Abstract
Duck circovirus disease (DuCVD), as an immunosuppressive disease, is a threat to the poultry industry. In order to diagnose this disease quickly and accurately, a real-time fluorescence-based recombinase-aided amplification (RF-RAA) method was established to detect duck circovirus (DuCV). The results showed that the quantity of amplification products was positively correlated with the value of fluorescence signal. Obvious detection results can be observed at 41°C after 15 min reaction. This method has good specificity and has no cross reaction with Muscovy duck parvovirus (MDPV), duck enteritis virus (DEV), fowl adenovirus (FAdV), porcine circovirus (PCV), and duck hepatitis A virus (DHAV). The sensitivity test showed that the minimum concentration of template detected by RF-RAA for DuCV was 10° copies/μL, and its sensitivity was 10 times higher than that of real-time fluorescence-based quantitative PCR (RFQ-PCR) and 10,000 times higher than that of polymerase chain reaction (PCR). Fifty-two clinical samples were detected by RF-RAA and RFQ-PCR, and the coincidence rate of the two methods was 98.08%. This method has the advantages of simple operation, good specificity and high sensitivity, and can be used for laboratory detection and clinical diagnosis of DuCV.
Key words: duck circovirus, recombinase-aided amplification, rapid detection
INTRODUCTION
Duck circovirus disease (DuCVD) is an infectious disease caused by duck circovirus (DuCV), and ducks of all ages and breeds are prone to infection by DuCV (Li et al., 2018). DuCV was first discovered in 2003 (Hattermann et al., 2003), and then cases of infection have been reported in many countries around the world. DuCV can damage the immune system of ducks (Li et al., 2020), cause immunosuppression (Liu et al., 2020), and lead to secondary infection or multiple infections (Yang et al., 2018). The diseased ducks showed loss of feather, loss of appetite, and developmental retardation. In severe cases, even shortness of breath, anemia, decreased production performance, and even death occur. The symptoms of DuCVD are similar to those of some bacterial, fungal infections and some nutritional diseases (Jiang et al., 2021). It is difficult to make an accurate diagnosis of DuCVD in clinic, so it is necessary to make laboratory differential diagnosis. In this study, a rapid and accurate RF-RAA method for the detection of DuCV was established, which is of positive significance for the prevention and control of DuCVD.
MATERIALS AND METHODS
Viral Nucleic Acid Extraction
The DNA of DuCV, Muscovy duck parvovirus (MDPV), duck enteritis virus (DEV), fowl adenovirus (FAdV), porcine circovirus (PCV), and duck hepatitis A virus (DHAV) (all the viruses were tested, isolated, identified, and preserved by our laboratory) were extracted according to the instructions of DNA/RNA nucleic acid extraction kit (Tiangen Biotech Co., Ltd., Beijing, China). The extracted RNA was reversely transcribed into cDNA.
Primer Screening and Probe Design for Detecting DuCV by RF-RAA
NCBI was used to find whole genome sequences of DuCV, and 3 pairs of specific primers were designed according to the principle of RAA primer design (Li et al., 2019) for the conserved region (rep gene). DuCV DNA was used as template to prepare 50 μL reaction: VI buffer (25 μL); purified water (17.1 μL), upstream primer, (10 μM, 2.1 μL), downstream primer (10 μM, 2.1 μL), template (1.2 μL), and magnesium acetate (2.5 μL). After reacting at 38°C for 30 min, the product was purified and the result was observed by 2% agarose gel electrophoresis. A pair of optimal primer combinations was selected, and a probe was designed between upstream and downstream primers. The primers and probes were synthesized and modified by a biochemical company (Sangon Biotech Co., Ltd., Shanghai, China) (Table 1).
Table 1.
Primers selected for DuCV reaction by RF-RAA.
| Primers | Sequence (5′-3′) | Gene localization |
|---|---|---|
| DuCV-1-F | CCTCTGATCTGGCCGAAGCGA CATCCGCTGT | 377-407 |
| DuCV-2-F | CTGTGATGGCTGGCGTCCCGC TGACTGAGGTG | 404-435 |
| DuCV-3-F | CCGCTGACTGAGGTGGCCCGG AAGTTCCCC | 421-450 |
| DuCV-1-R | TGCCGGGAGGACCAATCAG AACGATGACTT | 530-557 |
| DuCV-2-R | AATTCAAATGCATAACGGC TCTTTCCGGTG | 558-587 |
| DuCV-3-R | CCGCGTGGTTTGTAATACTTGT TTTCGGCG | 591-620 |
Abbreviations: DuCV, duck circovirus; RF-RAA, real-time fluorescence-based recombinase-aided amplification.
