Abstract
Apolipoprotein E (APOE) alleles impact pathogenesis and risk for multiple human diseases, making them primary targets for disease treatment and prevention. Previously, we and others reported an association between APOE alleles and the gut microbiome. Here, we evaluated effects of APOE heterozygosity and tested whether these overall results extended to mice maintained under ideal conditions for microbiome analyses. To model human APOE alleles, this study used APOE targeted replacement (TR) mice on a C57Bl/6 background. To minimize genetic drift, homozygous APOE3 mice were crossed to homozygous APOE2 or homozygous APOE4 mice prior to the study, and the resulting heterozygous progeny crossed further to generate the study mice. To maximize environmental homogeneity, mice with mixed genotypes were housed together and used bedding from the cages was mixed and added back as a portion of new bedding. Fecal samples were obtained from mice at 3-, 5- and 7-months of age, and microbiota analyzed by 16S ribosomal RNA gene amplicon sequencing. Linear discriminant analysis of effect size (LefSe) identified taxa associated with APOE status, depicted as cladograms to show phylogenetic relatedness. The influence of APOE status was tested on alpha-diversity (Shannon H index) and beta-diversity (principal coordinate analyses and PERMANOVA). Individual taxa associated with APOE status were identified by classical univariate analysis. Whether findings in the APOE mice were replicated in humans was evaluated by using published microbiome genome wide association data. Cladograms revealed robust differences with APOE in male mice and limited differences in female mice. The richness and evenness (alpha-diversity) and microbial community composition (beta-diversity) of the fecal microbiome was robustly associated with APOE status in male but not female mice. Classical univariate analysis revealed individual taxa that were significantly increased or decreased with APOE, illustrating a stepwise APOE2-APOE3–APOE4 pattern of association with heterozygous animals trending as intermediate in the stepwise pattern. The relative abundance of bacteria from the class Clostridia, order Clostridiales, family Ruminococacceae and related genera increased with APOE2 status. The relative abundance of Erysipelotrichia increased with APOE4 status, a finding that extended to humans. In this study, wherein mice were maintained in an ideal fashion for microbiome studies, gut microbiome profiles were strongly and significantly associated with APOE status in male APOE-TR mice. Erysipelotrichia are increased with APOE4 in both mice and humans. APOE allelic effects appeared generally intermediate in heterozygous animals. Further evaluation of these findings in humans, as well as studies evaluating the impact of the APOE-associated microbiota on disease-relevant phenotypes, will be necessary to determine if alterations in the gut microbiome represent a novel mechanism whereby APOE alleles impact disease.
Subject terms: Genetic association study, Microbiome
Introduction
Apolipoprotein E (APOE) alleles impact multiple facets of the human condition, ranging from Alzheimer’s disease (AD) to cardiovascular disease, metabolic syndrome, obesity, fertility and longevity (reviewed in Ref.1). The three primary APOE alleles include APOE3, which has a 78% minor allele frequency, as well as APOE4 and APOE2, with minor allele frequencies of 14 and 8%, respectively. Regarding AD, APOE2 reduces AD risk while APOE4 strongly increases AD risk, both relative to APOE3 (reviewed in Ref.2). This association has prompted intense evaluation of possible mechanism(s) underlying APOE effects in AD, resulting in APOE allelic association with amyloid-beta (Aβ) clearance, Aβ aggregation and astrocyte stress3–8. In the periphery, APOE2 is associated with decreased low-density lipoprotein (LDL) cholesterol, whereas APOE4 is associated with increased LDL cholesterol, relative to APOE3. While this may account for APOE association with cardiovascular disease, the mechanisms underlying APOE allelic effects on glucose metabolism, inflammation and innate immunity are less clear2. The effects of APOE alleles in heterozygous individuals are generally intermediate for phenotypes ranging from cholesterol to AD1,2. Elucidating these differential actions of APOE alleles may provide insights to these processes.
Several studies have suggested a relationship between APOE status, the gut microbiome and AD neuropathology. First, APOE-deficient mice display microbiome differences relative to wild-type mice9. Second, APOE4-targeted replacement (TR) mice were more resistant to gastrointestinal Cryptosporidium infection than APOE3 mice10. Third, the APOE4 allele in humans was associated with better defense against childhood diarrheal diseases in lower income countries11–13. Fourth, we and others have recently reported microbiome differences in a comparison of APOE3 and APOE4-TR mice14–16. Lastly, several reports have found that Aβ-burden in murine models is reduced in gnotobiotic mice or mice treated with antibiotics17–20. The mechanism(s) whereby APOE alleles influence the gut microbiome are unclear, although APOE4 has been associated with a greater inflammatory response to lipopolysaccharide (LPS), a microbiome product common to all gram-negative bacteria, in both humans and mice21,22.
To begin to evaluate whether APOE allelic effects are dominant, co-dominant, or recessive, we compared animals heterozygous and homozygous for APOE alleles. Additionally, we improved our study design for rigor and reproducibility by backcrossing the APOE-TR mice to obviate possible genetic drift, maintaining mice with mixed genotypes in the same cages to minimize possible cage effects, and mixing used bedding between cages to ensure a homogenous microbial environment among cages.
Materials and methods
Mice
APOE3-TR23,24 male mice were crossed to APOE4 and APOE2 female mice to produce APOE2/E3 and APOE3/E4 heterozygous offspring. These mice were then crossed to generate 76 experimental mice that included APOE2/E2 (N = 6), APOE2/E3 (N = 12), APOE3/E3 (N = 5), APOE3/E4 (N = 8), and APOE4/E4 (N = 4) female mice and APOE2/E2 (N = 5), APOE2/E3 (N = 7), APOE3/E3 (N = 13), APOE3/E4 (N = 11), and APOE4/E4 (N = 5) male mice. Genotypes were determined by TaqMan SNP assays (Thermo). At weaning, mice were separated by sex and housed as mixed genotypes, 2–5 mice per cage (average of 3.7 ± 1.4 (mean ± SD)). Mice were maintained on Teklad Global 18% Protein Rodent Diet. To minimize potential confounding effects of coprophagy (mice feeding partially on cage-mate feces)25, approximately 20% of the new bedding was a mixture of used bedding from all the cages. Feces were obtained from this cohort of mice at 3-, 5- and 7-months of age. To obtain feces, mice were temporarily removed from their cage and placed into a clean Styrofoam cup. Fresh fecal pellets were stored at − 80 °C until DNA isolation. All methods were approved by University of Kentucky Institutional Animal Care and Use Committee. This study was carried out in compliance with ARRIVE guidelines.
Microbiome analysis
Fecal DNA was isolated by using a QIAamp PowerFecal Pro DNA Kit (QIAGEN). Genomic DNA was polymerase chain reaction (PCR) amplified with primers CS1_515F and CS2_806R (modified from the primer set employed by the Earth Microbiome Project (EMP; GTGYCAGCMGCCGCGGTAA and GGACTACNVGGGTWTCTAAT) targeting the V4 regions of microbial small subunit ribosomal RNA genes were generated using a two-stage PCR amplification protocol as described previously26. The primers contained 5′ common sequence tags (known as common sequence 1 and 2, CS1 and CS2). First stage PCR amplifications were performed in 10 µl reactions in 96-well plates, using MyTaq HS 2× mastermix (Bioline). PCR conditions were 95 °C for 5 min, followed by 28 cycles of 95 °C for 30′′, 55 °C for 45′′ and 72 °C for 60′′.
