Abstract
Scope
Better understanding of factors contributing to inter-individual variability in biomarkers of vitamin K could enhance the understanding of the equivocal role of vitamin K in cardiovascular disease. Based on the known biology of phylloquinone, the major form of vitamin K, we hypothesized that plasma lipids contribute to the variable response of biomarkers of vitamin K metabolism to phylloquinone supplementation.
Methods and results
We examined the association of plasma lipids and 27 lipid-related genetic variants with the response of biomarkers of vitamin K metabolism in a secondary analysis of data from a 3-year phylloquinone supplementation trial in men (n=66) and women (n=85). Year 3 plasma triglycerides (TG), but not total cholesterol, LDL-cholesterol, or HDL-cholesterol, were associated with the plasma phylloquinone response (Men: β=1.01, p<0.001, R2=0.34; Women: β=0.61, p=0.008, R2=0.11; sex interaction p=0.077). Four variants and the TG-weighted genetic risk score were associated with the plasma phylloquinone response in men only. Plasma lipids were not associated with changes in biomarkers of vitamin K function (undercarboxylated osteocalcin and MGP) in either sex.
Conclusion
Plasma TG were an important determinant of the inter-individual response of plasma phylloquinone to phylloquinone supplementation, but changes in biomarkers of vitamin K carboxylation were not influenced by lipids.
Keywords: Lipids, Vitamin K
Graphical Abstract
A better understanding of factors that contribute to the inter-individual variability in biomarkers of vitamin K metabolism is needed. In a secondary analysis of data from a randomized trial in which phylloquinone, the major dietary and circulating form of vitamin K, was supplemented for three years, we found that plasma triglycerides were an important determinant of the inter-individual response of circulating phylloquinone, but changes in biomarkers of vitamin K carboxylation were not influenced by lipids.
1). INTRODUCTION
There are physiological functions of vitamin K-dependent proteins (VKDPs) found in vascular tissues that suggest the lipid-soluble vitamin K may have a protective role in cardiovascular disease (CVD).[1] One such VDKP is a known calcification inhibitor, yet associations of dietary and circulating vitamin K with vascular calcification in humans are equivocal.[2, 3] These inconsistencies may be related to the largely unexplained inter-individual variability in vitamin K biomarkers.[4, 5]
Phylloquinone is the primary form of vitamin K in the diet and in circulation.[6, 7] Circulating phylloquinone, which is carried on lipoproteins with other blood lipids, is a biomarker of vitamin K absorption.[8–10] Population means for circulating phylloquinone have been reported to range from 0.14 to 2.48 nM, with standard deviations up to 2.88 nM.[11] In two cohorts characterized for circulating phylloquinone, biologically plausible factors including lipids, explained approximately 20% of the variance in circulating phylloquinone.[4, 5] In both cohorts, triglycerides (TG) were a major predictor of circulating phylloquinone (explaining 2.8 to 15.7% of the variability, depending on the sex). In one cohort, LDL-cholesterol (LDL-C) and total cholesterol (TC) also accounted for small (~1%), but significant amounts of the variability in men and women, respectively.[4] Results of a genome-wide meta-analysis also provide evidence that circulating lipids contribute to variances in circulating phylloquinone.[12] A single nucleotide polymorphism (SNP) located on chromosome 11 near the APOA1/C3/A4/A5 gene cluster, consistently linked to TG, TC, LDL-C, and HDL-cholesterol (HDL-C) concentrations,[13, 14] was associated with circulating phylloquinone at p=5.91 × 10-8. However, large-scale meta-analyses of genome-wide association studies have identified many other lipid-related genes and it is plausible that some of them also influence vitamin K metabolism. [13, 14]
The classical role of vitamin K is as an essential cofactor for γ-glutamyl carboxylase, an enzyme that catalyzes the post-translational modification and activation of VKDPs.[15] Production of VKDPs is influenced by many factors including age and sex,[16] but the fractions of undercarboxylated VKDP in circulation have been shown to change when dietary phylloquinone changes and are considered functional indicators of vitamin K status of tissues utilizing the various proteins.[11, 17] This has been demonstrated for levels of undercarboxylated osteocalcin (OC), a VKDP synthesized in bone during bone formation,[18, 19] and uncarboxylated matrix gla protein (MGP), a calcification inhibitor in vascular tissue and cartilage.[20, 21] If lipids influence the availability of phylloquinone to tissues, then it is plausible that lipids could also influence the enzyme cofactor activity of vitamin K. The evidence relating lipids to the variability in biomarkers of vitamin K function is limited.[4]
Thus, we conducted a secondary data analysis from a randomized controlled trial designed to examine the effect of 3 years phylloquinone supplementation on age-related bone loss [19] and vascular calcification[2] to test the association of plasma TG, TC, LDL-C, HDL-C, and related SNPs, with changes in biomarkers of vitamin K metabolism. We hypothesized that lipids would contribute to the inter-individual variability in the biomarker response to phylloquinone supplementation.
2). EXPERIMENTAL SECTION
Study Design and Participants
Four hundred fifty-two free-living men and women, aged 60–80 years, enrolled in a 3-year, double-blind, randomized, controlled phylloquinone supplementation trial, as described in detail elsewhere.[19] Participants were primarily white (n=422, 93%, based on self-report). Prior to enrollment, participants completed a detailed medical history questionnaire. Participants were generally in good health, free of known coronary disease or osteoporosis, and not taking warfarin (a vitamin K antagonist). All participants provided written informed consent and the Institutional Review Board at Tufts University approved the protocol for this study. Equal numbers of participants were randomly assigned to either the treatment or nontreatment group, with separate randomization schemes for men and women. The trial was registered at clinicaltrials.gov as NCT00183001.
For this secondary analysis, only white participants randomized to the vitamin K treatment group who provided consent for genotyping were included.
Dosage Information
The treatment group received 500 ug of phylloquinone as part of a daily multivitamin formulation. During the 3-year intervention, each participant was instructed to take the multivitamin tablet each morning dissolved in a 5- to 6-ounce glass of water by mouth. Study participants also received a second daily tablet containing 600 mg calcium carbonate and 10 ug (400 IU) vitamin D as cholecalciferol. Adherence to the treatment protocol was 89.1% which was assessed by pill count.[19] For this analysis participants with protocol adherence greater than 85% were examined.
The supplemental dose of 500 μg/d phylloquinone exceeds the United States Institute of Medicine’s Adequate Intake for vitamin K which is currently 90 μg/d of phylloquinone for adult women and 120 μg/d for adult men.[22] However, there is large inter- and intra-individual variability in the daily intake of phylloquinone.[23] In a nationally representative group of approximately 4,700 men, women, and children that participated in a Market Research Corporation of America menu census survey, 14-day average intakes of men and women reached 613 μg/d and 495 μg/d, respectively with per-day maximums up to 1,650 ug/d.[24] Thus, 500 μg/d is within the range of reported dietary intakes of a community-dwelling population and is therefore attainable in the diet.
