Analysis of whole-exome sequencing data uncovers correlations between clonal hematopoiesis and lung cancer risk factors, identifies genetic variants correlated with clonal hematopoiesis, and highlights hundreds of potential novel clonal hematopoiesis mutations.
Abstract
Clonal hematopoiesis (CH) is a phenomenon caused by expansion of white blood cells descended from a single hematopoietic stem cell. While CH can be associated with leukemia and some solid tumors, the relationship between CH and lung cancer remains largely unknown. To help clarify this relationship, we analyzed whole-exome sequencing (WES) data from 1,958 lung cancer cases and controls. Potential CH mutations were identified by a set of hierarchical filtering criteria in different exonic regions, and the associations between the number of CH mutations and clinical traits were investigated. Family history of lung cancer (FHLC) may exert diverse influences on the accumulation of CH mutations in different age groups. In younger subjects, FHLC was the strongest risk factor for CH mutations. Association analysis of genome-wide genetic variants identified dozens of genetic loci associated with CH mutations, including a candidate SNP rs2298110, which may promote CH by increasing expression of a potential leukemia promoter gene OTUD3. Hundreds of potentially novel CH mutations were identified, and smoking was found to potentially shape the CH mutational signature. Genetic variants and lung cancer risk factors, especially FHLC, correlated with CH. These analyses improve our understanding of the relationship between lung cancer and CH, and future experimental studies will be necessary to corroborate the uncovered correlations.
Significance:
Analysis of whole-exome sequencing data uncovers correlations between clonal hematopoiesis and lung cancer risk factors, identifies genetic variants correlated with clonal hematopoiesis, and highlights hundreds of potential novel clonal hematopoiesis mutations.
Introduction
Clonal hematopoiesis (CH), also known as clonal hematopoiesis of indeterminate potential, is a phenomenon of asymptotic expansion of blood cells descended from a single mutated hematopoietic stem cell (HSC). In a healthy adult human, more than 500 billion mature blood cells are produced each day from only about 10,000 to 20,000 HSCs (1, 2). Hematopoietic stem or progenitor cells accumulate somatic mutations due to the increase of age, environmental exposures, or other reasons. While the majority of these somatic mutations are neutral or deleterious, some of them may contribute a competitive advantage to the host stem/progenitor cells during hematopoiesis. Consequently, a single HSC can produce a clonal population of blood cells that inherit the same set of somatic mutations.
The most clear clinical correlate with CH development is aging. CH was more common in older people as somatic mutations accumulate in HSCs with increased age (3, 4). The association between CH and age was first reported in the non-random X inactivation study: CH was observed in less than 5% of neonates and young healthy females, but 20%–25% in healthy women over 60 years old (5, 6). Subsequent large-scale analysis based on SNP microarray or DNA-sequencing data also observed low CH rate in young but over 10% in people older than 65 years (7–9). Exogenous stress such as chemical/radiotherapy and smoking also promotes CH mutations. In patients who had undergone chemotherapy, recurrent CH mutations were found in DNA damage–related genes (10, 11). Smoking was also highly related with CH mutations (8), affecting mutational signature of CH mutations (10).
While CH is not considered a hematologic disease, many CH mutations occur in genes that are frequently mutated in leukemia and other type of cancers, such as DNMT3A, TET2, ASXL1, and PPM1D (8, 9, 12). The presence of CH mutations has been associated with the increased risk of breast, ovarian, and hematologic cancer (12, 13), especially the therapy-induced acute myeloid leukemia (AML; ref. 14). Lung cancer is a major cause of cancer death worldwide accounting for over 1 million deaths each year (15). While CH mutations have been found associated with several solid tumors (10, 16), the connection between CH mutation and lung cancer remains largely unknown. A pan-cancer analysis found that patients with lung cancer tend to harbor more CH mutations than the average level across all tumor samples; however, this might be confounded by smoking history (10). Another large-scale whole-genome sequencing study detected weak association between CH mutations and lung adenocarcinoma (8); while the association between CH mutations and lung adenocarcinoma showed the lowest P- value among all cancer phenotypes, it do not reach the significance cutoff (8). Despite the limited number of cases that have previously been studied, lung cancer cases share similar risk factors associated with CH mutations, for example, age and smoking. Whether these risk factors contribute to the accumulation of CH mutations equally in patients with lung cancer and noncancer controls is unclear. In addition, germline genetic variants correlated with CH mutations are found at lung cancer susceptibility genes, such as TERT (8) and TRIM59 (16), suggesting potential connections between familial lung cancer and CH mutations. However, none of the previous studies has investigated the relationship between family history of lung cancer (FHLC) and CH mutations. The Integrative Analysis of Lung Cancer Etiology and Risk project of the International Lung Cancer Consortium (INTEGRAL-ILCCO) project (17) provided a comprehensive dataset from lung cancer and healthy cohorts, with additional clinical information such as age, sex, smoking status, and FHLC, providing the opportunity for us to uncover the linkages between CH mutations, lung cancer, and lung cancer risk factors. Here we utilized the whole-exome sequencing (WES) data from the INTEGRAL-ILCCO project to characterize the CH mutation status, its associated clinical impact in patients with lung cancer and/or lung cancer family history, and the inherited genetic causes of CH mutation status.
Patients and Methods
Human subjects
We utilized the clinical information, genotyping, and WES data from 1,958 samples in the INTEGRAL-ILCCO study (17). The study was approved by the Institutional Review Board of all sites accruing participants. The INTEGRAL-ILCCO project includes a total of 1,059 lung cancer cases and 899 controls from four sites: Harvard School of Public Health (HSPH), International Agency for Research on Cancer (IARC), University of Liverpool, and Mount Sinai Hospital and Princess Margaret Hospital in Toronto (Supplementary Table S1; ref. 17). Everyone in this study had not been treated prior to blood drawing. Lung cancer subjects that were patients with early-onset lung cancer with family history or with available tissues were preferred. Subjects without lung cancer diagnosis were defined as controls. Clinical information included sex, age, smoking history, and FHLC (Supplementary Table S1). Lung cancer samples were more likely to be smokers and associated with higher pack-years than controls (P < 0.0001), and were more likely to have FHLC (Supplementary Fig. S1).
Genotyping and WES
Genotyping, WES, and data processing were described by the previous study (17). Briefly, for the SNP array genotype data, DNA extracted from peripheral white blood cells was genotyped using the Human610-Quad BeadChip (Illumina), with low-quality SNPs removed. For WES data, paired-ended 125 bp WES was performed using the Agilent SureSelect v5 kit with additional custom capture targeted at known LC-GWAS (lung cancer genome-wide association study) regions. Sequence reads were mapped to the human reference GRCh37/hg19 using the Burrows-Wheeler Aligner. Potential PCR duplicates were filtered in subsequent analysis. Samples with abnormal heterozygosity rate, sex discordance, <95% completion rates, and unexpected relatedness (identity-by-state > 10%) were discarded. The median on-target coverage of all the samples was approximately 51×, with only less than 3% of on-target bases having a depth less than 10×. We also called SNPs from WES data using the GATK HaplotypeCaller pipeline. For both SNP array data and WES SNP calling results, we applied a χ2 Hardy-Weinberg equilibrium (HWE) test to remove SNPs that significantly deviated from HWE. For each SNP, we tested the significance in patients with lung cancer and controls separately. The SNPs with a P value larger than 5e-8 in both patients with lung cancer and controls were retained for further analysis.