Subsequently, a second PCR amplification was performed in 10 µl reactions in 96-well plates. A mastermix for the entire plate was made using MyTaq HS 2× mastermix. Each well received a separate primer pair with a unique 10-base barcode, obtained from the Access Array Barcode Library for Illumina (Fluidigm, South San Francisco, CA; Item# 100-4876). Cycling conditions were: 95 °C for 5 min, followed by 8 cycles of 95 °C for 30′′, 60 °C for 30′′ and 72 °C for 30′′. Samples were then pooled, purified, and sequenced on an Illumina MiniSeq platform employing paired-end 2 × 153 base reads. Fluidigm sequencing primers, targeting the CS1 and CS2 linker regions, were used to initiate sequencing. De-multiplexing of reads was performed on instrument. Library preparation, pooling, and sequencing were performed at the University of Illinois at Chicago Genome Research Core (GRC) within the Research Resources Center (RRC).
Forward and reverse reads were merged using PEAR27 and trimmed based on a quality threshold of p = 0.01. Ambiguous nucleotides and primer sequences were removed and sequences shorter than 225 bp were discarded. Chimeric sequences were identified and removed using the USEARCH algorithm with a comparison to the Silva 132_16S reference database28,29. Amplicon sequence variants (ASVs) were identified using DADA230 and their taxonomic annotations determined using the UCLUST algorithm and Silva 132_16S reference with a minimum similarity threshold of 90%28,29. Sequence processing and annotation was performed by the Research Informatics Core (RIC) within the RRC.
This sequencing effort yielded 10,162,042 reads. Raw sequence data files were submitted in the Sequence Read Archive (SRA) of the National Center for Biotechnology Information (NCBI) under the BioProject Identifier PRJNA787634. Two samples with fewer than 30,000 reads each were discarded. Since APOE effects may be sex dependent16, microbiomes from male and female mice were analyzed separately. Average read counts per sample for the 3-month males was 48,725, 3-month females was 49,499, 5-month males was 45,985, 5-month females was 49,939, in 7-month males was 50,975, in 7-month females was 51,931. Using MicrobiomeAnalyst31 (updated version February 2021), samples were rarified to the minimum library size, which for 3-month males was 36,421, 3-month females was 37,069, 5-month males to 30,738, 5-month females to 35,467, 7-month males to 35,452, and 7-month females to 36,730. Low abundance ASVs were removed, i.e., ASVs with < 3 counts in > 90% of the samples were removed, and low variance ASVs were also removed, i.e., ASVs whose inter-quantile range was in the lowest 10%31. These corrections reduced the number of ASVs from 263 to 63 ASVs in 3-month males and females, 67 in 5-month females, 69 in 5-month males, 68 in 7-month females and 66 in 7-month males. Count data were normalized with a centered log-ratio transformation. Regarding APOE genetics, the results were analyzed as separate genotypes or as pooled alleles, i.e., APOE2/E3 heterozygous mice were grouped with APOE2/E2 mice while APOE3/E4 mice were grouped with APOE4/E4 as described in other APOE studies32,33. This resulted in 12 APOE2, 13 APOE3 and 16 APOE4 male mice and 18 APOE2, five APOE3 and 12 APOE4 female mice.
Bacteria associated with APOE were identified by a linear discriminant analysis effect size (LefSe) approach34. Significance thresholds were set to 0.05 for the alpha values for Kruskal–Wallis/Wilcoxon tests and 2.0 for the logarithmic linear discriminant analysis (LDA) score, using a one-against-all multi-class analysis approach. These results were then plotted as a cladogram to document the phylogenetic relatedness of APOE allelic associations with the bacteria at each taxonomic level. Results are also presented as an LDA histogram.
Alpha-diversity was assessed using the Shannon H diversity index35 with APOE statistical significance determined by Kruskal–Wallis tests. Additional alpha-diversity tests included Margalef taxon richness, Pielou’s evenness and the Simpson index with APOE statistical significance determined by Jonckheere–Terpstra nonparametric tests. Beta-diversity was assessed using Principal Coordinates Analysis (PCoA) of Bray–Curtis matrices with statistical significance determined by Permutational Multivariate Analysis of Variance (PERMANOVA)36. Taxonomic levels that associate with APOE status were determined using a classical univariate analysis with a Kruskal–Wallis test. A false discovery rate (FDR) approach was used to correct for multiple testing31. Heatmaps of family-level bacterial relative abundances were generated for male and female mice as a function of APOE status by using the Ward analysis of variance clustering algorithm, which is based on Pearson Correlation Coefficient distance measures.
