Abstract
The purpose of our experiment was to explore how stochastic (inter‐individual variation) gut microbiome composition may link to inflammatory bowel disease (IBD) susceptibility and guide the development of a perinatal preventative probiotic. Dextran sodium sulfate (DSS) was introduced to C57BL/BJ mice to induce acute colitis as a model of IBD. Potentially protective bacteria were identified using a discovery‐validation cohort approach toward stochastic DSS susceptibility. Lactobacilli (two different cocktails of L. reuteri and L. johnsonii strains) or control media were supplemented by mouth to dams prior to delivery and during lactation (i.e., perinatal probiotic). The pups were evaluated for DSS susceptibility at young adulthood. Fecal Lactobacillus was increased in the DSS‐resistant mice in both the discovery and validation cohorts. Maternal supplementation of female offspring with an L. reuteri cocktail (strains 6798‐1, 6798‐jm, and 6798‐cm) induced progressive microbiome separation and protection against colitis by young adulthood. Maternal supplementation of L. reuteri could confer protection against DSS colitis in young adult female mice. This work is the first to exploit stochastic mammalian microbiome variation to guide microbial therapeutic identification. Our findings underscore neonatal microbiome plasticity and set the stage for the potential development of perinatally deliverable protective probiotics against human IBD.
Keywords: inflammatory bowel disease, Lactobacillus, microbiota, pediatric, probiotic, stochastic
Abbreviations
- CD
Crohn's disease
- DSS
dextran sulfate sodium
- DSS‐R
DSS‐resistant
- DSS‐S
DSS‐sensitive
- IBDs
inflammatory bowel diseases
- LJ‐PS
L. johnsonii probiotic supplementation
- LR‐PS
Lactobacillus reuteri probiotic supplementation
- PCoA
principal coordinate analysis
- PX
postnatal day X
- UC
ulcerative colitis
1. INTRODUCTION
Inflammatory bowel diseases (IBDs) are categorized into two major subtypes: ulcerative colitis (UC) and Crohn's disease (CD). These disorders are chronic inflammatory conditions of the digestive tract with unknown etiology. The peak onset of IBD occurs during young adulthood (second or third decade of life). 1 IBD incidence has increased in developed countries over the second half of the 20th century and more recently in populations adopting a westernized lifestyle. 2 , 3 This increasing incidence, along with the difficulty to treat the diseases has led to a large healthcare and economic burden worldwide, including more than $5.5 billion annual cost in the USA alone. 4 Therefore, novel preventative and therapeutic measures that could reduce the morbidity and economic burden of these diseases are sorely needed.
Genetic predisposition, environmental influences, the immune system, and the gut microbiome are essential components in the development of IBD. 5 An altered or dysbiotic gut microbial system has been implicated in the pathogenesis of IBD. 6 , 7 Monozygotic twin discordance in IBD is as high as 60% in CD and 85% in UC (reviewed in Ref. [4]). Monozygotic twins (genetically identical) are most commonly exposed to the same environment, and have similar microbiomes 8 ; theoretically a concordance rate close to 100% should be observed. 9 , 10 Since this is not the case, stochastic variation in individual biological systems must play an important role in the etiology of IBD. 6 , 11 , 12 Therefore, the gut microbiome is a highly plausible target where stochastic, environmentally responsive changes may induce susceptibility toward IBD, and stochastic interactions may even be at play after the onset of the disease. Intriguingly, one of the largest undertakings on the human gut microbiome to date has recently concluded that dysbiotic periods in IBD patients are “potentially stochastic”. 13 Even more recently, Clooney et al. 14 recognized that 90.3% of microbiome compositional variance is stochastic or unexplained in 531 IBD patients and 161 controls from Ireland and Canada.
In humans, early microbiome development has been found to impact disease susceptibility. 15 Studies by Yatsunenko et al. 16 and Zhou et al. 10 found that inter‐individual variation was greater in younger children compared to older individuals. Yatsunenko et al. 16 observed greater microbiome variation in children younger than 3 years of age, compared to adults in Malawi, Venezuela, and the United States, suggesting that this observation is independent of geography. Zhou et al. 10 found the gut microbiome to stabilize after 1 year of age, and highlighted a change in microbial metabolic genes when comparing infants younger than 1 with infants older than 1. These observations suggest an age‐related metabolic link that influences the variation in microbiome diversity. Therefore, it may be within the first few years of life that the microbiome undergoes stochastic selectivity. This random selection is influenced by genetic 17 and epigenetic 18 predisposition, as well as postnatal environmental influences (such as early antibiotic exposure 19 ), which can lead to a microbiome‐gut interactome 5 that is more, or less susceptible toward developing IBD later in life. 20
In this study, we aimed to examine how stochastic microbiome composition may link to IBD susceptibility. We did this to determine if such compositional associations may provide a means to modulate early microbiome development in order to decrease subsequent susceptibility to intestinal inflammation. Inbred mice, although imperfect models of IBD, 21 are uniquely advantageous to examine stochastic microbiome variation, but they are less commonly recognized for this purpose. In particular, due to their genetically identical nature and their identical nurture within research facilities, they are optimal for studying random variations within the microbiome, largely independently from genetics and environment. In mice, inter‐individual variation, or stochasticity, has been found to have a significant effect on the microbiome. 22 In this particular study by Hildebrand, et al., 22 inter‐individual variation was found to contribute to 45.5% of the variance in the gut microbiome. Previous research indicated that inter‐individual microbiome variation is greater in some mouse strains like C57BL6/J compared to others such as C3HRI, DBAJR, and WSB. 23 , 24 In a study by McCafferty et al., 25 stochastic changes within each cage of mice were found to be the main cause of cage effects. Hufeldt et al. 26 observed that inter‐individual variation in the gut microbiome was decreased when the female breeders were closely related. Another study found that wild type (WT) C57BL/6 mice from vendors were more susceptible to developing colitis than WT C57BL/6 animals maintained and bred within a smaller animal facility. 27 Even though random inter‐individual variation has been observed in the mammalian microbiome, this variation has surprisingly not been studied in the context of intestinal inflammation, specifically in respect to IBD.
Here, we examined how random microbiome variation at different stages of development associates with early adulthood (when the incidence of IBD peaks in humans 1 ) susceptibility to dextran sulfate sodium (DSS) induced colitis. Our primary outcome was weight loss as an inherent surrogate marker of colitis severity in this model. Indeed, we have recently underscored that weight loss is a sufficient and economically feasible single outcome measure of murine DSS colitis. 28 Our goal was to identify potential probiotics that, if given during early postnatal development (even in a transient fashion) may prevent/decrease microbiome evolution toward an IBD prone composition.