Identification of CH mutations
We designed a set of hierarchical filtering criteria to optimize the sensitivity and accuracy for CH mutation detection. We only kept bases with quality score >30 and processed aligned bam files with mpileup command of samtools to detect as many potential CH mutations as possible. We implemented a binomial error model to improve CH calling as described previously (18). Briefly, we estimated the mean sequencing error rate (0.032%) from duplicated reads by dividing the number unmatched bases with total bases, and then we used a binomial model to test whether the detected mutated reads were actually due to sequencing error. The following criteria were used to retain mutations: (i) sites with coverage > = 20; (ii) variant allele fraction (VAF) <35%; (iii) binomial model FDR-adjusted P < 0.001; (iv) sites were reported in Catalogue of Somatic Mutations in Cancer (COSMIC) version 92 (19).
Previous research has highlighted 34 leukemia/lymphoma-related genes (ASXL1, CBL, DNMT3A, GNAS, JAK2, NRAS, SF3B1, TP53, U2AF1, BCOR, PPM1D, TET2, IDH1, IDH2, SRSF2, RUNX1, SH2B3, ZRSR2, STAT3, KRAS, MYD88, ATM, CALR, CEBPA, ETV6, EZH2, FLT3, KIT, MPL, NPM1, STAG2, WT1, SETD2, CREBBP; ref. 10) as frequently associated with CH. More than 70% of reported CH mutations were reported in these genes (20). Thus, we considered any mutations located in those genes were likely to be true CH mutations. We relaxed the FDR cutoff to 0.01 on the previously reported mutations (20) to detect more potential mutated samples. For novel CH mutations, we removed any sites that overlap with dbSNP v151 (21) to eliminate noise from as many potential SNPs as possible.
We then applied more strict filtering criteria to genomic regions other than the 34 known CH-related genes to detect potential novel CH mutations. Because of the close relationship between CH, leukemia, and other types of cancer, functionally important CH mutations may also occur in other cancer genes. We further filtered CH mutations in 689 COSMIC cancer genes (excluding 34 leukemia genes; ref. 19) under the following criteria: (i) keep mutations with at least five reads supported the alternate allele; (ii) keep mutations with VAF no more than 0.1; (iii) if mutations had not been previously reported then remove sites that overlap with dbSNP v151. For the other genomic regions, we applied stricter filtering criteria to ensure the accuracy of CH identification. Only the mutations with (i) reads supported the alternate allele > = 10; (ii) VAF<0.1; (iii) not overlapped with dbSNP v151 sites were retained. All the sites were then annotated by ANNOVAR (22).
Mutational signature analysis
All the sites that remained, including exonic, intronic, and intergenic mutations were combined to estimate mutational signatures. Because of the limited number of mutations in each sample, we estimated the mutational signatures in pooling samples. For estimation of overall mutational signatures, we merged the mutations from all the samples. For correlation between mutational signatures and clinical factors, we merged the mutations from samples in each clinical factor group, randomly sampled 1,000 mutations and resampled 100 times. Then we assigned the mutations as well as their 3′ and 5′ nucleotide context into 96 trinucleotide mutational signatures. We assigned 30 previously described signatures (23) to our signatures using the decomposition algorithm developed by Coombs and colleagues (10). Each signature was assigned a weight that corresponded to the percentage of mutations explained by each given signature. We compared the weights of mutational signatures between the trait groups by Wilcoxon rank-sum tests.
Statistical analyses
Spearman correlation test (age) and Wilcoxon rank-sum tests (other traits) were used to test the relevance between CH mutations and traits. We also used Fisher exact tests to compare the number of samples with/without CH mutation between trait groups. Multivariate logistic regression analysis was used to examine the association between the prevalence of CH mutations and FHLC in both younger (age < 50) and older (age ≥ 50) samples separately, with age, disease status, smoking, and sex as covariates. For mediation test, we first constructed two linear/logistic regression models: independent variant—mediator and independent variant + mediator—dependent variant. Then we calculated the effect and significance of average causal mediation effects and proportion of the mediation effect by R function “mediate” of package “mediation.” Benjamini–Hochberg method (24) was used for multiple testing correction, with the significance cutoff of FDR as 0.1.
For the germline variation association, we obtained SNP array and WES SNP calling data for all of the samples. For WES SNP calling data, SNPs overlapped with SNP array were removed. We applied a linear regression model, with the number of CH mutations in each sample as the dependent variable, genotype of each SNP as independent variables. Sex, age, disease status, smoking, batch, sampling sites, and the top three principal components were included in the model as covariates. We used the “stepAIC” function from the MASS package to stepwisely optimize the model by AIC, and calculated the correlation between CH mutations and each SNP. To improve the statistical power, we required the sample size for each genotype ≥ 3, minimum minor allele frequency >0.01 and the total sample size ≥ 30. To correct for multiple testing, the P values were assessed using the Benjamini–Hochberg correction (24) to obtain the FDR. The significance cutoff was set to FDR < 0.1. Differences between CERES score and 0 were tested by one-sample t test. Because the number of blood/lymphocytes cell lines (78) was much less than other cell lines (912), to make significance level comparable between two kinds of cell lines, we randomly sampled 78 cell lines and calculated P values, shuffled 10,000 times, then used the mean P value as significance level of other cell lines.
Ethics approval and consent to participate
All research participants contributing clinical and genetic samples to this study provided written informed consent, subject to oversight by the University of Liverpool, Harvard University School of Public Health, University of Toronto, and Lunenfeld - Tanenbaum Research Institute or IARC review boards. The study was conducted according to the principles of the Declaration of Helsinki.
Availability of data and materials
Genotyping and WES data are available from Transdisciplinary Research Into Cancer of the Lung study (dbGaP Study Accession: phs000876.v2.p1), which is a part of INTEGRAL-ILCCO project. Data that support the differential expression of OTUD3 and survival status are available from Pediatric Acute Myeloid Leukemia (TARGET, 2018) and Study of origin Pediatric Acute Lymphoid Leukemia - Phase II (TARGET, 2018) datasets in cBioPortal database (https://www.cbioportal.org/). Data that support the pan-cancer expression landscape of OTUD3 are also available from The Cancer Genome Atlas (TCGA) PanCancer Atlas Studies dataset in cBioPortal database (https://www.cbioportal.org/). Gene expression and eQTL data of whole-blood are available at GTEx Portal (https://gtexportal.org/).
