Abstract
The study of multi-cell-type (MCT) interactions has the potential to significantly impact our understanding of tissue and disease biology. Such studies require innovative culture tools for unraveling the contributions of each cell type. Micro- and macro-scale platforms for MCT culture each have different advantages and disadvantages owing to their widely different capabilities, availability, and ease-of-use. However, as evidenced in the literature, there are very few examples of MCT studies and culture platforms, suggesting both biological and technical barriers. We have developed an open multi-culture platform to promote more rapid progress by integrating advantages of both micro- and macro-scale culture devices. The proposed open multi-culture platform addresses technical barriers by allowing easy customization, independent control of basic physical culture parameters, and incorporation of multiple culture modalities (e.g., 2D, 3D, transwell, and spheroid). The design also permits the user to obtain independent endpoints for each culture region. We demonstrate use of the platform in two example studies where we evaluated how cell ratio and cell types influence the response of triple negative breast cancer cells to heat damage and Hedgehog signaling. We also show that the platform can improve soluble factor transport between cell types compared to compartmentalized macro- and micro-scale alternatives. Last, we examine current and future challenges of the platform. We envision simple, yet flexible and customizable, platforms such as this will be important for advancing in vitro study of tissue and tumor biology.
Graphical Abstract
INTRODUCTION
In vitro cell culture allows researchers near complete control over the cellular microenvironment for advanced manipulation and interrogation while also offering a cost effective alternative to in vivo studies. However, a fundamental challenge for in vitro model development is to adequately recapitulate the in vivo environment to enable translation of results to in vivo application. For this reason, a variety of methods for incorporating tissue-specific 3D matrices and organotypic structures have been developed.1-5 Similarly, there is also focus on developing methods for moving beyond the study of co-culture interactions (i.e., between two cell types) to the study of multi-cell-type (MCT) interactions as these additional interactions can be seen to significantly impact important aspects of tissue and disease biology. For example, in vitro 3D tetra-culture (i.e., a model involving four cell-types)6 and tri-culture models7-8 have been shown to recapitulate critical cell-cell interactions. Engineered MCT culture models like these offer the potential to experimentally control the influence of each culture component and elucidate the complex molecular underpinnings of these important processes.
Examples of MCT studies in the literature utilize a variety of approaches that range from basic mixing of cells in traditional macroscale well-plates9-11 to advanced microfluidic embodiments that provide 3D culture 12, perfusion 13-15 and compartmentalization 16,17. However, there are some common limitations. Simple mixing approaches offer limited control over the nature of interactions (e.g., via soluble factor or cell-cell contact) or patterning of cell-types or structures (e.g., ducts or lumens). Likewise, it becomes inherently difficult to quantify changes occurring between individual cell-types within the complex mixture. Microscale approaches can offer improved control over the cellular microenvironment 18 but often add experimental complexity and additional expertise or specialized equipment such as microfluidic pumps, valves, and controls.19 Similarly, it is often difficult to isolate individual cell-types from within closed microfluidic devices for downstream analysis, which can limit the potential for new insights.
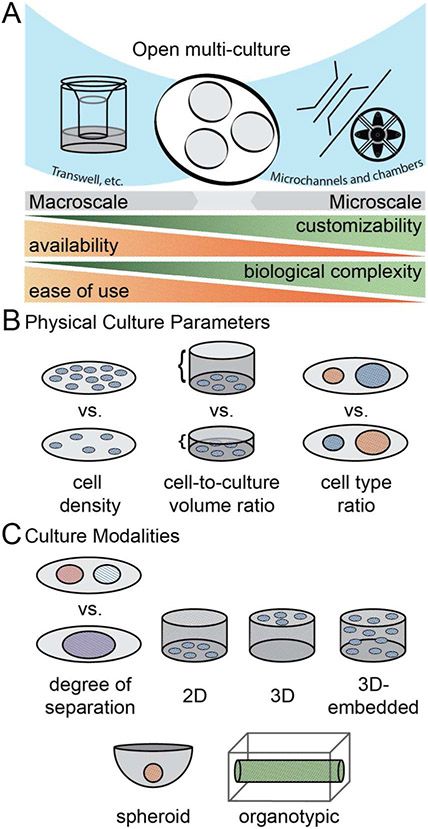
In addition to these limitations, in vitro studies of MCT interactions also present numerous experimental challenges.20,21 Different cell types often require significantly different culture conditions such as 2D vs 3D, ECM composition, or even geometry (e.g., monolayer vs. spheroid vs. lumen) yet integrating different culture modalities within a single culture model can be technically challenging (Fig 1C). Inclusion of additional cell types also requires one to choose appropriate ratios of cell-types (i.e., different ratios of cell types could lead to different conclusions). Similarly, basic variables of in vitro culture like the number of cells per unit volume of media (cell-to-volume ratio) chosen for each cell type can affect relative rates of nutrient depletion and waste accumulation, which then impact soluble factor signaling and response (Fig 1B). The ability to address each of these considerations depends upon the choice of a culture platform and comes with tradeoffs in terms of ease-of-use and technical complexity (Fig 1A). Tools that can balance these trade-offs by offering simple ways to isolate and control important culture parameters while enabling integration of different culture modalities will be important for advancing MCT studies.
Figure 1. General categories of multi-culture platforms.
(A) The open-multi-culture platform incorporates advantageous elements of both macroscale and microscale culture, creating a balance between customizability, availability, biological complexity, and ease of use. (B) Physical culture parameters, such as cell density, cell-to-volume ratio, and cell type ratio should be considered when forming models of inter-population interactions. (C) Culture modalities alter the degree of separation between cell populations and matrix interaction.
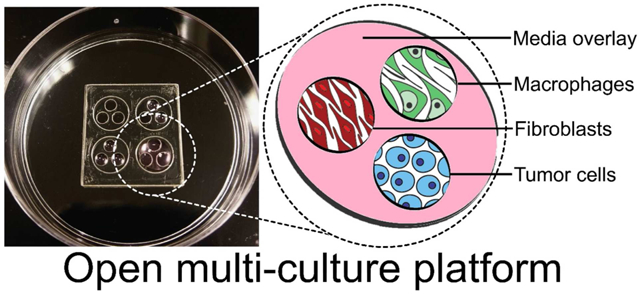
Here, we describe and characterize an open multi-culture methodology that blends the advantages of macro- and micro-scale approaches to balance biological, experimental, and device complexity (Fig 1A). The device consists of a series of wells (culture regions) that exist within a larger well (parent well) to allow use of a fluid or gel overlay to enable soluble factor communication or transmigration between cultures (Fig 1A and 2). The approach improves upon a simple macroscale mixing approach by providing discrete compartments that can be joined or separated at different times to allow cell-type-specific culture conditions and parameters while enabling independent access for harvesting the separate populations for downstream analysis. The approach simplifies micro-culture while enabling ultra-rapid customization by embracing an “open” microscale device design22-27 that can be fabricated via razor-printing. Although we introduced the concept of the open device design as part of a recent study28, the previous study focused on demonstrating the biocompatibility of the razorprinting fabrication method. Here, we study and characterize the specific advantages of the razor-printed device itself for the emerging and important area of MCT studies. More specifically, we demonstrate the ability to isolate effects of basic culture parameters (e.g., cell density, cell-type-ratio, cell-to-volume ratios), integrate multiple culture modalities, facilitate rapid soluble factor communication between culture compartments, and leverage independent access to each compartment to quantify the influence of MCT interactions.