2. MATERIALS AND METHODS
2.1. Animals in discovery‐validation cohort
Our novel approach is outlined in Figure S1. A discovery‐validation cohort method was used to investigate the impact of stochastic microbiome variation on dextran sodium sulfate (DSS) susceptibility. A discovery cohort of newly purchased 8‐week‐old C57BL/6J mice (n = 30) was studied after cage mixing and acclimation to our vivarium (i.e., vendor‐based discovery experiment). Stool in the discovery cohort was collected before experimental colitis induction at postnatal day 70 (P70, i.e., young adult mice). For the validation cohort, three breeding trios (one male, two females) of C57BL/6J mice were set up in our vivarium, and pups (n = 31) were studied (i.e., vivarium‐based validation experiment). Stool in the validation cohort was collected at P21 (weaning) and prior to experimental colitis induction at P70. For both cohorts, only male mice were used to limit potential gender‐based microbial and DSS sensitivity variation. 27
2.2. Administration of dextran sodium sulfate
At P70, 2.5% (wt/vol) dextran sodium sulfate (DSS) (MW = 36,000–50,000, MP Biomedicals, LLC) was administered to the mice through drinking water. Weight was monitored daily and DSS was stopped once mice started losing 5% of their original body weight.
2.3. 16S rRNA sequencing
The fecal microbiomes were analyzed by high throughput sequencing of the bacterial 16S rRNA gene at the Alkek Center for Metagenomics and Microbiome Research, and data was analyzed through ATIMA (Agile Toolkit for Incisive Microbial Analyses) developed by the Center for Metagenomics and Microbiome Research at Baylor College of Medicine, identical to Ihekweazu, et al. 29
2.4. Lactobacillus culture
The second portion of the study, as a proof of concept, examined the impact of early life Lactobacillus supplementation on subsequent DSS susceptibility in young adulthood (Figure S2). We selected L. reuteri and L. johnsonii species because they have been previously found to promote gut barrier function. 30 L. reuteri strains 6798‐1, 6798‐jm, and 6798‐cm and L. johnsonii strains 4901, 4903, 4931 were previously identified from mouse feces. 31 Only these two Lactobacillus species were used due to available expertise with the specific species at Baylor College of Medicine and previous postnatal studies in humans indicating early postnatal effects from Lactobacilli (see Section 4). Both cocktails were created from Lactobacillus strains originating from mice without colitis (Swiss Webster and inducible nitric oxide synthetase‐deficient C57BL/6 mice). 31 These Lactobacilli were cultured in an anaerobic workstation (Anaerobe Systems AS‐580) in a mixture of 5% CO2, 5% H2, and 90% N2. Colonies were grown in de Man, Rogosa and Sharpe (MRS) agar (Difco) anaerobically at 37°C overnight. Single colonies were used to inoculate MRS medium and grown anaerobically at 37°C overnight. After growth, Lactobacilli were pelleted by centrifuging at 5000 g for 5 min. Bacterial cells were washed three times with sterile anaerobic PBS to wash away residual MRS, and the bacterial pellet was suspended in anaerobic phosphate‐buffered saline (PBS) at 109 colony forming units (CFUs) per ml.
2.5. Lactobacillus supplementation
To determine if transient maternal supplement of Lactobacilli transfers a protective microbiome, we orally gavaged C57BL/6 dams (n = 6, 2 per group) with phosphate‐buffered saline (PBS), L. reuteri cocktail (strains 6798‐1, 6798‐jm, and 6798‐cm) or L. johnsonii cocktail (strains 4901, 4903, 4931). Both male and female offspring were subsequently examined. There were 39 offspring mice given the L. reuteri cocktail (24 male, 15 female), 41 given the L. johnsonii cocktail (19 male, 22 female), and 36 given PBS as a control (20 male, 16 female). The L. reuteri (strains 6798‐1, 6798‐jm, and 6798‐cm) and L. johnsonii (strains 4901, 4903, 4931) 31 cocktails were administered to the dams 1 week prior to giving birth, every other day, based on a previous experiment in which effective colonization of Bifidobacterium was transferred to the pups by gavaging the dams. 32 After birth, dams were left alone for a week before restarting every other day oral gavage for a week until weaning. The goal was to supplement the pups the desired Lactobacillus strains 1 week prior to delivery, and as intensely (but practically and safely) as possible through the last 2 weeks of lactation, through maternal transmission. Pre‐ and postnatally (during last 2 weeks of lactation) supplemented pups were weaned to a standard diet of our vivarium (PicoLab Select Rodent 50 IF/6F) and were maintained in 4/cage setting, with gender and with infantile probiotic treatment‐based separation. DSS (2.5%) was started on P70 and continued for 6 days. Daily weights were collected. Weight loss was examined as the single, sufficient and economically feasible measure of DSS colitis severity, as underscored by us recently. 28 Pup stools were collected at various points (P21 (weaning), P30, and P70 (pre‐DSS)). Fecal pellets from a select number of female mice (since only those had a variation in colitis phenotype) at each time point were sent for analysis to the Alkek Center for Metagenomics and Microbiome Research at Baylor College of Medicine.
2.6. Statistical analysis
For analysis of genera separation, the Mann–Whitney U‐test was utilized for comparisons between two groups (denoted as direct comparison), while the Kruskal–Wallis test was used for comparisons of three or more groups. The level of significance was set at p < 0.05. The protocol was approved by the Institutional Animal Care and Use Committee of Baylor College of Medicine (AN‐5351).
Analysis of variance (ANOVA) with repeated measures was performed using SAS 9.4 (SAS Institute Inc., 2007) to assess the percent changes in body weight in response to DSS‐induced colitis in P70 female pups that were exposed to maternal supplementation with L.reuteri (n = 15) versus control media (n = 16). The ANOVA with repeated measures was performed using PROC MIXED to assess the percent changes in body weight with main effects for treatment (L. reuteri or control media) and days post‐DSS treatment (days 0–11) and the treatment by day interaction. The Fisher's Least Significance Difference procedure was used to assess the nature of the differences observed with the significant treatment by day interaction. Statistical significance was set at p < 0.05.
We identified transient weight loss in a subgroup of mice on the second day of the experiment due to accidental water loss and transient dehydration. In order to address if this incident potentially influenced the experimental outcome, post‐hoc analysis was conducted with the exclusion of mice that were affected (Figure S3). The results of the analysis were similar to the original observations (see Figure 3 later), indicating that the transient water loss did not affect the difference we observed between the L. reuteri supplemented and the control females.
FIGURE 3.
