Abstract
Opioid use disorder (OUD) constitutes a significant public health burden as opioid overdose deaths have continued to rise in the United States. Although treatment modalities are available to manage OUD, some patients experience challenges achieving their OUD management goals. Some of these challenges may be attributable to inherited genetic variations, or polymorphisms, on the genes that code for proteins impacting the pharmacokinetics or pharmacodynamics of medications used in OUD management. Clinical pharmacogenomics testing can elucidate these polymorphisms; however, a lack of real-world evidence for the use of pharmacogenomics in OUD management complicates the implementation process. We conducted a retrospective cohort study of 113 patients undergoing buprenorphine-based OUD management in Northeast Washington D.C to determine if clinical pharmacogenomics testing for CYP3A4 and CYP3A5 would impact treatment outcomes. Data were collected from the electronic medical record (EMR) from December 30, 2015 to December 31, 2016. Study outcomes were based on presence of withdrawal symptoms, instances of unauthorized substances in UDTs, and SBN dose with SOC dosing versus PGx-based dosing. Pearson correlation tests, Wilcoxon signed rank tests, Wilcoxon rank sum tests, and one-way ANOVA tests were used. Linear and logistic regression analyses was used to assess predictors of withdrawal symptomatology. Kaplan-Meier survival analyses were used to assess time to first withdrawal. Our research suggests that patients with at least one copy of the CYP3A4*1B allele exhibit an accelerated rate of metabolism compared to the wild-type allele CYP3A4*1.
1. INTRODUCTION
According to the Diagnostic and Statistical Manual for Mental Disorders, Fifth Edition (DSM-V), opioid use disorder (OUD) is a problematic pattern of opioid use leading to clinically significant impairment or distress, as manifested by at least two of 11 symptoms occurring within a 12-month period [1]. Presently, OUD constitutes a significant public health burden as opioid overdose deaths have continued to rise in the United States [2]. Of the 700,000 people that died of a drug overdose between 1997 and 2017, almost 400,000 people died from an overdose involving any opioid, including prescription and illicit opioids [2]. In 2017, the number of overdose deaths involving opioids (including prescription opioids and illegal opioids like heroin and illicitly manufactured fentanyl) was 6 times higher than in 1999 and approximately 130 Americans die of an opioid overdose every day [2].
OUD is managed via both nonpharmacological and pharmacological approaches [3–5]. Nonpharmacological approaches employ cognitive behavioral therapy (CBT), such as 12-step programs, and pharmacological approaches employ the use of medication-assisted treatment (MAT) with methadone, buprenorphine (BUP), or extended-release intramuscular naltrexone (XR-NTX) [3–5]. For the purposes of OUD management, methadone is made available through opioid treatment programs (OTPs) in methadone clinics, whereas BUP is available via office-based opioid treatment (OBOT) programs [4].
Despite the availability of the MAT options for OUD management, some patients experience disparate treatment outcomes that may be attributable to multiple factors, such as barriers to care and interindividual genetic variations, or polymorphisms [6]. Pharmacogenomics (PGx), which combines pharmacology and genomics to study how genetic polymorphisms impact drug response [7], can be incorporated into clinical practice to identify genetic targets of interest that impact MAT response [8–10]. Genetic polymorphisms can impact both the pharmacokinetics (PK) and the pharmacodynamics (PD) of a given medication [7]. Polymorphisms in PD genes can affect drug action at its target, such as a receptor, and polymorphisms in PK genes, such as the cytochrome P450 (CYP) family of metabolic enzymes, can affect blood and tissue drug levels [8]. Functional variants in the CYP3A4 gene impact the rate at which drugs are metabolized and correlate to four basic phenotypes: poor metabolizers (PMs), intermediate metabolizers (IMs), extensive metabolizers (EMs), and ultra-rapid metabolizers (UMs) [8]. BUP is metabolized by CYP3A4 and CYP3A5 [11] and also undergoes glucuronidation via multiple isoforms of the uridine-diphosphate glucuronosyltransferase (UGT) enzyme, including UGT1A1, UGT1A3, and UGT2B7 [12]. Furthermore, as BUP exerts its action through agonism of the mu-opioid receptor [11, 13], this receptor serves as a pharmacodynamic target of interest and genetic polymorphisms impacting this receptor can impact treatment outcomes.
Clinical pharmacogenomic testing can help to elucidate genetic polymorphisms impacting medications used for OUD management in a given patient and allow health care providers to individualize medication selection and dosing. However, multiple barriers challenge the implementation of this testing process [14, 15]. One such barrier is the availability of real-world evidence that health care providers can use to enhance the clinical decision-making process [15]. A published case-report by our research group showed that a patient with the cytochrome P450 3A4 ultrarapid metabolizer phenotype required a higher-than-recommended daily dose of buprenorphine (32 mg) for adequate OUD management [4]. It is particularly noteworthy that this finding was included in evidence that was used to reverse the DC Medicaid’s PBM Policy that stopped prior authorizations intended to secure SBN doses exceeding 24 mg/day from December 30, 2015 to June 30, 2016. After this six month period passed, the reversal allowed eligible physicians the flexibility to dose the patients based on their clinical presentation and pharmacogenomic profile.
In an effort to fill the existing knowledge gap on evidence of the importance of pharmacogenomic testing in MAT, this study explored the impact of clinical characteristics, as well as CYP3A4 and CYP3A5 polymorphisms, on outcomes of BUP dosing, withdrawal, and relapse.