Ethics approval
Animal studies were performed in compliance with the Institutional Animal Care and Use Committee at University of Kentucky.
Consent for publication
Each of the authors have reviewed the manuscript and approved it for publication.
Results
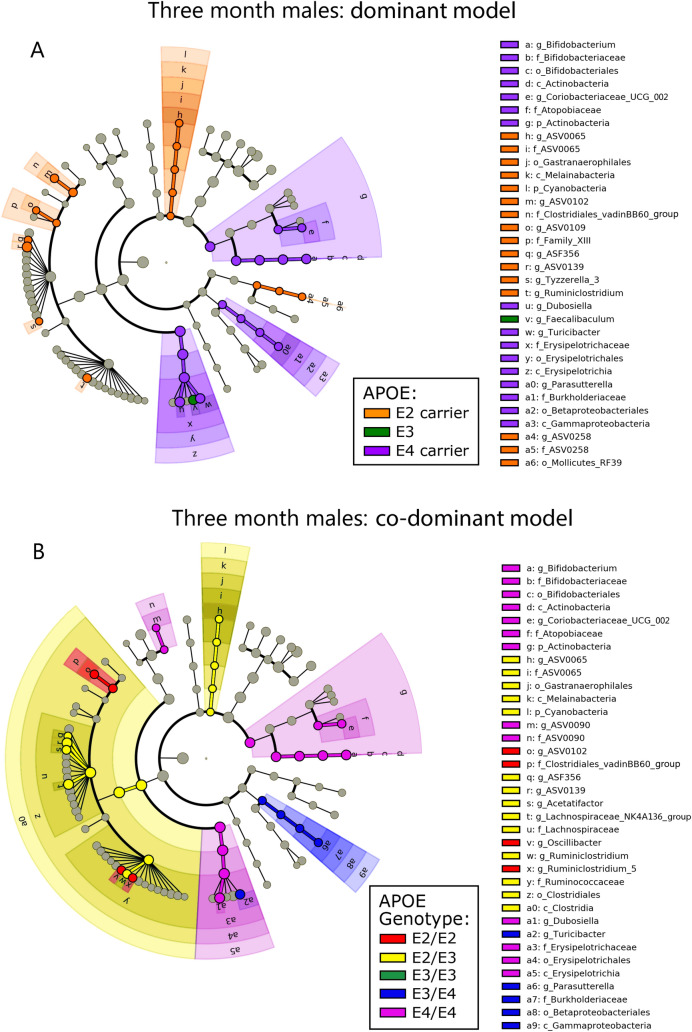
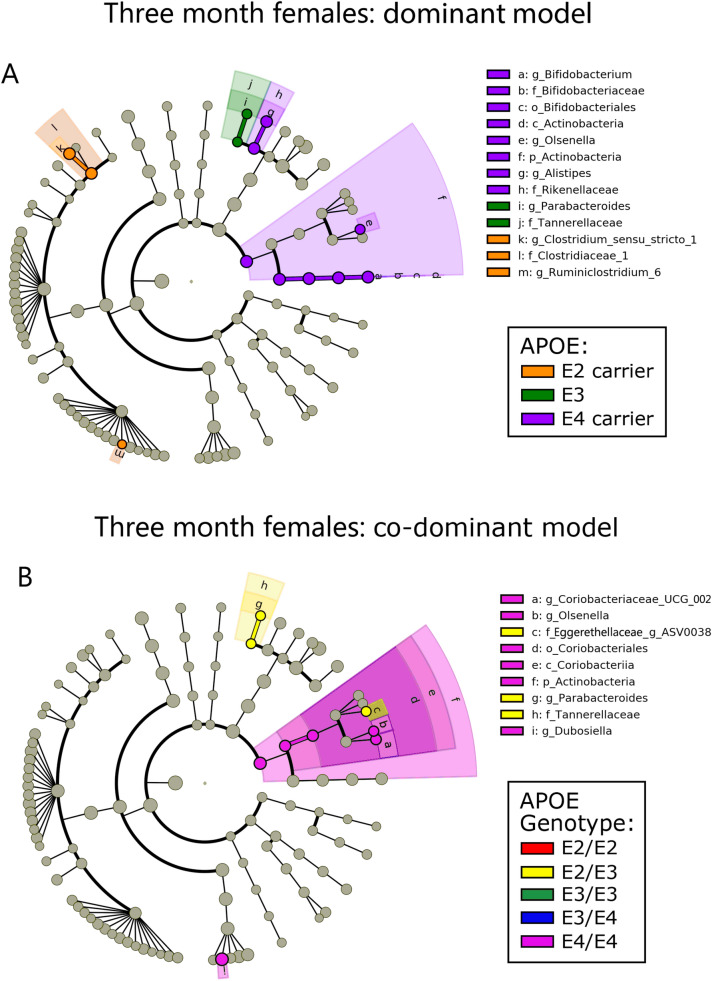
To investigate the hypothesis that APOE is associated with gut microbial community structure, we began with LefSe analysis and visualized the results with cladograms (Figs. 1, 2, Figs. S1, S2). This robust approach provides a visual means to identify statistically significant and phylogenetically-related taxa associated with APOE status34. These LefSe results are also presented as LDA histograms to provide a quantitative representation of the LefSe analyses (Fig. S3). Results are presented with APOE status stratified as APOE2 carriers, APOE3 and APOE4 carriers (pooled) (Figs. 1A, 2A) and with APOE status as separate genotypes (Figs. 1B, 2B). These two representations of the data provide insights into whether APOE allelic effects are dominant or co-dominant. Robust gut microbiome differences were observed in male mice compared to female mice at 3-months of age (Figs. 1, 2) with similar results found at 5- and 7-months of age (Figs. S1, S2). The microbiome of both male and female mice showed APOE4-associated increases in members of the Actinobacteria phylum (Figs. 1, 2). In contrast, only male mice showed APOE4-associated increases in the Erysipleotrichia and Gammaproteobacteria classes and APOE2-associated increases in the Cyanobacteria phylum. To gain further insights into these findings, we parsed the results into individual genotypes. This expanded our findings by showing that members of the Clostridia class were significantly associated with APOE2/E2 and APOE2/E3 (Fig. 1B). Overall, these results indicated that a subset of bacteria were consistently associated with APOE status, especially in males. The male population captured the majority of taxa significantly associated with APOE in the female population. In the following results, we present analyses of data from male mice at 3-months of age with analyses for all ages and sexes included within the Supplemental Files.
Figure 1.
Cladograms reveal microbial phylogenetic branches associated with APOE status in males. Taxa are represented as nodes and are connected by lines based on the phylogenetic relatedness of all taxa present in each experimental cohort. For example, the end node, a. represents the genus Bifodobacterium which is connected to other nodes representing higher level taxa related to Bifodobacterium including; b. the family Bifidobacteriaceae, c. the order Bifidobacteriales, and d. the class Actinobacteria. Many taxa are associated with APOE genetics, with node colors indicating the APOE associated with highest levels of that taxa. Statistical significance reflects both p < 0.05 for Kruskal–Wallis tests and a logarithmic LDA score > 2.0.
Figure 2.
Cladograms reveal microbial phylogenetic branches associated with APOE status in females. Taxa significantly associated with APOE are highlighted (p < 0.05 for Kruskal–Wallis tests and a logarithmic LDA score > 2.0).
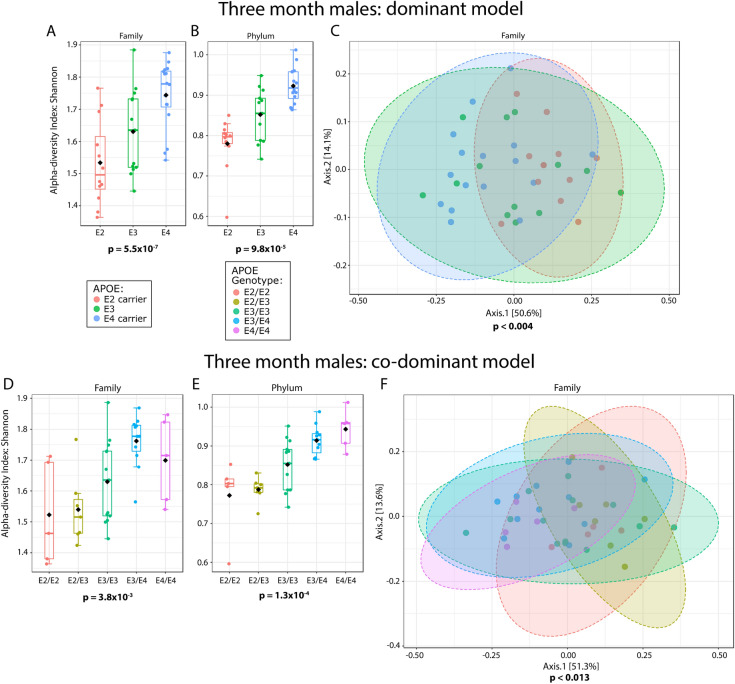
Microbiome alpha- (within sample) diversity was assessed by the Shannon H index and Simpson index, measures of taxon richness and evenness, as well the Margalef taxon richness index and evenness index37. A robust association between alpha-diversity and APOE was not detected in female mice (Shannon index p-values in Table 1, all measures of alpha-diversity in Supplemental Files). In contrast, male mice showed a stepwise trend towards higher alpha-diversity with APOE2-APOE3-APOE4 at the genus level, and this trend became more defined and statistically significant at every higher level through phylum (phylum and family depicted in Fig. 3, all levels presented in Table 1 and Supplemental Files, additional ages, and female data in Table S1). When the results are parsed into separate genotypes, the alpha-diversity of the APOE heterozygous animals tended to be intermediate relative to the homozygous animals. Hence, APOE was associated with alpha-diversity in male but not female mice and APOE allelic affects appeared co-dominant.
Table 1.
Microbiome alpha-diversity was significantly associated with APOE status in 3-month male but not female mice.
| Alpha-diversity (Shannon index) | ||
|---|---|---|
| Taxanomic level | 3 month p-values | |
| Males dominant | Males co-dominant | |
| Feature | 4.01E−01 | 1.02E−01 |
| Genus | 4.01E−01 | 1.02E−01 |
| Family | 5.49E−04 | 3.81E−03 |
| Order | 7.68E−05 | 5.05E−04 |
| Class | 7.68E−05 | 5.05E−04 |
| Phylum | 9.77E−05 | 1.28E−04 |
Figure 3.