Establishment of a RF-RAA Assay for Detecting DuCV
According to the instructions of RAA nucleic acid amplification kit (fluorescence method) (Qitian Gene Biological Technology Co., Ltd. Jiangsu. China), a RF-RAA reaction system (50 μL) was constructed: VI buffer (25 μL), forward primers (10 μM, 2.1 μL), reverse primers (10 μM, 2.1 μL), template (DuCV DNA) (1.2 μL), probe (0.6 μL), the final volume was made up to 47.5 μL with purified water (16.5 μL). The mixture was pipetted into the reaction unit, and 2.5 μL of magnesium acetate was added to the mixture. After blending in the thermostatic amplifier, it was immediately taken out and placed in the thermostatic nucleic acid amplification/detection device (Qitian Gene Biological Technology Co., Ltd. Jiangsu. China) for reaction for 25 min at 37°C, and slope ≥20 indicated a positive result.
Selection of Reaction Temperature and Time for Detecting DuCV by RF-RAA
When other conditions remained unchanged, only the reaction temperature (37, 38, 39, 40, 41, 42°C) was changed for test, and the best temperature was selected. Judging criterion for the best reaction time: a strong fluorescence signal can be observed in a short time.
Specificity Test of RF-RAA in Detecting DuCV
DuCV, MDPV, DEV, FAdV, and DHAV of susceptible ducks with similar clinical symptoms (depression, reduced feeding, and retardation) and PCV with the same circular structure were selected as templates. The same 50-μL reaction system as above was prepared according to the kit instructions. The reaction tube was placed in the instrument and RF-RAA was tested at the optimum temperature to verify the specificity of the reaction.
Sensitivity Test of RF-RAA in Detecting DuCV
Preparation of standard plasmids: using the conserved region of DuCV (rep gene) as a template, a pair of 18 to 27 bp PCR primers (F: ATCCTCTGATCTGGCCGAAG, R: AATTCAAATGCATAACGGCTCTTTC) were designed and synthesized near the probe. Reaction system: 2 × Taq Mix (25 μL) (Vazyme Biotech Co., Ltd., Nanjing, China), forward primers (10 μM, 1 μL), reverse primers (10 μM, 1 μL), template DNA (3 μL), the final volume was made up to 50 μL with purified water. Reaction conditions: pre-denatured at 94°C for 5 min; denatured at 94 °C for 30 s; annealed at 50°C for 30 s; extended at 72°C for 30 s, a total of 34 cycles; extended at 72°C for 5 min.
After purification, the product was ligated to pMD20-T vector (Takara Biomedical Technology Co., Ltd. Beijing. China) and introduced into DH5α competent cells. Positive single colonies were selected by blue and white screening, and plasmids were extracted after overnight culture, and then sent to a biochemical company (Sangon Biotech Co., Ltd.) for sequencing and verification. The copy number of DNA per unit volume of positive plasmids was calculated.
Plasmid copy number (copies/L) = [plasmid concentration (g.μL−1) × 6.02 × 1023]/[total fragment length (bp) × 660 g/mol].
Total fragment length = vector length (bp)+target fragment length (bp).
The plasmid concentration diluted to 109-10° copies/μL was used to analyze the sensitivities of RF-RAA, PCR, and RFQ-PCR.
RF-RAA sensitivity test: 1.0 μL of plasmid with a concentration of 1 × 104-1 × 10° copies/μL was used as the template, and the reaction was carried out according to the optimized conditions.
PCR sensitivity test: 2 × Taq Mix (12.5 μL) (Vazyme Biotech Co., Ltd), forward primers and reverse primers (10 μM, 0.5 μL) (primers same as above for standard plasmid preparation), template (plasmid with a concentration of 1 × 107-1 × 10° copies/μL) (1.0 μL), the final volume was made up to 25 μL with purified water. The reaction conditions were the same as those in the preparation of standard plasmids. The amplification products were observed by electrophoresis in 2% agarose gels.