2. MATERIALS AND METHODS
2.1. Study Design
A retrospective cohort study design was conducted among patients undergoing OBOT with sublingual BUP/naloxone (SBN) in a physician’s office located in Northeast Washington D.C. Data were collected from the electronic medical record (EMR) from December 30, 2015 to December 31, 2016. Patients were eligible for inclusion in the cohort if (1) they were at least 18 years of age, (2) they were undergoing BUP-based MAT, (3) and had a PGx test on file prior to the study period.
2.2. Study Variables
There were three outcome variables evaluated in this study including (1) SBN dosing, (2) withdrawal symptoms, and (3) occurrence of positive urine drug test (UDT) results. Other study variables encompassed patient demographics, medical insurance coverage, medical history (indicated by ICD-10 codes), medication history, UDT results, and PGx test information. Study outcomes were based on presence of withdrawal symptoms, instances of unauthorized substances in UDTs, and SBN dose with SOC dosing versus PGx-based dosing. Outcome variables corresponding to SBN dosing, withdrawal symptoms, and UDT results were characterized as both continuous and categorical variables to facilitate statistical analyses.
2.3. Operational Definitions
The three outcome variables in this study had to be operationalized in a manner that factored in policy-restricted dosing periods over the study period. Operationally, the period between December 30, 2015 and June 30, 2016 was defined as the SOC period when doses above the FDA approved maximum were not permitted. Conversely, the PGx period was defined as the time after June 2016 where higher dosing was permitted.
The SBN dosing was measured as a ratio of the average dose during the six-month PGx period over the average dose during the six-month SOC period [(average PGx dose) / (average SOC dose)] to account for variations in dosing over the respective time periods. Higher SBN ratios correspond to higher doses of SBN in the PGx period compared to the SOC period. This outcome was also operationalized as a categorical variable based on whether the average SBN dose in the PGx period was higher than 24 mg/day (1=yes, 0=no).
Withdrawal symptomatology was measured using the Subjective Opioid Withdrawal Scale (SOWS) assessment tool and was measured at each study visit. In order to account for the impact that (1) the mere presence of withdrawal symptoms and (2) the magnitude of each withdrawal symptom effect on OUD management outcomes; withdrawal symptomatology was represented using two variables. One variable characterized withdrawal symptoms as “withdrawal instances” without regard to the magnitude of the withdrawal symptom and the other variable captured the magnitude of the withdrawal symptom as the SOWS score. The withdrawal outcome variable was measured as the difference between the number of instances of withdrawal in the SOC period divided by the number of instances during the PGx period. Based upon this definition, a higher difference indicated fewer instances of withdrawal during the PGx period.
UDT data was collected as instances of unauthorized substances per urine screening. For example, if one urine screen identified three unauthorized substances, that would constitute three instances. The UDT outcome variable was measured as the difference between the number of instances that unauthorized substances were found in UDTs during the SOC period and the number of similar instances during the PGx period. Based upon this definition, a higher difference indicated fewer instances of unauthorized substances identified during the PGx period.
2.4. Statistical Analysis
Descriptive statistics including means and percentages were used to describe the study population. To examine bivariable associations between study outcomes and study variables, Pearson correlation tests, Wilcoxon signed rank tests, Wilcoxon rank sum tests, and one-way ANOVA tests were used. Linear and logistic regression analyses was used to assess predictors of withdrawal symptomatology. Kaplan-Meier survival analyses were used to assess time to first withdrawal. All analyses were conducted using SAS version 9.4 at an alpha of 0.05.
3. RESULTS
Description of Patient Characteristics
There was a total of 113 patients in the cohort, and of them, 86 were male and 27 were female. Most of the patients were single (n=80), Non-Hispanic African Americans (n=111) with an average age of 60 years (SD=8.5 years). DC Medicaid was the predominant primary insurance (n=62) followed by DC Medicare (n=28); there were 18 patients who were dually covered with DC Medicare as their primary insurance and DC Medicaid as their secondary insurance.
Approximately 50% of the patients lived independently (n=56) and 27 patients lived with a family member. The majority of the patients (n=92) were able to perform activities of daily living (ADLs) independently. Fourteen patients were employed, while 25 were unemployed, 58 were disabled, and three were retired. Sixty-eight (60.18%) patients had been incarcerated at least once in their lives with 43 patients reporting the actual years of incarceration; the average time of incarceration was 11.30 years (SD = 9.93 years).
In regard to the utilization of CBT, 44.25% (n=50) used Alcoholics Anonymous (AA) at least once, 59.29% (n=67) used Narcotics Anonymous (NA) at least once, and 73.45% (n=83) used counseling at least once.
The most frequently encountered medications aside from SBN were naloxone (n=67), amlodipine (n=38), hydrochlorothiazide (HCTZ) (n=28), and lisinopril (n=27); the latter three medications are commonly prescribed to manage hypertension.
Comorbid conditions were characterized as substance use disorders (SUDs), mood disorders, and infectious diseases. Among the SUDs, nicotine use was the most prevalent (n=88), followed by cocaine use (n=19), alcohol use (n=6), and cannabis use (n=4). Depression was the most frequently encountered mood disorder (n=44). There was a low HIV prevalence among the patients (n=6); however, 42 patients (37.2%) either had been exposed to the hepatitis C virus (HCV) in the past or were currently infected with HCV.
3.1. Description of Clinical Characteristics
The average SBN daily dose per patient was 23 mg/day during the SOC period (SD = 2.8 mg/day) and 26 mg/day (SD=4.6 mg/day) during the PGx period, with an overall max of 32 mg/day. According to the package insert for SBN, the FDA-approved maximum daily dose is 24 mg/day; 64.6% (n=73) of the patients had an average SBN daily dose that exceeded 24 mg/day during the PGx period.
