Abstract
Spermatogonial stem cells (SSCs) are able to undergo both self-renewal and differentiation. Unlike self-renewal, which replenishes the SSC and progenitor pool, differentiation is an irreversible process committing cells to meiosis. Although the preparations for meiotic events in differentiating spermatogonia (Di-SG) are likely to be accompanied by alterations in chromatin structure, the three-dimensional chromatin architectural differences between SSCs and Di-SG, and the higher-order chromatin dynamics during spermatogonial differentiation, have not been systematically investigated. Here, we performed in situ high-throughput chromosome conformation capture, RNA-seq, and chromatin immunoprecipitation-sequencing analyses on porcine undifferentiated spermatogonia (which consist of SSCs and progenitors) and Di-SG. We identified that Di-SG exhibited less compact chromatin structural organization, weakened compartmentalization, and diminished topologically associating domains in comparison with undifferentiated spermatogonia, suggesting that diminished higher-order chromatin architecture in meiotic cells, as shown by recent reports, might be preprogrammed in Di-SG. Our data also revealed that A/B compartments, representing open or closed chromatin regions respectively, and topologically associating domains were related to dynamic gene expression during spermatogonial differentiation. Furthermore, we unraveled the contribution of promoter-enhancer interactions to premeiotic transcriptional regulation, which has not been accomplished in previous studies due to limited cell input and resolution. Together, our study uncovered the three-dimensional chromatin structure of SSCs/progenitors and Di-SG, as well as the interplay between higher-order chromatin architecture and dynamic gene expression during spermatogonial differentiation. These findings provide novel insights into the mechanisms for SSC self-renewal and differentiation and have implications for diagnosis and treatment of male sub-/infertility.
Keywords: spermatogonia, 3D chromatin, A/B compartment, topologically associating domain, promoter-enhancer interaction
Abbreviations: 3D, three-dimensional; ChIP-seq, chromatin immunoprecipitation-sequencing; DI, directional index; Di-SG, differentiating spermatogonia; DPBS, Dulbecco's phosphate-buffered saline; DSBs, double-strand breaks; FACS, fluorescence-activated cell sorting; GO, Gene Ontology; Hi-C, high-throughput chromosome conformation capture; PCA, principal component analysis; PEIs, promoter-enhancer interactions; REs, regular enhancers; RP, regulatory potential; SEs, super enhancers; SSC, Spermatogonial stem cell; TADs, topologically associating domains; TSS, transcription start site; Un-SG, undifferentiated spermatogonia
With every heartbeat a man produces over 1000 spermatozoa, each in theory capable of generating a new-born child (1). The highly efficient production of spermatozoa is reliant on spermatogenesis, an intricate process occurring in the testis during which the primitive spermatogenic cells, that is, spermatogonial stem cells (SSCs), develop into mature spermatozoa (2). Spermatogonial stem cells are the only adult stem cells in males with the ability to transmit genetic information to the next generation, and thus they have a series of desirable attributes, with some shared by other stem cell categories. First, they strike a balance between self-renewal and differentiation to preclude exhaustion while simultaneously safeguarding the ongoing production of gametes. Second, being located at a specific place in the mammalian testis called “niche”, SSCs are orchestrated by a host of intrinsic and extrinsic factors with well-defined roles in SSC fate determination and behaviors (3, 4). Third, SSCs are capable of relocating to the basement membrane and reestablishing donor-derived spermatogenesis after transplantation into the allogenic recipient testis, being an appealing target for treatment of male infertility (5, 6).
Despite the crucial roles of SSCs in maintenance of male fertility, distinct models regarding the SSC property and cellular hierarchy have been proposed. Specifically, although traditional models, which are principally based on histological observations, propose that only the most primitive undifferentiated spermatogonia (Un-SG), that is, single spermatogonia (As), have stem cell characteristics (3), most data from later studies are more in favor of a dynamic stem cell model illustrating context-dependent and plastic stemness (4, 7). Nonetheless, it has generally been accepted that stem cell potential is typically limited to a rare subpopulation of Un-SG. Intriguingly, the recent boom of studies using single-cell RNA-seq methodology have uncovered the remarkable heterogeneity of SSCs (8, 9, 10), and with other omics approaches, the transcriptome, metabolome, DNA methylome, histone modification profiles, and chromatin accessibility of SSCs have been revealed (11, 12, 13, 14, 15, 16, 17, 18). Despite all these, the molecular mechanisms for SSC maintenance and development remain incompletely understood.
Spermatogonial stem cells are able to undergo both self-renewal and differentiation. Unlike the self-renewal that replenishes the SSC and progenitor pool, the differentiation is an irreversible process committed to meiosis, which is stringently modulated by the stages of the seminiferous epithelium in the testis (19, 20). Typically, when they start to differentiate, SSCs and progenitors are gradually preparing their genome to later undergo a series of events in meiosis, such as initiation of double-strand breaks (DSBs), alignment, pairing, and synapsis of homologous chromosomes, homologous recombination, and formation of crossovers (2). It has thus been assumed that the preparations for these meiotic events in differentiating spermatogonia (Di-SG) are accompanied by dramatic alterations in chromatin structure. Traditional histological studies have revealed that Di-SG are equipped with increasing amount of condensed chromatin, namely heterochromatin, that rims the nucleus (21, 22). Despite this, the three-dimensional (3D) chromatin architecture of SSCs and Di-SG, and the higher-order chromatin dynamics during spermatogonial differentiation, have not been systematically studied.
The recently developed high-throughput chromosome conformation capture (Hi-C) technique enables detection and visualization of the dynamic chromatin, providing desirable means to study the higher-order chromatin architecture and the key principles of genome packaging at the molecular level (23, 24). The higher-order chromatin can be spatially packaged into a hierarchy of the 3D genome, including A/B compartments, topologically associating domains (TADs), and chromatin loops (25, 26), further influencing numerous DNA-related biological processes such as transcription, DNA replication and repair, mitotic and meiotic cell cycle progress, etc. (27, 28). Here, by using in situ Hi-C, RNA-seq and chromatin immunoprecipitation-sequencing (ChIP-seq), we systematically investigated the 3D chromatin architecture of Un-SG (which consist of SSCs and progenitors) and Di-SG, with an aim to unravel the higher-order chromatin dynamics during spermatogonial differentiation, as well as the regulation in gene transcription. We performed the studies on pigs, because pigs are an increasingly prevalent animal model in fundamental and translational research due to the resemblance to humans concerning anatomy, physiology, genetics, and reproductive maturation (29, 30). Moreover, it is feasible to obtain a vast number of spermatogonial subpopulations from porcine testes with large size for subsequent advanced bioinformatic analyses. We gained novel insights into the changing chromatin dynamics during spermatogonial differentiation that have so far not been reported. For instance, our data suggest that the diminished higher-order chromatin architecture in meiotic cells, as shown by recent reports, might be preprogramed in Di-SG. Furthermore, we unraveled the contribution of promoter-enhancer interactions (PEIs) to premeiotic transcriptional regulation, which has not been accomplished in previous studies due to limited cell input and resolution. Together, our study uncovered the 3D chromatin structure of SSCs/progenitors and Di-SG, as well as the interplay between higher-order chromatin architecture and dynamic gene expression during spermatogonial differentiation, which is expected to better the biological understanding of SSC self-renewal and differentiation and have implications for diagnosis and treatment of male sub-/infertility.
Results
Dynamic 3D chromatin architecture during spermatogonial differentiation
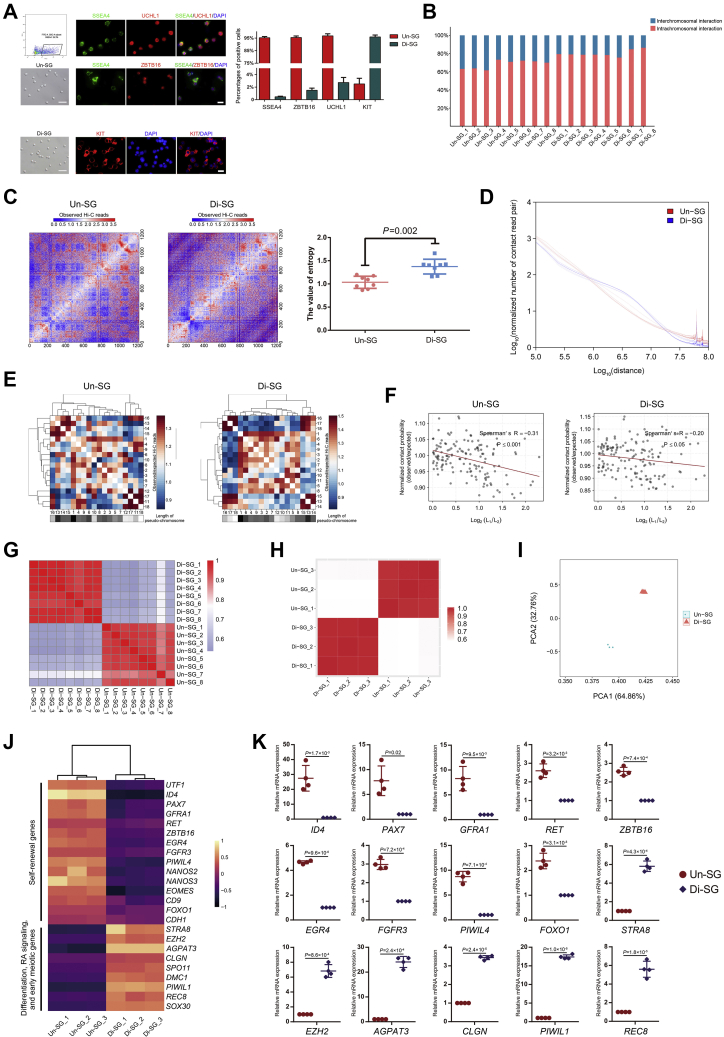
To uncover the 3D chromatin structure of SSCs/progenitors and Di-SG and the higher-order chromatin dynamics during spermatogonial differentiation, we first collected Un-SG and Di-SG from porcine testes. Our recent study has shown that SSEA4 is a surface marker of porcine Un-SG and that it can be used to enrich porcine Un-SG including transplantable SSCs with unprecedented efficiency (31). Hence, Un-SG were isolated from 90-day-old juvenile porcine testes and enriched by fluorescence-activated cell sorting (FACS) using an antibody against SSEA4, whereas Di-SG were isolated from 150-day-old pubertal porcine testes and enriched with a velocity sedimentation approach (STA-PUT) (32, 33). The high purity of collected spermatogonial subpopulations was validated by immunofluorescence staining and subsequent quantification of cells positive for stage-specific markers (Fig. 1A). Then, we performed in situ Hi-C and RNA-seq analyses on the collected spermatogonial samples. For Hi-C analysis, we generated high quality datasets from sufficient biological samples (eight for Un-SG and eight for Di-SG) and obtained around 6.3 billion valid interactions for the overall 16 samples, with an average of 392 million valid interactions per sample (Table S1). For RNA-seq analysis, we constructed transcriptomic libraries from six samples (biological triplicates for each spermatogonial subtype), with approximately 76 million paired reads per sample (Table S1).
Figure 1.
Dynamic 3D chromatin architecture during spermatogonial differentiation.A, enrichment and characterization of spermatogonial subpopulations. Un-SG were enriched by FACS using an antibody against SSEA4, and both cell populations were subjected to immunofluorescence staining and quantification of cells positive for stage-specific markers (SSEA4, ZBTB16, and UCHL1 for Un-SG and KIT for Di-SG). The bar represents 50 μm (brightfield) or 10 μm (immunofluorescence). The data are presented as the mean ± SEM of eight biological samples, and at least 300 cells were analyzed in each group. B, the interchromosomal and intrachromosomal interaction ratios in all Un-SG and Di-SG samples. C, the entropy difference between Un-SG and Di-SG. The intrachromosome log2 Hi-C matrices are shown at 100 kb resolution for chromosome 7. The data are presented as the mean ± SD of eight biological samples. P: Mann-Whitney U test, one-tailed. D, the P(s) curves of Un-SG and Di-SG showing the interaction probability patterns between bin pairs at defined genomic distances. E, the observed/expected number of contacts between any pair of 18 autosomes. The plaids with differential gray scale indicate the length of each chromosome. F, the observed/expected number of interactions between any pair of 18 autosomes plotted against the length difference of these chromosomes. L1 or L2 refers to the length of chromosome (L1 > L2), and length difference is indicated by log2 (L1/L2). The dotted line represents the linear trend for the obtained value. G, high-throughput chromosome conformation capture-Rep analysis illustrating the correlation of normalized Hi-C interaction matrices between Un-SG and Di-SG samples. H, Pearson correlation analysis illustrating the correlation of transcriptomic data between Un-SG and Di-SG samples. I, principal component analysis plot showing the transcriptomic profiles of Un-SG and Di-SG samples. J, a heatmap showing the representative downregulated and upregulated genes in Di-SG in relation to Un-SG. K, qPCR analysis of the mRNA expression of 15 genes in Un-SG and Di-SG. The data are presented as the mean ± SD of four independent experiments. P: paired Student’s t test, two-tailed. Di-SG, differentiating spermatogonia; FACS, fluorescence-activated cell sorting; Hi-C, high-throughput chromosome conformation capture; Un-SG, undifferentiated spermatogonia.