RFQ-PCR sensitivity test: TB Green Premix DimerEraser (2×) (12.5 μL) (Takara Biomedical Technology Co., Ltd.), forward primers and reverse primers (10 μM, 0.5 μL) (primers ibid), template (plasmid with a concentration of 1 × 106-1 × 10° copies/μL) (1.0 μL), the final volume was made up to 25 μL with purified water. Reaction conditions: pre-denatured at 95°C for 30 s; denatured at 95°C for 5 s; annealed at 55°C for 30 s; extended at 72°C for 30 s, a total of 40 cycles.
The PCR and RFQ-PCR methods used in this experiment have been verified by our laboratory for many times.
Robustness
The plasmid samples at different concentrations (104, 102, and 10° copies/μL) were selected for replication, and the experiment for each concentration was repeated for 5 times. According to the judging criterion of RAA-nucleic acid amplification kit (fluorescence method) (when the slope k ≥20, the result was positive), the time taken for the fluorescence curve to reach the criterion for a positive result was recorded, and the coefficient of variation (CV) within the group was calculated; at the same time, the above tests were carried out every 7 d for 3 consecutive times, and the CV between groups was calculated to evaluate the repeatability and stability of this method.
Clinical Sample Detection
RF-RAA and RFQ-PCR were used to detect the bursa of Fabricius in 52 ducks clinically diagnosed with DuCVD (samples from 5 duck farms in northern China, with clinical manifestations of shortness of breath, retarded growth, and messy feathers), and the viral nucleic acid was extracted. The specific operation was carried out according to the instructions of the viral DNA/RNA nucleic acid extraction kit. The detection results of RF-RAA and RFQ-PCR were compared, and the coincidence rate of the two methods was calculated.
RESULTS
Primer Screening and Probe Design for Detecting DuCV by RF-RAA
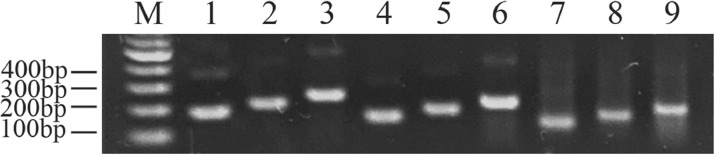
As can be seen from the figure, reaction product no.8 had only one highlighted band with correct fragment size (Figure 1). The corresponding combination was selected as the best primer and a probe is designed in it. DuCV-T (458–507 bp): ATGTTCTTTGGGCGTGGCCTGGAACGCC/i6FAMdT//idSp/CG/iBHQ1dT/CACCTGATCGTTGAG.
Figure 1.
Primer screening for detecting DuCV by RF-RAA. M: marker. 1: DuCV-1-F and DuCV-1-R are combined. 2: DuCV-1-F and DuCV-2-R are combined. 3: DuCV-1-F and DuCV-3-R are combined. 4: DuCV-2-F and DuCV-1-R are combined. 5: DuCV-2-F and DuCV-2-R are combined. 6: DuCV-2-F and DuCV-3-R are combined. 7: DuCV-3-F and DuCV-1-R are combined. 8: DuCV-3-F and DuCV-2-R are combined. 9: DuCV-3-F and DuCV-3-R are combined. Abbreviations: DuCV, duck circovirus; RF-RAA, real-time fluorescence-based recombinase-aided amplification.