Results
CH mutations in leukemia-associated genes
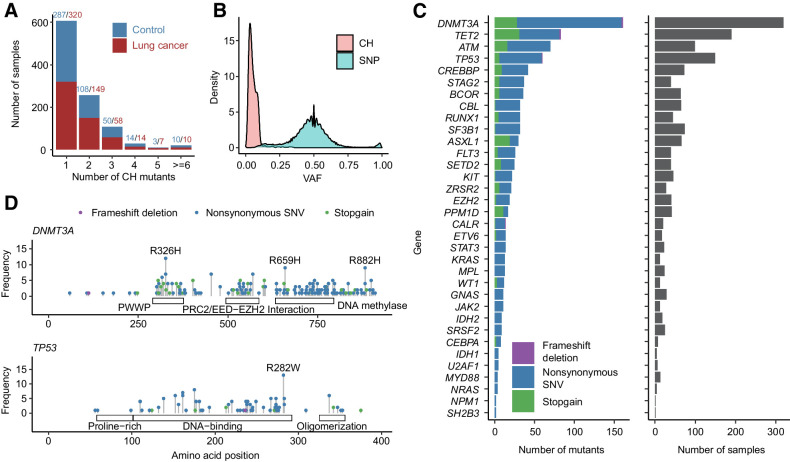
Previous study has identified a panel of leukemia-associated genes that are CH mutation hotspots (10). More than 70% of reported CH mutations were located in those genes (20). Thus, we first selected CH mutations located in those genes as the most robust dataset for subsequent analysis. We examined blood WES data to identify the prevalence of CH in 1,958 samples from the INTEGRAL-ILCCO project, including 1,059 from patients with lung cancer and 899 from controls (Supplementary Table S1). From these samples, we identified a total of 977 CH mutations located at 34 CH hotspot genes (Fig. 1A). Of the 1,958 subjects, 1,030 (52.6%) harbored at least one CH mutation. The majority of them (607 samples) have only one CH mutation with the maximum number per subject being 12 (Fig. 1A). The frequency of samples harbored at least one CH mutation in patients with lung cancer (558/1,059, 52.7%) and controls (472/899, 52.5%) do not have significantly differences (Fig. 1A). As expected, CH mutations have significantly lower VAFs compared with germline mutations, enabling us to correctly discriminate these two types of mutations (Fig. 1B). As shown, the median VAF for CH mutations was 0.047, with 98.9% of mutations having a VAF less than 0.2. In the 34 CH hotspot genes, DNMT3A had the largest number of mutated sites, followed by TET2, ATM, and TP53 (Fig. 1C). These top four genes accounted for 77.9% CH mutations in samples harboring at least one CH mutation.
Figure 1.
Overall distribution of CH mutations. A, Distribution of number of CH mutations in each sample. Most of the samples had 1–2 CH mutations. Red and blue denote number of CH mutations in patients with lung cancer and controls, respectively. B, VAF distribution of CH mutations and SNP. Most of CH mutations had VAF < 0.1. C, Number and type of CH mutations in 34 CH hotspot genes. D, CH mutation sites in DNMT3A and TP53. Mutations with samples ≥ 8 were labeled.
We therefore examined the pattern of mutation sites in two of the most frequently mutated genes, DNMT3A and TP53. In DNMT3A, we identified several high-frequency CH mutation sites including the most well-known R882H mutation (Fig. 1D; ref. 20). In TP53, the most frequent CH mutation was R282W (Fig. 1D). These frequent CH mutation sites are located at the functional domains of TP53 protein. In DNMT3A, the most frequent mutation R326H in DNMT3A is located in the Pro-Trp-Trp-Pro (PWWP) domain, and a less frequent mutation R659H is located in the DNA methylase domain (Fig. 1D). In TP53, the most frequent mutation R282W is located in the DNA-binding domain.
Association of CH mutations with age and lung cancer risk factors
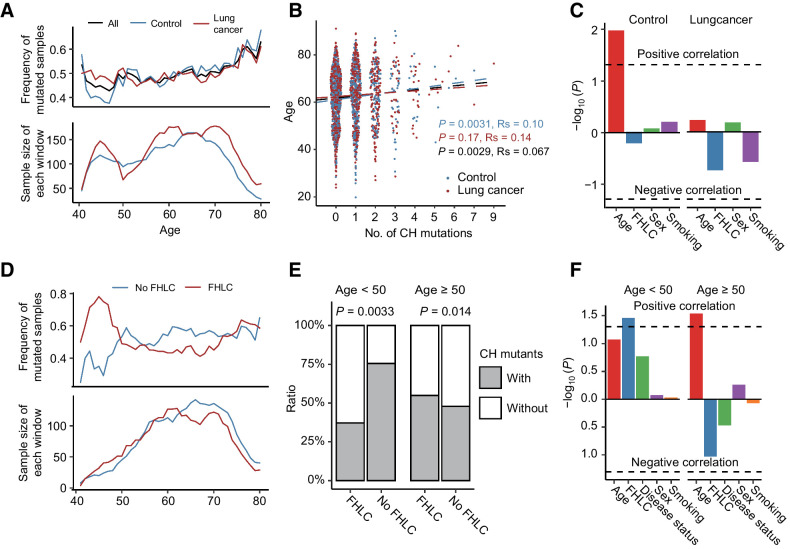
We investigated the association between number of CH mutations and available clinical variables, including age, sex, smoking history, disease status (lung cancer vs. control), and FHLC. Consistent with previous studies, we observed a continuously increase of CH mutation frequency with the increase of age (Fig. 2A; refs. 5, 6, 10, 25). Spearman correlation test suggest CH demonstrated a significant association with age (P = 0.0029; Fig. 2B). Both Spearman correlation and linear regression showed the association was more significant in control samples (P = 0.0031 and 0.011; Fig. 2B and C) than in lung cancer samples (P = 0.17 and 0.59), presumably due to the impact of other factors. Hence, we investigated the association between the number of CH mutations and other clinical traits, but observed no significant association without subject stratification. Notably, we observed in subjects younger than 50, lung cancer samples tend to have more CH mutations than control samples (Fig. 2A). Thus, we divided all subjects into a younger group (age <50) and an older group (age ≥50). We observed significant associations in subjects younger than 50. For example, we found that smoking has a much stronger impact on younger subjects in terms of CH mutations. In the young group, smokers had significantly higher CH mutation frequency than the nonsmokers (P = 0.025), while such a difference was not observed in the old group (P > 0.1; Supplementary Fig. S1A). Similarly, in the young group subjects with FHLC (regardless of their own cancer status) tend to have significantly more CH mutations than those without (P = 0.0033; Fig. 2D and E). We observed the opposite but nonsignificant trend among older subjects (Fig. 2E).
Figure 2.
CH mutations associated with age and lung cancer risk factors. A, Sliding window approach showed the mean frequency of mutated samples increasing with age. Mean frequency of mutated samples were calculated in each 5-year-old window with 1-year step. B and C, Spearman correlation (B) and linear regression (C) demonstrated a statistically significant association between CH and increased age, either in all samples or control samples. However, in lung cancer samples, the correlation between age and CH is not significant. D and E, In younger age group, subjects with family history of lung cancer have significantly fewer CH mutations than those without, while the opposite trend was observed in the older age group. F, Logistic regression found FHLC was the most significant trait associated with CH mutations in young samples. In older age samples, increasing age contributed the most to frequency of CH mutations.
It is well known that both smoking and FHLC are risk factors for lung cancer (26). In our dataset, we also observed that samples with smoking history and/or FHLC are more likely to be patients with lung cancer (Supplementary Fig. S1B and S1C). Thus, we further divided samples into subgroups by considering multiple traits, and then made comparisons in the younger (age < 50) and older age groups separately to characterize more effects of factors influencing lung cancer risk according to age groups. In younger subjects, FHLC was associated with more CH mutations, regardless of their lung cancer status, smoking history, and sex (Supplementary Fig. S1D). In older subjects, FHLC was associated with fewer CH mutations (Supplementary Fig. S1E). We further performed multivariate logistic regression analysis to examine the association between CH mutations and FHLC, while adjusting the effects of age, disease status, smoking, and sex as covariates. The result confirmed that FHLC is the most significant factor that associated with CH mutations (P = 0.035) in subjects with age < 50 (Fig. 2F). Instead, in old subjects, age is the most significant factor that associated with CH mutations (P = 0.029), followed by FHLC (P = 0.093; Fig. 2F). Thus, family history may contribute most to the accumulation of CH mutations in younger subjects; while in older individuals normal aging is the most important risk factor of CH mutations.