Biochemical Measurements
Blood samples were drawn after a minimum of a 10-hour fast at baseline and study completion (year 3). Samples were stored in individual cryogenic tubes at −80C and protected from light until the time of analysis. Unless noted, laboratory analyses were performed within 12 months of completion of the supplementation trial.
The measures of vitamin K metabolism analyzed included plasma phylloquinone, serum percent undercarboxylated osteocalcin (%ucOC), and plasma dephosphorylated-undercarboxylated Matrix Gla Protein (dp-ucMGP). Serum %ucOC and plasma dp-ucMGP reflect vitamin K cofactor activity in bone and vascular tissues and cartilage, respectively.[11, 25] Biomarkers were measured at Tufts University Human Nutrition Research Center on Aging (HNRCA), unless otherwise indicated. Plasma phylloquinone was analyzed by high-performance liquid chromatography (HPLC), followed by fluorometric detection.[26] Serum undercarboxylated osteocalcin and total osteocalcin were measured with a radioimmunoassay.[18] Total serum matrix gla protein (MGP) was assayed at the University of California, San Diego using a radioimmunoassay, as previously described.[27–29] Dp-ucMGP was measured by VitaK in Maastricht, Netherlands from stored samples of citrated plasma using a sandwich ELISA.[21, 30]
Automated enzymatic methods were used to determine TC and TG concentrations.[31] HDL-C was measured after precipitation of LDL and VLDL using a dextran sulfate-magnesium procedure.[32] LDL-C was calculated using the Friedewald equation for individuals with TG concentrations not greater than 400 mg/dl.[33]
SNP Selection and Genotyping
Twenty-seven SNPs was selected for analysis. Several loci were included based on their primary association with TG (n=7), TC (n=2), LDL-C (n=3), or HDL-C (n=2) in meta-analyses of genome-wide association studies (GWAS) conducted by the Global Lipids Genetics Consortium[13, 14] Six SNPs previously associated with plasma lipid concentrations and located within the APOA5 (n=3), APOA4 (n=2), or APOC3 (n=1) genes were included to examine the APOA1/C3/A4/A5 cluster with greater resolution.[34–44] SNPs in LPL (n=4) and APOE (n=3) were also investigated based on the biological roles of these genes in TG-rich lipoprotein metabolism.[45, 46]
For this secondary analysis, DNA was isolated from blood and purified for PCR analysis using the QIAamp DNA Mini Kit (Qiagen Inc., Chatsworth, CA). Genotyping for 20 SNPs was performed with the TaqMan® minor groove-binding method using the QuantStudio 6 Flex Real-Time PCR System (Applied Biosystems, Foster City, CA). The QuantStudio Real-Time PCR Software v1.1 was used for allele discrimination. SNPs in LPL and APOE were previously analyzed with TaqMan 5’ nuclease allelic discrimination assays designed by Applied Biosystems (Foster City, CA) using the Applied Biosystems Prism 7900.[47]
Genetic Risk Score
Weighted genetic risk scores (wGRS) were constructed to examine whether a genetic load of TG-, TC-, LDL-C-, or HDL-C- related variants is associated with the response of biomarkers of vitamin K metabolism. The methodology was consistent with the method reported by Renstrom et al.,[48] Ahmad et al.,[49] and Justesen et al.,[50] which has consistently produced wGRS associated with the respective lipids. Weights corresponded to reported effect sizes from the meta-analyzed (n >188,000 individuals of European ancestry) association of each SNP with the corresponding lipid.[13] For TG, TC, and LDL-C, weighted lipid increasing alleles were summed for each participant. Each wGRS was divided by the maximum wGRS for that lipid and multiplied by the total number of risk alleles to scale the wGRS to the range of an unweighted GRS. For the wHDL-GRS, lipid-decreasing alleles were added together. SNPs included in our wGRS were limited to the SNPs selected for genotyping in this study. If the direction of the association of a SNP with a lipid in this supplementation trial was not consistent with the reported direction, the SNP was not included in the wGRS (Supporting Information Table S1).
Statistical Analyses
Measures of plasma TG, plasma phylloquinone, and plasma dp-ucMGP were log-transformed to satisfy statistical assumptions of normality. Other exposures and biomarkers of vitamin K considered (TC, LDL-C, HDL-C, %ucOC) were normally distributed. Baseline characteristics of the individuals randomized to the treatment group were compared to similar individuals in the nontreatment group using a t-test for continuous variables or a Chi-square test for categorical variables. Within the group of individuals randomized to the treatment group, differences by sex were examined using the same methods.
Measures of vitamin K metabolism were reported to have changed in response to three years of phylloquinone supplementation in this trial.[19, 21, 51] In the present analyses, linear mixed effect models were used to examine the treatment effect (time x treatment group). Whether the changes in measures of vitamin K metabolism differed by sex or lipid-lowering medication use (Yes/No/Started during the trial (Initiated)) were examined in the treatment group with interaction terms (time x sex, and time x lipid-lowering medication use). Significant interactions of time with treatment, sex, and lipid-lowering medication use (p < 0.05) were examined further with stratification. Within each subgroup of men and women or lipid-lowering medication use, a Bonferroni-corrected p < 0.05 was used to identify significant changes in biomarkers of vitamin K metabolism. Models were additionally adjusted for percent body fat and smoking status at baseline and a random intercept for participant was included to account for intra-individual variability. Similar methods using linear mixed effect models were used to determine whether the plasma lipids changed from baseline to year 3.
Multiple linear regression models were used to test if plasma lipids were associated with the 3-year response of biomarkers of vitamin K metabolism. Outcomes included year 3 measures of plasma phylloquinone (nmol/L), serum %ucOC (%), and plasma dp-ucMGP (pmol/L). The primary exposures were year 3 concentrations of plasma TG, TC, LDL-C, and HDL-C. A separate model was used for each outcome-exposure pair. First, a crude model was used to examine the association of each lipid with the measures of vitamin K metabolism at year 3 adjusting for the baseline concentration of the vitamin K measures only. Covariates, including lipid-lowering medication use, current smoking, and percent body fat, were adjusted for in a second model. A third model was additionally adjusted for TG (primary exposures: TC, LDL-C, and HDL-C) or TC (primary exposure: TG). Like with OC,[52] circulating dp-ucMGP also depends on the circulating total MGP. The percent or fraction of undercarboxylated to total protein is considered a more robust indicator of vitamin K status than the concentration of the uncarboxylated protein itself. The assays for total MGP and dp-ucMGP were not compatible to create a %dp-ucMGP ratio due to use of different antibodies. Therefore we were unable to express the dp-ucMGP relative to total MGP, but we included the 3-year total MGP concentration as a covariate in all of the dp-ucMGP models. Interaction terms were applied to the fully adjusted model. We also performed sensitivity analyses excluding participants with TG ≥ 300 mg/dL and where appropriate combined men and women to examine whether associations were significantly different in men and women.
