Visual Abstract
Keywords: genetics, autosomal recessive Alport syndrome, genotype-phenotype correlation, hematuria, benign familial, heterozygous mutation, kidney failure, chronic, nephritis, hereditary, prognosis, prognostic predicting factor, retrospective studies, urinary abnormalities
Abstract
Background
Autosomal recessive Alport syndrome (ARAS) is an inherited renal disorder caused by homozygous and compound heterozygous mutations in COL4A3 or COL4A4, but the prognostic predictors for this disorder are not yet fully understood. Recently, the magnitude of the clinical spectrum of the COL4A3 and COL4A4 heterozygous state has attracted attention. This spectrum includes asymptomatic carriers of ARAS, benign familial hematuria, thin basement membrane disease, and autosomal dominant Alport syndrome.
Methods
We retrospectively analyzed 49 patients with ARAS from 41 families with a median age of 19 years to examine the clinical features and prognostic factors of ARAS, including the associated genotypes.
Results
The median age of patients with ARAS at ESKD onset was 27 years. There was no significant association between the presence or absence of hearing loss or truncating mutations and renal prognosis. However, there was a statistically significant correlation between renal prognosis and heterozygous variants that cause urinary abnormalities. Where the urinary abnormality–causing variant was absent or present in only one allele, the median age of ESKD onset was 45 years, whereas the same variant present on both alleles was associated with an age of onset of 15 years (P<0.001).
Conclusions
This study was the first to demonstrate the clinical importance in ARAS of focusing on variants in COL4A3 or COL4A4 that cause urinary abnormalities in both the homozygous or heterozygous state. Although heterozygous mutation carriers of COL4A3 and COL4A4 comprise a broad clinical spectrum, clinical information regarding each variant is important for predicting ARAS prognosis.
Introduction
Alport syndrome is an inherited renal disorder accompanied by hearing loss and eye lesions. Autosomal recessive Alport syndrome (ARAS), in particular, is caused by either homozygous or compound heterozygous mutations in COL4A3 (NM: 000091) or COL4A4 (NM: 000092), and the proportion of patients with ARAS among the total number of patients with Alport syndrome is estimated to be as low as 15%. The prognostic predictors of ARAS have not yet been sufficiently clarified, whereas the phenotype-genotype correlation in male X-linked Alport syndrome is relatively established; missense mutations exhibit milder phenotypes compared with truncating mutations (1–7). Storey et al. (2) reported that patients with ARAS carrying truncating mutations on at least one allele tend to show early onset of ESKD compared with patients without truncating mutations. Savige et al. (8) reported that renal failure tended to occur at a younger age in patients with two truncating variants compared with no truncating variants. In a systematic review, Lee et al. (9) reported that the median age at onset of ESKD in patients without missense mutations was earlier compared with patients with at least one missense mutation. However, we previously analyzed 30 patients with ARAS and found no association between the presence of truncating mutations and the age at onset of ESKD (10). Patients with ARAS have been reported to develop ESKD on average in their early 20s, although some patients preserve their renal function until middle age (2,8,9,11,12). The development of genetic testing methods such as next generation sequencing (NGS) in recent years has increased opportunities for genetic diagnosis and thereby, increased the number of patients diagnosed with ARAS at earlier stages with mild symptoms. Therefore, to better manage this syndrome, it is necessary to elucidate the prognostic predictors of ARAS.
Recently, the magnitude of the clinical spectrum of the COL4A3 and COL4A4 heterozygous state has attracted attention; some patients with heterozygous states are asymptomatic (carriers of ARAS), some may have benign familial hematuria or thin basement membrane disease, and yet others have autosomal dominant Alport syndrome and develop ESKD (13–15). Moreover, it is known that COL4A3, COL4A4, and COL4A5 mutations can also affect the severity of concomitant kidney diseases caused by mutations in other podocyte-related genes such as NPHS2 and MYH9 (16–18). In this study, we focused on variants in COL4A3 and COL4A4 in patients with ARAS and their parents and determined whether these variants are associated with urinary abnormalities in the heterozygous state. Although heterozygous mutation carriers of COL4A3 and COL4A4 have a broad clinical spectrum, clinical information regarding these variants is potentially important in predicting the prognosis of patients with ARAS.