The chromosomal conformation profiles revealed that compared with Un-SG, Di-SG exhibited increased intrachromosomal interaction ratio (68.3% in Un-SG versus 80.2% in Di-SG) but decreased interchromosomal interaction ratio (31.7% in Un-SG versus 19.8% in Di-SG, Fig. 1B and Table S1). Then, based on the normalized 100 kb intrachromosomal contact matrices, we applied entropy as a measurement of the chromatin structural organization (34, 35). Entropy serves as a means to quantify the uncertainty within a system, in which higher entropy corresponds to less structural organization. From a biological perspective, genomic regions with high entropy are typically correlated with high proportions of euchromatin (36, 37). We found that Di-SG had higher entropy (Fig. 1C), suggestive of less compact chromatin structural organization in Di-SG. Next, we conducted a P(s) analysis to gain the interaction probability patterns between bin pairs at defined genomic distances (38) and identified that Di-SG exhibited higher interaction probabilities than Un-SG at the distances between 1 and 10 Mb, but that the trend was reversed at long distances (Fig. 1D), in line with previous reports in mice and rhesus monkeys that the more advanced pachytene spermatocytes also displayed stronger interactions at short distances (between 1 and 5 Mb) but weaker interactions at long distances (>10 Mb) than spermatogonia (39, 40).
We further analyzed the interchromosomal interaction. As expected, Un-SG and Di-SG exhibited similar nonrandomly distributed chromosomal positions. Longer chromosomes preferentially interacted with each other and the same for shorter ones (Fig. 1E). The negative correlation between the interchromosomal interaction probability and the chromosomal length also applied to both spermatogonial populations (Fig. 1F), in accordance with recent findings in adipocytes and myoblasts (41).
Subsequently, we studied the normalized Hi-C interaction matrices with 100 kb bin size for all biological samples. By using HiC-Rep, we detected a substantially lower correlation coefficient between Un-SG and Di-SG (r = 0.60), in contrast with high correlation coefficients between ingroup samples (r = 0.93 for Un-SG and r = 0.96 for Di-SG, Fig. 1G). These patterns can be validated by using QuASAR-Rep (Fig. S1A), GenomeDISCO (Fig. S1B), the Pearson correlation (Fig. S1C), or principal component analysis (PCA, Fig. S1D), corroborating distinct 3D chromatin organizations in two spermatogonial subgroups. In addition, we observed difference in transcriptomes between Un-SG and Di-SG, as reflected by a low correlation between Un-SG and Di-SG (r = 0.58), in spite of the high correlation between ingroup samples (r = 0.988 for Un-SG and r = 0.997 for Di-SG, Fig. 1, H and I). As shown by the RNA-seq data, the markers for Un-SG and SSC self-renewal genes, such as UTF1, ID4, PAX7, GFRA1, RET, ZBTB16, EGR4, FGFR3, PIWIL4, NANOS2, NANOS3, EOMES, CD9, FOXO1, and CDH1, were markedly downregulated, whereas the genes involved in spermatogonial differentiation, retinoic acid (RA) signaling, and early meiosis, such as STRA8, EZH2, AGPAT3, CLGN, SPO11, DMC1, PIWIL1, REC8, and SOX30, were substantially upregulated in Di-SG (Fig. 1J), corroborating efficient isolation and enrichment of two spermatogonial subpopulations. The expression changes of some genes were validated by the qPCR analysis (Fig. 1K). Hence, these data indicate that alterations of chromatin configuration are accompanied by transcriptomic variations during spermatogonial differentiation.
A/B compartment switches and changes during spermatogonial differentiation
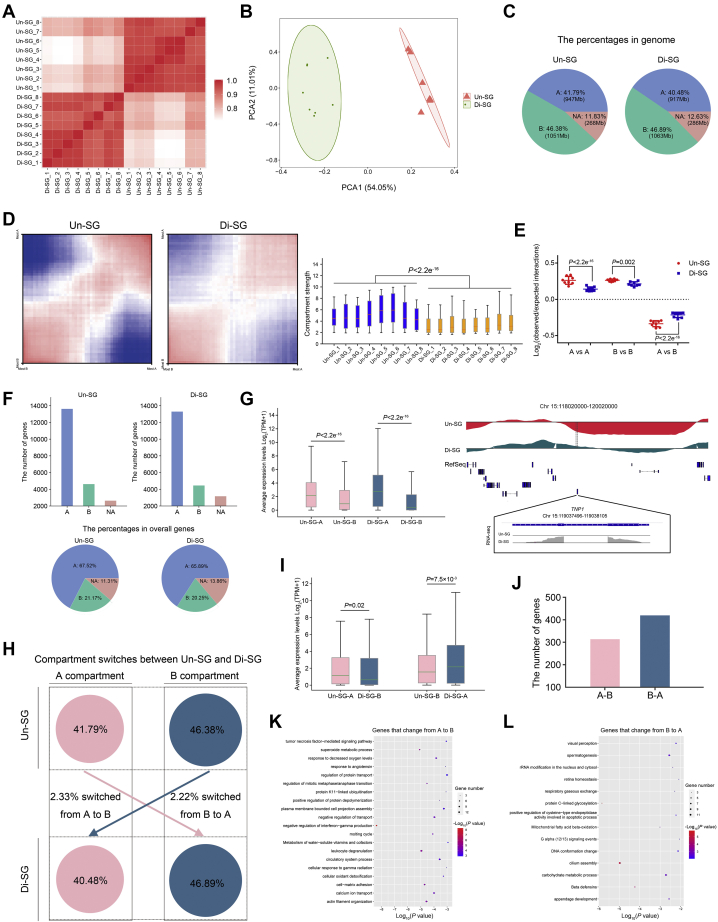
Higher-order chromatin can be divided into A and B compartments, representing open chromatin regions with active genes and closed chromatin regions with inactive genes, respectively (23). We then explored the A/B compartment index (20 kb bin size) in autosomes of Un-SG and Di-SG. Pearson correlation analysis illustrated a low correlation of global A/B compartment states between Un-SG and Di-SG (r = 0.78), in contrast with high correlations between ingroup samples (r = 0.96 for Un-SG and r = 0.95 for Di-SG, Fig. 2A), which was validated by PCA analysis (Fig. 2B). Despite the similar A/B compartment organization between Un-SG and Di-SG (Fig. 2C), the compartment strength was decreased in Di-SG (Fig. 2, D and E), indicative of weakened compartmentalization in Di-SG.
Figure 2.
A/B compartment switches during spermatogonial differentiation.A, Pearson correlation analysis illustrating the correlation of A/B indices between Un-SG and Di-SG samples. B, principal component analysis plot showing the A/B index profiles of Un-SG and Di-SG samples. C, the proportions and lengths of A/B compartments in genome. D, left, saddle plot showing the compartment strength in chromosome 9. Right, the compartment strength in all Un-SG and Di-SG samples, defined as the A-A and B-B compartment interaction strength relative to the A-B compartment interaction strength. P: Mann-Whitney U test, one-tailed. E, the interaction strength between A-A, B-B, or A-B compartments. The data are presented as the mean ± SD of eight biological samples. P: Mann-Whitney U test, one-tailed. F, the numbers (upper panel) and proportions (lower panel) of genes in A/B compartments. G, left, the average expression levels of genes in A/B compartments in Un-SG or Di-SG. P: Mann-Whitney U test, one-tailed. Right, PCA1 (the first eigenvalues, the upper part) and RefSeq view (the middle part) of chromosome 15, 118020000 to 120020000, as well as RNA-seq coverage track of TNP1 (chromosome 15, 119037496–119038105, the lower part) showing that TNP1, which was upregulated during spermatogonial differentiation, was located in the B compartment in Un-SG but switched to the A compartment in Di-SG. Principal component analysis 1 was calculated via eigenvector decomposition on the observed/expected intrachromosomal interaction matrices. H, a schematic overview illustrating the proportions of genomic regions subjected to A/B compartment switches (A to B or B to A) between Un-SG and Di-SG. I, the average expression levels of genes that changed from A to B or from B to A. P: Mann-Whitney U test, one-tailed. J, the numbers of genes that changed from A to B or from B to A. K and L, gene ontology-biological process (GO-BP) analysis of genes that changed from A to B (K) or from B to A (L). Di-SG, differentiating spermatogonia; PCA, principal component analysis; Un-SG, undifferentiated spermatogonia.
In both cell populations, the A compartments harbored the majority of genes (Fig. 2F), and genes in the A compartments showed higher expression levels than those in the B compartments (Fig. 2G). Previous studies have reported the correlation of the A/B compartment switch with transcriptional regulation (23). We thus probed the occurrence of the A/B compartment switch during spermatogonial differentiation. We found that 52.88 Mb and 50.22 Mb, making up 2.33% and 2.22% of the autosomal genome, underwent the A-B and B-A switch, respectively (Fig. 2H). Genes that changed from compartment A to B during spermatogonial differentiation tended to show lower expression levels in Di-SG than in Un-SG, whereas genes that changed from B to A tended to be upregulated (Fig. 2I). Specifically, 314 genes that changed from compartment A to B (Fig. 2J and Table S2) were enriched in cell-matrix adhesion, regulation of mitotic metaphase/anaphase transition, and response to decreased oxygen levels (Fig. 2K and Table S3), whereas 420 genes that changed from B to A (Fig. 2J and Table S2) fell in terms such as carbohydrate metabolic process, spermatogenesis, and DNA conformation change (Fig. 2L and Table S3). For instance, ATM, a protein involved in DNA damage response and located in the A compartment in Un-SG, was downregulated (FDR < 0.05, fold change > 2) in Di-SG where it was located in the B compartment. TNP1, which plays important roles in spermiogenesis and was significantly upregulated during spermatogonial differentiation, was located in the B compartment in Un-SG but switched to the A compartment in Di-SG (Fig. 2G and Table S2).
Compartments experiencing the A-A or B-B change can refer to those correlated with increasingly open or closed chromatin, respectively. We found that 44.2 Mb and 65.2 Mb, accounting for 1.95% and 2.88% of the autosomal genome, were subjected to the A-A and B-B change, respectively (Fig. S2A). Genes that underwent the compartment A-A change during spermatogonial differentiation tended to show higher expression levels in Di-SG than in Un-SG, whereas genes that underwent the B-B change tended to be downregulated (Fig. S2B). Specifically, 739 genes that underwent the compartment A-A change (Fig. S2C and Table S2) were enriched in male gamete generation, regulation of mitotic/meiotic cell cycle, and chromosome organization (Fig. S2D and Table S3), whereas 244 genes that underwent the B-B change (Fig. S2C and Table S2) fell in terms such as cellular response to hormone stimulus, regulation of cell adhesion, and Notch signaling pathway (Fig. S2E and Table S3). Genes that underwent the compartment A-A change included HSPA2, STRA8, SOX30, MLH1, and METTL3, all of which have been reported to be involved in spermatogenesis and showed higher expression levels in Di-SG than in Un-SG (FDR < 0.05, fold change > 2). YTHDC2, an N6-methyladenosine (m6A)-binding protein playing regulatory roles in spermatogenesis, underwent the B-B change and was downregulated (FDR < 0.05, fold change > 2) during spermatogonial differentiation (Table S2). Together, our data suggest that switches and changes of A/B compartments play important regulatory roles in dynamic gene expression during spermatogonial differentiation.