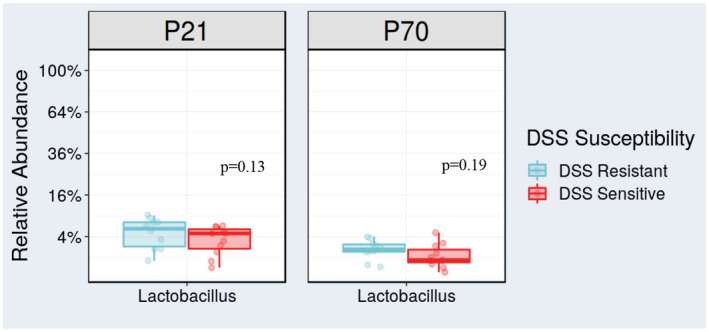
The validation cohort supported that increased Lactobacillus pre‐DSS may protect against colitis. Lactobacillus was increased in the DSS‐R group (mean = 2.07%) versus the DSS‐S group (mean = 1.40%) at P70. Although the difference in Lactobacillus was not significant (p = 0.19), it was one of the distinguishing genera between DSS‐R and DSS‐S in the validation cohort, and the results were consistent with the discovery cohort observations. Additionally, at P21, the DSS‐R group (mean = 5.28%) had higher abundance of Lactobacillus compared to DSS‐S (mean = 3.8%), which also did not reach statistical significance but supported the consistent trend (p = 0.13) [Colour figure can be viewed at wileyonlinelibrary.com]
3. RESULTS
3.1. Discovery cohort
In the initial discovery cohort, we created arbitrary groups of DSS‐resistant (DSS‐R) and DSS‐sensitive (DSS‐S) animals based on separation by weight loss during the experimentally induced colitis (Figure 1). These arbitrary groups were designated after the experiment, based on the theoretical approach described in Figure S1B. When comparing the microbiomes between the DSS‐S and DSS‐R mice at P70 (pre‐DSS), a distinct separation was found by Principal Coordinate Analysis (PCoA) (Figure 2A; p = 0.001). Three genera were found to significantly differ in abundance between the DSS‐R and the DSS‐S classified microbiomes (Figure 2B). Lactobacillus and Blautia were increased in DSS‐R mice, while Faecalibaculum was increased in DSS‐S animals. Therefore, Lactobacillus and Blautia were associated with protection against colitis in the discovery cohort. A validation cohort observing the DSS‐R and DSS‐S microbiomes at P21 (and other time points) was conducted to search for a developmental probiotic candidate.
FIGURE 1.
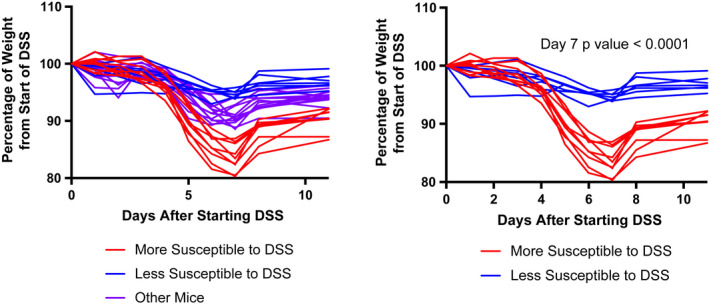
Discovery cohort weights. Weight graphs of all mice in discovery cohort (left graph) and comparison of mice we arbitrarily identified as most (n = 9)/least (n = 7) susceptible to DSS after experiment (right graph). The p value between these two groups at day 7 was <0.0001 [Colour figure can be viewed at wileyonlinelibrary.com]
FIGURE 2.
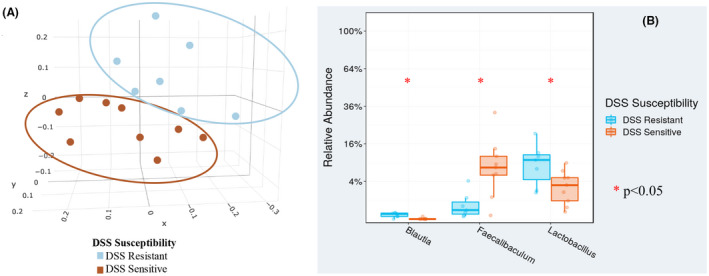
Principal coordinate analysis (PCoA) indicated microbiome separation and increased abundance of Lactobacillus pre‐DSS. (A) Weighted PCoA indicated that there was a distinct microbial separation (p = 0.001) between the mice that lost the most weight (DSS‐sensitive: DSS‐S) and the mice that lost the least weight (DSS‐resistant: DSS‐R) after start of DSS. (B) Blautia, Faecalibaculum, and Lactobacillus separated most at pre‐DSS between the DSS resistant and DSS sensitive microbiomes (Blautia: p = 0.003; Faecalibaculum: p = 0.01; Lactobacillus: DSS‐S mean = 3.53%, DSS‐R mean = 9.32%, p = 0.04) [Colour figure can be viewed at wileyonlinelibrary.com]
3.2. Validation cohort
In the validation cohort, DSS sensitivity‐based separation was not as distinct as in the discovery cohort (Figure S4). Correspondingly, beta diversity was not significantly different between DSS‐R and DSS‐S microbiomes (not shown). Lactobacillus was the only genus from the discovery and validation cohorts that was consistently associated with protection against colitis, although this separation between DSS‐R and DSS‐S groups was not significant either at P21 or P70 in the validation cohort (Figure 3). Nevertheless, the novelty of our approach and the consistent findings (despite the relatively small number of mice studied) with respect to Lactobacilli between our discovery and validation experiments, provided enough evidence for us to pursue this genus as a potential developmental probiotic in a proof‐of‐concept manner.
3.3. Lactobacillus supplementation
Lactobacillus reuteri (perinatal supplementation: LR‐PS) and L. johnsonii (LJ‐PS) cocktails of three strains or media only (control: C) were supplemented to dams according to the schematic in Figure S2. Although there was no difference between the weights at any time post‐DSS when comparing any of the groups to each other as a whole (i.e., LR‐PS vs. LJ‐PS vs. C), there was a significant difference in weight loss at day 6 post‐DSS when comparing LR‐PS and C female pups (Figure 4). Therefore, we analyzed microbiome responses to peri‐, and early postnatal supplementation of Lactobacilli in the female offspring. There was a progressive microbiome separation by beta diversity with aging (Figure 5A). At P21 (weaning), C separated the most from both LR‐PS and LJ‐PS (p = 0.056). By P30, microbiome separation between the groups became significant (p = 0.017), which progressed by P70 (pre‐DSS, p = 0.001) with LR‐PS separating the most.
FIGURE 4.
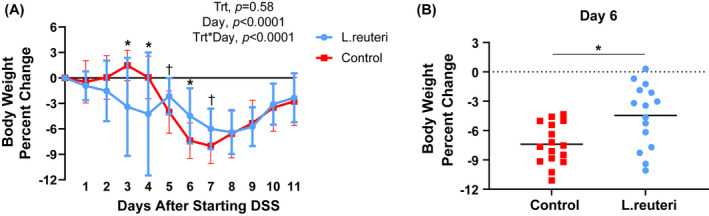
Female offspring may be protected by maternal L. reuteri supplementation. Statistical analysis included a two‐way ANOVA with repeated measures. (A) The P70 female pups supplemented with L. reuteri (n = 15) lost significantly less weight (were more DSS resistant) at day 6 compared to the female pups just given control media (n = 16) after the start of DSS. Values are means ± SD. (B) Individual data representation of P70 female pups supplemented with L. reuteri (n = 15), which lost significantly less weight (were more DSS resistant) at day 6 than control. Bars represent the means. *p < 0.05, † p < 0.10 [Colour figure can be viewed at wileyonlinelibrary.com]
FIGURE 5.
