Abstract
Arginine methylation is a posttranslational modification that affects many cellular processes in eukaryotes. The malaria parasite Plasmodium falciparum encodes three conserved protein arginine methyltransferases (PfPRMTs). We have determined that PfPRMT1 had authentic type I PRMT activity to form monomethylarginines and asymmetric dimethylarginines. Compared to mammalian PRMT1s, PfPRMT1 possesses a distinctive, ~50 amino acids longer N-terminal sequence, which is essential for enzyme activity. Recombinant PfPRMT1 methylated histones H4 and H2A, and several conserved substrates involved in RNA metabolism, including fibrillarin, poly(A) binding protein II, ribosomal protein S2, and a putative splicing factor. Using synthetic peptides and mass spectrometry, we determined target arginines in several substrates and studied the enzyme kinetics. While the kinetic parameters of recombinant PfPRMT1 on an H4 peptide and S-adenosylmethionine were similar to those of mammalian PRMT1s, PfPRMT1 had much higher substrate turnover rates. In the histone H4 N-terminus, PfPRMT1 could only methylate arginine 3, a mark for transcription activation. Western blots detected dynamic dimethylation of H4R3 during parasite development, suggesting that histone arginine methylation may play a conserved role in chromatin-mediated gene regulation. Consistent with the presence of potential substrates in both the cytoplasm and nucleus, green fluorescent protein-tagged PfPRMT1 and PfPRMT1 were localized in both cellular compartments with the majority in the cytoplasm. In vitro assays showed that PfPRMT1 could be inhibited by several small molecule inhibitors with 50% inhibitory concentrations in the sub-micromolar range. Most of these compounds also effectively inhibited parasite growth, suggesting that parasite PRMTs are promising targets for developing antiparasitic drugs.
Keywords: arginine methylation, enzyme activity, histones, transcription regulation, chemical inhibitors
INTRODUCTION
Protein arginine (R) methylation is a widespread posttranslational modification in eukaryotes, and its significance in regulating many cellular processes such as signal transduction, RNA processing and transport, and transcription has been increasingly appreciated [1–3]. Arginine methylation is catalyzed by a family of protein arginine methyltransferases (PRMTs), which add one or two methyl groups from S-adenosylmethionine (AdoMet) to the guanidine nitrogens of arginine, resulting in three major forms of methylated arginines: ω-NG, monomethylarginine (MMA), ω-NG, NG-asymmetric dimethylarginine (aDMA), or ω-NG, NG-symmetric dimethylarginine (sDMA). Four types of PRMTs have been described with type I and II enzymes as the main types in mammalian cells. Type I enzymes catalyze the formation of MMA and aDMA, while type II enzymes catalyze the formation of MMA and sDMA [3]. The human genome encodes at least 11 PRMTs, and PRMT1 (type I) and PRMT5 (type II) are evolutionarily conserved in most, if not all, eukaryotes [4, 5].
The many substrates identified for the type I enzymes often contain GAR (glycine and arginine-rich) motifs in the sequences of RGG, RG, or RXG [2, 6]. Among them, the best known RNA-binding proteins include hnRNPs, fibrillarin, nucleolin, and poly(A)-binding protein II (PABPII) which are involved in various aspects of RNA processing and transport [3]. Recently, histone arginine methylation has been recognized as an important means of transcription regulation. Three mammalian PRMTs have been identified to catalyze histone methylation. PRMT1 methylates R3 in histone H4, and to a lesser extent R3 in H2A [7]. Another type I enzyme PRMT4 or CARM1 (cofactor associated arginine methyltransferase 1) methylates R2, 17, 26 and several arginines in the C-terminus of H3 [8]. The type II enzyme PRMT5 methylates R8 in H3 and R3 in H4 [9]. In vivo, PRMTs often interact with other cofactors such as histone acetyltransferases and chromatin remodelers to regulate transcription [10, 11]. In mammals, the asymmetric arginine dimethylation in histones catalyzed by PRMT1 and CARM1 is associated with active genes, whereas PRMT5-mediated histone symmetric dimethylation is often linked to transcription repression [12].
The malaria parasite Plasmodium falciparum is responsible for more than one million deaths annually. Like other eukaryotes, P. falciparum genomic DNA is organized into nucleosomes involving both core and variant histones [13, 14]. In addition to general conservation of the basal transcription machinery in the parasite [15], the presence of a large number of chromatin-modifying proteins suggests a prominent role of chromatin-mediated mechanisms in transcription regulation [16]. While a deficiency of specific transcription factors in the parasite genome has been considered [17], the recent identification of a large family of AP2-domain proteins as transcription activators or repressors in several apicomplexan genomes has changed this view [18, 19]. Therefore, it is very likely that the malaria parasite uses both chromatin-based mechanisms and conventional DNA-binding transcriptional factors to regulate gene expression [20, 21].
The malaria parasite histones have many covalent modifications that play evolutionarily conserved roles in transcription regulation [14, 22]. Histone acetylation is generally associated with active transcription, whereas the H3K9 trimethylation is negatively correlated with global gene expression [23]. Specifically, in the control of monoallelic expression of the var gene families, H3K4 di- and tri-methylation and the heterochromatin marker H3K9 trimethylation are associated with active and silent var genes, respectively [24, 25]. Tandem mass spectrometry analysis of P. falciparum histones has revealed the presence of arginine methylation on H3 and H4 [14]. Besides, several PRMTs have recently been characterized in the protozoan parasite Trypanosoma brucei and Toxoplasma gondii [26–29]. In particular, H3R17 methylation, catalyzed by the CARM1-like PRMT in T. gondii has been attributed to gene activation [27]. These findings have prompted us to characterize the PRMTs in the malaria parasite and study their functions in transcription.
Here we report the identification and characterization of the PRMT1 homologue in P. falciparum, referred to as PfPRMT1. PfPRMT1 is an evolutionarily conserved protein with type I PRMT activity towards histone H4 and a number of GAR-motif proteins as potential substrates. PfPRMT1 is expressed in all asexual erythrocytic stages of the parasite and the protein is localized in both cytoplasm and nucleus. The inhibitory effect of general methyltransferase inhibitors on the parasite suggests that PRMTs could serve as potential targets for developing antimalarial drugs.
EXPERIMENTAL
Identification of potential PfPRMTs and substrates in the parasite genome
To identify PRMTs in the Plasmodium genomes, we used the BLASTP algorithm to search the malaria database (http://www.plasmoDB.org) using the protein sequences of human PRMTs [4]. Protein pattern and architecture were further researched using a Hidden Markov Model (HMM) embedded in the SMART program (http://smart.embl.de). To identify the protein homologues for known PRMT substrates, we used the human fibrillarin (CAA39935), PABPII (AAH10939), and ribosomal protein S2 (rpS2) (NP_002943), and Saccharomyces cerevisiae Gar1p (AAB68929) to search PlasmoDB using BLASTP.
Protein expression and purification
Full-length cDNA encoding PfPRMT1 was amplified by polymerase chain reaction (PCR) with primer pairs PfPRMT1-F1/PfPRMT1-R1 (Supplemental Table 1) and cloned at the BamHI and SalI sites of the vector pMAL-c2x (New England Biolabs) and pGEX-6P-1 to produce maltose binding protein (MBP)-PfPRMT1 and glutathione S-transferase (GST)-PfPRMT1, respectively. Recombinant protein was expressed in bacterial strain BL21 after induction with 0.3 mM of IPTG for 3 h at 37°C. For purification of recombinant MBP-PfPRMT1, bacteria were resuspended in amylose column buffer (20 mM Tris, pH 7.5, 200 mM NaCl, 2 mM dithiothreitol [DTT], 1 mM PMSF) and lysed by sonication (four pulses of 40 s each). The lysate was clarified by centrifugation at 14,000 × g for 20 min at 4°C. The supernatant was diluted (1:5 with column buffer) and loaded to an amylose resin column. The column was washed with 20 column volumes of amylose column buffer. The MBP-PfPRMT1 fusion protein was eluted with column buffer containing 10 mM maltose. Similarly, GST-PfPRMT1 was purified using a glutathione Sepharose 4B column (GE Healthcare) as described previously [22]. PfPRMT1 truncations corresponding to amino acids (aa) 35-401, 56-401, and 1 to 348 were amplified with primer pairs PfPRMT1-F7/PfPRMT1-R1, PfPRMT1-F4/PfPRMT1-R1, and PfPRMT1-F1/PfPRMT1-R7, respectively (Supplemental Table 1), and cloned into the pGEX-6P-1 at the BamHI and SalI sites to produce GST fusion proteins. Tag-free PfPRMT1 was cleaved off the column by using PreScission Protease (GE Healthcare). All purified recombinant proteins were dialyzed twice in 1 L of 50 mM sodium phosphate buffer (pH 8.0) containing 2 mM DTT and 10% glycerol at 4°C. When necessary, recombinant proteins were concentrated by using MICROCON Centrifugal Filter Devices YM-10 (Millipore). Purified recombinant PfPRMT1 was kept at −70°C for binding and enzyme activity assays. For antibody production, full-length PfPRMT1 was cloned into the pET28a (+) vector (Novagen) at BamHI and SalI sites. Recombinant PfPRMT1 was purified with Ni-NTA resin (Qiagen), and dialyzed in PBS. The purified recombinant PfPRMT1 was used to produce polyclonal rabbit anti-PfPRMT1 antiserum (Proteintech Group Inc). This antiserum was used directly in subsequent experiments without purification of the antibodies.