The mean withdrawal instances during the SOC period was 19.12 (SD = 43.23), with a minimum of zero and a maximum of 270 instances. This was higher than the mean withdrawal instances during the PGx period, which was 3.73 (SD = 9.98), with a minimum of zero and a maximum of 48. The mean SOWS score during the SOC period was 2.64 (SD = 3.76) with a minimum of zero and a maximum of 24.77, while the mean SOWS score during the PGx period was 1.21 (SD = 1.40) with a minimum of zero and a maximum of eight. Wilcoxon signed rank analyses resulted in a statistically significant difference in both the withdrawal instances (p <.0001) and SOWS scores (p <.0001) between the SOC and PGx periods. The heat maps below depict the average SOWS scores per month, stratified by CYP3A4 phenotype and primary insurance provider, respectively. There is a notable decrease in the instances of withdrawal symptoms from July 2016 onward—the beginning of the PGx-guided dosing period.
The mean number of UDT instances per patient decreased from 10.71 (SD = 14.09) during the SOC period to 5.23 (SD = 7.04) in the PGx period. The heat map showing the instances of replase over time In regard to pharmacogenomics, we identified four CYP3A44 genotypes across the patient cohort corresponding to three phenotypes: UM (n=93), EM (n=17), and IM (n=3). The CYP3A5 gene had a more polymorphic presentation across the patient cohort with 16 genotypes corresponding to three phenotypes: PM (n=42), IM (n=58), and EM (n=13). The most frequently encountered CYP3A4/CYP3A5 phenotype combination was CYP3A4 UM with a CYP3A5 IM (n=49). Table 2 below shows the descriptive summary of the pharmacogenomic profile of the 113 patients.
Table 2.
MULTIPLE LOGISTIC REGRESSION
| Parameter | Beta Coefficient | Odds Ratio Estimate | 95% Confidence Limits | p-value | |
|---|---|---|---|---|---|
| Ever Diagnosed with Nicotine Use Disorder? (1=Yes, 0=No) | 1.1450 | 3.143 | 0.813 | 12.151 | 0.0970 |
| DC Medicaid as Primary Insurance? (1=Yes, 0=No) | 1.6608 | 5.264 | 1.223 | 22.656 | 0.0257 |
| Gender (1=Female, 2=Male) | 0.7491 | 2.115 | 0.597 | 7.488 | 0.2455 |
| Ever Prescribed Lisinopril? (1=Yes, 0=No) | −0.6960 | 0.499 | 0.161 | 1.547 | 0.2282 |
| Ever Prescribed Naloxone? (1=Yes, 0=No) | −0.3902 | 0.677 | 0.259 | 1.766 | 0.4252 |
| Age | −0.0235 | 0.977 | 0.919 | 1.038 | 0.4518 |
| Ever Been Incarcerated? (1=Yes, 0=No) | 0.3181 | 1.375 | 0.515 | 3.671 | 0.5256 |
| Ever Diagnosed With Depression? (1=Yes, 0=No) | −0.3889 | 0.678 | 0.266 | 1.724 | 0.4142 |
| DC Medicare as Primary Insurance? (1=Yes, 0=No) | 0.8567 | 2.355 | 0.445 | 12.454 | 0.3133 |
| Presence of at Least One Copy of CYP3A4*1B? (1=Yes, 0=No) | 1.8940 | 6.646 | 1.208 | 36.553 | 0.0294 |
| CYP3A5 Extensive Metabolizer? (1=Yes, 0=No) | 0.2504 | 1.285 | 0.298 | 5.544 | 0.7372 |
3.2. Bivariable associations with withdrawal symptoms
There was a statistically-significant correlation between the difference in withdrawal instances and the difference in instances of unauthorized substances (Pearson correlation coefficient = 0.42, p <.0001). There was also a statistically-significant correlation between the difference in withdrawal instances and the SBN ratio (Pearson correlation coefficient = −0.50, p <.0001). There was a statistically-significant correlation between the difference in unauthorized substance instances and age (Pearson correlation coefficient = 0.26, p = .0061) as well as the difference in SOWS scores (Pearson correlation coefficient = −0.26, p = .0283). Wilcoxon rank sum analyses resulted in statistically-significant differences in mean SOWs scores between patients with DC Medicaid as their primary insurance and those without. A similar finding was seen with the difference in withdrawal instances and DC Medicaid as the primary insurance (p = 0.0018) as well as with the difference in withdrawal instances and the presence of at least one copy of the CYP3A4*1B allele (p = 0.0438). Simple linear regression analyses revealed that the use of amlodipine (B = 17.48, p = 0.0413), the average SBN daily dose during the PGx period (B = −4.84, p <.0001), incidence of unauthorized substances during the PGx period (B = −1.25, p = 0.0358), and incidence of unauthorized substances during the SOC period (B = −1.48, p <.0001).
3.3. Survival Analysis on Time to First Onset of Withdrawal Symptoms
The Kaplan-Meier curve indicates the time in which the first instance of withdrawal symptoms manifested in each patient and is stratified by CYP3A4 phenotype. The shaded area on the left indicates the SOC period and the area to the right indicates the PGx period. There is a statistically-significant difference in time until presence of withdrawal symptoms (p=0.015).