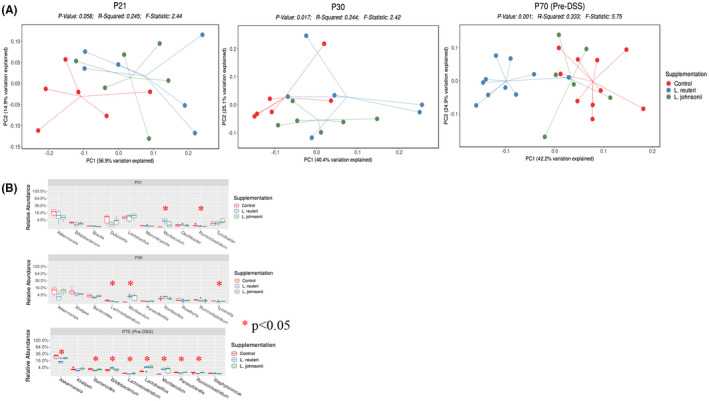
Differences in female gut microbiome based on supplementation. (A) Principal coordinate analysis (PCoA) of female pups’ microbiome samples was done at different time points: P21, P30, and P70. At P21, the control group microbiomes separated from Lactobacillus‐supplemented groups (p = 0.056). L. reuteri, L. johnsonii, and control groups progressively separated with aging (P30; p = 0.017 and P70; p = 0.001). By P70, L. reuteri supplemented female pups separated from L. johnsonii and control groups. (B) Comparisons of the female pups’ microbiomes were done at the genus level. The top 10 genera of microbial differences between L. reuteri, L. johnsonii, and control groups are shown at each time point. For P21, Muribaculum (p = 0.0038) and Ruminiclostridium (p = 0.0314) were the only genera with significant separation as p < 0.05. For P30, Lachnoclostridium (p = 0.0087), Muribaculum (p = 0.0131), and Tyzzerella (p = 0.0491) were significant. Finally, at P70, eight out of the top 10 genera separated significantly [Colour figure can be viewed at wileyonlinelibrary.com]
The top 10 genera of microbial differences between LR‐PS, LJ‐PS, and C females (based on p‐value) were then analyzed at P21, P30, and P70 (Figure 5B). P21 had two genera (Muribaculum (p = 0.0038) and Ruminiclostridium (p = 0.0314)) that significantly differed among the groups. P30 had three genera (Lachnoclostridium (p = 0.0087), Muribaculum (p = 0.0131), and Tyzzerella (p = 0.0491) that separated significantly. At P70, eight genera separated significantly between the female groups: Lachnoclostridium (p = 0.0038; LR‐PS decreased vs. C; 0.01% vs. 0.1%), Akkermansia (p = 0.0097; LR‐PS decreased vs. C; 13.52% vs. 26.95%), Lactobacillus (p = 0.0102; LR‐PS increased vs. C; 4.08% vs. 0.82%), Muribaculum (p = 0.0120; LR‐PS increased vs. C; 2.45% vs. 0%), Parasutterella (p = 0.0027; LR‐PS decreased vs. C; 0.12% vs. 0.3%), Ruminiclostridium (p = 0.0190; LR‐PS decreased vs. C; 0.06% vs. 0.25%), Bacteroides (p = 0.0233; LR‐PS decreased vs. C; 1.13% vs. 2.35%), Bifidobacterium (p = 0.0330; LR‐PS increased vs. C; 3.59% vs. 1.43%).
Interestingly, only three genera, Akkermansia, Bacteroides, and Ruminiclostridium, differentiated female LR‐PS from both female C and LJ‐PS groups (Figure S5A). Akkermansia stood out the most by being decreased in LR‐PS at P70 when compared directly to both C (p = 0.0087; 13.52% vs. 26.95%) and LJ‐PS (p = 0.0152; 13.52% vs. 21.98%) due to its relatively high abundance. Bacteroides was also decreased in LR‐PS at P70 when compared directly to both C (p = 0.0152; 1.13% vs. 2.35%) and LJ‐PS (p = 0.0260; 1.13% vs. 2.12%). Finally, Ruminiclostridium was decreased in LR‐PS at P70 when compared directly to C (p = 0.0301; 0.06% vs. 0.25%) and LJ‐PS (p = 0.0129; 0.06% vs. 0.24%).
Although the male LR‐PS group was not protected by the maternal L. reuteri cocktail supplementation, we examined how their microbiomes were affected at P70 (pre‐DSS) compared to control. Male LR‐PS separated out from control by beta‐diversity (Figure S6A). There were less significant genus level differences in male LR‐PS compared to control, however, than in females (Figure S6B). Notably, pre‐DSS Lactobacillus was significantly increased in female LR‐PS (4.08% abundance in LR‐PS vs 0.82% in C, p = 0.0260 with direct comparison) while there was no difference in the abundance of this genus between male LR‐PS offspring and controls (2.79% abundance in LR‐PS vs. 2.73% in C, p = 1.0 with direct comparison). Similarly, Akkermansia was not significantly decreased in male LR‐PS compared to controls (Figure S5B; 17.71% abundance in LR‐PS vs. 24.29% in C, p = 0.3429 with direct comparison), while the genus was separated in females (Figure S5A; 13.52% abundance in LR‐PS vs. 26.95% in C, p = 0.0087 with direct comparison). Lachnoclostridium (0.01% abundance in LR‐PS vs. 0.1% in C, p = 0.0059 with direct comparison) and Bacteroides (1.13% abundance in LR‐PS vs. 2.35% in C, p = 0.0152 with direct comparison) were the other two genera that significant separated in females, but not in males (Lachnoclostridium: LR‐PS = 0.03%, C = 0.07%, p = 0.1720; Bacteroides: LR‐PS = 0.76%, C = 2.74%, p = 0.0571). Of note, Alistipes was significantly decreased in male LR‐PS compared to male C (1.14% vs. 3.22%, p = 0.0286) while this separation was not found when comparing female LR‐PS to female C (1.62% abundance in LR‐PS vs. 2.24% in C, p = 0.2403 with direct comparison).
Akkermansia and Bacteroides were both decreased in LR‐PS females (in association with DSS resistance [i.e., protection against colitis]) as opposed to C and LJ‐PS females, and also LR‐PS males (ineffective Lactobacillus) (Figure 5). Therefore, these two genera significantly differentiated the perinatally supplemented groups in respect to early adulthood protection against DSS colitis secondary to maternal L. reuteri supplementation.