Given the association between CH and lung cancer risk factors, we wonder whether CH was a mediator between risk factors and lung cancer, or independently influenced by lung cancer risk factors. First, we test whether CH was a mediator between a risk factor and cancer. Either across all the samples or in young/old groups, none of the correlation between risk factor and cancer was significantly mediated by CH (Supplementary Fig. S2A). Because age and FHLC showed significant correlations with CH, we further tested whether these correlations could be mediated by another risk factor or cancer status. Consistent with Fig. 2F, in young samples, although the correlation between FHLC and CH could not be significantly mediated by any other risk factors, age has the lowest P values (P = 0.156) than all the other risk factors (Supplementary Fig. S2B); in old group, the correlation between age and CH could be significantly mediated by FHLC (P = 0.028; Supplementary Fig. S2C). In together, CH was more likely independently influenced by lung cancer risk factors than a mediator between risk factors and lung cancer, although we could not exclude the possibility that limited number of CH mutations reduced the statistic power.
Genetic variants associated with CH mutations
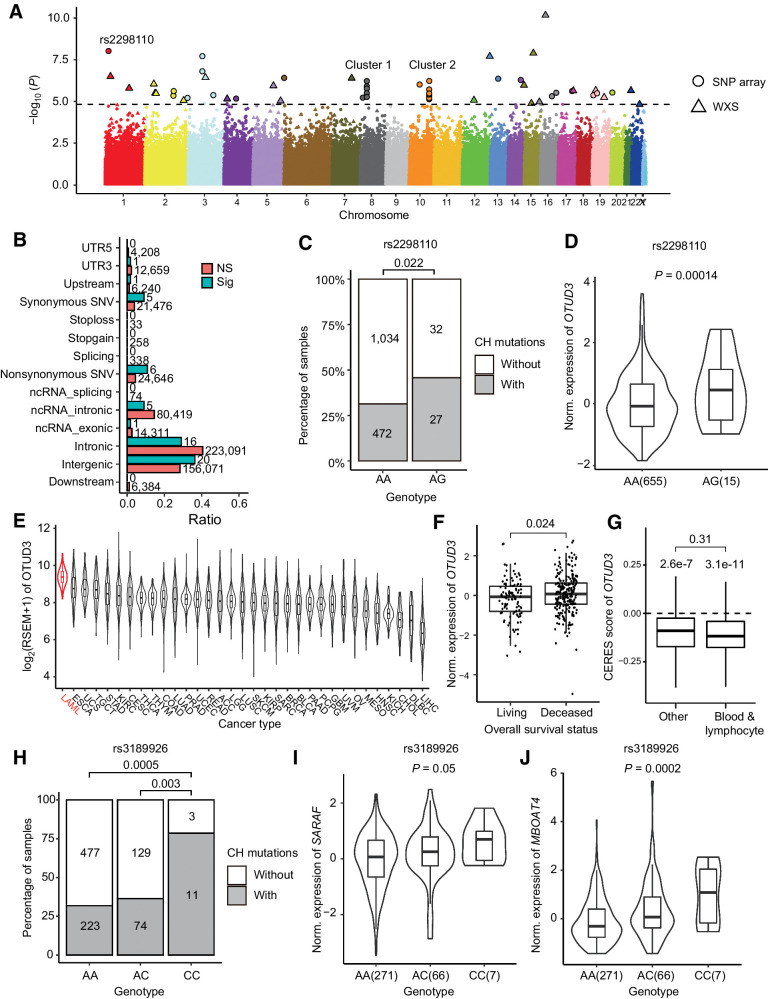
The association between FHLC and CH mutations suggested potential genetic effects. Thus, we performed a single-variant genome-wide association analysis by applying a linear regression model to all the samples. After removing SNPs that significantly deviated from HWE, we examined 407,635 SNPs from SNP array data and 150,292 SNPs called from the WES data. We discovered 55 sites (32 from SNP array data, 23 from WES data) significantly associated with the number of CH mutations at the significance level of 0.1 (FDR < 0.1; Fig. 3A; Supplementary Table S2).
Figure 3.
Genetic association of CH mutations. A, Manhattan plot shows 117 SNPs significantly associated with number of CH mutations. B, Category of SNPs associated with CH mutations. Numbers on each bar denote the number of SNPs in each category. Note that the total number of “Sig” SNPs were larger than 55, because a SNP might belong to multiple categories. C, Number of samples with CH mutations correlated with genotype of rs2298110. Samples with heterozygous genotype AG tend to have more CH mutations. D, In GTEx whole blood samples, expression of OTUD3 was positively correlated with heterozygous genotype AG of rs2298110. E, Expression of OTUD3 among TCGA cancers. Acute myeloid leukemia had the highest expression of OTUD3. F, Higher expression of OTUD3 was correlated with poor overall survival status in TARGET leukemia data (31). G, DepMap data suggested knockout OTUD3 broadly reduced the proliferation rate in various cell lines. While there were no significant CERES score differences between blood/lymphocyte cell lines and other cell lines (P = 0.31), both kind of cell lines had CERES scores significantly lower than zero (P = 3.1e-11 in blood/lymphocyte and 2.6e-7 in other cell lines, respectively). H, Number of samples with CH mutations correlated with genotype of rs3189926. Samples with homozygous genotype CC tend to have more CH mutations than genotype AA and AC. I and J, In GTEx whole blood data, samples with homozygous genotype CC at rs3189926 had higher expression of SARAF (I) and MBOAT4 (J) than other samples.
In total, we detected six nonsynonymous SNVs significantly correlated with CH mutations (Fig. 3B). Of these SNPs, rs2298110 located in OTUD3 showed the most significant correlation with CH mutations and the second lowest P value among all the SNPs (Fig. 3A; Supplementary Table S2). Samples with heterozygous genotype AG at rs2298110 tend to have more CH mutations than homozygous genotype AA (Fig. 3C). The A-to-G mutation leads to an asparagine to serine amino acid change at position 321. Despite the potential protein function change, rs2298110 is also an expression quantitative trait locus (eQTL). By investigating the eQTL data from whole blood samples of Genotype-Tissue Expression (GTEx) project (27), we found genotype AG at rs2298110 was correlated with higher expression of OTUD3 (Fig. 3D; Supplementary Table S3). As a deubiquitinase, the OTUD3 protein plays bifunctional roles in multiple cancers, which can be either a tumor suppressor by stabilizing PTEN protein in breast, colon, liver, and cervical cancer (28), or promote tumorigenesis by stabilizing the GRP78 protein in lung cancer (29). We investigated the expression of OTUD3 in TCGA dataset (30), and found leukemia had the highest expression of OTUD3 among all the cancer types (Fig. 3E). We found that higher expression of OTUD3 was associated with poor overall survival status (Fig. 3F) in TARGET leukemia data (31). These results suggested that OTUD3 may promote tumorigenesis in leukemia. In addition, we utilize the CRISPR-Cas9 knockout data from DepMap database (32, 33) to investigate whether knowckout OTUD3 will influence cell proliferation rate. In cell lines both derived from blood/lymphocyte and other tissues, the CERES scores (32) were significantly lower than 0 (Fig. 3G), suggesting knockout of OTUD3 would broadly reduce the proliferation rate of various cell lines. While there was no significant differences between CERES score of blood/lymphocyte cell lines and other cell lines (P = 0.31), the differences between CERES score and zero were more significant in blood/lymphocyte cell lines (P = 3.1e-11) than in other cell lines (P = 2.6e-7; Fig. 3G), indicating that OTUD3 may played more important role in the proliferation of blood/lymphocyte cells than in other tissues. We hypothesized that A-to-G mutation at rs2298110 may also gain the proliferation rate of hematopoietic stem cell by increasing the expression of OTUD3, and further promoting CH.