Methods to test the associations of each lipid-related genetic variant and wGRS with changes in each measure of vitamin K metabolism in response to supplementation mirrored those described for the plasma lipids. Crude models were adjusted for baseline concentrations of the vitamin K outcome measure, model 2 was adjusted for lipid-lowering medication use, current smoking, and percent body fat, model 3 was additionally adjusted for year 3 TG and TC concentrations, and interaction terms were applied to model 3.
Collinearity was assessed by calculating a variance inflation factor for each predictor in each model. All statistical tests were two-tailed. The significance of findings for the four major blood lipids and wGRS was detected at p < 0.05 after a Bonferroni correction. Due to the large number of lipid-related SNPs examined, the proportion of false discoveries for these analyses was controlled by using the Benjamini-Hochberg correction for multiple comparisons.[53] This portion of the analyses was designed to be hypothesis-generating, so the FDR threshold was set at Q < 0.3. All analyses were carried out in R version 3.4.1.
3). RESULTS
Equal numbers of subjects were randomly assigned to either the treatment (n=229) or the nontreatment (n=223) group. Of the 229 participants in the treatment group, 151 white individuals were greater than 85% adherent to the study protocol and provided consent for genotyping. At baseline, characteristics, biomarkers of vitamin K, and plasma lipids for these participants were not different from similar participants randomized to the nontreatment group (n=151, all p > 0.103 for Chi-square or t-test). Vitamin K biomarker and plasma lipid data at baseline and year 3 are shown in Table 1 along with participants’ baseline characteristics.
Table 1:
Characteristics and biomarkers of white individuals participating in a 3-year phylloquinone supplementation (500 μg/d) trial who were > 85% adherent to the study protocol and had DNA available for genotyping.
| Men (n=66) | Women (n=85) | By Sex | |||
|---|---|---|---|---|---|
| Mean | SD | Mean | SD | p-value | |
| Age (years) | 68 | 5.2 | 68 | 5.6 | 0.516 |
| BMI (kg/m2) | 28.1 | 4 | 28.3 | 5.4 | 0.824 |
| Body Fat (%) | 29 | 7.1 | 42.6 | 7.3 | <0.001 |
| Lipid-lowering medication use (%) | 0.468 | ||||
| No | 66.7 | 54.1 | |||
| Yes | 21.2 | 27.1 | |||
| Initiated | 12.1 | 14.1 | |||
| Current smoker (%) | 1.000 | ||||
| No | 95.5 | 94.1 | |||
| Yes | 4.5 | 5.9 | |||
| Dietary Phylloquinone (ug/d) | 139.9 | 81.2 | 196.9 | 128.9 | 0.003 |
| Biomarkers of vitamin K metabolism | |||||
| Plasma Phylloquinone (nmol/L) | |||||
| Baseline | 1.58 | 2.44 | 1.17 | 1.55 | 0.197 |
| 3-Year | 3.13ab | 2.93 | 3.62ab | 2.84 | 0.061 |
| % ucOC (%) | |||||
| Baseline | 35.42 | 14.35 | 43.33 | 16.47 | 0.002 |
| 3-Year | 16.32ab | 12.22 | 24.38ab | 16.27 | 0.001 |
| dp-ucMGP (pmol/L) | |||||
| Baseline | 482.12 | 312.78 | 551.68 | 220.29 | 0.010 |
| 3-Year | 120.64ab | 136.41 | 166.82ab | 169.58 | 0.023 |
| Plasma Lipids | |||||
| TG (mg/dL) | |||||
| Baseline | 118.1 | 84.2 | 118.5 | 75.5 | 0.648 |
| 3-Year | 114.1 | 90.7 | 114.9 | 52.4 | 0.305 |
| TC (mg/dL) | |||||
| Baseline | 189.8 | 34.2 | 214.2 | 40 | <0.001 |
| 3-Year | 181.3c | 35.2 | 204.9c | 39.9 | <0.001 |
| HDL-C (mg/dL) | |||||
| Baseline | 51.8 | 13.9 | 61.3 | 16.4 | <0.001 |
| 3-Year | 49.6 | 13.7 | 59.1b | 17.0 | <0.001 |
| LDL-C (mg/dL) | |||||
| Baseline | 114.8 | 31.1 | 128.3 | 29.3 | 0.008 |
| 3-Year | 109.3c | 28.9 | 123.2c | 32.9 | 0.008 |
Significant treatment effect (time × treatment interaction) at p <0.05 using linear mixed effect analysis
Significantly different from baseline (corrected p < 0.05) using linear mixed effect analysis
3-year change differed by lipid-lowering medication (corrected p for interaction < 0.05) use using linear mixed effect analysis
Significant treatment effects were observed for the biomarkers of vitamin K (all interaction p < 0.001), but not the lipids (all interaction p > 0.327). In the treatment group, plasma phylloquinone increased in both men and women (corrected p < 0.001), but the increase was greater in women than men (interaction p=0.048). Changes in biomarkers of vitamin K carboxylation did not differ in men and women. None of the changes in biomarkers of vitamin K differed by lipid-lowering medication (all interaction p > 0.100). Because the change in plasma phylloquinone was different in men and women and because there were separate randomization schemes, men and women were analyzed separately in subsequent analyses.
There were no significant changes in TG in men or women (corrected p > 0.506). Changes in TC and LDL-C differed by lipid-lowering medication use (interaction p < 0.001). TC and LDL-C did not change in participants with consistent use or non-use of lipid-lowering medication (corrected p > 1.0, Men: n=58, Women: n=69). TC and LDL-C declined in men and women who started taking lipid-lowering medication during the trial (corrected p< 0.001, n=8 and n=12, respectively). HDL-C did not change in men (corrected p=0.110), but it declined in women (corrected p=0.023). Changes in lipids, with relevant stratification by lipid-lowering medication use, can be viewed in Supporting Information Figure S2.