3.4. Factors Predicting Suboxone Dosing Ratio, Withdrawal Instance Ratio and Unauthorized Substance Ratio
Statistically-significant predictors for lower withdrawal instances on PGx-guided dosing were having DC Medicaid as primary insurance and having at least one copy of the CYP3A4*1B allele. Patients with DC Medicaid as their primary insurance had 5.3 times the odds of having a lower instance of withdrawal symptoms with PGx-guided dosing compared to those without DC Medicaid as their primary insurance. Patients with at least one copy of the CYP3A4*1B allele, either CYP3A4*1B homozygotes or heterozygotes, had 6.6 times the odds of having a lower instance of withdrawal symptoms on PGx-guided dosing compared to patients that did not.
4. DISCUSSION
The results from our research demonstrate that multiple factors can make a significant impact on OUD management outcomes, particularly regarding PGx, insurance, comorbid conditions, and social determinants of health. Firstly, it is notable that most of the patients in our cohort are African-American, have at least one copy of the CYP3A4*1B allele, and expressed a need for SBN doses exceeding the FDA-recommended 24 mg per day to effectively manage their OUD. In the literature, there is conflicting evidence regarding the phenotypic characterization of the CYP3A4*1B allele. For example, some research suggests that it has no effect on metabolism or predisposes an individual to certain cancers.
Our research suggests that patients with at least one copy of the CYP3A4*1B allele exhibit an accelerated rate of metabolism compared to the wild-type allele CYP3A4*1. Furthermore, as there is some substrate cross-specificity between CYP3A4 and CYP3A5, it was notable to see some evidence of a difference in treatment outcomes among patients with the CYP3A5 EM compared to CYP3A5 IM and PM. Therefore, this may demonstrate that both CYP3A4 and CYP3A5 need to be considered when determining how CYP enzymes impact BUP-based OUD management. Due to a relatively small sample size and the exploratory nature of the research, further studies are needed to develop robust clinical decision-making tools for pharmacogenomic testing implementation in the clinical setting. Furthermore, other genes of interest, such as the gene coding for the mu-opioid receptor, need to be interrogated to gain a more comprehensive understanding about how PGx impacts treatment outcomes for BUP-based OUD management.
