Abstract
Background and Objectives
To study human leukocyte antigen (HLA) allele associations in anti-leucine–rich glioma-inactivated 1 (LGI1) encephalitis.
Methods
A multiethnic cohort of 269 patients with anti-LGI1 encephalitis and 1,359 controls was included. Four-digit HLA sequencing and genome wide association single-nucleotide polymorphism typing imputation (0.99 concordance) were used for HLA typing. Significance of primary and secondary associations was tested using χ2, Fisher exact tests, or logistic regression with the control of population stratification covariates when applicable.
Results
DRB1*07:01 and DQA1*02:01, 2 alleles in strong linkage disequilibrium, were associated with the disease (90% vs 24%, OR = 27.8, p < 10e−50) across ethnicity independent of variation at DRB3 and DQB1, 2 flanking HLA loci. DRB1*07:01 homozygosity was associated with a doubling of risk (OR = 2.1, p = 0.010), suggesting causality. DRB1*07:01 negative subjects were younger (p = 0.003) and more frequently female (p = 0.015). Three patients with malignant thymomas did not carry DRB1*07:01, whereas patients with other tumors had high DRB1*07:01 frequency, suggesting that the presence of tumors other than thymomas may be coincidental and not causal. In both DRB1*07:01 heterozygous individuals and DRB1*07:01 negative subjects, DRB1*04:02 was associated with anti-LGI1 encephalitis, indicating an independent effect of this allele (OR = 6.85, p = 4.57 × 10−6 and OR = 8.93, p = 2.50 × 10−3, respectively). DRB1*04:02 was also independently associated with younger age at onset (β = −6.68, p = 9.78 × 10−3). Major histocompatibility complex peptide-binding predictions using LGI1-derived peptides revealed divergent binding propensities for DRB1*04:02 and DRB1*07:01 alleles, suggesting independent pathogenic mechanisms.
Discussion
In addition to the established primary DRB1*07:01 association in anti-LGI1 encephalitis, we observe a secondary effect of DRB1*04:02 with lower age at onset. Our study provides evidence for secondary effects within HLA locus that correlate with clinical phenotypes in anti-LGI1 encephalitis.
Anti-leucine–rich glioma-inactivated 1 (LGI1) encephalitis is one of the most common autoimmune encephalitis with approximate incidence of 0.83/million/year in Europe.1 The classic presentation is in men (66%) of around 65 years. Faciobrachial dystonic seizures (FBDSs) are a characteristic and often presenting clinical feature of the disease, with other focal seizures in addition to amnesia, disorientation, and psychiatry features following.1-4 Despite an initial good response to most immunotherapies,5,6 cognitive sequelae after the disease are common and disabling.1,7,8
LGI1 is a secreted neuronal protein preferentially expressed in the hippocampus, which forms a trans-synaptic complex between Kv1.1 potassium channels and AMPA receptors through its partners ADAM23 and ADAM22.9 Human antibodies against LGI1 block these interactions,10,11 although they have different properties depending on the location of their epitopes, for example, if targeting the leucine-rich repeat (LRR) domain, which forms LGI1 homodimers, or the epitempin (EPTP) domain that binds to ADAM22/23.12,13 No consistent environmental triggers have been described, and anti-LGi1 cases are only rarely paraneoplastic, with a few cases associated with thymoma.1,14
Strikingly, anti-LGI1 encephalitis is strongly associated with the human leukocyte antigen (HLA) allele DRB1*07:01 in ∼90% of Whites15-17 and Koreans,18 although a recent study found an association with DRB1*03:01 in 11 Chinese cases.19 Additional weaker effects have been suggested in HLA-DPB1 and HLA class I genes, although these remain unconfirmed.15 In this article, we extended the analysis of HLA genotypes in approximately 200 patients across 3 ethnic groups, confirming and extending results and their clinical relevance.
Methods
Subjects
A total number of 269 patients (246 Whites and 23 of Asian or African descent) diagnosed with anti-LGI1 encephalitis and referred to (1) the Oxford Autoimmune Neurology Group in England with the contribution of the University of California, San Francisco, and Mayo Clinic, Rochester, MN (n = 121), and (2) the French Reference Center for Paraneoplastic Neurological Syndromes and Autoimmune Encephalitis in Lyon, France (n = 148), were included. Detection of anti-LGI1 antibodies in serum and/or CSF was performed, as previously reported.2,20 HLA typing data on a subset of 68 patients recruited through Oxford and 72 patients recruited through Lyon have been previously published.8,16 The presence of FBDS and clinical severity measured by the modified Rankin score (mRS) at different stages of the disease (onset, maximum, and last follow-up) and tumoral status were determined from clinical or notes review.
Patients and controls recruited in Lyon were genotyped using the Affymetrix PMRA array, whereas patients recruited in Oxford were genotyped on Illumina GSAMD v1.0 and v2.0. A subset of controls were drawn from a different cohort and were genotyped with Illumina Infinium PsychArray-24 and had high-resolution HLA sequencing. All genotype data operations were performed using PLINK. All cohorts were imputed to the 1000 Genome Phase III21 after haplotype phasing and merged using QCTOOL. High-quality imputed calls (R2 ≥ 0.9) were used to extract genetic principal components using PLINK. A Euclidean distance-based measure was used to automatically match patients to the closest controls in a 1:5 ratio.