To identify potential methylation substrates for PfPRMT1, we have selected nine GAR-motif-containing P. falciparum genes and expressed the full-length or truncated proteins fused to either GST or MBP as described above using primers listed in Supplemental Table 1. These are PABPII (PFI1175c), fibrillarin (PF14_0068), putative PABP (MAL13P1.303), Gar1-like homologue (PF13_0051), rpS2 (PF14_0448), and four splicing factor-like proteins (PFF1135w, PF10_0068, PF10_0217, and PFE0865c). Purified recombinant proteins were used in methylation assays with MBP-PfPRMT1.
In vitro dimerization and AdoMet binding of recombinant PfPRMT1
To determine whether PfPRMT1 forms dimers and oligomers, purified recombinant protein (1 μM) was incubated without or with 0.025% glutaraldehyde in 50 mM sodium phosphate buffer (pH 8.0) at room temperature for 3 or 5 min [30]. The reactions were stopped by the addition of an equal volume of 2 × Laemmli sample buffer, and the proteins were separated by 10% SDS–polyacrylamide gel electrophoresis (PAGE), and then detected by immunoblotting with the polyclonal rabbit anti-PfPRMT1 antiserum (1:5000) and horse radish peroxidase-conjugated goat anti-rabbit IgG (1:3000).
To determine whether PfPRMT1 binds to AdoMet as the substrate, ~3 μg (1.2 μM) of recombinant MBP-PfPRMT1 or PfPRMT1 were incubated with 1.6 μM of [3H]AdoMet (82.0 Ci/mmol, GE Healthcare) in 30 μl of cross-linking buffer (50 mM sodium phosphate [pH 8.0], 5 mM MgCl2, 2 mM DTT) at 4°C overnight [26]. For competition experiments, a 1000-fold excess of unlabelled AdoMet or ATP (Sigma) was included the reaction. The reactions were placed on ice and exposed to UV irradiation (254 nm) for 60 min at a distance of 5 cm from the UV light. Afterwards, the proteins were resolved by 10% SDS-PAGE, stained with Coomassie blue, and processed for fluorography using an autoradiographic enhancer (Perkin Elmer). The gel was dried and exposed to X-ray film for five weeks.
In vitro methylation assay
In vitro methylation assay of PfPRMT1 activity was carried out in 20 μl of methylation assay buffer (50 mM sodium phosphate [pH 8.0], 5 mM MgCl2, 2 mM DTT), 1 μCi [3H]AdoMet, 2-4 μg of substrates, and 1.5 μg (0.6 μM) of recombinant MBP-PfPRMT1 at 30°C for 1 h. The reactions were stopped by the addition 4 × SDS-PAGE sample buffer and boiled for 5 min. Proteins were resolved by 10% or 15% SDS-PAGE, stained and processed for fluorography as described above.
Mass spectrometry analysis of substrate peptides and enzyme kinetics
To further determine the methylated arginines in potential substrates, two histone H4 peptides PfH4-21 (Ac-SGRGKGGKGLGKGGAKRHRKI) and PfH4-21R3K (Ac-SGKGKGGKGLGKGGAKRHRKI), and two GAR-motif peptides PFI1175c-16 (156-171) (KISPFRGRGMKSALGV) and PF14_0068-15 (44-58) (GRGGGGGGRGGGGG) were synthesized (GenScript Corporation). Methylation reaction was performed in 10 μl of methylation buffer, 250 μM AdoMet, 20 μM of each peptide, and 3 μg (1.2 μM) of recombinant MBP-PfPRMT1 at 37°C. Reactions were stopped at different time points by adding 0.5% trifluoroacetic acid (TFA) and methylation products were detected by mass spectrometry using the Micromass Matrix Assisted Laser Desorption Ionization Time of Flight (MALDI-TOF) mass spectrometer (Waters).
Enzyme kinetics was determined for the recombinant PfPRMT1 using a similar method as described [31]. To determine the kinetic parameters for the PfH4-21 peptide, the methylation reaction was performed with 1.1 μM of MBP-PfPRMT1, a constant amount of [3H]AdoMet (50 μM), and varying concentrations (2 - 20 μM) of PfH4-21 at 37°C for 5 min. For the substrate AdoMet, methylation reaction was performed with a constant amount of PfH4-21 peptide (50 μM) and varying concentrations of [3H]AdoMet (0.2 - 2 μM) at 37°C for 10 min. All reactions were performed in duplicate in a final volume of 10 μl, stopped by addition of 0.5% TFA, and spotted onto Whatman P-81 cation exchange filter. The radiolabeled products were processed for quantification using a liquid scintillation counter (LSC) as previously described [32]. Data for each point were fitted to the Michaelis–Menten plot to obtain the binding constant Km and catalytic turnover kcat values.
Parasite culture
The P. falciparum 3D7 clone was cultured as described [33]. For time-course studies, parasites were synchronized by two rounds of treatment of ring-stage parasites with 5% sorbitol, and then collected at ring, early trophozoite, late trophozoite and schizont stages [34]. Parasites were released by lysing the erythrocyte membrane with 0.05% saponin and washed twice with PBS.
In vivo PfPRMT1 expression and activity
Total RNA was extracted from the parasites using Trizol Reagent (Invitrogen) and treated with RNase-free DNase I [35]. The 5’ and 3’ ends of the PfPRMT1 mRNA were determined by using the FirstChoice RNA ligase-mediated rapid amplification of cDNA ends (RLM-RACE) kit (Ambion) with PfPRMT1-specific primers (Supplemental Table 2) [36]. RACE products were cloned in TOPO cloning vector (Invitrogen) and sequenced. The relative levels of PfPRMT1 expression were studied by real-time reverse transcriptase (RT)-PCR analysis at four stages of the P. falciparum asexual erythrocytic cycle using HotStart-IT SYBR Green One-Step qRT-PCR Master Mix Kit (USB) with primers PfPRMT1-F3 and PfPRMT1-R3 (Supplemental Table 2). The constitutively expressed seryl-tRNA synthetase gene (PF07_0073) was used as the internal reference. Data analysis and determination of the threshold cycle (Ct) values were performed as described previously [14].
To study PfPRMT1 protein expression during intraerythrocytic development cycle (IDC), synchronized parasites were lysed by sonication (three pulses of 10 sec each). Equal amounts of the parasite lysates (30 μg) at each stage were separated by 10% SDS–PAGE and transferred to nitrocellulose membranes. Western blots were performed using a standard procedure [37] with rabbit anti-PfPRMT1 antiserum (1:2000) as the primary antibodies and horse radish peroxidase-conjugated goat anti-rabbit IgG (1:3000) as the secondary antibodies. Blots were developed using the DAB substrate (Roche).
To determine PfPRMT1 activity in vivo, the endogenous PfPRMT1 was obtained by immunoprecipitation (IP) assay as described [40]. Briefly, 50 μl of protein A agarose (Sigma) was washed three times with an IP buffer (150 mM NaCl, 1% Triton X-100, 50 mM Tris, pH 8.0, 1 mM DTT, 0.2 mM PMSF), and then bound with 50 μl of preimmune or anti-PfPRMT1 antiserum. After washing with the IP buffer, the agarose beads were mixed with 300 μl of precleared lysate from 2 × 108 parasites. Afterwards, the beads were washed three times with the IP buffer, twice with the methylation assay buffer, and resuspended in methylation buffer to a total of 50% slurry. The agarose beads were directly assayed for methylation activity using recombinant PfH4 as the substrate.
Plasmid construction and parasite transfection
To generate green fluorescent protein (GFP)-tagged PfPRMT1, a construct containing PfPRMT1 amino acids 1-401 fused with a downstream GFP tag and pDT3 terminator was made in pBluescript. The resulting PfPRMT1-GFP-pDT3 cassette was then cloned into pHD22Y [38] at BamHI and NotI sites to obtain the final transfection construct pHD22Y-PfPRMT1-GFP. Parasite transfection was carried out by culturing the late stage schizonts with RBCs preloaded with 100 μg of plasmid DNA [39]. Parasites containing the plasmid were selected with 2.5 nM of WR99210 for about three weeks and replenished weekly with fresh RBCs. Parasites with stable chromosomal integration of the plasmid were enriched by two cycles of drug on-off selection, and single clones of transformed parasites were obtained by limiting dilution [40]. Integration of the plasmid at the PfPRMT1 locus was screened by integration-specific PCR and western blots. Primary antibodies were either anti-PfPRMT1 antiserum at 1:2000 or monoclonal anti-GFP antibody at 1:1000 (Roche).
Histone arginine methylation in parasites
To study histone arginine methylation, histones were isolated from the three IDC stages of synchronized parasites as described previously [14]. Equal amounts of proteins were separated by 18% SDS-PAGE analysis. Western blots were performed using anti-dimethylated H4R3 and anti-dimethylated H3R17 (Millipore). These antibodies do not react with recombinant P. falciparum histones expressed in E. coli (data not shown). As a loading control, the blot was probed with anti-H4 antibodies.
Subcellular localization of PfPRMT1
Immunofluorescence assay (IFA) was performed as described [41]. Briefly, infected RBCs were washed once with PBS buffer and cell pellet (~100 μl) was fixed with 1 ml of 4% paraformaldehyde and 0.0075% glutaraldehyde in PBS for 30 min. Fixed cells were washed twice with PBS and treated with 0.5% Triton X-100 in PBS for 10 min. Cells were washed twice with PBS and blocked in 3% BSA for one hour at room temperature. The polyclonal anti-PfPRMT1 antiserum (1:300) was added and incubated for another two hours. After washing the cells three times with PBS, FITC-conjugated anti-rabbit IgG antibodies (Sigma) were added at 1:100 in 3% BSA containing 4-6-diamidino-2-phenylindole (DAPI) and incubated for one hour. Images were captured using a Nikon Eclipse E600 epifluorescent microscope. Localization of GFP-tagged PfPRMT1 was determined similarly.
