Abstract
Lignocellulose is the most abundant biomass available on earth, including wood and agricultural wastes such as rice straw, corn cobs, and oil palm empty bunches. The biopolymer content in lignocellulose has a great potential as feedstock for producing industrial raw materials such as glucose, sorbitol, xylose, xylitol, and other pharmaceutical excipients. Currently, scientists and governments agree that the enzymatic delignification method is an environmentally friendly green method to be applied. This review attempts to explain the proper preparation of the enzymes laccase, lignin peroxidase, and manganese peroxidase, as well as the important factors influencing their activity. The recent applications of the enzymes for detoxification of hazardous substances, proper enzyme immobilization technique, and future prospect combination with DESs extraction of lignin are also discussed.
Keywords: Lignocellulose, Biopolymer, Laccase, Manganese peroxidase, Lignin peroxidase, Enzyme immobilization, Detoxification
Lignocellulose, Biopolymer, Laccase, Manganese peroxidase, Lignin peroxidase, Enzyme immobilization, Detoxification.
1. Introduction
Lignocellulose is the most abundant biomass on earth as the main component of wood and various agricultural residues (such as rice straw, wheat straw, and corn cobs) [1, 2, 3]. Biological deconstruction of plant cell walls is becoming an increasingly important research topic related to future bioeconomics that depends on the supply of biomass raw materials to produce various bioproducts and bioenergy [4].
Lignocellulose is a raw material that has many benefits where the main constituents are biopolymers of cellulose, hemicellulose, and lignin, which form complex structures that are resistant to decomposition [3, 5]. Cellulose is the main structural polysaccharide of plant cell walls weighing up to 30–50% of the dry weight of lignocellulose [6, 7]. Cellulose has unique optical, mechanical, and rheological properties and is easily chemically reconfigured as a natural polymer. Modifying cellulose by chemical processes can form cellulose esters and ethers commonly used as pharmaceutical excipients [8]. The presence of cellulose in plants binds closely to lignin, while lignin in the structure of lignocellulose is difficult to destroy. Therefore, it is necessary to carry out a delignification process (pretreatment) to obtain cellulose with high purity [9].
Lignocellulosic biomass is a raw material that is cheap, abundant, renewable and is widely produced by various industries [5]. In Indonesia, lignocellulosic materials can be obtained from various sources, both as a byproduct and as wastes, including agricultural industry waste (such as straw, rice husks, corn cobs, grass), plantations waste (such as oil palm empty bunches, bagasse, sugarcane pruning, cocoa pod husks, coffee pod skins), wood and forestry waste (wood stalks, saws leftovers, paper mill waste), and post-harvest processing, as well as organic waste (market and household waste) [9]. The main constituent structures of lignocellulose are cellulose, hemicellulose, and lignin. The exact composition of the constituents depends on the origin of the plant and its species. In general, lignocellulosic materials from hardwood sticks contain 40–55% cellulose, 24–40% hemicellulose, and 18–25% lignin, while softwood stick-based materials contain 45–50% cellulose, 25–35% hemicellulose, and 25–35% lignin [10].
So far, the existing pretreatment methods are physical, chemical, physicochemical, and biological pretreatment [5]. Today's leading pre-treatment methods are based on physicochemical processes (steam-explosion, ammonia fiber explosion, and wet oxidation) which in most cases involve high energy requirements and high capital investment. There are some degradations of sugars and also the formation of inhibitor compounds that affect subsequent processes. In addition, the use of chemicals in this process is expensive and also contributes to environmental problems due to toxic waste produced [11, 12].
Therefore, the biological pretreatment method was chosen as a solution to the above problems and is often known as biodelignification. Biodelignification can be done using microbes and enzymes. Many microorganisms in nature that can degrade lignin include fungi and bacteria. These microorganisms produce enzymes to attack, depolymerize, and degrade polymers in lignocellulosic substrates [13]. The microorganisms that have been reported as the most effective decomposers of lignin are white-rot fungi [14, 15].
The white-rot fungi (WRF, see Figure 1) basidiomycetes are able to degrade lignin efficiently [16]. They produce various extracellular ligninolytic enzymes, such as laccase, manganese peroxidase, and lignin peroxidase [17]. The degree of delignification of white-rot fungi varies greatly, depending on the type of fungi and the carbohydrate content in the lignocellulose raw material. Some white-rot fungi have been reported to have the potential to produce all cell wall components, while others are more selective [18]. Several types of white-rot fungi are Phanerochaete chrysosporium, Pleurotus ostreatus, Trametes versicolor, Cyathus stercoreus, and Ceriporiopsis subvermispora [19].
Figure 1.
White rot fungi at its natural habitat (A), and in PDA medium (B). (Source: personal documentation)
In biodelignification, the study on the optimal production process for each enzyme is essential to know and needs to explore [20, 21]. In this review article, the discussion focuses on a good preparation and optimum utilization of ligninolytic enzymes produced by white-rot fungi.
2. Main text
2.1. Cellulose, hemicellulose, and lignin
Cellulose is the most abundant renewable organic material on earth and is widely distributed in plants, bacteria, marine algae, and other biomass. Cellulose is rarely found in a pure state in nature. It permanently binds to other polymers such as lignin and hemicellulose [22]. Cellulose is a linear polymer of glucose units connected via 1,4-β-glucoside bonds and is insoluble in water, dilute acid solutions, and dilute alkaline solutions at room temperature. This poor solubility property is due to the solid intramolecular and intermolecular hydrogen bonds between the individual chains [23]. Although the structure and composition of plant cell walls vary widely, cellulose content usually makes up to 35–50% of the dry weight of biomass and, in particular, almost 100% for cotton [24]. Its linear structure causes cellulose to be crystalline and not easily degraded chemically or mechanically and is resistant to enzymatic decomposition [25].
The varying chemical structure of cellulose (α, β, γ) has a big influence on its reactivity. The hydroxyl groups contained in amorphous regions are very easy to reach and easy to react, while the hydroxyl groups that are present in crystalline regions with dense beams and strong inter-chain bonds may not be achieved at all [26]. Based on the degree of polymerization (DP) and solubility in the 17.5% sodium hydroxide (NaOH), there are three types of cellulose [30], namely alpha, beta, and gamma cellulose. Alpha cellulose is a long chain cellulose, insoluble in 17.5% NaOH solution or a strong alkaline solution with a DP of 600–1500. Alpha cellulose is used to determine the level of purity of cellulose. Alpha cellulose is the highest quality (pure) cellulose. The higher the alpha cellulose content, the better the quality of the cellulose. Beta cellulose is short-chain cellulose, dissolves in 17.5% NaOH or a strong base with a polymerization degree of 15–90, and can precipitate when neutralized. Gamma cellulose is the same as beta cellulose, but the degree of polymerization is less than 15 with the main content being hemicellulose [27].
The second polysaccharide component of lignocellulose is hemicellulose, which makes up to 15–30% of the plant cell wall. Hemicellulose is embedded in the plant cell walls, and its main function is to bind between cellulose and lignin. Hemicellulose with lignin forms covalent bonds between ether and ester, forming a carbohydrate-lignin complex. Hemicellulose also works by binding to cellulose microfibrils to strengthen the cell walls. Unlike cellulose, hemicellulose has an amorphous structure, consisting of several heteropolymers including polyxylose, glucomannan (Glu-Man), and galactoglucomannan (Gal-Glu-Man). Hemicellulose, as already mentioned has a low degree of polymerization, therefore it can be hydrolyzed with a dilute base or acid [15, 24].
The main hemicelluloses in softwood are galactoglucomannan, glucomannan, and arabinoglukuronoxy, while the main hemicellulose in hardwood is xylan (3). In general, they fall into four classes: (i) unbranched chains such as (1–4) xylan and mannan; (ii) helical chains such as (1–3) xylan; (iii) branched chains such as (1–4) -Galactoglucomannans; and (iv) pectic substances such as polyrhamnogalacturonans [27].
Lignin is the third most abundant polymer in nature. These substances are present in plant cell walls and provide strong resistance to microbial attack and oxidative stress. The composition of lignin varies among plant species. Hardwood has a higher lignin content (>30%) when compared to softwood or herbaceous plants [15, 28]. Compared with other constituents of the cell wall, lignin is hydrophobic and prevents water penetration into the cell wall. Lignin provides strength to the cell walls of cellulosic materials as well as protects the cell walls from biochemical stresses by inhibiting the enzymatic degradation of other components.
Lignin strengthens plant cell walls through the adhesion of cellulosic microfibrils which allows plants to significantly grow in size, improves water transport, resistance to pathogens, and slows down wood degradation by microorganisms [29]. Lignin has several benefits such as being antioxidant and antimicrobial. Lignin is often available in large quantities as a byproduct of industrial waste and is environmentally friendly [10]. In nature, lignin is broken down through the action of peroxidases, laccases, and additional oxidative enzymes from fungi and bacteria.
Lignin consists of three distinct phenylpropane units, namely, p-coumaryl, coniferyl, and synapyl alcohol. Lignin molecules are arranged in three-dimensional forms and cross-linked to each of the constituent molecules with different types of chemical bonds [30]. These monomers are linked by ether bonds (C = O) or bonds between carbons (C–C) which are connected randomly and non-linearly to form very complex polymers. The complex structure causes lignin to be recalcitrant to be degraded by chemical exposure and organisms [15, 28]. Lignin also contains various secondary metabolites such as hydroxycinnamyl alcohols (or monolignols), coniferyl alcohol, and synapyl alcohol with small amounts of p-coumaryl alcohol [31].
The constituents of lignin and hemicellulose are different so that the cross-linking between these polymers differs from plant to plant and from tissue to tissue [32]. Based on its chemical composition, the structure of lignin varies depending on the type and species of biomass [22]. For example, softwood mainly contains guaiacil (G) type lignin, while hardwood contains glucuronosilan type and guaiacyl-syrup (GS) lignin [4]. Wood from gymnosperms has a greater lignin concentration than wood from angiosperms and is generally composed of propyl guaiacyl units. Therefore, lignin biodegradation can be significantly influenced by, the amount and type of lignin present in lignocellulose [4]. Lignin is the most abundant renewable raw material, apart from cellulose, and plays an important role in the biosphere carbon cycle, namely by preventing the degradation of accessible carbohydrate microbes within the cell walls [31]. Lignin degrades poorly under anaerobic conditions but is extensively degraded by white-rot fungi under conditions of availability of oxygen & moisture [27].
2.2. Delignification method of lignocellulose
In order to obtain cellulose, the first step in utilizing lignocellulosic biomass is lignin degradation through a process known as pretreatment so that cellulose polymers can be exposed to further hydrolysis [1, 4, 33]. The four main objectives of pretreatment are (1) modifying the crystalline index of cellulose and degree of polymerisation (2), disrupting the lignin-carbohydrate bond (3), removal of lignin and/or hemicellulose and (4) increasing in porosity [34]. Delignification can be carried out by physical, chemical, physicochemical, and biological methods [35, 36]. The four methods have their respective advantages and disadvantages. Physical treatment has the advantage of being environmentally friendly by not producing toxic compounds, but the main disadvantage is that this method does not remove or release lignin, so it is necessary to futher treat the mass with other methods afterward [11, 36].
Next, chemical treatment has the advantage that this method is very effective in degrading lignin and the chemical solution used can be recovered for use in the next delignification process. However, the disadvantage of this method is that the chemical solution used in some processes is very expensive, the recovery of the solution needs to be done as much as possible which takes a long time and energy, and not all solutions can be recovered, where the chemical solution is toxic and wastes contribute to environmental pollution. The flammability and high volatility of organic solvents as chemical solutions make pre-treatment carried out under controlled conditions, which require more costs to maintain these conditions [35, 36].
Physicochemical treatment is the most commonly used method for delignifying lignocellulosic biomass. The advantages of this method are the effectiveness of high lignin degradation and time efficiency, but the disadvantages are high energy consumption, cellulose degradation, and the formation of inhibitor compounds in the process which can be detrimental in the next cellulose processing stage [11, 12].
Biological pretreatment or biodelignification can reduced those disadvantages of chemical and physicochemical treatments. It can be categorized into pretreatment that utilizes microorganisms, such as filamentous fungi and certain bacteria directly on the raw material, and enzymatic pretreatment that uses enzymes excreted by these lignin-decomposing microorganisms [22, 40, 41, 42]. Many microorganisms play a role in lignin degradation but only the white-rot fungi (WRF) are capable of depolymerizing lignin effectively [11, 18]. But despite its various advantages, biological pretreatment still faces several obstacles such as long processing times, large space requirements, and the need for continuous monitoring of microorganism growth [40].
