Abstract
Purpose:
Genetic differences in immunity may contribute to toxicity and outcomes with immune checkpoint inhibitor (CPI) therapy, but these relationships are poorly understood. We examined the genetics of immune-related thyroid dysfunction (thyroid irAEs).
Experimental Design:
In patients with NSCLC treated with CPIs at MSK and VUMC, we evaluated thyroid irAEs. We typed germline DNA using genome-wide single nucleotide polymorphism (SNP) arrays and imputed genotypes. Germline SNP imputation was also performed in an independent DFCI cohort. We developed and validated polygenic risk scores (PRS) for hypothyroidism in non-cancer patients using the UK and VUMC BioVU biobanks. These PRSs were applied to thyroid irAEs and CPI response in NSCLC patients at MSK, VUMC, and DFCI.
Results:
Among 744 patients at MSK and VUMC, thyroid irAEs occurred in 13% and were associated with improved outcomes (PFS aHR=0.68, 95%CI: 0.52–0.88). The PRS for hypothyroidism developed from UK Biobank predicted hypothyroidism in the BioVU dataset in non-cancer patients (OR per standard deviation [SD]=1.33, 95%CI: 1.29–1.37; AUROC= 0.6). The same PRS also predicted development of thyroid irAEs in both independent cohorts of patients treated with CPIs (HR per SD=1.34, 95%CI: 1.08–1.66; AUROC 0.6). The results were similar in the DFCI cohort. However, PRS for hypothyroidism did not predict CPI benefit.
Conclusions:
Thyroid irAEs were associated with response to anti-PD-1 therapy. Genetic risk for hypothyroidism was associated with risk of developing thyroid irAEs. Additional studies are needed to determine if other irAEs also have shared genetic risk with known autoimmune disorders and the association with treatment response.
Keywords: immune checkpoint inhibitors, Immune-related adverse events, polygenic risk scores, spontaneous autoimmune hypothyroidism, genome wide association studies
Introduction:
Immune checkpoint inhibitor (CPI) based therapy, which targets the adaptive immune system, is associated with remarkable long-term responses in a subset of many cancers (1–3). Benefit from CPI therapy is affected by tumor-specific features, such as PD-L1 expression (4,5) or tumor mutational burden (6) and host-specific features associated with underlying immunity such as the microbiome (7–9) and, possibly the germline genetics of the host (10–14).
The dual role of CPIs in promoting T cell activation, but also autoimmunity leads to a variety of clinically significant systemic autoinflammatory responses (immune related adverse events, irAEs) in a subset of individuals(15). The presentations of irAEs often mimic autoimmune conditions that occur spontaneously, but it is unclear if the underlying mechanism is shared or distinct despite the phenotypic similarity. Furthermore, it is uncertain whether and which irAEs are associated with CPI benefit. While some studies show an association between an irAE and improved outcomes(16–19) others do not(20,21), and some show worse outcomes(22,23). These conflicting findings may be due to factors such as (a) survivor bias (patients who respond to therapy and are on therapy longer are more likely to develop irAEs) and (b) heterogeneous cohorts (combining cancer types, irAEs with different pathophysiology, ranges of presentation/ severity and different types of treatment of irAEs). We hypothesized that examining a specific irAE in a single cancer type that is routinely observed early in the CPI treatment course would yield clarity on the relationship between irAE development, immunity, and CPI benefit.
Thyroid irAEs occur early in the course of CPI exposure(24,25) and are among the most common irAE, with a cumulative incidence of approximately 10% of patients on PD-1 blockade therapy(26,27). Spontaneous autoimmune hypothyroidism is common and appears to be at least partially heritable(28,29). Genome-wide association studies (GWAS) have identified many common variants contributing to risk(30). It is unclear whether hypothyroidism that occurs following PD-1 blockade therapy is genetically similar to hypothyroidism that occurs in the general population. Additionally, whether this genetic risk impacts PD-1 blockade benefit is unknown.
To further understand the relationships between underlying autoimmunity, thyroid irAEs, and immunotherapy outcomes, we examined patients with non-small cell lung cancer (NSCLC) receiving PD-1 blockade-based therapy in cohorts from 3 academic medical centers. To investigate the genetics of thyroid irAEs, we first developed a genetic predictor of spontaneous hypothyroidism in non-cancer patients. We first used the UK Biobank, an open access resource containing genetic and health related traits from ~500,000 volunteers in the UK and built a polygenic risk score (31) which is a risk score that integrates the information from many polymorphisms in the genome. We then validated that score in non-cancer patients in the Vanderbilt University biobank (BioVU). Finally, we evaluated whether genetic risk of sporadic hypothyroidism by GWAS is associated with incident hypothyroidism following PD-1 blockade and also evaluated the association between genetic risk and survival.
Materials and Methods:
Patients
This retrospective study was approved by the institutional review board at MSK, VUMC, and DFCI. All patients at MSK and DFCI provided written informed consent for blood banking. Patients in this study had advanced NSCLC and received PD-1 blockade-based therapy (either PD-1 blockade monotherapy or in combination with CTLA-4 blockade). In the MSK cohort, 551 patients who received CPIs between 2011 – 2018 and had an available baseline blood sample were included (Table 1). In the VUMC population, 195 individuals who had received CPIs between 2009 – 2019 were identified from BioVU, VUMC’s DNA biobank linked to de-identified electronic health records(32). At DFCI, 561 patients with NSCLC who received CPIs between 2013 – 2020 were examined.
Table 1:
Baseline patient characteristics and treatment details
| Characteristic | MSKCC | VUMC | DFCI |
|---|---|---|---|
| (n=551) | (n=193) | (n=561) | |
| Median age [IQR] - yr | 67 [59–73] | 63 [57–69] | 67 [60–74] |
| Biological sex - no. (%) | |||
| Female | 291 (53%) | 74 (38%) | 313 (56%) |
| Male | 260 (47%) | 119 (62%) | 248 (44%) |
| Race, self-reported - no./total no. (%) | |||
| White | 458/533 (86%) | 176/191 (92%) | 506/551 (92%) |
| Black | 39/533 (7%) | 11/191 (6%) | 22/551 (4%) |
| Asian | 34/533 (6%) | 2/191 (1%) | 17/551 (3%) |
| Other | 2/553 (<1%)* | 2/191 (1%) | 6/551 (1%) |
| Unknown | 18 | 2 | 10 |
| Ethnicity, self-reported. - no./total no. (%) | |||
| Hispanic or Latino | 18/545 (3%) | 2/186 (1%) | 10 (2%) |
| Non-Hispanic or Latino | 527/545 (96%)* | 184/186 (99%) | 551 (98%) |
| Unknown | 6 | 7 | 0 |
| Cigarette smoking status - no. (%) | |||
| Former or current | 472 (86%) | 166/188 (88%) | 363/421 (86%) |
| Never | 79 (14%) | 22/188 (12%) | 58/421 (14%) |
| Unknown | 0 | 5 | 140 |
| Histology - no. (%) | 5 | 140 | |
| Adenocarcinoma | 427 (78%) | 120 (62%) | 416 (74%) |
| Non-adenocarcinoma | 124 (22%) | 73 (38%) | 145 (26%) |
| Treatment - no. (%) | |||
| Anti-PD-(L)1 monotherapy | 480 (87%) | 179 (93%) | 527 (94%) |
| Anti-PD-(L)1+CTLA-4 combination | 71 (13%) | 14 (7%) | 34 (6%) |
Percentages may not add up to 100 due to rounding.
hyperthyroidism (n=2), thyroid cancer or metastasis (n=4);
IQR = interquartile range, PD-(L)1 = programmed cell death protein (ligand) 1, TPS = tumor proportion score, CTLA-4 = cytotoxic T-lymphocyte-associated protein 4
For the germline genetic analysis, patients from MSK and VUMC (MSK and VUMC cohort) were analyzed together as they were both genotyped using the same GWAS array. Patients from DFCI were analyzed separately as an independent cohort.
