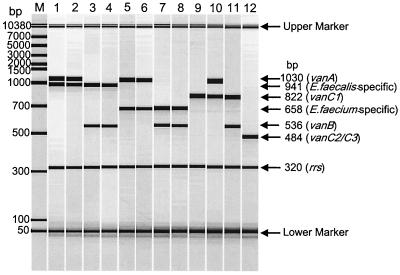
Kariyama and colleagues (5) recently described a multiplex PCR method for the detection of the vanA, vanB, vanC1, and vanC2/C3 genes, in addition to primers for the detection of the ddl genes of Enterococcus faecalis and E. faecium and of 16S rRNA (PCR control). However, the vanB primers used by these authors were derived from the sequence of the vanB1 subtype (1) and are incapable of detecting vanB2 or vanB3 subtypes, as previously described (2, 4, 7) and recently demonstrated in our laboratories.
Calgary Laboratory Services (CLS) recently implemented the PCR scheme described by Kariyama et al. (5). Three enterococcal strains which displayed a vancomycin-resistant enterococci phenotype were negative for the van genes using this methodology even though the ddlE. faecalis and 16S rRNA genes were amplified. PCR using the primer set for vanB alone yielded an amplicon of 1.1 kb for all three strains, which was larger than the expected 433 bp. The isolates were subsequently referred to the National Microbiology Laboratory (NML), Winnipeg, Manitoba, for further analysis. The NML has implemented a multiplex assay which uses novel, unpublished primers for the detection of vanB genes, the ddl genes of E. faecalis and E. faecium, and a published set of primers for the vanA gene (3). Using this assay, all three strains produced the E. faecalis-specific product and the vanB product. In order to explain the discordant results, the NML sequenced the 1.1-kb product produced using the Kariyama et al. (5) primer set. Sequence analysis revealed that the reverse primer bound correctly. However, the forward primer did not appear to anneal at the site 5′ GTG ACA AAC CGG AGG CGA GGA 3′ since the corresponding sequence of the vanB2 gene was 5′ GTG ACA AGC CGG AGG CGG GTG 3′. There are four nucleotide changes, with three of them at the 3-prime end, which likely results in no primer binding. This is predicted to also occur with the vanB3 gene (5′ GTG ACA AGC CGG AGA CGG GTG 3′). Further sequence analysis revealed that the forward primer is binding to the upstream vanH gene at the following site: 5′ GGA TGT GTT GGA GGG CGA GGA 3′. Note that the last 8 bp of this region and the forward primer used by Kariyama et al. (5) are identical. This primer binding would result in an amplicon of 1,086 bp, which is consistent with the PCR results.
The NML currently uses in-house primers developed from consensus vanB sequences (forward, 5′ AAG CTA TGC AAG AAG CCA TG 3′, and reverse, 5′ CCG ACA ATC AAA TCA TCC TC 3′) capable of detecting all vanB subtypes, with an amplicon size of approximately 536 bp. The multiplex assay described by Kariyama and colleagues (5) was easy to perform and reliably detected vanA, vanC1, vanC2/vanC3, and species-specific genes for E. faecalis and E. faecium from other isolates tested at both CLS and NML. However, if one were to substitute the vanB primers used by Kariyama et al. (5) with those developed by NML, the multiplex assay would be capable of detecting all vanB subtypes. Other vanB consensus primers have been developed (2); however, these primers cannot be used in the multiplex assay described by Kariyama et al. (5) since the vanC2 or vanC3 amplicon sizes would be identical.
In summary, the presence of vanB subtypes necessitates the use of consensus PCR primers for rapid and reliable detection. While the work was in progress Kawalec et al. (6) described similar results in a vanB2 E. faecium strain and determined that mispriming within vanH was responsible, though they did not precisely determine the anomalous primer binding site.
REFERENCES
- 1.Clark N C, Cooksey R C, Hill B C, Swenson J M, Tenover F C. Characterization of glycopeptide-resistant enterococci from U.S. hospitals. Antimicrob Agents Chemother. 1993;37:2311–2317. doi: 10.1128/aac.37.11.2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dahl K H, Simonsen G S, Olsvik O, Sundsfjord A. Heterogeneity in the vanBgene cluster of genomically diverse clinical strains of vancomycin-resistant enterococci. Antimicrob Agents Chemother. 1999;43:1105–1110. doi: 10.1128/aac.43.5.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dutka-Malen S, Evers S, Courvalin P. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J Clin Microbiol. 1995;33:24–27. doi: 10.1128/jcm.33.1.24-27.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gold H S, Unal S, Cercenado E, Thauvin-Eliopoulos C T, Eliopoulos G M, Wennersten C B, Moellering R C., Jr A gene conferring resistance to vancomycin but not teicoplanin in isolates of Enterococcus faecalis and Enterococcus faecium demonstrates homology with vanB, vanA, and vanCgenes of enterococci. Antimicrob Agents Chemother. 1993;37:1604–1609. doi: 10.1128/aac.37.8.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kariyama R, Mitsuhata R, Chow J W, Clewell D B, Kumon H. Simple and reliable multiplex PCR assay for surveillance isolates of vancomycin-resistant enterococci. J Clin Microbiol. 2000;38:3092–3095. doi: 10.1128/jcm.38.8.3092-3095.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kawalec M, Gniadkowski M, Zielinska U, Klos W, Hryniewicz W. Vancomycin-resistant Enterococcus faecium strain carrying the vanB2gene variant in a Polish hospital. J Clin Microbiol. 2001;39:811–815. doi: 10.1128/JCM.39.2.811-815.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patel R, Uhl J R, Kohner P, Hopkins M K, Steckelberg J M, Kline B, Cockerill F R., III DNA sequence variation within vanA, vanB, vanC-1, and vanC-2/3 genes of clinical Enterococcusisolates. Antimicrob Agents Chemother. 1998;42:202–205. doi: 10.1128/aac.42.1.202. [DOI] [PMC free article] [PubMed] [Google Scholar]