Abstract
The Drosophila GAGA factor (GAF) is a multifunctional protein implicated in nucleosome organization and remodeling, activation and repression of gene expression, long distance enhancer–promoter communication, higher order chromosome structure, and mitosis. This broad range of activities poses questions about how a single protein can perform so many seemingly different and unrelated functions. Current studies argue that GAF acts as a “pioneer” factor, generating nucleosome-free regions of chromatin for different classes of regulatory elements. The removal of nucleosomes from regulatory elements in turn enables other factors to bind to these elements and carry out their specialized functions. Consistent with this view, GAF associates with a collection of chromatin remodelers and also interacts with proteins implicated in different regulatory functions. In this review, we summarize the known activities of GAF and the functions of its protein partners.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00018-021-03776-z.
Keywords: GAGA, GAF, Pioneer factor, Promoter, Architectural protein, Promoter regulation, Boundary protein, Insulator, Polycomb, Chromatin silencing, Transcription regulation
Introduction
When chromosomal DNA is wrapped around nucleosomes, it can become inaccessible for interactions with transcription factors including activators, TFIID and RNA polymerase. For this reason, active or potentially active regulatory elements are typically located in nucleosome-free regions of chromatin and are hypersensitive to chemical agents or to cleavage by nucleases such as DNase I or MNase. Accessibility depends upon a local configuration of nucleosome–DNA interactions that is permissive for the assembly of a variety of different, often large-scale regulatory complexes. In Drosophila, these regulatory complexes include the combinations of general transcription factors and RNA polymerase that are found at promoter sequences, the different Polycomb group complexes that are assembled on polycomb response elements (PREs), the protein complexes that are associated with boundary elements (insulators), the complexes that assemble on X-linked chromatin entry sites (CES) and the multiple factors that interact with enhancer sequences. In most all of these cases, there are specialized DNA-binding factors that are able to mediate alterations in the configuration of nucleosomes in the immediate neighborhood and help create and maintain nucleosome-free regions that are accessible to other functionally specific factors.
One of the DNA-binding proteins in Drosophila that helps generate nucleosome-free regions of chromatin is the GAGA factor (GAF). GAF binds to (GA)n sequences throughout the genome [1] and is encoded by the Trithorax-like (Trl) gene [2]. It was first identified as a factor that binds to GAGAG motifs located in the upstream promoter regions of the Ultrabithorax (Ubx) [3] and engrailed (en) [4] genes and stimulates their transcription in nuclear extracts. Subsequent studies showed that GAGAG motifs are present in the upstream regions of promoters in many different genes including kruppel, actin5C, ecdysone E74 as well as several heat shock genes (e.g. hsp70, hsp26, hsp27) and implicated these motifs in their transcriptional activity either in vitro or in vivo [5, 6]. The localization of GAF to promoter regions was further documented in genome-wide studies which showed that it associates with ~ 20% of the Pol II-bound promoters, and that this association correlates with the presence of a paused Pol II at the promoter [7–9].
It was initially thought that GAF acts as a classical activator; however, studies by Kadonaga and colleagues demonstrated that GAF enhances transcription in vitro indirectly by countering the repressive effects of histone H1 [10, 11]. Their findings led to idea that GAF might function by counteracting the inhibitory effects of chromatin on transcription and this view was reinforced by two different lines of evidence. One came from in vitro chromatin assembly experiments. When chromatin was assembled in early embryonic extracts on plasmid templates containing the hsp70 heat shock gene, the promoter region was inaccessible to nuclease cleavage; however, if assembly took place in the presence of GAF, or GAF was added (together with ATP) after assembly was completed, the promoter region, but not elsewhere in the hsp70 gene, became accessible to nuclease cleavage [12]. These experiments suggested that GAF helped displace nucleosomes from the promoter region. A somewhat different result was obtained for the hsp26 promoter. In vivo, this promoter has two hypersensitive sites containing binding sequences for GAF and the heat shock factor, HSF, separated by a positioned nucleosome. When chromatin was assembled on a plasmid containing the hsp26 promoter region in vitro, a nucleosome was positioned in the promoter so that GAF and HSF could bind to their recognition sequences in the absence of added ATP; however, when ATP was include in combination with GAF or HSF, the nucleosome position was altered by what appeared to be a sliding mechanism [13]. GAF can also facilitate the remodeling of the fushi-tarazu (ftz) promoter and its transcriptional activity in vitro. In this case, remodeling activity depends upon four GAGA motifs in the promoter [14].
The second line of evidence came from in vivo studies on the hsp26 and hsp70 heat shock genes. In the case of the hsp26 gene, the requirements for forming a “native” chromatin structure appeared to be more stringent than those in vitro. The GAF recognition sequences in the proximal and distal nuclease hypersensitive sites in the hsp26 promoter were found to be critical not only for heat induction, but also for generating the two nuclease hypersensitive sites. In contrast, the HSF factor recognition sequences were critical for induction, but not for hypersensitivity [15–17]. While mutations in the TATA box led to only a minor reduction of hypersensitivity, more extensive sequence alterations that eliminate TFIID binding in vitro had a much greater effect on hypersensitivity without apparently compromising GAF association with the promoter [18, 19].
The GAF recognition sequences in the upstream region of the hsp70 promoter have a similar function. Lee et al. found that mutations in the GAF sequences in the hsp70 promoter significantly reduce the level of paused polymerase under non-heat shock conditions and compromise the response of the promoter to heat induction [20]. Subsequent studies showed that GAF association with the upstream hsp70 promoter region is important for generating nuclease hypersensitivity and for the association of TFIID (TBP), the NELF pausing complex and HSF [9, 21, 22]. Mutations in the TATA box that reduce the binding of TFIID by at least 20-fold [18] and also deplete NELF do not affect GAF binding [9] indicating that GAF is associated with the promoter prior to these factors. The role of GAF in generating nucleosome-free regions is also supported by genome-wide MNase-seq experiments, which show that many GAF associated promoters and intergenic regions become much less accessible upon GAF RNAi knockdown [7]. Additionally, there is a decrease in Pol II (Rpb3) and CBP histone acetyltransferase [23] association at a subset of promoters after GAF RNAi knockdown.
The association of GAF with promoter regions dovetailed with the discovery that Trl mutations dominantly enhanced the loss-of-function phenotypes seen in Ubx/ + heterozygous animals. Trl mutations were also found to enhance position effect variegation of the white gene in the inversion in(1)wm4h. However, these were not the only phenotypes observed. Adult survivors of a hypomophic Trl allele, Trl13C, have loss-of-function transformations in segment A6 (parasegment 11: PS11). Like A5 and A7 (PS10 and PS12), A6 identity is specified by Abdominal-B (Abd-B); however, both of these segments appear normal suggesting that misspecification of A6 (PS11) was due to a defect is in the functioning of the A6 regulatory domain, iab-6, and not in the activity of the Abd-B promoter [2]. Other findings also pointed to functions beyond ensuring that promoters are accessible to general transcription factors and transcriptional activators. For example, GAGA motifs are found in many polycomb response elements (PREs). Consistent with GAF being important for the silencing function of these elements, mutations either in Trl or in the GAGA motifs weaken or disrupt Polycomb silencing [24–29]. Additional evidence that GAF is required for polycomb-dependent silencing comes from chromatin immunoprecipitation (ChIP) experiments, which showed that GAF is associated with PREs in vivo [30–32]. GAF is also important for the functioning of fly chromatin boundary elements (insulators). GAF localizes to many known boundaries [33] and mutations in their GAGA motifs or in the Trl gene can impair insulator functions [34–38].
GAF association with promoters, PREs, boundary elements, and probably also many enhancers [39–41] is connected to chromatin remodeling and the process of zygotic genome activation (ZGA) in early embryos. The first indication of a possible role in the earliest steps of embryogenesis came from studies on the maternal effect allele Trl13C [42]. Even though maternal deposition of Trl mRNA in embryos from homozygous Trl13C is reduced, but not eliminated, only a small percentage of the embryos produced by homozygous Trl13C mothers hatch and most of the progeny arrest development prior to cellularization. Transcription of two genes, fushi taratzu (ftz) and engrailed (en), that are normally turned on during nuclear cycles 12–14 is disrupted. The severity of this disruption in individual nuclei correlates with the extent of reduction in the amount of GAF protein in the nucleus. Even more striking than the defects in transcription are abnormalities in the pre-blastoderm nuclear division cycles in Trl13C embryos. The defects include asynchrony, incomplete chromosome segregation, chromosome fragmentation, and nuclear disintegration. As was observed for transcription, the severity of the disruptions in mitosis is correlated with the extent of depletion of GAF [42]. Interestingly, GAF is found to associate with heterochromatic regions during mitosis, suggesting that it may contribute to centrosome function [42, 43].
Similar though somewhat less severe nuclear division defects have been reported in embryos produced by germline clone mothers mutant in the ZGA gene Zelda (zld) [44, 45] raising the possibility that GAF could also play a prominent role in ZGA. Several findings are consistent with this suggestion. The first indication that GAF could have a genome-wide role in ZGA was the discovery that many of the genes that are first transcribed at high levels during nuclear cycle 14 have paused polymerases [46]. Consistent with studies on fly heat shock genes, which showed that GAF plays an important role in polymerase pausing [7, 20, 47, 48], many of these ZGA genes have GAF motifs in their promoters [46]. Further suggesting that GAF is likely important for Pol II recruitment in pre-cellular blastoderm embryos, Blythe and Weischaus found that reducing Zld or GAF activity suppresses the mitotic defects in mei-41 mutant embryos. Since the mitotic defects in mei-41 are thought to arise from conflicts between the replication machinery and Pol II, limiting Pol II activity would be expected to reduce the severity of the defects [49]. Subsequent work by Schultz et al. showed that there are two classes of Zld sites in early embryos, those that become inaccessible after Zld depletion, and those that remain open. Most of the Zld sites in the latter class are marked by GAGA motifs and thus could be bound by GAF [50]. Taken together these findings implicate GAF in ZGA, and suggest that in this context, it likely has complementary and sometimes overlapping functions with the pioneer factor Zld. The relationship between Zld and GAF has been further explored by Gaskill et al. [51]. They found that the earliest transcribed genes depend more on Zld than GAF, while genes that are activated during the major wave of transcription in NC14 tend to require GAF. This correlation fits with the genome-wide distribution of these two proteins. Zld sites are enriched in the vicinity of genes activated in earlier nuclear division cycles, while GAF occupancy is enriched near genes activated in the major wave of transcription. As was found for Zld, there are thousands of sites in the genome in early embryos whose accessibility in chromatin depends upon GAF [51]. With some exceptions, most of the GAF-dependent sites do not depend on Zld. Likewise, most sequences that require Zld for accessibility, do not also depend on GAF.
How does GAF perform such a diverse array of functions ranging from mitosis to transcriptional activation and PcG dependent silencing? One common thread linking these different functions is the establishment of regions of chromatin that are nucleosome free so that other factors, which have dedicated activities, are able to access their binding sites. Another is the existence of numerous partners that are implicated in different GAF functions [52]. In this review, we have summarize our current understanding of how GAF functions and the role of its different protein partners.