Several parameters for comparison of the biodelignification method compared to the chemical delignification method are listed in the following table.
The main advantage of biological delignification is the technology that is safe and environmentally friendly by using no chemicals, does not involve acids, alkalis, or any reactive substances thus reducing the output of waste streams and reducing downstream processing costs. Minimum inhibitor formation, as well as byproducts that will not inhibit or hinder the hydrolysis process, are also advantages of biological pretreatment (37,38, see Table 1). Then, the use of microbial enzymes for hydrolysis causes high specificity of enzyme reactions and the absence of lost substrates or toxicity due to chemical modification [38, 39]. The opposite occurs in chemical delignification, which requires high energy costs, high chemical requirements, and the eluent used is classified as hazardous waste due to its toxicity [40].
Table 1.
Comparison of delignification methods and some criteria/condition.
| Type of Delignification | Parameter | Used Agent or Condition | References |
|---|---|---|---|
| Biodelignification | Active agent | microbial cell (mold or bacteria) or enzymes | [22] |
| Temperature/Pressure | Low/light, | [5] | |
| Energy used | Lower | [5] | |
| Cost/value of process | Cheap/safe and environmentally friendly | [37] | |
| Inhibitor formation | Low | [38, 39] | |
| Chemical Delignification- | Active agent | Basic and Acid chemicals agent & hazardous waste | [1, 40] |
| Temperature/Pressure | High | [3] | |
| Inhibitor/waste formation | High | [19] |
The strategy of using enzymes in lignocellulosic biomass degradation is considered more beneficial because it can overcome problems arising from the direct use of fungi, such as difficulties in meeting growth requirements on a large scale, long incubation processes, penetration of mycelia into the substrate, excessive consumption of polysaccharides by fungi that cause excessive degradation of carbohydrates, and contamination problems [1]. The use of ligninolytic enzymes for direct lignocellulose treatment minimizes the problems encountered in fungal pretreatment and represents a simpler and more effective method. Therefore, this method is an interesting topic for further exploration because of its ability to operate in a relatively short reaction time, only requires a few hours, and requires lower nutrient requirements for enzymatic reactions [19, 39]. The effect of enzymes on lignocellulosic biomass depends on the type of enzyme and the composition of the biomass itself. This is due to the specificity of the enzyme related to the type of reaction being catalyzed [43, 44].
However, there is a drawback to using enzymes compared to the direct use of fungal biomass, namely that enzymes are still more expensive to produce on an industrial scale. This may be overcome by the advances in enzyme immobilization technology that increase the efficiency in reusing enzymes both in quantity and activity may be greater than that of fungal biomass [18].
2.3. White-rot fungi as source of ligninolytic enzymes
White-rot fungi (WRF) are classified into the Agaricomycetes class. This type of fungi can be found growing on live trees that are still growing well or dead wood from temperate to tropical environments. The term white-rot is associated with the whitish (white) appearance often observed in wood overgrown by this type of fungi. The general characteristics of white-rot fungi include being able to degrade all the main components of plant cell walls: cellulose, hemicellulose, and lignin. Lignocellulose decomposition is carried out through a series of enzymatic processes (hydrolase and oxidoreductase) and non-enzymatic mechanisms [45].
There are about 10,000 species of white-rot fungi, with varying capacities to degrade lignin, cellulose, and hemicellulose. However, only a few types have been carried out further research. The most studied species of white-rot fungi are divided into six families: Phanerochaetaceae (Phanerochaete chrysosporium), Poliporaceae (Trametes versicolor and Pycnoporus sanguineus), Ganodermataceae (Ganoderma lucidum and Ganoderma applanatum) Marasmiaceae (Lentinula edodes), Pleurotus mushrooms, Hymenochaetaceae (Inonotus hispidus and Phellinus igniarius), Meruliaceae (Bjerkandera adusta, Irpex lacteus, and Phlebia radiate) [45].
Some white-rot fungi are able to degrade lignin selectively and some are non-selective. Examples of selective white-rot fungi are Ceriporiopsis subvermispora, Dichomitus squalens, Phanerochaete chrysosporium, Phlebia radiata whose mechanisms degrade lignin and hemicellulose more than cellulose, while examples of non-selective white-rot fungi are Trametes versicolor and Fomes fomentarius which can degrade all components. in the same amount. White-rot fungi are the only organisms that are believed to be able to completely degrade lignin into water-soluble products and CO2 [46]. The carbon source of white-rot fungi comes from lignin. Lignin is degraded by white-rot fungi into carbon dioxide as a whole to enter wood polysaccharides which are protected by lignin-carbohydrate complexes [47].
White-rot fungi produce extracellular oxidative enzymes during the lignin degradation process. These microorganisms produce enzyme secretions that function as biodegradation agents to break down lignocellulosic materials into simpler molecules. The secretion of enzymes produced by white-rot fungi is also able to degrade pesticide compounds and toxic waste. This is because white-rot fungi have a non-specific oxidative system to depolymerize and mineralize lignin, including some extracellular oxidoreductases, low molecular weight metabolites, and very effective oxygen action. In addition, white-rot fungi can depolymerize lignin and metabolize lignin into CO2 and H2O [46].
2.4. Ligninolytic enzymes
Ligninolytic enzymes are enzyme complexes that produce delignification abilities in various types of organisms such as bacteria, insects, and fungi. However, the most effective lignin-decomposing organisms are fungi, especially white-rot fungi. This enzyme is produced by white-rot fungi as a secondary metabolic product because lignin degradation does not produce energy in mushrooms [48]. The ligninolytic enzymes known so far are extracellular, not specific, and break down the aromatic structure of lignin and the bonds between lignocellulosic units through different oxidative reactions [45].
Ligninolytic enzymes are categorized into two major groups, namely heme peroxidase and phenol oxidase [14, 18]. Enzymes included in phenol oxidase are laccase (benzenediol: oxygen oxidoreductase; EC 1.10.3.2) while those included in heme peroxidase are lignin peroxidase (LiP, diarylpropane: oxygen oxidoreductase and hydrogen peroxide; EC 1.11.1.14), and manganese peroxidase (MnP)., Mn (II): hydrogen peroxide oxidoreductase; EC 1.11.1.13) [18,41,145]. These three enzymes form an enzymatic system that can degrade lignin effectively.
2.4.1. Laccase
The first laccase identified and reported came from the Japanese lacquer tree Toxicodendron vernicifluum (Rhus vernicifera). In general, the laccase produced from these plants has lower laccase. Plant lacquer is concerned with lignin biosynthesis and polymerization, elongation, and stress response in plants. In fungi, laccase appears more than plants, such as in Basidiomycetes such as Phanerochaete chrysosporium, Theiophora terrestris, and Lenzites true [49] and other white-rot fungi such as Phlebia radiate, Pleurotus ostreatus, and Trametes versicolor [50]. Many Trichoderma species such as T. atroviridae, T. harzianum, and T. longibrachiatum are sources of laccase [51].
In plants, laccase plays a role in the lignification process only, while in fungi it plays a role in delignification, sporulation, pigment production, fruit body formation, and plant pathogenesis [52]. Although most of the purified laccase is extracellular enzymes, white-rot fungi also contain intracellular laccase. Laccase exists in a variety of structures, the majority consisting of monomeric structures, but some in homodimeric, heterodimeric, and multimeric forms.
The molecular mass of laccase produced by fungi ranges from 50 to 140 kDa, depending on the type of fungi [53]. Laccase from fungi usually undergoes a glycosylation reaction, with a mass increase of 10–25%. The carbohydrate portion of the laccase has been shown to maintain conformational stability and to protect the enzyme from proteolysis and inactivation by radicals [54].
The redox potential (E °) of the laccase is the energy required to remove electrons from the reducing substrate. Laccase from fungi can oxidize substrates with high E ° (E °> 400 mV) [55]. Filamentous fungi are usually more attractive hosts for heterologous protein production because of their faster microbial growth, ease of gene manipulation, their ability to excrete large amounts of protein into the growth medium, and the ability to carry out post-translational modifications [56].
Laccase from bacteria has shown greater tolerability from neutral to alkaline pH, and high concentrations of sodium chloride. It was still active in concentrations of 1 M or higher, as in the laccase produced by Marinomonas mediterranea and Bacillus halodurans [57]. Some of the laccase produced by bacteria shows a high tolerance to different solvents, including ethanol, methanol, dimethylformamide, acetonitrile, acetone, and dimethylsulfoxide, as observed in the laccase produced by Bacillus pumilus, which generally retains more than 50% of its activity in a mixture of water and water solvent [58]. Although the laccase from bacteria is generally more stable than the fungal laccase, the laccase from bacteria has a low redox potential (E ° T1 <+460 mV) so it is less suitable to be applied as a lignin-degrading lignocellulose [59].
2.4.1.1. Factors affecting the production and activity of laccase
1.Temperature
The temperature effect is limited in terms of laccase production. The optimal temperature of lacquer differs greatly from one type to another. It has been found that 25 °C is the optimal temperature for the production of laccase with light but in the dark, the optimal temperature is 30 °C [52]. The optimal temperature range for laccase production is between 25 °C and 30 °C [60]. In a study conducted by Farnet et al., it was found that the enzyme preincubation at 40 °C and 50 °C was able to increase laccase activity drastically [61]. Laccase is almost completely active in the temperature range 40 °C – 60 °C, with maximum activity at 50 °C. Activity remained unchanged after prolonged incubation at 40 °C for more than 4 h [62].
-
2.
pH
The optimal pH varies according to the field because different media causes different reactions to laccase. Cordi et al. using syringaldazine as a substrate and determining the effect of pH on enzyme activity in the range of pH 3.0–8.0 [63]. Laccase was extracted from Trametes versicolor which showed high enzyme activity over a wide pH range and temperature range but optimal activity was found at pH 3.0 and 50 °C. Laccase extracted from Stereum ostrea showed the highest activity at pH 6.0 and temperature 40 °C [64]. When fungi are grown in pH 5.0 media, laccase is overproduced and most studies have shown that a pH between 4.5 and 6.0 is suitable for enzyme production [52].
-
3.
Inducer
Laccase production has been shown to be highly dependent on the cultivation of the fungi. During the secondary metabolic phase, the ligninolytic system is activated and triggered by nitrogen concentration [65]. Laccase is generally produced in low concentrations by laccase-producing fungi, but higher concentrations can be obtained by adding various supplements such as xenobiotic compounds to the media [66]. The addition of aromatic compounds such as 2,5-xylidine, lignin, and veratril alcohol is known to increase and induce laccase activity [67]. Veratril alcohol is an aromatic compound, its addition to the culture medium resulted in increasing laccase production.
The addition of 2,5-xilidine after 24 h of cultivation gave the highest induction of laccase activity and increased laccase activity ninefold. Higher concentrations of 2,5-xylidine has a reducing effect due to toxicity [68]. The addition of an inducer increases the concentration of the specific laccase enzyme [69]. The supply of nutrients such as carbon and nitrogen affects the synthesis of laccase by fungi, but copper and other metal ions are also important laccase inducers. Fungal species influence the choice of metal inducers. Laccase activity was achieved by adding different metal ions (Cu2+, Mn2+, Cd2+, and Fe2+) to the Pleurotus ostreatus and found that Cu2 + stimulated the production of the laccase enzyme from fungi most strongly. New Basidiomycetes, Trametes sp. 420, producing laccase in glucose medium and cellobiose medium with induction of 0.5 mM and 6 mM o-toluidine [70]. Five mM EDTA is known to be able to inhibit total laccase activity [71].
In a study conducted by Arora and Gill (2001), better enzyme production was obtained in media containing a mixture of salt broth (MSB) and malt extract compared to MSB alone. Malt extract contains a lot of the aromatic amino acids tryptophan and tyrosine. The tryptophan-derived metabolite (3- hydroxy-2-aminobenzoate) acts as a mediator in laccase-catalyzed oxidation reactions in the white-rot fungi Pycnoporus cinnabarinus. Additionally, laccase is known to react with 4-hydroxyindole, a tryptophan derivative. P. radiata is known to provide maximum laccase production (6.18 U/mL) in MSB-malt extract which is the best nutritional medium for high laccase producers [72]. The difference in laccase enzyme activity produced by several inducers on the media can be seen in Table 2.