Insurance coverage, particularly with DC Medicaid as the primary insurance, consistently made a significant impact on OUD management outcomes in our cohort. DC Medicaid had a policy that allowed the physician to obtain a prior authorization to cover SBN doses exceeding the FDA-recommended maximum dose of 24 mg per day. This allowed the prescribing physician to provide Medicaid patients with SBN dosing consistent with their pharmacogenomic profile. However, DC Medicare did not provide the same policy during our study period and as a result, DC Medicare patients could not obtain higher doses of SBN even if their pharmacogenomic profile suggested that a higher dose was necessary. This meant that DC Medicare patients exhibiting a CYP3A4 ultrarapid metabolizer phenotype may present with a higher incidence of withdrawal symptoms and increased relapse to unauthorized substances compared to patients with DC Medicaid as their primary insurance. These findings suggest that an explicit policy which allowed prescribers to adjust dosing beyond the FDA guidelines when the clinical presentation and evidence suggests may lead to better outcomes. Prior authorization and other stop gap measure could be replaced by a Pharmacist working with a prescriber in the medical home to advance these decisions in a more efficient manner thus improving treatment outcomes.
4.1. Policy Recommendations
Because of our work, we would like to propose several recommendations to improve the delivery of OUD management services. Firstly, we would like to emphasize the need to take a bio-psycho-social approach to OUD management. Although the combined use of CBT and MAT are the cornerstone of OUD management, we recommend expanding the “assisted” aspect of MAT by addressing social determinants of health, increased psychological support, and increased disease management services. For example, in addition to providing SBN and enrolling a patient in counseling services, the patient may also need housing, transportation, employment, legal, or medication therapy management (MTM) services. Utilizing a patient-centered medical home model with an interdisciplinary team comprised of prescribers, pharmacists, social workers, nurses, dentists, and other key professionals would serve as the best means of providing comprehensive care.