We also observed two SNP clusters located on chromosome 8 and 10, respectively (Fig. 3A). Cluster 1 included four SNPs (rs4733102, rs9656754, rs3189926, rs16876489) on 8p12, across a approximately 77 kb region (8:29893911-29971290). Two protein-coding genes are located in this region, SARAF and LEPROTL1. By investigated the eQTL data from whole blood samples of GTEx (27), we found that all the four SNPs were eQTLs, which were significantly correlated with the expression of SARAF and a nearby downstream gene MBOAT4 (Supplementary Table S3). For example, SNP rs3189926 was located in the 3′-untranslated region of SARAF. The homozygous genotype CC was correlated with more CH mutations and higher expression of SARAF and MBOAT4 than genotype AA and AC (Fig. 3H–J). As a negative regulator of store-operated calcium entry, SARAF might be related with abnormal calcium homeostasis of various cancers (34). MBOAT4 as well as LEPROTL1 were involved in the regulation of lipid metabolism (35). Cluster 2 of nine SNPs (rs78452361, rs1696819, rs1696820, rs1696821, rs17544933, rs4752586, rs17102481 and two novel sites) was located at chromosome 10q26.13, over a 26 kb intergenic region. While the potential function of these SNPs remain unknown, they are located immediately downstream of FGFR2, a FGFR that has been reported as a risk gene in breast cancer (36), gastric cancer (37), and leukemia (38). In addition, GWAS analysis has found a risk loci rs35837782 for childhood acute lymphoblastic leukemia at 10q26.13 (39). Overall, SNPs in these regions might play important regulation roles in tumor cell proliferation and survival, and might potentially promote CH via similar mechanisms.
CH mutations in other genes
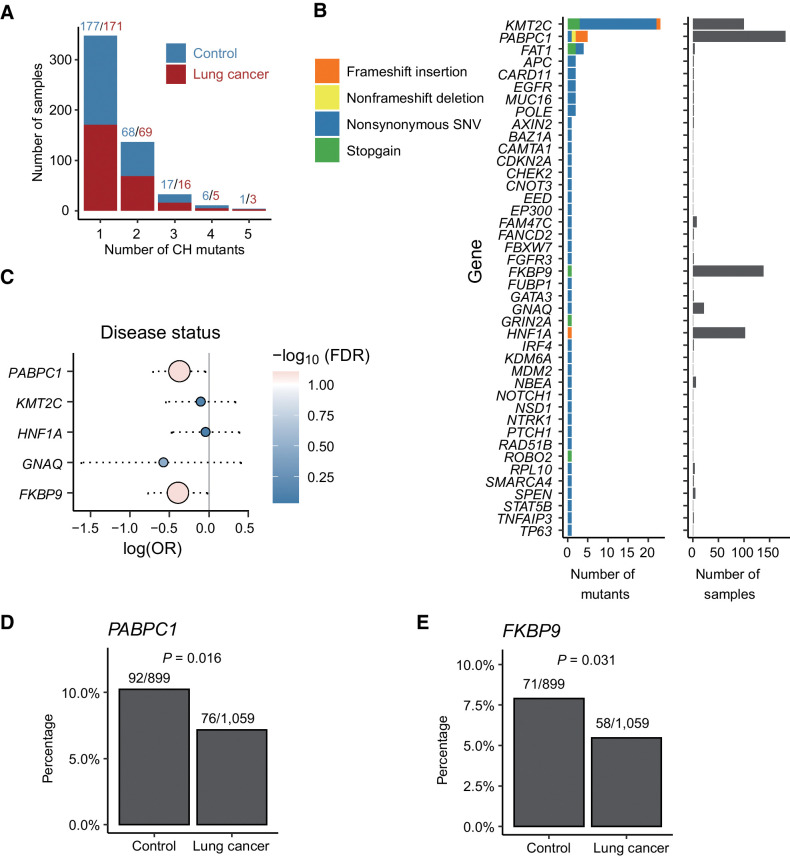
To detect potential novel CH mutations, we extended our analysis to other genes but applied a set of stringent selection criteria. First, we investigated 689 COSMIC cancer genes (19), and from them we identified a total of 85 different mutations located in 48 genes of 533 samples (Fig. 4A). While most genes are mutated in only a small number of samples with a unique mutation site, several genes, including KMT2C, PABPC1, FKBP9, and HNF1A, were mutated in a significantly large number of samples (Fig. 4B). Specifically, KMT2C was mutated in 101 subjects and harbored 23 different mutations. In contrast, PABPC1, FKBP9, and HNF1A were associated with only a few different mutation sites but these mutations presented in more than 80 samples.
Figure 4.
Potential novel CH mutations identified in other genomic regions and clinical associations. A, Distribution of number of CH mutations in each sample. Most of the samples had 1–2 CH mutations. Red and blue denote number of CH mutations in patients with lung cancer and controls, respectively. B, Number and type of CH mutations in COSMIC cancer genes. Nonsynonymous SNVs were most common. Most genes had only one CH mutation in a few samples. KMT2C and PABPC1 had the largest number of CH mutations; KMT2C, PABPC1, FKBP9, and HNF1A had the highest frequency of CH mutations. C, Comparison of mutation frequency of genes that were mutated in more than 10 samples in the patients with lung cancer versus the controls. D and E,PABPC1 (D) and FKBP9 (E) were less frequently mutated in subjects with lung cancer compared with controls.
We compared the mutation frequency of these genes in the patients with lung cancer versus the controls (Fig. 4C). We found that PABPC1 and FKBP9 mutations were significant correlated with disease status. As shown in Fig. 4D and E, PABPC1 and FKBP9 were less frequently mutated in subjects with lung cancer compared with controls. Following that, we further extended CH mutation identification to all genes, again, using the stringent selection criteria. This analysis resulted in 46 mutations located in 30 additional genes (Supplementary Fig. S3A and S3B) across 477 samples. Of them, PABPC3 and USP17L11 were the two most frequently mutated genes, with a mutation K254fs (a 1-bp frame-shifting insertion) in in PABPC3 and a mutation T360I (a point mutation) in USP17L11 presenting in 129 and 39 samples, respectively. By correlated these genes with sample subgroups stratified on the basis of different clinical features, we found that the USP17L11 mutation was negatively associated with FHLC and the PABPC3 mutation was positively associated with smoking (Supplementary Fig. S3C–S3F). These genes are not annotated as cancer-related genes according to COSMIC and their relevance with lung cancer or leukemia is largely unknown. Nevertheless, they may have cancer-related functions. For example, PABPC3 belongs to the poly(A)-binding protein (PABP) family, and post-transcription regulation mediated by PABPs was extensively altered in tumor and cancer cell lines (40, 41). In fact, we discovered another poly(A)-binding protein gene PABPC1 (Fig. 4B and E), which was included as a cancer gene in COSMIC. Reduced expression of PABPC1 has been reported to be associated with shorter postoperative survival time in esophageal cancer (42).