Clinical Variables
MSK medical records and pharmacy records were reviewed for age, self-reported demographics, smoking status, lung cancer histology, past medical history, and CPI treatment history. These elements were abstracted and entered into a clinical data sheet. VUMC data were extracted using MedEx or from structured tables within existing de-identified medical records (29). All treatment dates underwent manual chart review by a trained thoracic oncology nurse, and data were entered into a REDCap database.
Thyroid irAE event
A thyroid event after the start of CPI therapy was defined as either (1) incident hypothyroidism or (2) transient incident hyperthyroidism followed by incident hypothyroidism. Incident hypothyroidism was defined as (a) a TSH of ≥ 10 mU/L or (b) TSH of ≥ 5 mU/L with a new prescription of levothyroxine ≥ 50 mcg. Incident hyperthyroidism was defined as TSH < 0.05 mU/L. Incident hyperthyroidism without subsequent hypothyroidism (n=2) was excluded from the analysis due to testing shortly before patients transitioned to hospice with no additional follow up thereafter. A baseline history of hypothyroidism or hyperthyroidism was defined as documentation of a thyroid condition prior to the start of CPI therapy. We excluded these patients from the analysis due to the challenge in defining whether a thyroid irAE occurred.
In the MSK cohort, medical records were manually reviewed to identify cases of thyroid events. Extracted laboratory and medication data were used to identify thyroid events in the VUMC population, with manual chart review confirmation. In the DFCI cohort, extracted laboratory and medication data were similarly used to identify thyroid events.
Response assessment
In the MSK cohort, best overall response (BOR) was assessed by investigator-assessed Response Evaluation Criteria in Solid Tumors (RECIST) v1.1 from the start of CPI therapy to the date of tumor progression or death due to any cause. Progression-free survival (PFS) and overall survival (OS) was assessed from the date of start of CPI therapy.
In the DFCI cohort, overall survival (OS) was assessed from the date of start of CPI therapy, with linkage to the National Death Index (NDI) up to 12/2019. Patients treated after the NDI check were censored at last contact at DFCI. The VUMC and DFCI cohorts did not have response assessment available.
Genotyping of MSK and VUMC samples
Blood DNA from MSK and VUMC were genotyped on the Affymetrix Axiom Precision Medicine Diversity Array. Imputation was performed on the Michigan Imputation Server using the 1000 Genomes phase 3 v5 reference panel. Standard quality control measures were implemented to remove poorly genotyped samples (call rate < 95%), SNPs (genotyping rate < 95%), and rare variants (minor allele frequency < 0.005) prior to imputation. After dropping samples with missing genotypes or low quality genotypes we had a total of N=729 (N=536 from MSKCC and N=193) in the genetics dataset.
Genotype imputation of DFCI samples from tumor sequencing
Samples from DFCI had tumor panel sequencing as part of routine clinical care from which germline variants were imputed using off-target reads. OncoPanel, a custom targeted hybrid capture sequencing platform, was used to assay genomic variation from tumor biopsies. Germline variant imputation was performed across all samples from raw sequence reads using the STITCH imputation software, which leverages ultra-low coverage read data together with the 1000 Genomes phase 3 v5 reference panel to infer probabilistic germline calls for the autosomal chromosomes. Quality control was performed to remove poorly imputed variants (INFO < 0.4) and rare variants (minor allele frequency < 0.01).
Polygenic Risk Assessment
Summary statistics were obtained from previous genome-wide association studies of 2 thyroid-related phenotypic outcomes in the UK Biobank - self-reported hypothyroidism (22,500 individuals, 436,818 controls)(33), and thyroid medication use (H03A medication category; 24,835 individuals, 280,750 controls)(34). Polygenic risk scores (PRS) were constructed using LDpred, which estimates posterior mean effect sizes using Bayesian priors and linkage disequilibrium information(35). PRS weights are available at http://doi.org/10.5281/zenodo.5034793.
Quality control of PRS in DFCI samples
A partially overlapping set of 833 patients with a variety of solid tumors seen at the DFCI had both OncoPanel next generation sequencing (as part of the same clinical cohort) and direct germline genotyping (as part of an orthogonal biobank) were used to validate the tumor imputed PRS. DNA samples were processed from whole blood and genotyped on the Illumina Multi-Ethnic Genotyping Array (MEGA), the Expanded Multi-Ethnic Genotyping Array (MEGA Ex) array, and the Multi-Ethnic Global (MEG) BeadChip; imputed to the Haplotype Reference Consortium reference panel; and then restricted to ~1.1 million HapMap3 variants that typically exhibit high imputation accuracy across genotyping platforms. PRSs were inferred using the imputed variants that passed quality control in each respective study (i.e. no harmonization across the two platforms was imposed).
External Validation of Polygenic Risk Score
External validation of the PRS was performed in a population of 51,070 individuals of European descent with no cancer diagnosis in BioVU. European ancestry was determined using principal components analysis (PCA). Individuals in this cohort were genotyped on the Illumina Expanded Multi-Ethnic Genotyping Array and subjected to standard quality control. Imputation was performed with the Haplotype Reference Consortium reference panel on the Michigan Imputation Server(36). PRS were calculated using the previously derived weights from LDpred and the score function in PLINK 1.9(37,38). Spontaneous hypothyroidism cases and controls were defined using phecodes, which aggregate similar ICD-9-CM and ICD-10-CM. Individuals must have at least 2 ICD codes for hypothyroidism to be assigned a phecode, and individuals with other thyroid diseases were excluded from the control set.
Ancestry analysis
We determined genetic ancestry using PCA in PLINK. PCA was conducted in each genotype data separately: BioVU dataset without cancer and CPI (MEGA array), the MSK and VUMC cohorts (Affymetrix PMDA array) and DFCI dataset which included imputed SNPs from low coverage sequencing.
Statistical Analyses
De-identified data on duration of treatment and vital status at the time of the database lock on October 1, 2020 for the MSK cohort was used for the survival analysis. Patients who did not experience the event of interest at the database lock were censored at the time of the last follow up date. Survival analyses for PFS and OS were performed using Kaplan-Meier time-to-event estimates. We also determined the association between incident thyroid events and PFS and OS using multivariate Cox regression. We ran the Cox regression encoding thyroid events as a dichotomous variable counting any thyroid event during therapy. In addition, to account for potential survival bias, we also conducted analyses in which thyroid events were tested as time-dependent covariates. As a sensitivity analysis, landmark analysis in patients with OS of at least 90 days was performed in the MSK cohort. Ninety days was chosen as it includes more than half of patients with a thyroid irAE event (n=37/65, 57%). All models included age, sex, and concurrent use of anti-CTLA-4 therapy as covariates.
To test the association between prevalent hypothyroidism and PRS in BioVU (non-cancer patients), we used logistic regression models and adjusted for age at last visit, sex, and the first 10 principal components to account for genetic ancestry.