Figure 1:
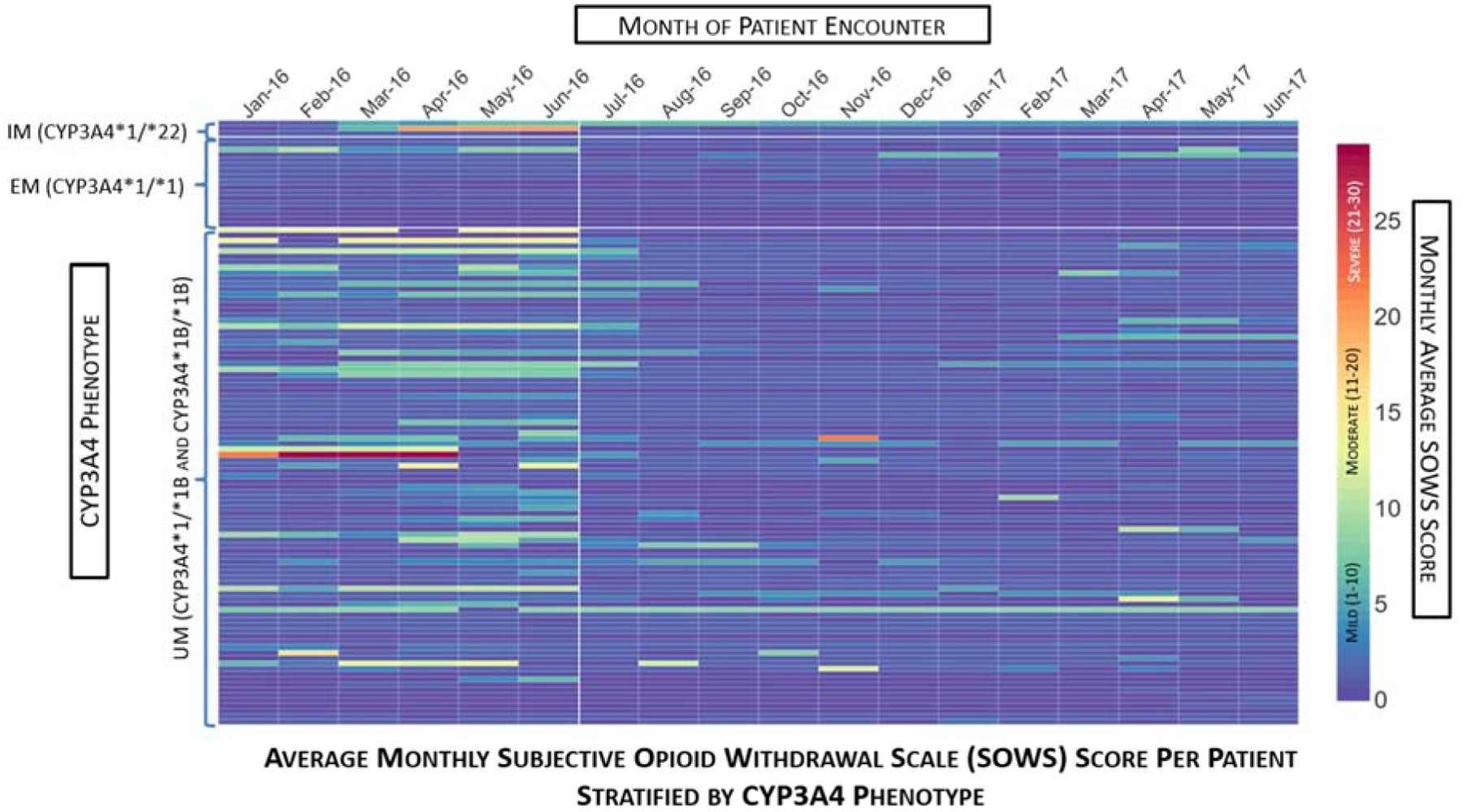
Average Monthly Subjective Opioid Withdrawal Scale (SOWS) Score per Patient Stratified by CYP3A4 Phenotype. This heat map depicts the average monthly SOWS score per month over an 18-month span of patient encounters. Each column represents the month of patient encounter, each row depicts an individual patient, and the color of the rectangle depicts the monthly average SOWS score. The patient rows are stratified by CYP3A4 phenotype, with white lines demarcating the IM, EM, and UM groupings. The color bar on the right indicates the relationship between the color of the rectangle and the SOWS score. The first six months (January 2016 – June 2016) comprise the SOC period in which patients were limited to a SBN maximum daily dose of 24 mg. The remaining 12 months (July 2016 – June 2017) comprise the PGx period in which patients could receive SBN doses exceeding 24 mg/day. The white vertical line after June 2016 demarcates the end of the SOC period. Note that within the SOC period, there is a tendency towards higher monthly average SOWS scores among the UMs compared to the EMs and IMs. Also note that during the PGx period, there is an overall observable reduction in monthly average SOWs scores compared to the SOC period.
Figure 2:
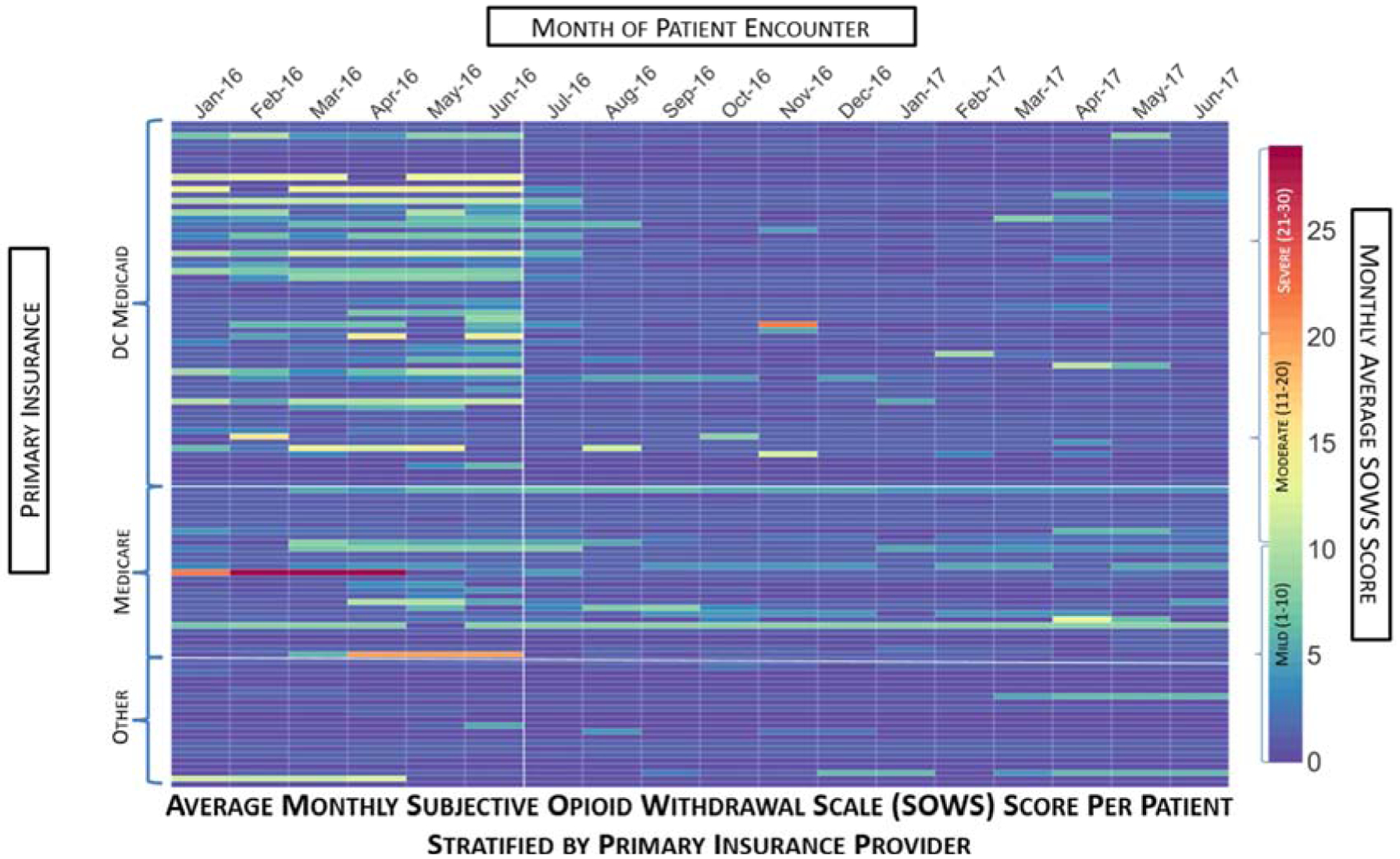
Average Monthly Subjective Opioid Withdrawal Scale (SOWS) Score per Patient Stratified By Primary Insurance Provider. This heat map depicts the average monthly SOWS score per month over an 18-month span of patient encounters. Each column represents the month of patient encounter, each row depicts an individual patient, and the color of the rectangle depicts the monthly average SOWS score. The patient rows are stratified by the primary insurance provider type, with white lines separating the DC Medicaid, Medicare, and Other Insurance groupings. The color bar on the right indicates the relationship between the color of the rectangle and the SOWS score. The first six months (January 2016 – June 2016) comprise the SOC period in which patients were limited to a SBN maximum daily dose of 24 mg. The remaining 12 months (July 2016 – June 2017) comprise the PGx period in which patients could receive SBN doses exceeding 24 mg/day. The white vertical