4. DISCUSSION
4.1. Overall findings
This work contains the first preclinical experimental approach to a developmental probiotic by uniquely employing stochastic‐microbiome‐variation directed identification in order to protect against IBD.
4.2. Large vendor versus small vivarium
Our mouse model‐based discovery‐validation cohort design allowed us to make several important observations. In the discovery cohort (when mice were sourced from a large vendor), there were obvious microbiome differences prior to exposure of DSS between groups classified as DSS‐R and DSS‐S, notably, increased Lactobacillus, likely driven by random, unpredictable differentiation in early life. In the validation cohort, when mice were reared in the same room of our small vivarium, stochastic microbiome variation influencing DSS susceptibility was significantly reduced, importantly lacking Faecalibaculum. Faecalibaculum has been rarely associated with DSS sensitivity in certain mouse strains. 33 These findings corroborate prior observations on small vivarium‐induced relative DSS resistance in mice, compared to large vendor bred animals 27 and implicate Faecalibaculum to have participated in the vendor‐originated increase in colitis severity variation of our work. Our results also showed that the convergence of gut microbiomes in a smaller vivarium compared to a large vendor is associated with a reduction in random murine DSS sensitivity. These observations support the prediction that stochastic microbiome variation in mammals is sensitive to environmental influences. Namely, larger environmental variation‐related microbiome divergence is inherently present in large breeding facilities, compared to a single room of a small vivarium. Large vendor‐based microbiome variation in mice may actually be more translatable to humans living in “microbially less tight” communities than coprophagic experimental mice (which share identical diet and environment (i.e., in the same room of a small vivarium)).
4.3. Importance of Lactobacilli
Lactobacillus was a genus in our discovery cohort that significantly associated with protection against DSS colitis (Figure 2B, DSS‐R group). We only uncovered trends for increased DSS‐R Lactobacillus abundance in our validation cohort, both at P21 (p = 0.13) and P70 (p = 0.20) (Figure 3). We took into consideration, however, the existing literature on Lactobacillus, and that our validation study cohort was likely underpowered to detect significant microbiome variation toward colitis protection in the setting of the same room in our small vivarium. Therefore, as a proof of concept, we decided to further examine Lactobacillus as a genus, which could furnish a developmentally protective probiotic against IBD in our DSS model system.
A study by Je et al. 34 found that a cocktail comprised of Lactobacillus johnsonii IDCC9203, Lactobacillus plantarum IDCC3501, and Bifidobacterium animalis subspecies lactis IDCC4301 was able to reduce symptoms of DSS‐induced colitis in mice. This study also found that the administration of this probiotic cocktail affected these symptoms in a dose‐dependent manner, and also the reduced expression of pro‐inflammatory cytokines such as tumor necrosis factor‐α, interleukin (IL)‐1β, and IL‐6 34 . Kim et al. 35 found that Lactobacillus acidophilus reduced the severity of DSS‐induced colitis and increased the lifespan of the mice by decreasing endoplasmic reticulum stress. Current literature on probiotics affecting human IBD, however, is limited and needs further research. 36 A review by Coquiero et al. 37 found that probiotic supplementation, especially with Lactobacillus, presented a possible therapeutic option for induction, maintaining remission, or decreasing symptoms in UC. No human or animal studies, however, have examined perinatally delivered probiotics as a preventative against the development of IBD or experimental murine colitis later in life.
4.4. Perinatal administration of a probiotic
A study by Atarashi et al. 38 observed that oral inoculation of Clostridium in infant mice led to an increase in regulatory T cells and colitis resistance, but they started supplementation two weeks after birth, and did not identify any changes with Lactobacillus supplementation. Liu et al. 39 observed that early supplementation (from P8 to P20) of L. reuteri DSM 17938 in mice promoted microbial diversity, increased immune tolerance through a higher proportion of Foxp3+ regulatory T cells in the intestine, and stimulated plasma metabolites that enhance tolerance to inflammatory stimuli. In humans, a randomized controlled trial by Schmidt et al. 40 found that supplementation of a probiotic containing Lactobacillus rhamnosus and Bifidobacterium animalis subsp lactis during late infancy decreased the incidence of eczema compared to a control group. Canova et al. 41 conducted a prospective study that discovered early life exposure to antibiotics increased the risk of childhood‐onset IBD, supporting the idea that early microbial influences may modulate the risk of subsequent IBD development. Therefore, based on our results in our discovery and validation cohorts, we decided to test two probiotic cocktails, containing three Lactobacillus strains each, as a perinatal/early postnatal intervention to decrease subsequent DSS colitis susceptibility in young adult mice. This approach allowed us to determine whether a transiently delivered, developmental probiotic could induce protection against mammalian colitis susceptibility.
4.5. Sexual dimorphism in probiotic response
Maternal L. reuteri cocktail supplementation successfully transferred DSS resistance to young adult LR‐PS female offspring (Figure 4). Intriguingly, we observed a progressively increasing microbiome separation from P21 to P70 (pre‐DSS) in the pups following maternal L. reuteri administration (Figure 5A). Male LR‐PS microbiomes also separated from control, but to a lesser degree than females (Figure S6). We speculate that estradiol driven 42 or other sex‐specific differences in gut mucosal biology 43 may be the reason for this gender‐based separation in mice. Sexual dimorphism has been previously observed to factor in the effectiveness of probiotic use in mice affecting intestinal inflammation and bone density. 44
4.6. Notable microbes
Lactobacillus, Muribaculum, and Bifidobacterium were significantly associated with DSS protection in the female LR‐PS group while Lachnoclostridium, Akkermansia, Parasutterella, Ruminiclostridium, and Bacteroides were associated with DSS susceptibility (i.e., increased abundance in female C vs. female LR‐PS). When comparing the female LR‐PS (DSS protected group) to female LJ‐PS and male LR‐PS groups (i.e., maternally Lactobacillus supplemented, but not protected against DSS), the relative decrease of Akkermansia and Bacteroides stood out most. In humans, decreased amounts of Akkermansia have been associated with UC at diagnosis (i.e. during the state of inflammation 45 ). Akkermansia and Bacteroides harbor extensively glycosyl hydrolases, which allow them to systematically degrade the protective mucus layer. 46 Mucolytic bacteria have been shown to be increased in the mucosa of IBD patients 47 , 48 , 49 and the degradation of the mucus layer has been hypothesized to contribute to the inflammatory milieu. It has been speculated that the degradation of mucus by mucolytic microbes in IBD patients provides a nutrient source for non‐mucolytic mucosa‐associated bacteria. In mice, mucin‐degrading microbes have been shown to promote the expansion of enteric pathogens and viruses, 46 , 50 , 51 and thus may inadvertently promote inflammation. Therefore, a decreased amount of Akkermansia may be protective against IBD by allowing for a more pronounced and efficient mucus barrier layer at the colonic mucosa. Recent work on Bacteroides‐supplementation found Akkermansia to be associated with therapeutic failure in respect to DSS protection, supporting this idea. 29 Conversely, in a SH2‐domain‐containing inositol 5′‐phosphatase (SHIP)‐deficient mouse model (a model of Crohn's disease), Bacteroides species were enriched prior to inflammation, 52 which is consistent with the decreased abundance of Bacteroides in our DSS resistant LR‐PS female offspring of our study. Additionally, in a study by Wu et al. 53 investigating the impact of triptolide on DSS‐induced colitis, decreased amount of Bacteroides was associated with anti‐inflammatory effects, including significantly increased microbial diversity. Based on our data, we speculate that supplementation of L. reuteri promoted a more anti‐inflammatory pro‐host microbiome, which in our mouse model consisted of decreased Akkermansia and Bacteroides.