To estimate the distribution of PfPRMT1 in the cytoplasmic and nuclear compartments of the parasite, ~100 μl of parasite pellet was resuspended in 300 μl of a hypotonic buffer (10 mM HEPES, pH7.9, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, 0.5 mM EDTA and 1% protease inhibitor cocktail) and incubated on ice for 10 min. The parasites were mechanically lysed by 30~40 strokes in a Dounce homogenizer with pestle B and then centrifuged at 2,700 × g for 10 min at 4°C. The supernatant was used as the cytoplasmic extract and the pellet was resuspended in 300 μl of PBS as the nuclear extract. Equal proportions (1/15) of the protein extracts (20 μl) were resolved by 15% SDS-PAGE, stained by Coomassie blue and detected by immunoblotting using the anti-PfPRMT1 antiserum (1:2000) and anti-histone H4 antibodies (1:1000, Millipore) as a control. For a cytoplasmic protein, we used a rabbit antiserum against the recombinant His-tagged P. falciparum histone-like protein (PfHLP, PFI0230c), which was tested specific for PfHLP in the parasite (data not shown).
Small molecule inhibitors
To evaluate the potential effect of small molecule inhibitors on recombinant PfPRMT1 and parasite growth, the following compounds were selected: sinefungin, 5-deoxy-5-methylthioadenosine [MTA], AMI-5 (eosin), chaetocin, and adenosine dialdehyde (AdoX). To estimate the IC50 values of these compounds for PfPRMT1, 0.8 μM of MBP-PfPRMT1 and different concentrations of the inhibitors were pre-incubated at room temperature for 15 min in the methylation buffer. Subsequently, 6 μM of recombinant PfH4 and 0.3 μM of [3H]AdoMet were added and incubated at 30°C for 30 min. Reactions were stopped by incubating at 80°C for 10 min and spotted onto P-81 filter for LSC. Assays were performed in duplicates. To determine the effect of these chemicals for parasite growth, a luciferase-expressing strain 3D7-luc was used [42]. Late ring-stage parasites (100 μl) were seeded in triplicate in 96-well plates at 2% parasitemia and 2% hematocrit and incubated at 37°C for 48 h with different concentrations of the chemicals. IC50s were calculated by linear regression analysis [42].
RESULTS
Plasmodium encodes conserved PRMTs
BLASTP search of the P. falciparum genome in PlasmoDB has identified three PRMT candidates (Fig. 1). Putative PfPRMT1 (PF14_0242) and PfPRMT5 (PF13_0323) have been identified previously [4, 26]. PF08_0092 encodes a putative PRMT with all conserved methyltransferase motifs. It is probably a homologue of the mammalian CARM1 protein, since its homologue in T. gondii could methylate R17 in histone H3 [27]. No additional domains were identified in the PfPRMT proteins. Phylogenetic analysis clearly identified PF14_0242 and PF13_0323 as PRMT1 and PRMT5 homologues, respectively (Fig. 1B). These three PRMTs are conserved in all Plasmodium genomes sequenced so far.
Figure 1.
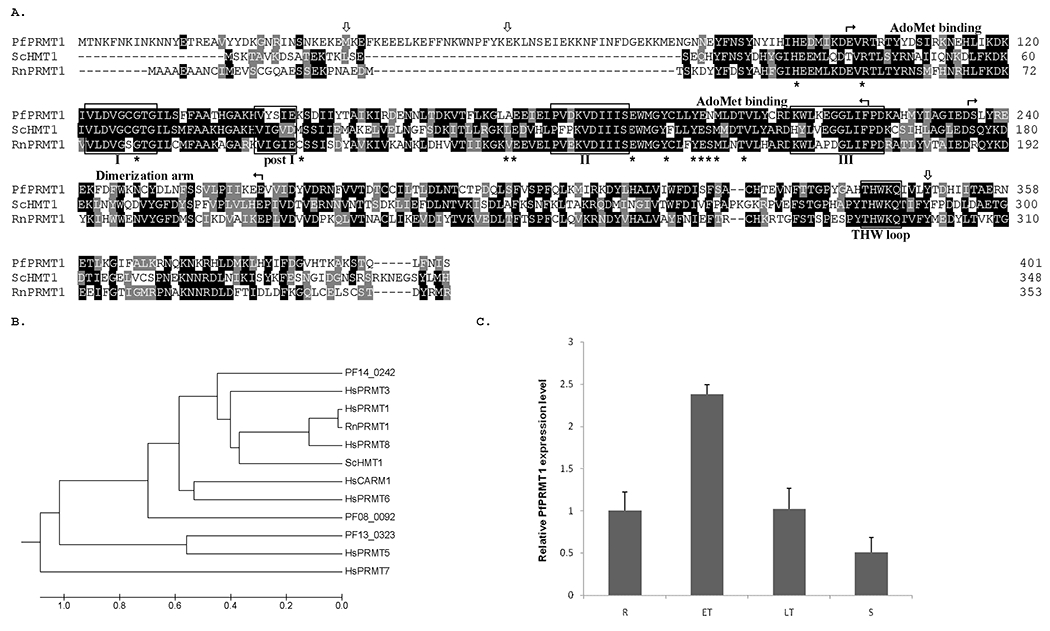
A. Alignment of PfPRMT1 with RnPRMT1 (NP_077339) and ScHMT1 (NP_009590). Identical and similar amino acids are shadowed in black and grey, respectively. The methyltransferase signature motifs I, post-I, II, III and the THW loop are boxed. Borders of the cofactor binding domain and dimerization arm are marked by arrows. Asterisks below the sequences indicate residues involved in AdoMet binding. Downward arrows indicate the sites of the truncation in PfPfPRMT1 for activity analysis. B. Phylogenetic analysis of Homo sapiens (Hs) PRMT1 (NP_001527), HsPRMT3 (AAH37544), HsCARM1 (Q86X55), HsPRMT5 (O14744), HsPRMT6 (AAH73866), HsPRMT7 (NP_061896), HsPRMT8 (AAH22458), ScHMT1, RnPRMT1 and three P. falciparum PRMTs. Shown here is the UPGMA tree constructed using MEGA program (http://www.megasoftware.net). C. Real-time RT-PCR determination of the relative expression levels of PfPRMT1 during the IDC of P. falciparum. R – ring, ET – early trophozoite, LT – late trophozoite, S – schizont.
PfPRMT1 has 401 amino acids with a predicted molecular weight of 47.4 kDa. Compared with the Rattus norvegicus (Rn) PRMT1 and the Saccharomyces cerevisiae (Sc) HMT1 (heterogeneous nuclear ribonucleoprotein methyltransferase 1), of which the structures have been solved [43, 44], PfPRMT1 is ~50 amino acids longer in the N-terminus (Fig. 1A). PfPRMT1 contains the conserved methyltransferase motifs I, post-I, II, III and THW loop. Most of the residues (10/13) involved in AdoMet binding are conserved [44]. In addition, the essential G68 for the yeast HMT1 activity, and the RnPRMT1 E144 and E153 involved in binding the guanidine group in the arginine substrate are also conserved in PfPRMT1. The dimerization domain shows a lesser degree of conservation with 62% amino acid identity.
Microarray data show that PfPRMT1 is expressed throughout the intraerythrocytic stages. Real-time RT-PCR analysis using RNA from synchronized parasites yielded a similar result, with the highest PfPRMT1 mRNA level being detected in early trophozoites and the lowest level in schizonts (Fig. 1C). To determine the transcription initiation site and polyadenylation sites of PfPRMT1 mRNA, RLM-RACE and 3’ RACE analyses were performed with RNA from asexual blood stage parasites. From a total of 18 clones sequenced for the 5’ RLM-RACE, the 5’ ends were mapped to −560 (8), −528 (6), −527 (1), −529 (1), −533 (1), and −537 bp (1) (number in parenthesis indicates number of clones sequenced) upstream of the putative ATG codon, indicating that PfPRMT1 has a relatively long 5’ untranslated region (5’UTR). Sequencing of 13 clones from 3’ RACE analysis detected two clusters of polyadenylation sites at 37 (1), 40 (6), 49 (1), and 363 (1), 364 (2), 365 bp (2) downstream of the stop codon. Therefore, the predicted PfPRMT1 mRNA is approximately 1.8-2.1 kb.
PfPRMT1 has conserved type I PRMT activity
Since PfPRMT1 has the conserved AdoMet-binding and dimerization motifs, we first tested whether these motifs are functional. Recombinant PfPRMT1 was expressed in bacteria and used in UV cross-linking experiments with radiolabeled AdoMet. The result showed that recombinant PfPRMT1 could bind to AdoMet (Fig. 2A). This binding was not affected by the addition of excessive amounts of non-substrate ATP, but was reduced to an undetectable level when 1000-fold excess of cold AdoMet was added in the cross-linking reaction. This suggests that the recombinant PfPRMT1 binds specifically to the cofactor AdoMet. The presence of the MBP fusion did not affect AdoMet binding.
Figure 2.