The association of PRS with incident hypothyroidism in the context of CPI therapy was tested using multivariate Cox regression with covariates for age, sex, and the first 10 principal components, and Hazard Ratios (HRs) were computed per standard deviation of the PRS. In the DFCI cohort, additional technical covariates were included for sequencing panel version, whether the patient was sequenced after the start of therapy, and number of prior therapy lines.
Receiver operating characteristic (ROC) curves were used to visualize how well the PRS model discriminated hypothyroidism events and area under the ROC curve (AUROC) was estimated to quantify the overall prediction accuracy of the PRS. All p-values were two-sided. Statistical analysis and data visualization was performed using R v4.0.3 (R foundation for Statistical Computing, Vienna, Austria) with RStudio v1.3.1093.
A summary of the methods is illustrated in Supplementary Figure 1.
Individual Variant Testing
To better understand the shared biology between thyroid irAEs and spontaneous hypothyroidism, we analyzed the association with individual SNPs and thyroid irAEs. We focused on 16,751 SNPs that were noted to be genome wide significant (p<5×10−8) from the UK Biobank dataset in the GWAS for self-reported hypothyroidism since the UK Biobank is so well-powered. Of these, we filtered on minor allele frequency >0.01 and imputation quality R2>0.5 in the MSKCC and VUMC GWAS dataset, leaving 16,132 SNPs. We tested the association of these SNPs with thyroid irAEs using Cox models, adjusted for age, sex, and ancestry as estimated by principal components (Supplementary Table 1). To determine the appropriate level for multiple hypothesis testing correction, we used the LD clump feature in PLINK and used a threshold of R2>0.5 to identify the number of effectively independent regions tested, with a result of n=1,057. Therefore, we used a multiple hypothesis testing correction of 0.05/1057 or 4.73 × 10−5 to account for multiple testing. All p-values were two-sided. To characterize potential function of a variant we used an online resource which includes distance from transcriptional start site, expression quantitative trait locus (eQTL) data and promoter Hi-C results (https://genetics.opentargets.org/)(39) and the GTEx consortium data (https://www.gtexportal.org/home/)(40).
Results:
Study population for analysis
We identified 3 cohorts of patients with NSCLC who received CPI for our analysis: 551 individuals from MSK (MSK cohort), 193 individuals from VUMC (VUMC cohort), as well as 561 individuals from DFCI (DFCI cohort). The median follow up time was 18.7 months (IQR: 7.5–44.4 months) for the MSK cohort, 11.1 months (IQR: 4.1–31.8 months) for the VUMC cohort, and 12.0 months (IQR: 4.3–26.2 months) for the DFCI cohort. Thyroid dysfunction, as defined by either hypothyroidism or hyperthyroidism with progression to hypothyroidism, occurred in 12% (n=65/551) of patients in the MSK cohort and was found to be an early event after CPI start, occurring within a median of 2.4 months (IQR: 1.4–4.2 months). Thyroid dysfunction occurred in 16% (n= 30/193, median of 4.1 months, IQR: 1.6–7.9 months) and 7% (n=42/561, median of 5.8 months, IQR: 4.8–13.1 months) of the patients in the VUMC and DFCI cohorts, respectively.
Age (median, interquartile range [IQR]: 67, 59–73 MSK; 63, 57–69 VUMC; 67, 60–74 DFCI), race and ethnicity, and smoking status (former or current: 86% MSK, 88% VUMC, 86% DFCI of patients where information is available) was similar across sites (Table 1). There were slight differences in gender (Female: 53% MSK, 38% VUMC, 56% DFCI). Adenocarcinoma was the most common histology (78% MSK, 62% VUMC, 74% DFCI) at the three sites, and most patients received anti-PD-(L)1 monotherapy (87% MSK, 93% VUMC, 94% DFCI). A small fraction of patients (13% at MSK, 7% at VUMC, 6% at DFCI) received anti-PD-(L)1 and anti-CTLA-4 combination therapy.
Thyroid irAEs are an early event and associated with longer survival
We first examined the association between thyroid irAEs and response in the MSK (BOR, PFS, and OS) and VUMC (PFS) cohorts, as these cohorts had manually abstracted outcomes data. We did this by comparing patients with thyroid irAEs and those who neither had a history of hypothyroidism nor thyroid irAEs.
Compared to those without a thyroid event, individuals with thyroid irAEs had a higher objective response rate (MSK cohort: 38/65, 58% vs 110/486, 23%, p<0.0001 Fisher’s exact) longer PFS (Combined MSK+VUMC cohort: 54% vs 19% progression-free at 1 year, 13% vs 3% progression-free at 3 years) and OS (MSK cohort: 85% vs 56% alive at 1 year, 35% vs 14% alive at 3 years) (Figure 1). The adjusted hazard ratios for PFS for thyroid irAEs in the combined cohort were 0.42 (95% CI: 0.33–0.54) and 0.68 (95% CI: 0.52–0.88) in our standard Cox and time-dependent analysis, respectively (Table 2). As a sensitivity analysis, this result did not change when including the two patients with hyperthyroidism without subsequent hypothyroidism 0.68 (95% CI: 0.52–0.88). We found that 57% (n=37/65) of thyroid dysfunction events occurred within 90 days of CPI start in the MSK cohort. As a sensitivity analysis, we examined whether the same relationship held by only examining the patients who had OS longer than 90 days in the MSK cohort (86%, 475/551). Thyroid irAEs remained significantly associated with PFS (49% vs 20% at 1 year, 14% vs 4% at 3 years, HR: 0.50, 95% CI: 0.33–0.74) and OS (73% vs 62% alive at 1 year, 30% vs 15% alive at 3 years, HR: 0.59, 95% CI: 0.38–0.94) (Supplementary Figure 2). In individuals with available PD-L1 expression data (n = 320), thyroid irAEs remained significantly associated with PFS (HR: 0.39, 95% CI: 0.26–0.58) and OS (HR: 0.36, 95% CI: 0.22–0.58) when adjusting for PD-L1 expression.