Structural features of the GAF protein
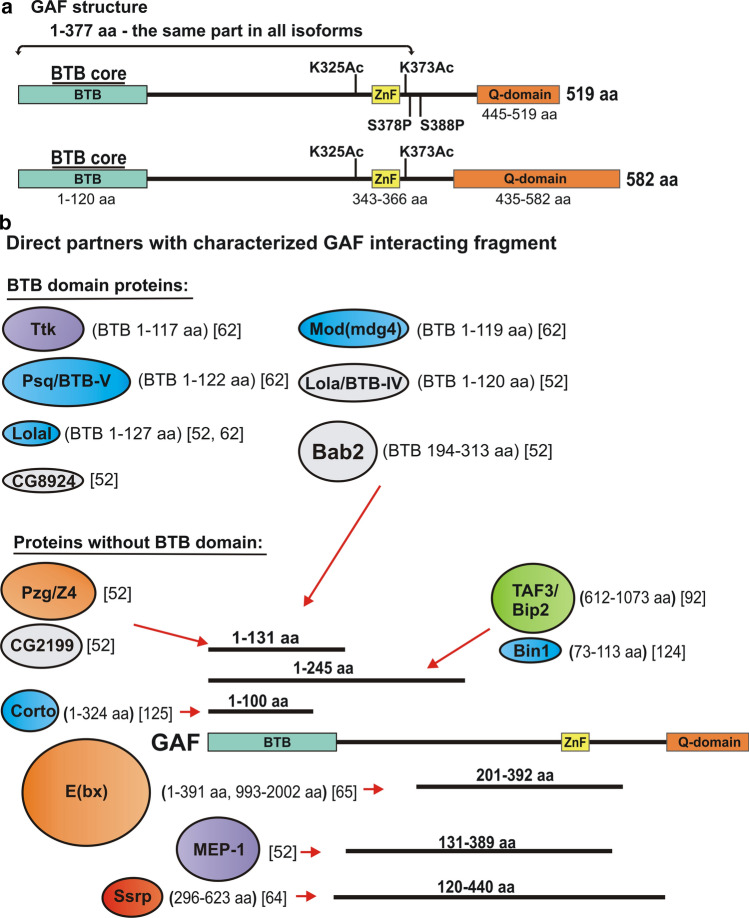
GAF has three distinguishable domains: an N-terminal BTB/POZ domain (Bric a brac, Tramtrack, Broad-complex/Poxvirus, Zinc finger), a central C2H2-type zinc finger and finally several alternative glutamine rich (Q) C-terminal domains [53, 54] (Fig. 1a). The two most abundant GAF isoforms, 519aa (GAF519) and 582aa (GAF582) share the BTB/POZ and zinc finger domains, but have distinct glutamine rich C-terminal domains of 142 and 205 amino acids (aa) in length respectively [55].
Fig. 1.
a Structure of GAF519 and GAF582 isoforms. Both isoforms share a 1-377 aa N-terminal domain that contains BTB/POZ domain and the C2H2-type zinc finger, while they have alternative C-terminal domains that are rich poly Q sequences. Amino acids K325 and K373 are targets for acetylation, while S378 and S388 may be phosphorylated in GAF519. b Proteins that interact directly with GAF. Proteins are shown as colored ovals. The size of the oval indicates the relative size of the proteins. The sequences for each protein that mediate interactions with GAF are indicated in the round brackets, while the corresponding references are indicated in the square brackets. Proteins that interact with GAF via their BTB domain are: Ttk, Mod(mdg4), Psq/BTB-V, Lola/BTB-IV, Lolal, Bab2, CG8924. Protein partners that interact with GAF but lack a BTB domain are: Pzq/Z4, CG2199, Corto, E(bx), Mep-1, Ssrp, TAF3/Bip2, Bin1. Red arrows indicate the GAF sequences that are required for interaction
The single zinc finger domain is responsible for DNA binding. A minimal trinucleotide sequence GAG is sufficient for recognition and binding [56], while the GAGAG pentanucleotide appears to be optimal for interaction [57, 58]. However, in vivo not all of the potential binding sites are occupied by GAF. Instead, GAF localization correlates with the number and density of GAGA-like motifs and sequences that are most highly enriched among GAF-bound fragments have multiple motifs [1, 30]. These findings suggest that interaction of GAF with its cognate recognition sequences in vivo is facilitated by cooperative interactions. Consistent with this possibility there is good evidence for cooperative binding in vitro [59, 60]. Cooperativity in DNA binding depends upon the zinc finger and the N-terminal BTB/POZ domain and the number of GAF-binding sites in the DNA substrate. When only a single GAGAG motif (1xGAGA) is present the BTB/POZ domain actually inhibits binding by the full-length GAF519 protein. In this case, truncated versions of GAF—either a BTB/POZ domain (∆POZ) deletion or a protein with only the DNA-binding domain (DBD)—binds to the 1xGAGA sequence with six–ninefold greater affinity than GAF519. A different result is obtained with a 5xGAGA oligonucleotide or natural DNA sequences, such as the Ubx, hsp26 and hsp70 promoters, that have multiple GAGA-like motifs. For these sequences, GAF519 binds with about tenfold greater affinity than either the ∆POZ or DBD proteins. Gel shift experiments indicate that this difference in affinity arises because GAF519 binds cooperatively to DNAs containing multiple GAGA-like motifs while the ∆POZ and DBD proteins do not. EM analysis shows that GAF assembles into large multi-subunit complexes on these natural templates, while such complexes can’t be detected with the ∆POZ protein. Moreover, complexes appear to be partially pre-assembled in solution in the absence of a DNA substrate as gel filtration experiments indicate that bacterially expressed GAF fractionates with a rather broad size distribution from a 60 kD monomer to multimers of up to 600 kD [60].
Other studies have also implicated the GAF BTB/POZ domain in homo and heterotypic protein–protein interactions [52, 61–63]. As was observed for the full-length GAF519 protein, the BTB/POZ domain self-assembles into a series of oligomers [62]. In gel filtration experiments, the BTB/POZ domain elutes in a broad distribution with a peak corresponding to a hexamer or octamer. In crosslinking experiments, the predominant species at low concentrations of crosslinker are monomers and dimers; while at higher concentrations several different multimeric species are evident. By contrast, the BTB/POZ domains of other proteins appear to assemble into more stable complexes. For example, a single multimeric species is observed for the Mod(mdg4) BTB/POZ domain and this multimer appears to correspond to an octamer [62]. The GAF BTB/POZ domain also mediates protein:protein interactions with other proteins that have a BTB/POZ domain [52, 61–63]. These GAF interacting BTB/POZ domain proteins include: Tramtrack (Ttk) [61–63], Lolal (Batman) [52, 62], Mod(mdg4), Pipsqueak (Psq/BTB-V) [62], Lola/BTB-IV, Bab2/BTB-II and CG8924 [52] (Fig. 1b). While the GAF BTB/POZ domain is promiscuous in its interactions with the BTB/POZ domains of other proteins, this is not the case for its interaction partners. Bonchuk et al. found that though the BTB/POZ domains of Mod(mdg4), Ttk and Psq all interact with GAF, Batman and themselves, they do not interact with each other [62].
While these findings indicate that GAF likely assembles into a variety of distinct complexes via interactions between its BTB/POZ domain and the BTB/POZ domain of other proteins, this is not the only protein:protein interaction domains. For example, other studies have suggested that the central part of the protein, which contains the zinc finger, might also facilitate interactions with other proteins [52, 64, 65] (Fig. 1b). The C-terminal poly Q domains could also have specialized protein interaction functions. The two primary isoforms, GAF519 and GAF582, have quite different developmental profiles. mRNAs encoding GAF519 are deposited in the developing egg during oogenesis and this isoform is present at high levels during the early stages of embryogenesis. In contrast, the larger GAF582 isoform only appears around 6 h of development [53]. In spite of the different developmental profiles, both isoforms co-localize in polytene chromosomes and behave indistinguishably in in vitro DNA binding and in tissue culture transient-transfection experiments [53]. On the other hand, while in vivo functional studies indicate that the two isoforms have overlapping functions, their activities are clearly not equivalent. The differences in activity may be related to distinct functional requirements at different stages of development [66]. Transgenes expressing both isoforms can rescue the zygotic lethality of Trl mutant flies; however, the extent of rescue is greater for GAF582, which is first expressed in the zygote at 6 h. The opposite result is obtained for the maternal effect lethality of the Trl13C allele. Transgenes expressing GAF519 partially rescue the maternal effect lethality, while transgenes expressing GAF582 do not. While this is consistent with their distinct expression patterns, the reason for this difference is not clear. Both isoforms associate with centromeric heterochromatin during mitosis and rescue the nuclear division defects in pre-cellular blastoderm embryos from Trl13C mothers. Both also rescue the defects in transcription of the ftz gene. Thus, there must be some other GAF target(s) in early embryos that requires the 519 aa and not the 582 aa isoform [66].
Another factor that is likely to impact GAF functionality is post-translational modification. The known modifications include O-glycosylation [67], phosphorylation [68] and acetylation [69]. Although the role and extent of the post-translational modifications are not completely understood, they are likely be important in modulating GAF activities through changes in DNA binding and protein:protein interactions. Consistent with this suggestion, phosphorylation and acetylation were shown to reduce the GAF DNA-binding activity [68, 69] suggesting that they could modulate GAF interactions with its target sequences.
GAF and chromatin remodelers
Eukaryotic gene expression in vivo takes places in the context of a chromatinized template, in which the DNA is assembled into nucleosomes and associated non-histone chromosomal proteins. Consequently, critical components of the transcriptional machinery (transcription factors, factors required for enhancer or silencer activity, RNA polymerases) cannot interact with their target sequences unless they are accessible. Accessible sequences are typically nucleosome free and can be detect as regions of DNA that are hypersensitive to nuclease cleavage [70, 71]. Nucleosome-free regions can extend over several hundred base pairs and are flanked by nucleosomes that are marked by highly dynamic H3.3 and H2A.Z histone variants [72]. One mechanism for generating accessible regions in chromatin involves a class of special “pioneer” factors that can interact with nucleosomal DNA and induce a change in the configuration of nucleosomes in the region [73]. As noted above, it is thought that the ZGA factor Zld is a pioneer protein [74, 75]. In the case of GAF, it has not only been implicated in ZGA, but there is also direct evidence that it can facilitate nucleosome displacement. Moreover, unlike Zld, GAF has been shown to associate with multiple remodeling complexes (see below).
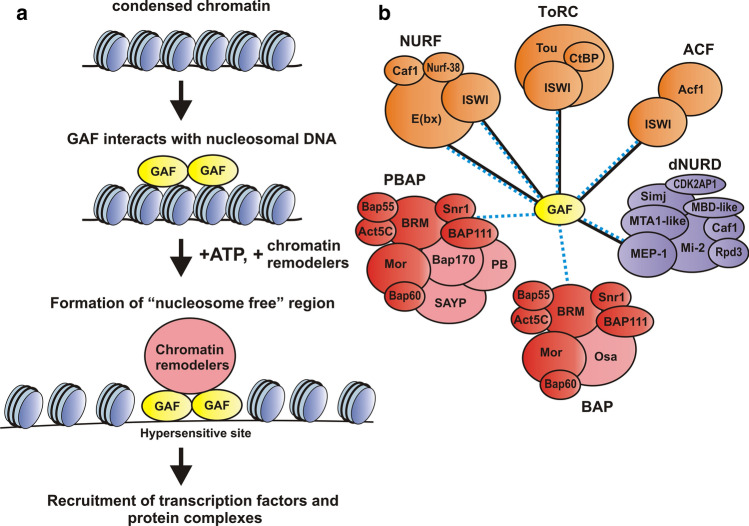
As discussed above, a role for GAF in chromatin remodeling and the establishment of nucleosome-free region is supported by in vitro chromatin assembly experiments [12–14]. In these experiments, the promoter regions containing GAF-binding sites become accessible when GAF is included in the nucleosome assembly reaction mix. Nucleosomes-free promoter regions could also be generated de novo when GAF was added to purified nucleosome templates that had been pre-assembled in the absence of exogenous GAF [12]. However, addition of GAF by itself was not sufficient to remodel the nucleosomes around the promoter. Instead, generating a nucleosome-free promoter required not only GAF, but also the assembly extract and ATP. Since GAF does not have an ATPase activity, these requirements imply that the extract must contain an ATP-dependent co-factor that can function with GAF to remodel nucleosomes [12]. Tsukiyama and Wu were able to purify an ATP-dependent chromatin remodeler called NURF using this assay [76]. NURF consists of four proteins: ISWI which has ATPase activity and can translocate DNA; a large 300 kD multi-domain (including a Bromodomain and 3 PHD fingers) protein E(bx) (NURF301); a histone binding protein, Caf1-55 and Nurf-38 which is a member of the inorganic pyrophosphatase protein family [76, 77]. When incubated with a pre-assembled chromatin template, GAF and ATP, NURF can displace nucleosomes from the hsp70 promoter, making it accessible to MNase or restriction enzyme digestion [76] (Fig. 2a).