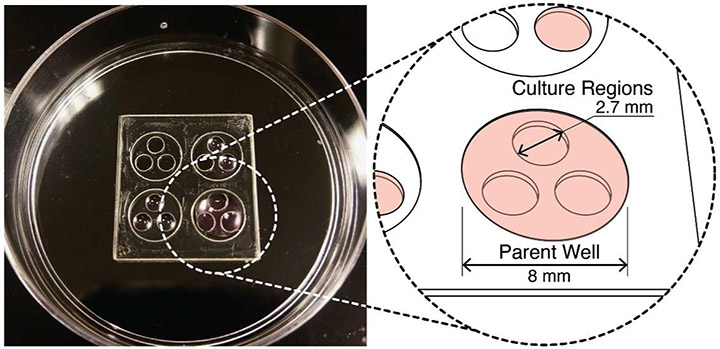
Figure 2. Open multi-culture device.
(A) Photograph of a 50 mm Petri dish with an array of 4 open-culture devices each at different stages of filling/seeding. Each device has 3 culture regions (2.7 mm dia. each) contained by a parent well (8 mm dia.). The pink color represents culture medium.
EXPERIMENTAL METHODS
Razor-printing (Xurography) -
Razor-printed culture platform components were made from either PDMS or polystyrene sheets and biomedical-grade adhesive tape as done previously.28 Briefly, open-multi culture platform preparation consists in cutting 125 μm (300711566, GoodFellow) and 190 μm (300711235, GoodFellow) polystyrene films bound to double-sided medical-grade adhesive tape (90106, Adhesives Research Inc., Glen Rock, PA, USA) into the desired geometries. The 125 μm sheet was used for the culture regions while the parent wells were constructed from 190 μm sheets. Once cut, the parent wells were manually aligned and bound over the sheet of culture regions, then placed on sterile tissue culture treated omnitrays or petri dishes. Assembled devices were placed under ultraviolet light for 20 min in a laminar flow hood for sterilization and washed twice with PBS prior to cell seeding.
Culture platform design and operation -
The simple multi-culture platform is composed of multiple adjacent culture regions contained within a larger parent well. Each cell type is seeded independently into a culture well using 5 μl and allowed to attach to the bottom culture surface. A 50-80 μl volume of cell culture medium is then dispensed over the top of the culture regions to interconnect the compartments. The 3-regions have a center-center connection that forms an equilateral triangle with side 3.5 mm. The parental well is recessed 125 μm while the culture regions are recessed an additional 600 μm through to the culture substrate.
Cell culture -
MDA-MB-231, MDA-MB-468, HS-578t, THP-1, and NIH-3T3 were purchased from ATCC. MDA-MB-231, MDA-MB-468, HS-578t and NIH-3T3 were sustained by DMEM high glucose media with L-Glutamine (D5796, Sigma-Aldrich) supplemented with 10% heat inactivated fetal bovine serum (F6765, Sigma-Aldrich) and 1% penicillin/streptomycin (P4333, Sigma-Aldrich). THP-1 cells were kept in RPMI 1640 (SH30027.01, GE Life Sciences) supplemented with 0.05 nM 2-mercaptoethanol (M3148, Sigma-Aldrich) with 10% heat inactivated fetal bovine serum and 1% penicillin/streptomycin. All cells were maintained at 37°C in a 5% CO2 incubator. Passages were performed at 75-80% confluence using 0.5% trypsin (59418C, Sigma-Aldrich) for adherent cells. Viable cells were counted using CBA Visio Image Cytometer (Nexcelom Bioscience LLC, Lawrence, MA, USA) with the Trypan Blue exclusion method (T8154, Sigma-Aldrich).
Immunofluorescent staining -
The staining process was carried out at room temperature (25°C). NIH-3T3 were fixed with 4% paraformaldehyde for 15-20 min. Cell permeabilization was performed using 0.5% triton (T8787, Sigma-Aldrich). Devices were washed twice with PBS 1X and incubated with 3% BSA in PBS 1X solution for 1-2 hrs. Primary antibody anti-alpha smooth muscle actin (ab7817, Abcam) was incubated at 1:200 ratio in 3% BSA + 0.1% Tween 20 in PBS 1X solution for 1 hr. Devices were washed three times 0.1% tween 20 in PBS 1X solution. Secondary antibody Alexa 488 anti-mouse (ab150117, Abcam) was incubated at a ratio of 1:500 in 3% BSA + 0.1% tween 20 in PBS 1X solution for 1 hr. Devices were washed three times 0.1% tween 20 in PBS 1X solution and stained with Hoechst 33342 nuclear dye for 30 min. Fluorescent images were obtained with an 3i-Spinning disk Confocal microscope at 10X.
Heat damage
MDA-MB-231 cells were used as the tumor cell model. Tumor cells were co-cultured adjacent to normal or cancer-associated stromal groups. Stromal cells within the normal group were modeled using THP-1 M1, THP-1 M1/M2, and NIH-3T3 cell lines. The cancer-associated stromal group consisted of THP-1 M2 and TGF-β treated NIH-3T3 cell lines. Preparation and seeding of normal stroma - NIH-3T3 (4,000 cells) and THP-1 (10,000 cells) cells were seeded simultaneously in adjacent culture regions. THP-1 were treated with 36 nM phorbol 12-myristate 13-acetate (PMA, 594400, Millipore) for 48hrs to induce differentiation into macrophages (mixed M1/M2 phenotype). For M1 polarization, differentiation consisted of an additional 48-hr incubation period in the same culture medium supplemented with 20 nM IFN-γ (300-02, PeproTech) and 10 ng/mL LPS (L4391, Sigma-Aldrich). Preparation and seeding of cancer-associated stroma - Seeding and treatment began the same as for the normal stromal group. However, NIH-3T3 and THP-1 M1/M2 cells underwent additional treatments for 3 days to induce cancer-associated phenotypes. NIH cells were treated with 20 nM TGF-β in DMEM HG (D5796, Sigma-Aldrich).29 THP-1 M1/M2 cells were treated with RPMI 1640 media supplemented with 20 nM IL-4 (200-04, PeproTech) and IL-13 (200-13, PeproTech) to induce an M2 phenotype.30 Preparation and seeding of tumor cells - The third adjacent culture region was seeded with 4,000 MDA-MB-231 cells.
Stromal groups were independently seeded adjacent to tumor cells within each 3-region device. After overnight incubation, 50 μL of RPMI media was added to each device, effectively interconnecting all three regions. As a positive control for hyperthermia temperature, cells were exposed to a temperature of 41°C using a water bath (66566, Precision Scientific, Chicago, IL, USA) for 30 min. Incubation at 37°C inside the cell culture incubator was used as a baseline control. Culture medium was replaced for all groups immediately after treatment as media components may have been negatively affected by the increased temperature.
After 48 hrs of culture, tumor proliferation was assessed using a Click-IT 594 Imaging Kit (C10639, Invitrogen) according to the manufacturer’s recommendations. Hoechst 33342 (56198, BD Biosciences) nuclear staining (1:1000 dilution) was done to obtain total cell counts. Images at 20X magnification were taken using a ZOE Fluorescent Cell Imager (Bio-Rad, Hercules, CA, USA) and cell counts were processed with ImageJ Processing and Analysis Software.31 Proliferation fraction was determined by dividing EDU positive cell counts by the total cell counts in the tumor culture region. 1-2 images were taken per culture region (30-60% of the total surface culture area) to determine fraction of proliferating cells. Student-t test was performed to report significance (p < 0.05).