Figure 1:
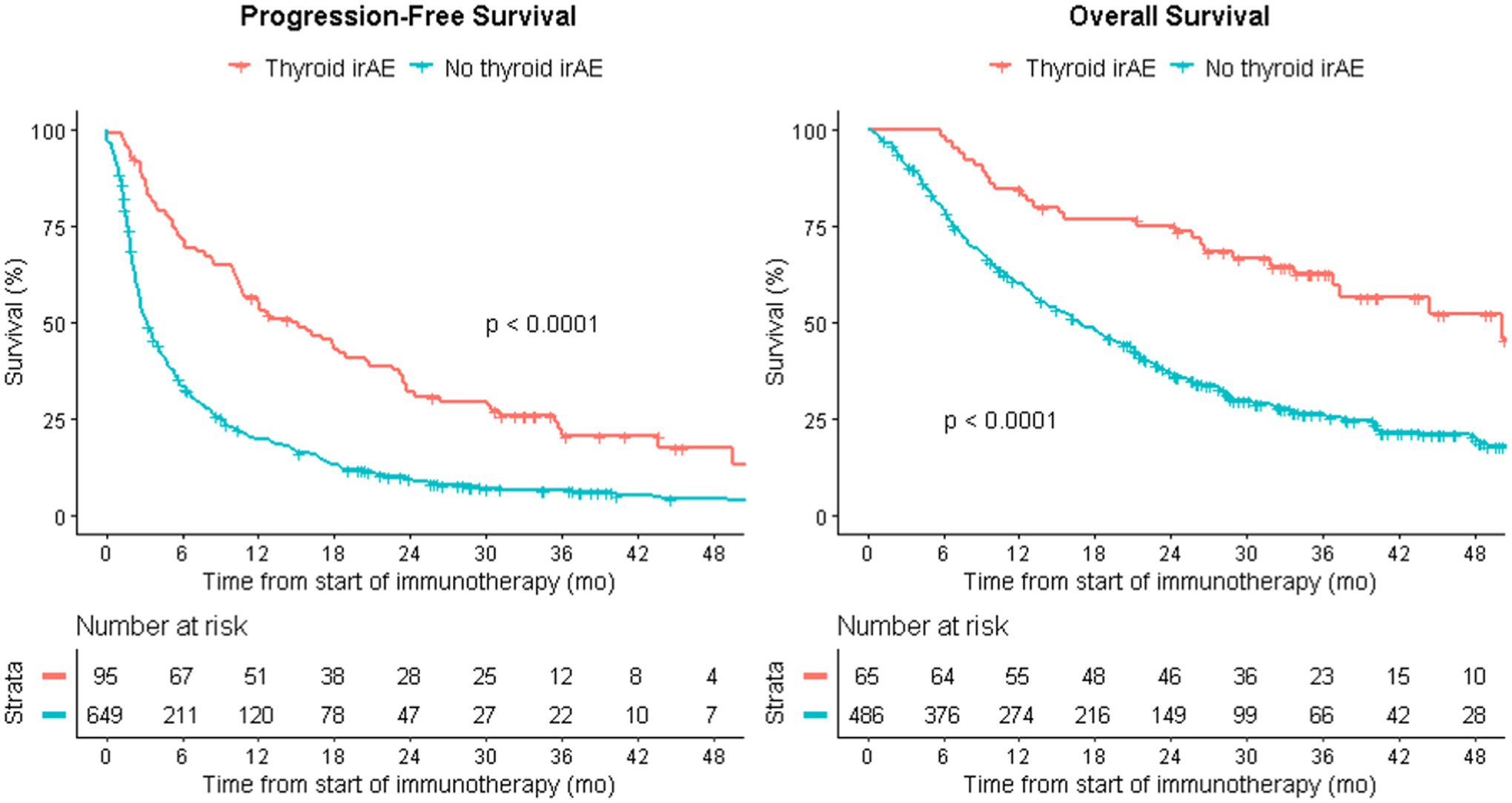
Thyroid irAEs as a predictor of PFS in the combined MSK+VUMC cohort and OS in the MSK cohort. Kaplan-Meier survival curves are unadjusted and compare those who had a thyroid irAE to those who did not have a thyroid irAE. The x-axis reflects time from start of CPI therapy.
Table 2: HRs of the effect of thyroid irAEs on PFS and OS in the combined MSK+VUMC cohort.
Models were adjusted for age, sex, and combined anti-PD-(L)1 + anti-CTLA-4 therapy. aHR = adjusted hazard ratio
| PFS | |||
|---|---|---|---|
| Analysis | aHR | 95% CI | p-value |
| Standard Cox regression analysis | 0.42 | 0.33–0.54 | 6×10–12 |
| Time-dependent analysis | 0.68 | 0.52–0.88 | 4×10–3 |
Given this highly significant relationship between development of thyroid irAEs and multiple objective measures of response demonstrating clinical benefit of CPIs, we evaluated whether a hereditary predisposition to development of hypothyroidism could predict thyroid irAEs as well as CPI benefit.
Polygenic risk score for thyroid disorders is associated with developing thyroid irAEs
We next asked whether predisposition to a baseline thyroid condition is associated with developing thyroid irAEs. We developed a polygenic risk score (PRS) for thyroid disorders using the UK Biobank. We used two different phenotypic proxies for thyroid disease: self-reported hypothyroidism and thyroid medication use and calculated the PRS for each of these using LDpred. The SNP weights for these PRSs were highly correlated (Pearson correlation = 0.9 for each pair of PRS). We validated these PRSs using participants who were not included in our VUMC lung cancer and immunotherapy cohort but had genotypes available in the VUMC BioVU population (non-cancer patients). Since UK Biobank is predominantly of European ancestry, we restricted the VUMC BioVU to those who were of European ancestry. Overall, there was a strong association of each individual PRS with hypothyroidism (AUROC = 0.6 for both) (Supplementary Figure 3A) with an odds ratio/standard deviation of 1.33 (95% CI 1.29–1.37) for the PRS for self-reported hypothyroidism and an increased relative risk by decile (Supplementary Figure 3B). We then applied these PRSs to our discovery cohort. Since the effect sizes for these 2 PRSs were highly correlated, we focused primarily on the PRS for self-reported hypothyroidism (hypothyroidism PRS) from the UK Biobank.
The hypothyroidism PRS was significantly associated with development of thyroid irAEs in the MSK+VUMC cohort (Figure 2A), and it had similar performance for predicting thyroid irAEs (AUROC = 0.6) in comparison with its performance in the VUMC BioVU cohort (spontaneous hypothyroidism). The thyroid medication PRS from UK Biobank had similar performance (Supplementary Figure 4). In a Cox regression model adjusted for age, sex, and the first ten principal components, the HR per standard deviation for the hypothyroidism PRS was 1.34 (95% CI: 1.08–1.66; p = 8.73×10−3), with similar effect sizes seen for the other PRS (Table 3). The hypothyroidism PRS results remained significant after removing individuals who received combination CPI therapy (HR: 1.34, 95% CI: 1.07–1.69, p = 0.01) and only including individuals of European ancestry (HR: 1.27, 95% CI: 1.02–1.59, p = 0.03). Additionally, when examining the PRS by tertile, individuals in the highest tertile PRS scores had higher rates of hypothyroidism events than the first and second tertile (Figure 2A, Supplementary Figure 4).