Associations of plasma lipids with the inter-individual vitamin K response to supplementation
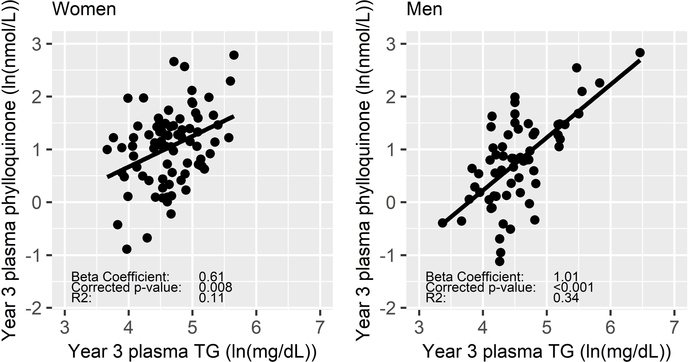
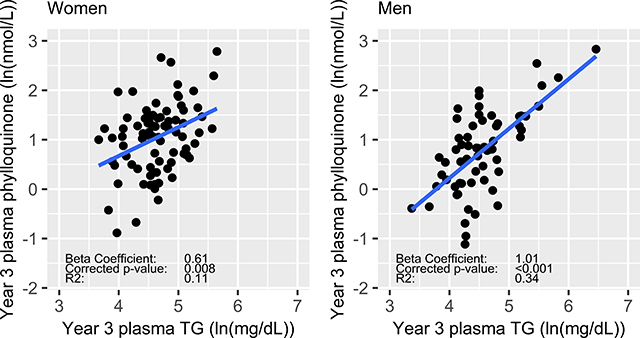
The TC-adjusted association of plasma TG with the inter-individual response of plasma phylloquinone to supplementation was significant in men and women (corrected p < 0.001 and =0.008, respectively) (Figure 1). The effect sizes and explained variability were not markedly changed with year 3 TG < 300 mg/dL (Men: β=1.0, p < 0.001, R2=0.29, Women: β=0.61, p=0.002, R2=0.11). Differences in effect sizes, variability explained, and level of significance are notable, but when men and women were combined the association of TG with the plasma phylloquinone response was not significantly different in men compared to women (interaction p=0.077). TC, LDL-C, and HDL-C were not associated with the response of any biomarker of phylloquinone supplementation in either sex.
Figure 1:
Natural-log transformed year 3 plasma phylloquinone (ln(nmol/L)) and year 3 plasma TG (ln(mg/dL) in women (n=85) and men (n=66).
Associations of SNPs at lipid-related genes with the inter-individual vitamin K response to supplementation
Twenty-seven lipid-related SNPs were genotyped in this group. For the APOE locus, rs7412 and rs429358 were analyzed together to be interpreted as the common APO-ε2, APO-ε3, APO-ε4 alleles. APO-ε4 was assigned as the risk. Two loss of function (LOF) variants in LPL, rs1801177 and rs268, had low MAF (2% and 0.5%, respectively) so their risk alleles were added together to construct an LPL LOF variant. Twenty-five of the 27 SNPs were in HWE. FADS1 (rs174546) and LPL (rs1800590, T-93G) were not (p < 0.02) and were not analyzed further. In total, 23 distinct variants were examined as exposures for the response of vitamin K biomarkers with supplementation.
Lipid-related SNPs and the inter-individual plasma phylloquinone response to supplementation
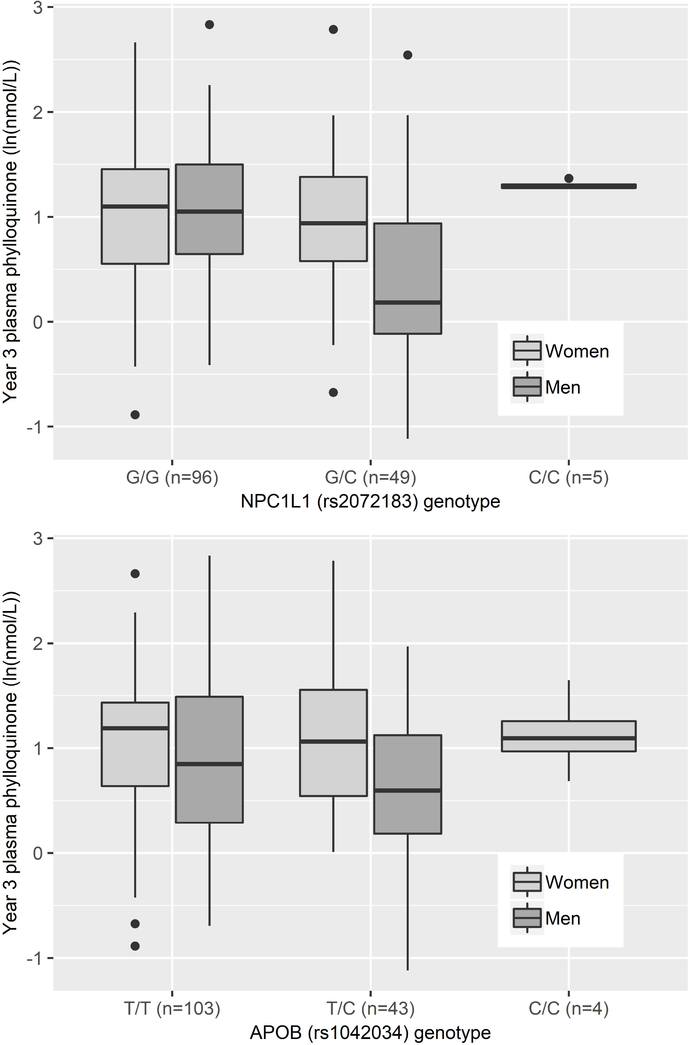
Lipid-related variants were associated with the inter-individual plasma phylloquinone response in men only. In covariate-adjusted models, variants in APOA5 (rs3135506, 56C>G), APOB (rs1042034), NPC1L1, (rs2072183), and ZPR1 (rs964184) were associated with the plasma phylloquinone response at FDR Q < 0.24. After adjusting for TG and TC, the associations of variants in APOA5 and ZPR1 were attenuated while associations of variants in the APOB (rs1042034) and the NPC1L1 (rs2072183) genes were associated with the plasma phylloquinone response at Q < 0.07 (APOB: R2=0.07, NPC1L1: R2=0.05). For both variants, the lipid increasing allele was inversely associated with the plasma phylloquinone response (Figure 2). The other 19 variants were not associated with the plasma phylloquinone response in men in covariate- or lipid-adjusted models. None of the 23 variants were associated with the plasma phylloquinone response in women.
Figure 2:
Association of variants in NPC1L1 (rs2072183, lipid increasing allele=C) and APOB (rs1042034, lipid increasing allele=C) with natural log-transformed year 3 plasma phylloquinone concentrations in men and women.