Soluble factor paracrine signaling
Mono-culture was performed by seeding 4,000 MDA-MB-468 in adjacent culture regions, allowing cells to attach overnight. For co-culture, 5,000 HS-578T cells were seeded at t = 0 hrs and treated with 10 μg/ml of TGF-β (100-21, PeproTech, Rocky Hill, NJ, USA) in cell culture media for 72 hrs (replenished at 48 hrs). TGF-β induces epithelial mesenchymal transition in HS-578T to a myofibroblast phenotype, termed HS-578T-myo. After 72 hrs of TGF-β treatment, culture regions were independently washed 3 times with 10 μl of 1X PBS to remove any remaining TGF-β. Approximately 4,000 MDA-MB-468 cells were seeded at t = 55 hrs in culture regions adjacent to the HS-578T-myo cells and allowed to attach overnight prior to overlaying to initiate co-culture at t = 72 hrs. At t = 72 hrs for both mono- and co-culture, cell culture media was replaced with low serum (0.5% FBS) cell culture media with 1X PBS (vehicle) or 5 nM Shh (100-45, PeproTech) +/− 5 μM cyclopamine (S1146, Selleckchem, Houston, TX, USA) using a volume of 50 μl loaded above all adjacent culture regions (contained within the outer ring). Low serum is required for optimal Shh signaling as shown in a previous publication.32 Cells were co-cultured for 96 hrs (from t = 72 to 168 hrs). Half of the total cell culture volume was replaced after 48 hrs of co-culture (t = 120 hrs) for metabolic waste removal and nutrient/Shh replenishment. Tumor cell growth was monitored during the last 2 hrs of co-culture using the Click-IT 594 flow cytometry assay according to the manufacturer’s recommendations (Molecular Probes, Eugene, OR). Following EDU incubation, MDA-MB-468 samples were independently collected using Accutase and prepared for flow cytometry analysis in the Accuri C6 instrument. For gene expression assays, cells were lysed independently in each compartment as facilitated by the use of adjacent culture regions and an open device design. mRNA was isolated directly from cell lysates using Dynabeads mRNA DIRECT Micro Kit (61021, Invitrogen, Carlsbad, CA, USA). Total mRNA (20–30 ng) was reverse transcribed to generate cDNA and quantified using One-Step SYBR Primescript kit (RR086A, Clontech Laboratories, Inc., San Francisco, CA) in the StepOne Real-time PCR System (Applied Biosystems). Primer sequences for mouse and human GAPDH, GLI1, and PTCH1 genes have been described before.33,34 Gene expression levels were normalized to the expression of GAPDH. Results were normalized again to the TGF-β treated condition for HS-578T or vehicle for MDA-MB-468 to calculate “normalized relative expression”.
Culture configurations
See Fig S1 for the methods of the Fig 3.
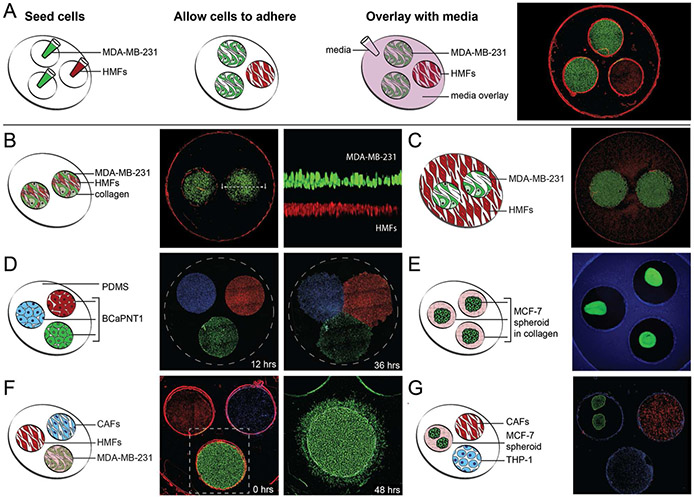
Figure 3. Configurations.
(A) The sequence of events needed to prepare the multi-culture platform. (A-image) Co-culture of HMFs (red, nuclear stain), and MDA-MB-231s (green, expressing GFP). (B) A “transwell” configuration, and a magnified horizontal section in which HMFs (red) and MDA-MB-231s (green) are separated vertically by a collagen layer (black) within each culture region. (C) The parent well area is utilized for cell-culture of HMFs (red) while culture regions contain monolayers of MDA-MB-231s (green). (D) The patterning of BCaPNT1 cells via razor printed PDMS templates. Cells are stained with Hoechst (blue), Cell Tracker Red (red) and Calcein AM (green), and seeded into culture regions of a razor-printed PDMS template within a 24-well plate. The PDMS template was removed at 12 hrs. Cell spreading and migration was observed at 36 hrs. (E) Spheroids of MCF-7s (green, expressing GFP), generated off-chip, are transferred into a three-region device, which was false-colored blue for visualization clarity. (F) The migration of MDA-MB-231s (green) towards culture regions containing HMFs (red, left) and CAFs (blue, right). The images were taken at 0 hrs and 48 hrs after seeding. (G) The tri-culture of the, CAFs (red), two MCF-7 spheroids (green), and the THP-1 immune cells (blue). (scale) All culture regions are 2.7 mm in diameter.
Diffusion Simulations
Modeling was performed in exactly the same manner as we have done previously in order to allow direct comparison of results.35 Briefly, modeling was performed using COMSOL Multiphysics. A diffusion coefficient, D, of 100 μm2/s is modeled, corresponding to proteins on the order to ~10 kDa36; however, given the time for diffusive transport scales linearly with D, results can be easily scaled for a particular protein or diffusion coefficient of interest. Transport within the closed device is diffusion dominant37 whereas transport in the open device is facilitated by convection when a fluid media overlay is used.38 Therefore, to provide an equal comparison, transport is modeled as purely diffusive for both the open and closed device, representing a scenario where gel is used to overlay the open multi-culture device instead of fluid media. The source of factors is modeled in two ways. First, a constant concentration boundary condition (normalized to 1) is used at the interface between the gel within the culture region and gel overlay (constant source). Second, the volume within the source culture region set to an initial normalized concentration of 1 and allowed to disperse without any additional factor production (non-constant source). A constant source approximates a co-culture scenario where source cells maintain a concentration of soluble factor in the source-well, while a non-constant source approximates a scenario where gel laden with soluble factor is seeded in the source well instead of cells and depletes over time. These two scenarios are used as they can be directly compared with previously published results for the closed multi-culture device and because they provide insight into (i) how rapidly cells can condition their environment to match local set-points (constant source) and (ii) how rapidly factors within a source region can disperse evenly throughout the device (non-constant source).
RESULTS
The open multi-culture device design helps to address basic challenges of studying MCT interactions
The device design enables a range of custom configurations for studying MCT interactions. Here we present seven different configurations of the multi-culture platform, using six distinct cell types related to the breast and prostate cancer microenvironments (Fig 3). The multi-culture platform is typically operated in three steps: 1) seeding of individual culture regions, 2) incubation to allow for cell settling or cell adhesion, and gel polymerization; and 3) overlaying the parent well with media in order to begin co-culture (Fig 3A). For a pictorial protocol and details of each configuration, see Fig S1.