Topologically associating domain dynamics during spermatogonial differentiation
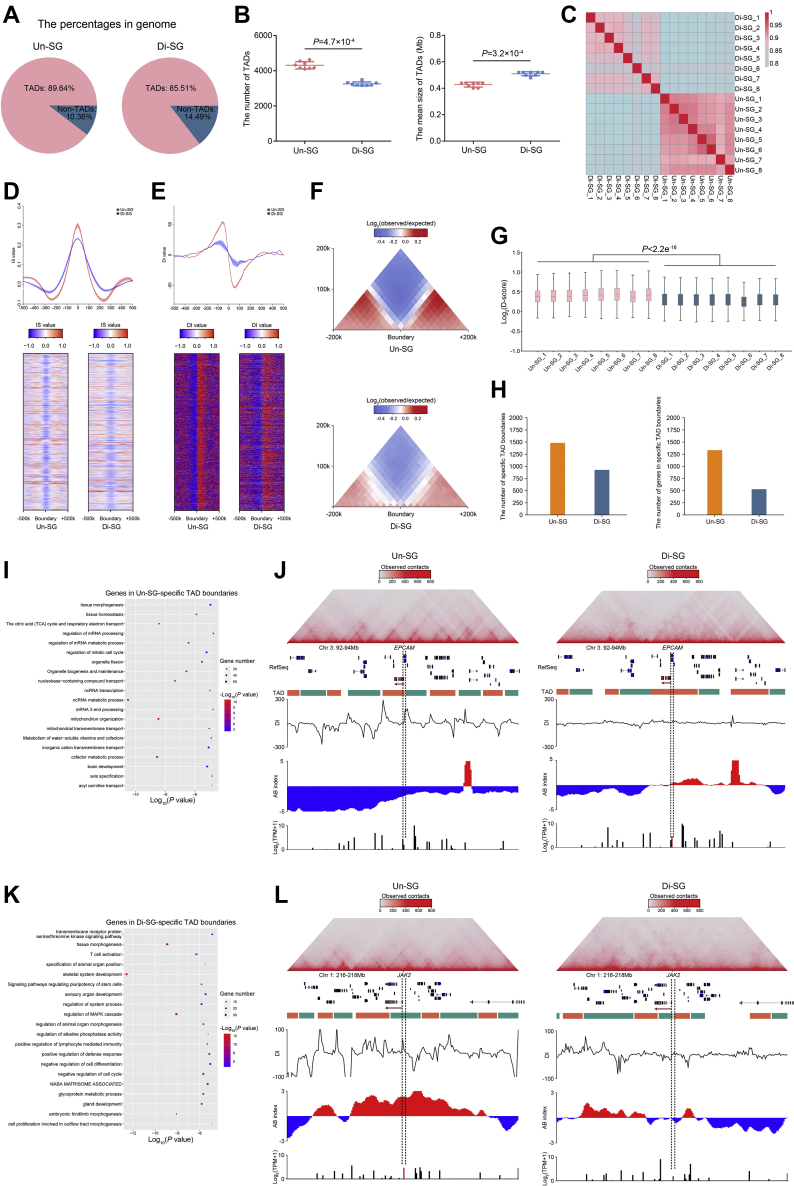
Topologically associating domains have been reported to be generally conserved among distinct cell types (41, 42, 43, 44, 45). Nevertheless, recent articles reported reorganization of TADs during spermatogenesis (39, 40, 46, 47, 48). Drastic alterations of TADs have also been identified in oogenesis, that is, TADs undergo gradual attenuation and then vanish during oogenesis, and it is only until the two-cell or even later embryo developmental stage that TADs reemerge and gradually restore (49, 50, 51), suggesting distinctive TAD dynamics in gametogenesis and early embryo development. Yet, whether TADs are conserved or subjected to alterations during spermatogonial differentiation remains to be explored. To this end, we analyzed the TAD architecture at 20 kb resolution in both spermatogonial subtypes. We identified that TADs constituted the majority of the genome (Fig. 3A), with a decrease of the TAD number but an increase of the mean TAD size (Fig. 3B) during spermatogonial differentiation. We observed a slightly lower correlation between Un-SG and Di-SG (0.80), in comparison with high correlations of TAD architecture between ingroup samples (0.91 for Un-SG and 0.86 for Di-SG), as reflected by Jaccard indices (Fig. 3C). Then, we used the insulation score (Fig. 3D), the directional index (DI, Fig. 3E), as well as aggregate Hi-C maps (Fig. 3F) to measure the strengths of TAD boundaries and found all of them declined in Di-SG. Moreover, the domain score (D-score), which is defined by the ratio of intra-TAD interactions in the overall intrachromosomal interactions (52) and able to quantify the tendency of TADs to self-interact (53), was decreased in Di-SG (Fig. 3G), further suggesting that TADs are weakened during spermatogonial differentiation. Thus, our data complement previous studies by showing that TAD attenuation already initiates at the premeiotic spermatogonial differentiation stage, toward the TAD dissolution occurring in subsequent meiosis (39, 40, 46, 47, 48).
Figure 3.
Topologically associating domain dynamics during spermatogonial differentiation.A, the proportions of TADs and non-TADs in genome. B, the numbers (left) and mean sizes (right) of TADs in Un-SG and Di-SG. The data are presented as the mean ± SD of eight biological samples. P: Mann-Whitney U test, one-tailed. C, Jaccard indices illustrating the correlation of TAD architecture between Un-SG and Di-SG samples. D and E, the mean IS (D) and DI (E) value of TADs and the flanking regions (±500k) in Un-SG and Di-SG. F, the aggregate Hi-C map showing the average observed/expected chromatin interaction frequencies at TADs and the flanking regions (±200k) in Un-SG and Di-SG. G, the D-score in all Un-SG and Di-SG samples. P: Mann-Whitney U test, one-tailed. H, the numbers of specific TAD boundaries (left) and their harbored genes (right) in Un-SG and Di-SG. I, gene ontology-biological process analysis of genes in Un-SG-specific TAD boundaries. J, views of the observed/expected chromatin interaction frequencies (the upper panel), RefSeq (the middle panel), DI, A/B index, and RNA-seq coverage (the lower panel) at chromosome 3, 92 to 94 Mb, revealing that EPCAM was harbored in Un-SG-specific TAD boundaries, in B-A switching compartments and upregulated in Di-SG. K, gene ontology-biological process analysis of genes in Di-SG-specific TAD boundaries. L, views of the observed/expected chromatin interaction frequencies (the upper panel), RefSeq (the middle panel), DI, A/B index, and RNA-seq coverage (the lower panel) at chromosome 1, 216 to 218 Mb, revealing that JAK2 was harbored in Di-SG-specific TAD boundaries, in A-B switching compartments and downregulated in Di-SG. DI, directional index; Di-SG, differentiating spermatogonia; GO-BP,gene ontology-biological process; Hi-C, high-throughput chromosome conformation capture; IS, insulation score; TADs, topologically associating domains; Un-SG, undifferentiated spermatogonia.
Later, we investigated whether TAD attenuation contributes to dynamic gene expression during spermatogonial differentiation. There were 1482 Un-SG-specific TAD boundaries harboring 1333 genes (Fig. 3H and Table S4) that are related to mitochondrion organization, regulation of mRNA metabolic process, and mitotic cell cycle (Fig. 3I and Table S5). These genes included spermatogonial markers (e.g., TSPAN33 and EPCAM, Fig. 3J), those involved in spermatogonial self-renewal (e.g., FOXO1), in differentiation (e.g., BMP4, DAZL, and WNT3A), and in meiosis (e.g., SPO11, Table S4). By contrast, 529 genes, which were embedded in 926 Di-SG-specific TAD boundaries (Fig. 3H and Table S4), were implicated in important biological processes during spermatogonial development, such as regulation of MAPK cascade, transmembrane receptor protein serine/threonine kinase signaling pathway, and regulation of cell cycle and cell differentiation (Fig. 3K and Table S5). Genes falling in this group included ZBTB16, a pivotal transcriptional regulator of spermatogonial self-renewal, those involved in pluripotency maintenance (e.g., LIF), and the JAK/STAT signaling component JAK2 (Fig. 3L and Table S4).
Long-range interacting TADs have been reported to be able to form TAD interaction networks, namely TAD cliques, to influence lineage-specific differentiation (54). It would be intriguing to explore whether the TAD interaction network, other than the TAD itself, is also attenuated during spermatogonial differentiation. Hence, we defined a TAD clique as a cluster of five or more interacting TADs in our Hi-C data. We observed high correlations of TAD cliques between ingroup samples (Pearson’s r = 0.77 for Un-SG and r = 0.78 for Di-SG, Fig. S3A). In accordance with the variation of TAD boundaries, the formation of TAD cliques (reflected by the number of TAD cliques, Fig. S3B), the genome coverage by TAD cliques (Fig. S3C), and the percentage of TADs in cliques (Fig. S3D) were diminished in Di-SG, indicating attenuation of the TAD interaction network during spermatogonial differentiation. Besides, we found that TAD cliques were enriched in B compartments relative to A compartments in both spermatogonial populations (Fig. S3E). Thus, the attenuation of TAD cliques in Di-SG suggests the facilitated transcription during spermatogonial differentiation.
Identification of PEIs and their regulation in gene expression during spermatogonial differentiation
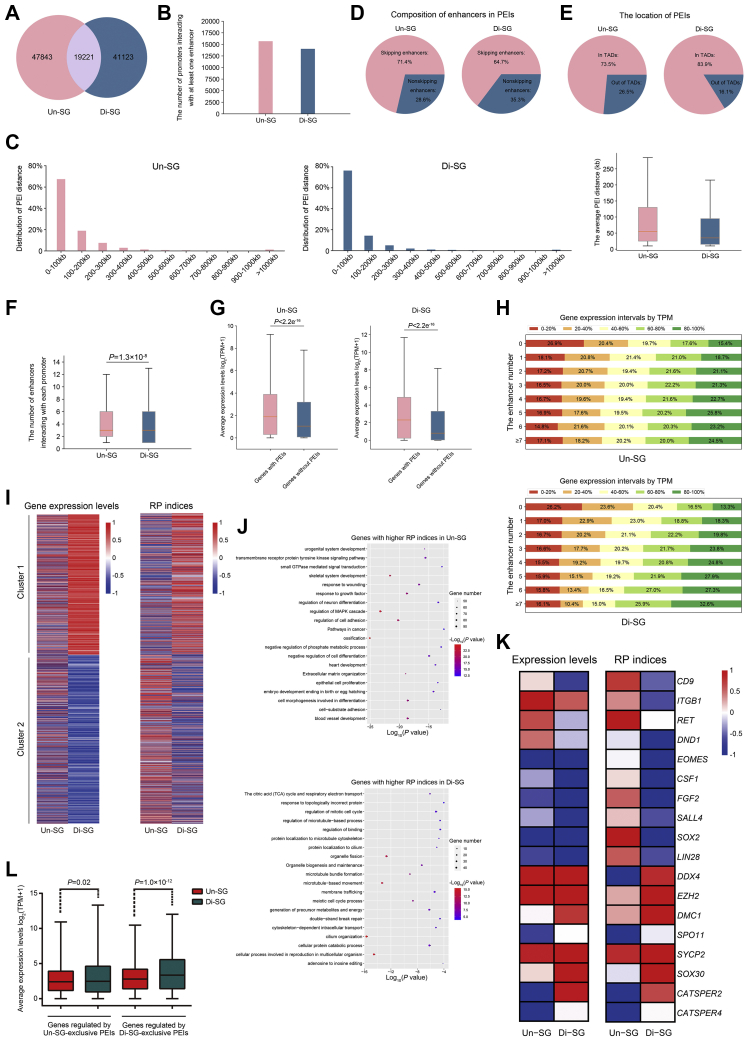
Chromatin can be spatially packaged into the 3D genome architecture chromatin loops, facilitating the interactions between promoters and distant DNA regulatory elements. In this way, long-range enhancers are able to physically contact with the target promoters, modulating the temporal and spatial expression of target genes (55, 56). To gain knowledge about the potential PEIs and their regulation in dynamic gene expression during spermatogonial differentiation, we combined the reads from eight biological samples of Un-SG or Di-SG into single sets of Hi-C data (to reach the resolution of 5 kb). We identified overall 67,064 PEIs in Un-SG and 60,344 in Di-SG (Fig. 4A), and that the 15,679 and 14,032 promoters interacted with at least one enhancer in Un-SG and Di-SG, respectively (Fig. 4B). The majority of PEIs were within 100 kb (Fig. 4C) were implicated skipping enhancers (enhancers that are not the closest to promoters, Fig. 4D) and were, as expected, within TADs (Fig. 4E) in both spermatogonial subtypes. Besides, we detected slightly more enhancers that interact with each promoter in Un-SG than in Di-SG (Fig. 4F).
Figure 4.
Promoter-enhancer interaction regulation in gene expression during spermatogonial differentiation.A, the numbers of PEIs in Un-SG and Di-SG samples. The number in the overlapped region refers to PEIs present in both populations. B, the numbers of promoters that interact with at least one enhancer in Un-SG and Di-SG samples. C, left, distribution of PEI distance in Un-SG and Di-SG samples. Right, the average PEI distance in Un-SG and Di-SG samples. D, composition of skipping and nonskipping enhancers in PEIs. E, the proportions of PEIs in or out of TADs. F, the average numbers of enhancers that interact with each promoter in Un-SG and Di-SG samples. P: Mann-Whitney U test, one-tailed. G, the average expression levels of genes with or without PEIs. P: Mann-Whitney U test, one-tailed. H, more interacting enhancers are associated with higher proportions of genes in top gene expression intervals. The numbers in columns refer to the proportions of genes in each gene expression interval. I, heatmaps of expression levels and RP indices of two clusters of genes that were expressed in either or both spermatogonial subpopulations (TPM > 1) and that exhibited a fold change of ≥4. J, gene ontology-biological process analysis of genes with differential RP indices during spermatogonial differentiation. K, heatmaps of representative genes, with differential RP indices and expression levels during spermatogonial differentiation. L, the average expression levels of genes regulated by Un-SG- or Di-SG-exclusive PEIs. P: Mann-Whitney U test, one-tailed. Di-SG, differentiating spermatogonia; PEI, promoter-enhancer interaction; RP, regulatory potential; TADs, topologically associating domains; TPM, transcripts per million; Un-SG, undifferentiated spermatogonia.
As expected, genes with PEIs exhibited generally higher expression levels than those without PEIs (Fig. 4G), and more interacting enhancers were also associated with higher gene expression in both Un-SG and Di-SG (Fig. 4H), suggesting cumulative effects of enhancers on the transcriptional levels of target genes. Because multiple enhancers that interact with promoters vary in interacting intensity, we then introduced a regulatory potential (RP) index that combines both the number and intensity of the interacting enhancers to quantify their potential for transcriptional regulation of target genes. We identified that alterations of the RP index are generally consistent with gene transcriptional changes (Fig. 4I), suggesting that PEIs orchestrate transcription during spermatogonial differentiation. Later, we detected that the 3985 genes showed significantly higher RP indices in Un-SG than in Di-SG (RPUn-SG-RPDi-SG > 3, fold change > 2, Table S6). These genes were related to regulation of cell adhesion, response to growth factor, and cell morphogenesis involved in differentiation (Fig. 4J and Table S7). Examples in this case are Un-SG markers (CD9 and ITGB1), genes important to spermatogonial self-renewal (RET, DND1, EOMES, CSF1, FGF2, and SALL4) and pluripotency (SOX2 and LIN28, Fig. 4K and Table S6) that exhibited both higher RP indices and expression levels (FDR < 0.05, fold change > 2) in Un-SG than in Di-SG. By contrast, 2741 genes, falling in terms such as cellular process involved in reproduction in multicellular organism, meiotic cell cycle process, regulation of mitotic cell cycle, and DSB repair (Fig. 4J and Table S7), showed significantly lower RP indices in Un-SG than in Di-SG (RPUn-SG-RPDi-SG < −3, fold change > 2, Table S6). Illustrations of this point are the pan-germ cell marker DDX4, genes involved in spermatogonial differentiation (EZH2), in meiosis (DMC1, SPO11, SYCP2, and SOX30), and in sperm motility (CATSPER2 and CATSPER4, Fig. 4K and Table S6) that displayed both lower RP indices and expression levels in Un-SG than in Di-SG, lending further support to the positive correlation between the RP index and gene expression.