Microbiome alpha- and beta-diversity as a function of APOE. Alpha-diversity is depicted as boxplots (A,B,D,E) and beta-diversity as PCoA plots (C,F). These results are from male mice at 3-months of age. Statistical significance for the findings is indicated below each graph. Ellipses in C and F represent 95% confidence intervals. Dominant model (C) R2 = 0.171 and co-dominant model (F) R2 = 0.206. Beta-diversity was also analyzed using a PERMDISP, which had no significant p-values, indicating that variances were not significantly different as a function of APOE.
Beta-diversity is a measure of between sample microbial communities based on their composition. Beta-diversity was visualized by using PCoA based on Bray–Curtis distance matrices38–41, and analyzed using a PERMANOVA. We found that APOE status was significantly associated with microbiome beta-diversity in male mice (Fig. 3, Table 2, additional ages and female in Table S2). When the data were analyzed with each separate APOE genotype, beta-diversity was still significant at each of the same taxonomic levels. Overall, these results demonstrate that the microbiome is robustly associated with APOE genetics in male mice.
Table 2.
Microbiome beta-diversity significantly associated with APOE status in male, but not female, mice.
| Beta-diversity (PERMANOVA) | |||
|---|---|---|---|
| Taxanomic level | 3 month | ||
| Males -dominant | Males co-dominant | ||
| Feature | p-value | < 0.002 | < 0.004 |
| R2 | 0.14 | 0.20 | |
| Genus | p-value | < 0.001 | < 0.008 |
| R2 | 0.15 | 0.18 | |
| Family | p-value | < 0.004 | < 0.013 |
| R2 | 0.17 | 0.21 | |
| Order | p-value | < 0.004 | < 0.015 |
| R2 | 0.21 | 0.24 | |
| Class | p-value | < 0.004 | < 0.015 |
| R2 | 0.21 | 0.24 | |
| Phylum | p-value | < 0.035 | < 0.112 |
| R2 | 0.12 | 0.15 | |
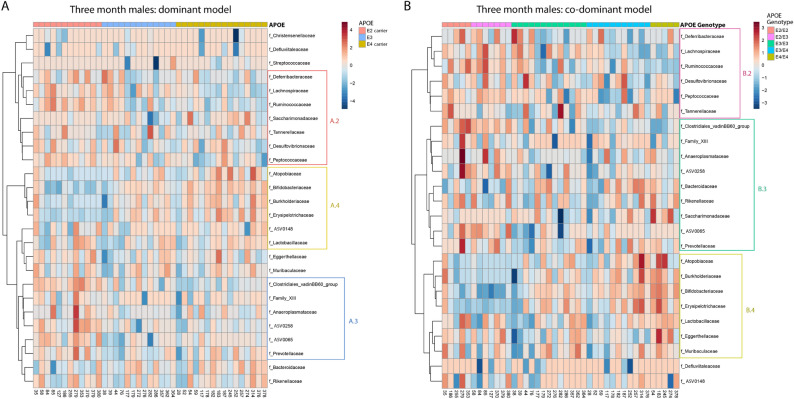
Variation in bacterial relative abundance per sample was visualized using heatmaps, as seen in Fig. 4. While cladograms identify bacterial phylogenetic branches that correlate with high abundance in association with a specific APOE status, heatmaps provide a per sample depth of information for each APOE status. Inspection of the heatmaps suggests that the data are relatively complex although some patterns of taxa association with APOE status are discernible, e.g., Ruminococcaceae and Erysipelotrichaceae in male mice (Fig. 4).
Figure 4.
Heatmaps depict overall microbiota profiles grouped by APOE status. These heatmaps depict per-sample relative abundance for family-level bacteria in female and male mice. Columns were grouped by APOE status, rows were grouped by the Ward clustering algorithm using Pearson Correlation Coefficient distance measures. Colored boxes highlight groups of taxa that follow either an APOE2- (A.2 and B.2), APOE3- (A.3 and B.3) or APOE4-associated pattern (A.4 and B.4).
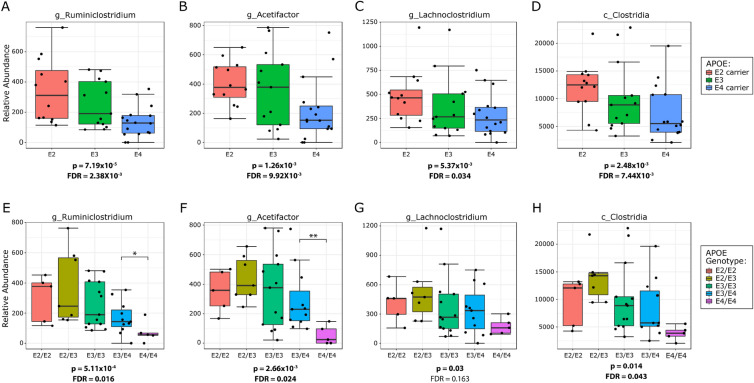
Although the use of LefSe analysis and cladograms provides insights into the microbiome and have been optimized for this purpose34, another perspective is provided by taxon-by-taxon classical univariate analysis using a Kruskal–Wallis test for significance and an FDR correction for multiple testing. To highlight the taxa most robustly associated with APOE, we applied classical univariate analysis to identify results that were significant with both approaches. Classical univariate analysis of the 3-month old female mice found no ASVs, genera, families, orders, classes, or phyla that were significantly associated with APOE in either APOE model. However, this approach applied to 3-month old male mice in the dominant model found 12 APOE-associated ASVs, as well as 12 genera, six families, five orders, five classes, and one phylum (statistics for all taxa in each age group are listed in Supplementary Tables S1.1–S3.4). In the co-dominant model, this approach found eight ASVs, eight genera, three families, four orders, four classes, and no phyla that were significantly associated with APOE. A graphical representation of the findings in 3-month old male mice is depicted in Fig. 5. Several bacteria showed stepwise associations with APOE on multiple taxonomic levels and were overall increased with APOE2. For example, the Clostridia class, Clostridiales order and two major families within this phylogenic branch, Ruminococcaceae and Lachnospiraceae showed an increase in their relative abundance from APOE4 to APOE3 to APOE2 (Fig. 5). The most abundant genera within the Ruminococcaceae family significantly associated with APOE were Ruminiclostridium (Fig. 5A,E), Ruminiclostridium_5 and Ruminiclostridium_9, which in aggregate represent approximately half of the Ruminococcaceae family. At 5- and 7-months of age, other genera within Ruminococcaceae were associated with APOE (Supplemental Tables S5.0–S7.5). Genera within the other major family, Lachnospiraceae, that increased with APOE2 were Acetifactor and Lachnoclostridium (Fig. 5B,C,F,G).
Figure 5.
The phylogenetic branch defined by Clostridia and its lower taxa shows a significant association with APOE in male mice. The relative abundance of each depicted bacteria was significantly associated with APOE status. The relative abundance of these bacteria decreased in a stepwise fashion from APOE2 to APOE3 to APOE4. P values have been corrected using an FDR approach. (A–D) Are the plots depicted using the dominant model representation, while (E–H) are the plots depicted using the co-dominant model representation. These data are derived from the 3-month male mice with data for all ages provided in Supplementary Tables S5.0–SS7.5.