Figure 2:
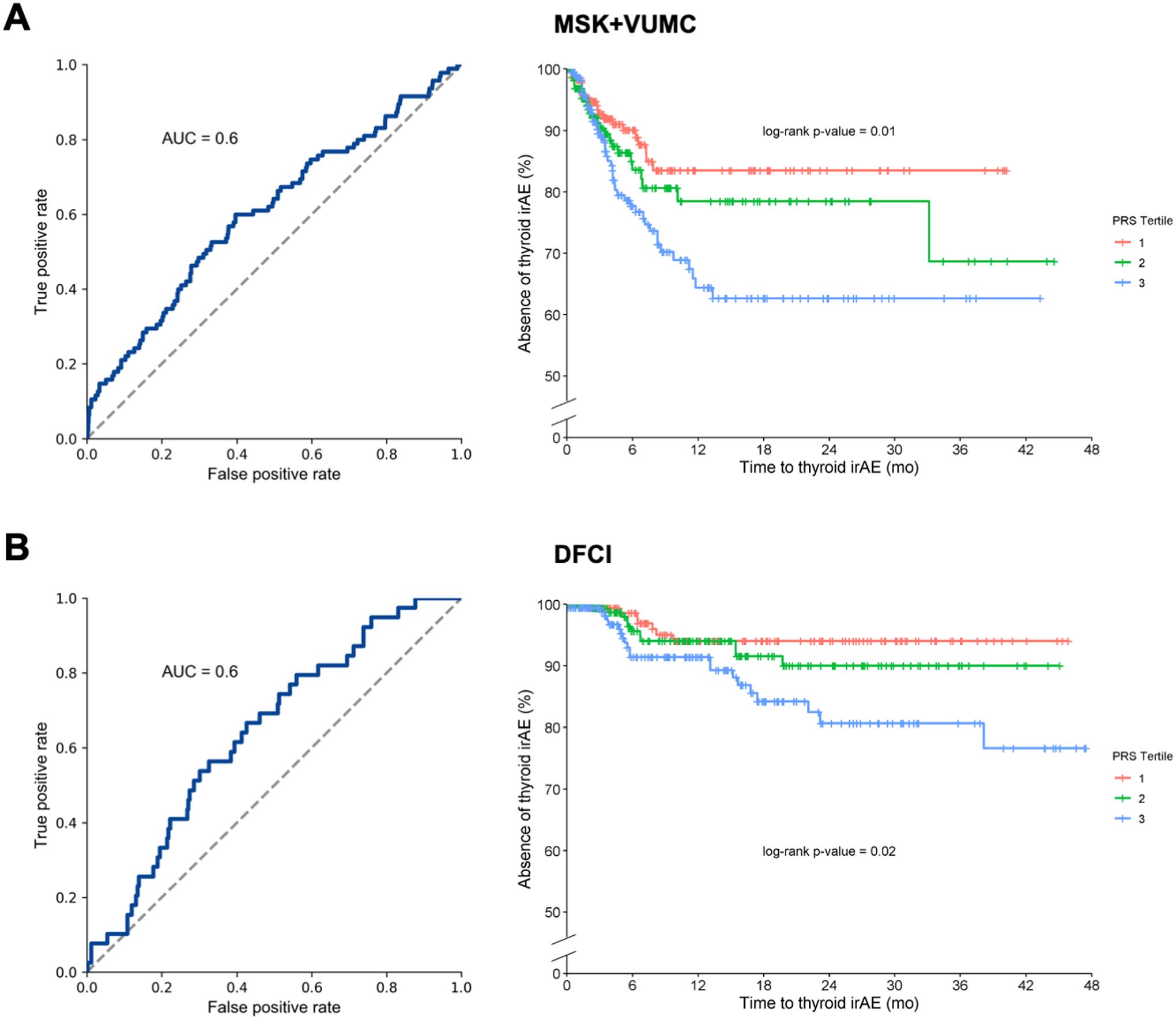
Hypothyroidism PRS (using self-reported hypothyroidism) as a predictor of CPI-induced thyroid irAEs in the (A) MSK+VUMC cohort and (B) the DFCI cohort. The left panel shows the ROC curve and right panel shows time to event by PRS tertile. P-values for the three curves are calculated using a log-rank test.
Table 3: PRS as a predictor of CPI-induced thyroid irAEs in the MSK+VUMC cohort.
HRs of the effect of PRS as a predictor of thyroid irAEs in the combined MSK+VUMC cohort. Cox regression model was adjusted for age, sex, and the first ten principal components
| PRS Phenotype | aHR | 95% CI | p-value |
|---|---|---|---|
| Hypothyroidism | 1.34 | 1.08–1.66 | 8.73×10–3 |
| Thyroid medications | 1.32 | 1.07–1.63 | 9.98×10–3 |
We sought to replicate these PRS associations in the DFCI cohort. First, we confirmed that the PRS was imputed accurately from tumors using a separate set of 833 benchmark individuals with both tumor sequencing and germline SNP array data available (see Methods): the correlation between the tumor imputed PRS and the germline ground truth PRS was >0.87 for all PRSs evaluated, with no visible outliers (Supplementary Figure 5). Next, we tested each PRS for association with the time to thyroid irAE: both PRSs were significantly associated, with the PRS for self-reported hypothyroidism yielding an HR of 1.39 (95% CI 1.07–1.82; p=0.01 by Cox regression) (AUROC=0.64) with similar effect sizes seen for the other PRS (Figure 2B, Supplementary Figure 6).
Analysis of individual loci associated with thyroid irAEs
As an exploratory analysis, we evaluated whether any of the genome-wide significant associations identified in the UK Biobank for self-reported hypothyroidism are also individually associated with thyroid irAEs. After filtering on minor allele frequency and imputation quality, we tested each of 16,132 significant UK Biobank SNPs for association with hypothyroidism irAEs in the MSK+VUMC cohort (Supplementary Table 1). Of these, 1,502 of were nominally associated (p<0.05) with thyroid irAEs and exhibited significant sign consistency (1,143/1,502 were associated in the same direction) underscoring the shared genetic effect observed through the PRS. One SNP, rs9268543 (p=7.5×10−7), surpassed stringent Bonferroni correction, even though the MSK+VUMC cohort was orders of magnitude smaller than the UK Biobank and thus underpowered for individual variant discovery. This SNP falls within the HLA locus and has been associated with numerous other autoimmune traits(41,42). This variant is also an expression quantitative trait locus (eQTL) for several genes at this locus, most strongly for HLA-DQA2 (40).
PRS for hypothyroidism is not associated with PFS or OS
Lastly, we assessed whether PFS for hypothyroidism was associated with CPI response. Despite the association between both PRSs and thyroid irAEs, neither PRS was significantly associated with PFS or OS in the MSK cohort (Supplementary Table 2, Supplementary Figure 7A). Likewise, in the DFCI cohort no significant association between PRS and OS was observed (Supplementary Figure 7B).
Discussion:
We examined the associations between hereditary predisposition for spontaneous hypothyroidism, thyroid irAEs, and benefit from CPI treatment with the goal of inferring whether there may be shared biology. Thyroid irAEs occurred early during treatment, and those who developed this irAE were more likely to have a beneficial initial response and to achieve a longer duration of response to CPI therapy compared to those who did not. We developed a PRS for hypothyroidism that performed similarly in predicting development of spontaneous hypothyroidism in the general population and thyroid irAEs in multiple patient cohorts. However, the PRS did not predict benefit from immunotherapy.
We saw a rapid onset of thyroid irAEs in the MSK and VUMC datasets. In the DFCI dataset, thyroid events occurred on average later, but this could have been due to the later and less frequent TSH measurements in this cohort and despite this the PRS performed similarly in the DFCI cohort. The rapid onset of thyroid irAEs in the MSK dataset and VUMC datasets in a genetically predisposed population and our previous work on finding shared auto-antibodies(24), suggests that the checkpoint inhibitors unmasked a pre-existing subclinical autoimmune condition suppressed by immune checkpoints.
A potential limitation of our study is survivor bias, or time from CPI start to development of thyroid irAEs. However, this irAE consistently occurred early after CPI start and survivor bias was adjusted for in time-dependent and landmark analyses. We found thyroid irAEs significantly associated with multiple markers of response: BOR, PFS, and OS. Additionally there is plausibility that tipping immunologic homeostasis of immune tolerance with CPI therapy not only leads to indirect cancer cell cytotoxicity but also potentiates (or drives) development of anti-thyroid antibodies leading to thyroid irAEs. Therefore, we infer there exists a shared immunologic mechanism that drives both thyroid irAEs and CPI benefit leading to improved survival. Furthermore, there are other cancer therapy settings in which development of thyroid dysfunction is associated with response to treatment (e.g. IL-2)(43).