line after June 2016 demarcates the end of the SOC period. During this 18-month period, Medicare did not allow patients to receive SBN doses exceeding 24 mg/day. Therefore, during the PGx period, patients with Medicare as their primary insurance were unable to obtain SBN doses exceeding 24 mg/day and some of these patients exhibited slightly higher average monthly SOWS scores compared to the rest of the patients.
Figure 3:
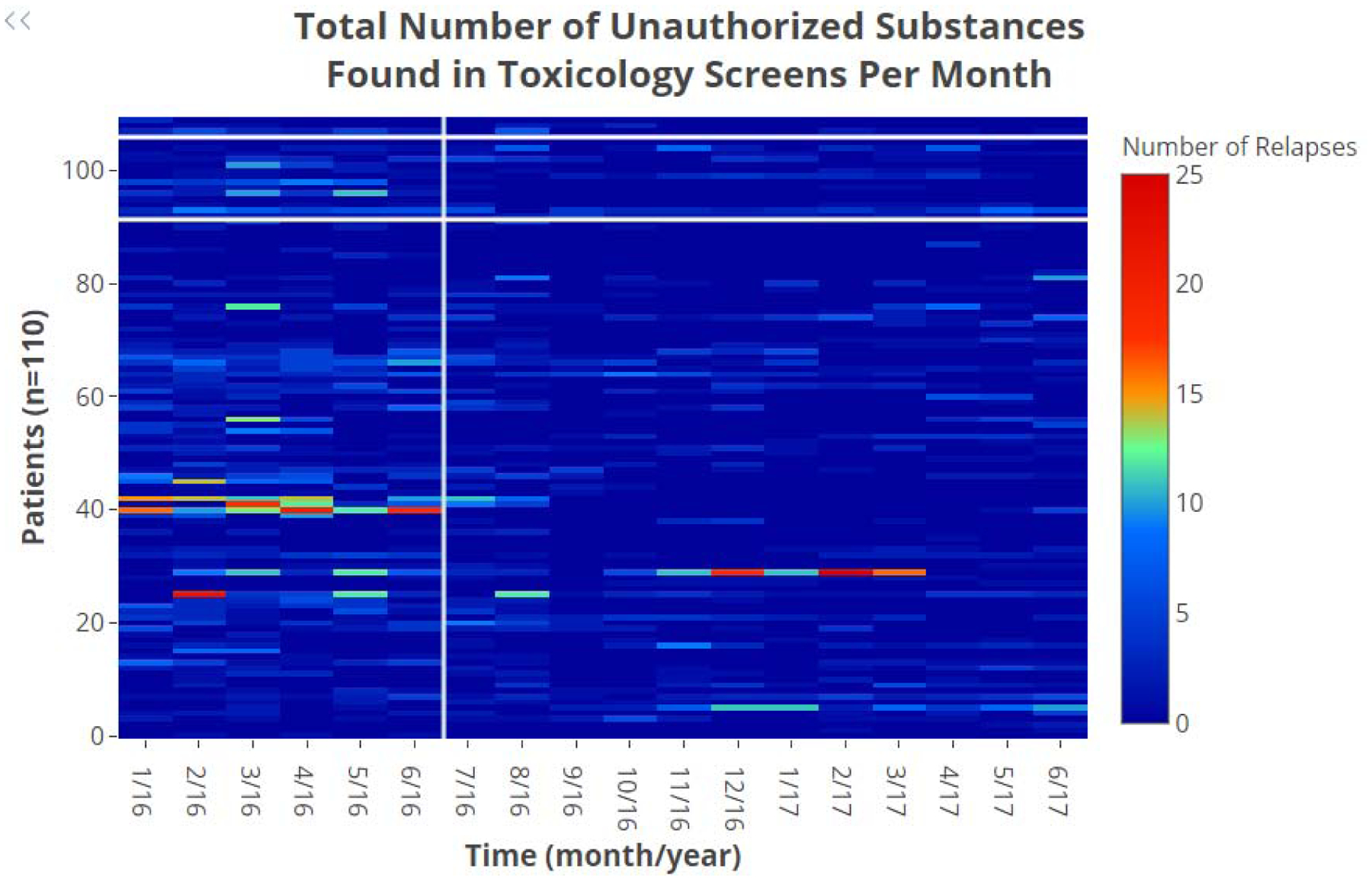
Total Number of Unauthorized Substances Found in Toxicology Screens per Month. This heat map depicts the total number of unauthorized substances found in toxicology screens per month over an 18-month span of patient encounters. Each column represents the month of patient encounter, each row depicts an individual patient, and the color of the rectangle depicts the total number of unauthorized substances per month. The patient rows are stratified by CYP3A4 phenotype, with IM on top, EM in the middle, and UM on the bottom. The color bar on the right indicates the number of unauthorized substances found in toxicology screens per month. The first six months (January 2016 – June 2016) comprise the SOC period in which patients were limited to a SBN maximum daily dose of 24 mg. The remaining 12 months (July 2016 – June 2017) comprise the PGx period in which patients could receive SBN doses exceeding 24 mg/day. The white vertical line after June 2016 demarcates the end of the SOC period. Note that within the SOC period, there is a tendency towards higher total numbers of unauthorized substances found in toxicology screens per month with UMs compared to the EMs and IMs.
Figure 4:
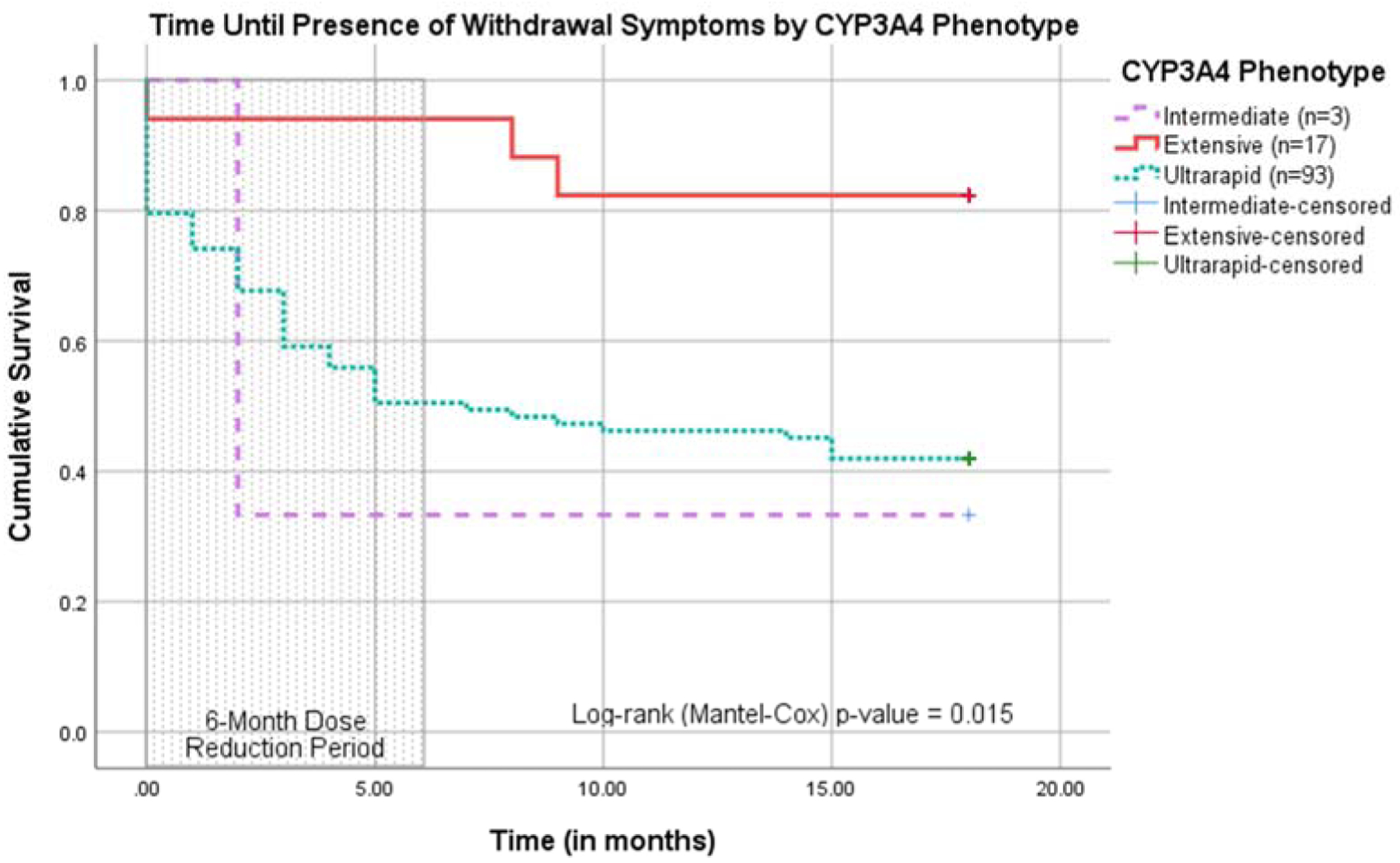
Survival Plot of Time until Presence of Withdrawal Symptoms by CYP3A4 Phenotype. This survival plot illustrates the time until the first instance of withdrawal symptoms over an 18-month span of patient encounters. The first six months (indicated by the shaded area) comprise the SOC period in which patients were limited to a SBN maximum daily dose of 24 mg. The remaining 12 months comprise the PGx period in which patients could receive SBN doses exceeding 24 mg/day. Each curve represents a CYP3A4 phenotype (IM, EM, or UM). According to this survival plot, UMs experienced withdrawal symptoms more frequently during the SOC period than IMs and EMs.