HLA imputation was performed using HLA Genotype Imputation with Attribute Bagging.22,23 Overall HLA imputation concordance was evaluated within a subset of 111 Oxford-recruited patients16 and 71 patients recruited in Lyon8 who were next-generation HLA-typed and found DRB1 concordance to be 99.4% and 99.6%, respectively. No differences were found between the Oxford and Lyon cohorts; thus, the results were analyzed as a single group after matching control subjects using principal component analysis (PCA). Genotypes with an imputation probability lower than 0.3 were removed.
Standard Protocol Approvals, Registrations, and Patient Consents
This study was approved by local ethics committees, and written informed consent was obtained from all the patients for the storage and use of biological samples and clinical information for research purposes.
Statistical Analyses
Categorical variables are presented as percentages and quantitative variables as means with SD. Although we report uncorrected p values, our primary analyses were Bonferroni or false discovery rate (FDR) corrected per allele over 5% carrier frequency so that only findings corrected for multiple comparisons were discussed and pursued. These primary analyses included (1) comparisons of HLA frequencies in all subjects (HLA phenotype carrier analysis) and (2) effects of the “other alleles” in DRB1*07:01 heterozygous subjects (compared with heterozygous subjects and expected allele counts derived from the control population). After these findings, we subsequently analyzed the effects of DRB1*07:01 homozygosity, differential effects of DRB3∼DRB1*07:01∼DQA1*02:01∼DQB1 haplotypes, and effects in DRB1*07:01 negative cases, both within the White subgroup using χ2 statistics and across ethnic groups using general logistic regressions, with successive conditioning of the various HLA-associated alleles and using population principal component matching when necessary. These secondary analyses were not Bonferroni-corrected or FDR-corrected because these were considered confirmatory. The analytical plan followed was similar to that reported for narcolepsy by Ollila et al.24 A significance level was set at p < 0.05. Analyses were performed using R.
Data Availability
Raw data are available on request to the corresponding author.
Results
Primary Association of DRB1*07:01 in Anti-LGI1 Encephalitis
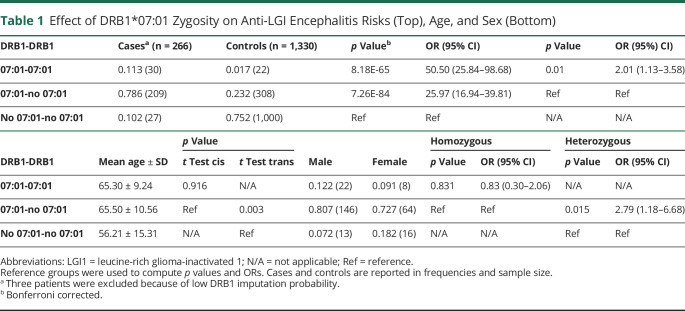
Consistent with previous studies,8,15-17,19 88.9% of the cases (239/266, 3 patients removed because of low imputation probability) carried HLA-DRB1*07:01 (Table 1), independent of ethnicity. DRB1*07:01 homozygosity was associated with an approximate doubling of risk in homozygous cases vs heterozygous without the effects of homozygosity on disease onset age or sex ratio (Table 1).
Table 1.
Effect of DRB1*07:01 Zygosity on Anti-LGI Encephalitis Risks (Top), Age, and Sex (Bottom)
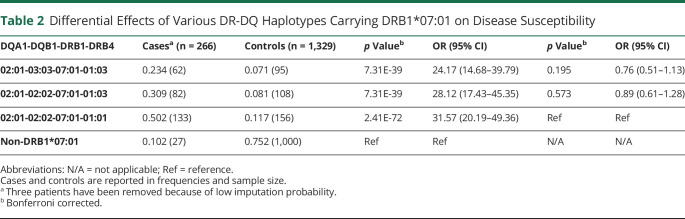
Extending on a previous study that has shown that the association fades in the DQ region,16 we next explore the influence of polymorphisms closely linked to DRB1*07:01 in the DRB3 and DQB1 loci. Because DRB1*07:01 haplotypes come in 3 frequent categories,25 DQB1*03:03∼DQA1*02:01∼DRB1*07:01∼DRB3*01:03N, DQB1*02:02∼DQA1*02:01∼DRB1*07:01∼DRB3*01:01, and DQB1*02:02∼DQA1*02:01∼DRB1*07:01∼ DRB3*01:03, we explored whether these haplotypes confer differential predisposition. As given in Table 2, these haplotypes had identical effects on disease predisposition. Because both DQA1 and DQB1 variations contribute to the HLA binding of the DQ molecule, this result suggests a primary effect of DRB1*07:01 on disease predisposition.
Table 2.