Table 2.
Laccase activity in several species of white-rot fungi using different media inducers.
| Inducer medium | Laccase activity of few species of white-rot fungi (U/mL) |
|||
|---|---|---|---|---|
| D. flavida [73] | P. brevispora [74, 75] | P. radiata [72, 76] | P. sanguineus [60, 64] | |
| Mineral salt broth (MSB), rice straw | 9.25 | 8.90 | 9.120 | 2.740 |
| Malt Extract, rice straw | 8.49 | 5.885 | 5.320 | 8.330 |
| MSB, malt extract, guaiacol | 1.24 | 5.700 | 11.590 | 1.617 |
| Malt Extract, sugarcane residues | 3.30 | 8.560 | 8.90 | 8.99 |
| MSB | 0.060 | 0.350 | 0.045 | 0.130 |
| Malt Extract | 0.050 | 1.370 | 0.415 | 0.672 |
| MSB, Malt Extract | 0.660 | 4.045 | 6.180 | 0.502 |
| Malt Extract | 0.150 | 0.165 | 9.470 | 0.055 |
| Malt Extract | 0.332 | 2.835 | 0.045 | 1.745 |
| MSB, Malt Extract, veratril alcohol | 0.64 | 4.175 | 11.185 | 0.235 |
| MSB, guaiacol | 0.217 | 0.605 | 10.475 | 0.647 |
| Malt Extract, guaiacol | 1.042 | 5.695 | 2.490 | 4.765 |
(Source: listed in the table).
Induction of ligninolytic activity can be triggered by high levels of lignin from natural lignocellulosic substrates which function as a natural inducer and a source of carbon for the enzyme production process. In the research conducted by Ningrum (2018), it can be seen that a significant increase in the activity of the laccase enzyme produced by the white-rot fungi, Cerrena sp. The highest laccase enzyme activity was obtained from corncob media on the 7th day after incubation, which was 24,237 U/mL. Whereas in the media of sago pile, rice husks and oil palm empty bunches also showed high activity, those respectively were 18,743 U/mL (day 6), 20,227 U/mL (day 5), and 16,274 U./mL (day 3). The substrate of corncob produced the highest enzyme activity even though it had lower lignin levels than rice husk. It means that the activity of the laccase enzyme produced is not only influenced by the amount of lignin present in the substrate, but also by the type of lignin. The three monolignol monomer structures (coniferyl, p-cumaryl alcohol, and synapyl alcohol) are the main constituents of lignin found in monocotyl plants such as corncob. It is different with the structure of lignin in grasses such as rice husks which only contain guaiacil, a derivative of coniferyl alcohol [77].
The production of the laccase enzyme from the species Pleurotus ostreatus has also been widely studied. Agricultural waste can also be used as an inducer in the laccase enzyme production process. The differences in the activity of the laccase from various agricultural wastes and various carbon/nitrogen sources can be seen in Table 3.
Table 3.
Laccase activity of few species of rot fungi in lignocellulose substrate.
| Species rot fungi | Substrate | Laccase Activity | Reference |
|---|---|---|---|
| Cerrena unicolor Han 849 | Populus beijingensis | 295.96 ± 4.85 U/L | [78] |
| Firmiana platanifolia | 625.98 ± 24.08 U/L | ||
| Sorghum bicolor | 371.71 ± 5.69 U/L | ||
| Oryza sativa | 102.17 ± 3.55 U/L | ||
| Ganoderma lucidum | Pineapple leaves | 472.31 + 41.2 IU/mL | [79] |
| Wheat straw | 100.4 IU/mL | [80] | |
| Banana stalk | 249.7 IU/mL | ||
| Sugarcane bagasse | 192.1 IU/mL | ||
| Rice straw | 338.4 ± 2.8 IU/mL | ||
| Trametes versicolor | Corn stalk | 1241.07 U/g | [81] |
| Steam-explode corn stalk | 2600.33 U/g | ||
| Olive leaves | 276.62 U/g | [82] | |
| Coriolopsis caperata | Wheat bran | 1623.55 U/g | [83] |
| Pleurotus ostreatus | ABTS | 774 U/L | [84] |
| Schizophyllum commune NI-07 | Polyurethane foam cubes (PUF) | 7307 U/mL | [85] |
| Coriolopsis gallica | Sawdust | 1480 U/L | [86] |
| Filter paper | 1300 U/L | ||
| Pleurotus ostreatus | Peel of mandarin Orange | 4,8 ± 0,08 U/L | [87] |
| Wood powder | 19,42 ± 0,14 U/g | ||
| 26 ± 0,98 U/g | |||
| Wood powder of grape tree | 6,9 ± 0,4 U/L | ||
| Sugarcane waste | 151,6 U/g | ||
| 9,942 U/g | |||
| 1079,8–1139,8 U/L |
(Source: listed in the table).
2.4.2. Lignin peroxidase (LiP)
Lignin peroxidase (LiP) was first discovered in 1983 in the ligninolytic culture of P. chrysosporium where it is one of the main components of the ligninolytic system. After further investigation, only about 40% of white-rot fungi produce Lignin Peroxidase, some other examples of microorganisms are T. versicolor, Phanerochaete sordida, Phlebia radiata, Pleurotus ostreatus, and Phlebia tremellosa. In most fungi, lignin peroxidase is present as a series of isoenzymes encoded by different genes [88].
Lignin peroxidase is a heme-containing enzyme that catalyzes the hydrogen peroxide-dependent oxidative degradation of lignin. LiP is a glycoprotein with an average molecular weight of 38–46 kDa. Lignin peroxidase purified from T.versicolor was found as a homogeneous monomer protein with a molecular weight of 30 kDa [89]. Lignin peroxidase also has high redox potential (700–1,400 mV), an optimum pH of 3–4.5, and the ability to catalyze the degradation of a large number of aromatic structures such as veratryl alcohol (3,4-dimethoxybenzyl) and methoxybenzenes.
2.4.2.1. Factors affecting LiP enzyme production and activity
-
1.
Substrate Selection
Several compounds were tested as potential substrates for lignin peroxidase at a concentration of 100 mM. The rate of substrate oxidation is measured by determining the increase in absorbance at the respective wavelengths. These substrates include veratryl alcohol (310 nm), guaiacol (436 nm), catechol (392 nm), butyl alcohol (310 nm), vanillic acid (390 nm), amyl alcohol (310 nm), and pyrogallol (450 nm). Lignin peroxidase can oxidize a number of aromatic phenolic compounds. Among all the mediator compounds tested, veratryl alcohol, which is known to act as an inducer in most white-rot fungi, causes increased lignin peroxidase activity. While other compounds tested failed to influence lignin peroxidase activity [31].
Another experiment by Parshetti (2011) used substrates commonly used for lignin peroxidase activity tests, namely L-Dopa, 8-hydroxyquinone, mimosine, veratryl alcohol, and xylidine. Of the various substrates tested, veratryl alcohol produced the highest activity compared to other substrates, followed by 8-hydroxyquinone, L-Dopa, mimosine, and the last one, xylidine [90]. Lignin peroxidase activity was measured using n-propanol as a substrate. The reaction mixture contains 40 mM n-propanol, 2 mM hydrogen peroxide, and 50 mM tartaric acid. One unit of enzyme activity is defined as the amount of enzyme required to oxidize 1 μmol of substrate per minute. This was measured by reading the absorbance at 310 nm for 1 min. The activity was found to be maximum with n-propanol followed by guaiacol, catechol, and veratryl alcohol. No activity was detected with the hydroxyquinone substrate [91].
-
2.
pH
Vandana et al (2019) tested lignin peroxidase activity using 100 mM veratryl alcohol at different pH values ranging from 3 to 10 with intervals of 0.5 using the appropriate buffer. Buffers include citric acid sodium phosphate buffer with a pH of 5–6.5, a sodium phosphate buffer with a pH of 7–8.5, a sodium tartrate buffer with a pH of 3–4.5, and a carbonate buffer with a pH of 9–10, all of which are prepared at a concentration of 0.5 M. The pH stability of pure lignin peroxidase was determined by exposing the enzymes to pH values ranging from 3 to 10 at intervals of 0.5 and estimating activity after 30 s and 15 min. In this study, lignin peroxidase activity with veratryl alcohol substrate had an optimal pH of 5.5 with lignin peroxidase activity of 400u/ml. Most activity was then followed by pH 5 (360u/ml); 4.5 (355 u/ml); 4 (350 u/ml); 3.5 (340u/ml); and 3 (310u/ml). At pH 6, the lignin peroxidase activity decreased significantly (280u/ml), then a drastic decrease starting from pH 6.5 to 10 where the lignin peroxidase activity was only in the range of 50u/ml to 150u/ml [31].
In Nature's research (2009), the method used to test lignin peroxidase activity was the same, but the results obtained were slightly different from those done by Vandana et al (2019). After 30 min of incubation, Lignin peroxidase activity was found to be stable at pH 5.0. Lignin peroxidase activity is lower at pH below 3 and higher below pH 7. More than 70% of enzyme activity is lost at pH 7–11 [16]. While in this research, Hariharan and Nambisan (2013), tested lignin peroxidase activity at pH 3–8. The highest activity reached 1300 IU/ml at pH 5. While the lowest activity occurred at pH 8, followed by pH 3, 7, 4, and 6 [79].
-
3.
Temperature
Lignin peroxidase was incubated with 100 mM veratryl alcohol in 0.5 M sodium tartrate buffer with a pH of 5.5. The temperature ranges from 25 °C to 40 °C with intervals of 0.5 °C, then the optimal temperature is determined. Subsequently, the enzymes were incubated for various time intervals ranging from 0 min to 10 min with 2-minute intervals at temperatures ranging from 25 °C to 85 °C with 10 °C intervals to test the effect of incubation temperature on temperature. enzyme activity. Enzyme thermostability is determined by incubating the enzymes at 60 °C, 65 °C, 70 °C, 75 °C, and 80 °C for periods ranging from 20 min to 120 min with 20-minute intervals. The tube is then immediately cooled in an ice bath and the activity is determined [31].
From this experiment, lignin peroxidase in P. chrysosporium had optimal activity at 30 °C. In contrast to the research of Vandana et al (2019), in a study conducted by Zeng et al (2010), it was found that the optimal culture temperature of P. chrysosporium is around 35 °C, where at this temperature microorganisms are more conducive to degrade lignin than inoculation at other times [31, 92]. In both studies, it was found that lignin peroxidase activity with temperatures above 60 °C was drastically lost after 10 min of exposure. As for the research conducted by Padma (2013), it was found that the activity of ligninolytic enzymes was highest at 27 °C, while at temperatures above that the activity decreased significantly [79]. Alam (2009), tested the effect of temperature on enzyme stability from 25 °C to 75 °C at pH 5. The results showed that lignin peroxidase activity was still 100% up to 55 °C. Lignin peroxidase activity decreased to more than 75% at temperatures above 55 °C and activity was lost by 66% at 75 °C [93]. Hariharan and Nambisan (2013) tested the activity of the lignin peroxidase enzyme in a temperature range of 23–33 °C. The highest activity occurs at 27 °C while the lowest occurs at 33 °C [79].
-
4.
Incubation time
The effect of contact time (incubation) was tested on the stability of LiP which was carried out together with the optimum pH and temperature. The enzymes were incubated in 1 ml 50 mM sodium acetate buffer (pH 5.0) at 55 °C for 1, 3, 6, 12, 24, 48, 72, 96, and 120 h. After each incubation period, the enzymes were immediately cooled in an ice bath and the residual activity was determined. Enzyme activity is expressed as the percentage with the highest activity recorded as 100%. LiP activity was maintained at 100% for the first 3 h of the incubation period. Activity decreased to less than 80% after 12 h of incubation. After 48 h, LiP activity remained stable at around 60% and remained constant for up to 120 h of incubation time. The effect of the incubation period on the activity of the enzyme lignin peroxidase was examined by incubating the inoculum at a temperature range of 24–192 h with an inoculum volume of 4 ml. Maximum ligninase production was obtained after 120 h of incubation, with LiP activity of 1203.7 IU/mL [79].