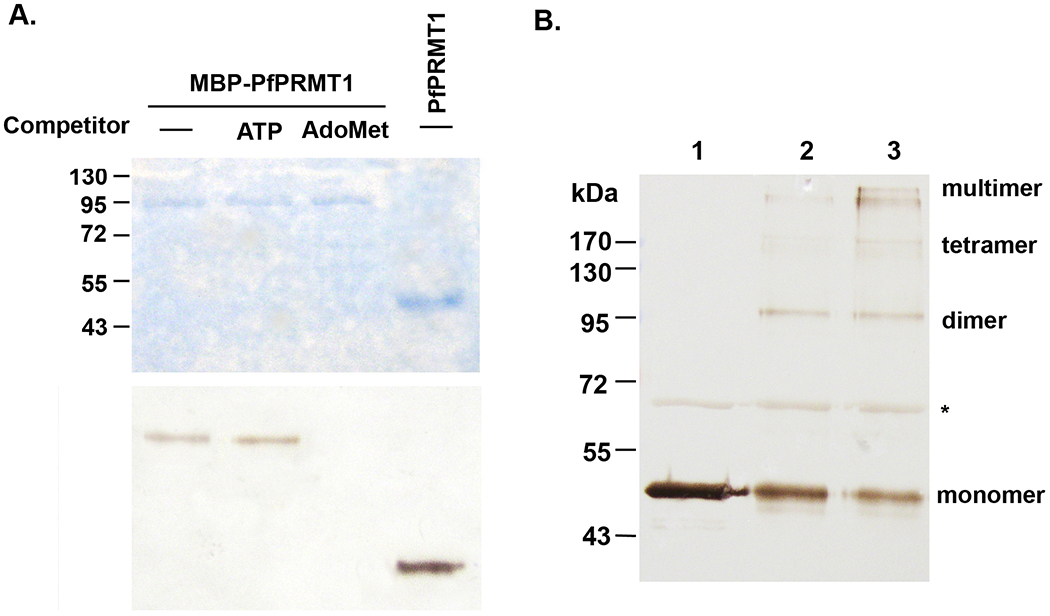
A. UV-crosslinking to detect specific binding of PfPRMT1 to AdoMet. Either recombinant MBP-PfPRMT1 (upper bands) or PfPRMT1 (lower band) was used in the binding reactions. 1000-fold excess of unlabeled ATP or AdoMet was included as the competitor. Upper panel shows Coomassie blue-stained gel, and lower panel shows the fluorograph. B. Oligomerization of recombinant PfPRMT1. Recombinant PfPRMT1 was incubated without (lane 1) or with 0.025% glutaraldehyde for 3 min (lane 2) and 5 min (lane 3), respectively. After cross-linking the proteins were separated in 10% SDS-PAGE and probed with the anti-PfPRMT1 antiserum. Monomers, dimers, tetramers, and multimers are indicated. The asterisk indicates a bacterial protein present in the purified PfPRMT1, which does not show detectable crosslinking under these conditions.
Oligomerization is important for enzymatic activity of PRMT1, 3 and 5 [44–46]. To determine whether PfPRMT1 could form homo-oligomers, purified PfPRMT1 was incubated with or without the crosslinking reagent glutaraldehyde. SDS-PAGE analysis indicated that untreated protein migrated at the predicted size of 47 kDa, corresponding to the monomer (Fig. 2B). In contrast, the treated samples showed proteins bands that migrated at sizes corresponding to homo-dimers, -tetramers, and -oligomers, respectively (Fig. 2B). As an internal control, a bacterial protein present in the sample did not form any oligomers. This suggests that despite the lesser degree of homology in the dimerization arm between PfPRMT1 and the mammalian PRMT1s, it has retained the ability to oligomerize.
Mammalian PRMT1 displays enzymatic activity towards histone H4 and H2A [7]. To determine whether the recombinant PfPRMT1 has arginine methyltransferase activity, we used purified recombinant P. falciparum core histones H2A, H2B, H3, and H4 in the methylation assay [22]. We have expressed and purified PfPRMT1 using different tags (GST, six histidines and MBP) and found that these tags did not interfere with the methylase activity (data not shown). In comparison to the MBP protein as the control, MBP-PfPRMT1 displayed robust methylase activity towards H4, and to a lesser extent, H2A (Fig. 3A). We then used specific antibodies against the monomethylated and asymmetrically dimethylated H4R3 and showed that recombinant PfPRMT1 indeed catalyzed H4R3 monomethylation and asymmetric dimethylation (Fig. 3B). To determine if PfPRMT1 from the parasite has a similar activity on histone H4, we performed IP of parasite lysate using preimmune serum or polyclonal antiserum against recombinant PfPRMT1. Precipitated proteins with the PfPRMT1 antiserum had authentic methyltransferase activity to methylate recombinant PfH4, whereas this activity was not observed with proteins precipitated with the preimmune serum (Fig. 3C).
Figure 3.

PfPRMT1 enzymatic activity on histones. A. Methylase activity of MBP-PfPRMT1 on histones. MBP-PfPRMT1 or MBP was used in methylation assays with recombinant P. falciparum core histones, and the reactions were separated by 15% SDS-PAGE. Left panel shows the Coomassie blue-stained gel, while the right panel shows the fluorograph. B. MBP-PfPRMT1 methylates H4R3. The same methylation reaction was performed using non-radioactive AdoMet and the blot was probed with polyclonal antibodies against asymmetrically dimethylated H4R3 (H4R3m2) and monomethylated H4R3 (H4R3m1). C. PfPRMT1 expressed in the parasite methylates H4. Parasite lysates were immunoprecipitated with anti-PfPRMT1 antiserum or preimmune serum, and the precipitated proteins were used in methylation reactions using recombinant PfH4 as the substrate. H4 methylation was detected by fluorography.
PRMT1 exhibits significant sequence variations in the first 40 amino acids at the N-terminus. In the rat PRMT1, deletion of the first 14 amino acids does not affect the enzymatic activity, whereas truncation to amino acid 38 abolishes enzymatic activity [44]. In the yeast HMT1, the first 15 amino acids are not essential, but deletion of the first 20 amino acids results in markedly reduced enzyme activity [43]. PfPRMT1 has ~50 extra amino acids in the N-terminus comparing with the yeast and rat homologues (Fig. 1A). To determine whether this sequence is important for the methyltransferase activity, we generated several truncations of PfPRMT1 and tested their enzyme activity using recombinant H4 as the substrate. We found that deletion of the first 34 amino acids had no effect on methyltransferase activity (Fig. 4). However, truncation to amino acid 56 completely abolished the enzyme activity. Similarly, the C-terminal 54 amino acids were also critical for the enzyme activity.
Figure 4.

The effect of terminal truncations on PfPRMT1 activity. Recombinant PfPRMT1s (full-length and different terminal truncations) were incubated with PfH4 in methylation reactions and the proteins were separated by SDS-PAGE. Upper panel shows the Coomassie blue-stained gel, and the lower panel the fluorograph.
PfPRMT1 has broad substrate specificity
In model eukaryotes, PRMT1 methylates a wide variety of substrates, including histones, transcription factors, and a number of proteins involved in RNA processing, transport and translation [1]. Among them, fibrillarin [47], PABPII [48, 49], Gar1p homologue [50], and rpS2 [51] are evolutionarily conserved in P. falciparum. In addition, we have also identified several potential splicing factors, which have putative RGG, RG, or RXG boxes. We wanted to determine whether these P. falciparum proteins could serve as the PfPRMT1 substrates in vitro. These proteins or their GAR-motif containing domains were expressed in E. coli as either GST- or MBP-fusion proteins (Supplemental Table 1), and were tested in methylation reactions using [3H]-labeled AdoMet. The results showed that fibrillarin (PF14_0068), PABPII (PFI1175c), rpS2 (PF14_0448), and a putative splicing factor (PFE0865c) could be methylated in vitro, indicating that they are potential physiological substrates for PfPRMT1 (Fig. 5). It is noteworthy that PfPRMT1 methylated the N-terminal polypeptides of fibrillarin (1-100 aa) and rpS2 (1-60 aa), which have extensive GAR motifs. Among the putative splicing factor-like proteins, PFE0865c could be methylated by PfPRMT1, suggesting that PfPRMT1 may affect pre-mRNA splicing events. PFE0865c is an arginine- and serine-rich protein with two RNA recognition motifs (RRMs). One typical RGG motif and three RG motifs are located in the N-terminus between the two RRMs, while the C-terminal 100 amino acids are highly enriched with serine (36/100 amino acids). To determine which region was methylated by PfPRMT1 in PFE0865c, we expressed the N-terminal RG-containing fragment (1-112 aa) and the C-terminal Ser-rich domain (199-298 aa) separately. The results showed that both regions could be methylated by PfPRMT1. Since the C-terminal domain contains only one RSG sequence, which was shown to be a very poor substrate for PRMT1, this result supported the notion that PRMT1 could methylate arginines beyond the RG motifs [52].
Figure 5.