Selection of Reaction Temperature and Time for Detecting DuCV by RF-RAA
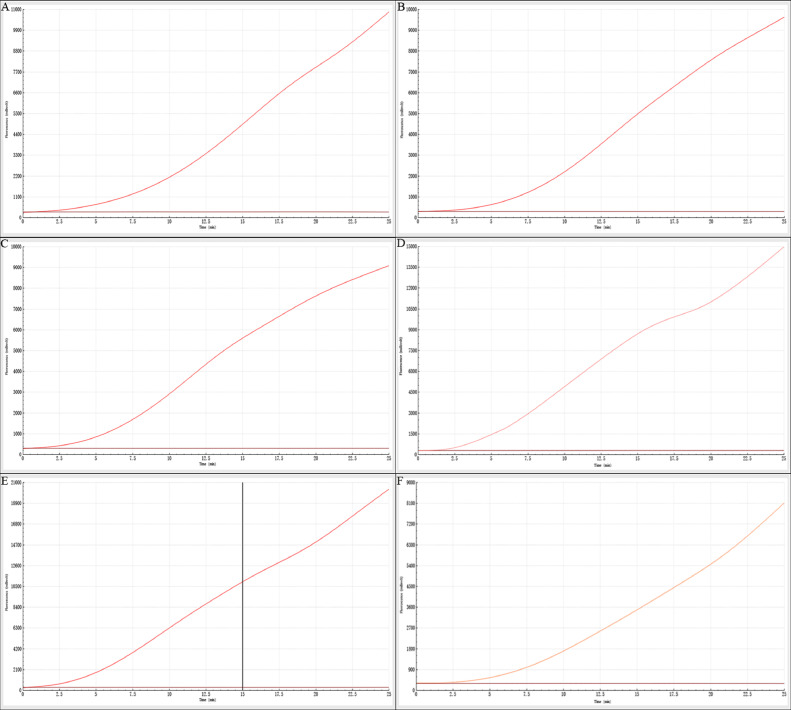
The fluorescence curve peaked at 41°C, and the value of fluorescence signal increased gradually with the reaction, but the growth rate decreased in the later stage of reaction. At 15 min, the value of fluorescence signal was high and the reaction time was short (Figure 2E). Therefore, 41°C and 15 min were chosen as the best reaction conditions for RF-RAA.
Figure 2.
Selection of reaction temperature and time for detecting DuCV by RF-RAA. (A–F) shows the fluorescence curves under the reaction conditions of 25 min, 37–42°C. According to the peak value and growth rate of fluorescence curve, 41°C and 15 min were determined as the best reaction conditions. Abbreviations: DuCV, duck circovirus; RF-RAA, real-time fluorescence-based recombinase-aided amplification.
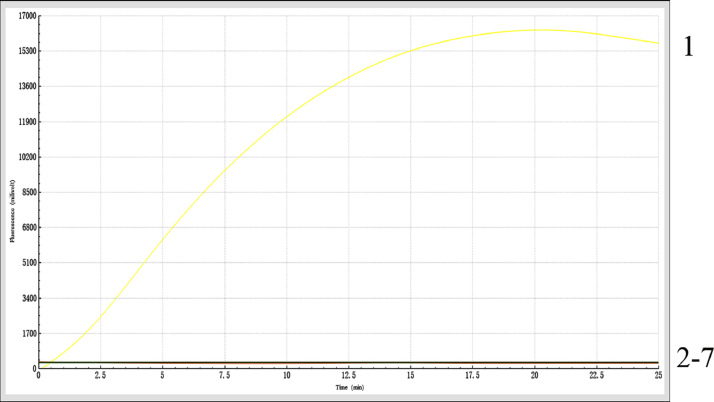
Verification of Specificity of RF-RAA in the Detection of DuCV
The results showed that there was no fluorescence curve with other viral nucleic acids as templates except DuCV (Figure 3), which proved that RF-RAA had good specificity and no cross reaction with other viral nucleic acids.
Figure 3.
Verification of Specificity of RF-RAA in the Detection of DuCV. 1: DuCV. 2: MDPV. 3: DEV. 4: FAdV. 5: DHAV. 6: PCV. 7: negative control. Fluorescence amplification curve was visible only when DuCV was used as template, indicating that RF-RAA has good specificity. Abbreviations: DuCV, duck circovirus; DEV, duck enteritis virus; DHAV, duck hepatitis A virus; FAdV, fowl adenovirus; MDPV, Muscovy duck parvovirus; PCV, porcine circovirus; RF-RAA, real-time fluorescence-based recombinase-aided amplification.