2.4.2.2. Screening method of LiP enzyme activity
To test the presence of lignin-degrading enzymes (ligninase), previously a spot test can be performed by dropping a 1.0% pyrogallol solution mixed with 0.4% H2O2 (composition 1:1) on the edge of the microbial culture being tested (which is still actively growing). The culture was then observed for 3 h, 24 h, and 72 h after dropping. The brownish-yellow color on the area where the pyrogallol reagent drops indicate the activity of the lignin peroxidase enzyme.
Lignin peroxidase activity was then determined spectrophotometrically (Shimadzu UV 1800 spectrophotometer) by measuring the oxidation rate of veratryl alcohol to veratraldehyde in the presence of H2O2 [94]. The veratryl alcohol test contains 0.8 mM veratryl alcohol in 0.1 M sodium tartrate buffer (pH = 3.0). 1 mL of culture buffer filtrate was added at 150 mM H2O2. The absorbance was determined at 310 nm for 1 min at 30 °C. One unit of lignin peroxidase activity is defined as 1 μmol veratraldehyde formed/minute and expressed as U/mL (molar extinction coefficient, ε max = 9300 M-1 cm-1) [31].
The second method is based on the oxidation of azure B dye. The reaction mixture contains 1 ml of 125 mM sodium tartrate buffer (pH 3.0), 500 μl 0.160 mM azure B, 500 μl culture filtrate and 500 μl 2 mM hydrogen peroxide. The reaction begins by adding hydrogen peroxide and one unit of enzyme activity has been denoted as OD. reduction of 0.1 units per minute per ml of culture filtrate [75].
Enzyme activity is measured by the following equation:
where:
At = Absorbance at the 30th minutes
A0 = Absorbance at zero minute
ϵmaks = Molar absorptivity of veratryl alcohol (9300 M−1 cm−1)
d = Cuvet width (cm)
t = Time (30 min)
Vtot = Total volume of solution (ml)
Venzyme = Volume of enzyme solution (ml)
The level of activity of lignin peroxidase can be influenced by several things such as the influence of the substrate, pH, temperature, and incubation time. These can be seen from the following Table 4.
Table 4.
Comparison of lignin peroxidase activity from several studies.
| Species | Temp (oC) | pH | Substrate | Activity (U/ml) | Reference |
|---|---|---|---|---|---|
| Phanerochaete chrysosporium | 30 | 5.5 | Veratryl Alcohol |
417 | [31] |
| Phlebia chrysochreas | 30 | 4.5 | Veratryl Alcohol |
45 | [95] |
| Ganoderma lucidum | 27 | 5 | Veratryl Alcohol | 1263 | [79] |
| Phanerochaete chrysosporium | 35 | 5 | Veratryl Alcohol | 744 | [93] |
| Ganoderma lucidum | 40 | 3 | Veratryl Alcohol | 766 | [96] |
|
Schyzophyllum Commune |
35 | 4 | Veratryl Alcohol | 468 | [97] |
| Kocuria rosea | 50 | 3 | Veratryl Alcohol |
64 | [90] |
| Phanerochaete chrysosporium | 40 | 2 | n-propanol | 1571 | [98] |
| Phlebia radiate | 35 | 4 | Veratryl Alcohol | 260 | [99] |
| Phanerochaete chrysosporium | 30 | 4.5 | Veratryl Alcohol | 370 | [100] |
| Phanerochaete chrysosporium | 35 | 5 | Veratryl Alcohol | 798 | [92] |
(Source: listed in the table)
2.4.3. Manganese peroxidase (MnP)
The enzyme manganese peroxidase (MnP, or Mn (II): H2O2 oxidoreductase, EC 1.11.1.13) is an extracellular glycoprotein containing heme as a prosthetic group [101]. MnP has a molecular weight ranging from 32 kDa - 75 kDa [102] and shows different characteristics depending on the source and isoform. MnP together with the H2O2 generating system are the main components of the lignin degradation system [103].
2.4.3.1. Sources of MnP
MnP is included in the heme peroxidase or non-animal peroxidase enzyme, namely peroxidase enzymes produced by microorganisms, and/or plants. The non-animal peroxidase enzyme group has three classes/subdivisions based on localization and cellular function, where MnP is included in class II, namely the peroxidase enzyme group secreted in the extracellular tissue of fungi [103, 104]. Fungi, especially white-rot fungi (WRF), are the largest producers of ligninase, precisely 90% of all wood-degrading species [29].
Apart from fungi, lignin-degrading enzymes can also be found in bacteria and algae [28, 97]. The first MnP was discovered by Glenn & Gold and Paszcznski et al., (1986) on Phanerochaete chrysosporium which then attracted researchers to study MnP further to date [15, 29].
The WRF group of fungi is a group of fungi under the Basidiomycota division that can completely degrade the lignin component of the lignocellulosic substrate. This group of fungi degrades lignin by producing extracellular ligninases, namely laccase, LiP, MnP, and VP [105]. Members of the order Polyporales and Agaricales, such as Ganoderma sp., Phlebia radiate, Lentinula edodes, and Pleurotus sp., Mostly represent the WRF group [29]. Table 5 shows several MnP sources that have been isolated and characterized from fungi.
Table 5.
Sources and characteristics of MnP from several fungi and bacteria.
| Species | MW (kDa)/A (U/L) | Substrate Of Cultivation | Method | Optimum Temp. (°C) | Stability Temp. (°C) | Optimum pH | Reference |
|---|---|---|---|---|---|---|---|
|
Ganoderma lucidum IBL-05 |
43 | Wheat husk | SSF | 40 | <45 | 5.0 | [106] |
|
Echinodont ium taxodii 2538 |
53.4 | Moso bamboo | SSF | 55 | <45 | 3.5 | [107] |
|
Trametes polyzona MnP1 dan MnP2 |
44 | Remazol brilliant red F3B gran |
SmF agitation 130 rpm | 90 | 20–37 | 4.5 | [108] |
| 42 | 70 | 20–50 | 4.5 | ||||
| Fusarium sp. | - | Rice straw, wood | SmF agitation 180 rpm | 30 | 20–50 | 4.0 | [101] |
|
Cerrena unicolor BBP6 |
45 | SmF agitation 150 rpm | 60 | 20–50 | 4.5 | [109] | |
|
Bjerkander a adusta CX-9 |
30 | o-dianisidine | SmF agitation 150 rpm | 70 | - | 3.0 | [110] |
|
Trametes versicolor IBL-04 |
43 | Corn cob | SSF | 50 | ≤65 | 5.0 | [111] |
|
Irpex lacteus CD2 |
42 | - | SmF agitation 150 rpm | 70 | 40–60 | 4.5 | [112] |
| Bacillus pumilus | [113] | ||||||
| Lysinibacillus sp. | -/76,4 | SmF | 30 | 7 | [114] |
In addition to fungi, soil bacteria and lumen bacterial of termite are composting microorganisms that also work in the process of lignin degradation [29, 115]. Study on bacterial-MnP has been published recently, such as using Bacillus pumilus, Paenibacillus sp. [113], Azospirillum brasilense [116]. The activity of MnP in those studies was performed with and without the addition of an inducer. A recent study reporting activity of MnP from bacterial isolated from the digestive tract of Coptotermes curgnathus, especially Bacillus sp., Lysinibacillus sp., and Acinetobacter sp. Lysinibacillus showed the highest MnP activity after 7 days of cultivation [114], and followed by Bacillus aryabhattai that isolated from a liquid waste of pulp and paper fabrics.
2.4.3.2. Screening methods of MnP activity
In conducting in vitro MnP enzyme activity assays, various phenolic and non-phenolic substrates can be used. MnP can oxidize phenolic substrates (ex. ABTS, DMP, vanililasetone, ferulic acid, syringol, guaiacol, isoeugenol, p-metoxyphenol, phenol red, catechol, hydroquinone, etc.) and non-phenolic (eg vanilyl alcohol, veratryl alcohol, and benzyl alcohol) in the presence of H2O2. Guaiacol is the most commonly used substrate [35]. Just like other parameters, each MnP has a different level of substrate specificity such as MnP from B.adusta has the highest substrate specificity with 2,6-DMP while from T. polyzone with ABTS [108, 110].
2.4.3.3. Factors affecting MnP production and activity
In the process of enzyme production from microorganisms, special production conditions need to be put in place so that they can grow and the enzymes are produced optimally. Factors that influence the production and activity of ligninolytic enzymes include cultivation methods, sources and ratios of carbon and nitrogen, pH, temperature, and metal ion inducers.
-
1.
Cultivation methods
The method of cultivating microorganisms plays an important role in the production of ligninolytic enzymes. The cultivation of microorganisms can be carried out using the Solid-State Fermentation (SSF) and Submerged Fermentation (SmF) methods. SSF is defined as fermentation involving solids in the absence of free water, but the substrate must have sufficient moisture to support the growth and metabolism of microorganisms [117]. SmF is a fermentation that is carried out in the presence of excess free water, a more conventional method, and is commonly involved with agitation during production [118].
There are four main ways to grow microorganisms in SmF, namely batch culture, fed-batch culture, perfusion batch culture, and continuous culture. In batch culture, the microorganisms are inoculated in a fixed volume medium. In the case of fed-batch culture, the concentrated nutritional component is gradually added to the batch culture. In a perfusion batch culture, the culture was added and the same volume was withdrawn from the cell-free media that had been used. In continuous culture, fresh media is added to the batch system in an exponential phase of microorganism growth by withdrawing the media containing the product. Sustainable cultivation provides nearly balanced growth, with little fluctuation in nutrients, metabolites, cell count, or biomass [119]. Table 5 shows some of the microorganisms and cultivation methods that have been applied.
-
2.
Source of Carbon and Nitrogen
Carbon and nitrogen play an important role in the growth of microorganisms and enzyme production [97]. Carbon acts as a building block for organic matter in cells of all organisms, and also serves as a source of energy. As a result, most of this carbon enters energy-producing metabolic pathways and is eventually secreted from cells as CO2. Carbohydrates are an excellent source of carbon, oxygen, hydrogen, and metabolic energy. The availability of carbohydrates for microorganisms usually depends on the complexity of the molecules. In general, they can be ranked as follows:
| Hexose > Disaccharides > Pentose > Polysaccharides (94) |
Lignocellulosic substrates are also often used as a carbon source in the form of polysaccharides [120]. Akpinar & Urek (2012) used grape stalks, skins, and seeds from the waste of grape juice production, which carbohydrate content was analyzed and used as a carbon source in the production of ligninase from Pleurotus eryngii. Nitrogen can be obtained from various sources with the commonly used extracts of yeast or peptone [26, 121].
Nitrogen is the second most abundant compound in the fermentation medium. Nitrogen is used for the anabolic synthesis of nitrogen-containing cellular compounds, such as amino acids, purines, DNA, and RNA. Many algae and fungi use ammonium nitrate and sodium nitrate as nitrogen sources, but yeast and bacteria have problems utilizing nitrogen in these forms due to decreasing pH soon after growing. Some organisms are capable of assimilating nitrites. Sources of organic nitrogen in synthetic media are specific amino acids, purines, pyrimidines, and urea. Urea, depending on the buffer capacity of the system, will increase the pH value of the medium. Ammonium sulfate produces acidic conditions because ammonia is quickly used and free acids are then released [120]. Some complex organic nitrogen sources such as corn, dry distillate, yeast, fish or bone meal, cottonseed, milk protein, are often used in commercial fermentation. These nitrogen sources provide carbon and many other nutrients and are quite cheap and affordable [94].
The ratio of carbon to nitrogen (C: N) plays an important role because microorganisms need a good balance of carbon and nitrogen to remain active [122]. A good balance of carbon and nitrogen needs to be maintained in the cells and it has been stated that a C: N ratio of 20–30 with an optimal ratio of 24 in the "diet" can maintain this balance [123].
-
3.
Temperature, pH, and metal ions
The effect of temperature and pH affects the activity and stability of the MnP enzyme. MnP from each species and strain of microorganisms gave varying reactions concerning temperature and pH. Adjustment to the pH of the growth medium can be done by adding a strong acid/base and then stabilizing it using a pH buffer.
Metal ions essential for fungal growth include Cu2 +, Fe2 +, Mn2 +, Mo2 +, Zn2 +, Ni2 + and common non-essential metals including Cr3+, Cd2 +, Pb2 +, Hg2 +, and Ag+. Essential metals are relatively less toxic than heavy metals and increase the growth rate of fungi when present at low concentrations [124]. However, although these metal ions are important for fungal growth, not all of them have a significant or positive effect on MnP activity.