In addition, we identified that some genes were regulated by spermatogonial subtype-exclusive PEIs, that is, PEIs present in only Un-SG or Di-SG. To be more precise, 2730 genes, including those involved in spermatogenesis, such as BMP4, DMRT1, RXRG, SYCP1, PRM1, PRM2, METTL3, BRCA2, WNT3A, FTO, NEDD4, and GDNF (Table S8), were regulated by Un-SG-exclusive PEIs, whereas 1671 genes, such as CXCR4, LHX1, LIF, RARA, RARB, SPO11, ATR, SMC4, TNP2, PRM3, and CATSPER4 (Table S8) that are also spermatogenesis-related, were with Di-SG-exclusive PEI regulation. As expected, genes regulated by cell type-exclusive PEIs were associated with elevated expression levels (Fig. 4L). Taken together, these data indicate that PEIs act as an important element of transcriptional regulation during spermatogonial differentiation.
Characterization of H3K27ac-marked active enhancers and their regulation in gene expression during spermatogonial differentiation
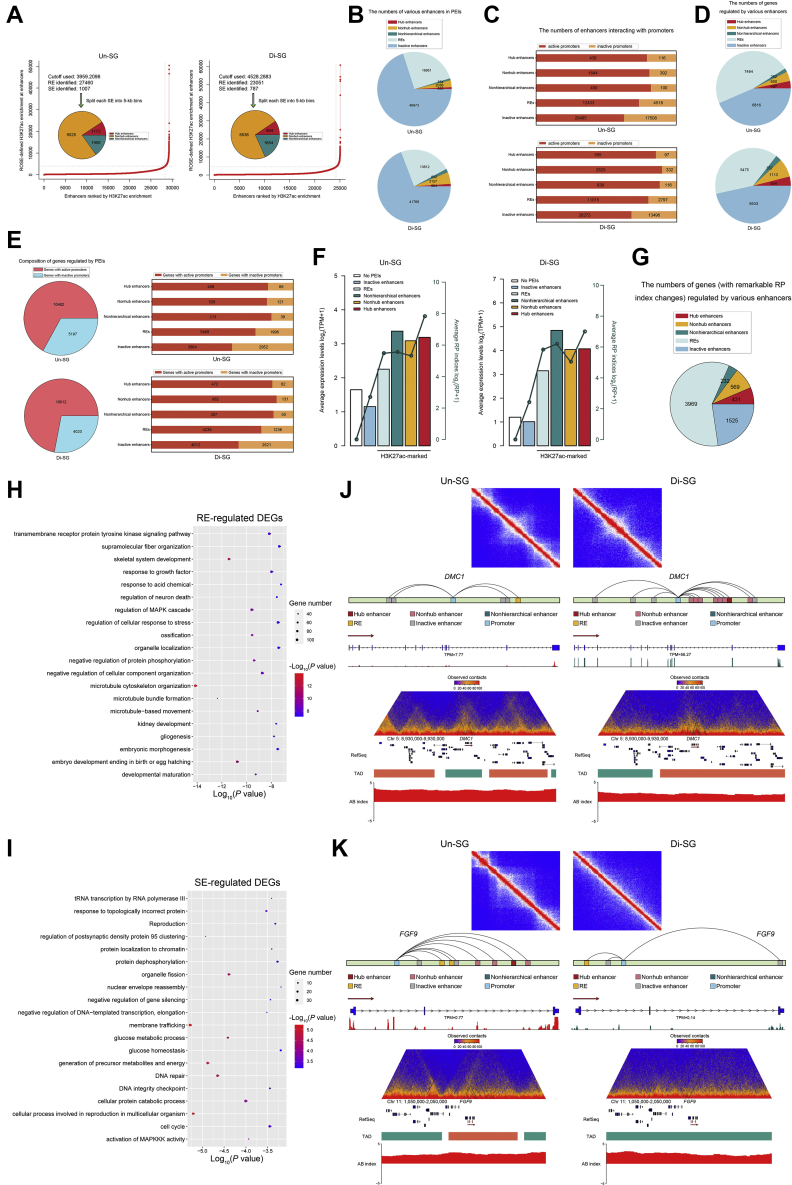
To delve into the role for enhancers in PEIs and further in gene expression, we performed ChIP-seq analyses on the collected spermatogonial samples, using antibodies against H3K27ac (an active enhancer marker) or H3K4me3 (an active promoter marker). By combining Hi-C chromatin interactions and H3K27ac ChIP-seq data and applying the ROSE algorithms (57), we defined regular enhancers (REs) and super enhancers (SEs), and by splitting each SE into 5 kb bins, we further classified SEs into hierarchical (which are composed of hub and nonhub enhancers) and nonhierarchical enhancers, as previously reported (58). Arguably, substantially more REs than SEs were identified in both spermatogonial populations (Fig. 5A). Then, we looked into the enhancers within PEIs and found that in both subgroups, only a small fraction of enhancers fell in the scope of SEs (Fig. 5B). Various enhancers tended to interact with active promoters (marked by H3K4me3, Fig. 5C). Consistently, when looking into genes with PEIs, it could be found that only a minor fraction of genes was regulated by SEs (Fig. 5D), even though most genes had active promoters interacting with enhancers (Fig. 5E).
Figure 5.
Regulation of H3K27ac-marked active enhancers in gene expression during spermatogonial differentiation.A, definition and numbers of REs and hierarchically organized SEs in two spermatogonial populations. B, the numbers of different categories of enhancers within PEIs. C, the numbers of different categories of enhancers that interact with active or inactive promotors. D, the numbers of PEI genes regulated by different categories of enhancers. E, left, the numbers of PEI genes with active or inactive promoters. Right, the numbers of PEI genes regulated by various enhancers and promoters. F, average expression levels and RP indices (indicated by bars and lines, respectively) of genes regulated by different categories of enhancers. G, the numbers of genes (with remarkable RP index changes) regulated by different categories of enhancers. H and I, gene ontology-biological process analysis of RE (H) and SE (I)-regulated genes with differential RP indices and expression levels during spermatogonial differentiation. J and K, the upper panel, the contact matrix showing stripes at the DMC1 (J) and FGF9 (K) SE loci. The middle panel, models for PEI regulation of DMC1 (J) and FGF9 (K), as well as RNA-seq coverage at their genomic loci. The lower panel, views of the observed/expected chromatin interaction frequencies, RefSeq, TAD, and A/B index at chromosome 5: 8,930,000 to 9,930,000 (J) and chromosome 11: 1,050,000 to 2,050,000 (K). PEIs, promoter-enhancer interactions; REs, regular enhancers; RP, regulatory potential; SEs, super enhancers; TAD, topologically associating domain.
It has been reported that various enhancers differentially contribute to gene expression (58, 59). As expected, we found that in both spermatogonial populations, genes with higher RP indices and expression levels tended to be regulated by active enhancers that are marked by H3K27ac (i.e., REs and SEs), and that genes regulated by SEs showed generally higher expression levels than those targeted by REs (Fig. 5F). Regular enhancer and SE-associated genes were also prone to have active promoters (marked by H3K4me3, Fig. 5E, right), together acting as hallmarks of active transcription. Further analysis revealed that of the genes showing remarkable RP index changes during spermatogonial differentiation (RPUn-SG-RPDi-SG > 3 or < −3, fold change > 2), 59.0% (3969/6726) were regulated by REs (Fig. 5G), of which 54.3% (2154/3969) showed differential expression levels (FDR < 0.05, fold change > 2) in two spermatogonial populations (Table S9). These genes were enriched with regulation of MAPK cascade, transmembrane receptor protein tyrosine kinase signaling pathway, response to growth factor, and developmental maturation (Fig. 5H and Table S10). By contrast, 18.3% (1232/6726) of the genes with remarkable RP index changes were regulated by SEs (Fig. 5G), of which 55.0% (677/1232) were differentially expressed (Table S9). These genes fell in terms such as cellular process involved in reproduction in multicellular organism, generation of precursor metabolites and energy, DNA repair, and activation of MAPKKK activity (Fig. 5I and Table S10). Hence, these data suggest that both REs and SEs are important regulatory elements underlying the dynamic transcriptome during spermatogonial differentiation.
Of the active enhancers that are marked by H3K27ac, SEs particularly appealed to us, because of their substantially high levels of activity, enrichment of active chromatin characteristics, and pivotal roles in cell fate determination (57, 59, 60). To gain more insights into SE-mediated transcriptional regulation during spermatogonial differentiation, we inspected 1232 SE-associated genes with remarkable RP index changes. Of these, 35.0% (431/1232), 46.2% (569/1232), and 18.8% (232/1232) were regulated by hub enhancers, nonhub enhancers, and nonhierarchical enhancers, respectively (Fig. 5G and Table S9). Intriguingly, we found that hub enhancer-targeted genes harbored some with well-known roles in spermatogenesis. For instance, DMC1, a gene essential for DSB repair and meiotic homologous recombination (61), was targeted by hub enhancers only in Di-SG and also upregulated in Di-SG (FDR < 0.05, fold change > 2, Fig. 5J and Table S9). Another illustration of this point is CATSPER2, a member of the CATSPER gene family with functions in sperm motility (62) (Fig. S4A and Table S9). FGF9 is a downstream gene of SOX9 that plays crucial roles in testicular development. Recently, a novel role for FGF9 in promoting SSC self-renewal has been reported (63). Interestingly, this gene was targeted by hub enhancers only in Un-SG and showed higher expression levels in Un-SG (Fig. 5K and Table S9). Genes with hub enhancer regulation also included DND1, a gene encoding DND1 which associates with NANOS2 to promote spermatogonial self-renewal (64) (Fig. S4B and Table S9), as well as EZH2, an epigenetic factor capable of modulating spermatogonial differentiation and apoptosis (65) (Fig. S4C and Table S9). These results thus corroborate that SEs, in particular hub enhancers, act as important regulatory elements of dynamic gene transcription during spermatogonial differentiation.
Discussion
Several recent articles reported reorganized chromatin architecture during mammalian spermatogenesis. As reported, compartmentalization, TADs, or loops underwent dissolution and reestablishment with spermatogenic cell development, and gene transcription seemed to be independent of the chromatin structure at certain stages such as the pachytene stage (39, 40, 46, 47, 48). Here, by integrating Hi-C, RNA-seq, and ChIP-seq data, we delved into the higher-order chromatin structural dynamics and their influences upon transcriptional regulation during spermatogonial differentiation. Our findings complement previous studies by showing that the dynamic alterations in 3D chromatin organization already initiate at the premeiotic spermatogonial differentiation stage: Di-SG exhibited less compact chromatin structural organization, weakened compartmentalization, and TADs in comparison with Un-SG. Our results also suggest that A/B compartments and TADs are related to dynamic gene expression during spermatogonial differentiation. Moreover, because it is feasible to obtain a vast number of spermatogonial subpopulations from porcine testes with large size, we proceeded to explore the contribution of PEIs to premeiotic transcriptional regulation, which has not been accomplished in previous studies due to limited cell input and resolution.
One of the most striking findings in this study might be the dynamic 3D chromatin structure during spermatogonial differentiation, which is in contrast with a recent article describing minimal alterations in higher-order chromatin architecture between primitive type A spermatogonia and type A spermatogonia in mice (39). From our perspective, the discrepancy could be ascribed to several respects. First, because peers and us have demonstrated that SSEA4 is a conserved surface marker of Un-SG and that it can be used to enrich nonhuman primate, human, and porcine Un-SG including transplantable SSCs efficiently (11, 31, 66), here, we exploited an antibody against SSEA4 in conjunction with FACS to enrich Un-SG with high purity for subsequent bioinformatic analyses, distinct from Luo et al. (39) who used a STA-PUT procedure to collect spermatogonial subpopulations with relatively lower purity. Second, as reported in that article, primitive type A spermatogonia were isolated from 6-day-old mice (39). Indeed, mouse male germ cells at this developmental stage consist of not only Un-SG but also a fraction of Di-SG likely committed to the first wave of spermatogenesis (67, 68, 69, 70), which are morphologically indistinguishable and difficult to be separated by velocity sedimentation approaches such as STA-PUT, and due to this heterogeneity, the reported minimal alterations in higher-order chromatin architecture between the collected spermatogonial subgroups might be underrepresented for the likely changing chromatin dynamics during spermatogonial differentiation. Third, although it has traditionally been acknowledged that spermatogenesis is a generally conserved process among mammalian species, recent single-cell RNA-seq analyses of testicular cells from mice, human, and nonhuman primates disclosed some divergent characteristics during mammalian spermatogenesis (71, 72). Hence, the possibility remains that the 3D chromatin dynamics during spermatogonial differentiation per se differ between mice and pigs.