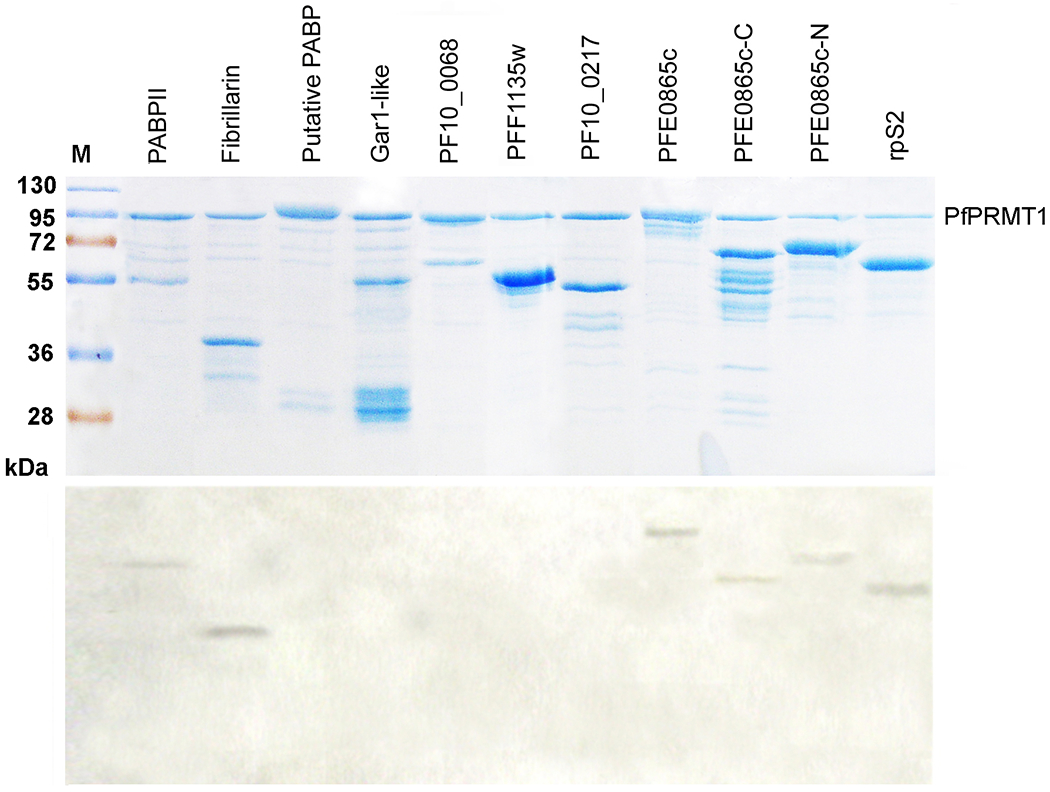
PfPRMT1 activity on potential substrates. Nine P. falciparum proteins were expressed in E. coli as either GST- or MBP-fusion proteins, purified, and incubated with MBP-PfPRMT1 in methylation assays. Upper panel shows the Coomassie blue stained gel, and lower panel is the fluorograph. GST-fusion proteins include PABPII (1-202), fibrillarin (1-100), putative PABP (1-383), Gar1-like (1-209), PF10_0068 (1-246), PFF1135w (128-347), and PF10_0217 (391-538). MBP-fusion proteins include PFE0865c (1-298), FE0865c-C (199-298), PFE0865c-N (1-112), and rpS2 (1-60).
PfPRMT1 enzyme kinetics with substrate peptides
Having determined that P. falciparum histone H4, PABPII, and fibrillarin were in vitro substrates of recombinant PfPRMT1, we wanted to identify the substrate arginines, and determine their methylation status and dynamics. To confirm that H4R3 is the only arginine in the N-terminus that is methylated by PfPRMT1, we synthesized the wild-type H4 peptide (1-21 aa) and H4 peptide with the R3K mutation (PfH4-21R3K). Despite that the PfH4-21 peptide contains three arginines, only R3 is in the RG context. The results showed that only the wild-type PfH4-21 peptide but not PfH4-21R3K was methylated by PfPRMT1, demonstrating the specificity of PfPRMT1 for R3 (Fig. 6A). In addition, the PABPII peptide (PFI1175c-16) with two RG motifs was also methylated by PfPRMT1 (Fig. 6A). To determine the methylation status of the peptides and its dynamics, time-course studies were performed with these peptides and the methylation products were analyzed by mass spectrometry. The spectra showed that the two RG motifs in PABPII peptide and the two RGG motifs in fibrillarin were both methylated by PfPRMT1 (Fig. 6B). For both peptides, the predominant forms of the methylation products at 60 min contained two methyl groups. For the H4 peptide, the presence of only one arginine for methylation allowed us to analyze the dynamics of the methylation reactions. By 20 min of the reaction, approximately equamolar amounts of MMA and DMA were detected (Fig. 6B, C), which conforms to the partially processive mechanism of the reaction proposed for PRMT1 and suggests that mono- and di-methylation occur sequentially without the release of the monomethylated product [31]. In two hours, the R3 in the H4 peptide was predominantly dimethylated (Fig. 6C).
Figure 6.
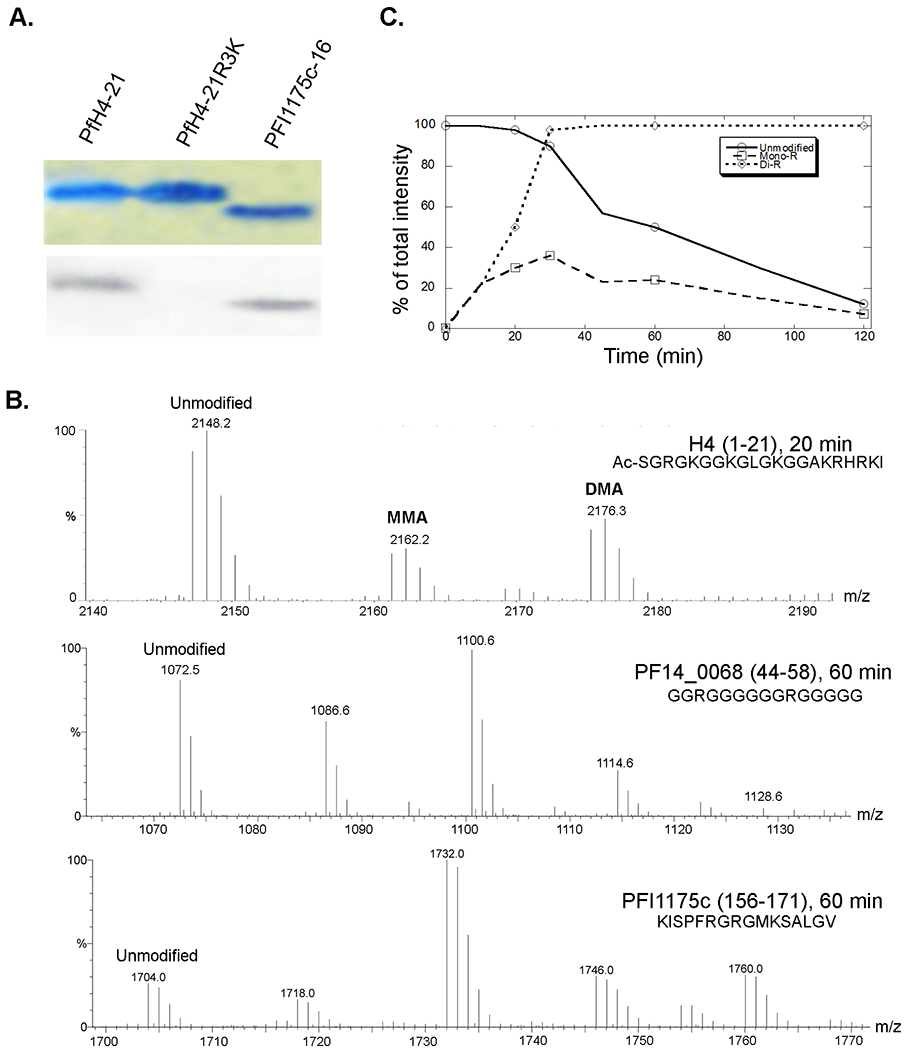
PfPRMT1 activity on peptides. A. Recombinant MBP-PfPRMT1 activity on peptides. Peptides PfH4-21, PfH4-21R3K, and PFI1175c-16 were incubated with MBP-PfPRMT1, separated in a 20% SDS-PAGE gel and stained by Coomassie blue (upper panel), while methylation of the peptides was detected by fluorography (lower panel). B. Spectra of the MALDI-TOF analysis of methylation of peptides PfH4, PF14_0068, and PFI1175c by recombinant MBP-PfPRMT1 to indicate the unmodified and methylated peptides (with an increment of a mass of 14). H4 peptide has only one substrate arginine, while the other two peptides both contain two substrate arginines. C. A time-course analysis of methylation of PfH4-21 by MBP-PfPRMT1. The graph is a representative of two experiments to show the relative quantities of the unmodified substrate and the products with MMA (mono-R) and aDMA (di-R).
The kinetics of the H4R3 methylation in the PfH4-21 peptide was characterized and compared with the mammalian homologues. While the kinetic parameters were generally agreeable with those of the rat PRMT1 [31], PfPRMT1 had >5 and ~2 times higher kcat/Km value than the mammalian homologue for the peptide and AdoMet substrates, respectively (Table 1).
Table 1.
Characterization of the kinetics for the recombinant PfPRMT1
| Substrates | Km (μM) | kcat (min−1) | kcat/Km (M−1. min−1) | |||
|---|---|---|---|---|---|---|
| PfRMT1 | rPRMT1* | PfRMT1 | rPRMT1* | PfRMT1 | rPRMT1* | |
| AcH4-21 | 1.36±0.21 | 3.55±0.68 | 0.77±0.07 | 0.39±0.01 | 5.66 x 105 | 1.10 x 105 |
| AdoMet | 1.44±0.10 | 2.52±0.54 | 0.85±0.08 | 0.84±0.03 | 5.90 x 105 | 3.36 x 105 |
Parameters for the rat PRMT1 (rPRMT1) were from reference [31].
PfPRMT1 protein expression in the parasite
To detect PfPRMT1 expression in the parasite, we produced polyclonal rabbit antiserum against recombinant PfPRMT1. We first assessed PfPRMT1 expression by western blots using equal amounts of protein lysates from synchronized parasites. Western blots detected a protein band of ~47 kDa, consistent with the predicted molecular weight of PfPRMT1 (Fig. 7A). PfPRMT1 was detected throughout the IDC with higher levels detected in trophozoites. In addition, a protein band of ~70 kDa cross-reacted with the PfPRMT1 antiserum. Preimmune serum did not detect any of these protein bands (data not shown). To further demonstrate that the 47 kDa protein is PfPRMT1, we tagged the endogenous PfPRMT1 with GFP at the C-terminus. Chromosomal integration-specific PCR confirmed the correct targeting of the endogenous PfPRMT1 locus (data not shown). Western blots using both anti-GFP and anti-PfPRMT1 antibodies detected a protein band of ~72 kDa in parasite lysates of two transgenic parasite clones, consistent with the predicted size of the PfPRMT1-GFP fusion protein (Fig. 7B). Meanwhile, the 47 kDa band disappeared from the GFP-tagged clones.