2.5. Application of ligninolytic enzymes
Neither laccase nor peroxidases (LiP and MnP) are strongly selective of substrates. The characteristics are advantageous for industrial application due to the wide range of substrates they can work. Laccase will act on the removal of the phenolic compound of wine, development of fuel cell and biosensor, and also be used for bioremediation processes of dyes and effluents in the pharmaceutical industry, as well as chemical, cosmetics, and textile industries. Laccase and both peroxidase enzymes have been applied in few delignification processes, such as fungal (biomass) delignification, enzymatic delignification [125, 126, 137, 138].
In the beginning, only the Laccase enzyme has been thoroughly studied for its singular ability. Hattaka et al. (2003) have reported several studies regarding the single ability of MnP to delignify lignocellulose, including writing that pure MnP from P. radiata has been reported to convert pine wood chips into small molecular weight fragments in the presence of Tween 80 and glucose-glucose oxidase complexes. Another study by Masarin, et al. (2016) carried out the delignification process of Eucalyptus grandis wood chips using the partially purified MnP enzyme from C. subvermispora, showing that MnP was able to degrade lignin as much as 23.44%. The reaction conditions contained the equivalent of 200 IU MnP, Tween 60, MnSO4, and glucose - glucose oxidase complex at pH 4.5, temperature 27 °C for 72 h, and agitated at 120 rpm [127].
Recently, ligninolytic enzymes especially MnP and LiP have been used singularly for the detoxication function of hazardous compounds [128]. MnP can degrade and detoxify aflatoxin B1 [129], persistent organic pollutants (PAHs) [130], and many toxic synthetic dyes, such as Poly R-478, Reactive Red 195A, Reactive Blue 21, Reactive Yellow 145A [131], Indigo carmine, Remazol Brilliant Blue R, Remazol Brilliant Violet 5R, and Methyl Green [132]. LiP has the ability to degrade and detoxify halogenated phenol [133], endocrine-disrupting pollutants like bisphenol A, estrone, ethinylestradiol [134], and also synthetic dyes [135, 136].
In its application, MnP is frequently present together with other ligninolytic enzymes in the form of crude enzyme extracts. However, research by Zeng et al. (2013) who compared the treatment of rice straw with crude extract and a pure mixture of MnP and LiP from P. chrysosporium showed that the pure mixture of enzymes provided an almost higher rate of lignin degradation (33%) compared to crude extract (17%) [21]. Ahmad et al., (2016) then deepened research on pure enzyme mixtures by testing the effect of the ratio of the LiP, MnP, and Lac enzymes on the effectiveness of lignin degradation in wheat straw, rice straw, and bagasse. It was found that wheat straw and bagasse were delignified optimally with a 2: 1: 2 MnP: LiP: Lac ratio, with 58.5% and 55% degradation rate respectively [20]. Meanwhile, rice straw was optimally delignified at a ratio of 1: 2: 2 with a degradation rate of 52 %. Although the use of pure enzymes shows higher effectiveness (almost 2 times), the use of pure enzymes is not cost-effective and not economical, especially when using commercial enzymes for large processes with lignocellulosic substrates from nature [20].
Solution for more cost-effective and economical use of enzyme was possibly by technique of enzyme immobilization, as well as an answer for better performance. Immobilization can be performed by using suitable supports, such as mesoporous silica, hydrogel, and smart polymer. There are several techniques of enzymes immobilization onto supports which range from reversible physical adsorption and ionic linkages, to the irreversible stable covalent bonds. Such techniques produce immobilized enzymes with different varying stability [140, 141].
Another study by Kong et al., (2016) isolated MnP from E. taxodii and tested the ability of purified MnP and Lac in oxidizing 3 models of phenolic lignin compounds and 3 models of non-phenolic compounds. It was found that MnP was able to degrade the lignin phenolic compound model; 4-Hydroxy-3-methoxy cinnamic acid, 4-Hydroxy cinnamic acid, and 4-Hydroxy-3,5-dimethoxy cinnamic acid as much as 97.79%, 28.35%, and 99.92%, respectively. MnP was also able to degrade the model compounds of the lignin non-phenolic model; cinnamic acid, 3-Methoxy cinnamic acid, and 3,5-Dimethoxy cinnamic acid as much as 26.19%, 30.03%, and 44.84%. Compared to Lac, the ability of MnP to degrade phenolic compounds was lower but for non-phenolic compounds, Lac was not able to degrade it at all [107]. Although the effectiveness of MnP looks very good on phenolic lignin but overall the delignification process has not been completed. Most likely the delignification process can be completed with the help of hydrolytic enzymes/non-oxidative enzyme system [146].
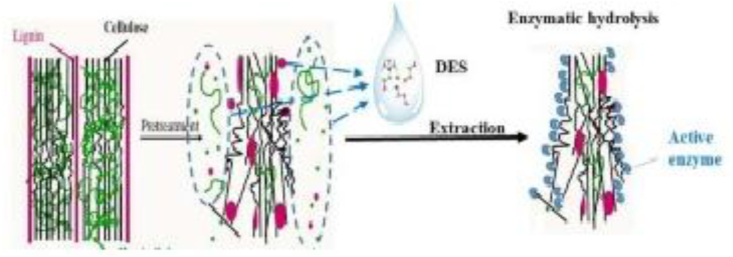
On the other hand, there has been an encouraging development of lignin extraction using DESs (deep eutectic solvents). Natural DESs or DESs are eutectic mixture of liquid solvent between hydrogen-bond acceptors (HBA) and hydrogen bond donors (HBD). These solvents gained much interest due to its ultimate capability of solubilizing lignin from lignocellulosic-biomass, as illustrated in Figure 2 [142,143]. Chen et al. (2020) reported that DESs also have favorable features such as easy synthesis, low vapor pressure, tunable physicochemical properties, lignin depolymerization and functionalization reaction, and biodegradability or green solvents [144]. These features open the way for combination with enzymatic methods, especially to separate non-phenolic lignin that difficult to remove by enzymatic method.
Figure 2.
Pretreatment of lignocellulosic biomass with DESs [142]. (Reprinted with permission from Ref. 142, Fang C. et al.,(2017), ACS).
3. Conclusion
The high activity of ligninolytic enzymes can result in a high level of delignification, and white-rot fungi Pleurotus ostreatus and Cerrena sp. are potential to produce high laccase activity compared to others, while Phanerochaete chrysosporium and Trametes versicolor are potential to produce lignin peroxidase and manganese peroxidase with high activity.
Some Factors and conditions can affect the optimal activity and production of ligninolytic enzymes including substrate, temperature, pH, incubation time, carbon and nitrogen sources, the addition of surfactants, mediators, and certain ions. Therefore, it is important to optimize each factors and conditions of cultivation before a large scale enzyme production.
The use of the two or three ligninolytic enzyme in a combination is the most effective way of biodelignification of lignocellulose biomass rather than microbial cell biodelignification or individual enzymatic biodelignification. However further studies need to be made to find the optimal ratio of the enzymes to be able to produces the highest lignin degradation, and the use of enzyme immobilization technique has to be considered for a more economical method.
Recent development in lignin extraction by DES solvent paving the way and a promising prospects for the combination of both methods, enzymatic delignification and DESs-extraction of lignin for complete delignification and better utilization of lignin.
Declarations
Author contribution statement
All authors listed have significantly contributed to the development and the writing of this article.
Funding statement
This work was supported by the Directorate of Research and Development UI (Hibah PUTI Q2, No:NKB1460/UN2.RST/HKP.05.00/2020).
Data availability statement
Data included in article/supp. material/referenced in article.
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
The authors would like to thanks the Directorate of Research and Development UI for supporting our research with grant Hibah PUTI Q2, No:NKB-1460/UN2.RST/HKP.05.00/2020.
References
- 1.Masran R., Zanirun Z., Bahrin E.K., Ibrahim M.F., Lai Yee P., Abd-Aziz S. Harnessing the potential of ligninolytic enzymes for lignocellulosic biomass pretreatment. Appl. Microbiol. Biotechnol. 2016;100(1):5231–5246. doi: 10.1007/s00253-016-7545-1. [DOI] [PubMed] [Google Scholar]
- 2.Shanmugapriya M., Lakshmiprabha M. Biobleaching and delignification of hard wood kraft pulp by white rot fungi. Int. J. Pharm. Chem. Sci. 2013;2(2):932–937. www.ijpcsonline.com [Google Scholar]
- 3.Shirkavand E., Baroutian S., Gapes D.J., Young B.R. Combination of fungal and physicochemical processes for lignocellulosic biomass pretreatment - a review. Renew. Sustain. Energy Rev. 2016;54:217–234. [Google Scholar]
- 4.Singh D., Chen S. The white-rot fungi Phanerochaete chrysosporium: conditions for the production of lignin-degrading enzymes. Appl. Microbiol. Biotechnol. 2008;81(3):399–417. doi: 10.1007/s00253-008-1706-9. [DOI] [PubMed] [Google Scholar]
- 5.Mood S.H., Golfeshan A.H., Tabatabaei M., Jouzani G.S., Najafi G.H., Gholami M., et al. Lignocellulosic biomass to bioethanol, a comprehensive review with a focus on pretreatment. Renew. Sustain. Energy Rev. 2013;27:77–93. [Google Scholar]
- 6.Foyle T., Jennings L., Mulcahy P. Compositional analysis of lignocellulosic materials: evaluation of methods used for sugar analysis of waste paper and straw. Bioresour. Technol. 2007;98(16):3026–3036. doi: 10.1016/j.biortech.2006.10.013. [DOI] [PubMed] [Google Scholar]
- 7.Harris D., DeBolt S. Synthesis, regulation and utilization of lignocellulosic biomass. Plant Biotechnol. J. 2010;8(3):244–262. doi: 10.1111/j.1467-7652.2009.00481.x. [DOI] [PubMed] [Google Scholar]
- 8.Su Y., Yu X., Sun Y., Wang G., Chen H., Chen G. Evaluation of screened lignin-degrading fungi for the biological pretreatment of corn stover. Sci. Rep. 2018;8(1):1–11. doi: 10.1038/s41598-018-23626-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Asmoro N.W., Afriyanti A., Ismawati I. Ekstraksi selulosa batang tanaman jagung (zea mays) metode basa. J. Ilm. Teknosains. 2018;4(1):24. [Google Scholar]
- 10.Thakur V.K., Thakur M.K. Recent advances in green hydrogels from lignin: a review. Int. J. Biol. Macromol. 2015;72:834–847. doi: 10.1016/j.ijbiomac.2014.09.044. [DOI] [PubMed] [Google Scholar]
- 11.Zabed H.M., Akter S., Yun J., Zhang G., Awad F.N., Qi X., et al. Recent advances in biological pretreatment of microalgae and lignocellulosic biomass for biofuel production. Renew. Sustain. Energy Rev. 2019:105. (June 2018):105–28. [Google Scholar]
- 12.Rajendran K., Drielak E., Sudarshan Varma V., Muthusamy S., Kumar G. Updates on the pretreatment of lignocellulosic feedstocks for bioenergy production–a review. Biomass Conv. Bioref. 2018;8(2):471–483. [Google Scholar]