Another key finding was that our L. reuteri cocktail increased the levels of Bifidobacterium. Similar to our study, oral administration of Bifidobacterium breve M1 and M2 was found to ameliorate DSS‐induced colitis while reducing the abundance of Bacteroides simultaneously. 54 In mice, Bifidobacteria beneficially modulate the protective mucus layer and B. lactis, B. bifidum, and B. longum have been shown to attenuate colitis and inflammation. 55 , 56 , 57 , 58 , 59 , 60 The importance of Bifidobacteria is also highlighted by the fact that Bifidobacteria are depleted in IBD. 61 , 62 We speculate that our Lactobacilli cocktail may be working in synergy with Bifidobacteria to inhibit colitis. Therefore, in our study, maternal supplementation of an L. reuteri cocktail most likely transferred colitis protection by modulating the evolution of the gut microbiome as a whole toward a less colitis prone state.
4.7. Study limitations
We acknowledge several limitations to our work, and highlight the proof‐of‐concept, paradigm‐shifting intention of it. Since we were aiming to study the difference of the two extremes on the distribution curve for DSS susceptibility (Figure S1), an ideal scenario would have been to include about 1000 mice (based on a power calculation) in our experiment and compare the most and least protected 10–15 animals’ microbiomes in the DSS‐R and DSS‐S groups from that large cohort. Due to the obvious cost and feasibility constraints, our experiment was limited to a much smaller sample size. A discovery‐validation cohort approach was utilized in an attempt to decrease the small group size limitations on stochastic microbiome variation.
Another constraint was in the depth of taxonomic identification in this study using 16S rRNA sequencing. Therefore, we did not confirm specifically the transfer of the experimental Lactobacillus strains into the dams and pups, but rather focused on the secondary functional (i.e. DSS resistance) and genus level microbiome changes that our intervention induced. This approach was used due to cost‐benefit considerations for our specific experiment since shotgun sequencing (or other means of detailed sequencing) is significantly more expensive for an arguably limited payoff. Namely, our goal was to introduce a novel conceptual approach toward the identification of a developmental probiotic against human IBD in an imperfect animal model, where specific microbiome associations may bare limited translational value. We also recognize that our probiotic identification was done in males, yet the beneficial effect from those occurred in females only. This finding repeatedly underscores the unpredictable nature of microbiome responses, whereby such approaches as ours may only provide taxonomic targets for manipulation, but not necessarily the means by which those targets can be achieved (i.e. increased Lactobacilli at P70 in males was not achieved by perinatal [maternal] supplementation of those, for example).
We only examined a single, but objective outcome measure of DSS colitis, namely weight loss. However, we do not find this a limitation. We intentionally aimed to further a paradigm shift toward the mouse DSS colitis model. We have recently underscored that in more than 3,000 peer‐reviewed publications that used the DSS model, weight loss has been the most economic, most consistent, and objective measurement of colitis severity. 28 Consequently, we wished to demonstrate the importance of the judicious use or research resources, especially during the COVID‐19 pandemic, and show that weight loss is a sufficient single marker of DSS colitis at current state of art.
In spite of the limitations, we are confident that our findings in addition to those by Schmidt et al. 40 (see earlier, where infantile Lactobacillus rhamnosus and Bifidobacterium animalis subsp lactis protected against eczema in a clinical trial) position Lactobacilli as outstanding candidates for translational studies in humans as a developmental (perinatally delivered) probiotic against IBD. Importantly, genetic predisposition to eczema and IBD is shared, 63 and a bidirectional clinical association between IBD and eczema (i.e., atopic dermatitis [AD]) appears to exist. 64 Furthermore, some similarities between AD‐ and CD‐associated microbiomes have been observed. 65 In the meantime, only a very large cohort, placebo‐controlled prospective trial with prolonged (i.e., through the first 30 years of life for the participants at minimum) follow‐up could provide answers for perinatal/early postnatal microbial therapeutics to prevent against IBD.
CONFLICT OF INTEREST
The authors have no conflicting interests to declare.
AUTHOR CONTRIBUTIONS
Mahesh Krishna performed data collection, data analysis, and drafted the manuscript. Melinda Engevik performed data collection and contributed new reagents. Karen Queliza, Savini Britto, Wenly Ruan, and Hongtao Wang performed data collection. Rajesh Shah performed data analysis. James Versalovic contributed to conceptual design, with reagents, and manuscript review. Richard Kellermayer performed the conceptual design, data analysis and manuscript writing, and obtained funding.
Supporting information
Fig S1‐S6
ACKNOWLEDGMENTS
The authors thank the Gutsy Kids Fund, including philanthropic leadership from the Brock Wagner family and donation from the Frugoni and other generous families (to RK); the Klaasmeyer family funds for PSC research (to RK); and the Crohn’s and Colitis Foundation ProKiids Network (for grant/research time support to RK). Additionally, the Alkek Center for Metagenomics and Microbiome Research at Baylor College of Medicine was very helpful in aiding microbiome data analysis. Bridget Stroup was also essential with her support of statistical analysis as well as critical review of the revised manuscript.