Differential Effects of Various DR-DQ Haplotypes Carrying DRB1*07:01 on Disease Susceptibility
Secondary Association of DPB1*03:01 and DRB1*04:02 in Anti-LGI1 Encephalitis
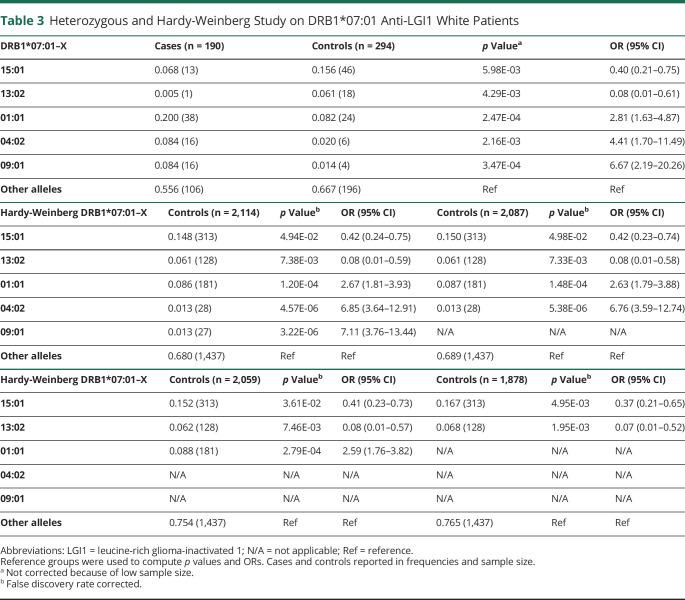
We next studied the effects of DRB1 alleles in trans of DRB1*07:01. To do so, we compared non-DRB1*07:01 DRB1 allele counts in trans of DRB1*07:01 positive cases (1) vs non-DRB1*07:01 DRB1 allele counts in trans of DRB1*07:01 positive controls (Table 3, top) and (2) vs all non-DRB1*07:01 allele counts in controls, a comparison which should be identical to that of non-DRB1*07:01 allele counts in controls considering the Hardy-Weinberg equilibrium (Table 3, middle and bottom, this comparison has more statistical power). Any identified allelic association in the Hardy-Weinberg study was sequentially and iteratively removed to explore whether another, yet unidentified allele remained significant until no alleles remained significant. To avoid issues with population structure matching in each stratum, this analysis was performed in Whites only. DRB1*04:02, an allele found in only 1.9% of controls, was significantly associated with the disease (OR = 4.41, p = 2.16 × 10−3 in DRB1*07:01 positive controls, and OR = 6.85, p = 4.57 × 10−6 in all controls' allele counts) in the DR7 heterozygous patients. Other effects included the susceptibility effects of DRB1*09:01 and DRB1*01:01 and protective effects of DRB1*15:01 and DRB1*13:02.
Table 3.
Heterozygous and Hardy-Weinberg Study on DRB1*07:01 Anti-LGI1 White Patients
Similar results were observed using logistic regression analysis across all ethnic groups in DRB1*07:01 heterozygous only (eTable 1, links.lww.com/NXI/A694) and all loci with controlling PCA (eTable 2, links.lww.com/NXI/A694), with an addition of DPB1*03:01, another rare allele. DPB1*03:01 was present in 218/1,358 controls and 8/269 patients in the context of the DPA1*01:02∼DPB1*03:01 (3% vs 16%, OR = 0.16, p = 3.56 × 10−04), and the effect was independent of other DRB1 effects. Unlike previously reported,16 we could not confirm any independent effects in HLA-A, B, or C loci, although expected class I alleles associated with DR7 haplotypes were enriched in cases vs controls when not controlled for the presence of DR7 (data not shown).
Association of DRB1*04:02 in Non-DRB1*07:01 Positive Anti-LGI1 Encephalitis Cases
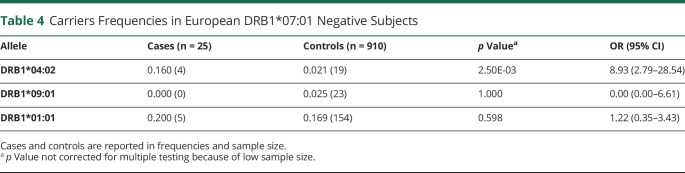
We next explored the possibility of a residual HLA association in the rare DRB1*07:01 negative patients with European ancestry (Table 4). Strikingly, DRB1*04:02 was also overrepresented in these cases, indicating the effects of this allele both in DR7 positive and negative cases (OR = 8.93, p = 0.003). By contrast, DRB1*09:01 and DRB1*01:01 were not associated with the disease.
Table 4.