Fig. 2.
GAF functions in conjunction with ATP-dependent chromatin remodelers to establish nucleosome-free regions of chromatin. a GAF interacts with nucleosomal DNA and recruits chromatin remodelers. In an ATP-dependent reaction, remodelers translocate nucleosomes and establish “nucleosome-free” regions of chromatin that are hypersensitive to various nucleases. This facilitates the recruitment of other transcription factors. b GAF interacts with ATP-dependent chromatin remodelers of different subfamilies: SWI/SNF (PBAP and BAP), ISWI (NURF, ACF and ToRC), CHD (dNURD). Contacts established by indirect methods are indicated by dotted blue lines; direct partners (E(bx), ISWI and MEP-1) are indicated by solid black lines
The formation of nucleosome-free regions of chromatin by the combination of GAF and NURF in vitro is not simply dependent on the ability of GAF to bind to DNA and block nucleosome occupancy. Instead, there are specific protein:protein interactions between GAF and the NURF subunits that likely mediate the recruitment of the NURF complex either to free GAF or to chromatin associated GAF. One interaction is between GAF and the E(bx) subunit (Fig. 2b). Xiao et al. found that the 300 kD E(bx) subunit co-IPs GAF519 in embryonic nuclear extracts [65]. Using recombinant GAF, they showed that DBD domain and the immediately flanking amino acids mediate interactions with E(bx). Conversely, two regions in E(bx) are responsible for binding to GAF. One is a 1–391 aa N-terminal fragment while the other is a ~ 1000 aa sequence located between 993aa-2002aa. Both E(bx) sequences also mediate interactions with nucleosomes. The second direct interaction is between GAF and the ISWI subunit [65] (Fig. 2b). The interactions detected in these experiments appear to be sufficient for the formation of stable complexes between GAF and NURF. In reciprocal purification, when GAF is immunoprecipitated from embryonic nuclear extracts, all four NURF subunits are detected by mass spectrometry [52]. In addition to NURF, there are two other remodeling complexes that utilize the DEAD/H-helicase ATPase ISWI, the ACF [78] and ToRC [79] complexes (Fig. 2b). The ACF complex has ISWI and Acf1, while ToRC has ISWI plus two other subunits, CtBP and Tou. Since ISWI can interact directly with GAF, one might expect that both ACF and ToRC would associate with GAF in vivo. This seems to be the case as Acf1, as well as the two ToRC subunits CtBP and Tou are detected by mass spectrometry in GAF co-IPs [52].
Although GAF together with NURF forms nucleosome-free regions of chromatin on hsp70 promoter in vitro [12, 76], it seems likely that the SWI/SNF family of ATP-dependent chromatin remodelers is primarily responsible for GAF-dependent remodeling of promoters and perhaps also boundary elements and PREs (Fig. 2a). There are two fly SWI/SNF complexes, PBAP and BAP (Fig. 2b). They share seven subunits—the Brahma helicase (Brm), Bap111, Bap55, Bap60, Snr1, Act5C and Mor. The unique subunits that specify PBAP complex are Polybromo, Bap170 and SAYP, while the BAP complex contains OSA [80, 81]. Although direct interactions between GAF and SWI/SNF subunits have not yet been demonstrated, Nakayama et al. [82] found that a FLAG-tagged GAF519 protein expressed in a wild type Trl background is associated with several common SWI/SNF remodeling complex subunits and Polybromo specific to the PBAP in nuclear extracts. Supporting the idea that this interaction is functionally significant, they found that GAF and the PBAP subunits Polybromo, Brm, and Bap60 are associated with Fab-7 and d1 boundaries and the bxdPRE in vivo, and that the association of SWI/SNF subunits with these elements is significantly reduced in Trl mutants [82]. As would be predicted from the findings of Nakayama et al. [82] all of the PABP subunits are detected by mass spectrometry in proteins that are co-immunoprecipitated with GAF from nuclear extracts [52]. The idea that that there is an intimate functional connection between GAF and PBAP has received support from recent study by Judd et al. [83]. They found that GAF acts synergistically with the PBAP complex at a majority of GAF-regulated promoters. RNAi knockdown experiments of GAF and the PBAP subunit Bap170 indicate that both are required to generate hypersensitive regions at the same set of promoters in vivo. As GAF binds weakly to chromatin in the absence of PBAP, it likely interacts with target sequences prior to PBAP; however, when PBAP is present, GAF binding is substantially enhanced as is general accessibility. At the same time, this study showed that GAF functions with NURF at a subset of promoters to position the + 1 nucleosome and that this relationship seems to be important in facilitating the release of paused polymerases [83]. While GAF functions in the establishment of hypersensitive (nucleosome free) regions of chromatin, other factors are expected to help maintain these regions as open chromatin. For example, studies by Gilchrist et al. [84] suggested that the presence of paused polymerases just beyond the transcription start site helps to occlude nucleosomes from the promoter region.
PBAP and NURF are not the only GAF associated ATP-dependent remodeler complexes. Experiments by Lomaev et al. showed that GAF also co-immunoprecipitates the BAP specific OSA protein (together with all of the other SWI/SNF subunits [52]. Nakayama et al. [82] also found one of the subunits, Mi-2 of the Drosophila dNURD complex, suggesting that this chromatin remodeler might also be associated with GAF [82] (Fig. 2b). Consistent with this suggestion, all of the subunits of the dNURD complex were detected by mass spectrometry in the immunoprecipitation experiments of Lomaev et al. [52].
While these findings demonstrate physical linkages between GAF and the main chromatin remodelers, many important questions remain unanswered. One is the extent of context specificity between different remodelers and GAF. The studies of Judd et al. [83] indicate that GAF recruits PABP to promoters, while there is evidence from Nakayama et al. [82] that that this interaction may also take place at some boundary elements and PREs. Does the GAF-dependent remodeling of nucleosomes at promoters, boundaries and PREs depend only on PBAP, or do some of the other remodeling complexes also function at these elements or at least at a subset of these elements? What about NURF? Are the interactions of NURF with GAF limited to regions downstream of the transcription start site? If there is specificity with respect to the recruitment of remodelers to different classes of GAF-dependent elements, what determines this specificity?
GAF and formation/maintenance of nucleosome-free regions of chromatin
Physical interactions between GAF and at least five different chromatin remodeling complexes would be expected to coordinate GAF binding to DNA with local nucleosome remodeling, and this would provide a mechanism for generating nucleosome-free regions. The experiments with pre-assembled chromatin hsp70 template and NURF [12, 76] indicate that GAF can function as a classical “pioneer” protein and displace pre-existing nucleosomes. Although similar studies have not been performed for GAF and PBAP or other associated remodelers, one imagines that when combined with GAF they would also be able to remodel pre-assembled chromatin templates. On the other hand, while pioneer proteins are thought to be capable of displacing pre-existing nucleosomes, it is not entirely clear to what extent such an activity is relevant in vivo. In flies, transcriptional activation (ZGA) takes place in a two-step process with a minor wave of transcription during nuclear division cycles 8–13 and a major wave during nuclear cycle 14 [74, 85, 86]. During these and the earlier division cycles, the chromosomal DNA is duplicated before each division and the daughter chromosomes must be assembled into chromatin before mitosis commences. It is entirely possible that GAF or GAF plus a remodeling complex could interact with its cognate recognition sequences after the replication fork passes but before these sequences are fully assembled into nucleosomes. This would only require a pre-existing pool of free GAF, which at this stage of development could be provided by the substantial deposition of maternal Trl mRNA. At stages in development in which GAF (or GAF plus a remodeling complex) is already bound to its recognition motifs in nucleosome-free regions of chromatin, the pre-existing complexes could potentially help template the re-association of GAF (or GAF plus the remodeling complex) with its recognition motifs following the passage of the replication fork ensuring that the nucleosome-free regions are inherited by both daughter chromosomes. In either of these cases, displacement of pre-existing nucleosomes would be unnecessary. On the other hand, there are instances in which nucleosome-free regions of chromatin are generated by displacing pre-existing nucleosomes. A classic example would be the SUC2 promoter in the yeast S. Cerevisiae, which is remodeled by the SWI/SNF under inducing conditions [87, 88]. In this case, induction is rapid, and thus nucleosome remodeling is not coupled to the passage of the replication fork.
The interaction of transcriptions factors with chromatin remodeling complexes is not in itself sufficient to generate nucleosome-free regions of chromatin. For example, like GAF, HSF physically interacts with the E(bx) subunit of NURF [65]. Moreover, when incubated with NURF and ATP it can also displace nucleosomes from the hsp70 promoter [76]. While HSF appears to function as a “pioneer” factor when combined with NURF in these in vitro assays, HSF recognition sequences are not sufficient to generate a nucleosome-free promoter in vivo [16–18] and GAGA elements contribute significantly to HSF binding in vivo [22]. A possible reason for this is that under normal growth conditions, HSF is localized in the cytoplasm and thus would not be able to prevent nucleosome encroachment even if it could promote nucleosome displacement under heat shock conditions [89].
In fact, most of the well-studied nucleosome-free regions of chromatin in flies appeared to be maintained for extended periods of time, in some cases throughout much of the life cycle. While HSF is an extreme example, interactions between DNA-binding proteins and their target sequences have limited half-lives, and once a protein dissociates from its target sequence, it would not be able to block nucleosome encroachment. One mechanism that appears to be deployed by GAF in maintaining nucleosome-free regions is cooperative binding. The well-characterized GAF-dependent nucleosome-free regions typically extend over sequences of 100–400 bp and contain several GAGA-like motifs. Consistent with this being a general feature of GAF-dependent nucleosome-free regions, genome-wide studies indicate that GAF is most frequently found associated with sequences that contain multiple GAGA-like motifs [1, 30]. Based on in vitro biochemical experiments, sequences with multiple GAGA-like motifs are likely occupied in vivo by multimeric GAF complexes. As noted above, the BTB domain of GAF mediates the assembly of recombinant GAF into a spectrum of multimeric complexes, and this ability to multimerize substantially augments binding to natural DNA sequences such as the hsp70 or Ubx promoters [60]. The notion that a single protein and its cognate binding site might not be sufficient to maintain open regions of chromatin is supported by recent work by Kyrchanova et al. on several insulator proteins [90]. They found that a truncated 106 bp Fab-8 boundary that has two binding sites for the fly dCTCF zinc finger protein is not sufficient for dCTCF binding or for insulator function in the context of BX-C. Instead, at least four dCTCF binding sites are required for function, while three binding sites have no boundary activity and significantly reduced dCTCF association [90]. Similar results were obtained for two other polydactyl zinc finger DNA-binding proteins, Pita and Su(Hw). These studies also showed that boundary elements which have only a single binding site for Pita, dCTCF, or Su(Hw) require surrounding sequences (> 100 bp) for functionality [90]. This may also be true for pioneer proteins like Zld. While Zld occupancy has been observed at single sites, Zld is usually found in DNA segments that have a cluster of Zld binding motifs or other relevant motifs such as GAGAG [91]. It clearly will be of interested to determine whether single Zld sites in isolation can be accessed by Zld and are functional.