Krishna M, Engevik M, Queliza K, et al. Maternal Lactobacillus reuteri supplementation shifts the intestinal microbiome in mice and provides protection from experimental colitis in female offspring. FASEB BioAdvances. 2022;4:109–120. doi: 10.1096/fba.2021-00078
REFERENCES
- 1. Loftus EV. Clinical epidemiology of inflammatory bowel disease: Incidence, prevalence, and environmental influences. Gastroenterology. 2004;126:1504‐1517. [DOI] [PubMed] [Google Scholar]
- 2. Ananthakrishnan AN. Epidemiology and risk factors for IBD. Nat Rev Gastroenterol Hepatol. 2015;12:205‐217. [DOI] [PubMed] [Google Scholar]
- 3. Malik TA. Inflammatory bowel disease: historical perspective, epidemiology, and risk factors. Surg Clin North Am. 2015;95(6):1105‐1122. [DOI] [PubMed] [Google Scholar]
- 4. Kellermayer R. Challenges for epigenetic research in inflammatory bowel diseases. Epigenomics. 2017;9:527‐538. [DOI] [PubMed] [Google Scholar]
- 5. de Souza HSP, Fiocchi C, Iliopoulos D. The IBD interactome: an integrated view of aetiology, pathogenesis and therapy. Nat Rev Gastroenterol Hepatol. 2017;14:739‐749. [DOI] [PubMed] [Google Scholar]
- 6. Fofanova TY, Petrosino JF, Kellermayer R. Microbiome‐epigenome interactions and the environmental origins of inflammatory bowel diseases. J Pediatr Gastroenterol Nutr. 2016;62:208‐219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Alipour M, Zaidi D, Valcheva R, et al. Mucosal barrier depletion and loss of bacterial diversity are primary abnormalities in paediatric ulcerative colitis. J Crohns Colitis. 2016;10:462‐471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Goodrich JK, Waters JL, Poole AC, et al. Human genetics shape the gut microbiome. Cell. 2014;159:789‐799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Castillo‐Fernandez JE, Spector TD, Bell JT. Epigenetics of discordant monozygotic twins: implications for disease. Genome Med. 2014;6:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhou S, Xu R, He F, et al. Diversity of gut microbiota metabolic pathways in 10 pairs of Chinese infant twins. PLoS ONE. 2016;11:e0161627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kellermayer R. Probability in gene expression. Genet Med. 2006;8:263‐264. [DOI] [PubMed] [Google Scholar]
- 12. Kellermayer R. Genetic drift. Physiologic noise obscures genotype‐phenotype correlations. Am J Med Genet A. 2007;143A: 1306‐1307. [DOI] [PubMed] [Google Scholar]
- 13. Lloyd‐Price J, Arze C, Ananthakrishnan AN, et al. Multi‐omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature. 2019;569:655‐662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Clooney AG, Eckenberger J, Laserna‐Mendieta E, et al. Ranking microbiome variance in inflammatory bowel disease: a large longitudinal intercontinental study. Gut. 2021;70: 499‐510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Milani C, Duranti S, Bottacini F, et al. The first microbial colonizers of the human gut: composition, activities, and health implications of the infant gut microbiota. Microbiol Mol Biol Rev. 2017;81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yatsunenko T, Rey FE, Manary MJ, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222‐227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tong M, McHardy I, Ruegger P, et al. Reprograming of gut microbiome energy metabolism by the FUT2 Crohn's disease risk polymorphism. ISME J. 2014;8:2193‐2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mir SA, Nagy‐Szakal D, Dowd SE, Szigeti RG, Smith CW, Kellermayer R. Prenatal methyl‐donor supplementation augments colitis in young adult mice. PLoS ONE. 2013;8:e73162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kronman MP, Zaoutis TE, Haynes K, Feng R, Coffin SE. Antibiotic exposure and IBD development among children: a population‐based cohort study. Pediatrics. 2012;130:e794‐e803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kellermayer R, Zilbauer M. The gut microbiome and the triple environmental hit concept of inflammatory bowel disease pathogenesis. J Pediatr Gastroenterol Nutr. 2020;71(5):589‐595. [DOI] [PubMed] [Google Scholar]
- 21. DeVoss J, Diehl L. Murine models of inflammatory bowel disease (IBD): challenges of modeling human disease. Toxicol Pathol. 2014;42:99‐110. [DOI] [PubMed] [Google Scholar]
- 22. Hildebrand F, Nguyen TL, Brinkman B, et al. Inflammation‐associated enterotypes, host genotype, cage and inter‐individual effects drive gut microbiota variation in common laboratory mice. Genome Biol. 2013;14:R4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Friswell MK, Gika H, Stratford IJ, et al. Site and strain‐specific variation in gut microbiota profiles and metabolism in experimental mice. PLoS ONE. 2010;5:e8584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Campbell JH, Foster CM, Vishnivetskaya T, et al. Host genetic and environmental effects on mouse intestinal microbiota. ISME J. 2012;6:2033‐2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. McCafferty J, Mühlbauer M, Gharaibeh RZ, et al. Stochastic changes over time and not founder effects drive cage effects in microbial community assembly in a mouse model. ISME J. 2013;7:2116‐2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hufeldt MR, Nielsen DS, Vogensen FK, Midtvedt T, Hansen AK. Family relationship of female breeders reduce the systematic inter‐individual variation in the gut microbiota of inbred laboratory mice. Lab Anim. 2010;44:283‐289. [DOI] [PubMed] [Google Scholar]
- 27. Chassaing B, Aitken JD, Malleshappa M, Vijay‐Kumar M. Dextran sulfate sodium (DSS)‐induced colitis in mice. Curr Protoc Immunol. 2014;104:15.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lanka Britto S, Krishna M, Kellermayer R. Weight loss is a sufficient and economical single outcome measure of murine dextran sulfate sodium colitis. FASEB Bioadv. 2019;1:493‐497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ihekweazu FD, Fofanova TY, Queliza K, et al. ATCC 8483 monotherapy is superior to traditional fecal transplant and multi‐strain bacteriotherapy in a murine colitis model. Gut Microbes. 2019;10:504‐520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu HY, Roos S, Jonsson H, et al. Effects of Lactobacillus johnsonii and Lactobacillus reuteri on gut barrier function and heat shock proteins in intestinal porcine epithelial cells. Physiol Rep. 2015;3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Peña JA, Li SY, Wilson PH, Thibodeau SA, Szary AJ, Versalovic J. Genotypic and phenotypic studies of murine intestinal lactobacilli: species differences in mice with and without colitis. Appl Environ Microbiol. 2004;70:558‐568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Luk B, Veeraragavan S, Engevik M, et al. Postnatal colonization with human "infant‐type" Bifidobacterium species alters behavior of adult gnotobiotic mice. PLoS ONE. 2018;13:e0196510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang L, Tang H, Wang C, Hu Y, Wang S, Shen L. Aquaporin 4 deficiency alleviates experimental colitis in mice. FASEB J. 2019;33:8935‐8944. [DOI] [PubMed] [Google Scholar]
- 34. Je IG, Lee DG, Jeong DG, et al. The probiotic, ID‐JPL934, attenuates dextran sulfate sodium‐induced colitis in mice through inhibition of proinflammatory cytokines expression. J Med Food. 2018;21:858‐865. [DOI] [PubMed] [Google Scholar]