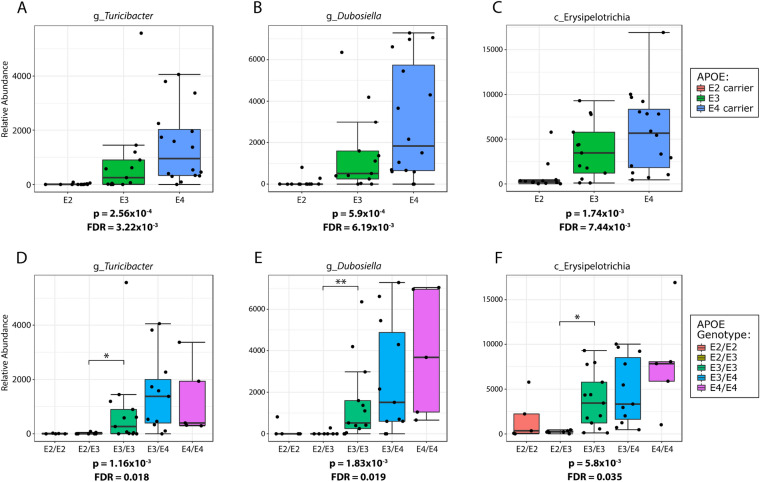
In contrast, the relative abundance of other taxa was increased in APOE4 mice, most notably bacteria within the class Erysipelotrichia class, confirming the findings from the LefSe cladograms (Fig. 1). Bacteria within the Erysipelotrichia branch that were significantly associated with APOE included the order Erysipelotrichiales, its family Erysipelotrichaceae and its genera Turicibacter and Dubosiella (Fig. 6, data for all ages shown in Supplementary Tables S5.0–S7.5). Consistent within this branch, bacterial relative abundance was near zero in the APOE2 mice, moderate in APOE3 and highly enriched in APOE4 (Fig. 6). Hence, both the LefSe and classical approaches identified members of the Clostridia class as enriched in APOE2 mice while members of the Erysipelotrichia class were enriched in APOE4 mice.
Figure 6.
The phylogenetic branch defined by Erysipelotrichia and its lower taxa shows a significant association with APOE in male mice. All depicted bacteria were significantly associated with APOE status. The relative abundance of these bacteria increased in a stepwise fashion from APOE2 to APOE3 to APOE4. P values have been corrected using an FDR approach. (A–C) Are the plots depicted using the dominant model representation, while (D–F) are the plots depicted using the co-dominant model representation. These results are derived from 3-month male mice.
To discern whether the Clostridiales and Erysipelotrichiales phylogenetic branches associated with APOE in this murine APOE-TR model are also associated with APOE in humans, we turned to a recent genome wide association study (GWAS) that evaluated the relationship between the gut microbiome and human polymorphisms42. This meta-analysis included data from as many as 18,340 individuals42. The only genetic locus that reached genome wide statistical significance was rs182549, which is associated with lactose intolerance. Interestingly, this SNP is modestly associated with the risk of Alzheimer’s disease (p = 0.003, N = 445,779)43, consistent with the possibility that the gut microbiome may influence AD risk. Focusing on APOE, the alleles of APOE2, APOE3 and APOE4 are defined by two SNPs, rs7412 and rs429358. The minor allele of rs7412 defines APOE2 while the minor allele of rs429358 determines APOE4 status. The Clostridiales and Erysipelotrichiales phylogenetic branches were not significantly associated with rs7412 (APOE2) at any phylogenetic level. However, the class Erysipelotrichia, the order Erysipelotrichales and the family Erysipelotrichaceae were nominally associated with rs429358 (Table 3). For each of these taxa, the minor APOE4 allele was associated with an increase in the relative abundance of these bacteria, reproducing the findings observed in the murine APOE-TR model.
Table 3.
Bacteria in the Erysipelotrichia phylogenetic branch are nominally associated with rs429358 in humans.
| Bacteria | SNP | Reference Allele | Effect Allele | Beta | SE | p value | N |
|---|---|---|---|---|---|---|---|
| Class: Erysipelotrichia | rs429358 | T | C | 0.032 | 0.015 | 0.035 | 18,097 |
| Order: Erysipelotrichales | rs429358 | T | C | 0.032 | 0.015 | 0.035 | 18,097 |
| Family: Erysipelotrichaceae | rs429358 | T | C | 0.032 | 0.015 | 0.035 | 18,097 |
| Genus: Turicibacter | rs429358 | T | C | 0.001 | 0.021 | 0.87 | 8921 |
The positive beta values reflect that the bacterial taxa are increased with the minor APOE4 allele of rs429358. These results combine data from men and women and are supplemental data from a large microbiome genetics study42.
Significance p values are in bold font.
Discussion
The primary finding reported here is that murine gut microbiome profiles are significantly associated with APOE status and APOE alleles appeared to act in a co-dominant fashion in heterozygous mice. These findings are strengthened given that the APOE-TR mice were maintained in an optimized fashion for microbiome analyses. The microbiome association with APOE was observed in alpha- and beta-diversity, encompasses multiple bacterial lineages and was predominately observed in male mice. Both LefSe and classical univariate analyses identified specific taxa that were associated with APOE. This association occurred in a stepwise fashion in the mice with the progression from APOE2-APOE3-APOE4. The stepwise association between indices of the gut microbiome and APOE2-APOE3-APOE4 reported here are reminiscent of APOE allelic association with other phenotypes ranging from LDL-cholesterol to AD risk1,2. Additionally, at least one of these associations, an increase in the relative abundance of Erysipelotrichia with APOE4, also has been observed in the human gut microbiome42. Moreover, the stepwise trends were also seen in the heterozygous mice, which reflected resulting intermediate effects of APOE on the gut microbiome. Overall, these findings confirm and extend prior reports that APOE genetics are associated with the gut microbiome14–16.
To identify the impact of APOE alleles on the microbiome, we used several approaches in this study, including alpha-diversity, beta-diversity, LefSe and classical univariate analyses. Alpha- and beta-diversity analyses aggregate multiple variables to provide an assessment of overall microbiome diversity and of microbiome profile similarity, respectively. In contrast, LefSe and classical univariate analyses provide an indication of differences in the relative abundance of specific taxa between experimental groups. These analyses were done by pooling the genotypes in a way representative of a dominant model of allelic effect and as individual genotypes representative of a co-dominant model of allelic effect. In this discussion, we will highlight the primary significant findings from these various analyses.
APOE4 was associated with increased alpha-diversity as assessed by the Shannon H index. A stepwise progression was observed in the dominant model representation with lowest alpha-diversity in APOE2, moderate in APOE3 and highest in APOE4. The co-dominant model representation showed an intermediate effect in the heterozygous animals following the stepwise APOE trend. Alpha-diversity is a measure of the number of distinct taxa and the evenness of these numbers across taxa. High alpha-diversity in the gut microbiome has been associated with improved gut health and microbiome homeostasis (reviewed in Ref.44). The APOE4 association with increased alpha-diversity observed here is consistent with prior observations that APOE4 is associated with better response to diarrheal infections in a third-world environment12,13. Indeed, the enrichment of APOE4 in people indigenous to Amazonian basin has been proposed to be a result of evolutionary selection in this environment with insufficient sanitation45.