- 13.Bolaji O.J. Thesis Newcastle University; 2019. Fungal Treatment of Selected Matured Forages to Improve Their Value for Ruminant Feeding. [Google Scholar]
- 14.Saini V.K., Thapliyal B.P., Sanjay N. Improved Delignification and Fiber Accessibility of Dendrocalamus Strictus by white Rot Fungi. 2013. Chapter 3 study on effect of variables for enhanced delignification by white rot fungi in mechanically destructured and non destructured bamboo (dendrocalamus strictus) p. 70.http://hdl.handle.net/10603/6212 [Google Scholar]
- 15.Solbiati J.O., Abdel-Hamid A.M., Cann I.K.O. Vol. 82. Elsevier; 2013. Chapter one - insights into lignin degradation and its potential industrial applications; pp. 1–28. (Adv. Appl. Microbiol.). [DOI] [PubMed] [Google Scholar]
- 16.Wong D.W.S. Structure and action mechanism of ligninolytic enzymes. Appl. Biochem. Biotechnol. 2009;157:174–209. doi: 10.1007/s12010-008-8279-z. [DOI] [PubMed] [Google Scholar]
- 17.Nagai M., Sakamoto Y., Nakade K., Sato T. Isolation and characterization of the gene encoding a manganese peroxidase from Lentinula edodes. Mycoscience. 2007;48(2):125–130. [Google Scholar]
- 18.Rodríguez-Couto S. Industrial and environmental applications of white-rot fungi. Mycosphere. 2017;8(3):456–466. [Google Scholar]
- 19.Wan C., Li Y. Fungal pretreatment of lignocellulosic biomass. Biotechnol. Adv. 2012;30:1447–1457. doi: 10.1016/j.biotechadv.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 20.Ahmad Z., Bajwa M.A., Hussain F., Randhawa M.A. A novel approach to delignify lignocellulosic materials by using ligninolytic enzyme consortium. Bioresources. 2016;11(4):10511–10527. [Google Scholar]
- 21.Zeng G.M., Zhao M.H., Huang D.L., Lai C., Huang C., Wei Z., et al. Purification and biochemical characterization of two extracellular peroxidases from Phanerochaete chrysosporium responsible for lignin biodegradation. Int. Biodeterior. Biodegrad. 2013;85:166–172. [Google Scholar]
- 22.Tsegaye B., Balomajumder C., Roy P. Microbial delignification and hydrolysis of lignocellulosic biomass to enhance biofuel production: an overview and future prospect. Bull. Natl. Res. Cent. 2019;43(1):1–16. [Google Scholar]
- 23.Kamel S., Ali N., Jahangir K., Shah S.M., El-Gendy A.A. Pharmaceutical significance of cellulose: a review. Express Polym. Lett. 2008;2(11):758–778. www.expresspolymlett.com [Google Scholar]
- 24.Chen H. 2014. Biotechnology of Lignocellulose: Theory and Practice. Biotechnology of Lignocellulose: Theory and Practice; pp. 1–511. [Google Scholar]
- 25.Nosya M.A. Universitas Lampung; 2016. Pembuatan Mikrokristal Selulosa Dari Tandan Kosong Kelapa Sawit. [Google Scholar]
- 26.Prasher I., Chauhan R. Effect of carbon and nitrogen sources on the growth, reproduction and ligninolytic enzymes activity of dictyoarthrinium synnematicum somrith. Adv. Zool. Bot. 2015;3(2):24–30. [Google Scholar]
- 27.Banik N., Dey V., Sastry G.R.K. An overview of lignin & hemicellulose effect upon biodegradable bamboo fiber composites due to moisture. Mater. Today Proc. 2017;4:3222–3232. [Google Scholar]
- 28.Ishak N., Sari A., Kassim M., Aripin A.M., Sharifah M., Oluwatosin A.F., et al. 2019. A Review on Lignin and Biodelignification; pp. 41–72. [Google Scholar]
- 29.Janusz G., Pawlik A., Sulej J., Swiderska-burek U., Jarosz-wilkołazka A., Paszczy A. Lignin degradation : microorganisms , enzymes involved , genomes analysis and evolution. FEMS Microbiol. Rev. 2017;41(6):941–962. doi: 10.1093/femsre/fux049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Gonzalo G., Colpa D.I., Habib M.H.M., Fraaije M.W. Bacterial enzymes involved in lignin degradation. J. Biotechnol. 2016;236:110–119. doi: 10.1016/j.jbiotec.2016.08.011. [DOI] [PubMed] [Google Scholar]
- 31.Vandana T., Kumar S.A., Swaraj S., Manpal S. Purification, characterization, and biodelignification potential of lignin peroxidase from immobilized phanerochaete chrysosporium. Bio. 2019;14(3):5380–5399. [Google Scholar]
- 32.Jeffries T.W. In: Biochemistry of Microbial Degradation. Ratledge C., editor. Kluwer Academic Publishers; Netherlands: 1994. 8.Biodegradation of lignin and hemicelluloses; pp. 233–277. [Google Scholar]
- 33.Sasmitaloka K.S., Suryani A., Mangunwidjaja D. Microbial delignification of corn stover by phanerochaete chrysosporium in solid state fermentation. IOSR J. Pharm. 2016;6(1):12–19. www.iosrphr.org [Google Scholar]
- 34.Moreno A.D., Tomás-Pejó E., Ballesteros M., Negro M.J. 2019. Pretreatment technologies for lignocellulosic biomass deconstruction within a biorefinery perspective. Biomass, Biofuels, Biochem Biofuels Altern Feed Convers Process Prod Liq Gaseous Biofuels; pp. 379–399. [Google Scholar]
- 35.Chandra R., Kumar V., Yadav S. I. 2017. Extremophilic ligninolytic enzymes; pp. 115–154. (Extremophilic Enzymatic Processing of Lignocellulosic Feedstocks to Bioenergy). [Google Scholar]
- 36.Baruah J., Nath B.K., Sharma R., Kumar S., Deka R.C., Baruah D.C., et al. Recent trends in the pretreatment of lignocellulosic biomass for value-added products. Front. Energy Res. 2018;6(DEC):1–19. [Google Scholar]
- 37.Nurjanah L., Falah S., Azhari A., Suryani S., Artika I.M. Trametes versicolor as agent for delignification of rice husks. Curr. Biochem. 2014;1(1):37–44. [Google Scholar]
- 38.Ponnusamy V.K., Nguyen D.D., Dharmaraja J., Shobana S., Banu J.R., Saratale R.G., et al. A review on lignin structure, pretreatments, fermentation reactions and biorefinery potential. Bioresour. Technol. 2019;271:462–472. doi: 10.1016/j.biortech.2018.09.070. [DOI] [PubMed] [Google Scholar]
- 39.Rezania S., Oryani B., Cho J., Talaiekhozani A., Sabbagh F., Hashemi B., et al. Different pretreatment technologies of lignocellulosic biomass for bioethanol production: an overview. Energy. 2020;199:117457. [Google Scholar]
- 40.Nadir N., Ismail N.L., Hussain A.S. Biomass for Bioenergy - Recent Trends and Future Challenges [Intenet] Intechopen; 2019. Fungal pretreatment of lignocellulosic materials; pp. 1–19. Available from: https://www.intechopen.com/books/advanced-biometric-technologies/liveness-detection-in-biometrics. [Google Scholar]
- 41.Kumar A., Chandra R. Ligninolytic enzymes and its mechanisms for degradation of lignocellulosic waste in environment. Heliyon. 2020;6(2) doi: 10.1016/j.heliyon.2020.e03170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peral C. Biotransformation of Agricultural Waste and By-Products, the Food, Feed, Fibre, Fuel (4F) Economy. Elsevier; 2016. Chapter 5: biomass pretreatment strategies (technologies, environmental performance, economic considerations, industrial implementation) pp. 125–160. [Google Scholar]
- 43.Bhatia S.K., Jagtap S.S., Bedekar A.A., Bhatia R.K., Patel A.K., Pant D., et al. Recent developments in pretreatment technologies on lignocellulosic biomass: effect of key parameters, technological improvements, and challenges. Bioresour. Technol. 2020;300:1–13. doi: 10.1016/j.biortech.2019.122724. [DOI] [PubMed] [Google Scholar]
- 44.Hosseini Koupaie E., Dahadha S., Bazyar Lakeh A.A., Azizi A., Elbeshbishy E. Enzymatic pretreatment of lignocellulosic biomass for enhanced biomethane production-A review. J. Environ. Manag. 2019;233:774–784. doi: 10.1016/j.jenvman.2018.09.106. [DOI] [PubMed] [Google Scholar]
- 45.Peralta R.M., da Silva B.P., Gomes Côrrea R.C., Kato C.G., Vicente Seixas F.A., Bracht A. Elsevier; 2017. Enzymes from Basidiomycetes-Peculiar and Efficient Tools for Biotechnology; pp. 119–149. Biotechnology of Microbial Enzymes: Production, Biocatalysis and Industrial Applications. [Google Scholar]
- 46.Lestari E. Universitas Hasanuddin; 2013. Potensi Jamur Pelapuk Kayu Isolat Lokal Makassar Dalam Mendekomposisi Komponen Lignoselulosa Jerami Padi Oryza Sativa L. Makassar. [Google Scholar]
- 47.Boyd-Wilson K., Walter M. 2008. Development of a Biotechnology Tool Using New Zealand White-Rot Fungi to Degreade Pentachlorophenol; pp. 1–11. 1. Available from: https://www.wasteminz.org.nz/pubs/development-of-a-biotechnology-tool-using-new-zealand-white-rot-fungi-to-degrade-pentachlorophenol/ [Google Scholar]
- 48.Hatti-Kaul R., Ibrahim V. In: Bioprocessing Technologies in Biorefinery for Sustainable Production of Fuels, Chemicals, and Polymers. Yang S., El-Enshasy H.A., Thongchul N., editors. John Wiley & Sons; 2013. Lignin-degrading enzymes: an overview. [Google Scholar]
- 49.Viswanath B., Subhosh Chandra M., Pallavi H., Rajasekhar Reddy B. Screening and assessment of laccase producing fungi isolated from different environmental samples. Afr. J. Biotechnol. 2008;7(8):1129–1133. [Google Scholar]
- 50.Kiiskinen L.L., Rättö M., Kruus K. Screening for novel laccase-producing microbes. J. Appl. Microbiol. 2004;97(3):640–646. doi: 10.1111/j.1365-2672.2004.02348.x. [DOI] [PubMed] [Google Scholar]
- 51.Hölker U., Dohse J., Höfer M. Extracellular laccases in ascomycetes Trichoderma atroviride and Trichoderma harzianum. Folia Microbiol. 2002;47(4):423–427. doi: 10.1007/BF02818702. [DOI] [PubMed] [Google Scholar]
- 52.Thurston C.F. The structure and function of fungal laccases. Microbiology. 1994;1(140):19–26. [Google Scholar]
- 53.Baldrian P. Fungal laccases-occurrence and properties. FEMS Microbiol. Rev. 2006;30(2):215–242. doi: 10.1111/j.1574-4976.2005.00010.x. [DOI] [PubMed] [Google Scholar]
- 54.Maestre-Reyna M., Liu W.C., Jeng W.Y., Lee C.C., Hsu C.A., Wen T.N., et al. Structural and functional roles of glycosylation in fungal laccase from lentinus sp. PLoS One. 2015;10(4) doi: 10.1371/journal.pone.0120601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Agrawal K., Chaturvedi V., Verma P. Fungal laccase discovered but yet undiscovered. Biores. Bioproc. 2018;5(4):1–12. [Google Scholar]
- 56.Garcia L.F., Lacerda M.F.A.R., Thomaz D.V., de Souza Golveia J.C., Pereira M das G.C., de Souza Gil E., et al. Optimization of laccase–alginate–chitosan-based matrix toward 17 α-ethinylestradiol removal. Prep. Biochem. Biotechnol. 2019;49(4):375–383. doi: 10.1080/10826068.2019.1573195. [DOI] [PubMed] [Google Scholar]
- 57.Jimenez-Juarez N., Roman-Miranda R., Baeza A., Sánchez-Amat A., Vazquez-Duhalt R., Valderrama B. Alkali and halide-resistant catalysis by the multipotent oxidase from Marinomonas mediterranea. J. Biotechnol. 2005;117(1):73–82. doi: 10.1016/j.jbiotec.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 58.Guan Z.B., Song C.M., Zhang N., Zhou W., Xu C.W., Zhou L.X., et al. Overexpression, characterization, and dye-decolorizing ability of a thermostable, pH-stable, and organic solvent-tolerant laccase from Bacillus pumilus W3. J. Mol. Catal. B Enzym. 2014;101:1–6. [Google Scholar]
- 59.Mate D.M., Alcalde M. Laccase engineering: from rational design to directed evolution. Biotechnol. Adv. 2015;33(1):25–40. doi: 10.1016/j.biotechadv.2014.12.007. [DOI] [PubMed] [Google Scholar]