- 35. Kim DH, Kim S, Lee JH, et al. Lactobacillus acidophilus suppresses intestinal inflammation by inhibiting endoplasmic reticulum stress. J Gastroenterol Hepatol. 2018. [DOI] [PubMed] [Google Scholar]
- 36. Scarpato E, Russo M, Staiano A. Probiotics in pediatric gastroenterology: emerging Indications: Inflammatory Bowel Diseases. J Clin Gastroenterol. 2018, 52 Suppl 1, Proceedings from the 9th Probiotics, Prebiotics and New Foods, Nutraceuticals and Botanicals for Nutrition & Human and Microbiota Health Meeting, held in Rome, Italy from September 10 to 12, 2017:S7‐S9. [DOI] [PubMed] [Google Scholar]
- 37. Coqueiro AY, Raizel R, Bonvini A, Tirapegui J, Rogero MM. Probiotics for inflammatory bowel diseases: a promising adjuvant treatment. Int J Food Sci Nutr. 2018;1‐10. [DOI] [PubMed] [Google Scholar]
- 38. Atarashi K, Tanoue T, Shima T, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337‐341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liu Y, Tian X, He B, et al. DSM 17938 feeding of healthy newborn mice regulates immune responses while modulating gut microbiota and boosting beneficial metabolites. Am J Physiol Gastrointest Liver Physiol. 2019;317:G824‐G838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schmidt RM, Pilmann Laursen R, Bruun S, et al. Probiotics in late infancy reduce the incidence of eczema: a randomized controlled trial. Pediatr Allergy Immunol. 2019;30:335‐340. [DOI] [PubMed] [Google Scholar]
- 41. Canova C, Ludvigsson JF, Di Domenicantonio R, Zanier L, Barbiellini Amidei C, Zingone F. Perinatal and antibiotic exposures and the risk of developing childhood‐onset inflammatory bowel disease: a nested case‐control study based on a population‐based birth cohort. Int J Environ Res Public Health. 2020;17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bábíčková J, Tóthová Ľ, Lengyelová E, et al. Sex differences in experimentally induced colitis in mice: a role for estrogens. Inflammation. 2015;38:1996‐2006. [DOI] [PubMed] [Google Scholar]
- 43. Jasarevic E, Morrison KE, Bale TL. Sex differences in the gut microbiome‐brain axis across the lifespan. Philos Trans R Soc Lond B Biol Sci. 2016;371:20150122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. McCabe LR, Irwin R, Schaefer L, Britton RA. Probiotic use decreases intestinal inflammation and increases bone density in healthy male but not female mice. J Cell Physiol. 2013;228:1793‐1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shah R, Cope JL, Nagy‐Szakal D, et al. Composition and function of the pediatric colonic mucosal microbiome in untreated patients with ulcerative colitis. Gut Microbes. 2016;7: 384‐396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Engevik MA, Banks LD, Engevik KA, et al. Rotavirus infection induces glycan availability to promote ileum‐specific changes in the microbiome aiding rotavirus virulence. Gut Microbes. 2020;1‐24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Png CW, Lindén SK, Gilshenan KS, et al. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am J Gastroenterol. 2010;105:2420‐2428. [DOI] [PubMed] [Google Scholar]
- 48. Prindiville T, Cantrell M, Wilson KH. Ribosomal DNA sequence analysis of mucosa‐associated bacteria in Crohn's diseataske. Inflamm Bowel Dis. 2004;10:824‐833. [DOI] [PubMed] [Google Scholar]
- 49. Willing BP, Dicksved J, Halfvarson J, et al. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology. 2010;139:1844‐1854.e1841. [DOI] [PubMed] [Google Scholar]
- 50. Ng KM, Ferreyra JA, Higginbottom SK, et al. Microbiota‐liberated host sugars facilitate post‐antibiotic expansion of enteric pathogens. Nature. 2013;502:96‐99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pacheco AR, Curtis MM, Ritchie JM, et al. Fucose sensing regulates bacterial intestinal colonization. Nature. 2012;492: 113‐117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Dobranowski PA, Tang C, Sauvé JP, Menzies SC, Sly LM. Compositional changes to the ileal microbiome precede the onset of spontaneous ileitis in SHIP deficient mice. Gut Microbes. 2019;10:578‐598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wu H, Rao Q, Ma GC, Yu XH, Zhang CE, Ma ZJ. Effect of triptolide on dextran sodium sulfate‐induced ulcerative colitis and gut microbiota in mice. Front Pharmacol. 2019;10: 1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chen Y, Jin Y, Stanton C, et al. Alleviation effects of Bifidobacterium breve on DSS‐induced colitis depends on intestinal tract barrier maintenance and gut microbiota modulation. Eur J Nutr. 2021;60(1):369‐387. [DOI] [PubMed] [Google Scholar]
- 55. Philippe D, Favre L, Foata F, et al. Bifidobacterium lactis attenuates onset of inflammation in a murine model of colitis. World J Gastroenterol. 2011;17:459‐469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Engevik MA, Luk B, Chang‐Graham AL, et al. Bifidobacterium dentium fortifies the intestinal mucus layer via autophagy and calcium signaling pathways. mBio. 2019;10(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Alard J, Peucelle V, Boutillier D, et al. New probiotic strains for inflammatory bowel disease management identified by combining in vitro and in vivo approaches. Benef Microbes. 2018;9:317‐331. [DOI] [PubMed] [Google Scholar]
- 58. Schroeder BO, Birchenough GMH, Ståhlman M, et al. Bifidobacteria or fiber protects against diet‐induced microbiota‐mediated colonic mucus deterioration. Cell Host Microbe. 2018;23:27‐40.e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Srutkova D, Schwarzer M, Hudcovic T, et al. Bifidobacterium longum CCM 7952 promotes epithelial barrier function and prevents acute DSS‐induced colitis in strictly strain‐specific manner. PLoS ONE. 2015;10:e0134050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Abrantes FA, Nascimento BB, Andrade MER, et al. Treatment with Bifidobacterium longum 51A attenuates intestinal damage and inflammatory response in experimental colitis. Benef Microbes. 2020;11:47‐57. [DOI] [PubMed] [Google Scholar]
- 61. Duranti S, Gaiani F, Mancabelli L, et al. Elucidating the gut microbiome of ulcerative colitis: bifidobacteria as novel microbial biomarkers. FEMS Microbiol Ecol. 2016;92(12):fiw191. [DOI] [PubMed] [Google Scholar]
- 62. Gevers D, Kugathasan S, Denson LA, et al. The treatment‐naive microbiome in new‐onset Crohn's disease. Cell Host Microbe. 2014;15:382‐392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Paternoster L, Standl M, Waage J, et al. Multi‐ancestry genome‐wide association study of 21,000 cases and 95,000 controls identifies new risk loci for atopic dermatitis. Nat Genet. 2015;47:1449‐1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Shi X, Chen Q, Wang F. The bidirectional association between inflammatory bowel disease and atopic dermatitis: a systematic review and meta‐analysis. Dermatology. 2020;236(6):546‐553. [DOI] [PubMed] [Google Scholar]
- 65. Reddel S, Del Chierico F, Quagliariello A, et al. Gut microbiota profile in children affected by atopic dermatitis and evaluation of intestinal persistence of a probiotic mixture. Sci Rep. 2019;9:4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1‐S6