Carriers Frequencies in European DRB1*07:01 Negative Subjects
Clinical Associations
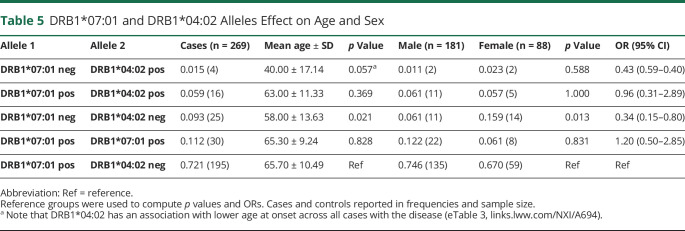
As reported previously,8 DRB1*07:01 negative subjects had a younger age at onset (56.7 vs 65.5 years, mean age, p = 0.003) and were more frequently female (53% vs 46%, p = 0.015). Intriguingly, the presence of DRB1*04:02 was also associated with reduced disease onset age (−6.7 years, p < 0.01, eTable 3, links.lww.com/NXI/A694). However, this finding could not be confirmed in DRB1*04:02 positive/DRB1*07:01 negative cases because of the small sample size (n = 4), despite a very low average age at onset (mean age ∼40 years old, p = 0.057) in comparison with DRB1*07:01 positive patients (Table 5).
Table 5.
DRB1*07:01 and DRB1*04:02 Alleles Effect on Age and Sex
Next, these striking findings led us to examine whether DRB1*04:02 could also associate with a different clinical presentation. To address this, we analyzed the effect of DRB1*07:01 and DRB1*04:02 on the mRS scores at onset, maximal mRS score, mRS score in the last visit, and the presence of FBDS (eTable 4, links.lww.com/NXI/A694). The results showed a moderate association of DRB1*04:02 with an increased mRS score at onset (p = 0.049).
Additionally, we studied the association of DRB1*07:01 and DRB1*04:02 in patients with tumors (n = 32) and matched controls. Of 32 patients, 3 were diagnosed with malignant thymoma, none of them carrying DRB1*07:01 or DRB1*04:02. Conversely, in patients with tumors other than malignant thymomas, we found that the association of DRB1*07:01 with the disease status was also present and of the same magnitude as in patients without tumor (eTable 5, links.lww.com/NXI/A694).
HLA Binding Predictions
As DRB1*07:01 and DRB1*04:02 share some residues (eFigure 1, links.lww.com/NXI/A694), we finally examined if specific LGI-derived peptides could bind uniquely to these subtypes. We parsed the full-length sequences of LGI1 from the UniProt database26 in the FASTA format (accession number O95970). We used NetMHCIIpan4.027 with offset correction for DRB1*07:01 and DRB1*04:02 HLA binding prediction in consecutive overlapping peptides with a length of 15 amino acids. We considered rank values ≤1 as strong binders, and >1 but ≤3 as medium binders and compared DRB1*07:01 and DRB1*04:02 binding spectrums with that of the remaining DRB1 (eTables 6 and 7, links.lww.com/NXI/A694).
Thirty-three LGI1-derived peptides with a length of 15 amino acids were predicted as medium/strong binders for DRB1*07:01, encompassing 12 core-peptides located in both the LRR and EPTP domains. This binding peptide-spectrum was shared in more than 50% with DRB1*01:01, DRB1*04:01, DRB1*09:01, and DRB1*16:01. However, only ∼18% of the peptides predicted as medium/strong binders for DRB1*07:01 (∼30% of the binding cores) were predicted as well for DRB1*04:02; thus, overlap with these predisposing subtypes was low. The common medium/strong binders between DRB1*07:01 and DRB1*04:02 were predicted to bind with other DRB1 as well (eTable 6, links.lww.com/NXI/A694).
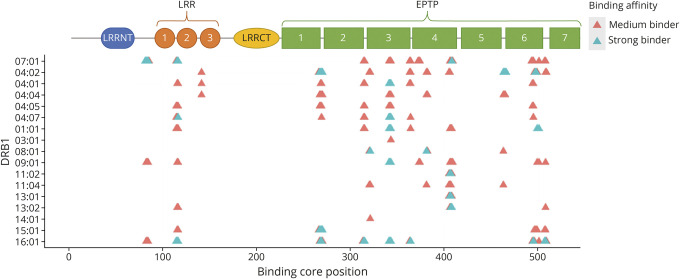
In the LRR domain, 4 peptides with core FLFTPSLQL were predicted to bind with DRB1*07:01 with ranks ≤1%, 3 of them also binding to DRB1*09:01 and DRB1*16:01 with ranks >1%. On the other hand, in the EPTP 6 domain, 4 peptides with binding core IQRMPSRGS were predicted as strong binders to DRB1*04:01 with ranks ≤1% (Figure 1), only sharing binding prediction with ranks >1%, with DRB1*04:04 (3 peptides), DRB1*08:01 (1 peptide), and DRB1*11:04 (1 peptide).
Figure 1. Binding Affinities of LGI1 Cores to DRB1 Alleles Associated With Anti-LGI1 Encephalitis and Other Common Alleles.
Strong binders: rank ≤1% and medium binders: rank >1% and ≤3%. EPTP = epitempin; LGI1 = leucine-rich glioma-inactivated 1; LRR = leucine-rich repeat.