Materials and Methods
Ethical Considerations
All procedures involving human participants in this study were performed in accordance with the ethical standards of the Institutional Review Board of Kobe University School of Medicine and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. Informed consent was obtained from patients and/or their parents.
Participants and Inclusion Criteria
Patients suspected of having Alport syndrome on the basis of their clinical symptoms, pathology finding, and/or family history were referred to Kobe University for genetic analysis between January 2006 and August 2019. After diagnosis, most patients were then followed in various local hospitals throughout Japan, but no further data were collected for the purpose of our study. For this study, we included all patients who were genetically defined as having ARAS, comprising a total of 49 patients from 41 families with compound heterozygous or homozygous mutations in COL4A3 or COL4A4. Among these, our group previously reported 30 patients from 24 families (10). We added newly diagnosed patients to the previously cohort and also analyzed data from other individuals possessing the heterozygous mutations of interest, such as the patients’ parents or patients reported in the literature. Only subjects who were found to have homozygous or compound heterozygous variants in either COL4A3 or COL4A4 were diagnosed with ARAS. The median age of the patients was 19 years. The men-to-women ratio was 20:29; 32 patients had COL4A3 variants, and 17 patients had COL4A4 variants.
Clinical Information
All clinical information was collected with an information sheet that was sent with the blood samples for genetic analysis. We collected information regarding age, sex, height, body weight, serum Cr, urinary protein-creatinine ratio, history of dialysis or kidney transplantation (if applicable), the existence of hearing loss or eye lesions, and the results of kidney biopsy (if applicable). With regard to family history, we requested information on the presence or absence of renal disease, dialysis, hearing loss, eye lesions, and the age at which ESKD developed. We defined the age at ESKD onset as the time that dialysis was introduced or kidney transplantation was performed. The eGFR (milliliters per minute per 1.73 m2) was calculated on the basis of serum creatinine using the equation reported by Uemura et al. (19) for patients aged 2–18 years and by Matsuo et al. (20) for patients aged 19 years and older.
Genetic Analyses
All patients were referred to our hospital for genetic testing between January 2006 and August 2019, and those confirmed to have ARAS were included in this study. Genomic DNA was isolated from patient peripheral blood leukocytes with the QuickGene-Mini80 system (Kurabo Industries, Tokyo, Japan) according to the manufacturer’s instructions. Genetic testing was conducted either with Sanger sequencing of COL4A3 and COL4A4 genes (before NGS was available in our laboratory, January 2006 to November 2015) or with NGS using a targeted sequencing panel containing causative genes of inherited kidney disease (November 2015 to August of 2019). NGS analyses were conducted as described previously (21). Where heterozygous mutations were not identified by direct sequencing, mRNA or multiplex ligation–dependent probe amplification was performed for confirmation.
Variant Evaluation
Variants were evaluated to determine whether they could cause urinary abnormalities or were likely asymptomatic in the heterozygous state. This was performed by obtaining clinical information from the patients’ parents who were heterozygous variant carriers. Urinary abnormalities were considered to be present when at least hematuria was noted. To mitigate the fact that not all of the patients’ parents had undergone urinalysis or where there was a lack in uniformity in the urinalysis procedure, we used variant information from the HGMD variant database (https://portal.biobase-international.com/hgmd/pro/start.php) to determine which variants could cause urinary abnormalities (autosomal dominant Alport syndrome, benign familial hematuria, or thin basement membrane) in the heterozygous state.
Statistical Analyses
All calculations were performed using standard statistical software (JMP version 11 for Windows; SAS Institute, Cary, NC). The occurrence of events (age at ESKD onset) was analyzed with the Kaplan–Meier method. To calculate the P value, we used the log-rank test. For the comparison of eGFR, we used analysis of covariance after adjusting for age. We considered an association to be significant when the P value was 0.05.