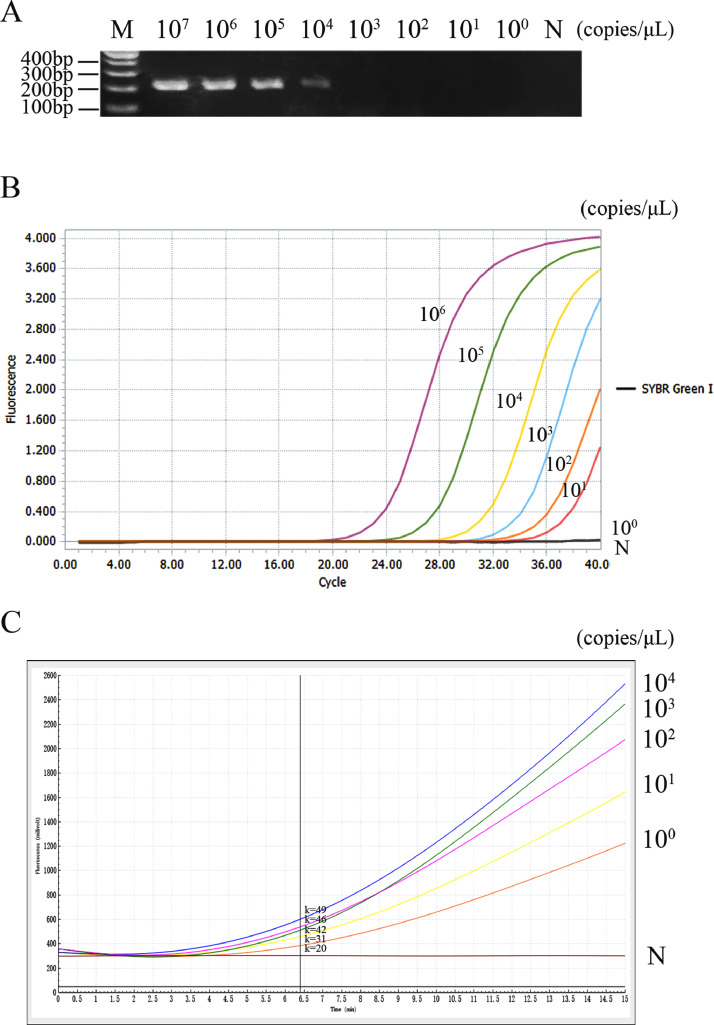
Comparison of Sensitivity of Three Methods for Detecting DuCV
The minimum concentration of template detected of DuCV was 104 copies/μL for PCR (Figure 4A), 101 copies/μL for RFQ-PCR (Figure 4B), and 10° copies/μL for RF-RAA (Figure 4C), indicating that the sensitivity of RF-RAA was 10 times higher than that of RFQ-PCR, 10,000 times higher than that of conventional PCR.
Figure 4.
Comparison of sensitivity of three methods in detecting DuCV. (A) Sensitivity of PCR in detecting DuCV. M: marker. N: negative control. (B) Sensitivity of RFQ-PCR in detecting DuCV. N: negative control. (C) Sensitivity of RF-RAA in detecting DuCV. N: negative control. K: slope. As can be seen from the figure, the minimum concentration of template detected of DuCV was 104 copies/μL for PCR, 101 copies/μL for RFQ-PCR and 10° copies/μL for RF-RAA. Abbreviations: DuCV, duck circovirus; RF-RAA, real-time fluorescence-based recombinase-aided amplification; RFQ-PCR, real-time fluorescence-based quantitative PCR.
Robustness
The results showed that the CV was 2.30 to 3.77% for intra-batch replication and 2.44 to 5.80% for inter-batch replication (both less than 10%) (Table 2), indicating that the established RF-RAA assay had good repeatability and stability.
Table 2.
RF-RAA robustness results.
| Concentration (copies/μL) | Intragroup replication |
Intergroup replication |
||
|---|---|---|---|---|
| Mean (X±SD) | CV (100%) | Mean (X±SD) | CV (100%) | |
| 104 | 2.22 ± 0.08 | 3.77 | 2.37 ± 0.06 | 2.44 |
| 102 | 2.62 ± 0.08 | 3.19 | 2.63 ± 0.15 | 5.80 |
| 100 | 4.96 ± 0.11 | 2.30 | 5.03 ± 0.21 | 4.14 |
Abbreviation: RF-RAA, real-time fluorescence-based recombinase-aided amplification.
Clinical Sample Detection
Two methods were used to detect 52 clinical samples, and 36 positive strains and 16 negative strains were detected by RFQ-PCR. Thirty seven positive strains and 15 negative strains were detected by RF-RAA. The coincidence rate of sample test results between RF-RAA and RFQ-PCR was 98.08% (Table 3).
Table 3.