Discussion
This study expands our understanding of the role of HLA in anti-LGI1 encephalitis. First, we confirmed that the disease is strongly associated with DRB1*07:01 across different ethnicities. Second, homozygosity for the main disease association allele was found to be linked with an increased risk of developing the disease with minor or no effects on disease presentation or age at onset, suggesting a causal effect. This effect is similar to that observed in other HLA class II autoimmune diseases associated with single HLA class II heterodimers, such as narcolepsy with DQA1*01:02-DQB1*06:02,24,28,29 celiac disease with DQA1*05-DQB1*02,30,31 and rheumatoid arthritis with DRB1*04.32 An explanation for these “allele dosage” effects may be that the encounter between the HLA-DRB1*07:01, the initial autoantigen trigger, and CD4+ autoreactive T cells is a rare event that can be modeled as a stochastic process proportional to HLA class II molecule amounts. This may contrast with effects on age at onset, which has been shown to involve different HLA associations in many instances, possibly reflecting regulatory CD4+ T cells or subsequent CD8+ T-cell or B-cell effects. For instance, DQB1*06:02/DQB1*03:01 heterozygosity but not DQB1*06:02 homozygosity is strongly associated with an earlier age at onset in narcolepsy.33
Another finding of this study is the observation of a strong secondary predisposing association with a rare DRB1*04 subtype, DRB1*04:02. This association was significant in both DRB1*07:01 heterozygous and DRB1*07:01 negative subjects. DRB1*04:02 carriers appeared with an earlier age at onset and a slightly increased severity. Younger age at onset was also observed in DRB1*07:01 negative subjects, which moreover showed a female predominance, confirming our previous results.8 Of interest, 3 rare cases with malignant thymoma did not carry DRB1*07:01 or DRB1*04:02, suggesting the absence of HLA association. This mirrors our finding in diseases with antibodies against contactin-associated protein-like 2 where only nonparaneoplastic cases were associated with DRB1*11:01.34 Patients with nonthymoma tumors did not differ in DRB1*07:01 carrier frequency compared with nonparaneoplastic cases, suggesting that the presence of these diverse tumors, unlike malignant thymoma, is coincidental.1,7,8 Of note, the association between DRB1*07:01 or DRB1*04:02 and anti-LGI1 encephalitis would increase in significance if these thymoma-associated cases are removed from the analysis.
Possible associations were found with DRB1*09:01, a subtype also associated with DQA1*03:01∼DQB1*03:03.25 Because DQB1*03:03 is also present in approximately one-third of DRB1*07:01 haplotypes associated with the disease (Table 2), this could suggest DQB1 effects additional to DRB1*07:01 effects. We, however, believe this to be unlikely because DRB1*07:01 predisposes to anti-LGI1 encephalitis, independently of DQB1*03:03 (Table 1). Furthermore, this association was present in Whites and Asians in trans of DRB1*07:01 (eTable 1, links.lww.com/NXI/A694) but not in DRB1*07:01 negative subjects (eTable 8). A predisposing effect of DRB1*01:01 was also evident in trans of DRB1*07:01 but was not present in DRB1*07:01 negative subjects. For these reasons, these weaker and less consistent associations will need confirmation alongside that of DRB1*04:02.
In addition to the above, we explored the possibility of residual effects in other HLA genes after controlling for DRB1 effects. A notable finding was a strong independent protective effect of DPA1*01:02∼DPB1*03:01, a finding distinct from previously reported DPA1*02:01∼DPB1*11:01 predisposing effects.16 Of interest, DPB1*03:01 is a high-expression allele35 and has been involved in susceptibility (not protection) to a number of other autoimmune diseases, such as multiple sclerosis,36-38 severe aplastic anemia,39 primary biliary cholangitis,40 and B27 negative ankylosing spondylitis.41 By contrast, although HLA class I alleles known to be associated with DRB1*07:01, such as B*57:01, B*44:03, C*06:02, and C*16:01, were increased in the sample uncorrected for DRB1*07:01 (data not shown) as previously reported,16 all HLA class I association disappeared after controlling for the main DR7 association (data not shown).
To explore whether specific motifs of LGI1 could be involved in these effects, we finally conducted peptide-binding prediction analysis for critical DRB1 alleles. In contrast to other DRB1*04 molecules, DRB1*04:02, like DRB1*07:01, shares I and D at positions 67 and 70 (eFigure 1, links.lww.com/NXI/A694), positions critical to HLA pocket binding 4 (P4), possibly suggesting that a similar LGI1 epitope could be involved. Nonetheless, HLA binding predictions of LGI1-derived peptides in addition to the differential association with age at onset and severity suggest a different mechanism. Although DRB1*07:01 binds most strongly and independently of other alleles to the beginning of the LRR 1 domain, DRB1*04:02 binds to higher affinity at the beginning of the EPTP 6 domain (Figure 1). These results suggest that the DRB1*04:02 association is related to an independent epitope leading to anti-LGI1 autoimmunity. Tetramer studies will be needed to explore these hypotheses.