Results
Clinical Features
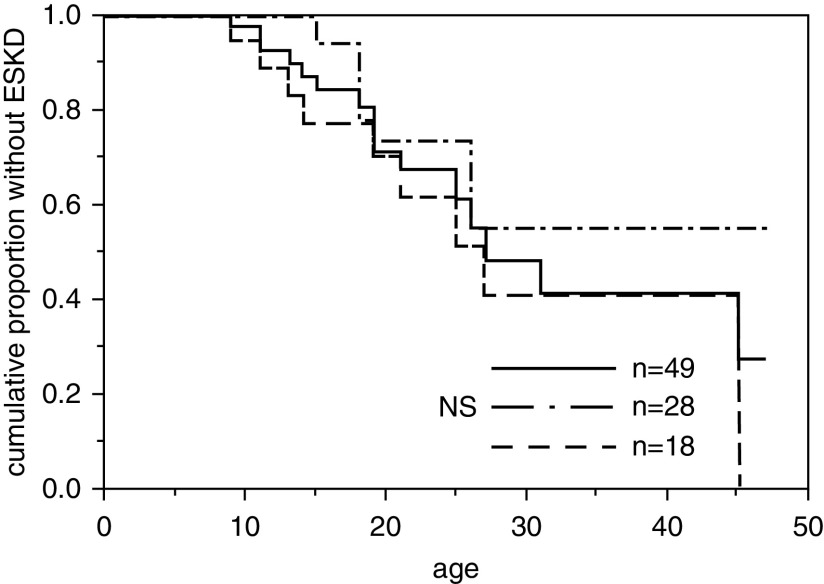
A total of 49 patients from 41 families were included in this study. The clinical and genetic information of the patients is shown in Table 1 and Supplemental Table 1. The median age of the patients when genetically diagnosed with ARAS was 19 (16–21) years. COL4A3 variants were detected in 32 patients from 25 families, and COL4A4 variants were detected in 17 patients from 16 families. Among these patients, 16 progressed to ESKD (14 patients had a COL4A3 mutation, and two patients had a COL4A4 mutation), and the median age for developing ESKD was 27 years (Figure 1). At the time of genetic testing, 18 patients had been diagnosed with hearing loss, 28 had not been diagnosed with hearing loss, and three had no information. The renal survival curves for patients with and without hearing loss at the time of genetic testing are shown in Figure 1. There was no statistically significant difference between these two groups. Only four patients had ocular lesions indicated at the time of genetic testing.
Table 1.
Patients’ clinical and allele information
| Patient Identification | Sex | Age, yr | ESKD Age (Creatinine-eGFR) | Urine Protein-Creatinine Ratio | Hearing Loss | Ocular Lesion | 5 Staining (Glomerular Basement Disease) | Gene | Truncating Allelea | Urinary Findings Alleleb |
| 108 | Men | 16 | (107.0) | 1.2 | − | − | Positive | COL4A3 | 2 | 1 |
| 115 | Women | 19 | (57.8) | 0.12 | + | − | Negative | COL4A3 | 2 | 0 |
| 155 | Men | 36 | 19 | ESKD | + | − | COL4A3 | 2 | 0 | |
| 155–1 | Women | 33 | 21 | ESKD | + | Band keratopathy | COL4A3 | 2 | 0 | |
| 143 | Women | 2 | (122.1) | 0.57 | Negative | COL4A3 | 1 | 1 | ||
| 165 | Men | 6 | (167.8) | 0.30 | − | − | Negative | COL4A3 | 1 | 2 |
| 166 | Women | 18 | 18 | ESKD | − | − | Negative | COL4A3 | 1 | 1 |
| 169 | Men | 19 | (107.8) | 6.3 | + | − | Negative | COL4A3 | 1 | 1 |
| 170 | Women | 7 | (119.5) | 0.80 | − | − | Negative | COL4A3 | 1 | 1 |
| 245 | Men | 41 | (91.5) | 7.4 | + | − | Negative | COL4A3 | 1 | 0 |
| 415 | Women | 17 | (107.7) | 0.35 | + | − | Negative | COL4A3 | 1 | 0 |
| 525 | Women | 11 | (129.8) | 0.35 | + | − | Negative | COL4A3 | 1 | 1 |
| 570 | Men | 45 | (53.8) | 2.2 | − | − | Positive | COL4A3 | 1 | 1 |
| 570–1 | Men | 47 | 31 | ESKD | COL4A3 | 1 | 1 | |||
| 412 | Men | 19 | (80.7) | 1.7 | − | − | COL4A3 | N/A | 1 | |