Another limitation of our analyses is that our PRS was developed in UK Biobank which includes participants who are predominantly of European ancestry. The cohorts of CPI-treated patients we tested were also predominantly of European ancestry. Since PRS may not generalize well across different ancestry populations(37) the PRS we tested may not work to predict irAEs in non-European ancestry populations. Larger studies of patients on CPI in non-European ancestry populations are needed to understand the genetics of treatment benefit and irAEs in these populations.
The underlying factor leading to thyroid irAEs and CPI benefit may have a hereditary origin, an environmental origin, or may be a combination of multiple factors. Our PRS developed for spontaneous hypothyroidism, a diagnosis that generally has an autoimmune etiology, was able to perform similarly well in predicting thyroid irAEs. This supports a shared biological mechanism encapsulated by the PRS driving autoimmune hypothyroidism and thyroid irAEs. In short, this suggests there exists genetic polymorphism-based hereditary factors that contribute to developing thyroid irAEs. Our results suggest that other irAEs may be predictable using analogous PRS for similar clinical phenotypes.
Our analyses may also be useful to help understand the mechanisms that underlie thyroid irAEs. To examine shared loci, we focused on the large, well-powered UK Biobank as the discovery GWAS, and investigated which SNPs replicated in our MSK+VUMC cohort. We found that one loci was associated with thyroid irAEs after stringent multiple test correction. Among these, was the HLA locus with the top variant (rs9268543) previously associated with many autoimmune diseases, including rheumatoid arthritis (41), inflammatory bowel disease (42), and with hypothyroidism and type 1 diabetes in the UK Biobank. The overlap between HLA genetic variants for spontaneous hypothyroidism and thyroid irAEs suggests that at least some of the antigens underlying the disorder overlap. This variant is an attractive target for downstream experimental analysis to understand the mechanisms of irAEs. Future studies with larger sample sizes may help to identify additional shared loci between spontaneous hypothyroidism and thyroid irAEs and possibly to identify new loci at which the genetics do not overlap.
Our hypothyroidism PRS did not distinguish between those benefiting and not benefiting from CPIs. This suggests that in contrast to inherited factors, metabolic, epigenetic or environmental factors (e.g. shared exogenous exposures among those who benefit/ do not benefit from CPI that shape the adaptome over time) play a larger role in driving the mechanism behind thyroid irAEs and CPI benefit. Additionally, the present analysis may be underpowered to detect the difference. Since the PRS for hypothyroidism had an AUROC of ~0.6, larger cohort sizes may be needed to overcome the heterogeneity of environmental factors. The association between thyroid irAEs and treatment response could also be due to inherited genetic factors that are not captured by the PRS. The PRS developed from UK Biobank was developed exclusively from common genetic variants and does not capture the effect of rare genetic variants. Since rare variants may contribute a large fraction of heritability for some complex traits(38,39) future studies using whole genome sequencing data may reveal a relationship between heritable genetic variation for thyroid irAEs and CPI benefit. An alternative explanation may be that, the genetic contribution captured by the PRS may solely reflect a “branch” off target effect of CPI therapy (e.g. cross presentation of shared antigens that are not associated with CPI benefit) decoupled from the adaptive immune mechanisms of CPI therapy that lead to improved outcomes. If the latter explanation is correct, it would suggest that the genetic component of an individual’s cancer-immune set point may not be the same factors that confer autoimmune thyroid disease risk.
Individual irAEs have distinct disease kinetics that differentially affect survival bias and varied severity ranging from subclinical to life-threatening(15) - unsurprisingly, not all are consistently associated with CPI benefit(20,44,45). Our finding that development of thyroid irAEs secondary to PD-1 blockade-based therapy is associated with CPI benefit is similar to what has been seen in the literature(46). Thyroid irAEs occur early, are common, and can be treated with thyroid hormone supplementation, making it an early clinical signal suggestive of long-term benefit from CPI therapy.
Genetic predisposition to irAEs has previously been examined in studies associating irAE risk with human leukocyte antigen (HLA) genes(47–50) and/or individual SNPs(51). Only one other study has examined these associations using PRSs, which is more reflective of underlying susceptibility to autoimmune disease(52,53). Our findings differ from this study of genetics of autoimmune skin conditions and survival(54). Khan et al. developed a PRS of autoimmune dermatologic conditions and applied the PRSs to a clinical trial of PD-1 blockade in bladder cancer. Similar to our results, they found an association between the autoimmune PRSs and irAE. In contrast to our findings where hypothyroid PRSs were not associated with CPI benefit, dermatologic PRSs were associated with overall survival in patients receiving CPI therapy. This suggests predisposition for specific autoimmune diseases differentially impacts immunotherapy benefit.
In summary, in this study of patients from three academic centers, we found that developing thyroid irAEs was robustly associated with benefit from CPI therapy. Genetic predisposition measured by PRSs for spontaneous - immune-mediated - hypothyroidism was associated with the irAE but did not predict CPI benefit, suggesting distinct immune pathways driving underlying genetic risk for hypothyroidism and benefit from CPI. Large scale and mechanistic studies are needed to elucidate underlying pathways linking genetic risk, specific irAEs, and therapeutic response. Understanding the relationships and biological underpinnings of these processes are critical for both advancement of precision immunotherapy and development of new therapies for our patients.
Supplementary Material
Translational relevance.
Understanding and predicting who experiences durable benefit from immune checkpoint inhibitor (CPI) therapy is key to advancing precision immuno-oncology and developing new immunotherapies. We observed immune-mediated thyroid dysfunction is a common, early, low-grade event in those receiving PD-1 blockade-based therapy. Building upon this observation we asked whether genetic risk for autoimmune (spontaneous) hypothyroidism is associated with thyroid irAEs. We developed and validated a polygenic risk score for spontaneous hypothyroidism. The PRS associated with thyroid irAEs but did not associate with CPI benefit. Mechanisms mediating immunotherapy benefit may be distinct from autoimmune genetic predisposition to thyroid dysfunction. These findings have implications in clarifying the genetic factors predisposing individuals to CPI benefit.
Funding
Supported by the National Institutes of Health [R01-CA227466 and K24-CA169004 to E Ziv; T32-CA009207 and K30-UL1TR00457 to J Luo; T32-GM007347, and F30-HL140756 to VL Martucci; U54CA217450-01, U01CA224276-01, P30-CA086485, and UG1CA233259 to CM Lovly; U01CA253560-01 to MC Aldrich; and R01-CA227237 and R01-CA244569 to A Gusev]; Memorial Sloan Kettering Cancer Center Support Grant/Core [P30-CA008748]; the Druckenmiller Center for Lung Cancer Research at Memorial Sloan Kettering Cancer Center; the Vanderbilt Institute for Clinical and Translational Research [UL1TR002243]; the American Diabetes Association Grant [1-19-PDF-131 to Z Quandt]; the Conquer Cancer Foundation [Young Investigator Award to J Luo]; the Damon Runyon Cancer Research Foundation [CI-98-18 to MD Hellmann]; the Louis B. Mayer Foundation [A Gusev]. MD Hellmann is a member of the Parker Institute for Cancer Immunotherapy.
Conflicts of Interest Statement
J.L. has received honoraria from Targeted Oncology and Physicians’ Education Resource.
M.A.G. receives institutional research funding from Celgene, Merck, Novartis, OncoMed Pharmaceuticals, and Roche; has been a compensated consultant for AstraZeneca, Beyond Spring Pharmaceuticals, Boehringer Ingelheim, Bristol-Myers Squibb, Genentech/Roche, Heron Therapeutics, and Takeda Pharmaceuticals.