A primary finding of this study was that both the LefSe and classical univariate analyses found that taxa within the Clostridia class were increased with APOE2 status, confirming results from our prior study14 and that of Tran et al.15. This phylogenetic branch included the Clostridiales order, Ruminococcaceae family and several genera within this family. The Clostridiales order was increased in APOE2 mice compared to APOE3 and APOE4 mice, with the heterozygous mice as intermediates. This was most robust in the 3-month males with similar findings at 5- and 7-months. The two major bacterial families within this order, Ruminococcaceae and Lachnospiraceae, were also both increased with APOE2. The stepwise fashion of the decline in Ruminnococcaceae relative abundance from APOE2 to APOE3 to APOE4 confirms the stepwise pattern seen previously14,16 and extends it along the phylogenetic branch from the Clostridia class to associated genera, such as Ruminiclostridium, Ruminiclostridium_5, Ruminiclostridium_9. Interestingly, Tran et al. also reported an increase in relative abundance of the Clostridiales order and Ruminococcaceae family in APOE2/E3 humans compared to APOE3/E4 and APOE4/E4 humans15. This suggests that this increase in Clostridiales and Ruminococcaceae with APOE2 may extend to humans. Two additional genera in the Clostridia class, within the Lachnospiraceae family, i.e., Acetifactor and Lachnoclostridium, also increased with APOE2 status in the current study. However, this finding was not replicated by Tran et al., who reported that Lachnospiraceae increased in APOE4 mice compared to APOE3 mice15.
Our study strengthens the associations between APOE status and the Clostridiales order, Ruminococcaceae family and related genera, and the Acetifactor and Lachnoclostridium genera by demonstrating a stepwise pattern with APOE allelic status across the entire phylogenetic branch from the Clostridia class down to related genera. Ruminococcaceae and Lachnospiraceae are bacterial families that highly express genes responsible for the metabolism of resistant starches in the large intestine, generating short chain fatty acids (SCFAs). The presence of SCFAs in the gut affect human health in general (reviewed in Refs.46,47) and have been reported to promote microglial maturation and function in particular48. Treatment with SCFAs has been shown to reduce microglial pro-inflammatory signals and promote a homeostatic profile that is neuroprotective49–52. Considering these findings relative to disease pathology associated with the stepwise APOE2-APOE3-APOE4 phenotype, we propose a tentative model wherein (i) APOE2 is associated with an increase in the relative abundance of microbiome bacteria Ruminococcaceae, Acetifactor and Lachnoclostridium, relative to APOE3 and APOE4, (ii) this shift in bacterial profile increases the production of SCFAs and (iii) this increase in SCFAs promotes microglial homeostasis and disease-ameliorating signaling, as suggested by robust genetic evidence53–61, (reviewed in Refs.62,63). While speculative, this model serves as a framework for future studies.
Another primary finding detected by both the LefSe and classical univariate analyses was that the Erysipelotrichia phylogenetic branch was significantly associated with APOE status in a stepwise APOE2-APOE3-APOE4 pattern with heterozygous mice acting as intermediates. This finding appeared to extend to humans and replicates the increase of the Erysipelotrichaceae family in APOE4 mice that we observed previously14. This parallels the association of the Erysipelotrichia class, Erysipelotrichales order and Erysipelotrichaceae family with the APOE4 minor allele rs429358 in human GWAS data. Our current study also extends this finding from the Erysipelotrichia phylogenetic order to its major genera, i.e., Turicibacter and Dubosiella. However, Tran et al. reported Erysipelotrichaceae were significantly increased in APOE3 compared to APOE4 mice. Erysipelotrichaceae has been shown to increase in animals fed a high-fat diet and to decrease in patients on a low-fat diet64,65. Hence, diet variation between the mice in our study and those of Tran et al. may account for the Erysipelotrichaceae difference, noting that our Teklad Global 18% (2018) chow has a fat content that accounts for 18% of total calories, whereas the RPM3, Special Diet Services chow used in the Tran et al. study has a fat content that accounts for 12% of total calories15. Since APOE genetics have been associated with BMI and obesity1, there may be a complex interplay between diet, APOE genotype and relative abundance of Erysipelotrichaceae in the gut.
Although the cladograms depict these results as phylogenetic trees, and the relatedness of taxa could imply functional relatedness, a limitation of this study is that findings were not extended to a functionality analysis. A future extension of these findings would be an in-depth analysis of the functional phenotypes of the significantly associated taxa. Future metabolomic studies would provide information regarding differences in the gut as a function of APOE genetics.
Conclusions
In this study in which mice were maintained with optimized conditions for microbiome analysis, we report a significant association between APOE status and gut microbiome profiles in 3-month male mice that reproduces at 5- and 7-months of age. The Clostridia class, Clostridiales order, its related family Ruminococcaceae, as well as related genera Ruminoclostridium, and Acetifactor and Lachnoclostridium of the Lachnopsiraceae family, increase with APOE2, which may reflect an increase in resistant starch metabolism with APOE2, and a possible impact on SCFA levels. The Erysipelotrichia class, Erysipelotrichiales order, Erysipelotrichaceae family, and Turicibacter and Dubosiella genera increase with APOE4. The findings with the Erysipelotrichia phylogenetic branch appear to extend to humans. Both the dominant model representation and the co-dominant model representation of the data showed stepwise APOE effects on the gut microbiome, and the comparison of the two provides a more robust understanding of the allelic effects. Understanding the effects of APOE genetics on the gut microbiome may provide novel approaches to counter deleterious APOE genetic effects on human disease.
Supplementary Information
Acknowledgements
The authors appreciate helpful discussion by Drs. Jamie Sturgill and Sarah D’Orazio.
Author contributions
D.J.Z. purified DNA, analyzed sequencing results, generated figures and contributed to writing the manuscript. S.J.G. contributed to data analysis and writing the manuscript, L.A.J. optimized the microbiome study design, and S.E. contributed to data analysis and writing the manuscript.
Funding
The authors acknowledge funding support from the NIH (R56-AG057589 and T32-GM118292).
Data availability
Raw sequence data (FASTQ files) were deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA), under the BioProject identifier PRJNA787634.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-05763-1.