Previous studies have suggested that chromatin reorganization is a characteristic event during stem cell differentiation and lineage specification (49, 54, 73). Here, we identified that also spermatogonial differentiation entailed dynamic alterations in 3D chromatin organization, characterized by loosened chromatin structural organization and attenuated compartmentalization and TADs. Spermatogonial differentiation has been known as a process that implicates pronounced transitions in cell-cycle, transcriptional, and metabolic regulators, separating the largely quiescent SSCs (which principally rely on glycolysis for energy supply) from the more proliferative Di-SG (which preferentially use oxidative phosphorylation to produce ATP) (9, 10, 11, 15, 74, 75). Loosened chromatin structural organization and weakened compartmentalization in Di-SG might therefore be related to cell-cycle transitions and metabolic shifts. Topologically associating domains have recently been reported to almost vanish in pachytene spermatocytes (39, 40, 46, 47, 48), even though extensive transcription occurs with dissolved TADs. Our data demonstrated that TAD attenuation already initiated at the premeiotic spermatogonial differentiation stage. This, along with the marked upregulation of many meiosis-related transcripts in Di-SG, as revealed by the present and previous studies (15, 16, 76), suggest that spermatogonial differentiation might be a transitional process that gradually prepares the genome for the subsequent meiotic events, which warrants systematic investigations in future.
Previous studies have also suggested the need to unravel the elusive and enigmatic relationship between transcription and chromatin configuration. Here, we identified that the dynamic gene expression during spermatogonial differentiation could be influenced by A/B compartment switches and changes, as well as TAD boundaries and cliques. To gain more knowledge about the contribution of delicate chromatin organization to gene transcription during this process, we probed PEIs in Un-SG and Di-SG under 5 kb bins. We introduced a RP index to quantify the potential of interacting enhancers for transcriptional regulation. As expected, the RP index was found positively correlated with gene expression during spermatogonial differentiation. Our findings thus provide direct evidence that apart from epigenetic modification and noncoding RNAs, also PEIs are an important element of transcriptional regulation in the process of male germline development. Further, we characterized REs and SEs and investigated the structural hierarchy of SEs on the basis of chromatin interactions in two spermatogonial populations. Intriguingly, we identified that a couple of genes with well-known roles in spermatogenesis were potentially regulated by hub enhancers within hierarchical SEs, suggesting a role for the structural hierarchy of SEs in transcriptional regulation during spermatogonial differentiation. Future perturbation studies by using, for example, the CRISPR-Cas9 strategy, will functionally validate the role of hub enhancers in SE structure and further in transcriptional regulation. Nevertheless, a prerequisite for this would be establishment of an optimized long-term culture system that enables stable propagation and induced differentiation of porcine SSCs, akin to their mouse counterparts that not only readily proliferate and differentiate in vitro but also seem amenable to CRISPR-Cas9-mediated genome editing (77, 78, 79).
To sum up, we systematically investigated the 3D genome organization and its correlation with transcriptional regulation during spermatogonial differentiation. We identified that diminished higher-order chromatin architecture in meiotic cells, as shown by recent reports, might be preprogramed in Di-SG, delineating unidirectional development of male germline and have also for the first time, to our knowledge, unraveled the contribution of PEIs to premeiotic transcriptional regulation. Recent studies exploiting the single-cell RNA-seq technique uncovered the transcriptomes of progressive spermatogenic subtypes during mammalian spermatogenesis (8, 9, 10). In future, development of single-cell Hi-C technology would help to unravel the finer 3D chromatin structural difference between SSCs and progenitors, enabling more comprehensive insights into the higher-order chromatin dynamics during male germline development and with functional perturbation analyses, the roles of 3D genome conformation in transcriptional activity could be validated. Overall, the present study greatly adds to the growing body of knowledge about chromatin configuration related to male fertility. As failure to maintain the proper chromatin architecture during early spermatogenesis, that is, the phase of SSC self-renewal and differentiation, can cause spermatogonial apoptosis (80), this study also provides a desirable reference catalog for diagnosis of male sub-/infertility, as well as for development of SSC therapy, for example, SSC auto-transplantation (6) and in vitro differentiation into sperm (81).
Experimental procedures
Testis samples
Testes were obtained from 90-day-old juvenile or 150-day-old pubertal Duroc pigs (Besun farm). After surgical castration, testes were placed in Dulbecco's phosphate-buffered saline (DPBS) supplemented with 2% penicillin/streptomycin (Hyclone) and transported to the laboratory on ice. All animal procedures were in accordance with and approved by the Institutional Animal Care and Use Committee of Northwest A&F University.
Isolation and enrichment of spermatogonial populations
Undifferentiated spermatogonia were isolated from 90-day-old juvenile porcine testes and enriched by FACS using an antibody against SSEA4. To obtain the single-cell suspension, the testis tunica albuginea and visible connective tissue were removed and then exposed to Type IV Collagenase (2 mg/ml; Thermo Fisher Scientific) at 35 °C for 20 min with periodic shaking. After three times of washing with DPBS to remove interstitial cells, the obtained seminiferous tubules were incubated with hemolytic fluid for 2 min to remove erythrocytes, followed by treatment with 0.25% trypsin-EDTA (Hyclone) at 37 °C for 5 min to obtain the single-cell suspension. After centrifugation, the cell pellet was resuspended in Dulbecco’s modified Eagle medium (DMEM, high glucose; Hyclone) supplemented with 5% fetal bovine serum (Hyclone) and subjected to differential plating, as previously reported (31). The suspension containing Un-SG was then collected and applied to FACS. In brief, the cells were washed with chilled FACS buffer (DPBS with 1% fetal bovine serum and 2 mM EDTA) and then incubated with the mouse anti-SSEA4 antibody (1: 50; 4755S, Cell Signaling Technology) on ice for 30 min, followed by washing and incubation with the Alexa fluor 488-conjugated donkey anti-mouse secondary antibody (1: 200, diluted in FACS buffer; Thermo Fisher Scientific) on ice for 20 min. After washing, the cells were subjected to FACS using a BD FACS AriaTM Ⅲ Flow Cytometer (BD Biosciences).
Differentiating spermatogonia were isolated from 150-day-old pubertal porcine testes and enriched with a velocity sedimentation approach (STA-PUT), following previously published protocols (32, 33). Different fractions of germ cells were examined, and only fractions with high purity of Di-SG were pooled.
Immunofluorescence
Immunofluorescence staining was performed on 4% paraformaldehyde-fixed cytospin slides of collected cells. Briefly, the cells were permeabilized with 0.1% triton-X (Sigma-Aldrich) for 10 min, followed by washing and blocking with 5% bovine serum albumin (MP Biomedicals) for 1 h. The cells were then incubated with the primary antibodies at 4 °C overnight. The primary antibodies used were as follows: mouse anti-SSEA4 (1: 200; 4755S, Cell Signaling Technology), rabbit anti-UCHL1 (1: 200; ab108986, Abcam), rabbit anti-ZBTB16 (1: 200; sc-22839, Santa Cruz Biotechnology), and rabbit anti-KIT (1: 200; 3074S, Cell Signaling Technology). The corresponding isotype IgGs in place of the primary antibodies were used as negative controls. The next day, the cells were washed and incubated with the Alexa fluor 488-conjugated donkey anti-mouse and/or 594-conjugated donkey anti-rabbit secondary antibodies (1: 400; Thermo Fisher Scientific) for 1 h, followed by nuclear counterstaining with DAPI (1: 1000; Bioworld Technology) for 5 min. After washing, the cells were visualized under a Nikon Eclipse 80i fluorescence microscope. The purity of collected spermatogonial populations was determined by the percentage of cells positive for stage-specific markers (SSEA4, ZBTB16, and UCHL1 for Un-SG and KIT for Di-SG) in the total cells (>300 cells analyzed in each group).
High-throughput chromosome conformation capture library construction
High-throughput chromosome conformation capture libraries were constructed with isolated Un-SG and Di-SG, after a previously published protocol, with minor modifications (26). In brief, 1.0 × 106 to 5.0 × 106 cells were crosslinked with 37% formaldehyde and then incubated with a glycine solution for 10 min to quench crosslinking. After washing with PBS, the cells were pelleted, snap-frozen, and stored at −80 °C. To construct Hi-C libraries, cell pellets were resuspended in lysis buffer and homogenized. DNAs were digested with 200 units of MboI for 1 h at 37 °C. Restriction fragment overhangs were filled and labeled with biotinylated nucleotides and ligated. Ligated DNAs were then purified and sheared to 300 to 500 bp. Ligation junctions were pulled down with streptavidin beads and subjected to Illumina NovaSeq 6000 sequencing in Novogene Co, LTD.
High-throughput chromosome conformation capture data processing
The clean Hi-C reads were mapped to the Sscrofa 11.1 genome, and the Hi-C contact frequency between genomic loci was computed using the Juicer pipeline (version 1.8.9). Low-quality alignments (defined as MAPQ < 30) and intrafragment reads were filtered from unique reads, thereby generating valid Hi-C contacts that were used for later analyses. All contact matrices used for further analyses were KR-normalized with Juicer. The value of matrices for different samples was standardized using the R software bnbc (version 1.12.0). Correlation in contact matrices was evaluated using HiCRep (version 1.14.0), QuASAR-Rep, or GenomeDISCO (82) in the default settings. The global interaction patterns of the whole chromosome were constructed by the scaled matrices with 100 kb or 20 kb bin size for biological samples. We selected 20 kb to show local interactions and perform TAD calling. To compare the high-resolution contact frequency, we merged the valid pairs from eight biological samples of different stages and attained the KR-normalized contact matrices with the resolution of 5 kb.
Von Neumann entropy of intrachromosomal contacts
The Von Neumann entropy was used to measure the chromatin structural organization, based on the normalized 100 kb intrachromosomal contact matrices, as previously described (34, 35).
P(s) analysis
P(s) analysis was performed on the normalized interaction matrices with 100 kb resolution, following previously reported methods (38). In brief, genome distances were first divided into 100 kb equal bins. Then, for each bin, the mean number of interactions at corresponding distances was counted. To obtain the P(s), the number of interactions in each bin was divided by the total number of possible region pairs.
RNA-seq library construction
Total RNAs were extracted from biological triplicates of Un-SG and Di-SG, using the RNeasy Mini Kit (Qiagen) and following the protocol provided by the manufacturer. After DNase (Qiagen) treatment, the poly A-mRNA-seq libraries were constructed with an Illumina TruSeq stranded RNA-seq library protocol.
RNA-seq data processing
RNA-seq libraries were quantified with the Qubit dsDNA High Sensitivity Assay Kit (Thermo Fisher Scientific) and sequenced on the Hiseq 4000 platform (Illumina), producing approximately 75.75 million 150 bp paired-end raw reads and 72.87 million high-quality reads for each library. The expression levels of protein-coding genes (gene annotation file from Ensembl Sscrofa 11.1 release 90) were quantified as transcripts per million using Kallisto (version 0.44.0). EdgeR (version 3.30.0) was used in the differential gene expression analysis. Genes with false discovery rate (FDR) ≤ 0.05 and log2 |fold change| > 1 were identified as differentially expressed genes.
Quantitative PCR analysis
Total RNAs were extracted from Un-SG and Di-SG, using the RNeasy Mini Kit (Qiagen) and following the protocol provided by the manufacturer. Total RNAs were then subjected to reverse transcription into cDNAs, using the Transcriptor First Strand cDNA Synthesis Kit (Roche). Real-time PCR reactions were performed in a 25-μl volume system with SYBR green (Roche) incorporation and were run in parallel triplicates from four independent experiments using an IQ5 platform (Bio-Rad). HPRT1 was used as a reference gene, and the data were analyzed with a 2−ΔΔCt method. Information on primers and on qPCR products is depicted in Table 1.
Table 1.