M.D.H. receives institutional research funding from Bristol-Myers Squibb; has been a compensated consultant for Merck, Bristol-Myers Squibb, AstraZeneca, Genentech/Roche, Nektar, Syndax, Mirati, Shattuck Labs, Immunai, Blueprint Medicines, Achilles, Arcus and Natera; received travel support/ honoraria from AstraZeneca, Eli Lilly, and Bristol-Myers Squibb; has options from Shattuck Labs, Immunai, and Arcus; has a patent filed by his institution related to the use of tumor mutation burden to predict response to immunotherapy (PCT/US2015/062208), which has received licensing fees from PGDx.
C.M.L. is a consultant/advisory board member for Pfizer, Novartis, AstraZeneca, Genoptix, Sequenom, Ariad, Takeda, Blueprints Medicine, Cepheid, Foundation Medicine, Roche, Achilles Therapeutics, Genentech, Syros, Amgen, EMD-Serono, and Eli Lilly and reports receiving commercial research grants from Xcovery, AstraZeneca, and Novartis.
The remaining authors declare no potential conflicts of interest.
References:
- 1.Robert C, Ribas A, Schachter J, Arance A, Grob J-J, Mortier L, et al. Pembrolizumab versus ipilimumab in advanced melanoma (KEYNOTE-006): post-hoc 5-year results from an open-label, multicentre, randomised, controlled, phase 3 study [Internet]. The Lancet Oncology. 2019. page 1239–51. Available from: 10.1016/s1470-2045(19)30388-2 [DOI] [PubMed] [Google Scholar]
- 2.Larkin J, Chiarion-Sileni V, Gonzalez R, Grob J-J, Rutkowski P, Lao CD, et al. Five-Year Survival with Combined Nivolumab and Ipilimumab in Advanced Melanoma. N Engl J Med. 2019;381:1535–46. [DOI] [PubMed] [Google Scholar]
- 3.Garon EB, Hellmann MD, Rizvi NA, Carcereny E, Leighl NB, Ahn M-J, et al. Five-year overall survival for patients with advanced non--small-cell lung cancer treated with pembrolizumab: results from the phase I KEYNOTE-001 study. J Clin Oncol. American Society of Clinical Oncology; 2019;37:2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garon EB, Rizvi NA, Hui R, Leighl N, Balmanoukian AS, Eder JP, et al. Pembrolizumab for the Treatment of Non–Small-Cell Lung Cancer. N Engl J Med. Massachusetts Medical Society; 2015;372:2018–28. [DOI] [PubMed] [Google Scholar]
- 5.Doroshow DB, Bhalla S, Beasley MB, Sholl LM, Kerr KM, Gnjatic S, et al. PD-L1 as a biomarker of response to immune-checkpoint inhibitors. Nat Rev Clin Oncol [Internet]. 2021; Available from: 10.1038/s41571-021-00473-5 [DOI] [PubMed] [Google Scholar]
- 6.Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, et al. Mutational landscape determines sensitivity to PD-1 blockade in non–small cell lung cancer. Science. American Association for the Advancement of Science; 2015;348:124–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peled JU, Devlin SM, Staffas A, Lumish M, Khanin R, Littmann ER, et al. Intestinal Microbiota and Relapse After Hematopoietic-Cell Transplantation. J Clin Oncol. 2017;35:1650–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baruch EN, Youngster I, Ben-Betzalel G, Ortenberg R, Lahat A, Katz L, et al. Fecal microbiota transplant promotes response in immunotherapy-refractory melanoma patients. Science. 2021;371:602–9. [DOI] [PubMed] [Google Scholar]
- 9.Davar D, Dzutsev AK, McCulloch JA, Rodrigues RR, Chauvin J-M, Morrison RM, et al. Fecal microbiota transplant overcomes resistance to anti–PD-1 therapy in melanoma patients. Science. American Association for the Advancement of Science; 2021;371:595–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med. 2015;372:2509–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Orrù V, Steri M, Sole G, Sidore C, Virdis F, Dei M, et al. Genetic variants regulating immune cell levels in health and disease. Cell. 2013;155:242–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lim YW, Chen-Harris H, Mayba O, Lianoglou S, Wuster A, Bhangale T, et al. Germline genetic polymorphisms influence tumor gene expression and immune cell infiltration. Proc Natl Acad Sci U S A. 2018;115:E11701–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shahamatdar S, He MX, Reyna MA, Gusev A, AlDubayan SH, Van Allen EM, et al. Germline Features Associated with Immune Infiltration in Solid Tumors. Cell Rep. 2020;30:2900–2908.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sayaman RW, Saad M, Thorsson V, Hu D, Hendrickx W, Roelands J, et al. Germline genetic contribution to the immune landscape of cancer. Immunity. 2021;54:367–386.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Postow MA, Sidlow R, Hellmann MD. Immune-Related Adverse Events Associated with Immune Checkpoint Blockade [Internet]. New England Journal of Medicine. 2018. page 158–68. Available from: 10.1056/nejmra1703481 [DOI] [PubMed] [Google Scholar]
- 16.Ricciuti B, Genova C, De Giglio A, Bassanelli M, Bello MGD, Metro G, et al. Impact of immune-related adverse events on survival in patients with advanced non-small cell lung cancer treated with nivolumab: long-term outcomes from a multi-institutional analysis [Internet]. Journal of Cancer Research and Clinical Oncology. 2019. page 479–85. Available from: 10.1007/s00432-018-2805-3 [DOI] [PubMed] [Google Scholar]
- 17.Fujimoto D, Yoshioka H, Kataoka Y, Morimoto T, Kim YH, Tomii K, et al. Efficacy and safety of nivolumab in previously treated patients with non-small cell lung cancer: A multicenter retrospective cohort study. Lung Cancer. 2018;119:14–20. [DOI] [PubMed] [Google Scholar]
- 18.Street S, Chute D, Strohbehn I, Zhao S, Rengarajan M, Faje A, et al. The positive effect of immune checkpoint inhibitor-induced thyroiditis on overall survival accounting for immortal time bias: a retrospective cohort study of 6,596 patients [Internet]. Annals of Oncology. 2021. Available from: 10.1016/j.annonc.2021.05.357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shankar B, Zhang J, Naqash AR, Forde PM, Feliciano JL, Marrone KA, et al. Multisystem Immune-Related Adverse Events Associated With Immune Checkpoint Inhibitors for Treatment of Non–Small Cell Lung Cancer. JAMA Oncol. American Medical Association; 2020;6:1952–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horvat TZ, Adel NG, Dang T-O, Momtaz P, Postow MA, Callahan MK, et al. Immune-related adverse events, need for systemic immunosuppression, and effects on survival and time to treatment failure in patients with melanoma treated with ipilimumab at Memorial Sloan Kettering Cancer Center. J Clin Oncol. American Society of Clinical Oncology; 2015;33:3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weber JS, Hodi FS, Wolchok JD, Topalian SL, Schadendorf D, Larkin J, et al. Safety Profile of Nivolumab Monotherapy: A Pooled Analysis of Patients With Advanced Melanoma. J Clin Oncol. 2017;35:785–92. [DOI] [PubMed] [Google Scholar]
- 22.Verheijden RJ, May AM, Blank CU, Aarts MJB, van den Berkmortel FWPJ, van den Eertwegh AJM, et al. Association of Anti-TNF with Decreased Survival in Steroid Refractory Ipilimumab and Anti-PD1–Treated Patients in the Dutch Melanoma Treatment Registry. Clin Cancer Res. American Association for Cancer Research; 2020;26:2268–74. [DOI] [PubMed] [Google Scholar]