GAF and context-dependent partners
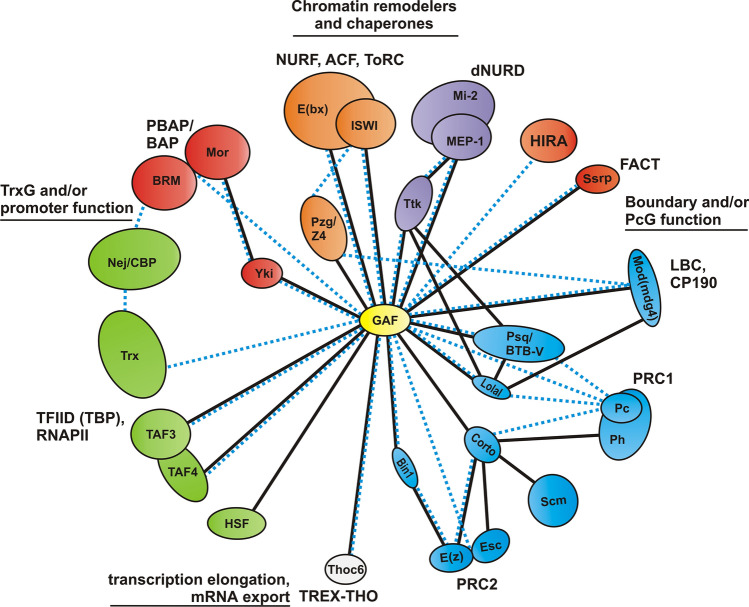
The formation and maintenance of nucleosome-free regions of chromatin by GAF is likely to involve not only GAF associated chromatin remodelers but also a collection of other proteins, some of which have functions that are dedicated to specific types of regulatory elements. In each regulatory context, the activity of GAF is augmented by these other protein partners. Some of these protein partners are part of larger complexes and may interact only indirectly with GAF, while other partners may involve direct physical interactions. Consistent with this later possibility, more than 20 proteins have found to interact directly with GAF (Fig. 3, Supplementary excel file).
Fig. 3.
GAF interactome. GAF interacts with chromatin remodelers, Trx and promoter-associated factors, proteins involved in boundary and PcG function, transcription elongation and mRNA export. Solid black lines—direct partners; dotted blue lines—partners identified by indirect methods
GAF promoter partners
Although functional studies have suggested that GAF interacts with NELF, the PolII subunit Rpb3 and CBP [7, 9, 23], none of these proteins are found stably associated with GAF in co-IP studies suggesting that they may interact with GAF indirectly. On the other hand, direct physical association has been shown for subunits of the TFIID complex [92, 93] and HSF [94] (Fig. 3). In the case of TFIID, GAF was shown to directly interact with TAF3 [92, 93] (Fig. 3). Both proteins could be co-purified in reciprocal co-IP experiments [92]. Another potential direct connection between GAF and TFIID is TAF4 (Fig. 3). It was identified in a yeast 2-hybrid (Y2H) screen as a GAF partner [93] and is present in GAF co-IP [52].
In addition to interactions with components of the general transcriptional machinery, GAF also associates with co-factors that have pathway or gene specific functions like HSF. For example, GAF was found to interact with the Yorkie (Yki) protein, a regulatory target of the Hippo signaling pathway (Fig. 3). Over 60% of Yki sites overlap with 49% of GAF sites genome wide in embryos and wing discs, and Yki and GAF co-purify from extracts of Drosophila S2 cells [95]. This interaction is functional—GAF is required for expression of dE2f1-Yki targets and depletion of GAF suppresses Yki-driven cell proliferation [95, 96]. Two other potential GAF interactors are the Atro/Gug and Gro co-repressor proteins. Atro and Gro proteins overlap with 24.3% and 21.3% (11.47% TSS) of GAF sites, respectively [97, 98]. Atro was shown to co-immunoprecipitate GAF in IP-western experiments [98], while Gro was found associated with GAF in IP/MS studies [52].
GAF boundary and PRE partners
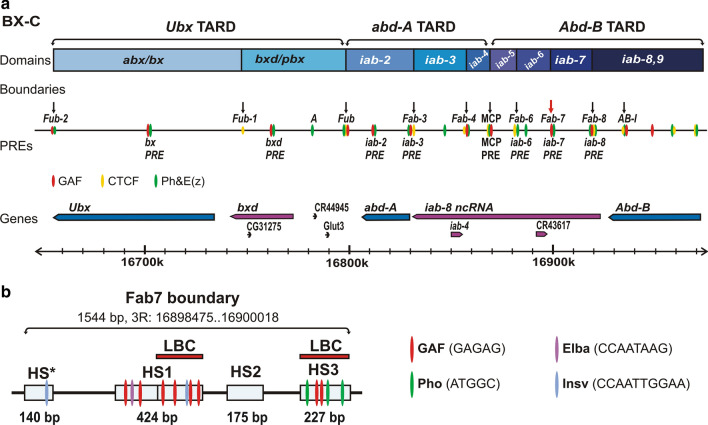
Many fly chromatin boundary elements (insulators) have multiple GAGAG motifs and are occupied by GAF in ChIP experiments [33]. Two classic examples of GAF associated boundaries are Fab-7 [99–104] and Fab-8 [105, 106] from the Drosophila Bithorax complex (BX-C) (Fig. 4a). These two boundaries flank the iab-7 regulatory domain, insulating it from the adjacent iab-6 and iab-8 regulatory domains. The iab-6, iab-7 and iab-8 regulatory domains, together with iab-5, are responsible for the parasegment (PS) specific expression of the BX-C homeotic gene Abdominal-B (Abd-B) in the posterior parasegments PS10 (iab-5), PS11 (iab-6), PS12 (iab-7) and PS13 (iab-8). In addition to blocking cross-talk between adjacent regulatory domains the boundaries in this region of BX-C also have bypass activity. Bypass activity is required so that more distal domains can bypass the intervening boundary elements and activate Abd-B expression. For example, the iab-6 regulatory domain must bypass Fab-7 and Fab-8 to regulate Abd-B in PS11. Likewise, the iab-7 regulatory domain has to bypass Fab-8 to regulate Abd-B in PS12 [107–110].
Fig. 4.
GAF is required for the functioning of chromatin boundaries and PREs in BX-C. a The diagram shows the BX-C TARDs, containing Ubx, abd-A, and Abd-B genes and their respective cis-regulatory domains. The positions of the boundaries and PREs are indicated above and below the sequence coordinate line correspondingly. The red arrow marks Fab-7 boundary. The red, yellow and green ovals are regions bound by GAF, CTCF or E(z) and Ph, respectively. b The Fab-7 boundary has four nuclease hypersensitive regions: one minor—HS* and three prominent—HS1, HS2, and HS3. The locations of the recognition motifs for GAF, Pho, Elba and Insv proteins known to be associated with Fab-7 are indicated. The LBC complex binds to distal half of HS1 (dHS1) and HS3
Both Fab-7 and Fab-8 map to nuclease hypersensitive regions in chromatin. Fab-7 has four nuclease hypersensitive regions, HS*, HS1, HS2 and HS3 [37, 99, 102, 111] (Fig. 4b). The two largest are HS1, which is about 400 bp in length and HS3, which is nearly 200 bp. In nuclear extracts, GAF interacts with large probes spanning the centromere distal half of HS1, dHS1, and with a similarly sized HS3 probe as part of a large > 1000 kD complex called the LBC (Large Boundary Complex) [35, 38]. The sequence recognition properties of the LBC are complex. The available evidence indicates that the preferred substrates for the LBC are 120–200 bp in length and contain one or more GAGAG-like motifs [35]. However, other than GAGAG-like motifs, the sequences that are known to be bound by the LBC share no obvious similarities. dHS1 has four GAGAG motifs, while HS3 has two and mutations in these motifs substantially weakened LBC binding in nuclear extracts and boundary function in vivo [35, 38] (Fig. 4b). The LBC also binds to a ~ 130 bp sequence on centromere proximal side of Fab-8 that has a single GAGAG; however, unlike dHS1 or HS3, mutations in this GAGAG motif have only a minimal impact on LBC binding. While most other known LBC recognition sequences have GAn motifs that are also important for LBC binding, there are examples of sequences that are bound by the LBC but lack GAGAG motifs [112].
Supershift EMSA experiments using gel filtration fractions containing the LBC with antibodies directed against known insulator proteins suggest that the LBC complex contains GAF, Mod(mdg4), E(y)2 and CLAMP [35, 38, 52, 113]. While Mod(mdg4) and E(y)2 are also detected by mass spectrometry in GAF co-IPs from nuclear extracts, surprisingly CLAMP is not [52]. Given this discrepancy, it is not clear at this point if CLAMP is a bona fide component of the LBC. While it conceivable that it fails to remain associated with GAF/LBC in co-IP experiments it is also possible that the two CLAMP antibodies used in the supershift experiments cross-react with an as yet unknown LBC associated protein. In this context, other boundary factors that do not appear to be components of the LBC are nevertheless found in GAF co-IPs. These include CP190, Su(Hw), CP60, and the boundary associated factors Pzg/Z4, Dref and Chro [52]. GAF was shown to directly interact with Pzg/Z4 protein [52] (Fig. 1b, Fig. 3), but not with CP190 or Su(Hw) proteins [52, 62, 114]. Since the CP190 and Su(Hw) proteins were shown not found to be associated with the LBC in EMSA supershift experiments [38], these boundary proteins appear to associate with GAF in complexes that are independent of the LBC.
Unlike CLAMP, the presence of Mod(mdg4) in the LBC is supported not only by the supershift and co-IP experiments, but also by the fact that its BTB/POZ domain is known to interact with the BTB/POZ domain of GAF [62] (Fig. 1b, Fig. 3). Since BTB/POZ domain of Mod(mdg4) forms a stable octamer [62], it seems possible that a Mod(mdg4) octamer could provide a scaffold for the assembly of GAF and other proteins into the LBC.
Mod(mdg4) also has another property which could help explain the seemingly promiscuous sequence recognition properties of the LBC. There are 31 predicted Mod(mdg4) isoforms. All share the N-terminal BTB/POZ domain but have different C-terminal domains. Of the 31 distinct C-terminal domains, 27 have unique FLYWCH zinc finger DNA-binding domains and each would be expected to have somewhat different sequence recognition specificities. The temporal/tissue-specific patterns of expression and the relative abundance of the different Mod(mdg4) isoforms are not known; however, 14 of the Mod(mdg4) isoforms were detected in GAF co-IPs with embryonic nuclear extracts [52]. Included in this group is the PT isoform [also known as Mod(mdg4)67.2 or 2.2] that has implicated genetically in boundary function [115, 116]. Based on what is known about Mod(mdg4) oligomerization, it is possible that eight different Mod(mdg4) isoform combinations could be present in each LBC. If this were the case, it could explain how the LBC is able to interact with seemingly quite different DNA sequences. The presence of an octamer in each LBC plus at least one other DNA-binding protein (GAF) could also help account for the large size (120–200 bp) of the sequences that are bound by the LBC in nuclear extracts. In this context, it also worth noting the LBC recognition sequences that have been characterized by DNAase or MNase digests of chromatin span sequences of similar size [99, 102].
The functions of the LBC recognition sequences in Fab-7 and Fab-8 are somewhat different. In Fab-8, the primary function of the LBC recognition sequence is to provide bypass activity, while the sequences on the distal side of the hypersensitive region function to provide blocking activity [112]. Not unexpectedly, these distal sequences contain two binding sites for the fly dCTCF protein [112]. In the case of the Fab-7, Kyrchanova et al. found that a combination of dHS1 (the LBC recognition sequence in HS1), and HS3 is sufficient to fully reconstitute the blocking and bypass activity of the much larger WT Fab-7 boundary in boundary replacement experiments [35]. This finding was a surprise as previous studies suggested that HS3 is not part of the Fab-7 boundary but rather is a PRE (Polycomb Response Element) for the BX-C iab-7 regulatory domain, the “iab-7 PRE” [26, 103]. HS3 has two GAGAG motifs and three recognition sequences for the PcG group DNA-binding protein Pleiohomeotic [35] (Pho, [117]) (Fig. 4b). Mutants in the two GAGAG motifs or in the Pho binding sites eliminate the silencing activity of HS3 [28]. Silencing activity is also dependent upon the GAF gene Trl and the pho gene [26, 118]. As noted above, LBC binding to HS3 is disrupted by mutations in the two GAGAG motifs; however, it is not affected by mutations in the three binding sites for Pho [35]. This is also true for boundary function of HS3; it is eliminated by mutations in the GAGAG motifs, but not by mutations in the Pho binding sites [35]. Thus, HS3 has two distinct functions. One is a boundary function, which depends on the LBC, while the other is a silencing function, which requires both the LBC and Pho.