Mutational signatures associated with CH mutations
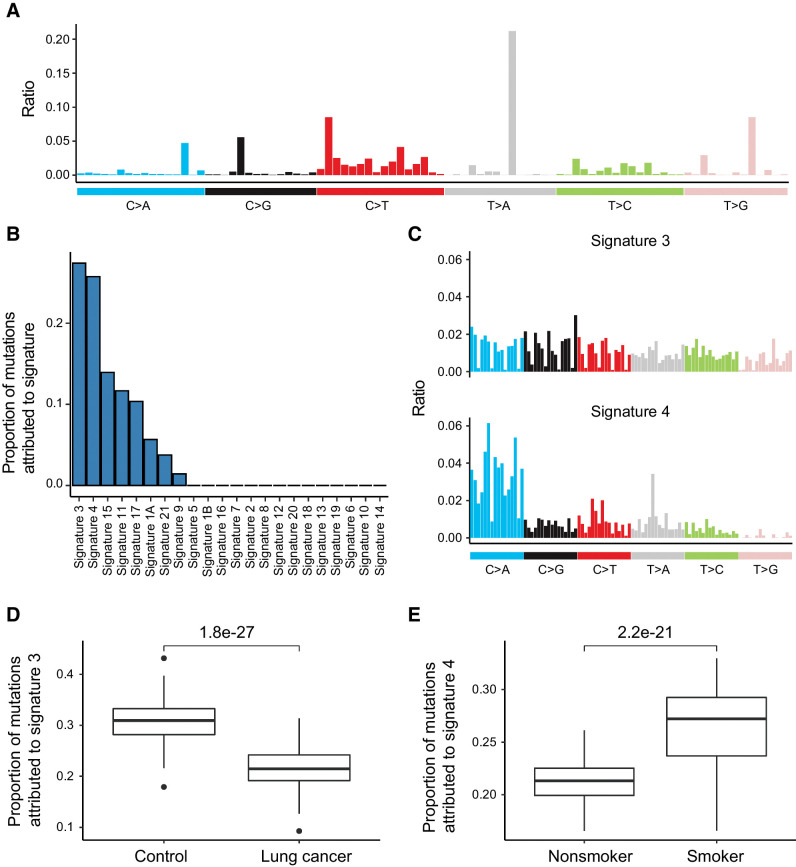
Mutational signature analysis has been widely used to characterize mutation patterns in tumors to gain insight on mutagenesis processes associated with tumorigenesis (23). Alexandrov and colleagues has defined a catalog of mutational signatures with mutational profile and associated etiology in multiple cancer types (23). To determine what types of mutations are enriched in CH mutations, here we pooled the CH mutations identified from all samples and defined the overall mutation profile (Fig. 5A; refs. 10, 23). Then we performed signature deconvolution using the established mutational signatures. Among all mutational signatures, we found Signature 3 and 4 to be the most informative ones, each accounting for more than 25% of CH mutation counts (Fig. 5B and C). According to annotation, Signature 3 was associated with BRCA1/2 mutations and proposed as a predictor of homologous recombination-based DNA repair deficiency (23). Signature 4 was known to be associated with tobacco smoking (23). Together, our results suggest that both genetic and environmental factors might contribute to the accumulation of CH mutations in the blood samples from our cohort.
Figure 5.
Signatures of CH mutations. A, Overall mutational signature of CH mutations. B, Proportion of mutations attributed to signatures, which were assigned previously by Alexandrov and colleagues (23). Signature 3 and Signature 4 contributed most to the total mutational signature. C, Rates of nucleotide substitution of Signature 3 and Signature 4. Data came from Alexandrov and colleagues (23). D, Patients with lung cancer had lower proportion of Signature 3. E, Smoker had higher fraction of Signature 4.
We then correlated the two mutational signatures with several clinical factors, including sex, age, smoking, disease status, and FHLC. We identified Signature 3 has the most significant negative correlation with lung cancer diagnosis (Fig. 5D). This result suggested that defective homologous recombination-based DNA repair might contribute to CH mutations more in controls than in patients with lung cancer. For Signature 4, we observed that it provides significantly higher contribution to the CH mutations in smokers than in non-smokers (Fig. 5E), consistent with the tobacco-related etiology of this signature.
Discussion
In this study, we examined blood WES data to identify the prevalence of CH in 1958 INTEGRAL-ILCCO project samples, investigated known CH mutations in 34 leukemia genes, and identified potential novel CH mutations in 65 other genes. In addition to the well-known age association of CH mutations, we found that CH mutations are associated with FHLC and smoking, especially in young samples. We investigated genetic variants associated with CH mutations, and discovered 55 sites significantly associated with the number of CH mutations. We found a SNP, rs2298110. That may promote CH by regulating the expression of OTUD3. We also observed that smoking significantly shaped the CH mutational signature. Overall, we uncovered a correlation between CH and lung cancer risk factors especially FHLC, identified potential genetic variants correlated with CH, and highlighted hundreds of potential novel CH mutations.
Prior studies have uncovered the blood-specific mutations in cancer-associated genes, identified recurrently mutated leukemia and lymphoma-associated genes (5, 6, 10, 25). Our analysis benefited from the CH mutation sites identified in these study. We also found CH mutations positively correlated with age, which was consistent with these studies. However, these studies usually lacked the comparison between patients with cancer and healthy individuals, had limited number of patients with lung cancer, or lacked the clinical information of lung cancer such as family history. INTEGRAL-ILCCO project provided us an opportunity to investigate the association between CH mutations and lung cancer and lung cancer risk factors. Interestingly, we found that CH mutations might have stronger associations with smoking and/or FHLC in the younger age group. While prior research had uncovered the association between CH mutations and smoking status, the variation in risk by age group has not been identified. Among known risk factors for lung cancer, FHLC showed the most significant association with CH mutations, suggesting that potential genetic factors may contribute to the accumulation of CH mutations. Indeed, a GWAS in individuals of European ancestry identified a germline polymorphism, which associated with a higher likelihood of having CH in TERT, a gene encoding a component of the telomerase complex (8). Another recent study uncovered three genetic loci associated with CH status, including one African ancestry specific locus, which disrupted a TET2 distal enhancer, resulted in increased self-renewal of hematopoietic stem cells (16). In our study, we identified 55 potential risk locus of CH status. As one of the most significant examples, our work highlighted rs2298110 as a potential genetic locus associated with CH; mutations at rs2298110 might promote CH by affecting the expression of OTUD3. We also observed two SNP clusters at 8p12 and 10q26.13, which might promote CH by regulating expression of tumor-associated genes. While we also utilized the CRISPR-Cas9 knockout data from DepMap database to validate the roles of these genes in regulating cell proliferation rate, the lack of experimental data was a limitation of our study. Future experimental studies would investigate how these genetic variants affect the expressions of candidate genes and promote CH, which could further corroborate our findings. In the younger age group, we also observed that smoking and lung cancer diagnosis correlated with CH mutations. Smoking is responsible for a variety of cancers, including lung cancers and myeloid leukemia (43). Tobacco smoke contains more than 60 carcinogens, which can directly induce cancer-related mutations and shape the distinct smoking-related mutational signatures (43, 44). In our study, we found the CH mutational signature was significantly associated with smoking, which was consistent with a previous study (10).