Clinical test results of two methods.
| Methods | Positive cases | Negative cases | Positive coincidence rate (%) | Negative coincidence rate (%) | Total consistent rate (%) |
|---|---|---|---|---|---|
| RFQ-PCR | 36 | 16 | - | - | - |
| RF-RAA | 37 | 15 | 100 | 93.75 | 98.08 |
Abbreviations: RF-RAA, real-time fluorescence-based recombinase-aided amplification; RFQ-PCR, real-time fluorescence-based quantitative PCR.
DISCUSSION
Existing laboratory diagnostic methods for DuCV have their own advantages and disadvantages. PCR needs high temperature denaturation, low temperature annealing, and moderate temperature extension (Li et al., 2015), so it is inseparable from the use of thermocycler; RFQ-PCR is sensitive, but the instrument is expensive; there are some problems such as nonspecific reaction in the detection of antibodies by enzyme-linked immunosorbent assay (Yang et al., 2019); loop-mediated isothermal amplification is highly specific and sensitive, but the primer design is complex and the reaction can only be carried out at 60 to 65°C (Zhao et al., 2012).
Recombinase-aided amplification (RAA) is a new isothermal in vitro nucleic acid amplification technique that uses recombinase, single-stranded DNA-binding protein (SSBP), and DNA polymerase instead of conventional thermally stable enzymes for rapid amplification of target genes at a constant temperature in 15 to 30 min (Zheng et al., 2019). Since the advent of RAA, it has been widely used in the detection of pathogenic microorganisms, including bacteria (Zhang et al., 2019a), viruses (Wang et al., 2021), parasites (Zhang et al., 2019b), and other pathogens.
In this experiment, specific primers and probes were designed on the basis of RAA. The probe contained a tetrahydrofuran (THF) residue, the T bases on both sides were labeled with a fluorophore and a quencher, respectively, and a blocking group (C3-spacer) was connected to the 3′ end of the probe. When the probe was single-stranded, the fluorescence signal emitted by the fluorophore was absorbed by the quencher. Once the target sequence was identified to be double-stranded, the THF was cut off, the fluorophore was in a free state, and the fluorescence signal can be detected by the instrument, thus achieving a positive correlation between the accumulation of amplification products and the value of fluorescence signal.
The detection of DuCV by RF-RAA requires only 15-min reaction under the constant temperature of 41°C to complete the detection, which is significantly shorter than the PCR or RFQ-PCR methods commonly used in clinical practice. And the lowest template concentration that can be detected by RF-RAA can reach 10° copies/μL, and the sensitivity is 10 times that of RFQ-PCR and 10,000 times that of PCR. In addition, the instrument used in this method is smaller in size and lower in price, and the detection results are judged by the instrument, which is more accurate and objective. The operation steps of detecting DuCV by RF-RAA method are simpler and easier to operate, and the reaction reagent is stored in the form of freeze-dried powder, which is easier to preserve and transport than the liquid reagent, and more conducive to promotion. These advantages make RF-RAA is likely to be widely used in the detection market as an important detection method in the future.
CONCLUSIONS
In this study, specific primers and probes were designed for the conserved region (REP gene) of DuCV, and a RF-RAA method was established for the detection of DuCV. The accumulation of amplification products was positively correlated with the intensity of fluorescence signal, and obvious results can be observed by instrument under reaction conditions of 15 min, 41°C. RF-RAA is expected to be a supplement to conventional PCR and RFQ-PCR in clinical detection, and has a positive significance for epidemic prevention and control.
DISCLOSURES
The authors declare that they have no competing interests, the manuscript has not been published or submitted to other journals previously.
ACKNOWLEDGMENTS
This work was supported by Key R&D Plan of Science and Technology Department of Hebei Province (20326621D) from Science and Technology Department, Hebei Province, China, and Open Fund of State Key Laboratory of Veterinary Etiological Biology (BYSWX2021KFKT03) from Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences, China.