Figure 7.
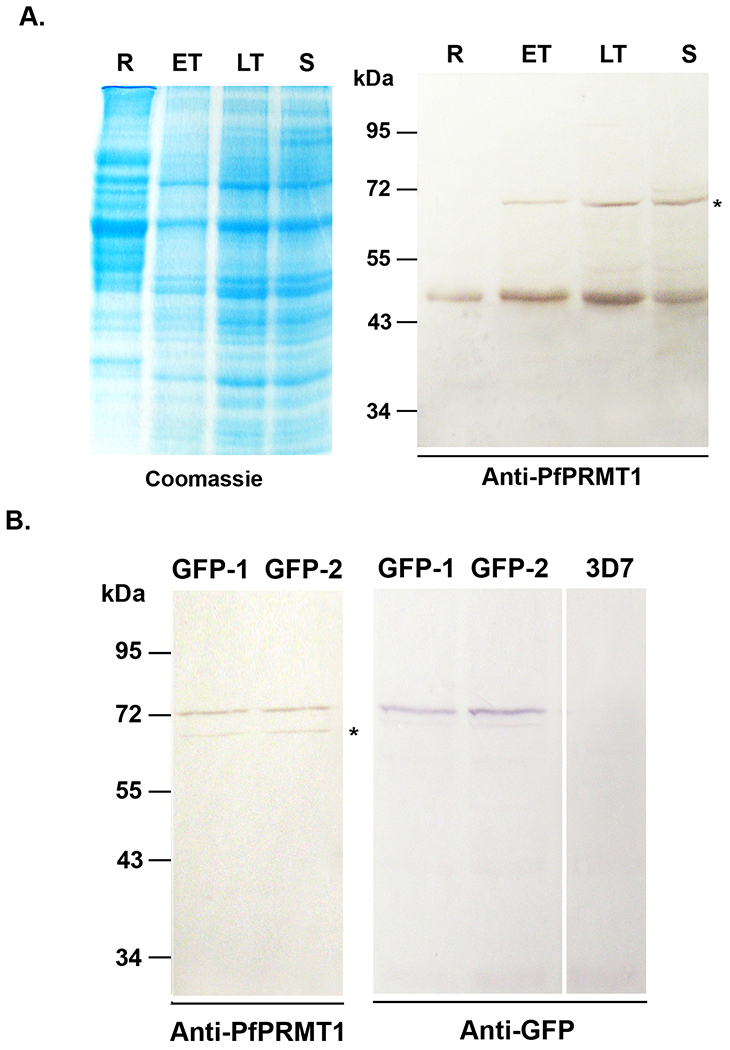
PfPRMT1 expression in the parasite. A. Time course expression of PfPRMT1 during IDC. Equal amounts of parasite lysate (~30 μg) were separated in 10% SDS-PAGE and probed with polyclonal anti-PfPRMT1 antiserum. Left panel is the Coomassie blue-stained gel to indicate approximately equal loading. Asterisk indicates the cross-reacting protein. R – ring, ET – early trophozoite, LT – late trophozoite, S – schizont. B. Confirmation of the C-terminal GFP tagging of the endogenous PfPRMT1 locus. Two clones (GFP-1 and GFP-2) were probed with the anti-PfPRMT1 antiserum (left panel) or the anti-GFP antibody (right panel). Lysate from wild-type 3D7 parasite was included as a GFP-negative control.
Histone arginine methylation during parasite development
With H4 being determined as a PfPRMT1 substrate in vitro, we next wanted to determine the presence and dynamics of H4R3 dimethylation during IDC. The result showed that H4R3 dimethylation was present in all asexual erythrocytic stages with much higher levels found in late trophozoites and schizonts (Fig. 8). In comparison, the level of dimethylated H3R17, possibly modified by the CARM1-like PRMT in the parasite, was prevalent throughout IDC.
Figure 8.

Histone arginine methylation during IDC of the parasite. Histones from synchronized parasites were purified, separated and probed with anti-dimethylated H4R3 (H4R3m2) and anti-dimethylated H3R17 (H3R17m2) antibodies. Immunoblotting with anti-H4 antibodies was included to indicate equal loading. R – ring, ET – early trophozoite, LT – late trophozoite, S – schizont.
Subcellular localization of PfPRMT1
In the parasite line with GFP-tagged PfPRMT1, strong GFP fluorescence was detected in both the cytoplasm and the nucleus (Fig. 9A). Similar results were obtained with wild-type 3D7 parasites by IFA using anti-PfPRMT1 antiserum (data not shown). Further analysis of the nuclear-cytoplasmic distribution of PfPRMT1 was performed using fractionated parasite lysates. Parasites were fractioned into nuclear and cytoplasmic extracts and the proteins from each fraction were resolved on SDS-PAGE and subjected to immunoblotting. The correct fractionation of parasites was demonstrated by the predominant detection of histone H4 in the nuclear fraction and the apicoplast-specific protein PfHLP in the cytoplasmic fraction [53]. The results showed presence of PfPRMT1 in both cellular fractions with the majority localized in the cytoplasm (Fig. 9B). Only a minor fraction of PfPRMT1 was present in the nucleus, which is similar to the cellular distribution of the human PRMT1 [54]. This is consistent with the presence of both nuclear (e.g., fibrillarin and histones) and cytoplasmic (e.g., rpS2) substrates.
Figure 9.
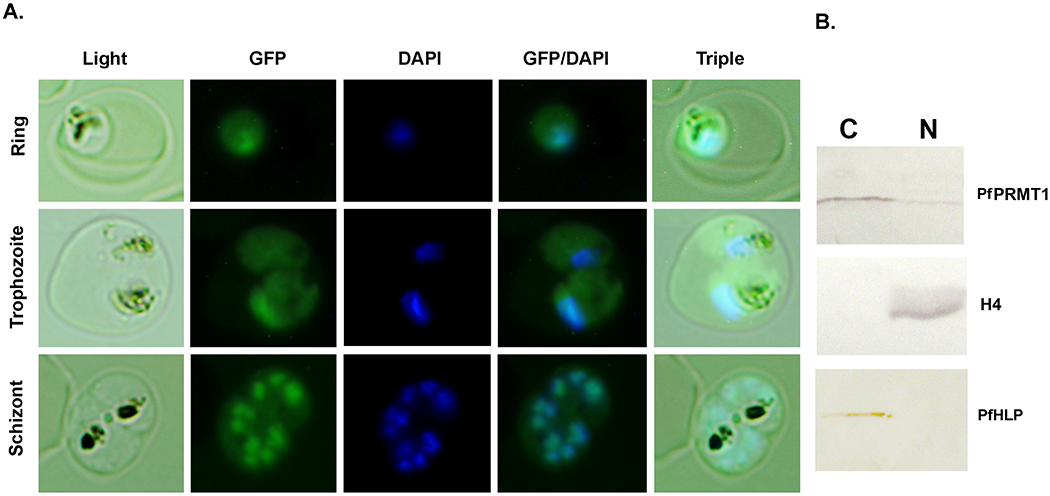
Subcellular localization of PfPRMT1 in ring, trophozoite and schizont stages. A. Localization of GFP-PfPRMT1. Representative images of GFP-PfPRMT1 in a ring, trophozoite, and schizont. Nuclei were stained with DAPI. Triple indicates merge of the light, GFP and DAPI images of the same cells. B. Western blots of the parasite nuclear (N) and cytoplasmic (C) fractions, separated by 10% SDS-PAGE, and probed with anti-PfPRMT1 antiserum (upper panel), anti-H4 antibodies (middle panel), and anti-PfHLP antiserum (lower panel).
The effect of small molecule inhibitors
AdoMet-dependent methyltransferases are inhibited by the reaction product, S-adenosyl-L-homocysteine (SAH). Among the two structural analogues of SAH, sinefungin was a potent inhibitor of PfPRMT1 with an IC50 of <1 μM, whereas MTA was a poor inhibitor (Table 2). These values are compatible with those for the recombinant human PRMT1 [31]. The recently identified AMI-5 as a general inhibitor for arginine and lysine methyltransferases also displayed strong inhibitory activity for PfPRMT1 [55]. In comparison, the fungal metabolite chaetocin, an inhibitor for lysine-specific histone methyltransferase Su(var)3-9 [56], displayed no obvious inhibition of PfPRMT1 in the lower micromolar concentration. When tested on the intraerythrocytic stages of the malaria parasites, all compounds except MTA had potent parasite growth inhibition activities with IC50s below 15 μM (Table 2). Adox, an inhibitor for SAH hydrolase, also showed similar inhibitory activity on parasite growth. Adox treatment may have affected all AdoMet-dependent methyltransferases, thus leading to parasite growth inhibition, since it can elevate the cellular level of SAH [57].
Table 2.