In hematologic malignancies, stem cells carrying CH mutations can be thought of as precursors to cancer stem cells (45). However, the relationship between CH and primary solid tumors is still unclear. In our study, we detected correlations between CH mutations and lung cancer risk factors rather than lung cancer status, indicated the connection between CH mutations may be indirectly. Mediation tests suggested that CH was more likely independently influenced by lung cancer risk factors than a mediator between risk factors and lung cancer, although we could not exclude the possibility that limited number of CH mutations reduced the statistic power. In some cases, the correlation between CH and cancer could be explained as a toxic effect of prior treatment with cytotoxic chemotherapy and/or radiation (10, 14, 46). Given the fact that CH mutations could alter the function of circulating immune cells, another hypothesis is CH may influence the immune response to tumors. Studies in mouse models suggested that deletion of DNMT3A in CD8+ T cells prevented T-cell exhaustion (47), while deletion of TET2 in murine myeloid cells increased numbers of effector T cells in the tumor and reduced tumor growth (48). In addition, we could also hypothesize that the correlation between CH mutations and lung cancer status or risk factors may be different between lung cancer subtypes, because different lung cancer subtypes varies greatly in genetic, expression profile, and pathology. Because of lack the subtype information, we could not analysis the correlation between CH and lung cancer subtypes. Future analysis on larger dataset with more cancer pathology information will improve our understanding of the relationship between lung cancer, smoking, and CH mutations, and the potential role of CH in tumor immunosurveillance.
Compared with targeted sequencing panels that usually have higher sequencing depth (>400×; refs. 10, 18, 46), the germline exome data for our CH analysis have much lower coverage (∼51×). In our cases, several hotspot mutations (e.g., DNMT3A p.R882H) were only supported by 1–2 read counts in many samples. Thus, we designed a hierarchical filtering criterion to balance the sensitivity and accuracy. For the hotspot mutations that were previously reported by high-coverage targeted sequencing analysis, we set a loose criterion and identified 977 high-confidential CH mutations in 34 leukemia genes. The VAF distribution of these CH mutations was similar to previous studies, which partially supported the accuracy of our filtering criteria. Compared with targeted sequencing panels, the whole-exome data provides the opportunity of detecting novel CH mutations. We identified 106 novel CH mutations in the other part of exome under the strict filtering criteria, highlighted several candidate genes. The CH dataset we defined here would be a valuable resource for further analysis of the mutation status and functional mechanism based on ultrasensitive-targeted sequencing.
Authors' Disclosures
C.I. Amos reports grants from NCI and Cancer Prevention Research Institute of Texas during the conduct of the study. No disclosures were reported by the other authors.
Supplementary Material
Supplementary Figures S1-S3, corresponded figure legends, and legends of supplementary table S1-S3
Table S1-S3
Acknowledgments
This study is supported by the Cancer Prevention Research Institute of Texas (CPRIT; RR180061 to C. Cheng; RR170048 to C.I. Amos; RR190104 to A. Li) and the National Cancer Institute of the National Institutes of Health (1R21CA227996 to C. Cheng; U19CA203654 to C.I. Amos). C. Cheng, C.I. Amos, and A. Li are CPRIT Scholars in Cancer Research.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors' Contributions
W. Hong: Investigation, visualization, methodology, writing–original draft, writing–review and editing. A. Li: Funding acquisition, methodology, writing–review and editing. Y. Liu: Resources, data curation, investigation, writing–review and editing. X. Xiao: Resources, data curation, methodology. D.C. Christiani: Resources, writing–review and editing. R.J. Hung: Resources, data curation, writing–review and editing. J. McKay: Resources, data curation, writing–review and editing. J. Field: Resources, data curation, writing–review and editing. C.I. Amos: Resources, data curation, funding acquisition, methodology, writing–review and editing. C. Cheng: Supervision, funding acquisition, writing–review and editing.
References
- 1. Abkowitz JL, Catlin SN, McCallie MT, Guttorp P. Evidence that the number of hematopoietic stem cells per animal is conserved in mammals. Blood 2002;100:2665–7. [DOI] [PubMed] [Google Scholar]
- 2. Fliedner TM, Graessle D, Paulsen C, Reimers K. Structure and function of bone marrow hemopoiesis: mechanisms of response to ionizing radiation exposure. Cancer Biother Radiopharm 2002;17:405–26. [DOI] [PubMed] [Google Scholar]
- 3. Blokzijl F, de Ligt J, Jager M, Sasselli V, Roerink S, Sasaki N, et al. Tissue-specific mutation accumulation in human adult stem cells during life. Nature 2016;538:260–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Machiela MJ, Zhou W, Sampson JN, Dean MC, Jacobs KB, Black A, et al. Characterization of large structural genetic mosaicism in human autosomes. Am J Hum Genet 2015;96:487–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Busque L, Mio R, Mattioli J, Brais E, Biais N, Lalonde Y, et al. Nonrandom X-inactivation patterns in normal females: lyonization ratios vary with age. Blood 1996;88:59–65. [PubMed] [Google Scholar]
- 6. Champion KM, Gilbert JGR, Asimakopoulos FA, Hinshelwood S, Green AR. Clonal haemopoiesis in normal elderly women: implications for the myeloproliferative disorders and myelodysplastic syndromes. Br J Haematol 1997;97:920–6. [DOI] [PubMed] [Google Scholar]
- 7. Genovese G, Kähler AK, Handsaker RE, Lindberg J, Rose SA, Bakhoum SF, et al. Clonal hematopoiesis and blood-cancer risk Inferred from blood DNA sequence. N Engl J Med 2014;371:2477–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zink F, Stacey SN, Norddahl GL, Frigge ML, Magnusson OT, Jonsdottir I, et al. Clonal hematopoiesis, with and without candidate driver mutations, is common in the elderly. Blood 2017;130:742–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Laurie CC, Laurie CA, Rice K, Doheny KF, Zelnick LR, McHugh CP, et al. Detectable clonal mosaicism from birth to old age and its relationship to cancer. Nat Genet 2012;44:642–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Coombs CC, Zehir A, Devlin SM, Kishtagari A, Syed A, Jonsson P, et al. Therapy-related clonal hematopoiesis in patients with non-hematologic cancers Is common and associated with adverse clinical outcomes. Cell Stem Cell 2017;21:374–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gibson CJ, Lindsley RC, Tchekmedyian V, Mar BG, Shi J, Jaiswal S, et al. Clonal hematopoiesis associated with adverse outcomes after autologous stem-cell transplantation for lymphoma. J Clin Oncol 2017;35:1598–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med 2014;371:2488–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ruark E, Snape K, Humburg P, Loveday C, Bajrami I, Brough R, et al. Mosaic PPM1D mutations are associated with predisposition to breast and ovarian cancer. Nature 2013;493:406–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wong TN, Ramsingh G, Young AL, Miller CA, Touma W, Welch JS, et al. Role of TP53 mutations in the origin and evolution of therapy-related acute myeloid leukaemia. Nature 2015;518:552–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394–424. [DOI] [PubMed] [Google Scholar]