REFERENCES
- Hattermann K., Schmitt C., Soike D., Mankertz A. Cloning and sequencing of Duck circovirus (DuCV) Arch Virol. 2003;148:2471–2480. doi: 10.1007/s00705-003-0181-y. [DOI] [PubMed] [Google Scholar]
- Jiang S.Y., Zhang T.T., Tian Q., Zhang M.H., Zhou B.J., Cheng Z.T., Wang K.G., Wen M. Research progress of duck circovirus disease. Guizhou J. Anim. Husbandry Vet. Med. 2021;45:52–55. [Google Scholar]
- Li P., Li J., Zhang R., Chen J., Wang W., Lan J., Xie Z., Jiang S. Duck "beak atrophy and dwarfism syndrome" disease complex: interplay of novel goose parvovirus-related virus and duck circovirus? Transbound. Emerg. Dis. 2018;65:345–351. doi: 10.1111/tbed.12812. [DOI] [PubMed] [Google Scholar]
- Li X.N., Shen X.X., Li M.H., Qi J.J., Wang R.H., Duan Q.X., Zhang R.Q., Fan T., Bai X.D., Fan G.H. Applicability of duplex real time and lateral flow strip reverse-transcription recombinase aided amplification assays for the detection of Enterovirus 71 and Coxsackievirus A16. Virol. 2019;16:166–176. doi: 10.1186/s12985-019-1264-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z.G., Wang X., Zhang R.H., Chen J.H., Xia L.L., Lin S.L., Xie Z.J., Jiang S.J. Establishment and application of double PCR method for detection of two genotypes of duck circovirus. Chin. J. Vet. Med. 2015;35:1060–1068. [Google Scholar]
- Li Z.L., Fu G.H., Feng Z.H., Chen J.H., Shi S.H., Liu R.C., Cheng L.F., Chen H.M., Wan C.H., Huang Y. Evaluation of a novel inactivated vaccine against duck circovirus in muscovy ducks. Vet. Microbiol. 2020;241 doi: 10.1016/j.vetmic.2019.108574. [DOI] [PubMed] [Google Scholar]
- Liu H., Li L.X., Sun W.C., Shi N., Sun X.T., Jin N.Y., Si X.K. Molecular survey of duck circovirus infection in poultry in southern and southwestern China during 2018 and 2019. BMC Vet. Res. 2020;16:130–138. doi: 10.1186/s12917-020-02301-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.H., Li P., Lin X., Jia H., Jiang Y.T., Wang X.J., Hou S.H. Application of portable real-time recombinase-aided amplification (rt-RAA) assay in the clinical diagnosis of ASFV and prospective DIVA diagnosis. Appl. Microbiol. Biot. 2021;105:3249–3264. doi: 10.1007/s00253-021-11196-z. [DOI] [PubMed] [Google Scholar]
- Yang J., Zhang R.T., Wang Z.Z., Yu X.L., Niu X.Y., Tang Y., Diao Y.X. Epidemiological investigation on co-infection of duck viral disease in some areas of China. Chin. J. Vet. Med. 2018;38:1872–1877. [Google Scholar]
- Yang Z.X., Zhu J.B., Xiao L., Gao L., Miao H.S., He Y.W., Meng J.X., Yang H., Li H.C. Establishment of antigen-captured ELISA for detecting epizootic haemorrhagic disease virus. Chin J. Vet. Sci. 2019;39:1–7. [Google Scholar]
- Zhang Q., Ding X., Wu X.M., Liu Y.H., Liu J.F., Xu X.Z., Ying Q.J., Cao J., Dai Y. Establishment and preliminary evaluation of recombinant enzyme-mediated isothermal amplification of specific nucleic acid of Clonorchis sinensis. Chin. J. Schi. Control. 2019;31:468–473. doi: 10.16250/j.32.1374.2019178. [DOI] [PubMed] [Google Scholar]
- Zhang R.Q., Li G.X., Li X.N., Shen X.X., Gao Y., Wang L., Fan T., Duan Q.X., Wang Y.K., Wang J., Feng Z.S., Ma X.J. A rapid and sensitive recombinase aided amplification assay incorporating competitive internal control to detect Bordetella pertussis using the DNA obtained by boiling. Int. J. Infect. Dis. 2019;86:108–113. doi: 10.1016/j.ijid.2019.06.028. [DOI] [PubMed] [Google Scholar]
- Zhao G.Y., Xie Z.X., Xie L.J., Pang Y.S., Deng X.W., Liu J.B., Fan Q. Establishment of LAMP visual detection method for duck circovirus. Chin. J. Anim. Quarant. 2012;29:24–26. [Google Scholar]
- Zheng X.C., Chen Y., Wen Z.Q., Ye W.T., Zhang J.X., Huang Q.J., Liang X.Q., Liu H. Establishment of a fluorescence RAA method for rapid detection of common carp edema virus. Chin. J. Prev. Vet. Med. 2019;41:721–725. [Google Scholar]