An as yet unanswered question is what role does the LBC play in PRE (and also TRE) function and how does this relate to its boundary activities? Recent Y2H experiments by Shokri et al. suggest that there is a direct interaction between GAF and Pho [119]. However, this association does not appear to be stable in Co-IP experiments as neither Pho nor its partner Stmbt are consistently detected by mass spectrometry [52]. This would suggest that one of the reasons why the GAGAG motifs in HS3 (iab-7 PRE) are required for PcG dependent silencing is that GAF (or in this case the LBC) is needed to open up the chromatin so that Pho can gain access. This possibility is supported by the studies of Mahmoudi et al. who showed that Pho could not gain access to chromatized Ubx PRE templates in the absence of added GAF [120].
On the other hand, other known components of the PcG silencing machinery form stable complexes with GAF in nuclear extracts. These include all of the components of the PcG complex PRC2 (Polycomb repressive complex), Zeste and Adf [52]. There are also several proteins that appear to interact directly with GAF. These include: Psq [62, 121], Batman [52, 62, 93, 122, 123], Bin1 [124] and Corto [125] (Fig. 1b, Fig. 3). GAF association in vivo has been shown for Psq [52, 121, 126], Batman [52, 122], Bin1 [52], while an association has not yet been found for the Corto protein.
As mentioned above, the Psq protein contains a BTB/POZ domain through which it can interact with GAF [62, 121] and DNA-binding domain. The DNA-binding domain consists of four tandem repeats of a conserved 50-amino acid sequence (Psq domain) and, like GAF, it recognizes GAGAG sequence motifs [127]. Both GAF and Psq could function together – they are colocalized on the polythene chromosomes [121] and bind similar sites genome-wide in S2 Drosophila cells as measured by ORGANIC profiling [128]. Psq co-purifies not only with GAF [52, 121], but also with the PcG protein Polycomb [129].
The Batman protein is only 127 aa in length. Most of the Batman protein corresponds to its BTB/POZ domain, which can interact directly with the GAF BTB/POZ domain [52, 62, 122, 123]. Batman and GAF are codistributed and both co-localize with about 50% of the Ph sites on polytene chromosomes [122]. It was shown that GAF association with Ph in embryos depends on externally provided Batman. On the other hand, in larval extracts, GAF can be co-IPd with Ph irrespective of Batman [123].
Another protein that might mediate GAF interactions with PcG complexes is the Bicoid interacting protein 1 (Bin1) also known as SAP18. While Bin1 lacks a BTB/POZ it nevertheless interacts with BTB/POZ domain of GAF [124]. Bin1 also interacts with the PRC2 subunit E(z) and they can be co-purified from nuclear extracts [130]. Apparently, Bin1 participates only in subset of GAF interacting associated sequences, since the GAF and Bin1 show only limited overlap in polytene chromosomes. However, both GAF and Bin1 co-localize at the region of the Bithorax complex [124].
Conclusions
GAF is unusual in that it is a participant in many different and seemingly unrelated regulatory contexts. In vivo, it is found associated with promoters, enhancers, boundary elements, PREs and TREs. Though not discussed here, GAF also co-localizes with CLAMP at a subset of the X-chromosome chromatin entry sites (CES) for the male specific lethal complex (MSL) [113]. In these different contexts, it facilitates promoter activity, transcriptional activation by enhancers including long distance enhancer–promoter interactions, insulating activity and transcriptional silencing. This diverse array of context-dependent activities poses two important questions. First, how does GAF carry out these different functions? Second, is there a common mechanism(s) of action or unifying theme or are these distinct functions completely “unrelated?” Probably the answer, at least in part, to the first question is that GAF has many different protein partners, either direct interactors, or proteins that are in complexes and interact with GAF only indirectly. In many instances, it is likely that these protein partners are largely responsible for carrying out distinct GAF functions. As for the second question, the common theme may be the ability of GAF to induce and then help maintain open regions of chromatin. In this respect, the cooperation of GAF with ATP-dependent chromatin remodelers could represent a conserved mechanism of action for “pioneer” proteins in other organisms. For example, it was shown that the OCT4 needs the BRG1/SMARCA4—SWI/SNF homologue, to support its binding and pioneering activity in mouse embryonic stem cells [131]. Similarly, chromatin association of the NANOG pioneer factor is facilitated by the BRG1 in mouse blastocysts [132]. Although two mammalian GAF homologs have been identified, their properties have not been explored in as great a detail as the Drosophila protein and it is not known whether they function in generating nucleosome-free regions of chromatin. One of the potential homologs is ZBTB3. It was computationally predicted to be the mammalian protein most structurally related to Drosophila GAF [133]. The other is the ThPOK (also known as c-KROX/ZBTB7B) protein, and it has been studied in more detail [134]. Like GAF, it has an N-terminal BTB/POZ domain, but instead of a single zinc finger, it has four C-terminal zinc fingers. ThPOK binds to GAGA sequences in vitro and in vivo and it functions in transcriptional activation of genes in different cellular contexts [135]. It has also been implicated in the differentiation of CD4 + progenitor cells where it is thought to activate the expression of the “suppressors of cytokine signaling” genes and repress expression of Blimp1 and Runx3 [136, 137]. ThPOK has also been found to contribute to the insulator function of an element located between the mouse Evx2 and Hoxd13 genes [138]. While it is not known whether ThPOK functions in generating nucleosome-free regions of chromatin, it has been shown to co-purify with the ACF and NurD chromatin remodeling complexes so this possibility remains open [139].
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We are grateful to the Center for Precision Genome Editing and Genetic Technologies for Biomedicine IGB RAS for computer equipment. This study was supported by the Russian Science Foundation (RSF) 18-74-10091 to D.C, by the National Institutes of Health (NIH) 5R35GM126975 to P.S.
Author contributions
All authors designed, wrote, and revised the review.
Funding
This study was supported by the Russian Science Foundation (RSF) 18-74-10091 to D.C, by the National Institutes of Health (NIH) 5R35GM126975 to P.S.
Data availability
All relevant data are within the paper and its Supporting Information file.
Compliance with ethical standards
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Darya Chetverina, Email: daria.chetverina@gmail.com.
Paul Schedl, Email: pschedl@Princeton.EDU.
References
- 1.van Steensel B, Delrow J, Bussemaker HJ. Genomewide analysis of Drosophila GAGA factor target genes reveals context-dependent DNA binding. Proc Natl Acad Sci USA. 2003;100(5):2580–2585. doi: 10.1073/pnas.0438000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Farkas G, Gausz J, Galloni M, Reuter G, Gyurkovics H, Karch F. The Trithorax-like gene encodes the Drosophila GAGA factor. Nature. 1994;371(6500):806–808. doi: 10.1038/371806a0. [DOI] [PubMed] [Google Scholar]
- 3.Biggin MD, Tjian R. Transcription factors that activate the Ultrabithorax promoter in developmentally staged extracts. Cell. 1988;53(5):699–711. doi: 10.1016/0092-8674(88)90088-8. [DOI] [PubMed] [Google Scholar]
- 4.Soeller WC, Poole SJ, Kornberg T. In vitro transcription of the Drosophila engrailed gene. Genes Dev. 1988;2(1):68–81. doi: 10.1101/gad.2.1.68. [DOI] [PubMed] [Google Scholar]
- 5.Granok H, Leibovitch BA, Shaffer CD, Elgin SC. Chromatin. Ga-ga over GAGA factor. Curr Biol. 1995;5(3):238–241. doi: 10.1016/S0960-9822(95)00048-0. [DOI] [PubMed] [Google Scholar]
- 6.Wilkins RC, Lis JT. Dynamics of potentiation and activation: GAGA factor and its role in heat shock gene regulation. Nucleic Acids Res. 1997;25(20):3963–3968. doi: 10.1093/nar/25.20.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fuda NJ, Guertin MJ, Sharma S, Danko CG, Martins AL, Siepel A, Lis JT. GAGA factor maintains nucleosome-free regions and has a role in RNA polymerase II recruitment to promoters. PLoS Genet. 2015;11(3):e1005108. doi: 10.1371/journal.pgen.1005108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li J, Gilmour DS. Distinct mechanisms of transcriptional pausing orchestrated by GAGA factor and M1BP, a novel transcription factor. EMBO J. 2013;32(13):1829–1841. doi: 10.1038/emboj.2013.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li J, Liu Y, Rhee HS, Ghosh SK, Bai L, Pugh BF, Gilmour DS. Kinetic competition between elongation rate and binding of NELF controls promoter-proximal pausing. Mol Cell. 2013;50(5):711–722. doi: 10.1016/j.molcel.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kerrigan LA, Croston GE, Lira LM, Kadonaga JT. Sequence-specific transcriptional antirepression of the Drosophila Kruppel gene by the GAGA factor. J Biol Chem. 1991;266(1):574–582. doi: 10.1016/S0021-9258(18)52474-1. [DOI] [PubMed] [Google Scholar]
- 11.Croston GE, Kerrigan LA, Lira LM, Marshak DR, Kadonaga JT. Sequence-specific antirepression of histone H1-mediated inhibition of basal RNA polymerase II transcription. Science. 1991;251(4994):643–649. doi: 10.1126/science.1899487. [DOI] [PubMed] [Google Scholar]
- 12.Tsukiyama T, Becker PB, Wu C. ATP-dependent nucleosome disruption at a heat-shock promoter mediated by binding of GAGA transcription factor. Nature. 1994;367(6463):525–532. doi: 10.1038/367525a0. [DOI] [PubMed] [Google Scholar]
- 13.Wall G, Varga-Weisz PD, Sandaltzopoulos R, Becker PB. Chromatin remodeling by GAGA factor and heat shock factor at the hypersensitive Drosophila hsp26 promoter in vitro. EMBO J. 1995;14(8):1727–1736. doi: 10.1002/j.1460-2075.1995.tb07162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okada M, Hirose S. Chromatin remodeling mediated by Drosophila GAGA factor and ISWI activates fushi tarazu gene transcription in vitro. Mol Cell Biol. 1998;18(5):2455–2461. doi: 10.1128/MCB.18.5.2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glaser RL, Thomas GH, Siegfried E, Elgin SC, Lis JT. Optimal heat-induced expression of the Drosophila hsp26 gene requires a promoter sequence containing (CT)n.(GA)n repeats. J Mol Biol. 1990;211(4):751–761. doi: 10.1016/0022-2836(90)90075-W. [DOI] [PubMed] [Google Scholar]
- 16.Lu Q, Wallrath LL, Allan BD, Glaser RL, Lis JT, Elgin SC. Promoter sequence containing (CT)n.(GA)n repeats is critical for the formation of the DNase I hypersensitive sites in the Drosophila hsp26 gene. J Mol Biol. 1992;225(4):985–998. doi: 10.1016/0022-2836(92)90099-6. [DOI] [PubMed] [Google Scholar]