With the open-multi culture device, basic physical parameters of the segregated cellular microenvironments (Fig 1B) can be independently controlled. This is demonstrated in Fig 3A for the parameters of cell number ratio, cell-to-culture volume ratio, and cell surface density (Fig S1A). Differentially stained MDA-MB-231 cells and HMFs are shown in a 3-region device at a ratio of 2:1 MDA-MB-231s to HMFs. By inverting the ratio to 1:2 using the same device, cell surface density and cell-to-culture volume ratio are maintained while cell number ratio is altered. As we have also recently shown, razor-printing can be used to easily alter the number of culture regions and their geometries to permit additional independent control of these three parameters,28 something that is particularly challenging with commercially available devices with fixed configurations. Further flexibility for varying these physical parameters can be achieved by recognizing that the parent well surface area can also be utilized for cell culture. This can be seen in Fig 3C where MDA-MB-231 cells were seeded into the culture regions while HMFs are restricted to the parent well outside the culture regions (Fig S1C).
Our approach allows the use of different culture modalities to control the ‘degree of separation’ between cell types. For example, in Fig 3B, we demonstrate that it is easy to incorporate matrices into the device to embody a “transwell” configuration, where a collagen layer separates the two cell types seeded in the same culture region (Fig S1B). The device design can also be used to pattern cells, controlling the degree of separation. Here, patterning is achieved using a razor-printed PDMS open-culture device to define separate culture regions and allow observation of cell spreading and migration (Fig 3D & S1D).
The platform also enables integration of different culture matrices and modalities to help recapitulate different extracellular environments present in vivo and meet the unique requirements of each cell type within a single culture device (Fig 3). Fig 3E and S1E illustrate use of spheroid culture. Spheroids are useful for recapitulating aspects of solid tumors such as boundary interactions with ECM and gradients of factors, nutrients, and differentiation. Fig 3G and S3G demonstrate that 2D and spheroid culture modalities can be integrated to construct a tumor:stroma:immune (MCF-7:HMF:THP-1) cell tri-culture model. In Fig 3F three cell types (3D-collagen-embedded MDA-MD-231 cells and 2D monolayer of HMFs and CAFs) are all overlaid with a matrix of collagen that allows for 2D and 3D cell migration (see also Fig S1F). The culture configurations of Fig 3 illustrate the flexibility of open multi-culture and how it can address basic challenges of studying MCT interactions by controlling physical culture parameters and integrating multiple culture modalities.
Open device designs can improve soluble factor communication during in vitro culture
Soluble factor communication in vitro is affected by multiple parameters such as the cell-to-volume ratio (cell number / culture volume) and rates of convection/diffusion. High cell-to-volume ratios cause more rapid accumulation and depletion of soluble factor concentrations which, in turn, allow cells to more readily influence one another. Likewise, convection can also aid transport of soluble factors relative to diffusion dominant environments. In microfluidic devices, cell-to-volume ratios are generally high and transport during culture is generally limited to diffusion whereas well-plates typically have lower cell-to-volume ratios and can have significant convection.38 The microscale open multi-culture device helps to provide a balance of these considerations for improving communication between cells relative to traditional well-plates and closed microchannel culture. To observe this, we compared cell-to-volume ratio for various culture devices and the effect of cell ratios in a model of paracrine Hedgehog signaling in breast cancer. To better describe this comparison, we refer to culture region where secretions are produced as the source region whereas the corresponding region where the factor will be received is the responding region.
First, cell-to-volume ratios in the open co-culture design are compared to traditional well-plates and closed microchannel-based devices (Tab 1). For 2D culture, the ratio is dictated by the culture-area-to-culture-volume (CACV) ratio, assuming similar surface cell densities. The open multi-culture design used here significantly increases the source CACV ratio compared to traditional transwell inserts and 96-well culture, thereby accumulating source paracrine factors more rapidly for more robust signaling. However, the previously published closed multi-culture device35 has the highest overall source CACV ratios and thus the fastest average accumulation of secretions. However, there are two potential caveats to this advantage. First, nutrient depletion will also occur most rapidly in the closed devices. Indeed, it has been estimated that media in such closed (non-perfused) microfluidic devices should likely be changed every ~8 hours to maintain nutrients due to the dramatically increased total CACV ratios.37 Second, like many microfluidic co-culture devices, the closed device uses a constriction between each culture region to prevent mixing of cell-types while allowing soluble factor communication. Therefore, although secretions will rapidly accumulate within each source region, constrictions may prevent efficient transport of factors to responding regions of the closed device to achieve successful co-culture. Given these caveats, the closed multi-culture device can provide significant advantages for short-term mono-culture, mixed-culture, or patterned-culture assays where rapid accumulation within a single-chamber is necessary; however, diffusion simulations are needed to better understand the relative performance of the open and closed multi-culture devices for co-culture applications (Fig 4).
Table 1.
Comparison of culture-area-to-culture-volume ratios for various devices when applied to co-culture of 2 cell types.
| Culture Device | Source Cell Culture Area [mm2] |
Total Cell Culture Area [mm2] |
Total Volume [μL] |
Source CACV Ratio [mm−1] |
Total CACV Ratio [mm−1] |
|---|---|---|---|---|---|
| 96-well* | 16 | 32 | 200 | 0.08 | 0.16 |
| 24-well / transwell insert | 190 / 33 | 223 | 700 | 0.27 / 0.047 | 0.32 |
| Open multi-culture** | 5.7 | 11.4 | 39 | 0.15 | 0.29 |
| Closed multi-culture** | 9.6 | 19.2 | 5.8 | 1.7 | 3.31 |
For a 96-well plate, cell types would need to be mixed, resulting in half of the the total culture surface covered by the source cell type.
Both the open and closed multi-culture device35 can be configured with different numbers of culture regions/chambers. Embodiments with just 2-regions are used in these calculations to allow cross-comparison with other devices for the purpose of co-culture
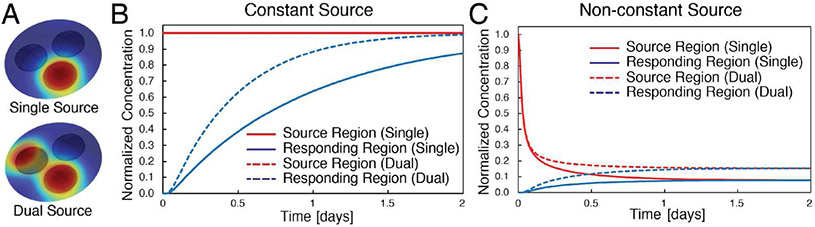
Figure 4.
Diffusion modeling of open multi-culture scenarios for comparison with previous closed multi-culture simulations.35 (A) 3D isometric view of 3-region device with either 1 or 2 source regions where soluble factor originates. Each well contains 3 μL of gel while the gel overlaying the three regions is 30 μL. (B) Simulations of source regions containing cells. The cells are presumed to maintain a constant normalized concentration of 1 within the well and is modeled as a constant concentration boundary condition at the interface between the gel within the culture region and gel overlay. (C) Simulations of the source regions initially loaded with gel containing soluble factor at a normalized concentration of 1. Concentration of the factor is depleted as it diffuses throughout the rest of the gel in the device with no additional production.