| 94 | Women | 17 | (64.1) | 0.24 | − | − | Positive | COL4A3 | 0 | 1 |
| 114 | Men | 20 | (34.0) | 1.6 | + | − | Negative | COL4A3 | 0 | 0 |
| 125 | Women | 22 | (138.1) | 0.46 | − | Retinal regeneration | Negative | COL4A3 | 0 | 1 |
| 125–1 | Men | 21 | (8.9) | 2.4 | − | − | Negative | COL4A3 | 0 | 1 |
| 125–2 | Men | 11 | (126.2) | 0.14 | − | − | COL4A3 | 0 | 1 | |
| 130 | Women | 16 | 15 | ESKD | − | Perimacular fleck | COL4A3 | 0 | 2 | |
| 130–1 | Women | 18 | 11 | ESKD | COL4A3 | 0 | 2 | |||
| 137 | Men | 20 | 13 | ESKD | + | COL4A3 | 0 | 1 | ||
| 137–1 | Women | 27 | 26 | ESKD | − | COL4A3 | 0 | 1 | ||
| 167 | Women | 21 | (158.7) | 2.1 | + | − | Negative | COL4A3 | 0 | 1 |
| 168 | Men | 19 | 19 | ESKD | − | − | Negative | COL4A3 | 0 | 2 |
| 171 | Men | 16 | 9 | ESKD | + | − | COL4A3 | 0 | 2 | |
| 171–1 | Women | 11 | 11 | ESKD | + | − | COL4A3 | 0 | 2 | |
| 173 | Men | 25 | 25 | ESKD | + | − | Negative | COL4A3 | 0 | 1 |
| 179 | Women | 45 | 45 | ESKD | + | − | Positive | COL4A3 | 0 | 1 |
| 473 | Women | 8 | (73.9) | 1.8 | − | − | COL4A3 | 0 | 2 | |
| 595 | Women | 19 | 18 | ESKD | − | − | COL4A3 | 0 | 2 | |
| 309 | Women | 2 | (126.7) | 0.78 | − | − | Negative | COL4A4 | 2 | 2 |
| 471 | Women | 41 | (29.5) | 2.9 | + | − | Positive | COL4A4 | 2 | 0 |
| 156 | Women | 7 | (136.6) | 0.43 | − | − | Negative | COL4A4 | 1 | 0 |
| 172 | Men | 16 | 14 | ESKD | + | − | Positive | COL4A4 | 1 | 2 |
| 174 | Men | 2 | (100.8) | 0.53 | − | − | Negative | COL4A4 | 1 | 1 |
| 218 | Men | 12 | (65.8) | 0.17 | − | Hyperopia | Negative | COL4A4 | 1 | 2 |
| 85 | Women | 23 | (98.4) | 1.6 | − | − | COL4A4 | 0 | 2 | |
| 145 | Women | 26 | (106.1) | 0.42 | − | − | Positive | COL4A4 | 0 | 0 |
| 204 | Men | 11 | (102.7) | 1.1 | − | − | Negative | COL4A4 | 0 | 1 |
| 257 | Women | 18 | (107.4) | 0.75 | − | − | COL4A4 | 0 | 1 | |
| 270 | Women | 47 | (45.5) | 1.9 | − | − | Positive | COL4A4 | 0 | 1 |
| 468 | Women | 4 | (153.9) | 0.22 | − | − | COL4A4 | 0 | 1 | |
| 476 | Women | 21 | (125.0) | 0.37 | − | − | Positive | COL4A4 | 0 | 2 |
| 601 | Women | 22 | (82.1) | 4.1 | − | − | Positive | COL4A4 | 0 | 1 |
| 741 | Women | 7 | (132.3) | 8.1 | − | − | COL4A4 | 0 | 2 | |
| 738 | Men | 37 | N/A | N/A | + | − | COL4A4 | N/A | 1 | |
| 738–1 | Women | 33 | 27 | ESKD | + | − | COL4A4 | N/A | 1 |
−, no indication at the time of genetic testing; +, it was pointed out at the time of genetic testing; N/A, not available.