- 17.Lu Q, Wallrath LL, Granok H, Elgin SC. (CT)n (GA)n repeats and heat shock elements have distinct roles in chromatin structure and transcriptional activation of the Drosophila hsp26 gene. Mol Cell Biol. 1993;13(5):2802–2814. doi: 10.1128/mcb.13.5.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leibovitch BA, Lu Q, Benjamin LR, Liu Y, Gilmour DS, Elgin SC. GAGA factor and the TFIID complex collaborate in generating an open chromatin structure at the Drosophila melanogaster hsp26 promoter. Mol Cell Biol. 2002;22(17):6148–6157. doi: 10.1128/MCB.22.17.6148-6157.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu Q, Wallrath LL, Emanuel PA, Elgin SC, Gilmour DS. Insensitivity of the present hsp26 chromatin structure to a TATA box mutation in Drosophila. J Biol Chem. 1994;269(22):15906–15911. doi: 10.1016/S0021-9258(17)40766-6. [DOI] [PubMed] [Google Scholar]
- 20.Lee H, Kraus KW, Wolfner MF, Lis JT. DNA sequence requirements for generating paused polymerase at the start of hsp70. Genes Dev. 1992;6(2):284–295. doi: 10.1101/gad.6.2.284. [DOI] [PubMed] [Google Scholar]
- 21.Li B, Weber JA, Chen Y, Greenleaf AL, Gilmour DS. Analyses of promoter-proximal pausing by RNA polymerase II on the hsp70 heat shock gene promoter in a Drosophila nuclear extract. Mol Cell Biol. 1996;16(10):5433–5443. doi: 10.1128/mcb.16.10.5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shopland LS, Hirayoshi K, Fernandes M, Lis JT. HSF access to heat shock elements in vivo depends critically on promoter architecture defined by GAGA factor, TFIID, and RNA polymerase II binding sites. Genes Dev. 1995;9(22):2756–2769. doi: 10.1101/gad.9.22.2756. [DOI] [PubMed] [Google Scholar]
- 23.Boija A, Mahat DB, Zare A, Holmqvist PH, Philip P, Meyers DJ, Cole PA, Lis JT, Stenberg P, Mannervik M. CBP regulates recruitment and release of promoter-proximal RNA polymerase II. Mol Cell. 2017;68(3):491–503 e495. doi: 10.1016/j.molcel.2017.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Americo J, Whiteley M, Brown JL, Fujioka M, Jaynes JB, Kassis JA. A complex array of DNA-binding proteins required for pairing-sensitive silencing by a polycomb group response element from the Drosophila engrailed gene. Genetics. 2002;160(4):1561–1571. doi: 10.1093/genetics/160.4.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Busturia A, Lloyd A, Bejarano F, Zavortink M, Xin H, Sakonju S. The MCP silencer of the Drosophila Abd-B gene requires both Pleiohomeotic and GAGA factor for the maintenance of repression. Development. 2001;128(11):2163–2173. doi: 10.1242/dev.128.11.2163. [DOI] [PubMed] [Google Scholar]
- 26.Hagstrom K, Muller M, Schedl P. A polycomb and GAGA dependent silencer adjoins the Fab-7 boundary in the Drosophila bithorax complex. Genetics. 1997;146(4):1365–1380. doi: 10.1093/genetics/146.4.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hodgson JW, Argiropoulos B, Brock HW. Site-specific recognition of a 70-base-pair element containing d(GA)(n) repeats mediates bithoraxoid polycomb group response element-dependent silencing. Mol Cell Biol. 2001;21(14):4528–4543. doi: 10.1128/MCB.21.14.4528-4543.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mishra RK, Mihaly J, Barges S, Spierer A, Karch F, Hagstrom K, Schweinsberg SE, Schedl P. The iab-7 polycomb response element maps to a nucleosome-free region of chromatin and requires both GAGA and pleiohomeotic for silencing activity. Mol Cell Biol. 2001;21(4):1311–1318. doi: 10.1128/MCB.21.4.1311-1318.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poux S, Horard B, Sigrist CJ, Pirrotta V. The Drosophila trithorax protein is a coactivator required to prevent re-establishment of polycomb silencing. Development. 2002;129(10):2483–2493. doi: 10.1242/dev.129.10.2483. [DOI] [PubMed] [Google Scholar]
- 30.Negre N, Hennetin J, Sun LV, Lavrov S, Bellis M, White KP, Cavalli G. Chromosomal distribution of PcG proteins during Drosophila development. PLoS Biol. 2006;4(6):e170. doi: 10.1371/journal.pbio.0040170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ogiyama Y, Schuettengruber B, Papadopoulos GL, Chang JM, Cavalli G. Polycomb-dependent chromatin looping contributes to gene silencing during Drosophila development. Mol Cell. 2018;71(1):73–88 e75. doi: 10.1016/j.molcel.2018.05.032. [DOI] [PubMed] [Google Scholar]
- 32.Strutt H, Cavalli G, Paro R. Co-localization of polycomb protein and GAGA factor on regulatory elements responsible for the maintenance of homeotic gene expression. EMBO J. 1997;16(12):3621–3632. doi: 10.1093/emboj/16.12.3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Negre N, Brown CD, Shah PK, Kheradpour P, Morrison CA, Henikoff JG, Feng X, Ahmad K, Russell S, White RA, Stein L, Henikoff S, Kellis M, White KP. A comprehensive map of insulator elements for the Drosophila genome. PLoS Genet. 2010;6(1):e1000814. doi: 10.1371/journal.pgen.1000814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Belozerov VE, Majumder P, Shen P, Cai HN. A novel boundary element may facilitate independent gene regulation in the Antennapedia complex of Drosophila. EMBO J. 2003;22(12):3113–3121. doi: 10.1093/emboj/cdg297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kyrchanova O, Kurbidaeva A, Sabirov M, Postika N, Wolle D, Aoki T, Maksimenko O, Mogila V, Schedl P, Georgiev P. The bithorax complex iab-7 polycomb response element has a novel role in the functioning of the Fab-7 chromatin boundary. PLoS Genet. 2018;14(8):e1007442. doi: 10.1371/journal.pgen.1007442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ohtsuki S, Levine M. GAGA mediates the enhancer blocking activity of the eve promoter in the Drosophila embryo. Genes Dev. 1998;12(21):3325–3330. doi: 10.1101/gad.12.21.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schweinsberg S, Hagstrom K, Gohl D, Schedl P, Kumar RP, Mishra R, Karch F. The enhancer-blocking activity of the Fab-7 boundary from the Drosophila bithorax complex requires GAGA-factor-binding sites. Genetics. 2004;168(3):1371–1384. doi: 10.1534/genetics.104.029561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wolle D, Cleard F, Aoki T, Deshpande G, Schedl P, Karch F. Functional requirements for fab-7 boundary activity in the bithorax complex. Mol Cell Biol. 2015;35(21):3739–3752. doi: 10.1128/MCB.00456-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kvon EZ, Stampfel G, Yanez-Cuna JO, Dickson BJ, Stark A. HOT regions function as patterned developmental enhancers and have a distinct cis-regulatory signature. Genes Dev. 2012;26(9):908–913. doi: 10.1101/gad.188052.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moshe A, Kaplan T. Genome-wide search for Zelda-like chromatin signatures identifies GAF as a pioneer factor in early fly development. Epigenetics Chromatin. 2017;10(1):33. doi: 10.1186/s13072-017-0141-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zabidi MA, Arnold CD, Schernhuber K, Pagani M, Rath M, Frank O, Stark A. Enhancer-core-promoter specificity separates developmental and housekeeping gene regulation. Nature. 2015;518(7540):556–559. doi: 10.1038/nature13994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bhat KM, Farkas G, Karch F, Gyurkovics H, Gausz J, Schedl P. The GAGA factor is required in the early Drosophila embryo not only for transcriptional regulation but also for nuclear division. Development. 1996;122(4):1113–1124. doi: 10.1242/dev.122.4.1113. [DOI] [PubMed] [Google Scholar]
- 43.Raff JW, Kellum R, Alberts B. The Drosophila GAGA transcription factor is associated with specific regions of heterochromatin throughout the cell cycle. EMBO J. 1994;13(24):5977–5983. doi: 10.1002/j.1460-2075.1994.tb06943.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liang HL, Nien CY, Liu HY, Metzstein MM, Kirov N, Rushlow C. The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature. 2008;456(7220):400–403. doi: 10.1038/nature07388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Staudt N, Fellert S, Chung HR, Jackle H, Vorbruggen G. Mutations of the Drosophila zinc finger-encoding gene vielfaltig impair mitotic cell divisions and cause improper chromosome segregation. Mol Biol Cell. 2006;17(5):2356–2365. doi: 10.1091/mbc.e05-11-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen K, Johnston J, Shao W, Meier S, Staber C, Zeitlinger J. A global change in RNA polymerase II pausing during the Drosophila midblastula transition. eLife. 2013;2:e00861. doi: 10.7554/eLife.00861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsai SY, Chang YL, Swamy KB, Chiang RL, Huang DH. GAGA factor, a positive regulator of global gene expression, modulates transcriptional pausing and organization of upstream nucleosomes. Epigenetics Chromatin. 2016;9:32. doi: 10.1186/s13072-016-0082-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang YV, Tang H, Gilmour DS. Identification in vivo of different rate-limiting steps associated with transcriptional activators in the presence and absence of a GAGA element. Mol Cell Biol. 2005;25(9):3543–3552. doi: 10.1128/MCB.25.9.3543-3552.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Blythe SA, Wieschaus EF. Zygotic genome activation triggers the DNA replication checkpoint at the midblastula transition. Cell. 2015;160(6):1169–1181. doi: 10.1016/j.cell.2015.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schulz KN, Bondra ER, Moshe A, Villalta JE, Lieb JD, Kaplan T, McKay DJ, Harrison MM. Zelda is differentially required for chromatin accessibility, transcription factor binding, and gene expression in the early Drosophila embryo. Genome Res. 2015;25(11):1715–1726. doi: 10.1101/gr.192682.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gaskill MM, Gibson TJ, Larson ED, Harrison MM (2020) The pioneer factor GAF is essential for zygotic genome activation and chromatin accessibility in the early <em>Drosophila</em> embryo. bioRxiv:2020.2007.2015.204248. doi:10.1101/2020.07.15.204248 [DOI] [PMC free article] [PubMed]
- 52.Lomaev D, Mikhailova A, Erokhin M, Shaposhnikov AV, Moresco JJ, Blokhina T, Wolle D, Aoki T, Ryabykh V, Yates JR, 3rd, Shidlovskii YV, Georgiev P, Schedl P, Chetverina D. The GAGA factor regulatory network: identification of GAGA factor associated proteins. PLoS ONE. 2017;12(3):e0173602. doi: 10.1371/journal.pone.0173602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Benyajati C, Mueller L, Xu N, Pappano M, Gao J, Mosammaparast M, Conklin D, Granok H, Craig C, Elgin S. Multiple isoforms of GAGA factor, a critical component of chromatin structure. Nucleic Acids Res. 1997;25(16):3345–3353. doi: 10.1093/nar/25.16.3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Soeller WC, Oh CE, Kornberg TB. Isolation of cDNAs encoding the Drosophila GAGA transcription factor. Mol Cell Biol. 1993;13(12):7961–7970. doi: 10.1128/mcb.13.12.7961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Adkins NL, Hagerman TA, Georgel P. GAGA protein: a multi-faceted transcription factor. Biochem Cell Biol = Biochimie et biologie cellulaire. 2006;84(4):559–567. doi: 10.1139/o06-062. [DOI] [PubMed] [Google Scholar]