In conclusion, our study confirmed a primary effect of DRB1*07:01 on disease susceptibility and revealed solid novel HLA associations with DRB1*04:02 and DPB1*03:01. The DRB1*04:02 association was notable because of a secondary effect on age at onset and a slight increase of severity in both DRB1*07:01 positive and negative patients. This suggests that DRB1*04:02 positive patients could represent a different subgroup or a modulation of the existing sporadic phenotype that should be further explored.
Acknowledgment
The authors thank NeuroBioTec Hospices Civils de Lyon BRC (France, AC-2013-1867, NFS96-900) for banking DNA, sera, and CSF samples.
Glossary
- EPTP
epitempin
- FBDS
faciobrachial dystonic seizure
- FDR
false discovery rate
- HLA
human leukocyte antigen
- LGI1
leucine-rich glioma-inactivated 1
- LRR
leucine-rich repeat
- mRS
modified Rankin score
- PCA
principal component analysis
Appendix. Authors
Contributor Information
Vicente Peris Sempere, Email: vsempereperis@gmail.com.
Sergio Muñiz-Castrillo, Email: ext-sergio.muniz@chu-lyon.fr.
Aditya Ambati, Email: ambati@stanford.edu.
Sophie Binks, Email: sophie.binks@st-hildas.ox.ac.uk.
Anne-Laurie Pinto, Email: anne-laurie.pinto@chu-lyon.fr.
Veronique Rogemond, Email: veronique.rogemond@chu-lyon.fr.
Sean J. Pittock, Email: pittock.sean@mayo.edu.
Divyanshu Dubey, Email: dubey.divyanshu@mayo.edu.
Michael D. Geschwind, Email: michael.geschwind@ucsf.edu.
Jeffrey Marc Gelfand, Email: jeffrey.gelfand@ucsf.edu.
Sonam Dilwali, Email: sonam.dilwali@ucsf.edu.
Soon-Tae Lee, Email: slee@snuh.org.
Julian Knight, Email: julian@well.ox.ac.uk.
Katherine S. Elliott, Email: kelliott@well.ox.ac.uk.
Sarosh Irani, Email: sarosh.irani@ndcn.ox.ac.uk.
Jérôme Honnorat, Email: jerome.honnorat@chu-lyon.fr.
Study Funding
This study was supported by research grants NIH U01-AI152590 and Fondation pour la recherche médicale (FRM, DQ20170336751). It has been developed within the frameworks of the LABEX CORTEX (reference ANR-11-LABX-0042) and the BETPSY project (reference ANR-18-RHUS-0012), both operated by the French National Research Agency (ANR). SRI was supported by the Wellcome Trust (104079/Z/14/Z), Medical Research Council (MR/V007173/1), BMA Research Grants—Vera Down grant (2013), Margaret Temple (2017), Epilepsy Research UK (P1201), the Fulbright UK-US commission (MS Society research award), and the NIHR Oxford Biomedical Research Centre. This research was funded in whole, or in part, by the Wellcome Trust (Grant number 104079/Z/14/Z). S. Binks has received support from the NIHR and Wellcome Trust. For the purpose of Open Access, the author has applied a CC BY public copyright license to any Author Accepted Manuscript version arising from this submission.
Disclosure
S.R. Israni is a coapplicant and receives royalties on patent application WO/210/046716 (U.K. patent no., PCT/GB2009/051441) entitled “Neurological Autoimmune Disorders”; the patent has been licensed for the development of assays for LGI1 and other VGKC-complex antibodies; has filed a patent (Diagnostic Strategy to improve specificity of CASPR2 antibody detection; Ref. JA94536P.GBA); has received research support from and/or consultancy with UCB, Immunovant, MedImmune, ADC therapeutics, CSL Behring and ONO Pharmaceuticals. The remaining authors report no disclosures relevant to the manuscript. Go to Neurology.org/NN for full disclosures.