Information on primers and on qPCR products
| Gene symbol | Primer sequences | Gene bank accession | Length of qPCR product (bp) |
|---|---|---|---|
| HPRT1 | F: GAAGAGCTACTGTAATGACCAGTCAACGG R: TCATTGTAGTCAAGGGCATAGCCTACC |
NM_001032376.2 | 285 |
| ID4 | F: CGACTACATCCTGGACCTGC R: AGAATGCTGTCGCCCTGCT |
NM_001123130.1 | 183 |
| PAX7 | F: GATCCTCTGCCGCTACCAA R: ATCACAGTGCCCGTCCTTC |
XM_021095458.1 | 178 |
| GFRA1 | F: CATCTGCAGATCTCGCCTGGC R: GCCAAAGGCTTGAATTGCATTTTTGAGAC |
XM_013983780.2 | 277 |
| RET | F: TGAAGAGGAGCAAGGGTCGGAT R: AGCATCAGACTGTACATCTCCTCGC |
XM_021072832.1 | 238 |
| ZBTB16 | F: GCGGAAGACCTGGATGACCT R: GTCGTCTGAGGCTTGGATGGT |
XM_021062868.1 | 105 |
| EGR4 | F: GCCACAAACCCTTCCAGTG R: GCTGTGCCGTTTCTTCTCAT |
NM_001285968.1 | 155 |
| FGFR3 | F: CAACCCCACTCCATCCAT R: GCTGCCAAACTTGTTCTCC |
XM_021100898.1 | 169 |
| PIWIL4 | F: ATGTACCAAATTGGACGGAA R: CTCACATCAGCACTAAACAGGA |
NM_001315802.1 | 137 |
| FOXO1 | F: CCATGCTACTCATTTGCGCC R: GAGTCCCGCTGCACAGTTAT |
NM_214014.3 | 177 |
| STRA8 | F: AGCCGTTTACTTTCACTCTGACC R: GCTGTTTGCATTCCCATCCT |
NM_001285970.1 | 148 |
| EZH2 | F: TGTGGACACTCCTCCCAGAA R: GTCGCAGGGCTGATAGTTG |
NM_001244309.1 | 121 |
| AGPAT3 | F: CCAAAGTCCTGGCTAAACG R: CTTCAGTCCTTCGACCACAG |
NM_001143700.1 | 128 |
| CLGN | F: TGAAAGCGAACCTGCTCA R: TGCCTCCCATTCTCCATC |
NM_001243207.1 | 142 |
| PIWIL1 | F: TGAGGGCATGGAATAGCTG R: CGTTCACTCGTTTCTTCACC |
NM_001194973.1 | 189 |
| REC8 | F: TTCCGGTCACTGTGCGGTCT R: ATCAGCAGGTCCAGGTCTCG |
XM_013978192.2 | 124 |
Analysis of A/B compartment
Identification of A/B compartments at 20 kb resolution was performed via two steps. First, PC1 vectors were generated by using PCA as previously described at 100 kb resolution (23). The o/e contact matrix was then generated by the first two principal components that were obtained by using the ‘prcomp’ function in R. The initial position of gene model was defined by transcription start site (TSS) of each gene, and gene density was calculated by the number of TSS in each 100 kb bin. Bins with positive Pearson’s correlation between PC1 value and gene density were defined as compartment A, otherwise compartment B. Second, the A-B index, which represents the comparative likelihood of a sequence interacting with A or B, was generated as previously described at 20 kb resolution (83). Bins at 20 kb resolution with the positive A-B index were considered as A compartment and vice versa. The compartment strength was calculated by using AA × BB/AB2 as previously described (84). AA/BB is the mean contact enrichment between pairs of bins with compartment A/B signals, whereas AB is the mean contact enrichment between pairs of bins with compartment A and B signals. To identify genome regions that switched the A/B compartment state between Un-SG and Di-SG, the 20 kb bin was defined as the A or B status in 1 cell type if it showed a compartment A or B signal in more than 85% of Hi-C libraries in this cell type. Genes with TSS located in A or B regions were considered as A or B genes.
Functional enrichment analysis
Functional enrichment analysis of Gene Ontology (GO) and pathway was performed using the Metascape (http://metascape.org) (85). The genes were mapped to their human orthologs, and the lists were submitted to Metascape for enrichment analysis of the significant representation of GO Biological Process, KEGG pathway, Reactome Gene Sets, and CORUM. All genes in the genome were used as the enrichment background. Cutoffs for significantly enriched terms were p < 0.01, minimum count of 3 and the enrichment factor >1.5. The terms were grouped into clusters based on their membership similarities.
Analysis of TADs
Based on the normalized 20 kb contact matrices, TADs were identified by using the DI, following a previously reported method (25). The DI was calculated up to 2 Mb flanking the center of each bin at 20 kb resolution, and the Hidden Markov model was then used to predict DI states for final TAD generation. The insulation score for each 20 kb bin was calculated as previously reported (86). The correlations of TAD architecture between samples were assessed by Jaccard indices (53), and aggregate Hi-C maps were constructed as previously reported (87). To quantify the tendency of TADs to self-interact, we calculated the D-score for each TAD, according to a previously described method (53). Topologically associating domain boundaries between TADs were smaller than 400 kb, and the regions over 400 kb were defined as unorganized chromatin. Cell type-specific TAD boundaries were identified as previously reported (25). To investigate TAD interaction networks, we defined TAD cliques as clusters of five or more interacting TADs in the Hi-C data, as previously reported (54).
Promoter-enhancer interactions identification and RP index calculation
To identify PEIs at the resolution of 5 kb, we generated aggregated Hi-C maps for each cell type. Promoter-enhancer interactions were identified by applying PSYCHIC based on the 5 kb contact matrices (88). The genome was divided into TADs, and similar neighboring domains were further merged into a hierarchical structure. Then, a domain-specific background model was built according to the fitted bilinear power-law model for each or merged TADs. High-confidence PEIs were identified by interaction intensity normalized by the background model with FDR value <0.001 and interaction distance ≥15 kb.
Later, we calculated the RP index that combines both the number and intensity of the interacting enhancers to quantify the potential of PEIs for transcriptional regulation of target genes. RP = Σn[log10 (normalized interaction intensity of PEIs)], where n refers to the number of interacting enhancers. The normalized interaction intensity of PEIs was calculated by the observed contact frequency minus the background contact frequency.
Chromatin immunoprecipitation-sequencing library construction
The ChIP assay was conducted as previously described (89). In brief, 1.0 × 106 to 5.0 × 106 cells were crosslinked with 37% formaldehyde and then incubated with a glycine solution for 10 min to quench crosslinking. After washing with PBS, the cells were pelleted and lysed. Chromatins were sonicated to obtain the sheared 200 to 500 bp DNA. Around 20 μl chromatin was saved as input DNA at −20 °C, and 100 μl chromatin was used for immunoprecipitation with the H3K4me3 antibody (9751, Cell Signaling Technology) or the H3K27ac antibody (ab4729, Abcam). Approximately, 5 μg antibody was used in each immunoprecipitation reaction at 4 °C overnight. The next day, 30 μl protein A/G beads were added and subjected to further incubation for 3 h. After washing, the binding materials were eluted from the beads. The immunoprecipitated DNAs were then used to construct the ChIP-seq library, following the protocol provided by the manufacturer and sequenced on Illumina Xten with the PE 150 method.
Chromatin immunoprecipitation-sequencing data processing
Trimmomatic (version 0.38) was used to filter out low-quality reads (90). The cleaned ChIP-seq reads were aligned to the pig genome (Sscrofa 11.1), using the BWA (version 0.7.15) with default settings. The biological samples of each cell type were pooled using SAMtools (version 0.1.19). To identify enriched regions of active markers (H3K4me3 and H3K27ac), peak calling was performed using SICER (version 1.1).
Annotation of PEIs with ChIP-seq data
To define active enhancer-involved PEIs, we first identified REs and SEs by using the ROSE algorithm (57, 91). Next, we divided all SEs into two categories as previously reported, to which we referred as hierarchical and nonhierarchical SEs (58). The hierarchical SEs were then divided into hub and nonhub enhancers by applying a threshold value of H-score, which corresponds to the 90th percentile of Z-score. The active enhancer-involved PEIs were defined if the 5 kb enhancer bin overlapped with the identified enhancer by at least 1 bp.
Statistics
Statistical comparisons were conducted by using the Mann-Whitney U test (one-tailed), unless otherwise stated. A difference was considered statistically significant when p < 0.05.
Data availability
The raw and processed datasets generated in this study have been submitted to the NCBI SRA database and are available under the accession number PRJNA743697.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
Author contributions
Y. Zh., L. Z., L. J., P. Z., M. L., and W. Z. conceptualization; Y. Zh., L. Z., L. J., P. Z., F. L., M. G., Q. G., and Y. Ze. investigation; Y. Zh., L. Z., L. J., P. Z., and M. L. formal analysis; Y. Zh. and L. Z. writing–original draft; Y. Zh. and L. J. writing–review and editing; M. L. and W. Z. supervision.
Funding and additional information
This study was supported by National Natural Science Foundation of China to W. Z. (Grant No. 31772605) and Y. Zh. (Grant No. 32002178), the First-class University and Academic Program from Northwest A&F University to W. Z. (Grant No. Z102021906), and the Open Fund of Farm Animal Genetic Resources Exploration and Innovation Key Laboratory of Sichuan Province to Y. Zh. (Grant No. SNDK-KF-201804).
Edited by Qi Qun Tang
Contributor Information
Mingzhou Li, Email: mingzhou.li@sicau.edu.cn.
Wenxian Zeng, Email: zengwenxian2015@126.com.
Supporting information
Hi-C and RNA-seq data summaries.
List of genes subjected to the A-B/B-A/A-A/B-B change.
GO-BP analysis of genes subjected to the A-B/B-A/A-A/B-B change.
List of genes harbored in Un-SG- or Di-SG-specific TAD boundaries.
GO-BP analysis of genes harbored in Un-SG- or Di-SG-specific TAD boundaries.
List of genes with differential RP indices.
GO-BP analysis of genes with differential RP indices.
List of genes with Un-SG- or Di-SG-exclusive PEI regulation.
List of RE- and hierarchically organized SE-associated genes.
GO-BP analysis of RE and SE-regulated genes with differential RP indices and expression levels.
The correlation of normalized Hi-C interaction matrices between Un-SG and Di-SG samples illustrated by QuASAR-Rep (A), GenomeDISCO (B), Pearson correlation analysis (C) and PCA plot (D).
A/B compartment changes during spermatogonial differentiation. A, a schematic overview illustrating the proportions of genomic regions subjected to A/B compartment changes (A-A or B-B) between Un-SG and Di-SG. B, the average expression levels of genes that underwent the A-A or B-B change. P: Mann-Whitney U test, one-tailed. C, the numbers of genes that underwent the A-A or B-B change. D and E, GO-BP analysis of genes that underwent the A-A (D) or B-B change (E).
TAD cliques in spermatogonial subgroups. A, Pearson correlation analysis illustrating the correlation of TAD cliques between Un-SG (left) and Di-SG (right) ingroup samples. B, the numbers of TAD cliques in Un-SG and Di-SG. Data points refer to eight biological samples. C, the genome coverage by TAD cliques in Un-SG and Di-SG. Data points refer to eight biological samples. D, the proportions of TADs in cliques and non-cliques. Data are presented as the mean ± SD of eight biological samples. E, the proportions of TAD cliques in A/B compartments. Data are presented as the mean ± SD of eight biological samples.
PEI regulation of CATSPER2 (A), DND1 (B) and EZH2 (C) in Un-SG and Di-SG. The upper panel, the contact matrix showing stripes at the CATSPER2 (A), DND1 (B) and EZH2 (C) SE loci. The middle panel, models for PEI regulation of CATSPER2 (A), DND1 (B) and EZH2 (C), as well as RNA-seq coverage at their genomic loci. The lower panel, views of the observed/expected chromatin interaction frequencies, RefSeq, TAD and A/B index at chromosome 1: 127,310,000 to 128,310,000 (A), chromosome 2: 141,890,000 to 142,890,000 (B) and chromosome 9: 108,885,000 to 109,885,000 (C).