Lipid-related SNPs and the inter-individual response of biomarkers of vitamin K function to supplementation
Of the 23 lipid-related variants examined, the four variants associated with the inter-individual response of plasma phylloquinone to supplementation in men were not associated with the response of functional markers of vitamin K activity in either sex. None of the genetic variants examined were significantly associated with the response of %ucOC or dp-ucMGP to phylloquinone supplementation in men.
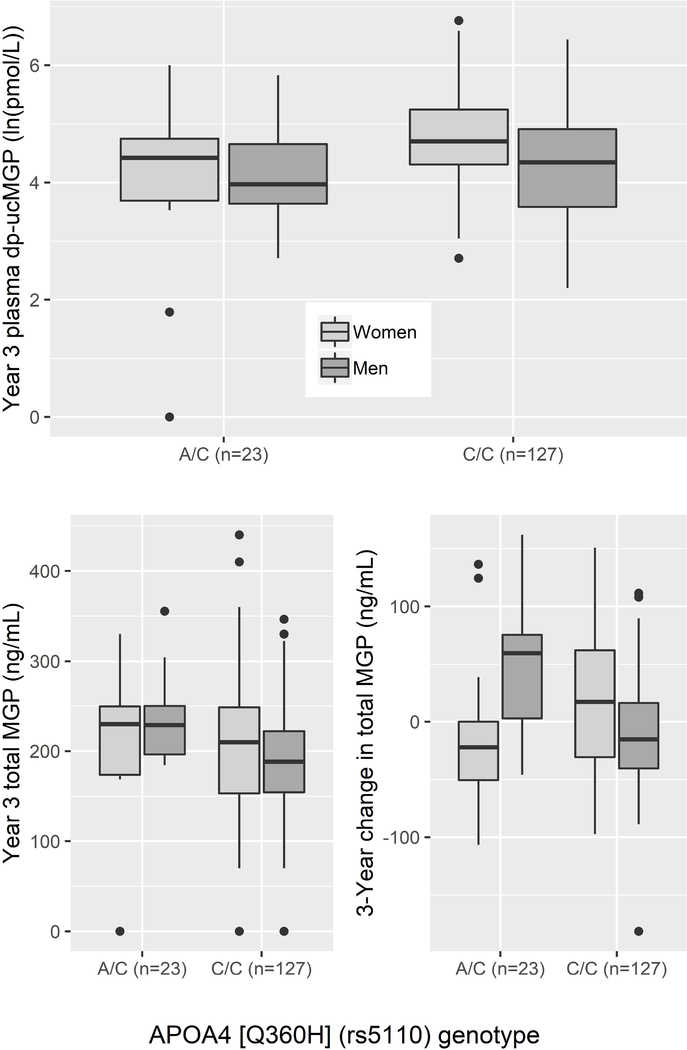
In women, two of the 23 lipid-related variants examined were associated with the dp-ucMGP response to supplementation. SNPs in APOA4 (rs5110, Q360H) (Figure 3) and SORT1 (rs629301) were associated with a smaller dp-ucMGP response (fully adjusted models Q < 0.16). The APOA4 and SORT1 variants explained 13.4% and 5.3% of the variability in the dp-ucMGP response, respectively. These associations were not related to changes in total MGP (p-values for paired t-tests by genotype > 0.15). These two SNPs were not associated with the plasma phylloquinone or %ucOC response to supplementation. While none of the main effects of the 23 variants for the %ucOC response to supplementation were found to be significant, the %ucOC response differed by lipid-lowering medication use in women for variants in LDLR (rs6511720,) GCKR (rs1260326), and TRIB1 (rs2954029) (Q-interaction < 0.07). In those taking lipid-lowering medication, the lipid increasing alleles of LDLR and GCKR were associated with a greater reduction in %ucOC (β =−1.2, p= 0.04 and β=−14.7, p=0.003 and). In those not taking lipid-lowering medication, the variant near TRIB1 was associated with a greater increase in %ucOC (β=10.7, p=0.01).
Figure 3:
Association of the APOA4 variant (rs5110, Q360H, lipid increasing allele=C) with the year 3 plasma dp-ucMGP (natural log-transformed) in men and women. Note: Based on a paired t-test, the change in total MGP from baseline to 3 year was not significantly different for any genotype in women (all p > 0.15). Total MGP declined in men for both genotypes (p=0.04). In men and women, for genotype A/A, n=0.
Lipid-related GRSs and the inter-individual response of biomarkers of vitamin K metabolism to supplementation
Six of the eight TG-associated SNPs[13] showed directionally consistent associations with TG and were included in the wTG-GRS (Supporting Information Table S1). In the covariate-adjusted model, the wTG-GRS was significantly associated with the plasma phylloquinone response in men (R2=0.143) (Supporting Information Table S3). This association was attenuated with adjustment for TG and TC. The wTG-GRS was not associated with the plasma phylloquinone response in women (all corrected p >1.0) nor was it associated with the %ucOC, or dp-ucMGP response. The other wGRS were not associated with the response of any biomarker of vitamin K metabolism.
4). DISCUSSION
Based on the known biology of vitamin K metabolism, specifically the lipoprotein transport of phylloquinone,[9, 10] and preliminary evidence of possible shared genetic architecture with lipids,[12] we hypothesized that lipids contribute to the variable response of biomarkers of vitamin K metabolism to phylloquinone supplementation. Leveraging data from a 3-year randomized, controlled, 500 μg/d phylloquinone supplementation trial, we tested the associations of blood lipids, 23 established lipid-related SNPs, and four wGRS with changes in biomarkers of vitamin K metabolism. On average plasma phylloquinone increased in response to supplementation, but plasma TG were an important contributor to the inter-individual variability in the change in plasma phylloquinone over the 3-year study. In our genetically-based analyses, the association between TG and the plasma phylloquinone response was only observed in men. Biomarkers of vitamin K function (%ucOC and dp-ucMGP) also responded to phylloquinone supplementation, but the inter-individual change in these measures did not differ by lipids, lipid-variants, or by sex.
