–558. doi: 10.1093/jac/dkz483. [DOI] [PubMed] [Google Scholar]
- 57.Knudsen BE, Bergmark L, Munk P, Lukjancenko O, Priemé A, Aarestrup FM, et al. Impact of sample type and DNA isolation procedure on genomic inference of microbiome composition. mSystems. 2016;1:5. doi: 10.1128/mSystems.00095-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17(1):3. [Google Scholar]
- 60.Clausen P, Aarestrup FM, Lund O. Rapid and precise alignment of raw reads against redundant databases with KMA. BMC Bioinform. 2018;19(1):307. doi: 10.1186/s12859-018-2336-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.R Core Team T. R: A Language and Environment for Statistical Computing. 2020.
- 62.McMurdie PJ, Holmes S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8(4):e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wickham H. ggplot2 Elegant Graphics for Data Analysis. Springer; 2016. [Google Scholar]
- 64.Lahti, L., & Shetty, S. Tools for microbiome analysis in R. 2017.
- 65.Shetty, S., & Lahti, L. Utilities for Microbiome Analytics. 2020.
- 66.Oksanen, J., et al. vegan Community Ecology Package. 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The 149 MSS and 64 ResCap sequencing files have been deposited in the European Nucleotide Archive repository under the study accession no. PRJEB45944 and PRJEB46230, respectively. R scripts to reproduce the analysis reported in this study can be found at; https://gitlab.com/PB_Stege/diet_microbiome_resistome/.