References
- 1.Johnson L., Petty C.S., Neaves W.B. A comparative study of daily sperm production and testicular composition in humans and rats. Biol. Reprod. 1980;22:1233–1243. doi: 10.1093/biolreprod/22.5.1233. [DOI] [PubMed] [Google Scholar]
- 2.Jan S.Z., Hamer G., Repping S., de Rooij D.G., van Pelt A.M.M., Vormer T.L. Molecular control of rodent spermatogenesis. Biochim. Biophys. Acta. 2012;1822:1838–1850. doi: 10.1016/j.bbadis.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 3.de Rooij D.G. The nature and dynamics of spermatogonial stem cells. Development. 2017;144:3022–3030. doi: 10.1242/dev.146571. [DOI] [PubMed] [Google Scholar]
- 4.Makela J.A., Hobbs R.M. Molecular regulation of spermatogonial stem cell renewal and differentiation. Reproduction. 2019;158:R169–R187. doi: 10.1530/REP-18-0476. [DOI] [PubMed] [Google Scholar]
- 5.Kubota H., Brinster R.L. Spermatogonial stem cells. Biol. Reprod. 2018;99:52–74. doi: 10.1093/biolre/ioy077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mulder C.L., Zheng Y., Jan S.Z., Struijk R.B., Repping S., Hamer G., van Pelt A.M. Spermatogonial stem cell autotransplantation and germline genomic editing: A future cure for spermatogenic failure and prevention of transmission of genomic diseases. Hum. Reprod. Update. 2016;22:561–573. doi: 10.1093/humupd/dmw017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lord T., Oatley J.M. A revised Asingle model to explain stem cell dynamics in the mouse male germline. Reproduction. 2017;154:R55–R64. doi: 10.1530/REP-17-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suzuki S., Diaz V.D., Hermann B.P. What has single-cell RNA-seq taught us about mammalian spermatogenesis? Biol. Reprod. 2019;101:617–634. doi: 10.1093/biolre/ioz088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tan K., Wilkinson M.F. Human spermatogonial stem cells scrutinized under the single-cell magnifying glass. Cell Stem Cell. 2019;24:201–203. doi: 10.1016/j.stem.2019.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tan K., Wilkinson M.F. A single-cell view of spermatogonial stem cells. Curr. Opin. Cell Biol. 2020;67:71–78. doi: 10.1016/j.ceb.2020.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo J., Grow E.J., Yi C., Mlcochova H., Maher G.J., Lindskog C., Murphy P.J., Wike C.L., Carrell D.T., Goriely A., Hotaling J.M., Cairns B.R. Chromatin and single-cell RNA-seq profiling reveal dynamic signaling and metabolic transitions during human spermatogonial stem cell development. Cell Stem Cell. 2017;21:533–546.e6. doi: 10.1016/j.stem.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hammoud S.S., Low D.H., Yi C., Carrell D.T., Guccione E., Cairns B.R. Chromatin and transcription transitions of mammalian adult germline stem cells and spermatogenesis. Cell Stem Cell. 2014;15:239–253. doi: 10.1016/j.stem.2014.04.006. [DOI] [PubMed] [Google Scholar]
- 13.Lesch B.J., Silber S.J., McCarrey J.R., Page D.C. Parallel evolution of male germline epigenetic poising and somatic development in animals. Nat. Genet. 2016;48:888–894. doi: 10.1038/ng.3591. [DOI] [PubMed] [Google Scholar]
- 14.Maezawa S., Yukawa M., Alavattam K.G., Barski A., Namekawa S.H. Dynamic reorganization of open chromatin underlies diverse transcriptomes during spermatogenesis. Nucleic Acids Res. 2018;46:593–608. doi: 10.1093/nar/gkx1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen W., Zhang Z., Chang C., Yang Z., Wang P., Fu H., Wei X., Chen E., Tan S., Huang W., Sun L., Ni T., Yang Y., Wang Y. A bioenergetic shift is required for spermatogonial differentiation. Cell Discov. 2020;6:56. doi: 10.1038/s41421-020-0183-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jan S.Z., Vormer T.L., Jongejan A., Roling M.D., Silber S.J., de Rooij D.G., Hamer G., Repping S., van Pelt A.M.M. Unraveling transcriptome dynamics in human spermatogenesis. Development. 2017;144:3659–3673. doi: 10.1242/dev.152413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharma S., Wistuba J., Pock T., Schlatt S., Neuhaus N. Spermatogonial stem cells: Updates from specification to clinical relevance. Hum. Reprod. Update. 2019;25:275–297. doi: 10.1093/humupd/dmz006. [DOI] [PubMed] [Google Scholar]
- 18.Cheng K., Chen I.C., Cheng C.E., Mutoji K., Hale B.J., Hermann B.P., Geyer C.B., Oatley J.M., McCarrey J.R. Unique epigenetic programming distinguishes regenerative spermatogonial stem cells in the developing mouse testis. iScience. 2020;23:101596. doi: 10.1016/j.isci.2020.101596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Rooij D.G., Griswold M.D. Questions about spermatogonia posed and answered since 2000. J. Androl. 2012;33:1085–1095. doi: 10.2164/jandrol.112.016832. [DOI] [PubMed] [Google Scholar]
- 20.de Rooij D.G., Russell L.D. All you wanted to know about spermatogonia but were afraid to ask. J. Androl. 2000;21:776–798. [PubMed] [Google Scholar]
- 21.Chiarini-Garcia H., Russell L.D. High-resolution light microscopic characterization of mouse spermatogonia. Biol. Reprod. 2001;65:1170–1178. doi: 10.1095/biolreprod65.4.1170. [DOI] [PubMed] [Google Scholar]
- 22.Chiarini-Garcia H., Russell L.D. Characterization of mouse spermatogonia by transmission electron microscopy. Reproduction. 2002;123:567–577. doi: 10.1530/rep.0.1230567. [DOI] [PubMed] [Google Scholar]
- 23.Lieberman-Aiden E., van Berkum N.L., Williams L., Imakaev M., Ragoczy T., Telling A., Amit I., Lajoie B.R., Sabo P.J., Dorschner M.O., Sandstrom R., Bernstein B., Bender M.A., Groudine M., Gnirke A., et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Belton J.M., McCord R.P., Gibcus J.H., Naumova N., Zhan Y., Dekker J. Hi-C: A comprehensive technique to capture the conformation of genomes. Methods. 2012;58:268–276. doi: 10.1016/j.ymeth.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dixon J.R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y., Hu M., Liu J.S., Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rao S.S.P., Huntley M.H., Durand N.C., Stamenova E.K., Bochkov I.D., Robinson J.T., Sanborn A.L., Machol I., Omer A.D., Lander E.S., Aiden E.L. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smallwood A., Ren B. Genome organization and long-range regulation of gene expression by enhancers. Curr. Opin. Cell Biol. 2013;25:387–394. doi: 10.1016/j.ceb.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gorkin D.U., Leung D., Ren B. The 3D genome in transcriptional regulation and pluripotency. Cell Stem Cell. 2014;14:762–775. doi: 10.1016/j.stem.2014.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Swindle M.M., Makin A., Herron A.J., Clubb F.J., Frazier K.S. Swine as models in biomedical research and toxicology testing. Vet. Pathol. 2012;49:344–356. doi: 10.1177/0300985811402846. [DOI] [PubMed] [Google Scholar]
- 30.Voigt A.L., Kondro D.A., Powell D., Valli-Pulaski H., Ungrin M., Stukenborg J.B., Klein C., Lewis I.A., Orwig K.E., Dobrinski I. Unique metabolic phenotype and its transition during maturation of juvenile male germ cells. FASEB J. 2021;35 doi: 10.1096/fj.202002799R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang P., Li F., Zhang L., Lei P., Zheng Y., Zeng W. Stage-specific embryonic antigen 4 is a membrane marker for enrichment of porcine spermatogonial stem cells. Andrology. 2020;8:1923–1934. doi: 10.1111/andr.12870. [DOI] [PubMed] [Google Scholar]
- 32.Bryant J.M., Meyer-Ficca M.L., Dang V.M., Berger S.L., Meyer R.G. Separation of spermatogenic cell types using STA-PUT velocity sedimentation. J. Vis. Exp. 2013 doi: 10.3791/50648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Y., Niu M., Yao C., Hai Y., Yuan Q., Liu Y., Guo Y., Li Z., He Z. Fractionation of human spermatogenic cells using STA-PUT gravity sedimentation and their miRNA profiling. Sci. Rep. 2015;5:8084. doi: 10.1038/srep08084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seaman L., Rajapakse I. 4D nucleome analysis toolbox: Analysis of Hi-C data with abnormal karyotype and time series capabilities. Bioinformatics. 2018;34:104–106. doi: 10.1093/bioinformatics/btx484. [DOI] [PubMed] [Google Scholar]
- 35.Lindsly S., Chen C., Liu S., Ronquist S., Dilworth S., Perlman M., Rajapakse I. 4DNvestigator: Time series genomic data analysis toolbox. Nucleus. 2021;12:58–64. doi: 10.1080/19491034.2021.1910437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.MacArthur B.D., Lemischka I.R. Statistical mechanics of pluripotency. Cell. 2013;154:484–489. doi: 10.1016/j.cell.2013.07.024. [DOI] [PubMed] [Google Scholar]
- 37.Rajapakse I., Groudine M., Mesbahi M. What can systems theory of networks offer to biology? PLoS Comput. Biol. 2012;8 doi: 10.1371/journal.pcbi.1002543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Naumova N., Imakaev M., Fudenberg G., Zhan Y., Lajoie B.R., Mirny L.A., Dekker J. Organization of the mitotic chromosome. Science. 2013;342:948–953. doi: 10.1126/science.1236083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luo Z.Y., Wang X.R., Jiang H., Wang R.Y., Chen J., Chen Y.S., Xu Q.L., Cao J., Gong X.W., Wu J., Yang Y.G., Li W.B., Han C.S., Cheng C.Y., Rosenfeld M.G., et al. Reorganized 3D genome structures support transcriptional regulation in mouse spermatogenesis. iScience. 2020;23:101034. doi: 10.1016/j.isci.2020.101034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Y., Wang H.B., Zhang Y., Du Z.H., Si W., Fan S.X., Qin D.D., Wang M., Duan Y.C., Li L.F., Jiao Y.Y., Li Y.Y., Wang Q.J., Shi Q.H., Wu X., et al. Reprogramming of meiotic chromatin architecture during spermatogenesis. Mol. Cell. 2019;73:547–561.e6. doi: 10.1016/j.molcel.2018.11.019. [DOI] [PubMed] [Google Scholar]
- 41.He M., Li Y., Tang Q., Li D., Jin L., Tian S., Che T., He S., Deng L., Gao G., Gu Y., Jiang Z., Li X., Li M. Genome-wide chromatin structure changes during adipogenesis and myogenesis. Int. J. Biol. Sci. 2018;14:1571–1585. doi: 10.7150/ijbs.25328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nora E.P., Lajoie B.R., Schulz E.G., Giorgetti L., Okamoto I., Servant N., Piolot T., van Berkum N.L., Meisig J., Sedat J., Gribnau J., Barillot E., Bluthgen N., Dekker J., Heard E. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fraser J., Ferrai C., Chiariello A.M., Schueler M., Rito T., Laudanno G., Barbieri M., Moore B.L., Kraemer D.C., Aitken S., Xie S.Q., Morris K.J., Itoh M., Kawaji H., Jaeger I., et al. Hierarchical folding and reorganization of chromosomes are linked to transcriptional changes in cellular differentiation. Mol. Syst. Biol. 2015;11:852. doi: 10.15252/msb.20156492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rubin A.J., Barajas B.C., Furlan-Magaril M., Lopez-Pajares V., Mumbach M.R., Howard I., Kim D.S., Boxer L.D., Cairns J., Spivakov M., Wingett S.W., Shi M., Zhao Z., Greenleaf W.J., Kundaje A., et al. Lineage-specific dynamic and pre-established enhancer-promoter contacts cooperate in terminal differentiation. Nat. Genet. 2017;49:1522–1528. doi: 10.1038/ng.3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Siersbaek R., Madsen J.G.S., Javierre B.M., Nielsen R., Bagge E.K., Cairns J., Wingett S.W., Traynor S., Spivakov M., Fraser P., Mandrup S. Dynamic rewiring of promoter-anchored chromatin loops during adipocyte differentiation. Mol. Cell. 2017;66:420–435.e5. doi: 10.1016/j.molcel.2017.04.010. [DOI] [PubMed] [Google Scholar]
- 46.Vara C., Paytuvi-Gallart A., Cuartero Y., Le Dily F., Garcia F., Salva-Castro J., Gomez-H L., Julia E., Moutinho C., Cigliano R.A., Sanseverino W., Fornas O., Pendas A.M., Heyn H., Waters P.D., et al. Three-dimensional genomic structure and Cohesin occupancy correlate with transcriptional activity during spermatogenesis. Cell Rep. 2019;28:352–367.e9. doi: 10.1016/j.celrep.2019.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Patel L., Kang R., Rosenberg S.C., Qiu Y.J., Raviram R., Chee S., Hu R., Ren B., Cole F., Corbett K.D. Dynamic reorganization of the genome shapes the recombination landscape in meiotic prophase. Nat. Struct. Mol. Biol. 2019;26:164–174. doi: 10.1038/s41594-019-0187-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alavattam K.G., Maezawa S., Sakashita A., Khoury H., Barski A., Kaplan N., Namekawa S.H. Attenuated chromatin compartmentalization in meiosis and its maturation in sperm development. Nat. Struct. Mol. Biol. 2019;26:175–184. doi: 10.1038/s41594-019-0189-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ke Y., Xu Y., Chen X., Feng S., Liu Z., Sun Y., Yao X., Li F., Zhu W., Gao L., Chen H., Du Z., Xie W., Xu X., Huang X., et al. 3D chromatin structures of mature gametes and structural reprogramming during mammalian embryogenesis. Cell. 2017;170:367–381.e20. doi: 10.1016/j.cell.2017.06.029. [DOI] [PubMed] [Google Scholar]
- 50.Hug C.B., Grimaldi A.G., Kruse K., Vaquerizas J.M. Chromatin architecture emerges during zygotic genome activation independent of transcription. Cell. 2017;169:216–228.e19. doi: 10.1016/j.cell.2017.03.024. [DOI] [PubMed] [Google Scholar]
- 51.Du Z., Zheng H., Huang B., Ma R., Wu J., Zhang X., He J., Xiang Y., Wang Q., Li Y., Ma J., Zhang X., Zhang K., Wang Y., Zhang M.Q., et al. Allelic reprogramming of 3D chromatin architecture during early mammalian development. Nature. 2017;547:232–235. doi: 10.1038/nature23263. [DOI] [PubMed] [Google Scholar]