In this study, plasma TG, but not TC, LDL-C, or HDL-C, were directly associated with the plasma phylloquinone response to supplementation. That only TG were associated with the plasma phylloquinone response is consistent with the predominance of circulating phylloquinone carried by the TG-rich lipoproteins.[8–10] In our study, TG accounted for 34% and 11% of the variability in the plasma phylloquinone response in men and women, respectively. Other biologically plausible factors, specifically lipid-lowering medication use, percent body fat, and smoking status at baseline, only explained an additional 7.6% in men and 1.8% in women. Though the association of TG with the plasma phylloquinone response was not significantly different in men compared to women, the differences in effect sizes, explained variability, and level of significance are notable. Furthermore, at the genetic level, the association of the wTG-GRS with the plasma phylloquinone response was only observed in men. These data suggest the relationship between TG and the plasma phylloquinone response to supplementation may be stronger in men than women. The association of lipids with circulating phylloquinone was analyzed in cross-sectional analyses of the Multi-Ethnic Study of Atherosclerosis (MESA) and Framingham Offspring Study (FOS) and the results were consistent with ours. In MESA, TG explained a large proportion of the variance (race/ethnicity: 12.1%, TG: 7.4%, total explained: 20.9%) in circulating phylloquinone in men and women combined. TC was not a contributing factor.[5] In FOS, where men and women were analyzed separately, TG explained the largest proportion of the variance in circulating phylloquinone in men (15.7% of 20.1% total) compared to other variables examined. In women, TG also contributed (2.8% of 12.3% total), but dietary intake of phylloquinone was a stronger predictor (7.4%).[4] LDL-C and TC only explained 0.4% and 1.5% of the variance in circulating phylloquinone in men and women, respectively.
To investigate the role of lipid genetics in the response of biomarkers of vitamin K metabolism to supplementation further, we identified and genotyped 27 SNPs that have been consistently linked to TG, TC, LDL-C, and/or HDL-C for a candidate SNP analysis.[13, 14, 34–46] No lipid-related SNPs were associated with the plasma phylloquinone response to supplementation in women. In men, variants in APOB (rs1042034), NPC1L1 (rs2072183), APOA5 (rs3135506), and ZPR1 (rs964184) were associated with the plasma phylloquinone response to supplementation in the covariate-adjusted model. After adjusting for plasma lipids, only the associations of SNPs in APOB and NPC1L1 remained significantly associated with the plasma phylloquinone response which suggests the associations with variants in APOA5 and ZPR1 were driven by plasma TG. In a genome-wide meta-analysis of circulating phylloquinone, the variant in ZPR1 (rs964184) was also associated with circulating phylloquinone in the covariate-adjusted model.[12] As in our study, the association with rs964184 was attenuated with adjustment for TG.
Associations of variants in NPC1L1 and APOB persisted after adjusting for TG and TC. This suggests these lipid genes may have roles in regulating phylloquinone metabolism that are independent of circulating lipid concentrations. NPC1L1 expression is enriched in the brush-border membrane of intestinal enterocytes, is recognized as a transporter of cholesterol and α-tocopherol,[54, 55] and preliminary data from cell and animal experiments and a retrospective analysis of clinical data suggest that NPC1L1 is involved in the intestinal absorption of vitamin K.[56] ApoB is highly expressed in the liver and the intestine. Since adjustment for plasma lipids strengthened the association between the genetic variants in APOB and the plasma phylloquinone response, we hypothesize that this association is related to apoB’s role in phylloquinone’s delivery to circulation. The sex-specific associations of these SNPs with the plasma phylloquinone response to supplementation needs to be validated in larger cohorts.
An important strength of this study is that in addition to the response of plasma phylloquinone, we analyzed the association of lipids and lipid-related genetic variants with the inter-individual response of biomarkers of vitamin K carboxylation. Whereas there was a consistent influence of lipids on plasma phylloquinone, surprisingly the same was not observed for the biomarkers, %ucOC and dp-ucMGP. For %ucOC, there was no significant association of any plasma lipids studied on the change in %ucOC in response to phylloquinone supplementation. Consistent with this observation, there was no genetic influence on this biomarker using the panel of 23 lipid-related candidate genetic variants. Whereas there was no observed influence of circulating lipids on dp-ucMGP, two (APOA4 [rs5110 Q360H], SORT1 [rs629301]) of the 23 variants were associated with the dp-ucMGP response to supplementation in women, not men. The association was significant even with adjustment for lipids suggesting the association was independent of lipids. With respect to vitamin K function, %ucOC and dp-ucMGP reflect vitamin K’s activity as an enzyme cofactor in specific tissues.[11] One could interpret these data to mean that vitamin K activity in tissues of interest is more reflected by carboxylation of these proteins and that the influence of circulating lipids is moot. However, neither of these biomarkers have yet to be calibrated to a specific physiologic response, hence the interpretation of how this relates to vitamin K function is not known.[22] Furthermore, there is growing evidence that vitamin K has physiological roles independent of an enzyme cofactor[57] so caution must be taken in assuming the response of biomarkers indicative of carboxylation are all-inclusive representation of vitamin K activity. These data however, do support the use of multiple biomarkers of vitamin K to obtain a robust measure of vitamin K status particularly as our data demonstrate that different biomarkers respond differently to lipids and their associated genetic variants.[11]
Additional strengths of our study are the measurement of plasma phylloquinone from fasting samples, hence the inter-individual variation in the postprandial response to dietary phylloquinone intake[23, 58, 59] which may vary with meal patterns,[60] was not introduced into our analysis. Historically, some variability in circulating phylloquinone was attributed to assay differences. However, with the implementation of an international standardized external quality assurance program,[61] this source of variation is minimized when the measures are conducted by laboratories that participate in the program, as we do. Nonetheless, large inter-individual variability remains[4, 5] and the source of this variation merits future study.
There are several important limitations of this study. All participants were white, which limits the generalizability to other racial/ethnic groups. A large percentage of participants were taking or started taking lipid-lowering medication during the study (43% of women and 33% of men). Most took the medication throughout the trial and TC and LDL-C did not change in these groups. However, the variable effects of these medications on TC and LDL-C may be considerable. To account for this, we adjusted for lipid-lowering medication use and tested whether the association of each lipid with the response of each biomarker of vitamin K metabolism differed by lipid-lowering medication use. Regardless, it is prudent to interpret the results related to TC and LDL-C cautiously. Finally, a large number of exposures were tested given the small size of the sample. Since the nature of this study was to provide preliminary evidence, we set a lenient FDR Q threshold of 0.3 to detect potential associations for the lipid-related SNPs. We also constructed four wGRSs which, compared to a single SNP, has a greater genetic load. Using a wGRS also consolidates the number of exposures (4 wGRS vs 23 SNPs). Both qualities increased power to detect genetic based associations.
We examined whether lipids and lipid-related genetic variants contribute to the inter-individual response of biomarkers of vitamin K metabolism with 3 years of daily phylloquinone supplementation. We observed that on average plasma phylloquinone increased, but plasma TG were an important determinant of the inter-individual change in plasma phylloquinone. A stronger influence in men than women was supported by our genetically-based analyses, but this requires further examination. The biomarkers of vitamin K function analyzed in this study also responded to supplementation, but the change in these measures were not related to lipids highlighting the need for a rigorous investigation of the relationships amongst the various biomarkers of vitamin K metabolism.