Inhibition of recombinant PfPRMT1 activity and P. falciparum 3D7-luc growth by small molecule inhibitors.
| Compounds | Structures | IC50s for PRMT1 activity (μM) | IC50s for 3D7 growth (μM) |
|---|---|---|---|
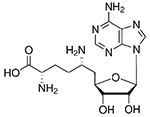
| Sinefungin |
|
0.84 ± 0.15 | 2.71 ± 0.04 |
| 5-Deoxy-5-methylthioadenosine (MTA) |
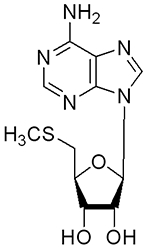
|
>360 | >100 |
| Eosin Y (AMI-5) |
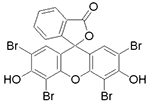
|
11.62 ± 0.08 | 14.79 ± 1.01 |
| Chaetocin |
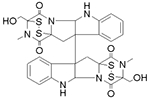
|
>120 | 14.00 ± 1.56 |
| Adenosine-2’,3’-dialdehyde (AdoX) |
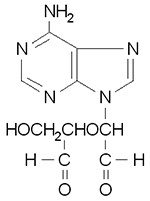
|
Not determined | 8.72 ± 0.73 |
DISCUSSION
Here, we present biochemical evidence to show that PfPRMT1 has intrinsic type I PRMT activity, catalyzing the formation of MMA and aDMA. Particularly, PfPRMT1 shares significant sequence homology and similar enzymatic activity on potential substrates to PRMT1 homologues in higher eukaryotes. Although the methyltransferase motifs in PfPRMT1are highly conserved, its extra 50 aa N-terminal sequence is not only highly divergent from the mammalian orthologues, but also essential for its enzymatic activity. Further, despite that enzyme kinetic parameters of the recombinant PfPRMT1 are similar to those of its mammalian enzymes, PfPRMT1 has a much higher turnover rates. These and other differences from the mammalian enzymes may serve the basis for developing specific inhibitors for the parasite enzyme.
Methylation of arginines in histones has profound effects on gene expression [12]. We showed that recombinant PfPRMT1 predominantly methylates H4 at R3, forming MMA and aDMA. Besides, both forms of H4R3 methylation are present during the entire IDC. Furthermore, both PfPRMT1 and asymmetric H4R3 dimethylation are dynamic during parasite development, exhibiting similar patterns of increases as the parasites mature (Fig. 7 and 8). We have also confirmed the presence of H3R17 methylation in the parasite, a potential substrate of the CARM1-like PRMT (Fig. 8). While methylation of H4R3 and R17 is involved in transcription activation in mammals [11, 58, 59], its significance in epigenetics of the malaria parasite awaits further elucidation.
Besides histones, a large number of cellular proteins has been identified as the physiological substrates of PRMT1s [1]. PfPRMT1 could methylate four of the nine selected P. falciparum proteins, including conserved PRMT1 substrates fibrillarin and PABPII. Fibrillarin is an evolutionarily conserved ribose-2’-O-methylase associated with C/D class small nucleolar RNAs, and involves in rRNA maturation and ribosome assembly [60]. PABPII is a nuclear protein involved in mRNA polyadenylation [49, 61]. In addition, PfPRMT1 could methylate a new GAR-motif-containing splicing factor-like protein (PFE0865c) in vitro. These results strongly indicate conserved functions for PfPRMT1 in RNA processing and metabolism. In higher eukaryotes, the evolution of many more PRMTs allows functional divisions of PRMTs. Malaria parasites have only three PRMTs; thus, each of these protozoan PRMTs may have a broader substrate range, which may represent their ancestral functions. Interestingly, we also found that PfPRMT1 could methylate rpS2 in vitro, a physiological substrate of PRMT3 [62], suggesting that PfPRMT1 might affect ribosome biogenesis and the translational machinery. In trypanosomes, PRMT1 also methylates proteins that affect the metabolism of specific mitochondrial mRNAs [29]. Therefore, investigations into the in vivo substrates of protozoan PRMT1s may reveal novel pathways where these enzymes play regulatory roles.
PRMTs have been considered promising pharmaceutical targets [4]. We have found that the inhibitor sinefungin as well as AMI-5 had both lower micromolar IC50s for the recombinant PfPRMT1 and the parasite growth. This result indicates that parasite methyltransferases could serve as potential targets for development of antimalarials. Since most of the compounds tested presumably affect all methyltransferase activities non-specifically, it is highly desirable to screen for specific inhibitors for different PRMTs [55]. The robust PRMT activity of the recombinant PfPRMT1 and its divergence from the mammalian PRMT1s may provide such a tool for screening specific inhibitors.
Supplementary Material
ACKNOWLEDGEMENTS
We also want to thank Dr. Hesheng Zhang for providing the PfHLP antiserum and Ms. Xiaolian Li for technical assistance.
FUNDING
This work was supported by a grant (R01 AI064553) from National Institute of Allergy and Infectious Diseases, NIH.
Abbreviations used:
- PRMT
protein arginine methyltransferases
- GAR
glycine and arginine-rich
- AdoMet
S-adenosylmethionine
- CARM1
cofactor associated arginine methyltransfease 1
- IDC
intraerythrocytic developmental cycle
- MBP
maltose-binding protein
- LSC
liquid scintillation counter
- GFP
green fluorescent protein
- MTA
5-deoxy-5-methylthioadenosine
- SAH
S-adenosyl-L-homocysteine
- AdoX
adenosine dialdehyde
- RLM-RACE
RNA ligase-mediated rapid amplification of cDNA end.
REFERENCES
- 1.Pahlich S, Zakaryan RP and Gehring H (2006) Protein arginine methylation: Cellular functions and methods of analysis. Biochim. Biophys. Acta 1764, 1890–1903 [DOI] [PubMed] [Google Scholar]
- 2.Bedford MT and Richard S (2005) Arginine methylation an emerging regulator of protein function. Mol. Cell 18, 263–272 [DOI] [PubMed] [Google Scholar]
- 3.Gary JD and Clarke S (1998) RNA and protein interactions modulated by protein arginine methylation. Prog. Nucleic Acid Res. Mol. Biol 61, 65–131 [DOI] [PubMed] [Google Scholar]
- 4.Krause CD, Yang ZH, Kim YS, Lee JH, Cook JR and Pestka S (2007) Protein arginine methyltransferases: evolution and assessment of their pharmacological and therapeutic potential. Pharmacol. Therapeutics 113, 50–87 [DOI] [PubMed] [Google Scholar]
- 5.Bachand F (2007) Protein arginine methyltransferases: from unicellular eukaryotes to humans. Eukaryot. Cell 6, 889–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McBride AE and Silver PA (2001) State of the Arg: protein methylation at arginine comes of age. Cell 106, 5–8 [DOI] [PubMed] [Google Scholar]
- 7.Strahl BD, Briggs SD, Brame CJ, Caldwell JA, Koh SS, Ma H, Cook RG, Shabanowitz J, Hunt DF, Stallcup MR and Allis CD (2001) Methylation of histone H4 at arginine 3 occurs in vivo and is mediated by the nuclear receptor coactivator PRMT1. Curr. Biol 11, 996–1000 [DOI] [PubMed] [Google Scholar]
- 8.Schurter BT, Koh SS, Chen D, Bunick GJ, Harp JM, Hanson BL, Henschen-Edman A, Mackay DR, Stallcup MR and Aswad DW (2001) Methylation of histone H3 by coactivator-associated arginine methyltransferase 1. Biochemistry 40, 5747–5756 [DOI] [PubMed] [Google Scholar]
- 9.Pal S, Vishwanath SN, Erdjument-Bromage H, Tempst P and Sif S (2004) Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol. Cell. Biol 24, 9630–9645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pal S and Sif S (2007) Interplay between chromatin remodelers and protein arginine methyltransferases. J. Cellular Physiol 213, 306–315 [DOI] [PubMed] [Google Scholar]
- 11.An W, Kim J and Roeder RG (2004) Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell 117, 735–748 [DOI] [PubMed] [Google Scholar]
- 12.Wysocka J, Allis CD and Coonrod S (2006) Histone arginine methylation and its dynamic regulation. Front Biosci. 11, 344–355 [DOI] [PubMed] [Google Scholar]
- 13.Cary C, Lamont D, Dalton JP and Doerig C (1994) Plasmodium falciparum chromatin: nucleosomal organisation and histone-like proteins. Parasitol. Res 80, 255–258 [DOI] [PubMed] [Google Scholar]
- 14.Miao J, Fan Q, Cui L, Li J, Li J and Cui L (2006) The malaria parasite Plasmodium falciparum histones: organization, expression, and acetylation. Gene 369, 53–65 [DOI] [PubMed] [Google Scholar]
- 15.Callebaut I, Prat K, Meurice E, Mornon JP and Tomavo S (2005) Prediction of the general transcription factors associated with RNA polymerase II in Plasmodium falciparum: conserved features and differences relative to other eukaryotes. BMC Genomics 6, 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aravind L, Iyer LM, Wellems TE and Miller LH (2003) Plasmodium biology: genomic gleanings. Cell 115, 771–785 [DOI] [PubMed] [Google Scholar]
- 17.Coulson RM, Hall N and Ouzounis CA (2004) Comparative genomics of transcriptional control in the human malaria parasite Plasmodium falciparum. Genome Res. 14, 1548–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Balaji S, Babu MM, Iyer LM and Aravind L (2005) Discovery of the principal specific transcription factors of Apicomplexa and their implication for the evolution of the AP2-integrase DNA binding domains. Nucleic Acids Res. 33, 3994–4006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Silva EK, Gehrke AR, Olszewski K, Leon I, Chahal JS, Bulyk ML and Llinas M (2008) Specific DNA-binding by apicomplexan AP2 transcription factors. Proc. Natl. Acad. Sci. USA. 105, 8393–8398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coleman BI and Duraisingh MT (2008) Transcriptional control and gene silencing in Plasmodium falciparum. Cell Microbiol. 10, 1935–1946 [DOI] [PubMed] [Google Scholar]
- 21.Chookajorn T, Ponsuwanna P and Cui L (2008) Mutually exclusive var gene expression in the malaria parasite: multiple layers of regulation. Trends Parasitol. 24, 455–461 [DOI] [PubMed] [Google Scholar]