- 16. Bick AG, Weinstock JS, Nandakumar SK, Fulco CP, Bao EL, Zekavat SM, et al. Inherited causes of clonal haematopoiesis in 97,691 whole genomes. Nature 2020;586:763–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang Z, Wei Y, Zhang R, Su L, Gogarten SM, Liu G, et al. Multi-omics analysis reveals a HIF network and hub gene EPAS1 associated with lung adenocarcinoma. EBioMedicine 2018;32:93–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Young AL, Challen GA, Birmann BM, Druley TE. Clonal haematopoiesis harbouring AML-associated mutations is ubiquitous in healthy adults. Nat Commun 2016;7:12484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sondka Z, Bamford S, Cole CG, Ward SA, Dunham I, Forbes SA. The COSMIC Cancer Gene Census: describing genetic dysfunction across all human cancers. Nat Rev Cancer 2018;18:696–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Watson CJ, Papula AL, Poon GYP, Wong WH, Young AL, Druley TE, et al. The evolutionary dynamics and fitness landscape of clonal hematopoiesis. Science 2020;367:1449–54. [DOI] [PubMed] [Google Scholar]
- 21. Sherry ST. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res 2001;29:308–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010;38:e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SAJR, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature 2013;500:415–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B 1995;57:289–300. [Google Scholar]
- 25. Xie M, Lu C, Wang J, McLellan MD, Johnson KJ, Wendl MC, et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med 2014;20:1472–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Malhotra J, Malvezzi M, Negri E, La Vecchia C, Boffetta P. Risk factors for lung cancer worldwide. Eur Respir J 2016;48:889–902. [DOI] [PubMed] [Google Scholar]
- 27. Aguet F, Barbeira AN, Bonazzola R, Brown A, Castel SE, Jo B, et al. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 2020;369:1318–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yuan L, Lv Y, Li H, Gao H, Song S, Zhang Y, et al. Deubiquitylase OTUD3 regulates PTEN stability and suppresses tumorigenesis. Nat Cell Biol 2015;17:1169–81. [DOI] [PubMed] [Google Scholar]
- 29. Du T, Li H, Fan Y, Yuan L, Guo X, Zhu Q, et al. The deubiquitylase OTUD3 stabilizes GRP78 and promotes lung tumorigenesis. Nat Commun 2019;10:2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Weinstein JN, Collisson EA, Mills GB, Shaw KRM, Ozenberger BA, Ellrott K, et al. The cancer genome atlas pan-cancer analysis project. Nat Genet 2013;45:1113–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Farrar JE, Schuback HL, Ries RE, Wai D, Hampton OA, Trevino LR, et al. Genomic profiling of pediatric acute myeloid leukemia reveals a changing mutational landscape from disease diagnosis to relapse. Cancer Res 2016;76:2197–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Meyers RM, Bryan JG, McFarland JM, Weir BA, Sizemore AE, Xu H, et al. Computational correction of copy number effect improves specificity of CRISPR–Cas9 essentiality screens in cancer cells. Nat Genet 2017;49:1779–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Behan FM, Iorio F, Picco G, Gonçalves E, Beaver CM, Migliardi G, et al. Prioritization of cancer therapeutic targets using CRISPR–Cas9 screens. Nature 2019;568:511–6. [DOI] [PubMed] [Google Scholar]
- 34. Palty R, Raveh A, Kaminsky I, Meller R, Reuveny E. SARAF inactivates the store operated calcium entry machinery to prevent excess calcium refilling. Cell 2012;149:425–38. [DOI] [PubMed] [Google Scholar]
- 35. Cai Y, Crowther J, Pastor T, Abbasi Asbagh L, Baietti MF, De Troyer M, et al. Loss of chromosome 8p governs tumor progression and drug response by altering lipid metabolism. Cancer Cell 2016;29:751–66. [DOI] [PubMed] [Google Scholar]
- 36. Hunter DJ, Kraft P, Jacobs KB, Cox DG, Yeager M, Hankinson SE, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet 2007;39:870–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kunii K, Davis L, Gorenstein J, Hatch H, Yashiro M, Di Bacco A, et al. FGFR2-amplified gastric cancer cell lines require FGFR2 and Erbb3 signaling for growth and survival. Cancer Res 2008;68:2340–8. [DOI] [PubMed] [Google Scholar]
- 38. Carll T, Patel A, Derman B, Hyjek E, Lager A, Wanjari P, et al. Diagnosis and treatment of mixed phenotype (T-myeloid/lymphoid) acute leukemia with novel ETV6-FGFR2 rearrangement. Blood Adv 2020;4:4924–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vijayakrishnan J, Kumar R, Henrion MYR, Moorman AV, Rachakonda PS, Hosen I, et al. A genome-wide association study identifies risk loci for childhood acute lymphoblastic leukemia at 10q26.13 and 12q23.1. Leukemia 2017;31:573–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xiang Y, Ye Y, Lou Y, Yang Y, Cai C, Zhang Z, et al. Comprehensive characterization of alternative polyadenylation in human cancer. J Natl Cancer Inst 2018;110:379–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Xia Z, Donehower LA, Cooper TA, Neilson JR, Wheeler DA, Wagner EJ, et al. Dynamic analyses of alternative polyadenylation from RNA-seq reveal a 3′-UTR landscape across seven tumour types. Nat Commun 2014;5:5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Takashima N, Ishiguro H, Kuwabara Y, Kimura M, Haruki N, Ando T, et al. Expression and prognostic roles of PABPC1 in esophageal cancer: correlation with tumor progression and postoperative survival. Oncol Rep 2006;15:667–71. [PubMed] [Google Scholar]
- 43. Hecht SS. Tobacco carcinogens, their biomarkers and tobacco-induced cancer. Nat Rev Cancer 2003;3:733–44. [DOI] [PubMed] [Google Scholar]
- 44. Alexandrov LB, Ju YS, Haase K, Van Loo P, Martincorena I, Nik-Zainal S, et al. Mutational signatures associated with tobacco smoking in human cancer. Science 2016;354:618–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Silver AJ, Jaiswal S. Clonal hematopoiesis: pre-cancer PLUS. Adv Cancer Res 2019;141:85–128. [DOI] [PubMed] [Google Scholar]
- 46. Mouhieddine TH, Sperling AS, Redd R, Park J, Leventhal M, Gibson CJ, et al. Clonal hematopoiesis is associated with adverse outcomes in multiple myeloma patients undergoing transplant. Nat Commun 2020;11:2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ghoneim HE, Fan Y, Moustaki A, Abdelsamed HA, Dash P, Dogra P, et al. De novo epigenetic programs inhibit PD-1 blockade-mediated T cell rejuvenation. Cell 2017;170:142–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pan W, Zhu S, Qu K, Meeth K, Cheng J, He K, et al. The DNA methylcytosine dioxygenase Tet2 sustains immunosuppressive function of tumor-Infiltrating myeloid cells to promote melanoma progression. Immunity 2017;47:284–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1-S3, corresponded figure legends, and legends of supplementary table S1-S3
Table S1-S3
Data Availability Statement
Genotyping and WES data are available from Transdisciplinary Research Into Cancer of the Lung study (dbGaP Study Accession: phs000876.v2.p1), which is a part of INTEGRAL-ILCCO project. Data that support the differential expression of OTUD3 and survival status are available from Pediatric Acute Myeloid Leukemia (TARGET, 2018) and Study of origin Pediatric Acute Lymphoid Leukemia - Phase II (TARGET, 2018) datasets in cBioPortal database (https://www.cbioportal.org/). Data that support the pan-cancer expression landscape of OTUD3 are also available from The Cancer Genome Atlas (TCGA) PanCancer Atlas Studies dataset in cBioPortal database (https://www.cbioportal.org/). Gene expression and eQTL data of whole-blood are available at GTEx Portal (https://gtexportal.org/).