References
- 1.Martinez-Martinez AB, Torres-Perez E, Devanney N, Del Moral R, Johnson LA, Arbones-Mainar JM. Beyond the CNS: The many peripheral roles of APOE. Neurobiol. Dis. 2020;138:104809. doi: 10.1016/j.nbd.2020.104809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu CC, Kanekiyo T, Xu H, Bu G. Apolipoprotein E and Alzheimer disease: Risk, mechanisms and therapy. Nat. Rev. Neurol. 2013;9(2):106–118. doi: 10.1038/nrneurol.2012.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rodriguez GA, Tai LM, LaDu MJ, Rebeck GW. Human APOE4 increases microglia reactivity at Abeta plaques in a mouse model of Abeta deposition. J. Neuroinflamm. 2014;11:111. doi: 10.1186/1742-2094-11-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Conejero-Goldberg C, Gomar JJ, Bobes-Bascaran T, Hyde TM, Kleinman JE, Herman MM, et al. APOE2 enhances neuroprotection against Alzheimer's disease through multiple molecular mechanisms. Mol. Psychiatry. 2014;19(11):1243–1250. doi: 10.1038/mp.2013.194. [DOI] [PubMed] [Google Scholar]
- 5.Tai LM, Mehra S, Shete V, Estus S, Rebeck GW, Bu G, et al. Soluble apoE/Abeta complex: Mechanism and therapeutic target for APOE4-induced AD risk. Mol. Neurodegener. 2014;9(1):2. doi: 10.1186/1750-1326-9-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ophir G, Meilin S, Efrati M, Chapman J, Karussis D, Roses A, et al. Human apoE3 but not apoE4 rescues impaired astrocyte activation in apoE null mice. Neurobiol. Dis. 2003;12(1):56–64. doi: 10.1016/s0969-9961(02)00005-0. [DOI] [PubMed] [Google Scholar]
- 7.Zhu Y, Nwabuisi-Heath E, Dumanis SB, Tai LM, Yu C, Rebeck GW, et al. APOE genotype alters glial activation and loss of synaptic markers in mice. Glia. 2012;60(4):559–569. doi: 10.1002/glia.22289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim J, Jiang H, Park S, Eltorai AE, Stewart FR, Yoon H, et al. Haploinsufficiency of human APOE reduces amyloid deposition in a mouse model of amyloid-beta amyloidosis. J. Neurosci. 2011;31(49):18007–18012. doi: 10.1523/JNEUROSCI.3773-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saita D, Ferrarese R, Foglieni C, Esposito A, Canu T, Perani L, et al. Adaptive immunity against gut microbiota enhances apoE-mediated immune regulation and reduces atherosclerosis and western-diet-related inflammation. Sci. Rep. 2016;6:29353. doi: 10.1038/srep29353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Azevedo OG, Bolick DT, Roche JK, Pinkerton RF, Lima AA, Vitek MP, et al. Apolipoprotein E plays a key role against cryptosporidial infection in transgenic undernourished mice. PLoS ONE. 2014;9(2):e89562. doi: 10.1371/journal.pone.0089562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oria RB, Patrick PD, Blackman JA, Lima AA, Guerrant RL. Role of apolipoprotein E4 in protecting children against early childhood diarrhea outcomes and implications for later development. Med. Hypotheses. 2007;68(5):1099–1107. doi: 10.1016/j.mehy.2006.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oria RB, Patrick PD, Oria MO, Lorntz B, Thompson MR, Azevedo OG, et al. ApoE polymorphisms and diarrheal outcomes in Brazilian shanty town children. Braz. J. Med. Biol. Res. 2010;43(3):249–256. doi: 10.1590/s0100-879x2010007500003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oria RB, Patrick PD, Zhang H, Lorntz B, de Castro Costa CM, Brito GA, et al. APOE4 protects the cognitive development in children with heavy diarrhea burdens in Northeast Brazil. Pediatr. Res. 2005;57(2):310–316. doi: 10.1203/01.PDR.0000148719.82468.CA. [DOI] [PubMed] [Google Scholar]
- 14.Parikh IJ, Estus JL, Zajac DJ, Malik M, Maldonado Weng J, Tai LM, et al. Murine gut microbiome association with APOE alleles. Front. Immunol. 2020;11:200. doi: 10.3389/fimmu.2020.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tran TTT, Corsini S, Kellingray L, Hegarty C, Le Gall G, Narbad A, et al. APOE genotype influences the gut microbiome structure and function in humans and mice: relevance for Alzheimer's disease pathophysiology. FASEB J. 2019;33(7):8221–8231. doi: 10.1096/fj.201900071R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maldonado Weng J, Parikh I, Naqib A, York J, Green SJ, Estus S, et al. Synergistic effects of APOE and sex on the gut microbiome of young EFAD transgenic mice. Mol. Neurodegener. 2019;14(1):47. doi: 10.1186/s13024-019-0352-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Minter MR, Zhang C, Leone V, Ringus DL, Zhang X, Oyler-Castrillo P, et al. Antibiotic-induced perturbations in gut microbial diversity influences neuro-inflammation and amyloidosis in a murine model of Alzheimer's disease. Sci. Rep. 2016;6:30028. doi: 10.1038/srep30028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harach T, Marungruang N, Duthilleul N, Cheatham V, Mc Coy KD, Frisoni G, et al. Reduction of Abeta amyloid pathology in APPPS1 transgenic mice in the absence of gut microbiota. Sci. Rep. 2017;7:41802. doi: 10.1038/srep41802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bonfili L, Cecarini V, Berardi S, Scarpona S, Suchodolski JS, Nasuti C, et al. Microbiota modulation counteracts Alzheimer's disease progression influencing neuronal proteolysis and gut hormones plasma levels. Sci. Rep. 2017;7(1):2426. doi: 10.1038/s41598-017-02587-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Minter MR, Hinterleitner R, Meisel M, Zhang C, Leone V, Zhang X, et al. Antibiotic-induced perturbations in microbial diversity during post-natal development alters amyloid pathology in an aged APPSWE/PS1DeltaE9 murine model of Alzheimer's disease. Sci. Rep. 2017;7(1):10411. doi: 10.1038/s41598-017-11047-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vitek MP, Brown CM, Colton CA. APOE genotype-specific differences in the innate immune response. Neurobiol. Aging. 2009;30(9):1350–1360. doi: 10.1016/j.neurobiolaging.2007.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gale SC, Gao L, Mikacenic C, Coyle SM, Rafaels N, Murray Dudenkov T, et al. APOepsilon4 is associated with enhanced in vivo innate immune responses in human subjects. J. Allergy Clin. Immunol. 2014;134(1):127–134. doi: 10.1016/j.jaci.2014.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sullivan PM, Mezdour H, Aratani Y, Knouff C, Najib J, Reddick RL, et al. Targeted replacement of the mouse apolipoprotein E gene with the common human APOE3 allele enhances diet-induced hypercholesterolemia and atherosclerosis. J. Biol. Chem. 1997;272(29):17972–17980. doi: 10.1074/jbc.272.29.17972. [DOI] [PubMed] [Google Scholar]
- 24.Strattan LE, Britsch DRS, Calulot CM, Maggard RSJ, Abner EL, Johnson LA, et al. Novel influences of sex and apoe genotype on spinal plasticity and recovery of function after spinal cord injury. eNeuro. 2021 doi: 10.1523/ENEURO.0464-20.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bo TB, Zhang XY, Kohl KD, Wen J, Tian SJ, Wang DH. Coprophagy prevention alters microbiome, metabolism, neurochemistry, and cognitive behavior in a small mammal. ISME J. 2020;14(10):2625–2645. doi: 10.1038/s41396-020-0711-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Naqib A, Poggi S, Wang W, Hyde M, Kunstman K, Green SJ. Making and Sequencing Heavily Multiplexed, High-Throughput 16S Ribosomal RNA Gene Amplicon Libraries Using a Flexible, Two-Stage PCR Protocol. Gene Expression Analysis. Humana Press; 2018. pp. 149–169. [DOI] [PubMed] [Google Scholar]
- 27.Zhang J, Kobert K, Flouri T, Stamatakis A. PEAR: A fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics. 2014;30(5):614–620. doi: 10.1093/bioinformatics/btt593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Silva GG, Green KT, Dutilh BE, Edwards RA. SUPER-FOCUS: A tool for agile functional analysis of shotgun metagenomic data. Bioinformatics. 2016;32(3):354–361. doi: 10.1093/bioinformatics/btv584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 30.Glockner FO, Yilmaz P, Quast C, Gerken J, Beccati A, Ciuprina A, et al. 25 years of serving the community with ribosomal RNA gene reference databases and tools. J. Biotechnol. 2017;261:169–176. doi: 10.1016/j.jbiotec.2017.06.1198. [DOI] [PubMed] [Google Scholar]
- 31.Dhariwal A, Chong J, Habib S, King IL, Agellon LB, Xia J. MicrobiomeAnalyst: A web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res. 2017;45:W180–W188. doi: 10.1093/nar/gkx295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu H, Tucker HM, Grear KE, Simpson JF, Manning AK, Cupples LA, et al. A common polymorphism decreases low-density lipoprotein receptor exon 12 splicing efficiency and associates with increased cholesterol. Hum. Mol. Genet. 2007;16(14):1765–1772. doi: 10.1093/hmg/ddm124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tao Q, Ang TFA, DeCarli C, Auerbach SH, Devine S, Stein TD, et al. Association of chronic low-grade inflammation with risk of Alzheimer disease in ApoE4 carriers. JAMA Netw. Open. 2018;1(6):e183597. doi: 10.1001/jamanetworkopen.2018.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12(6):R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hammer Ø, Harper DAT, Ryan PD. Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001;4(1):9. [Google Scholar]
- 36.Oksanen, J., Blanchet, F., Kindt, R., Legendre, P. & O'Hara, R. B. vegan: Community ecology package. R Package Version. 1:17–10 (2011).