- 23.Suresh K, Naidoo J. Lower Survival in Patients Who Develop Pneumonitis Following Immunotherapy for Lung Cancer. Clin. Lung Cancer. 2020. page e169–70. [DOI] [PubMed] [Google Scholar]
- 24.Osorio JC, Ni A, Chaft JE, Pollina R, Kasler MK, Stephens D, et al. Antibody-mediated thyroid dysfunction during T-cell checkpoint blockade in patients with non-small-cell lung cancer. Ann Oncol. 2017;28:583–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee H, Hodi FS, Giobbie-Hurder A, Ott PA, Buchbinder EI, Haq R, et al. Characterization of Thyroid Disorders in Patients Receiving Immune Checkpoint Inhibition Therapy. Cancer Immunol Res. 2017;5:1133–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quandt Z, Young A, Perdigoto AL, Herold KC, Anderson MS. Autoimmune Endocrinopathies: An Emerging Complication of Immune Checkpoint Inhibitors. Annu Rev Med. 2021;72:313–30. [DOI] [PubMed] [Google Scholar]
- 27.Robert C, Joshua AM, Kefford R, Joseph RW, Wolchok JD, Hodi FS, et al. Association of immune-related thyroid disorders with pembrolizumab (pembro, MK-3475) in patients (pts) with advanced melanoma treated in KEYNOTE-001. J Clin Orthod. American Society of Clinical Oncology; 2015;33:9050–9050. [Google Scholar]
- 28.Hansen PS, Brix TH, Sørensen TIA, Kyvik KO, Hegedüs L. Major genetic influence on the regulation of the pituitary-thyroid axis: a study of healthy Danish twins. J Clin Endocrinol Metab. 2004;89:1181–7. [DOI] [PubMed] [Google Scholar]
- 29.Panicker V, Wilson SG, Spector TD, Brown SJ, Falchi M, Richards JB, et al. Heritability of serum TSH, free T4 and free T3 concentrations: a study of a large UK twin cohort. Clin Endocrinol. 2008;68:652–9. [DOI] [PubMed] [Google Scholar]
- 30.Teumer A, Chaker L, Groeneweg S, Li Y, Di Munno C, Barbieri C, et al. Genome-wide analyses identify a role for SLC17A4 and AADAT in thyroid hormone regulation. Nat Commun. 2018;9:4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12:e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roden DM, Pulley JM, Basford MA, Bernard GR, Clayton EW, Balser JR, et al. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin Pharmacol Ther. Springer Nature; 2008;84:362–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loh P-R, Kichaev G, Gazal S, Schoech AP, Price AL. Mixed-model association for biobank-scale datasets. Nat Genet. 2018;50:906–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu Y, Byrne EM, Zheng Z, Kemper KE, Yengo L, Mallett AJ, et al. Genome-wide association study of medication-use and associated disease in the UK Biobank. Nat Commun. 2019;10:1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vilhjálmsson BJ, Yang J, Finucane HK, Gusev A, Lindström S, Ripke S, et al. Modeling Linkage Disequilibrium Increases Accuracy of Polygenic Risk Scores. Am J Hum Genet. 2015;97:576–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Das S, Forer L, Schönherr S, Sidore C, Locke AE, Kwong A, et al. Next-generation genotype imputation service and methods. Nat Genet. 2016;48:1284–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets [Internet]. GigaScience. 2015. Available from: 10.1186/s13742-015-0047-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ghoussaini M, Mountjoy E, Carmona M, Peat G, Schmidt EM, Hercules A, et al. Open Targets Genetics: systematic identification of trait-associated genes using large-scale genetics and functional genomics. Nucleic Acids Res. 2021;49:D1311–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Consortium GTEx. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369:1318–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stahl EA, Raychaudhuri S, Remmers EF, Xie G, Eyre S, Thomson BP, et al. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat Genet. 2010;42:508–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Lange KM, Moutsianas L, Lee JC, Lamb CA, Luo Y, Kennedy NA, et al. Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat Genet. 2017;49:256–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hamnvik O-PR, Larsen PR, Marqusee E. Thyroid Dysfunction from Antineoplastic Agents [Internet]. JNCI Journal of the National Cancer Institute. 2011. page 1572–87. Available from: 10.1093/jnci/djr373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beattie J, Rizvi H, Fuentes P, Luo J, Schoenfeld A, Lin I-H, et al. Success and failure of additional immune modulators in steroid-refractory/resistant pneumonitis related to immune checkpoint blockade. J Immunother Cancer [Internet]. 2021;9. Available from: 10.1136/jitc-2020-001884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moslehi JJ, Salem J-E, Sosman JA, Lebrun-Vignes B, Johnson DB. Increased reporting of fatal immune checkpoint inhibitor-associated myocarditis. Lancet. 2018;391:933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kotwal A, Kottschade L, Ryder M. PD-L1 Inhibitor-Induced Thyroiditis Is Associated with Better Overall Survival in Cancer Patients. Thyroid. 2020;30:177–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hasan Ali O, Berner F, Bomze D, Fässler M, Diem S, Cozzio A, et al. Human leukocyte antigen variation is associated with adverse events of checkpoint inhibitors. Eur J Cancer. 2019;107:8–14. [DOI] [PubMed] [Google Scholar]
- 48.Cappelli LC, Dorak MT, Bettinotti MP, Bingham CO, Shah AA. Association of HLA-DRB1 shared epitope alleles and immune checkpoint inhibitor-induced inflammatory arthritis. Rheumatology. Oxford Academic; 2018;58:476–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heaney AP, Sumerel B, Rajalingam R, Bergsneider M, Yong WH, Liau LM. HLA Markers DQ8 and DR53 Are Associated With Lymphocytic Hypophysitis and May Aid in Differential Diagnosis. J Clin Endocrinol Metab. 2015;100:4092–7. [DOI] [PubMed] [Google Scholar]
- 50.Quandt Z, Ziv E, Young A, Kang JH, Bluestone J, Anderson M. Investigation of the Predictive Utility of a Type 1 Diabetes Genetic Risk Score in Immune Checkpoint Inhibitor Induced Diabetes Mellitus.
- 51.Abdel-Wahab N, Diab A, Yu RK, Futreal A, Criswell LA, Tayar JH, et al. Genetic determinants of immune-related adverse events in patients with melanoma receiving immune checkpoint inhibitors. Cancer Immunol Immunother [Internet]. 2021; Available from: 10.1007/s00262-020-02797-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Concannon P, Rich SS, Nepom GT. Genetics of type 1A diabetes. N Engl J Med. 2009;360:1646–54. [DOI] [PubMed] [Google Scholar]
- 53.Tomer Y Genetic susceptibility to autoimmune thyroid disease: past, present, and future. Thyroid. 2010;20:715–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Khan Z, Di Nucci F, Kwan A, Hammer C, Mariathasan S, Rouilly V, et al. Polygenic risk for skin autoimmunity impacts immune checkpoint blockade in bladder cancer. Proc Natl Acad Sci U S A. 2020;117:12288–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