- 56.Wilkins RC, Lis JT. GAGA factor binding to DNA via a single trinucleotide sequence element. Nucleic Acids Res. 1998;26(11):2672–2678. doi: 10.1093/nar/26.11.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Omichinski JG, Pedone PV, Felsenfeld G, Gronenborn AM, Clore GM. The solution structure of a specific GAGA factor-DNA complex reveals a modular binding mode. Nat Struct Biol. 1997;4(2):122–132. doi: 10.1038/nsb0297-122. [DOI] [PubMed] [Google Scholar]
- 58.Pedone PV, Ghirlando R, Clore GM, Gronenborn AM, Felsenfeld G, Omichinski JG. The single Cys2-His2 zinc finger domain of the GAGA protein flanked by basic residues is sufficient for high-affinity specific DNA binding. Proc Natl Acad Sci USA. 1996;93(7):2822–2826. doi: 10.1073/pnas.93.7.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Espinas ML, Jimenez-Garcia E, Vaquero A, Canudas S, Bernues J, Azorin F. The N-terminal POZ domain of GAGA mediates the formation of oligomers that bind DNA with high affinity and specificity. J Biolo Chem. 1999;274(23):16461–16469. doi: 10.1074/jbc.274.23.16461. [DOI] [PubMed] [Google Scholar]
- 60.Katsani KR, Hajibagheri MA, Verrijzer CP. Co-operative DNA binding by GAGA transcription factor requires the conserved BTB/POZ domain and reorganizes promoter topology. EMBO J. 1999;18(3):698–708. doi: 10.1093/emboj/18.3.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bardwell VJ, Treisman R. The POZ domain: a conserved protein-protein interaction motif. Genes Dev. 1994;8(14):1664–1677. doi: 10.1101/gad.8.14.1664. [DOI] [PubMed] [Google Scholar]
- 62.Bonchuk A, Denisov S, Georgiev P, Maksimenko O. Drosophila BTB/POZ domains of "ttk group" can form multimers and selectively interact with each other. J Mol Biol. 2011;412(3):423–436. doi: 10.1016/j.jmb.2011.07.052. [DOI] [PubMed] [Google Scholar]
- 63.Pagans S, Ortiz-Lombardia M, Espinas ML, Bernues J, Azorin F. The Drosophila transcription factor tramtrack (TTK) interacts with Trithorax-like (GAGA) and represses GAGA-mediated activation. Nucleic Acids Res. 2002;30(20):4406–4413. doi: 10.1093/nar/gkf570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shimojima T, Okada M, Nakayama T, Ueda H, Okawa K, Iwamatsu A, Handa H, Hirose S. Drosophila FACT contributes to Hox gene expression through physical and functional interactions with GAGA factor. Genes Dev. 2003;17(13):1605–1616. doi: 10.1101/gad.1086803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xiao H, Sandaltzopoulos R, Wang HM, Hamiche A, Ranallo R, Lee KM, Fu D, Wu C. Dual functions of largest NURF subunit NURF301 in nucleosome sliding and transcription factor interactions. Mol Cell. 2001;8(3):531–543. doi: 10.1016/S1097-2765(01)00345-8. [DOI] [PubMed] [Google Scholar]
- 66.Greenberg AJ, Schedl P. GAGA factor isoforms have distinct but overlapping functions in vivo. Mol Cell Biol. 2001;21(24):8565–8574. doi: 10.1128/MCB.21.24.8565-8574.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jackson SP, Tjian R. O-glycosylation of eukaryotic transcription factors: implications for mechanisms of transcriptional regulation. Cell. 1988;55(1):125–133. doi: 10.1016/0092-8674(88)90015-3. [DOI] [PubMed] [Google Scholar]
- 68.Bonet C, Fernandez I, Aran X, Bernues J, Giralt E, Azorin F. The GAGA protein of Drosophila is phosphorylated by CK2. J Mol Biol. 2005;351(3):562–572. doi: 10.1016/j.jmb.2005.06.039. [DOI] [PubMed] [Google Scholar]
- 69.Aran-Guiu X, Ortiz-Lombardia M, Oliveira E, Bonet Costa C, Odena MA, Bellido D, Bernues J. Acetylation of GAGA factor modulates its interaction with DNA. Biochemistry. 2010;49(43):9140–9151. doi: 10.1021/bi1004427. [DOI] [PubMed] [Google Scholar]
- 70.Gross DS, Garrard WT. Nuclease hypersensitive sites in chromatin. Annu Rev Biochem. 1988;57:159–197. doi: 10.1146/annurev.bi.57.070188.001111. [DOI] [PubMed] [Google Scholar]
- 71.Weisbrod S. Active chromatin. Nature. 1982;297(5864):289–295. doi: 10.1038/297289a0. [DOI] [PubMed] [Google Scholar]
- 72.Talbert PB, Henikoff S. Histone variants on the move: substrates for chromatin dynamics. Nat Rev Mol Cell Biol. 2017;18(2):115–126. doi: 10.1038/nrm.2016.148. [DOI] [PubMed] [Google Scholar]
- 73.Iwafuchi-Doi M, Zaret KS. Pioneer transcription factors in cell reprogramming. Genes Dev. 2014;28(24):2679–2692. doi: 10.1101/gad.253443.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hamm DC, Harrison MM. Regulatory principles governing the maternal-to-zygotic transition: insights from Drosophila melanogaster. Open Biol. 2018;8(12):180183. doi: 10.1098/rsob.180183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McDaniel SL, Gibson TJ, Schulz KN, Fernandez Garcia M, Nevil M, Jain SU, Lewis PW, Zaret KS, Harrison MM. Continued activity of the pioneer factor zelda is required to drive zygotic genome activation. Mol Cell. 2019;74(1):185–195 e184. doi: 10.1016/j.molcel.2019.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tsukiyama T, Wu C. Purification and properties of an ATP-dependent nucleosome remodeling factor. Cell. 1995;83(6):1011–1020. doi: 10.1016/0092-8674(95)90216-3. [DOI] [PubMed] [Google Scholar]
- 77.Tsukiyama T, Daniel C, Tamkun J, Wu C. ISWI, a member of the SWI2/SNF2 ATPase family, encodes the 140 kDa subunit of the nucleosome remodeling factor. Cell. 1995;83(6):1021–1026. doi: 10.1016/0092-8674(95)90217-1. [DOI] [PubMed] [Google Scholar]
- 78.Ito T, Bulger M, Pazin MJ, Kobayashi R, Kadonaga JT. ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell. 1997;90(1):145–155. doi: 10.1016/S0092-8674(00)80321-9. [DOI] [PubMed] [Google Scholar]
- 79.Emelyanov AV, Vershilova E, Ignatyeva MA, Pokrovsky DK, Lu X, Konev AY, Fyodorov DV. Identification and characterization of ToRC, a novel ISWI-containing ATP-dependent chromatin assembly complex. Genes Dev. 2012;26(6):603–614. doi: 10.1101/gad.180604.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chalkley GE, Moshkin YM, Langenberg K, Bezstarosti K, Blastyak A, Gyurkovics H, Demmers JA, Verrijzer CP. The transcriptional coactivator SAYP is a trithorax group signature subunit of the PBAP chromatin remodeling complex. Mol Cell Biol. 2008;28(9):2920–2929. doi: 10.1128/MCB.02217-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mohrmann L, Langenberg K, Krijgsveld J, Kal AJ, Heck AJ, Verrijzer CP. Differential targeting of two distinct SWI/SNF-related Drosophila chromatin-remodeling complexes. Mol Cell Biol. 2004;24(8):3077–3088. doi: 10.1128/MCB.24.8.3077-3088.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nakayama T, Shimojima T, Hirose S. The PBAP remodeling complex is required for histone H3.3 replacement at chromatin boundaries and for boundary functions. Development. 2012;139(24):4582–4590. doi: 10.1242/dev.083246. [DOI] [PubMed] [Google Scholar]
- 83.Judd J, Duarte FM, Lis JT (2020) Pioneer factor GAF cooperates with PBAP and NURF to regulate transcription. bioRxiv:2020.2005.2010.087262. doi:10.1101/2020.05.10.087262 [DOI] [PMC free article] [PubMed]
- 84.Gilchrist DA, Dos Santos G, Fargo DC, Xie B, Gao Y, Li L, Adelman K. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell. 2010;143(4):540–551. doi: 10.1016/j.cell.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schulz KN, Harrison MM. Mechanisms regulating zygotic genome activation. Nat Rev Genet. 2019;20(4):221–234. doi: 10.1038/s41576-018-0087-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vastenhouw NL, Cao WX, Lipshitz HD. The maternal-to-zygotic transition revisited. Development. 2019;146(11):1. doi: 10.1242/dev.161471. [DOI] [PubMed] [Google Scholar]
- 87.Hirschhorn JN, Brown SA, Clark CD, Winston F. Evidence that SNF2/SWI2 and SNF5 activate transcription in yeast by altering chromatin structure. Genes Dev. 1992;6(12A):2288–2298. doi: 10.1101/gad.6.12a.2288. [DOI] [PubMed] [Google Scholar]
- 88.Schwabish MA, Struhl K. The Swi/Snf complex is important for histone eviction during transcriptional activation and RNA polymerase II elongation in vivo. Mol Cell Biol. 2007;27(20):6987–6995. doi: 10.1128/MCB.00717-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zandi E, Tran TN, Chamberlain W, Parker CS. Nuclear entry, oligomerization, and DNA binding of the Drosophila heat shock transcription factor are regulated by a unique nuclear localization sequence. Genes Dev. 1997;11(10):1299–1314. doi: 10.1101/gad.11.10.1299. [DOI] [PubMed] [Google Scholar]
- 90.Kyrchanova O, Maksimenko O, Ibragimov A, Sokolov V, Postika N, Lukyanova M, Schedl P, Georgiev P. The insulator functions of the Drosophila polydactyl C2H2 zinc finger protein CTCF: necessity versus sufficiency. Science Adv. 2020;6(13):eaaz3152. doi: 10.1126/sciadv.aaz3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Harrison MM, Li XY, Kaplan T, Botchan MR, Eisen MB. Zelda binding in the early Drosophila melanogaster embryo marks regions subsequently activated at the maternal-to-zygotic transition. PLoS Genet. 2011;7(10):e1002266. doi: 10.1371/journal.pgen.1002266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chopra VS, Srinivasan A, Kumar RP, Mishra K, Basquin D, Docquier M, Seum C, Pauli D, Mishra RK. Transcriptional activation by GAGA factor is through its direct interaction with dmTAF3. Dev Biol. 2008;317(2):660–670. doi: 10.1016/j.ydbio.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 93.Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, Li Y, Hao YL, Ooi CE, Godwin B, Vitols E, Vijayadamodar G, Pochart P, Machineni H, Welsh M, Kong Y, Zerhusen B, Malcolm R, Varrone Z, Collis A, Minto M, Burgess S, McDaniel L, Stimpson E, Spriggs F, Williams J, Neurath K, Ioime N, Agee M, Voss E, Furtak K, Renzulli R, Aanensen N, Carrolla S, Bickelhaupt E, Lazovatsky Y, DaSilva A, Zhong J, Stanyon CA, Finley RL, Jr, White KP, Braverman M, Jarvie T, Gold S, Leach M, Knight J, Shimkets RA, McKenna MP, Chant J, Rothberg JM. A protein interaction map of Drosophila melanogaster. Science. 2003;302(5651):1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- 94.Mason PB, Jr, Lis JT. Cooperative and competitive protein interactions at the hsp70 promoter. J Biol Chem. 1997;272(52):33227–33233. doi: 10.1074/jbc.272.52.33227. [DOI] [PubMed] [Google Scholar]
- 95.Oh H, Slattery M, Ma L, Crofts A, White KP, Mann RS, Irvine KD. Genome-wide association of Yorkie with chromatin and chromatin-remodeling complexes. Cell Rep. 2013;3(2):309–318. doi: 10.1016/j.celrep.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bayarmagnai B, Nicolay BN, Islam AB, Lopez-Bigas N, Frolov MV. Drosophila GAGA factor is required for full activation of the dE2f1-Yki/Sd transcriptional program. Cell Cycle. 2012;11(22):4191–4202. doi: 10.4161/cc.22486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kaul A, Schuster E, Jennings BH. The Groucho co-repressor is primarily recruited to local target sites in active chromatin to attenuate transcription. PLoS Genet. 2014;10(8):e1004595. doi: 10.1371/journal.pgen.1004595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yeung K, Boija A, Karlsson E, Holmqvist PH, Tsatskis Y, Nisoli I, Yap D, Lorzadeh A, Moksa M, Hirst M, Aparicio S, Fanto M, Stenberg P, Mannervik M, McNeill H. Atrophin controls developmental signaling pathways via interactions with Trithorax-like. eLife. 2017 doi: 10.7554/eLife.23084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Galloni M, Gyurkovics H, Schedl P, Karch F. The bluetail transposon: evidence for independent cis-regulatory domains and domain boundaries in the bithorax complex. EMBO J. 1993;12(3):1087–1097. doi: 10.1002/j.1460-2075.1993.tb05750.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gyurkovics H, Gausz J, Kummer J, Karch F. A new homeotic mutation in the Drosophila bithorax complex removes a boundary separating two domains of regulation. EMBO J. 1990;9(8):2579–2585. doi: 10.1002/j.1460-2075.1990.tb07439.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hagstrom K, Muller M, Schedl P. Fab-7 functions as a chromatin domain boundary to ensure proper segment specification by the Drosophila bithorax complex. Genes Dev. 1996;10(24):3202–3215. doi: 10.1101/gad.10.24.3202. [DOI] [PubMed] [Google Scholar]