The number of alleles with truncating mutation.
The number of alleles that can cause urinary findings with heterozygous mutation.
Figure 1.
Probability of developing ESKD was not associated with hearing loss. The solid line indicates all patients (n=49). The median age at ESKD onset was 27 years. The dashed line indicates patients with hearing loss at the time of genetic diagnosis (n=18). Their median age at ESKD onset was 27 years. The dashed-dotted line indicates patients without hearing loss at the time of genetic diagnosis (n=28). This group had not reached the median age of ESKD onset even at 40 years. NS, not significant.
Genotype-Phenotype Correlation Analyses of Truncating Mutations
Missense mutations, insertions, and deletions of bases in multiples of three and skipping of bases in exons in multiples of three were defined as nontruncating mutations, whereas nonsense mutations, insertions, and deletions of bases in multiples of other than three, exon skipping of bases in multiples other than three, and large deletions were defined as truncating mutations. Three patients (patient identifications 412, 738, and 738–1) had a mutation in the splicing consensus sequence that was expected to cause exon skipping; however, the transcript was not analyzed and could not be included in this analysis. Twenty-six patients possessed no alleles with a truncating mutation, whereas 14 patients possessed one such allele, and six patients possessed two such alleles. The median age of ESKD onset for these different groups was as follows: no allele, 26 years; one allele, 31 years; two alleles, 21 years; and overall, 31 years (Figure 2, Table 2). There were no statistically significant differences between the three groups. The eGFR at the timing of genetic testing of these patients who had not developed ESKD is shown in Table 1. There was no tendency of the decrease of the eGFR between the three groups (P=0.53).
Figure 2.
Probability of developing ESKD was not associated with truncating mutations. The solid line indicates patients with two alleles containing a truncating mutation (n=5), the dashed line indicates those with one such allele (n=13), and the dashed-dotted line indicates those with no such alleles (n=28). The median ages at ESKD onset of these three groups were 21, 31, and 26 years, respectively. NS, not significantot significant.
Table 2.
Distribution of patients on the basis of the number of alleles containing truncating mutations
| No. of Alleles with Truncating Mutation | Patients with ESKD (Median ESKD Age, yr) | Patients without ESKD |
| 0 | 10 (26) | 18 |
| 1 | 3 (31) | 10 |
| 2 | 2 (21) | 3 |
| Total | 15 (31) | 31 |
Urinary Abnormalities Associated with Heterozygous Variants