- 37.Hubálek Z. Measures of species diversity in ecology: An evaluation. Folia Zool. 2000;49(4):241–260. [Google Scholar]
- 38.McMurdie PJ, Holmes S. Waste not, want not: Why rarefying microbiome data is inadmissible. PLoS Comput. Biol. 2014;10(4):e1003531. doi: 10.1371/journal.pcbi.1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40(10):4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McMurdie PJ, Holmes S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE. 2013;8(4):e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kurilshikov A, Medina-Gomez C, Bacigalupe R, Radjabzadeh D, Wang J, Demirkan A, et al. Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat. Genet. 2021;53(2):156–165. doi: 10.1038/s41588-020-00763-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jansen IE, Savage JE, Watanabe K, Bryois J, Williams DM, Steinberg S, et al. Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer's disease risk. Nat. Genet. 2019;53:156. doi: 10.1038/s41588-018-0311-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rinninella E, Raoul P, Cintoni M, Franceschi F, Miggiano GAD, Gasbarrini A, et al. What is the healthy gut microbiota composition? A changing ecosystem across age, environment, diet, and diseases. Microorganisms. 2019;7(1):14. doi: 10.3390/microorganisms7010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vasunilashorn S, Finch CE, Crimmins EM, Vikman SA, Stieglitz J, Gurven M, et al. Inflammatory gene variants in the Tsimane, an indigenous Bolivian population with a high infectious load. Biodemogr. Soc. Biol. 2011;57(1):33–52. doi: 10.1080/19485565.2011.564475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tan J, McKenzie C, Potamitis M, Thorburn AN, Mackay CR, Macia L. The role of short-chain fatty acids in health and disease. Adv. Immunol. 2014;121:91–119. doi: 10.1016/B978-0-12-800100-4.00003-9. [DOI] [PubMed] [Google Scholar]
- 47.Dalile B, Van Oudenhove L, Vervliet B, Verbeke K. The role of short-chain fatty acids in microbiota-gut-brain communication. Nat. Rev. Gastroenterol. Hepatol. 2019;16(8):461–478. doi: 10.1038/s41575-019-0157-3. [DOI] [PubMed] [Google Scholar]
- 48.Erny D, de Angelis ALH, Jaitin D, Wieghofer P, Staszewski O, David E, et al. Host microbiota constantly control maturation and function of microglia in the CNS. Nat. Neurosci. 2015;18(7):965–977. doi: 10.1038/nn.4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang P, Zhang Y, Gong Y, Yang R, Chen Z, Hu W, et al. Sodium butyrate triggers a functional elongation of microglial process via Akt-small RhoGTPase activation and HDACs inhibition. Neurobiol. Dis. 2018;111:12–25. doi: 10.1016/j.nbd.2017.12.006. [DOI] [PubMed] [Google Scholar]
- 50.Patnala R, Arumugam TV, Gupta N, Dheen ST. HDAC inhibitor sodium butyrate-mediated epigenetic regulation enhances neuroprotective function of microglia during ischemic stroke. Mol. Neurobiol. 2017;54(8):6391–6411. doi: 10.1007/s12035-016-0149-z. [DOI] [PubMed] [Google Scholar]
- 51.Yamawaki Y, Yoshioka N, Nozaki K, Ito H, Oda K, Harada K, et al. Sodium butyrate abolishes lipopolysaccharide-induced depression-like behaviors and hippocampal microglial activation in mice. Brain Res. 2018;1680:13–38. doi: 10.1016/j.brainres.2017.12.004. [DOI] [PubMed] [Google Scholar]
- 52.Soliman ML, Puig KL, Combs CK, Rosenberger TA. Acetate reduces microglia inflammatory signaling in vitro. J. Neurochem. 2012;123(4):555–567. doi: 10.1111/j.1471-4159.2012.07955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang Y, Cella M, Mallinson K, Ulrich JD, Young KL, Robinette ML, et al. TREM2 lipid sensing sustains the microglial response in an Alzheimer's disease model. Cell. 2015;160(6):1061–1071. doi: 10.1016/j.cell.2015.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jonsson T, Stefansson H, Steinberg S, Jonsdottir I, Jonsson PV, Snaedal J, et al. Variant of TREM2 associated with the risk of Alzheimer's disease. N. Engl. J. Med. 2013;368(2):107–116. doi: 10.1056/NEJMoa1211103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jonsson T, Stefansson H, Ph DS, Jonsdottir I, Jonsson PV, Snaedal J, et al. Variant of TREM2 associated with the risk of Alzheimer's disease. N. Engl. J. Med. 2012;368:107. doi: 10.1056/NEJMoa1211103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guerreiro R, Wojtas A, Bras J, Carrasquillo M, Rogaeva E, Majounie E, et al. TREM2 variants in Alzheimer's disease. N. Engl. J. Med. 2012;368:117. doi: 10.1056/NEJMoa1211851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sutherland MK, Yu C, Lewis TS, Miyamoto JB, Morris-Tilden CA, Jonas M, et al. Anti-leukemic activity of lintuzumab (SGN-33) in preclinical models of acute myeloid leukemia. MAbs. 2009;1(5):481–490. doi: 10.4161/mabs.1.5.9288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bradshaw EM, Chibnik LB, Keenan BT, Ottoboni L, Raj T, Tang A, et al. CD33 Alzheimer's disease locus: Altered monocyte function and amyloid biology. Nat. Neurosci. 2013;16(7):848–850. doi: 10.1038/nn.3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Griciuc A, Serrano-Pozo A, Parrado AR, Lesinski AN, Asselin CN, Mullin K, et al. Alzheimer's disease risk gene CD33 inhibits microglial uptake of amyloid beta. Neuron. 2013;78(4):631–643. doi: 10.1016/j.neuron.2013.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Malik M, Simpson JF, Parikh I, Wilfred BR, Fardo DW, Nelson PT, et al. CD33 Alzheimer's risk-altering polymorphism, CD33 expression, and exon 2 splicing. J. Neurosci. 2013;33(33):13320–13325. doi: 10.1523/JNEUROSCI.1224-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang KL, Marcora E, Pimenova AA, Di Narzo AF, Kapoor M, Jin SC, et al. A common haplotype lowers PU.1 expression in myeloid cells and delays onset of Alzheimer's disease. Nat. Neurosci. 2017;20:1052. doi: 10.1038/nn.4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Malik M, Parikh I, Vasquez JB, Smith C, Tai L, Bu G, et al. Genetics ignite focus on microglial inflammation in Alzheimer's disease. Mol. Neurodegener. 2015;10:52. doi: 10.1186/s13024-015-0048-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Efthymiou AG, Goate AM. Late onset Alzheimer's disease genetics implicates microglial pathways in disease risk. Mol. Neurodegener. 2017;12(1):43. doi: 10.1186/s13024-017-0184-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hildebrandt MA, et al. High-fat diet determines the composition of the murine gut microbiome independently of obesity. Gastroenterology. 2009;137:1716–1724. doi: 10.1053/j.gastro.2009.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Turnbaugh PJ, Backhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 2008;3(4):213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequence data (FASTQ files) were deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA), under the BioProject identifier PRJNA787634.