- 102.Karch F, Galloni M, Sipos L, Gausz J, Gyurkovics H, Schedl P. Mcp and Fab-7: molecular analysis of putative boundaries of cis-regulatory domains in the bithorax complex of Drosophila melanogaster. Nucleic Acids Res. 1994;22(15):3138–3146. doi: 10.1093/nar/22.15.3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mihaly J, Hogga I, Gausz J, Gyurkovics H, Karch F. In situ dissection of the Fab-7 region of the bithorax complex into a chromatin domain boundary and a Polycomb-response element. Development. 1997;124(9):1809–1820. doi: 10.1242/dev.124.9.1809. [DOI] [PubMed] [Google Scholar]
- 104.Zhou J, Barolo S, Szymanski P, Levine M. The Fab-7 element of the bithorax complex attenuates enhancer-promoter interactions in the Drosophila embryo. Genes Dev. 1996;10(24):3195–3201. doi: 10.1101/gad.10.24.3195. [DOI] [PubMed] [Google Scholar]
- 105.Barges S, Mihaly J, Galloni M, Hagstrom K, Muller M, Shanower G, Schedl P, Gyurkovics H, Karch F. The Fab-8 boundary defines the distal limit of the bithorax complex iab-7 domain and insulates iab-7 from initiation elements and a PRE in the adjacent iab-8 domain. Development. 2000;127(4):779–790. doi: 10.1242/dev.127.4.779. [DOI] [PubMed] [Google Scholar]
- 106.Zhou J, Ashe H, Burks C, Levine M. Characterization of the transvection mediating region of the abdominal-B locus in Drosophila. Development. 1999;126(14):3057–3065. doi: 10.1242/dev.126.14.3057. [DOI] [PubMed] [Google Scholar]
- 107.Chetverina D, Aoki T, Erokhin M, Georgiev P, Schedl P. Making connections: insulators organize eukaryotic chromosomes into independent cis-regulatory networks. BioEssays News Rev Mole Cell Dev Biol. 2014;36(2):163–172. doi: 10.1002/bies.201300125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kyrchanova O, Mogila V, Wolle D, Magbanua JP, White R, Georgiev P, Schedl P. The boundary paradox in the Bithorax complex. Mech Dev. 2015;138(Pt 2):122–132. doi: 10.1016/j.mod.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Maeda RK, Karch F. The open for business model of the bithorax complex in Drosophila. Chromosoma. 2015;124(3):293–307. doi: 10.1007/s00412-015-0522-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mihaly J, Barges S, Sipos L, Maeda R, Cleard F, Hogga I, Bender W, Gyurkovics H, Karch F. Dissecting the regulatory landscape of the Abd-B gene of the bithorax complex. Development. 2006;133(15):2983–2993. doi: 10.1242/dev.02451. [DOI] [PubMed] [Google Scholar]
- 111.Schweinsberg SE, Schedl P. Developmental modulation of Fab-7 boundary function. Development. 2004;131(19):4743–4749. doi: 10.1242/dev.01343. [DOI] [PubMed] [Google Scholar]
- 112.Kyrchanova O, Wolle D, Sabirov M, Kurbidaeva A, Aoki T, Maksimenko O, Kyrchanova M, Georgiev P, Schedl P. Distinct elements confer the blocking and bypass functions of the Bithorax fab-8 boundary. Genetics. 2019;213(3):865–876. doi: 10.1534/genetics.119.302694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kaye EG, Kurbidaeva A, Wolle D, Aoki T, Schedl P, Larschan E. Drosophila dosage compensation loci associate with a boundary-forming insulator complex. Mol Cell Biol. 2017 doi: 10.1128/MCB.00253-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Pai CY, Lei EP, Ghosh D, Corces VG. The centrosomal protein CP190 is a component of the gypsy chromatin insulator. Mol Cell. 2004;16(5):737–748. doi: 10.1016/j.molcel.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 115.Gause M, Morcillo P, Dorsett D. Insulation of enhancer-promoter communication by a gypsy transposon insert in the Drosophila cut gene: cooperation between suppressor of hairy-wing and modifier of mdg4 proteins. Mol Cell Biol. 2001;21(14):4807–4817. doi: 10.1128/MCB.21.14.4807-4817.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ghosh D, Gerasimova TI, Corces VG. Interactions between the Su(Hw) and Mod(mdg4) proteins required for gypsy insulator function. EMBO J. 2001;20(10):2518–2527. doi: 10.1093/emboj/20.10.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Brown JL, Mucci D, Whiteley M, Dirksen ML, Kassis JA. The Drosophila Polycomb group gene pleiohomeotic encodes a DNA binding protein with homology to the transcription factor YY1. Mol Cell. 1998;1(7):1057–1064. doi: 10.1016/s1097-2765(00)80106-9. [DOI] [PubMed] [Google Scholar]
- 118.Dejardin J, Cavalli G. Chromatin inheritance upon Zeste-mediated Brahma recruitment at a minimal cellular memory module. EMBO J. 2004;23(4):857–868. doi: 10.1038/sj.emboj.7600108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Shokri L, Inukai S, Hafner A, Weinand K, Hens K, Vedenko A, Gisselbrecht SS, Dainese R, Bischof J, Furger E, Feuz JD, Basler K, Deplancke B, Bulyk ML. A Comprehensive Drosophila melanogaster transcription factor interactome. Cell Rep. 2019;27(3):955–970957. doi: 10.1016/j.celrep.2019.03.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Mahmoudi T, Zuijderduijn LM, Mohd-Sarip A, Verrijzer CP. GAGA facilitates binding of Pleiohomeotic to a chromatinized Polycomb response element. Nucleic Acids Res. 2003;31(14):4147–4156. doi: 10.1093/nar/gkg479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Schwendemann A, Lehmann M. Pipsqueak and GAGA factor act in concert as partners at homeotic and many other loci. Proc Natl Acad Sci USA. 2002;99(20):12883–12888. doi: 10.1073/pnas.202341499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Faucheux M, Roignant JY, Netter S, Charollais J, Antoniewski C, Theodore L. batman Interacts with polycomb and trithorax group genes and encodes a BTB/POZ protein that is included in a complex containing GAGA factor. Mol Cell Biol. 2003;23(4):1181–1195. doi: 10.1128/MCB.23.4.1181-1195.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Mishra K, Chopra VS, Srinivasan A, Mishra RK. Trl-GAGA directly interacts with lola like and both are part of the repressive complex of Polycomb group of genes. Mech Dev. 2003;120(6):681–689. doi: 10.1016/S0925-4773(03)00046-7. [DOI] [PubMed] [Google Scholar]
- 124.Espinas ML, Canudas S, Fanti L, Pimpinelli S, Casanova J, Azorin F. The GAGA factor of Drosophila interacts with SAP18, a Sin3-associated polypeptide. EMBO Rep. 2000;1(3):253–259. doi: 10.1093/embo-reports/kvd046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Salvaing J, Lopez A, Boivin A, Deutsch JS, Peronnet F. The Drosophila Corto protein interacts with Polycomb-group proteins and the GAGA factor. Nucleic Acids Res. 2003;31(11):2873–2882. doi: 10.1093/nar/gkg381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Guruharsha KG, Rual JF, Zhai B, Mintseris J, Vaidya P, Vaidya N, Beekman C, Wong C, Rhee DY, Cenaj O, McKillip E, Shah S, Stapleton M, Wan KH, Yu C, Parsa B, Carlson JW, Chen X, Kapadia B, VijayRaghavan K, Gygi SP, Celniker SE, Obar RA, Artavanis-Tsakonas S. A protein complex network of Drosophila melanogaster. Cell. 2011;147(3):690–703. doi: 10.1016/j.cell.2011.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lehmann M, Siegmund T, Lintermann KG, Korge G. The pipsqueak protein of Drosophila melanogaster binds to GAGA sequences through a novel DNA-binding domain. J Biol Chem. 1998;273(43):28504–28509. doi: 10.1074/jbc.273.43.28504. [DOI] [PubMed] [Google Scholar]
- 128.Kasinathan S, Orsi GA, Zentner GE, Ahmad K, Henikoff S. High-resolution mapping of transcription factor binding sites on native chromatin. Nat Methods. 2014;11(2):203–209. doi: 10.1038/nmeth.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Huang DH, Chang YL, Yang CC, Pan IC, King B. pipsqueak encodes a factor essential for sequence-specific targeting of a polycomb group protein complex. Mol Cell Biol. 2002;22(17):6261–6271. doi: 10.1128/MCB.22.17.6261-6271.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wang L, Ding L, Jones CA, Jones RS. Drosophila enhancer of zeste protein interacts with dSAP18. Gene. 2002;285(1–2):119–125. doi: 10.1016/s0378-1119(02)00418-3. [DOI] [PubMed] [Google Scholar]
- 131.King HW, Klose RJ. The pioneer factor OCT4 requires the chromatin remodeller BRG1 to support gene regulatory element function in mouse embryonic stem cells. eLife. 2017 doi: 10.7554/eLife.22631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hainer SJ, Boskovic A, McCannell KN, Rando OJ, Fazzio TG. Profiling of pluripotency factors in single cells and early embryos. Cell. 2019;177(5):1319–1329 e1311. doi: 10.1016/j.cell.2019.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kumar S. Remote homologue identification of Drosophila GAGA factor in mouse. Bioinformation. 2011;7(1):29–32. doi: 10.6026/97320630007029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Matharu NK, Hussain T, Sankaranarayanan R, Mishra RK. Vertebrate homologue of Drosophila GAGA factor. J Mol Biol. 2010;400(3):434–447. doi: 10.1016/j.jmb.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 135.Srivastava A, Kumar AS, Mishra RK. Vertebrate GAF/ThPOK: emerging functions in chromatin architecture and transcriptional regulation. Cell Mol Life Sci CMLS. 2018;75(4):623–633. doi: 10.1007/s00018-017-2633-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ciucci T, Vacchio MS, Gao Y, Tomassoni Ardori F, Candia J, Mehta M, Zhao Y, Tran B, Pepper M, Tessarollo L, McGavern DB, Bosselut R. The emergence and functional fitness of memory CD4(+) T cells require the transcription factor thpok. Immunity. 2019;50(1):91–105 e104. doi: 10.1016/j.immuni.2018.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Luckey MA, Kimura MY, Waickman AT, Feigenbaum L, Singer A, Park JH. The transcription factor ThPOK suppresses Runx3 and imposes CD4(+) lineage fate by inducing the SOCS suppressors of cytokine signaling. Nat Immunol. 2014;15(7):638–645. doi: 10.1038/ni.2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Vasanthi D, Anant M, Srivastava S, Mishra RK. A functionally conserved boundary element from the mouse HoxD locus requires GAGA factor in Drosophila. Development. 2010;137(24):4239–4247. doi: 10.1242/dev.058701. [DOI] [PubMed] [Google Scholar]
- 139.Srivastava A, Mishra RK. Interactome of vertebrate GAF/ThPOK reveals its diverse functions in gene regulation and DNA repair. J Biosci. 2020;45:1. doi: 10.1007/s12038-020-0014-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data are within the paper and its Supporting Information file.