Simulation results for the open multi-culture design are compared to previously published diffusion results for the closed multi-culture design (Fig S2).35 Results illustrate the impact of the constriction between the culture chambers in the closed multi-culture device. Despite the fact that the open culture device has a CACV ratio ~10 times lower than the closed device (Tab 1), soluble factor accumulation in the responding region is more rapid in the open device design. The responding region of the open culture device for constant source scenarios at 24 hrs reaches concentrations ~3 times higher than the analogous responding chamber of the closed device (Fig 4 & Tab 2). Thus, the constriction between the closed multi-culture chambers significantly hinders transport of factors from source chambers to responding chambers. Furthermore, the reduced rate of transport results in a ~2-fold increase in equilibration times for the closed device compared to the open device with a non-constant source (Tab 2). The rapid equilibration within the open-culture device can be seen by comparing Fig 4B and 4C and Movie S2 & S3 which contain animations of the 3D concentration profiles over the course of 48 hrs. To better understand the implications of these results, the relative timescale of nutrient depletion must also be considered. Given the total CACV for open culture is similar to 24-well transwells (Tab 1), similar media replenishment times can be expected (i.e., ~48 hours). However, as mentioned earlier, equivalent media replenishment in the closed device would occur on the order of ~8 hours.37 Therefore, results suggest the open device allows a ~2 to 3-fold increase in the rate of accumulation for a ~6-fold longer period of time, indicating a potential ~12 to 18-fold increase in the final responder region concentration prior to media replenishment in the open device relative to the closed device. If a fluid overlay were used in the open device, convection would be expected to further enhance communication between culture regions in the open-device. Results are similar for an open device with only two regions (Tab 1 & Fig S2).
Table 2.
Responding well normalized concentration after 24 hrs of co-culture for open and closed multi-culture devices. Concentration is normalized to the initial concentration of the source well.
| Culture Device | Constant Source |
Non-constant Source |
||
|---|---|---|---|---|
| Single Source | Dual Source | Single Source | Dual Source | |
| Open multi-culture | 63% | 88% | 7.5% | 15% |
| Closed multi-culture | 17% | 32% | 12.5% | 23% |
Demonstration Study I: Tumor-to-mesenchyme ratio impacts tumor proliferation
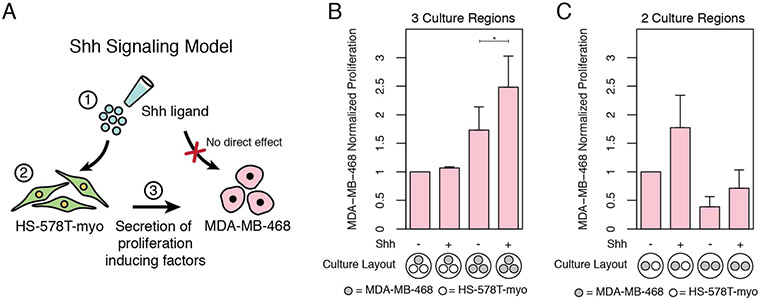
In order to demonstrate use of the platform to study paracrine signaling interactions, we utilized a model of Sonic Hedgehog (Shh) dependent cell-cell signaling involving HS-578T cells and MDA-MB-468 cells. This model parallels an analogous model we previously developed in prostate cancer but for breast cancer.32 Thus, the biological results of the experiments are not specifically the focus. Instead, the primary results of the experiments are the technical demonstrations of open-device capabilities. In this co-culture model, the cell lines must initially be cultured separately within the open culture device prior to overlaying with fluid for soluble factor communication. Prior to co-culture media overlay, the HS-578T cells are seeded and treated with TGF-β for 72 hrs to induce a mesenchymal phenotype (HS-578T-myo) and generate a confluent monolayer, both of which are necessary to make the HS-578T cells sensitive to Shh ligand. The MDA-MB-468 cells are seeded ~55 hrs after the HS-578T cells in the separate but adjacent culture regions of the device, thereby avoiding treatment with TGF-β. After first washing the individual cultures and then overlaying with fluid for 96 hrs of co-culture, the overlay is removed to allow each culture region to be separately addressed for different endpoints (see also Tab S1). The MDA-MB-468 breast cancer cells are detached and processed for flow cytometry analysis to quantify proliferation whereas the adjacent HS-578T-myo cells are lysed and processed for qRT-PCR analysis. As an additional demonstration of the advantages of customizability (vs. standard transwell plates), a 1:1 and 2:1 tumor:mesenchyme ratio are tested and compared without changing cell surface density to assure mesenchyme confluence for Shh signaling (Fig 5).
Figure 5. Shh signaling model and experimental results.
(A) Shh based cell-cell signaling model. Both HS-578T-myo cells and MDA-MB-468 tumor cells are treated with Shh ligand (Step 1) which causes the transcription of Hh-target genes, GLI1 and PTCH1, in the HS-578T-myo cells (Step 2) but not in the MDA-MB-468 cells (red ‘X’) (see also Fig S4). Secreted factors from the HS-578T-myo cells then cause proliferation of the MDA-MB-468 cells (Step 3). (B-C) MDA-MB-468 were cultured alone (mono-culture) or co-cultured with confluent monolayers of HS-578T that were previously treated with TGF-β to undergo epithelial-mesenchymal transition (HS-578T-myo). MDA-MB-468 and HS-578T-myo samples were simultaneously and independently collected from adjacent culture regions for proliferation and gene expression analysis. Fold change in proliferation rates of MDA-MB-468 in response to Hedgehog signaling at 1:1 and 1:2 culture region ratios. Co-culture proliferation rates were normalized to the analogous mono-culture rates (e.g., 3-region co-culture treated with Shh was normalized to 3-region mono-culture with Shh). Data represents average ± SE for 3 independent studies n=6 (2-region) and 2 independent studies with n=6 (3-region).
Results of co-culture show expected biological responses. Tumor cell proliferation is not affected by exogenous Shh alone but is enhanced in co-culture compared to mono-culture and is further enhanced when the mesenchyme is activated by exogenous Shh (Fig 5C-D & Fig S4). The presence of active Shh signaling during co-culture is confirmed via gene expression of GLI1 and PTCH1 in comparison with cyclopamine treated negative controls. In the additional experiment that compares co-culture response when using a 1:1 vs 1:2 cell ratio, we see that the co-culture system is sensitive to the relative quantity of tumor and mesenchyme despite surface cell densities being the same.