- 60.Pointing S.B., Jones E.B.G., Vrijmoed L.L.P. Optimization of laccase production by Pycnoporus sanguineus in submerged liquid culture. Mycologia. 2000;92(1):139–144. [Google Scholar]
- 61.Farnet A.M., Criquet S., Tagger S., Gil G., Le Petit J. Purification, partial characterization, and reactivity with aromatic compounds of two laccases from Marasmius quercophilus strain 17. Can. J. Microbiol. 2000;46(3):189–194. doi: 10.1139/w99-138. [DOI] [PubMed] [Google Scholar]
- 62.Palmeiri G., Giardina P., Marzullo L., Desiderio B., Nittii G., Cannio R., et al. Stability and activity of a phenol oxidase from the ligninolytic fungi Pleurotus ostreatus. Appl. Microbiol. Biotechnol. 1993;39(4–5):632–636. doi: 10.1007/BF00205066. [DOI] [PubMed] [Google Scholar]
- 63.Cordi L., Minussi R.C., Freire R.S., Durán N. Fungal laccase: copper induction, semi-purification, immobilization, phenolic effluent treatment and electrochemical measurement. Afr. J. Biotechnol. 2007;6(10):1255–1259. [Google Scholar]
- 64.Valeriano V.S., Silva A.M.F., Santiago M.F., Bara M.T.F., Garcia T.A. Production of laccase by Pycnoporus sanguineus using 2,5 - xylidine and ethanol. Braz. J. Microbiol. 2009;40(4):790–794. doi: 10.1590/S1517-83822009000400009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fu S.Y., Yu H.S., Buswell J.A. Effect of nutrient nitrogen and manganese on manganese peroxidase and laccase production by Pleurotus sajor-caju. FEMS Microbiol. Lett. 1997;147(1):133–137. [Google Scholar]
- 66.Lee I.-Y., Jung K.-H., Lee C.-H., Park Y.-H. Enhanced production of laccase in Trametes vesicolor by the addition of ethanol. Biotechnol. Lett. 1999;21(1):965–968. [Google Scholar]
- 67.Bollag J.M., Leonowicz A. Comparative studies of extracellular fungal laccases. Appl. Environ. Microbiol. 1984;48(4):849–854. doi: 10.1128/aem.48.4.849-854.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Eggert C., Temp U., Eriksson K.E.L. The ligninolytic system of the white rot fungi Pycnoporus cinnabarinus: purification and characterization of the laccase. Appl. Environ. Microbiol. 1996;62(4):1151–1158. doi: 10.1128/aem.62.4.1151-1158.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Robene-Soustrade I., Lung-Escarmant B. Laccase isoenzyme patterns of European Armillaria species from culture filtrates and infected woody plant tissues. Eur. J. For. Pathol. 1997;27(2):105–114. [Google Scholar]
- 70.Tong P., Hong Y., Xiao Y., Zhang M., Tu X., Cui T. High production of laccase by a new basidiomycete, Trametes sp. Biotechnol. Lett. 2007;29(2):295–301. doi: 10.1007/s10529-006-9241-1. [DOI] [PubMed] [Google Scholar]
- 71.Dubé E., Shareck F., Hurtubise Y., Daneault C., Beauregard M. Homologous cloning, expression, and characterisation of a laccase from Streptomyces coelicolor and enzymatic decolourisation of an indigo dye. Appl. Microbiol. Biotechnol. 2008;79(4):597–603. doi: 10.1007/s00253-008-1475-5. [DOI] [PubMed] [Google Scholar]
- 72.Arora D.S., Gill P.K. 2001. Effects of Various media and Supplements on Laccase Production by Some white Rot Fungi; pp. 89–91. 77. [DOI] [PubMed] [Google Scholar]
- 73.Meehnian H., Jana A.K., Jana M.M. Effect of particle size, moisture content, and supplements on selective pretreatment of cotton stalks by Daedalea flavida and enzymatic saccharification. 3 Biotech. 2016;6(2):1–13. doi: 10.1007/s13205-016-0548-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sharma R.K., Arora D.S. Biodegradation of paddy straw obtained from different geographic locations by means of Phlebia spp. for animal feed. Biodegradation. 2011;22(1):143–152. doi: 10.1007/s10532-010-9383-7. [DOI] [PubMed] [Google Scholar]
- 75.Arora D.S., Sharma R.K., Chandra P. Biodelignification of wheat straw and its effect on in vitro digestibility and antioxidant properties. Int. Biodeterior. Biodegrad. 2011;65(2):352–358. [Google Scholar]
- 76.Arora D.S., Chander M., Gill P.K. Involvement of lignin peroxidase, manganese peroxidase and laccase in degradation and selective ligninolysis of wheat straw. Int. Biodeterior. Biodegrad. 2002;50(2):115–120. [Google Scholar]
- 77.Ningrum A.C. Institut Pertanian Bogor; 2018. Produksi Dan Purifikasi Lakase (Lac) Cerrena Sp. B.Md.T.A.1 Pada Berbagai Media Limbah Lignoselulosa Sebagai Agen Pendegradasi Warna. [Google Scholar]
- 78.Han M.-L., An Q., Ma K.-Y., An W.-N., Hao W.-Y., Liu M.-Y., et al. 2021. A Comparative Study on the Laccase Activity of Four Basidiomycete Fungi with Different Lignocellulosic Residues via Solid-State Fermentation [Internet] pp. 1–15. Available from: https://bioresources.cnr.ncsu.edu/wp-content/uploads/2021/03/BioRes_16_2_3017_Han_AMAHLSYB_Compar_Study_Laccase_Activity_Fungi_Lignocell_Resid_18580.pdf. [Google Scholar]
- 79.Hariharan S., Nambisan P. Optimization of lignin peroxidase, manganese peroxidase and laccase enzyme production form Ganerdoma lucidum under solid state fermentation using pineapple leaf lignocellulosic substrate. Bio. 2013;8:250–271. (Sanchez 2009) [Google Scholar]
- 80.Asgher M., Sharif Y., Bhatti H.N. Enhanced production of ligninolytic enzymes by ganoderma lucidum IBL-06 using lignocellulosic agricultural wastes. Int. J. Chem. React. Eng. 2010;8 [Google Scholar]
- 81.Adekunle A.E., Zhang C., Guo C., Liu C.Z. Laccase production from Trametes versicolor in solid-state fermentation of steam-exploded pretreated cornstalk. Waste Biomass Valoriz. 2017;8(1):153–159. [Google Scholar]
- 82.Aydinoǧlu T., Sargin S. Production of laccase from Trametes versicolor by solid-state fermentation using olive leaves as a phenolic substrate. Bioproc. Biosyst. Eng. 2013;36(2):215–222. doi: 10.1007/s00449-012-0777-2. [DOI] [PubMed] [Google Scholar]
- 83.Nandal P., Ravella S.R., Kuhad R.C. Laccase production by Coriolopsis caperata RCK2011: optimization under solid state fermentation by Taguchi DOE methodology. Sci. Rep. 2013;3:1–7. doi: 10.1038/srep01386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Reksohadiwinoto B.S., Rosmalawati S. Enzim laccase Dari edible mushroom untuk pemutihan pati sagu ramah lingkungan laccase enzyme from edible mushroom for biobleaching sago starch with environmental friendly. J. Teknol. Lingkung. 2017;18(2):224–232. [Google Scholar]
- 85.Kumar V.P., Naik C., Sridhar M. Production, purification and characterization of novel laccase produced by Schizophyllum commune NI-07 with potential for delignification of crop residues. Appl. Biochem. Microbiol. 2015;51(4):432–441. [Google Scholar]
- 86.Daâssi D., Zouari-Mechichi H., Frikha F., Rodríguez-Couto S., Nasri M., Mechichi T. Sawdust waste as a low-cost support-substrate for laccases production and adsorbent for azo dyes decolorization. J. Environ. Heal. Sci. Eng. 2016;14(1) doi: 10.1186/s40201-016-0244-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang F., Terry N., Xu L., Zhao L., Ding Z., Ma H. Fungal laccase production from lignocellulosic agricultural wastes by solid-state fermentation: a review. Microorganisms. 2019;7(12) doi: 10.3390/microorganisms7120665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pérez J., Muñoz-Dorado J., De La Rubia T., Martínez J. Biodegradation and biological treatments of cellulose, hemicellulose and lignin: an overview. Int. Microbiol. 2002;5(2):53–63. doi: 10.1007/s10123-002-0062-3. [DOI] [PubMed] [Google Scholar]
- 89.Sahadevan L.D.M., Misra C.S., Thankamani V. Characterization of lignin-degrading enzymes (LDEs) from a dimorphic novel fungi and identification of products of enzymatic breakdown of lignin. 3 Biotech. 2016;6(1):1–16. doi: 10.1007/s13205-016-0384-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Parshetti G.K., Parshetti S., Kalyani D.C., Doong R.A., Govindwar S.P. Industrial dye decolorizing lignin peroxidase from Kocuria rosea MTCC 1532. Ann. Microbiol. 2012;62(1):217–223. [Google Scholar]
- 91.Singh T., Singh A.P. Fungal Applications in Sustaintable Evironmental Biotechnology [Internet] Springer International Publishing Switzerland; New Zealand: 2016. Chapter 9 white and Brown rot fungi as decomposers of lignocellulosic materials and their role in waste and pollution control; pp. 233–247. (6). Scion (Formerly, Forest Research Institute), Te Papa Tipu Innovation Park, 49 Sala Street, Rotorua 304. Available from: https://www.researchgate.net/publication/308086613. [Google Scholar]
- 92.Liang Y.S., Yuan X.Z., Zeng G.M., Hu C.L., Zhong H., Huang D.L., et al. Biodelignification of rice straw by Phanerochaete chrysosporium in the presence of dirhamnolipid. Biodegradation. 2010;21(4):615–624. doi: 10.1007/s10532-010-9329-0. [DOI] [PubMed] [Google Scholar]
- 93.Alam M.Z., Mansor M.F., Jalal K.C.A. Optimization of lignin peroxidase production and stability by Phanerochaete chrysosporium using sewage-treatment-plant sludge as substrate in a stirred-tank bioreactor. J. Ind. Microbiol. Biotechnol. 2009;36(5):757–764. doi: 10.1007/s10295-009-0548-5. [DOI] [PubMed] [Google Scholar]
- 94.Lundell T., Wever R., Floris R., Harvey P., Hatakka A., Brunow G., et al. 1993. Lignin Peroxidase L3 from Phlebia Radiata; pp. 391–402. 402. [DOI] [PubMed] [Google Scholar]
- 95.Mtui G., Nakamura Y. Lignocellulosic enzymes from Flavodon flavus, a fungi isolated from Western Indian Ocean off the coast of Dar es Salaam, Tanzania. Afr. J. Biotechnol. 2008;7(17):3066–3072. [Google Scholar]
- 96.Shaheen R., Asgher M., Hussain F., Bhatti H.N. Ac ce p t. Int. J. Biol. Macromol. 2017 doi: 10.1016/j.ijbiomac.2017.04.040. [DOI] [PubMed] [Google Scholar]
- 97.Chowdhary P., Shukla G., Raj G., Ferreira L.F.R., Bharagava R.N. Microbial manganese peroxidase: a ligninolytic enzyme and its ample opportunities in research. SN Appl. Sci. 2019;1(1):1–12. [Google Scholar]
- 98.Singh R., Shedbalkar U.U., Nadhe S.B., Wadhwani S.A., Chopade B.A. Lignin peroxidase mediated silver nanoparticle synthesis in Acinetobacter sp. Amb. Express. 2017;7(1) doi: 10.1186/s13568-017-0528-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Niku-Paavola M.-L., Karhunen E., Salola P., Raunio V. 1988. Ligninolytic of the white-rot Fungi Phlebia Radiata; pp. 877–883. 254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang P., Hu X., Cook S., Begonia M., Lee K.S., Hwang H.M. Effect of culture conditions on the production of ligninolytic enzymes by white rot fungi Phanerochaete chrysosporium (ATCC 20696) and separation of its lignin peroxidase. World J. Microbiol. Biotechnol. 2008;24(1):2205–2212. [Google Scholar]
- 101.Huy N.D., Tien N.T.T., Huyen L.T., Quang H.T., Tung T.Q., Luong N.N., et al. Screening and production of manganese peroxidase from Fusarium sp. on residue materials. MYCOBIOLOGY. 2017;45(1):52–56. doi: 10.5941/MYCO.2017.45.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chandra M.R.G.S., Madakka M. Recent Developments in Applied Microbiology and Biochemistry. Elsevier; 2018. Comparative biochemistry and kinetics of microbial lignocellulolytic enzymes; pp. 147–159. [Google Scholar]