References
- 1.Van Sonderen A, Thijs RD, Coenders EC, et al. Anti-LGI1 encephalitis: clinical syndrome and long-term follow-up. Neurology. 2016;87(14):1449-1456. doi: 10.1212/WNL.0000000000003173. [DOI] [PubMed] [Google Scholar]
- 2.Irani SR, Alexander S, Waters P, et al. Antibodies to Kv1 potassium channel-complex proteins leucine-rich, glioma inactivated 1 protein and contactin-associated protein-2 in limbic encephalitis, Morvan's syndrome and acquired neuromyotonia. Brain. 2010;133(9):2734-2748. doi: 10.1093/brain/awq213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Irani SR, Michell AW, Lang B, et al. Faciobrachial dystonic seizures precede Lgi1 antibody limbic encephalitis. Ann Neurol. 2011;69(5):892-900. doi: 10.1002/ANA.22307. [DOI] [PubMed] [Google Scholar]
- 4.Ariño H, Armangué T, Petit-Pedrol M, et al. Anti-LGI1-associated cognitive impairment. Neurology. 2016;87(8):759-765. doi: 10.1212/WNL.0000000000003009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thompson J, Bi M, Murchison AG, et al. The importance of early immunotherapy in patients with faciobrachial dystonic seizures. Brain. 2018;141(2):348-356. doi: 10.1093/brain/awx323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dubey D, Britton J, McKeon A, et al. Randomized placebo-controlled trial of intravenous immunoglobulin in autoimmune LGI1/CASPR2 epilepsy. Ann Neurol. 2020;87(2):313-323. doi: 10.1002/ana.25655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Binks S, Veldsman M, Easton A, et al. Residual fatigue and cognitive deficits in patients after leucine-rich glioma-inactivated 1 antibody encephalitis. JAMA Neurol. 2021;78(5):617-619. doi: 10.1001/jamaneurol.2021.0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muñiz-Castrillo S, Haesebaert J, Thomas L, et al. Clinical and prognostic value of immunogenetic characteristics in anti-LGI1 encephalitis. Neurol Neuroimmunol Neuroinflamm. 2021;8(3):e974. doi: 10.1212/NXI.0000000000000974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fukata Y, Adesnik H, Iwanaga T, Bredt DS, Nicoll RA, Fukata M. Epilepsy-related ligand/receptor complex LGI1 and ADAM22 regulate synaptic transmission. Science. 2006;313(5794):1792-1795. doi: 10.1126/science.1129947. [DOI] [PubMed] [Google Scholar]
- 10.Petit-Pedrol M, Sell J, Planagumà J, et al. LGI1 antibodies alter Kv1.1 and AMPA receptors changing synaptic excitability, plasticity and memory. Brain. 2018;141(11):3144. doi: 10.1093/BRAIN/AWY253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ohkawa T, Fukata Y, Yamasaki M, et al. Autoantibodies to epilepsy-related LGI1 in limbic encephalitis neutralize LGI1-ADAM22 interaction and reduce synaptic AMPA receptors. J Neurosci. 2013;33(46):18161. doi: 10.1523/JNEUROSCI.3506-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramberger M, Berretta A, Tan JMM, et al. Distinctive binding properties of human monoclonal LGI1 autoantibodies determine pathogenic mechanisms. Brain. 2020;143(6):1731-1745. doi: 10.1093/brain/awaa104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kornau HC, Kreye J, Stumpf A, et al. Human cerebrospinal fluid monoclonal LGI1 autoantibodies increase neuronal excitability. Ann Neurol. 2020;87(3):405-418. doi: 10.1002/ana.25666. [DOI] [PubMed] [Google Scholar]
- 14.Guasp M, Landa J, Martinez-Hernandez E, et al. Thymoma and autoimmune encephalitis: clinical manifestations and antibodies. Neurol Neuroimmunol Neuroinflamm. 2021;8(5):e1053. doi: 10.1212/NXI.0000000000001053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mueller SH, Färber A, Prüss H, et al. Genetic predisposition in anti-LGI1 and anti-NMDA receptor encephalitis. Ann Neurol. 2018;83(4):863-869. doi: 10.1002/ana.25216. [DOI] [PubMed] [Google Scholar]
- 16.Binks S, Varley J, Lee W, et al. Distinct HLA associations of LGI1 and CASPR2-antibody diseases. Brain. 2018;141(8):2263-2271. doi: 10.1093/brain/awy109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Sonderen A, Roelen DL, Stoop JA, et al. Anti-LGI1 encephalitis is strongly associated with HLA-DR7 and HLA-DRB4. Ann Neurol. 2017;81(2):193-198. doi: 10.1002/ana.24858. [DOI] [PubMed] [Google Scholar]
- 18.Kim TJ, Lee ST, Moon J, et al. Anti-LGI1 encephalitis is associated with unique HLA subtypes. Ann Neurol. 2017;81(2):183-192. doi: 10.1002/ana.24860. [DOI] [PubMed] [Google Scholar]
- 19.Hu F, Liu X, Zhang L, et al. Novel findings of HLA association with anti-LGI1 encephalitis: HLA-DRB1*03:01 and HLA-DQB1*02:01. J Neuroimmunol. 2020;344:577243. doi: 10.1016/j.jneuroim.2020.577243. [DOI] [PubMed] [Google Scholar]
- 20.Navarro V, Kas A, Apartis E, et al. Motor cortex and hippocampus are the two main cortical targets in LGI1-antibody encephalitis. Brain. 2016;139(4):1079-1093. doi: 10.1093/brain/aww012. [DOI] [PubMed] [Google Scholar]
- 21.Auton A, Abecasis GR, Altshuler DM, et al. A global reference for human genetic variation. Nature. 2015;526(7571):68-74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng X. Imputation-based HLA typing with SNPs in GWAS studies. In: Methods in Molecular Biology. Vol. 1802. Humana Press Inc.; 2018:163-176. doi: 10.1007/978-1-4939-8546-3_11. [DOI] [PubMed] [Google Scholar]