Demonstration Study II: Cell type compositions in tri-culture regulate therapy response
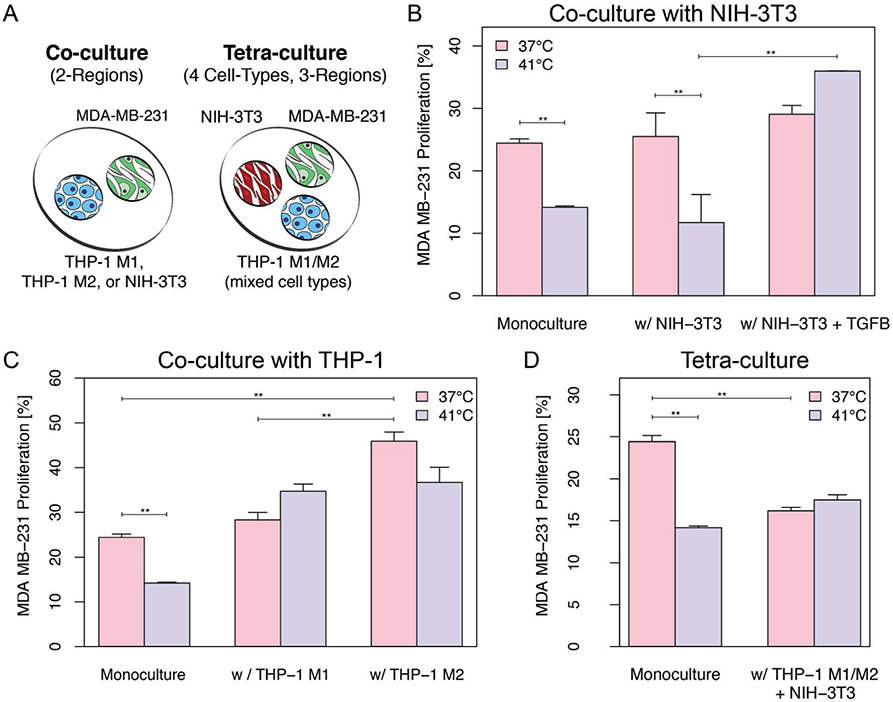
To demonstrate use of the platform for observing the influence of MCT interactions, we looked at the influence of cell neighbors on tumor cell recovery from heat damage (Fig 6). As expected, monoculture suffered a significant decrease in cell proliferation upon exposure to 41°C temperature for 30 min. The same effect was observed for co-cultures with NIH-3T3 cells. Although a slight increase in average proliferation can be observed for cocultures with NIH-3T3 treated with TGF-β, it was not statistically significant when compared with monoculture and NIH-3T3 co-culture at 37°C. A significant increase in tumor cell proliferation post heat-damage is clearly noticeable in MDA-MB-231 when co-cultured adjacent to TGF-β+NIH-3T3 (Fig 6B), indicating cancer-associated fibroblasts support MDA-MB-231 recovery after heat-stress culture conditions. M2 polarized macrophages increased proliferation of tumor cells when compared to its M1 phenotype counterpart and monoculture (Fig 6C). MDA-MB-231 cells cultured with M1 or M2 also show resistance to heat-stress, suggesting that both of the inflammatory cell subtypes individually support tumor cell recovery post heat-stress. Interestingly, the tetra-culture condition provided a strong protective effect against heat-therapy but also inhibited tumor cell proliferation relative to monoculture (Fig 6D). Results indicate that stromal composition of the tumor microenvironment plays a significant role in regulating tumor proliferation and response to therapy.
Figure 6. Proliferation levels in MDA-MB-231 after exposure to heat damage.
A) Schematic representation of cell seeding layout. MDA-MB-231 cells were cultured alone (monoculture) and with NIH-3T3 (B), THP-1 (C) or both (D). Cells were cultured at 37°C or exposed to 41°C for 30 min. Proliferation was monitored at 48 hrs post-heat damage. Co-culture data represents average of two independent experiments with n=8 devices. Tri-culture and monoculture data represents average of 3 independent experiments with n=8 and n=4 devices, respectively. Data represents mean data average +/− 1 SE. Student t-test, ** = p < 0.005.
More broadly, the co-culture experiment demonstrates how the open design facilitates complex protocols and experimental flexibility. The design allowed each culture region to be individually addressable both before and after co-culture to facilitate differential pretreatment and separate flow cytometry and gene expression endpoints (Fig S4 and Tab S1). Similarly, the customizability of the design allowed cell number ratio to be varied while maintaining identical local cell surface densities as required by the nature of the signaling mechanism.
DISCUSSION
Open device designs provide a balance of biological complexity and experimental control
The development of MCT models of disease and biology involves a multitude of potential variables. For example, as the number of cell types grow, the number and potential extent of interactions grows rapidly. In addition to biological complexity, there are also significant technical challenges associated with MCT models. To adequately support the requirements of different cell types, devices must often mix different culture modalities. The ability to adjust device geometry is also needed in order to adequately control basic physical parameters of culture such as cell surface densities, cell-to-volume ratios, and cell number ratios. Addressing these technical challenges requires fabricating custom device designs which generates additional barriers to studying MCT interactions in terms of expertise and cost.
The proposed open multi-culture platform helps to address these technical challenges by combining advantages of both macroscale and microscale assays. Often, these two general categories are considered to be at opposite ends of the spectrum. Macroscale platforms are commercially available and easy to use, while microscale approaches enable customization for better control over complex biological models but require additional design and fabrication expertise. Our proposed open multi-culture design provides a balance between those tradeoffs. The open device design offers simple pipette-based operation and modularity akin to macroscale wells. Using razor-printing to fabricate the device enables simple, affordable, ultra-rapid customization of device design and geometry.28,39-41 Similarly, the device also blends advantages of both macro- and micro-scale platforms to promote soluble factor communication. The open device increases source cell-to-volume ratios relative 96-well plates and 24-well transwell inserts, promoting co-culture communication. Further despite cell-to-volume ratios that are ~10-fold lower than closed multi-culture devices, the open device can provide significantly higher accumulation rates and final factor concentrations within responding regions of the device. This is because the open device eliminates constrictions that exist between chambers in the closed device. Therefore, although separated co-culture was more effective in open devices, closed devices would significantly outperform open devices in short-term (≲ 8 hr) mono-culture, mixed-culture, or patterned-culture applications where factors do not need to diffuse through constrictions to adjacent chambers. We illustrate that this the hybrid approach also enables individual control of basic physical culture parameters via rapidly customizable designs (Fig 3 & 5). We also demonstrate integration of different micro- and macro-scale culture modalities into the platform, including 2D, 3D, 3D-embedded, and spheroids (Fig 3). Thus, open multi-culture device design can provide advantages over both macroscale and closed microsale alternatives and are specifically well-adapted for studying soluble factor signaling.
The open device design provides some additional notable advantages over closed device alternatives for the broader biology community. An often unmentioned challenge of working with closed devices (e.g., interconnected microfluidic channels and chambers), is the issue of removing or avoiding trapped bubbles. Bubbles, if left unresolved, can block exchange of reagents and diffusion or expand during culture, killing cells over time. The lack of a ceiling and corners within an open culture device eliminates the problem of trapped bubbles. Thus, unlike closed microchannels made of the same materials, these open devices did not require any pretreatment to avoid bubble entrapment.28 Similarly, open designs also reduce device complexity, requiring fewer steps to assemble, fill, and use while increasing accessibility for pipetting (manual or automated) throughout the microscale device for added flexibility and user-friendly operation.23,42,43
Overall, the data illustrates that open microscale device designs can be both simple and highly functional for addressing complex problems such as systematic studies of MCT interactions.
The open multi-culture platform enables study of MCT interactions
We first used an established Shh paracrine signaling model of breast cancer to demonstrate use of the platform to study MTC interactions (Fig 5B). Results recapitulate what has been reported in the literature.32,44,45 Exogenous Shh had no significant effect on MDA-MB-468 proliferation in mono-culture, whereas the same treatment significantly increased proliferation when in co-culture with myo-differentiated HS-578T cells. In addition to supporting use of the platform for MCT studies, results also provided a baseline for examining the importance of basic culture parameters.