- 103.González-Rábade N., Del Carmen Oliver-Salvador M., Salgado-Manjarrez E., Badillo-Corona J.A. In vitro production of plant peroxidases - a review. Appl. Biochem. Biotechnol. 2012;166(7):1644–1660. doi: 10.1007/s12010-012-9558-2. [DOI] [PubMed] [Google Scholar]
- 104.Medina J.D.C., Woiciechowski A.L., Guimarães L.R.C., Karp S.G., Soccol C.R. Peroxidases. Curr. Dev. Biotechnol. Bioeng. Prod. Isol. Purif. Ind. Prod. 2016:217–232. [Google Scholar]
- 105.Moore D. 2014. David Moore ’ S World of Fungi : where Mycology Starts; pp. 1–5. (1972) [Google Scholar]
- 106.Asgher M., Wahab A., Bilal M., Nasir Iqbal H.M. Lignocellulose degradation and production of lignin modifying enzymes by Schizophyllum commune IBL-06 in solid-state fermentation. Biocatal. Agric. Biotechnol. 2016;6:195–201. [Google Scholar]
- 107.Kong W., Chen H., Lyu S., Ma F., Yu H., Zhang X. Characterization of a novel manganese peroxidase from white-rot fungi Echinodontium taxodii 2538, and its use for the degradation of lignin-related compounds. Process Biochem. 2016;51(11):1776–1783. [Google Scholar]
- 108.Lueangjaroenkit P., Teerapatsakul C., Sakka K., Sakka M., Kimura T., Kunitake E., et al. Two manganese peroxidases and a laccase of Trametes polyzona KU-RNW027 with novel properties for dye and pharmaceutical product degradation in redox mediator-free system. MYCOBIOLOGY. 2019;47(2):217–229. doi: 10.1080/12298093.2019.1589900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhang H., Zhang J., Zhang X., Geng A. Purification and characterization of a novel manganese peroxidase from white-rot fungi Cerrena unicolor BBP6 and its application in dye decolorization and denim bleaching. Process Biochem. 2018;66(September):222–229. [Google Scholar]
- 110.Bouacem K., Rekik H., Jaouadi N.Z., Zenati B., Kourdali S., El Hattab M., et al. Purification and characterization of two novel peroxidases from the dye-decolorizing fungi Bjerkandera adusta strain CX-9. Int. J. Biol. Macromol. 2018;106:636–646. doi: 10.1016/j.ijbiomac.2017.08.061. [DOI] [PubMed] [Google Scholar]
- 111.Asgher M., Ahmad Z., Iqbal H.M.N. Alkali and enzymatic delignification of sugarcane bagasse to expose cellulose polymers for saccharification and bio-ethanol production. Ind. Crop. Prod. 2013;44:488–495. [Google Scholar]
- 112.Qin X., Zhang J., Zhang X., Yang Y. Induction, purification and characterization of a novel manganese peroxidase from irpex lacteus CD2 and its application in the decolorization of different types of dye. PLoS One. 2014;9(11) doi: 10.1371/journal.pone.0113282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.de Oliveira P.L., Duarte M.C.T., Ponezi A.N., Durrant L.R. Purification and partial characterization of manganese peroxidase from Bacillus pumilus and Paenibacillus sp. Braz. J. Microbiol. 2009;40(4):818–826. doi: 10.1590/S1517-838220090004000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ayeronfe F., Kassim A., Hung P., Ishak N., Syarifah S., Aripin A. Production of ligninolytic enzymes by coptotermes curvignathus gut bacteria. Environ. Clim. Technol. 2019;23(1):111–121. [Google Scholar]
- 115.Scully E.D., Geib S.M., Hoover K., Tien M., Tringe S.G., Barry K.W., et al. Metagenomic profiling reveals lignocellulose degrading system in a microbial community associated with a wood-feeding beetle. PLoS One. 2013;8(9):1–22. doi: 10.1371/journal.pone.0073827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kupryashina M.A., Selivanov N.Y., Nikitina V.E. Isolation and purification of Mn-peroxidase from Azospirillum brasilense SP245. Appl. Biochem. Microbiol. 2012;48(1):17–20. [PubMed] [Google Scholar]
- 117.Kim D., Han G.D. Elsevier; 2014. Fermented Rice Bran Attenuates Oxidative Stress. Wheat and Rice in Disease Prevention and Health; pp. 467–480. [Google Scholar]
- 118.Elisashvili V., Kachlishvili E. Physiological regulation of laccase and manganese peroxidase production by white-rot Basidiomycetes. J. Biotechnol. 2009;144(1):37–42. doi: 10.1016/j.jbiotec.2009.06.020. [DOI] [PubMed] [Google Scholar]
- 119.Gupta R., Mehta G., Khasa Y.P., Kuhad R.C. 2011. Fungal Delignification of Lignocellulosic Biomass Improves the Saccharification of Cellulosics; pp. 797–804. [DOI] [PubMed] [Google Scholar]
- 120.Kinnunen A., Maijala P., Jarvinen P., Hatakka A. Improved efficiency in screening for lignin-modifying peroxidases and laccases of basidiomycetes. Curr. Biotechnol. 2016;6(2):105–115. [Google Scholar]
- 121.Asgher M., Wahab A., Bilal M., Nasir Iqbal H.M. Lignocellulose degradation and production of lignin modifying enzymes by Schizophyllum commune IBL-06 in solid-state fermentation. Biocatal. Agric. Biotechnol. 2016;6:195–201. [Google Scholar]
- 122.Akratos C.S., Tekerlekopoulou A.G., Vasiliadou I.A., Vayenas D.V. Elsevier; 2017. Cocomposting of Olive Mill Waste for the Production of Soil Amendments. Olive Mill Waste: Recent Advances for Sustainable Management; pp. 161–182. [Google Scholar]
- 123.Brust G.E. Elsevier; 2019. Management Strategies for Organic Vegetable Fertility; pp. 193–212. Safety and Practice for Organic Food. [Google Scholar]
- 124.Chauhan R. Nitrogen sources and trace elements influence Laccase and peroxidase enzymes activity of Grammothele fuligo. Vegetos. 2019;32(3):316–323. [Google Scholar]
- 125.Zapata-Castillo Patricia, Villalonga-Santana de M.L., Tamayo-Cortés J., Rivera-Muñoz G., Solís-Pereira S. Purification and characterization of laccase from Trametes hirsuta Bm-2 and its contribution to dye and effluent decolorization. Afr. J. Biotechnol. 2012;11(15):3603–3611. [Google Scholar]
- 126.Plácido J., Capareda S. Ligninolytic enzymes: a biotechnological alternative for bioethanol production. Biores. Bioproc. 2015;2(1) [Google Scholar]
- 127.Masarin F., Norambuena M., Ramires H.O., Demuner B.J., Pavan P.C., Ferraz A. Manganese peroxidase and biomimetic systems applied to in vitro lignin degradation in Eucalyptus grandis milled wood and kraft pulps. J. Chem. Technol. Biotechnol. 2016;91(5):1422–1430. [Google Scholar]
- 128.Wang X., Yao B., Su X. Linking enzymatic oxidative degradation of lignin to organics detoxification. Int. J. Mol. Sci. 2018;19(11) doi: 10.3390/ijms19113373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yehia R.S. Aflatoxin detoxification by manganese peroxidase purified from Pleurotus ostreatus. Braz. J. Microbiol. 2014;45(1):127–133. doi: 10.1590/S1517-83822014005000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Baborová P., Möder M., Baldrian P., Cajthamlová K., Cajthaml T. Purification of a new manganese peroxidase of the white-rot fungi Irpex lacteus, and degradation of polycyclic aromatic hydrocarbons by the enzyme. Res. Microbiol. 2006;157(3):248–253. doi: 10.1016/j.resmic.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 131.Bilal M., Asgher M., Shahid M., Bhatti H.N. Characteristic features and dye degrading capability of agar-agar gel immobilized manganese peroxidase. Int. J. Biol. Macromol. 2016;86:728–740. doi: 10.1016/j.ijbiomac.2016.02.014. [DOI] [PubMed] [Google Scholar]
- 132.Zhang H., Zhang S., He F., Qin X., Zhang X., Yang Y. Characterization of a manganese peroxidase from white-rot fungi Trametes sp.48424 with strong ability of degrading different types of dyes and polycyclic aromatic hydrocarbons. J. Hazard Mater. 2016;320:265–277. doi: 10.1016/j.jhazmat.2016.07.065. [DOI] [PubMed] [Google Scholar]
- 133.Ward G., Hadar Y., Dosoretz C.G. Lignin peroxidase-catalyzed polymerization and detoxification of toxic halogenated phenols. J. Chem. Technol. Biotechnol. 2003;78(12):1239–1245. [Google Scholar]
- 134.Wang J., Majima N., Hirai H., Kawagishi H. Effective removal of endocrine-disrupting compounds by lignin peroxidase from the white-rot fungi phanerochaete sordida YK-624. Curr. Microbiol. 2012;64(3):300–303. doi: 10.1007/s00284-011-0067-2. [DOI] [PubMed] [Google Scholar]
- 135.Alam M.Z., Mansor M.F., Jalal K.C.A. Optimization of decolorization of methylene blue by lignin peroxidase enzyme produced from sewage sludge with Phanerocheate chrysosporium. J. Hazard Mater. 2009;162(2–3):708–715. doi: 10.1016/j.jhazmat.2008.05.085. [DOI] [PubMed] [Google Scholar]
- 136.Rekik H., Nadia Z.J., Bejar W., Kourdali S., Belhoul M., Hmidi M., et al. Characterization of a purified decolorizing detergent-stable peroxidase from Streptomyces griseosporeus SN9. Int. J. Biol. Macromol. 2015;73(1):253–263. doi: 10.1016/j.ijbiomac.2014.11.021. [DOI] [PubMed] [Google Scholar]
- 137.Moreno A.D., Ibarra D., Alvira P., Tomás-Pejó E., Ballesteros M. A review of biological delignification and detoxification methods for lignocellulosic bioethanol production. Crit. Rev. Biotechnol. 2015;35(3):342–354. doi: 10.3109/07388551.2013.878896. [DOI] [PubMed] [Google Scholar]
- 138.Bak J.S., Ko J.K., Choi I.G., Park Y.C., Seo J.H., Kim K.H. Fungal pretreatment of lignocellulose by Phanerochaete chrysosporium to produce ethanol from rice straw. Biotechnol. Bioeng. 2009;104(3):471–482. doi: 10.1002/bit.22423. [DOI] [PubMed] [Google Scholar]
- 140.Sheldon R.A. Enzyme immobilization: the quest for optimum performance. Review. 2007 [Google Scholar]
- 141.Mohamad N.R., Marzuki N.H.C., Buang N.A., Huyop F., Abdul Wahab R. An overview of technologies for immobilization of enzymes and surface analysis techniques for immobilized enzymes. Biotech. Biotechnol. Equip. 2015;29(2):205–220. doi: 10.1080/13102818.2015.1008192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Fang C., Thomsen M.H., Frankaer C.G., Brudecki G.P., Schmidt J.E., Alnashef I.M. Reviving pretreatment effectiveness of deep eutectic solvents on lignocellulosic date palm residues by prior recalcitrance reduction. Ind. Eng. Chem. Res. 2017;56(12):3167–3174. [Google Scholar]
- 143.Kim K.H., Dutta T., Sun J., Simmons B., Singh S. Biomass pretreatment using deep eutectic solvent from lignin derived phenols. Green Chem. 2018;20:809–815. [Google Scholar]
- 144.Chen Z., Ragauskas A., Wan C. Lignin extraction and upgrading using deep eutectic solvents. Ind. Crop. Prod. 2020;147(112241):1–8. [Google Scholar]
- 145.Isroi Millati R., Syamsiah S., Niklasson C., Cahyanto M.N., Lundquist K., Taherzadeh M.J. Biological pretreatment of lignocelluloses with white-rot fungi and its application: a Review. Bio. 2011;6(4):5224–5259. [Google Scholar]
- 146.Andlar M., Rezic T., Mardetko N., Kracher D., Ludwig R., Santek B. Lignicellulose degradation: an overview of fungi and fungal enzymes involved in lignocellulose degradation. Eng. Life Sci. 2018;18:768–778. doi: 10.1002/elsc.201800039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data included in article/supp. material/referenced in article.