- 23.Zheng X, Shen J, Cox C, et al. HIBAG—HLA genotype imputation with attribute bagging. Pharmacogenomics J. 2014;14(2):192-200. doi: 10.1038/tpj.2013.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ollila HM, Ravel JM, Han F, et al. HLA-DPB1 and HLA class I confer risk of and protection from narcolepsy. Am J Hum Genet. 2015;96(1):136-146. doi: 10.1016/j.ajhg.2014.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Creary LE, Sacchi N, Mazzocco M, et al. High-resolution HLA allele and haplotype frequencies in several unrelated populations determined by next generation sequencing: 17th International HLA and Immunogenetics Workshop joint report. Hum Immunol. 2021;82(7):505-522. doi: 10.1016/j.humimm.2021.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bateman A, Martin MJ, Orchard S, et al. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021;49(D1):D480-D489. doi: 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reynisson B, Barra C, Kaabinejadian S, Hildebrand WH, Peters B, Nielsen M. Improved prediction of MHC II antigen presentation through integration and motif deconvolution of mass spectrometry MHC eluted ligand data. J Proteome Res. 2020;19(6):2304-2315. doi: 10.1021/ACS.JPROTEOME.9B00874. [DOI] [PubMed] [Google Scholar]
- 28.Ollila HM, Fernandez-Vina M, Mignot E. HLA-DQ allele competition in narcolepsy: a comment on Tafti et al.DQB1 locus alone explains most of the risk and protection in narcolepsy with cataplexy in Europe. Sleep. 2015;38(1):147-151. doi: 10.5665/sleep.4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van der Heide A, Verduijn W, Haasnoot GW, Drabbels JJM, Lammers GJ, Claas FHJ. HLA dosage effect in narcolepsy with cataplexy. Immunogenetics. 2015;67(1):1-6. doi: 10.1007/s00251-014-0808-z. [DOI] [PubMed] [Google Scholar]
- 30.Airaksinen L, Laurikka P, Huhtala H, et al. Influence of HLA-DQ2.5 dose on clinical picture of unrelated celiac disease patients. Nutrients. 2020;12(12):1-9. doi: 10.3390/nu12123775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ploski R, Ek J, Thorsby E, Sollid LM. On the HLA‐DQ(α1*0501, β1*0201)‐associated susceptibility in celiac disease: a possible gene dosage effect of DQB1*0201. Tissue Antigens. 1993;41(4):173-177. doi: 10.1111/j.1399-0039.1993.tb01998.x. [DOI] [PubMed] [Google Scholar]
- 32.Oka S, Furukawa H, Shimada K, et al. Association of HLA-DRB1 genotype with younger age onset and elder age onset rheumatoid arthritis in Japanese populations. Medicine (Baltimore). 2019;98(48):e18218. doi: 10.1097/MD.0000000000018218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Han F, Faraco J, Dong XS, et al. Genome wide analysis of narcolepsy in China implicates novel immune loci and reveals changes in association prior to versus after the 2009 H1N1 influenza pandemic. PLoS Genet. 2013;9(10):e1003880. doi: 10.1371/journal.pgen.1003880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muñiz-Castrillo S, Joubert B, Elsensohn MH, et al. Anti-CASPR2 clinical phenotypes correlate with HLA and immunological features. J Neurol Neurosurg Psychiatry. 2020;91(10):1076-1084. doi: 10.1136/JNNP-2020-323226. [DOI] [PubMed] [Google Scholar]
- 35.Shieh M, Chitnis N, Clark P, Johnson FB, Kamoun M, Monos D. Computational assessment of miRNA binding to low and high expression HLA-DPB1 allelic sequences. Hum Immunol. 2019;80(1):53-61. doi: 10.1016/j.humimm.2018.09.002. [DOI] [PubMed] [Google Scholar]
- 36.Field J, Browning SR, Johnson LJ, et al. A polymorphism in the HLA-DPB1 gene is associated with susceptibility to multiple sclerosis. PLoS One. 2010;5(10):e0013454. doi: 10.1371/journal.pone.0013454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anagnostouli M, Artemiadis A, Gontika M, et al. HLA-DPB1*03 as risk allele and hla-dpb1*04 as protective allele for both early-and adult-onset multiple sclerosis in a hellenic cohort. Brain Sci. 2020;10(6):1-13. doi: 10.3390/brainsci10060374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Watanabe M, Nakamura Y, Sato S, et al. HLA genotype-clinical phenotype correlations in multiple sclerosis and neuromyelitis optica spectrum disorders based on Japan MS/NMOSD Biobank data. Sci Rep. 2021;11(1):607. doi: 10.1038/s41598-020-79833-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Savage SA, Viard M, O’hUigin C, et al. Genome-wide association study identifies HLA-DPB1 as a significant risk factor for severe aplastic anemia. Am J Hum Genet. 2020;106(2):264-271. doi: 10.1016/j.ajhg.2020.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang C, Zheng X, Jiang P, et al. Genome-wide association studies of specific antinuclear autoantibody subphenotypes in primary biliary cholangitis. Hepatology. 2019;70(1):294-307. doi: 10.1002/hep.30604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reveille JD, Zhou X, Lee MJ, et al. HLA class I and II alleles in susceptibility to ankylosing spondylitis. Ann Rheum Dis. 2019;78(1):66-73. doi: 10.1136/annrheumdis-2018-213779. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Raw data are available on request to the corresponding author.