Often basic culture parameters such as cell number ratio, or cell-to-volume ratio can seem trivial or unimportant in comparison to other experimental variables; however, they can have significant influence on the interpretation of results. This can be seen in the results of Fig 5C, which were obtained at the same time as results for Fig 5B. Enhanced tumor growth rates in response to Shh were observed when cultured at a 1:2 ratio whereas the effect was not significant at a 1:1 ratio. Although surface cell densities were maintained constant to facilitate Shh signaling, it is possible that either cell-to-volume ratio or cell number ratio could be the reason that 2-region and 3-region devices showed significant differences. Questions such as this can be tested, if necessary, using the multi-culture platform by simultaneously altering cell-ratio while maintaining a constant cell confluence and cell-to-volume ratio.
In a final demonstration of the platform, we leveraged the compartmentalized nature of the open multiculture device to show that the composition of tumor-adjacent cell neighbors has important implications for models of cancer therapy response. In this application, we are modeling the use of heat which is studied as a potential adjuvant to drug therapy.46,47 Our study quantifies the sensitivity of tumor cells to heat damage in terms of cell growth and recovery rates in the presence of normal and cancer-associated stromal cell neighbors. Three different types of neighbors and their combination were studied. In agreement with observations by others, we see that MDA-MB-231 monoculture cell growth is negatively affected by heat damage (Fig 6). 48 However, we also see that fibroblasts and macrophages can individually influence tumor cell growth rates and speed up recovery post-heat damage. Interestingly, inclusion of all the cell-types together (tetra-culture) had a response not observed in co-culture with each cell type individually. Instead of maintaining or promoting growth at 37°C, growth at 37°C was inhibited relative to monoculture yet there was a strong protective effect against heat-stress, mitigating the effect of therapy (Fig 6D). As our platform supports MCT communication via soluble factors, is highly likely that this protective effect is driven by a set of secreted factors stimulated by an interaction among tumor cells, fibroblasts, and macrophages. This MCT interaction is analogous to what occurs in tissues but not considered in most in vitro models. Results such as these have significant implications for how the TME may perturb progression and therapeutic effects in vivo and underscore the need for engineered MCT models to unravel the complexity of these interactions and potentially offer new approaches to increasing therapeutic efficacy. The study also illustrates the value of an open and compartmentalized culture device, providing flexible configuration, a simple user interface, and individual endpoints for each culture region.
Future challenges
The challenge of biological complexity in MCT culture studies will persist, but technical challenges can continue to be addressed. As mentioned above, open device designs provide one approach that can help to address the technical complexity; however, there are also inherent challenges of “open” small-scale fluid systems that must still be considered. The effects of evaporation are increased in open microfluidic systems where the fluid is directly exposed to the air instead of closed in microfluidic tubing or channel. Currently, issues of evaporation are avoided by limiting air exposure time in biosafety hoods and multiple humidification chambers when culturing in an incubator.49,50 A potential not tested here is to use the sticker-like co-culture device in the bottom of standard well-plates (e.g., a 48-well plate), which is expected reduce convection and evaporation compared to a Petri dish. The decreased cell numbers in microscale culture also generally increase technical replicate variability due to the increasing influence of cell-cell variation and inherent challenges of working with small volumes.51 However, one goal of the proposed approach is to partially mitigate the increased potential for variability of small samples by reducing the potential for error through pipette-based operation and open access throughout the device. Last, although the ability to fabricate custom device designs in the laboratory can be enabling, quality control or reproducibility can be a concern for any laboratory-made reagent or device. For this reason, parameters of device preparation should be controlled like any other experimental parameter. This contrasts with purchased devices where one can typically rely on the quality control measures put in place by an established manufacturer. However, such practices are straightforward to implement and enable a significant leap forward in experimental flexibility and control.
CONCLUSION
Here, we present a simple and customizable open culture approach for MCT studies. The open design can be rapidly fabricated and customized and facilitates independent control over the influence of different cell types and basic cell culture parameters. We illustrate the flexibility of the platform by integrating different culture modalities such as 2D, 3D, and spheroid culture into 7 different culture configurations. We then successfully demonstrate the utility of the platform for MCT studies in two separate studies of cell-cell communication and response to therapy. The use of MCT culture approaches such as these will be of significant value for improving our understanding basic tissue and tumor biology, such as how different tumor microenvironments may impact therapy response of different individuals (i.e., personalized medicine). We envision that simple and customizable approaches to study MCT interactions will enable a broader community to explore new areas that would not otherwise be studied, empowering biologists themselves to engineer custom solutions and generate new applications and insights.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank the following individuals and organizations for important contributions and financial support. Theodorus de Groot and Patrick Ingram for editing and reviewing this manuscript. María Virumbrales-Munoz and Jose M. Ayuso for assistance with spheroid formation and David Lung, Jiaquan Yu, Molly M. Morgan and Karina Lugo Cintron for help acquiring human cells for the configuration experiments. We also thank Clare L. Procknow and Scotland Adkins for assistance with device fabrication. Support for DJB, JWW, YAG, & LES - NIH-NCI 4R01 CA155192. Support for LES - The National Human Genome Research Institute through the Genomic Science Training Program 5T32HG002760. Partial support for KPR, RJA & MD - NIH-NCI 1K01CA188167 and NSF-CREST 1345156 at UPR-Mayaguez. Adhesives Research, Inc. generously donated some of the ARCare 90106 tape for the studies. David J. Beebe holds equity in Bellbrook Labs LLC, Tasso Inc., Stacks to the Future LLC, Lynx Biosciences LLC, Onexion Biosystems LLC and Salus Discovery LLC.