Supplementary Material
ACKNOWLEDGEMENTS
This study was supported by the USDA Agricultural Research Service under Cooperative Agreement No. 58-1950-7-707. JMK, SLB, JMO, CES, GM, GSH, HSD conceived and designed research; RI Performed genotyping; JMK conducted statistical analyses; JMK, SLB, JMO, CES, GM, GSH interpreted results; JMK, SLB wrote manuscript; All authors read and revised and approved the final manuscript.
Abbreviations
- CVD
Cardiovascular Disease
- dp-ucMGP
Dephosphorylated-undercarboxylated Matrix Gla Protein
- FOS
Framingham Offspring Study
- GWAS
Genome-wide Association Study
- HDL-C
High-density lipoprotein-cholesterol
- HNRCA
Human Nutrition Research Center for Aging
- LOF
Loss of Function
- LDL-C
Low-density lipoprotein-cholesterol
- MGP
Matrix Gla Protein
- MESA
Multi-Ethnic Study of Atherosclerosis
- %ucOC
Percent Undercarboxylated Osteocalcin
- SNP
Single Nucleotide Polymorphism
- TC
Total Cholesterol
- TG
Triglycerides
- VKDPs
Vitamin K-Dependent Proteins
- wGRS
Weighted Genetic Risk Score
Footnotes
CONFLICT OF INTEREST STATEMENT
The authors have declared no conflict of interest.
Disclaimers: Any opinions, findings, conclusions, or recommendations expressed in this publication are those of the authors and do not necessarily reflect the views of the USDA.
Clinical Trial Registry number and website: This trial was registered at clinicaltrials.gov as NCT00183001
5) REFERENCES
- [1].Berkner KL, Runge KW, J. Thromb. Haemost. 2004, 2, 2118. [DOI] [PubMed] [Google Scholar]
- [2].Shea MK, O’Donnell CJ, Hoffmann U, Dallal GE, Dawson-Hughes B, Ordovas JM, Price PA, Williamson MK, Booth SL, Am J Clin Nutr 2009, 89, 1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Dalmeijer GW, van der Schouw YT, Booth SL, de Jong PA, Beulens JWJ, Arterioscler. Thromb. Vasc. Biol. 2014, 34, 1587. [DOI] [PubMed] [Google Scholar]
- [4].Shea MK, Benjamin EJ, Dupuis J, Massaro JM, Jacques PF, D’Agostino RBS, Ordovas JM, O’Donnell CJ, Dawson-Hughes B, Vasan RS, Booth SL, Eur. J. Clin. Nutr. 2009, 63, 458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Shea MK, Booth SL, Nettleton JA, Burke GL, Chen H, Kritchevsky SB, J. Nutr. 2012, 142, 1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Booth SL, Food Nutr. Res. 2012, 56, 5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Schurgers LJ, Vermeer C, Biochim. Biophys. Acta 2002, 1570, 27. [DOI] [PubMed] [Google Scholar]
- [8].Shearer MJ, Newman P, Thromb. Haemost. 2008, 100, 530. [PubMed] [Google Scholar]
- [9].Lamon-Fava S, Sadowski JA, Davidson KW, O’Brien ME, McNamara JR, Schaefer EJ, Am. J. Clin. Nutr. 1998, 67, 1226. [DOI] [PubMed] [Google Scholar]
- [10].Erkkila AT, Lichtenstein AH, Dolnikowski GG, Grusak MA, Jalbert SM, Aquino KA, Peterson JW, Booth SL, Metabolism. 2004, 53, 215. [DOI] [PubMed] [Google Scholar]
- [11].Shea MK, Booth SL, Nutrients 2016, 8, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Dashti HS, Shea MK, Smith CE, Tanaka T, Hruby A, Richardson K, Wang TJ, Nalls MA, Guo X, Liu Y, Yao J, Li D, Johnson C, Benjamin EJ, Kritchevsky SB, Siscovick DS, Ordovás JM, Booth SL, Am J Clin Nutr 2014, 100, 1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].C. J. Global Lipids Genetics Consortium, Willer, Schmidt EM, Sengupta S, Peloso GM, Gustafsson S, Kanoni S, Ganna A, Chen J, Martin L, Mora S, Beckmann JS, Bragg-gresham JL, Ehret GB, Esko T, Feitosa MF, Ferreira T, Papamarkou T, Pomilla C, Pouta A, Nat. Genet. 2013, 45, 1.23268125 [Google Scholar]
- [14].Teslovich T, Musunuru K, Smith A, Edmondson A, Stylianou I, Koseki M, Pirruccello J, Ripatti S, Chasman D, Willer C, Johansen C, Fouchier S, Isaacs A, Peloso G, Barbalic M, Ricketts S, Bis J, Aulchenko Y, Thorleifsson G, Nature 2010, 466, 707.20686565 [Google Scholar]
- [15].Shearer MJ, Newman P, J. Lipid Res. 2014, 55, 345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Vanderschueren D, Gevers G, Raymaekers G, Devos P, Dequeker J, Calcif. Tissue Int. 1990, 46, 179. [DOI] [PubMed] [Google Scholar]
- [17].Truong JT, Fu X, Saltzman E, Al Rajabi A, Dallal GE, Gundberg CM, Booth SL, J. Nutr. 2012, 142, 936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Gundberg CM, Nieman SD, Abrams S, Rosen H, J Clin Endocrinol Metab 1998, 83, 3258. [DOI] [PubMed] [Google Scholar]
- [19].Booth SL, Dallal G, Shea MK, Gundberg C, Peterson JW, Dawson-Hughes B, J. Clin. Endocrinol. Metab. 2008, 93, 1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Luo G, Ducy P, McKee MD, Pinero GJ, Loyer E, Behringer RR, Karsenty G, Nature 1997, 386, 78. [DOI] [PubMed] [Google Scholar]
- [21].Shea MK, Donnell CJO, Vermeer C, Magdeleyns EJP, Crosier MD, Gundberg CM, Ordovas J, Kritchevsky SB, Booth SL, Nutr J. 2011, 141, 1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Food and Nutrition Board. Institute of Medicine, 2001 Washington, DC, 2001. [Google Scholar]
- [23].Booth SL, Tucker KL, Mckeown NM, Davidson KW, Dallal GE, Sadowski JA, J Nutr 1997, 127, 587. [DOI] [PubMed] [Google Scholar]