Table 1:
CYP3A4 Phenotype Frequencies
| CYP3A4 | Corresponding Phenotype | Frequency | Percent |
|---|---|---|---|
| *1/*1 | EM | 17 | 15.04 |
| *1B/*1 | UM | 49 | 43.36 |
| *1B/*1B | UM | 44 | 38.94 |
| *1B/*22 | IM | 3 | 2.65 |
HIGHLIGHTS.
Opioid use disorder (OUD) is a significant public health issue in the United States
Sublingual buprenorphine/naloxone (SBN) is indicated for OUD management
Buprenorphine is metabolized by the CYP3A4 enzyme
CYP3A4 ultrarapid metabolizers (UMs) may metabolize buprenorphine at an accelerated rate compared to CYP3A4 extensive metabolizers
Pharmacogenomics testing can help improve OUD management outcomes
ACKNOWLEDGEMENTS
We would like to acknowledge the Howard University Research Centers in Minority Institutions (RCMI) program and the District of Columbia Department of Healthcare Finance for their continued support in our research endeavors.
FUNDING
This project was supported (in part) by the National Institute on Minority Health and Health Disparities of the National Institutes of Health under Award Number G12MD007597. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICT OF INTEREST
The authors have no conflicts of interest to disclose.
Howard University IRB approval number: 18-MED-46
REFERENCES
- 1.APA, DSM-5 Handbook of Differential Diagnosis, in Substance-Related and Addictive Disorders, APA, Editor. 2019, American Psychiatry Association: APA | Psychiatry Online | DSM Library. [Google Scholar]
- 2.CDC. Understanding the Epidemic | Drug Overdose | CDC Injury Center. Opioid Overdose 2018. 2018-12-19T06:41:54Z [cited 2019 January 31]; Available from: https://www.cdc.gov/drugoverdose/epidemic/index.html.
- 3.Schuckit MA and Longo DL, Treatment of Opioid-Use Disorders. 10.1056/NEJMra1604339, 2016. [DOI] [Google Scholar]
- 4.SAMHSA. Treatments for Substance Use Disorders. 2016. 2014-09-30T20:07–04:00; Available from: http://www.samhsa.gov/treatment/substance-use-disorders#opioid.
- 5.WHO. Treatment of opioid dependence. WHO 2011. 2011-02-08 15:04:15; Available from: http://www.who.int/substance_abuse/activities/treatment_opioid_dependence/en/.
- 6.Owusu Obeng A, e.a., Review of Opioid Pharmacogenetics and Considerations for Pain Management. - PubMed - NCBI. Pharmacotherapy, 2017. 37(9): p. 1105–1121. [DOI] [PubMed] [Google Scholar]
- 7.NIH. What is pharmacogenomics? Genetics Home Reference 2016 [cited 2016. December 13, 2016]; Available from: https://www.ncbi.nlm.nih.gov/pubmed/.
- 8.Ettienne EB, et al. , Pharmacogenomics-guided policy in opioid use disorder (OUD) management: An ethnically-diverse case-based approach. Addict Behav Rep, 2017. 6: p. 8–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Somogyi AA, e.a., Pharmacogenetics of opioid response. Clinical Pharmacology and Therapeutics, 2015. 97(2): p. 125–7. [DOI] [PubMed] [Google Scholar]
- 10.Bastami S, et al. , Influence of UGT2B7, OPRM1 and ABCB1 gene polymorphisms on postoperative morphine consumption. Basic Clin Pharmacol Toxicol, 2014. 115(5): p. 423–31. [DOI] [PubMed] [Google Scholar]
- 11.Ferrari A, et al. , Methadone--metabolism, pharmacokinetics and interactions. Pharmacol Res, 2004. 50(6): p. 551–9. [DOI] [PubMed] [Google Scholar]
- 12.Rouguieg K, et al. , Contribution of the different UDP-glucuronosyltransferase (UGT) isoforms to buprenorphine and norbuprenorphine metabolism and relationship with the main UGT polymorphisms in a bank of human liver microsomes. Drug Metab Dispos, 2010. 38(1): p. 40–5. [DOI] [PubMed] [Google Scholar]
- 13.Indivior, Suboxone Prescribing Information. 2015.
- 14.Shuldiner AR, et al. , The Pharmacogenomics Research Network Translational Pharmacogenetics Program: overcoming challenges of real-world implementation. Clinical pharmacology and therapeutics, 2013. 94(2): p. 207–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O’Donnell PH, et al. , The 1200 patients project: creating a new medical model system for clinical implementation of pharmacogenomics. Clinical pharmacology and therapeutics, 2012. 92(4): p. 446–9. [DOI] [PMC free article] [PubMed] [Google Scholar]