- 22.Cui L, Fan Q, Cui L and Miao J (2008) Histone lysine methyltransferases and demethylases in Plasmodium falciparum. Int. J. Parasitol 38, 1083–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cui L, Miao J, Furuya T, Li X, Su XZ and Cui L (2007) PfGCN5-mediated histone H3 acetylation plays a key role in gene expression in Plasmodium falciparum. Eukaryot. Cell 6, 1219–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chookajorn T, Dzikowski R, Frank M, Li F, Jiwani AZ, Hartl DL and Deitsch KW (2007) Epigenetic memory at malaria virulence genes. Proc. Natl. Acad. Sci. USA. 104, 899–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lopez-Rubio JJ, Gontijo AM, Nunes MC, Issar N, Hernandez Rivas R and Scherf A (2007) 5’ flanking region of var genes nucleate histone modification patterns linked to phenotypic inheritance of virulence traits in malaria parasites. Mol. Microbiol 66, 1296–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pelletier M, Pasternack DA and Read LK (2005) In vitro and in vivo analysis of the major type I protein arginine methyltransferase from Trypanosoma brucei. Mol. Biochem. Parasitol 144, 206–217 [DOI] [PubMed] [Google Scholar]
- 27.Saksouk N, Bhatti MM, Kieffer S, Smith AT, Musset K, Garin J, Sullivan WJ Jr., Cesbron-Delauw MF and Hakimi MA (2005) Histone-modifying complexes regulate gene expression pertinent to the differentiation of the protozoan parasite Toxoplasma gondii. Mol. Cell. Biol 25, 10301–10314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pasternack DA, Sayegh J, Clarke S and Read LK (2007) Evolutionarily divergent type II protein arginine methyltransferase in Trypanosoma brucei. Eukaryot. Cell 6, 1665–1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goulah CC, Pelletier M and Read LK (2006) Arginine methylation regulates mitochondrial gene expression in Trypanosoma brucei through multiple effector proteins. RNA 12, 1545–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsumoto M, Hwang SB, Jeng KS, Zhu N and Lai MM (1996) Homotypic interaction and multimerization of hepatitis C virus core protein. Virology 218, 43–51 [DOI] [PubMed] [Google Scholar]
- 31.Osborne TC, Obianyo O, Zhang X, Cheng X and Thompson PR (2007) Protein arginine methyltransferase 1: positively charged residues in substrate peptides distal to the site of methylation are important for substrate binding and catalysis. Biochemistry 46, 13370–13381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cui L, Miao J, Furuya T, Fan Q, Li X, Rathod PK, Su XZ and Cui L (2008) Histone acetyltransferase inhibitor anacardic acid causes changes in global gene expression during in vitro Plasmodium falciparum development. Eukaryot. Cell 7, 1200–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trager W and Jensen JB (1976) Human malaria parasites in continuous culture. Science 193, 673–675 [DOI] [PubMed] [Google Scholar]
- 34.Lambros C and Vanderberg JP (1979) Synchronization of Plasmodium falciparum erythrocytic stages in culture. J. Parasitol 65, 418–420 [PubMed] [Google Scholar]
- 35.Cui L, Rzomp KA, Fan Q, Martin SK and Williams J (2001) Plasmodium falciparum: differential display analysis of gene expression during gametocytogenesis. Exp. Parasitol 99, 244–254 [DOI] [PubMed] [Google Scholar]
- 36.Fan Q, An L and Cui L (2004) Plasmodium falciparum histone acetyltransferase, a yeast GCN5 homologue involved in chromatin remodeling. Eukaryot. Cell 3, 264–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fan Q, An L and Cui L (2004) PfADA2, a Plasmodium falciparum homologue of the transcriptional coactivator ADA2 and its in vivo association with the histone acetyltransferase PfGCN5. Gene 336, 251–261 [DOI] [PubMed] [Google Scholar]
- 38.Fidock DA and Wellems TE (1997) Transformation with human dihydrofolate reductase renders malaria parasites insensitive to WR99210 but does not affect the intrinsic activity of proguanil. Proc. Natl. Acad. Sci. USA. 94, 10931–10936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deitsch KW, Driskill CL and Wellems TE (2001) Transformation of malaria parasites by the spontaneous uptake and expression of DNA from human erythrocytes. Nucleic Acids Res. 29, 850–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rosario V (1981) Cloning of naturally occurring mixed infections of malaria parasites. Science 212, 1037–1038 [DOI] [PubMed] [Google Scholar]
- 41.Tonkin CJ, van Dooren GG, Spurck TP, Struck NS, Good RT, Handman E, Cowman AF and McFadden GI (2004) Localization of organellar proteins in Plasmodium falciparum using a novel set of transfection vectors and a new immunofluorescence fixation method. Mol. Biochem. Parasitol 137, 13–21 [DOI] [PubMed] [Google Scholar]
- 42.Cui L, Miao J, Wang J, Li Q and Cui L (2008) Plasmodium falciparum: development of a transgenic line for screening antimalarials using firefly luciferase as the reporter. Exp. Parasitol 120, 80–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weiss VH, McBride AE, Soriano MA, Filman DJ, Silver PA and Hogle JM (2000) The structure and oligomerization of the yeast arginine methyltransferase, Hmt1. Nature Struct. Biol 7, 1165–1171 [DOI] [PubMed] [Google Scholar]
- 44.Zhang X and Cheng X (2003) Structure of the predominant protein arginine methyltransferase PRMT1 and analysis of its binding to substrate peptides. Structure 11, 509–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang X, Zhou L and Cheng X (2000) Crystal structure of the conserved core of protein arginine methyltransferase PRMT3. EMBO J. 19, 3509–3519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rho J, Choi S, Seong YR, Cho WK, Kim SH and Im DS (2001) Prmt5, which forms distinct homo-oligomers, is a member of the protein-arginine methyltransferase family. J. Biol. Chem 276, 11393–11401 [DOI] [PubMed] [Google Scholar]
- 47.Najbauer J, Johnson BA, Young AL and Aswad DW (1993) Peptides with sequences similar to glycine, arginine-rich motifs in proteins interacting with RNA are efficiently recognized by methyltransferase(s) modifying arginine in numerous proteins. J. Biol. Chem 268, 10501–10509 [PubMed] [Google Scholar]
- 48.Smith JJ, Rucknagel KP, Schierhorn A, Tang J, Nemeth A, Linder M, Herschman HR and Wahle E (1999) Unusual sites of arginine methylation in Poly(A)-binding protein II and in vitro methylation by protein arginine methyltransferases PRMT1 and PRMT3. J. Biol. Chem 274, 13229–13234 [DOI] [PubMed] [Google Scholar]
- 49.Perreault A, Lemieux C and Bachand F (2007) Regulation of the nuclear poly(A)-binding protein by arginine methylation in fission yeast. J. Biol. Chem 282, 7552–7562 [DOI] [PubMed] [Google Scholar]
- 50.Xu C, Henry PA, Setya A and Henry MF (2003) In vivo analysis of nucleolar proteins modified by the yeast arginine methyltransferase Hmt1/Rmt1p. RNA 9, 746–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Swiercz R, Person MD and Bedford MT (2005) Ribosomal protein S2 is a substrate for mammalian PRMT3 (protein arginine methyltransferase 3). Biochem. J 386, 85–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wooderchak WL, Zang T, Zhou ZS, Acuna M, Tahara SM and Hevel JM (2008) Substrate profiling of PRMT1 reveals amino acid sequences that extend beyond the “RGG” paradigm. Biochemistry 47, 9456–9466 [DOI] [PubMed] [Google Scholar]
- 53.Ram EV, Naik R, Ganguli M, and Habib S (2008). DNA organization by the apicoplast-targeted bacterial histone-like protein of Plasmodium falciparum. Nucleic Acids Res. 36, 5061–5073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Herrmann F, Lee J, Bedford MT and Fackelmayer FO (2005) Dynamics of human protein arginine methyltransferase 1(PRMT1) in vivo. J. Biol. Chem 280, 38005–38010 [DOI] [PubMed] [Google Scholar]
- 55.Cheng D, Yadav N, King RW, Swanson MS, Weinstein EJ and Bedford MT (2004) Small molecule regulators of protein arginine methyltransferases. J. Biol. Chem 279, 23892–23899 [DOI] [PubMed] [Google Scholar]
- 56.Greiner D, Bonaldi T, Eskeland R, Roemer E and Imhof A (2005) Identification of a specific inhibitor of the histone methyltransferase SU(VAR)3-9. Nature Chem. Biol 1, 143–145 [DOI] [PubMed] [Google Scholar]
- 57.Li C, Ai LS, Lin CH, Hsieh M, Li YC and Li SY (1998) Protein N-arginine methylation in adenosine dialdehyde-treated lymphoblastoid cells. Arch. Biochem. Biophys 351, 53–59 [DOI] [PubMed] [Google Scholar]
- 58.Wang H, Huang ZQ, Xia L, Feng Q, Erdjument-Bromage H, Strahl BD, Briggs SD, Allis CD, Wong J, Tempst P and Zhang Y (2001) Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science 293, 853–857 [DOI] [PubMed] [Google Scholar]
- 59.Bauer UM, Daujat S, Nielsen SJ, Nightingale K and Kouzarides T (2002) Methylation at arginine 17 of histone H3 is linked to gene activation. EMBO Rep. 3, 39–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reichow SL, Hamma T, Ferre-D’Amare AR and Varani G (2007) The structure and function of small nucleolar ribonucleoproteins. Nucleic Acids Res. 35, 1452–1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mangus DA, Evans MC and Jacobson A (2003) Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 4, 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bachand F and Silver PA (2004) PRMT3 is a ribosomal protein methyltransferase that affects the cellular levels of ribosomal subunits. EMBO J. 23, 2641–2650 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