- [24].Booth SL, Webb R, Peters JC, J. Am. Diet. Assoc. 1999, 99, 1072. [DOI] [PubMed] [Google Scholar]
- [25].Schurgers LJ, Spronk HMH, Skepper JN, Hackeng TM, Shanahan CM, Vermeer C, Weissberg PL, Proudfoot D, J. Thromb. Haemost. 2007, 5, 2503. [DOI] [PubMed] [Google Scholar]
- [26].Davidson KW, Sadowski JA, Methods Enzymol. 1997, 282, 408. [DOI] [PubMed] [Google Scholar]
- [27].Otawara Y, Price PA, J. Biol. Chem. 1986, 261, 10828. [PubMed] [Google Scholar]
- [28].Price PA, Rice JS, Williamson MK, Protein Sci. 1994, 3, 822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Price PA, Williamson MK, J. Biol. Chem. 1985, 260, 14971. [PubMed] [Google Scholar]
- [30].Cranenburg ECM, Koos R, Schurgers LJ, Magdeleyns EJ, Schoonbrood THM, Landewé RB, Brandenburg VM, Bekers O, Vermeer C, Thromb. Haemost. 2010, 104, 811. [DOI] [PubMed] [Google Scholar]
- [31].McNamara JR, Schaefer EJ, Clin. Chim. Acta 1987, 166, 1. [DOI] [PubMed] [Google Scholar]
- [32].Warnick GR, Benderson J, Albers J, Baillie EE, Sexton B, Schaefer EJ, Carison D, Brewer HB, Clin Chem 1982, 28, 1379. [PubMed] [Google Scholar]
- [33].Friedewald WT, Levy RI, Fredrickson DS, Clin. Chem. 1972, 18, 499. [PubMed] [Google Scholar]
- [34].Ken-Dror G, Goldbourt U, Dankner R, J. Hum. Genet. 2010, 55, 300. [DOI] [PubMed] [Google Scholar]
- [35].Hubacek JA, Gene 2016, 592, 193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Aouizerat BE, Kulkarni M, Heilbron D, Drown D, Raskin S, Pullinger CR, Malloy MJ, Kane JP, J. Lipid Res. 2003, 44, 1167. [DOI] [PubMed] [Google Scholar]
- [37].Caussy C, Charrière S, Marçais C, Di Filippo M, Sassolas A, Delay M, Euthine V, Jalabert A, Lefai E, Rome S, Moulin P, Am. J. Hum. Genet. 2014, 94, 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Guardiola M, Ribalta J, Curr. Atheroscler. Rep. 2017, 19. [DOI] [PubMed] [Google Scholar]
- [39].Pennacchio LA, Olivier M, Hubacek JA, Cohen JC, Cox DR, Fruchart JC, Krauss RM, Rubin EM, Science. 2001. 169. [DOI] [PubMed] [Google Scholar]
- [40].Pennacchio LA, Hum. Mol. Genet. 2002, 11, 3031. [DOI] [PubMed] [Google Scholar]
- [41].Chandak GR, Ward KJ, Yajnik CS, Pandit AN, Bavdekar A, V Joglekar C, Fall CHD, Mohankrishna P, Wilkin TJ, Metcalf BS, Weedon MN, Frayling TM, Hattersley AT, BMC Med. Genet. 2006, 7, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Talmud PJ, Hawe E, Martin S, Olivier M, Miller GJ, Rubin EM, Pennacchio LA, Humphries SE, Hum. Mol. Genet. 2002, 11, 3039. [DOI] [PubMed] [Google Scholar]
- [43].Weinberg RB, Genet. Mol. Biol. 2002, 81, 125. [Google Scholar]
- [44].Dancer M, Marcais C, Caussy C, Di filippo M, Moulin P, Charrière S, Atherosclerosis 2016, 252, e74. [DOI] [PubMed] [Google Scholar]
- [45].Shearer MJ, Fu X, Booth SL, Adv. Nutr. 2012, 3, 182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Julve J, Martín-Campos JM, Escolà-Gil JC, Blanco-Vaca F, Clin. Chim. Acta 2016, 455, 134. [DOI] [PubMed] [Google Scholar]
- [47].Peter I, Crosier M, Yoshida M, Osteoporos. Int. 2011, 22, 1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Renstrom F, Shungin D, Johansson I, the MAGIC Investigators, Florez JC, Hallmans G, Hu FB, Franks PW, Diabetes 2011, 60, 345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Ahmad S, Mora S, Franks PW, Orho-melander M, Ridker PM, Hu FB, Chasman DI, Clin. Chem. 2018, 64, 231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Justesen JM, Allin KH, Sandholt CH, Borglykke A, Krarup NT, Grarup N, Circ Cardiovasc Genet 2015, 8, 465. [DOI] [PubMed] [Google Scholar]
- [51].Rajabi A, Peterson J, Choi SW, Suttie J, Barakat S, Booth SL, J Chromatogr B Anal. Technol Biomed Life Sci 2010, 878, 2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Lian JB, Gundberg CM, Clin. Orthop. Relat. Res.267. [PubMed] [Google Scholar]
- [53].Benjamini Y, Hochberg Y, Stat JR. Soc. 1995, 57, 289. [Google Scholar]
- [54].Davis HR, Zhu L, Hoos LM, Tetzloff G, Maguire M, Liu J, Yao X, Iyer SPN, Lam M, Lund EG, Detmers PA, Graziano MP, Altmann SW, J. Biol. Chem. 2004, 279, 33586. [DOI] [PubMed] [Google Scholar]
- [55].Narushima K, Takada T, Yamanashi Y, Suzuki H, Mol. Pharmacol. 2008, 74, 42. [DOI] [PubMed] [Google Scholar]
- [56].Takada T, Yamanashi Y, Konishi K, Yamamoto T, Toyoda Y, Masuo Y, Yamamoto H, Suzuki H, Sci. Transl. Med. 2015, 7, 1. [DOI] [PubMed] [Google Scholar]
- [57].Shearer MJ, Okano T, Annu. Rev. Nutr. 2018, 38, 127. [DOI] [PubMed] [Google Scholar]
- [58].Booth SL, O’Brien-Morse ME, Dallal GE, Davidson KW, Gundberg CM, Am. J. Clin. Nutr. 1999, 70, 368. [DOI] [PubMed] [Google Scholar]
- [59].Novotny JA, Kurilich AC, Britz SJ, Baer DJ, Clevidence BA, Br. J. Nutr. 2010, 104, 858. [DOI] [PubMed] [Google Scholar]
- [60].Jones KS, Bluck LJC, Wang LY, Stephen AM, Prynne CJ, Coward WA, Br. J. Nutr. 2009, 102, 1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Card DJ, Shearer MJ, Schurgers LJ, Harrington DJ, Biomed. Chromatogr. 2009, 23, 1276. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.