REFERENCES
- 1.Lamichhane SP, Arya N, Kohler E, Xiang S, Christensen J and Shastri VP, BMC Cancer, 2016, 16, 581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang K, Park H-J, Han S, Lee J, Ko E, Kim J, Lee JS, Yu JH, Song KY, Cheong E, Cho S-R, Chung S and Cho S-W, Biomaterials, 2015, 63, 177–188. [DOI] [PubMed] [Google Scholar]
- 3.Xu H, Li Z, Yu Y, Sizdahkhani S, Ho WS, Yin F, Wang L, Zhu G, Zhang M, Jiang L, Zhuang Z and Qin J, Sci. Rep, 2016, 6, 36670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Osaki T, Sivathanu V and Kamm RD, Sci. Rep, 2018, 8, 5168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zervantonakis IK, Hughes-Alford SK, Charest JL, Condeelis JS, Gertler FB and Kamm RD, Proc. Natl. Acad. Sci. U. S. A, 2012, 109, 13515–13520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DesRochers TM, Holmes L, O’Donnell L, Mattingly C, Shuford S, O’Rourke MA, Rippon MB, Edenfield WJ, Gevaert MR, Orr DE and Crosswell HE, Journal for ImmunoTherapy of Cancer, 2015, 3, P401. [Google Scholar]
- 7.Bray LJ, Binner M, Körner Y, von Bonin M, Bornhäuser M and Werner C, Haematologica, 2017, 102, 1215–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bruce A, Evans R, Mezan R, Shi L, Moses BS, Martin KH, Gibson LF and Yang Y, PLoS One, 2015, 10, e0140506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hatherell K, Couraud P-O, Romero IA, Weksler B and Pilkington GJ, J. Neurosci. Methods, 2011, 199, 223–229. [DOI] [PubMed] [Google Scholar]
- 10.Pagani S, Torricelli P, Veronesi F, Salamanna F, Cepollaro S and Fini M, J. Cell. Physiol, 2018, 233, 291–301. [DOI] [PubMed] [Google Scholar]
- 11.Nash CE, Mavria G, Baxter EW, Holliday DL, Tomlinson DC, Treanor D, Novitskaya V, Berditchevski F, Hanby AM and Speirs V, Oncotarget, 2015, 6, 13731–13741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shin Y, Han S, Jeon JS, Yamamoto K, Zervantonakis IK, Sudo R, Kamm RD and Chung S, Nat. Protoc, 2012, 7,1247–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Regier MC, Alarid ET and Beebe DJ, Integr. Biol , 2016, 8, 684–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DesRochers TM, Holmes L, O’Donnell L, Mattingly C, Shuford S, O’Rourke MA, Rippon MB, Edenfield WJ, Gevaert MR, Orr DE and Crosswell HE, Journal for ImmunoTherapy of Cancer, 2015, 3, P401. [Google Scholar]
- 15.Bruce A, Evans R, Mezan R, Shi L, Moses BS, Martin KH, Gibson LF and Yang Y, PLoS One, 2015, 10, e0140506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang I-NE, Bogdanowicz DR, Mitroo S, Shan J, Kala S and Lu HH, Connect. Tissue Res, 2016, 57, 476–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barkal LJ, Procknow CL, Álvarez-García YR, Niu M, Jiménez-Torres JA, Brockman-Schneider RA, Gern JE, Denlinger LC, Theberge AB, Keller NP, Berthier E and Beebe DJ, Nat. Commun, 2017, 8, 1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zervantonakis IK, Kothapalli CR, Chung S, Sudo R and Kamm RD, Biomicrofluidics, 2011, 5, 13406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sackmann EK, Fulton AL and Beebe DJ, Nature, 2014, 507, 181–189. [DOI] [PubMed] [Google Scholar]
- 20.Goers L, Freemont P and Polizzi KM, J. R. Soc. Interface, , DOI: 10.1098/rsif.2014.0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Warrick JW, Murphy WL and Beebe DJ, IEEE Rev. Biomed. Eng, 2008, 1, 75–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang D, Krasowska M, Priest C, Popescu MN and Ralston J, J. Phys. Chem. C, 2011, 115, 18761–18769. [Google Scholar]
- 23.Casavant BP, Berthier E, Theberge AB, Berthier J, Montanez-Sauri SI, Bischel LL, Brakke K, Hedman CJ, Bushman W, Keller NP and Beebe DJ, Proc. Natl. Acad. Sci. U. S. A, 2013, 110, 10111–10116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim HJ, Boedicker JQ, Choi JW and Ismagilov RF, Proc. Natl. Acad. Sci. U. S. A, 2008, 105, 18188–18193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oh S, Ryu H, Tahk D, Ko J, Chung Y, Lee HK, Lee TR and Jeon NL, Lab Chip, 2017, 17, 3405–3414. [DOI] [PubMed] [Google Scholar]
- 26.Berry SB, Zhang T, Day JH, Su X, Wilson IZ, Berthier E and Theberge AB, Lab Chip, 2017, 17, 4253–4264. [DOI] [PubMed] [Google Scholar]
- 27.Byrne MB, Leslie MT, Patel HS, Gaskins HR and Kenis PJA, Biomicrofluidics, 2017, 11, 054116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stallcop LE, Álvarez-García YR, Reyes-Ramos AM, Ramos-Cruz KP, Morgan MM, Shi Y, Li L, Beebe DJ, Domenech M and Warrick JW, Lab Chip, 2018, 18, 451–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Negmadjanov U, Godic Z, Rizvi F, Emelyanova L, Ross G, Richards J, Holmuhamedov EL and Jahangir A, PLoS One, 2015, 10, e0123046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Genin M, Clement F, Fattaccioli A, Raes M and Michiels C, BMC Cancer, 2015, 15, 577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schneider CA, Rasband WS and Eliceiri KW, Nat. Methods, 2012, 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Domenech M, Bjerregaard R, Bushman W and Beebe DJ, Integr. Biol , 2012, 4, 142–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fan L, Pepicelli CV, Dibble CC, Catbagan W, Zarycki JL, Laciak R, Gipp J, Shaw A, Lamm MLG, Munoz A, Lipinski R, Thrasher JB and Bushman W, Endocrinology, 2004, 145, 3961–3970. [DOI] [PubMed] [Google Scholar]
- 34.Zhang J, Lipinski RJ, Gipp JJ, Shaw AK and Bushman W, BMC Dev. Biol, 2009, 9, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Domenech M, Yu H, Warrick J, Badders NM, Meyvantsson I, Alexander CM and Beebe DJ, Integr. Biol , 2009, 1, 267–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saltzman WM, Radomsky ML, Whaley KJ and Cone RA, Biophys. J, 1994, 66, 508–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Young EWK and Beebe DJ, Chem. Soc. Rev, 2010, 39, 1036–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lindsay SM and Yin J, AIChE J, 2016, 62, 2227–2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martínez-Löpez JI, Mojica M, Rodríguez CA and Siller HR, Sensors , , DOI: 10.3390/s16050705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cosson S, Aeberli LG, Brandenberg N and Lutolf MP, Lab Chip, 2015, 15, 72–76. [DOI] [PubMed] [Google Scholar]
- 41.Renaud L, Selloum D and Tingry S, Microfluid. Nanofluidics, 2015, 18, 1407–1416. [Google Scholar]
- 42.Yang D, Krasowska M, Priest C, Popescu MN and Ralston J, J. Phys. Chem. C, 2011, 115, 18761–18769. [Google Scholar]
- 43.Berry SB, Zhang T, Day JH, Su X, Wilson IZ, Berthier E and Theberge AB, Lab Chip, 2017, 17, 4253–4264. [DOI] [PubMed] [Google Scholar]
- 44.Valenti G, Quinn HM, Heynen GJJE, Lan L, Holland JD, Vogel R, Wulf-Goldenberg A and Birchmeier W, Cancer Res., 2017, 77, 2134–2147. [DOI] [PubMed] [Google Scholar]
- 45.Gao L and Albarracin CT, Breast Diseases: A Year Book Quarterly, 2011, 22, 383–384. [Google Scholar]
- 46.Court KA, Hatakeyama H, Wu SY, Lingegowda MS, Rodríguez-Aguayo C, Löpez-Berestein G, Ju-Seog L, Rinaldi C, Juan EJ, Sood AK and Torres-Lugo M, Mol. Cancer Ther, 2017, 16, 966–976. [DOI] [PubMed] [Google Scholar]
- 47.Hervault A and Thanh NTK, Nanoscale, 2014, 6, 11553–11573. [DOI] [PubMed] [Google Scholar]
- 48.Thompson EA, Graham E, MacNeill CM, Young M, Donati G, Wailes EM, Jones BT and Levi-Polyachenko NH, Int. J. Hyperthermia, 2014, 30, 312–323. [DOI] [PubMed] [Google Scholar]
- 49.Berthier E, Warrick J, Yu H and Beebe DJ, Lab Chip, 2008, 8, 860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Berthier E, Warrick J, Yu H and Beebe DJ, Lab Chip, 2008, 8, 852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brehm-Stecher BF and Johnson EA, Microbiol. Mol. Biol. Rev, 2004, 68, 538–59, table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.