- 52.Krijger P.H., Di Stefano B., de Wit E., Limone F., van Oevelen C., de Laat W., Graf T. Cell-of-origin-specific 3D genome structure acquired during somatic cell reprogramming. Cell Stem Cell. 2016;18:597–610. doi: 10.1016/j.stem.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stadhouders R., Vidal E., Serra F., Di Stefano B., Le Dily F., Quilez J., Gomez A., Collombet S., Berenguer C., Cuartero Y., Hecht J., Filion G.J., Beato M., Marti-Renom M.A., Graf T. Transcription factors orchestrate dynamic interplay between genome topology and gene regulation during cell reprogramming. Nat. Genet. 2018;50:238–249. doi: 10.1038/s41588-017-0030-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Paulsen J., Liyakat Ali T.M., Nekrasov M., Delbarre E., Baudement M.O., Kurscheid S., Tremethick D., Collas P. Long-range interactions between topologically associating domains shape the four-dimensional genome during differentiation. Nat. Genet. 2019;51:835–843. doi: 10.1038/s41588-019-0392-0. [DOI] [PubMed] [Google Scholar]
- 55.Mifsud B., Tavares-Cadete F., Young A.N., Sugar R., Schoenfelder S., Ferreira L., Wingett S.W., Andrews S., Grey W., Ewels P.A., Herman B., Happe S., Higgs A., LeProust E., Follows G.A., et al. Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C. Nat. Genet. 2015;47:598–606. doi: 10.1038/ng.3286. [DOI] [PubMed] [Google Scholar]
- 56.Schoenfelder S., Furlan-Magaril M., Mifsud B., Tavares-Cadete F., Sugar R., Javierre B.M., Nagano T., Katsman Y., Sakthidevi M., Wingett S.W., Dimitrova E., Dimond A., Edelman L.B., Elderkin S., Tabbada K., et al. The pluripotent regulatory circuitry connecting promoters to their long-range interacting elements. Genome Res. 2015;25:582–597. doi: 10.1101/gr.185272.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Whyte W.A., Orlando D.A., Hnisz D., Abraham B.J., Lin C.Y., Kagey M.H., Rahl P.B., Lee T.I., Young R.A. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang J., Li K., Cai W., Liu X., Zhang Y., Orkin S.H., Xu J., Yuan G.C. Dissecting super-enhancer hierarchy based on chromatin interactions. Nat. Commun. 2018;9:943. doi: 10.1038/s41467-018-03279-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hnisz D., Abraham B.J., Lee T.I., Lau A., Saint-Andre V., Sigova A.A., Hoke H.A., Young R.A. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Parker S.C., Stitzel M.L., Taylor D.L., Orozco J.M., Erdos M.R., Akiyama J.A., van Bueren K.L., Chines P.S., Narisu N., NISC Comparative Sequencing Program, Black B.L., Visel A., Pennacchio L.A., Collins F.S., National Institutes of Health Intramural Sequencing Center Comparative Sequencing Program Authors, et al. Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants. Proc. Natl. Acad. Sci. U. S. A. 2013;110:17921–17926. doi: 10.1073/pnas.1317023110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kagawa W., Kurumizaka H. From meiosis to postmeiotic events: Uncovering the molecular roles of the meiosis-specific recombinase Dmc1. FEBS J. 2010;277:590–598. doi: 10.1111/j.1742-4658.2009.07503.x. [DOI] [PubMed] [Google Scholar]
- 62.Visser L., Westerveld G.H., Xie F., van Daalen S.K., van der Veen F., Lombardi M.P., Repping S. A comprehensive gene mutation screen in men with asthenozoospermia. Fertil. Steril. 2011;95:1020–1024.e1-9. doi: 10.1016/j.fertnstert.2010.11.067. [DOI] [PubMed] [Google Scholar]
- 63.Yang F., Whelan E.C., Guan X., Deng B., Wang S., Sun J., Avarbock M.R., Wu X., Brinster R.L. FGF9 promotes mouse spermatogonial stem cell proliferation mediated by p38 MAPK signalling. Cell Prolif. 2020;54 doi: 10.1111/cpr.12933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Niimi Y., Imai A., Nishimura H., Yui K., Kikuchi A., Koike H., Saga Y., Suzuki A. Essential role of mouse Dead end1 in the maintenance of spermatogonia. Dev. Biol. 2019;445:103–112. doi: 10.1016/j.ydbio.2018.11.003. [DOI] [PubMed] [Google Scholar]
- 65.Jin C., Zhang Y., Wang Z.P., Wang X.X., Sun T.C., Li X.Y., Tang J.X., Cheng J.M., Li J., Chen S.R., Deng S.L., Liu Y.X. EZH2 deletion promotes spermatogonial differentiation and apoptosis. Reproduction. 2017;154:615–625. doi: 10.1530/REP-17-0302. [DOI] [PubMed] [Google Scholar]
- 66.Fayomi A.P., Orwig K.E. Spermatogonial stem cells and spermatogenesis in mice, monkeys and men. Stem Cell Res. 2018;29:207–214. doi: 10.1016/j.scr.2018.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Law N.C., Oatley J.M. Developmental underpinnings of spermatogonial stem cell establishment. Andrology. 2020;8:852–861. doi: 10.1111/andr.12810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Culty M. Gonocytes, from the fifties to the present: Is there a reason to change the name? Biol. Reprod. 2013;89:46. doi: 10.1095/biolreprod.113.110544. [DOI] [PubMed] [Google Scholar]
- 69.Manku G., Culty M. Mammalian gonocyte and spermatogonia differentiation: Recent advances and remaining challenges. Reproduction. 2015;149:R139–R157. doi: 10.1530/REP-14-0431. [DOI] [PubMed] [Google Scholar]
- 70.Niedenberger B.A., Busada J.T., Geyer C.B. Marker expression reveals heterogeneity of spermatogonia in the neonatal mouse testis. Reproduction. 2015;149:329–338. doi: 10.1530/REP-14-0653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shami A.N., Zheng X., Munyoki S.K., Ma Q., Manske G.L., Green C.D., Sukhwani M., Orwig K.E., Li J.Z., Hammoud S.S. Single-cell RNA sequencing of human, macaque, and mouse testes uncovers conserved and divergent features of mammalian spermatogenesis. Dev. Cell. 2020;54:529–547.e12. doi: 10.1016/j.devcel.2020.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lau X., Munusamy P., Ng M.J., Sangrithi M. Single-cell RNA sequencing of the cynomolgus macaque testis reveals conserved transcriptional profiles during mammalian spermatogenesis. Dev. Cell. 2020;54:548–566.e7. doi: 10.1016/j.devcel.2020.07.018. [DOI] [PubMed] [Google Scholar]
- 73.Dixon J.R., Jung I., Selvaraj S., Shen Y., Antosiewicz-Bourget J.E., Lee A.Y., Ye Z., Kim A., Rajagopal N., Xie W., Diao Y., Liang J., Zhao H., Lobanenkov V.V., Ecker J.R., et al. Chromatin architecture reorganization during stem cell differentiation. Nature. 2015;518:331–336. doi: 10.1038/nature14222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Caldeira-Brant A.L., Martinelli L.M., Marques M.M., Reis A.B., Martello R., Almeida F., Chiarini-Garcia H. A subpopulation of human Adark spermatogonia behaves as the reserve stem cell. Reproduction. 2020;159:437–451. doi: 10.1530/REP-19-0254. [DOI] [PubMed] [Google Scholar]
- 75.Lord T., Nixon B. Metabolic changes accompanying spermatogonial stem cell differentiation. Dev. Cell. 2020;52:399–411. doi: 10.1016/j.devcel.2020.01.014. [DOI] [PubMed] [Google Scholar]
- 76.Zheng Y., Lei Q., Jongejan A., Mulder C.L., van Daalen S.K.M., Mastenbroek S., Hwang G., Jordan P.W., Repping S., Hamer G. The influence of retinoic acid-induced differentiation on the radiation response of male germline stem cells. DNA Repair. 2018;70:55–66. doi: 10.1016/j.dnarep.2018.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wu Y., Zhou H., Fan X., Zhang Y., Zhang M., Wang Y., Xie Z., Bai M., Yin Q., Liang D., Tang W., Liao J., Zhou C., Liu W., Zhu P., et al. Correction of a genetic disease by CRISPR-Cas9-mediated gene editing in mouse spermatogonial stem cells. Cell Res. 2015;25:67–79. doi: 10.1038/cr.2014.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sato T., Sakuma T., Yokonishi T., Katagiri K., Kamimura S., Ogonuki N., Ogura A., Yamamoto T., Ogawa T. Genome editing in mouse spermatogonial stem cell lines using TALEN and double-nicking CRISPR/Cas9. Stem Cell Rep. 2015;5:75–82. doi: 10.1016/j.stemcr.2015.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zheng Y., Jongejan A., Mulder C.L., Mastenbroek S., Repping S., Wang Y., Li J., Hamer G. Trivial role for NSMCE2 during in vitro proliferation and differentiation of male germline stem cells. Reproduction. 2017;154:181–195. doi: 10.1530/REP-17-0173. [DOI] [PubMed] [Google Scholar]
- 80.de Rooij D.G., de Boer P. Specific arrests of spermatogenesis in genetically modified and mutant mice. Cytogenet. Genome Res. 2003;103:267–276. doi: 10.1159/000076812. [DOI] [PubMed] [Google Scholar]
- 81.Lei Q., Lai X., Eliveld J., Chuva de Sousa Lopes S.M., van Pelt A.M.M., Hamer G. In vitro meiosis of male germline stem cells. Stem Cell Rep. 2020;15:1140–1153. doi: 10.1016/j.stemcr.2020.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yardimci G.G., Ozadam H., Sauria M.E.G., Ursu O., Yan K.K., Yang T., Chakraborty A., Kaul A., Lajoie B.R., Song F., Zhan Y., Ay F., Gerstein M., Kundaje A., Li Q., et al. Measuring the reproducibility and quality of Hi-C data. Genome Biol. 2019;20:57. doi: 10.1186/s13059-019-1658-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rowley M.J., Nichols M.H., Lyu X., Ando-Kuri M., Rivera I.S.M., Hermetz K., Wang P., Ruan Y., Corces V.G. Evolutionarily conserved principles predict 3D chromatin organization. Mol. Cell. 2017;67:837–852.e7. doi: 10.1016/j.molcel.2017.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Flyamer I.M., Gassler J., Imakaev M., Brandao H.B., Ulianov S.V., Abdennur N., Razin S.V., Mirny L.A., Tachibana-Konwalski K. Single-nucleus Hi-C reveals unique chromatin reorganization at oocyte-to-zygote transition. Nature. 2017;544:110–114. doi: 10.1038/nature21711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Conn S.J., Pillman K.A., Toubia J., Conn V.M., Salmanidis M., Phillips C.A., Roslan S., Schreiber A.W., Gregory P.A., Goodall G.J. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125–1134. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 86.Crane E., Bian Q., McCord R.P., Lajoie B.R., Wheeler B.S., Ralston E.J., Uzawa S., Dekker J., Meyer B.J. Condensin-driven remodelling of X chromosome topology during dosage compensation. Nature. 2015;523:240–244. doi: 10.1038/nature14450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bonev B., Mendelson Cohen N., Szabo Q., Fritsch L., Papadopoulos G.L., Lubling Y., Xu X., Lv X., Hugnot J.P., Tanay A., Cavalli G. Multiscale 3D genome rewiring during mouse neural development. Cell. 2017;171:557–572.e24. doi: 10.1016/j.cell.2017.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ron G., Globerson Y., Moran D., Kaplan T. Promoter-enhancer interactions identified from Hi-C data using probabilistic models and hierarchical topological domains. Nat. Commun. 2017;8:2237. doi: 10.1038/s41467-017-02386-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Han K., Ren R., Cao J., Zhao S., Yu M. Genome-wide identification of histone modifications involved in placental development in pigs. Front. Genet. 2019;10:277. doi: 10.3389/fgene.2019.00277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bolger A.M., Lohse M., Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Loven J., Hoke H.A., Lin C.Y., Lau A., Orlando D.A., Vakoc C.R., Bradner J.E., Lee T.I., Young R.A. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Hi-C and RNA-seq data summaries.
List of genes subjected to the A-B/B-A/A-A/B-B change.
GO-BP analysis of genes subjected to the A-B/B-A/A-A/B-B change.
List of genes harbored in Un-SG- or Di-SG-specific TAD boundaries.
GO-BP analysis of genes harbored in Un-SG- or Di-SG-specific TAD boundaries.
List of genes with differential RP indices.
GO-BP analysis of genes with differential RP indices.
List of genes with Un-SG- or Di-SG-exclusive PEI regulation.
List of RE- and hierarchically organized SE-associated genes.
GO-BP analysis of RE and SE-regulated genes with differential RP indices and expression levels.
The correlation of normalized Hi-C interaction matrices between Un-SG and Di-SG samples illustrated by QuASAR-Rep (A), GenomeDISCO (B), Pearson correlation analysis (C) and PCA plot (D).
A/B compartment changes during spermatogonial differentiation. A, a schematic overview illustrating the proportions of genomic regions subjected to A/B compartment changes (A-A or B-B) between Un-SG and Di-SG. B, the average expression levels of genes that underwent the A-A or B-B change. P: Mann-Whitney U test, one-tailed. C, the numbers of genes that underwent the A-A or B-B change. D and E, GO-BP analysis of genes that underwent the A-A (D) or B-B change (E).
TAD cliques in spermatogonial subgroups. A, Pearson correlation analysis illustrating the correlation of TAD cliques between Un-SG (left) and Di-SG (right) ingroup samples. B, the numbers of TAD cliques in Un-SG and Di-SG. Data points refer to eight biological samples. C, the genome coverage by TAD cliques in Un-SG and Di-SG. Data points refer to eight biological samples. D, the proportions of TADs in cliques and non-cliques. Data are presented as the mean ± SD of eight biological samples. E, the proportions of TAD cliques in A/B compartments. Data are presented as the mean ± SD of eight biological samples.
PEI regulation of CATSPER2 (A), DND1 (B) and EZH2 (C) in Un-SG and Di-SG. The upper panel, the contact matrix showing stripes at the CATSPER2 (A), DND1 (B) and EZH2 (C) SE loci. The middle panel, models for PEI regulation of CATSPER2 (A), DND1 (B) and EZH2 (C), as well as RNA-seq coverage at their genomic loci. The lower panel, views of the observed/expected chromatin interaction frequencies, RefSeq, TAD and A/B index at chromosome 1: 127,310,000 to 128,310,000 (A), chromosome 2: 141,890,000 to 142,890,000 (B) and chromosome 9: 108,885,000 to 109,885,000 (C).
Data Availability Statement
The raw and processed datasets generated in this study have been submitted to the NCBI SRA database and are available under the accession number PRJNA743697.