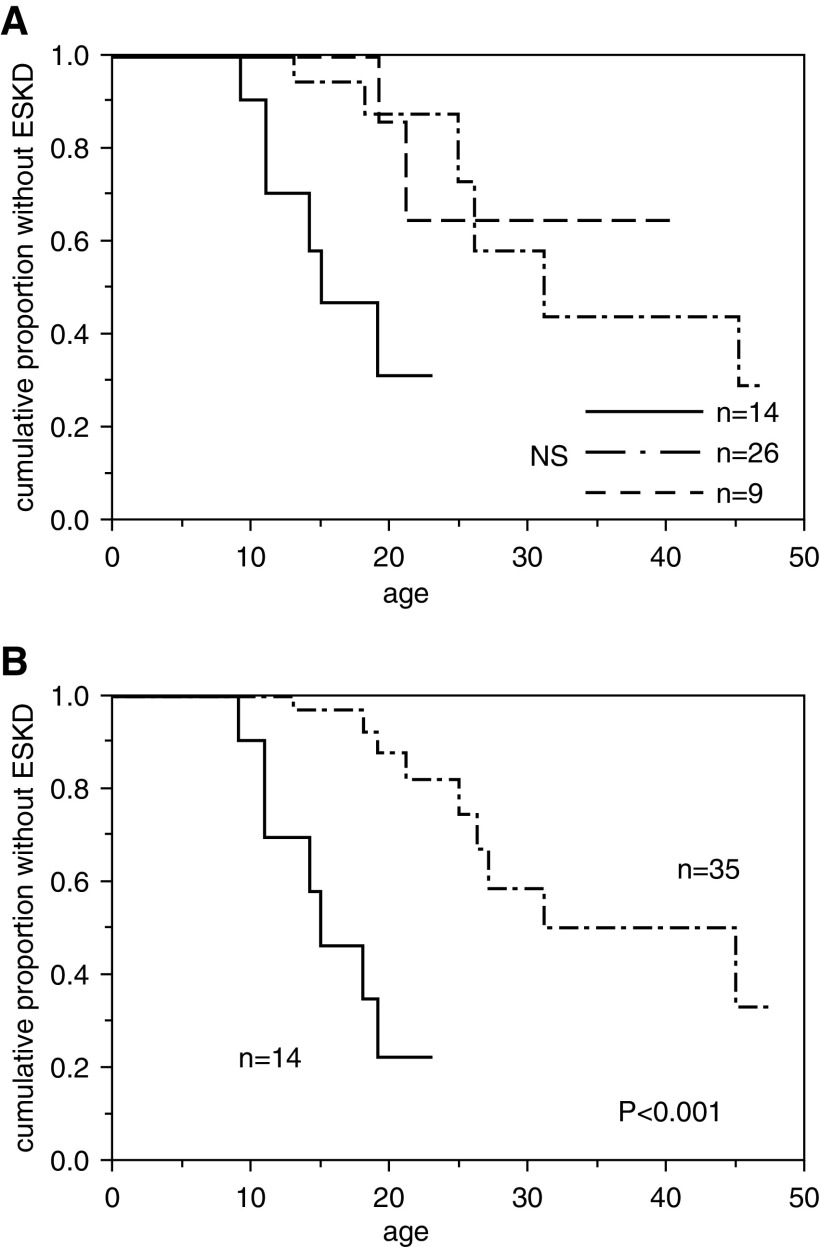
Participants were classified according to the number of COL4A3 and COL4A4 alleles that they had that may be associated with urinary abnormalities in the heterozygous state. As indicated in previous reports, even in cases where the patients’ parents have not been found to have an abnormal urinalysis result, the heterozygous patient may have urinary abnormalities (10,22,23). Such variants were classified as “variants that may be associated urinary abnormalities in the heterozygous state.” Nine patients possessed no alleles with mutations that could cause urinary abnormalities in the heterozygous state, whereas 26 patients possessed one such allele, and 14 patients possessed two such alleles (Table 3). The renal survival curves of these three groups are shown in Figure 3A. The median age at ESKD onset for the three groups was as follows: one allele, 31 years; two alleles, 15 years; and overall, 27 years. In the group with no alleles of interest, seven of nine patients had not developed ESKD at the time of genetic diagnosis, which may reflect a better prognosis; however, we could not compare the different prognoses statistically. Thus, we next divided all of the patients into two groups instead: a zero-allele and one-allele group (n=35) versus the two-allele group (n=14). The median ages of ESKD onset for these two groups were 45 and 15 years, respectively (Figure 3B). Age at onset of ESKD differed significantly between these two groups (P<0.001). The decrease of the eGFR tended to be faster (P=0.08) in the two-allele group (n=14) compared with the combined zero-allele and one-allele group (n=34).
Table 3.
Distribution of patients on the basis of the number of alleles containing variants that could cause urinary abnormalities in the heterozygous state
| No. of Alleles That Can Cause Urinary Findings with Heterozygous Mutation | Patients with ESKD (Median ESKD Age, yr) | Patients without ESKD |
| 0 | 2 (N/A) | 7 |
| 1 | 7 (31) | 19 |
| 2 | 7 (15) | 7 |
| Total | 16 (27) | 33 |
N/A, not available.
Figure 3.
Probability of developing ESKD was associated with heterozygous urinary abnormality–causing mutations. (A) Probability of patients with autosomal recessive Alport syndrome developing ESKD on the basis of the number of alleles containing heterozygous mutations associated with urinary abnormalities. The solid line indicates patients with two such alleles (n=14), the dashed-dotted line indicates those with one such allele (n=26), and the dashed line indicates those with no such alleles (n=9). The median ages at ESKD onset of these three groups were 15, 31 years, and undetermined, respectively. The group with no alleles had not reached the median age of ESKD onset even at 40 years. NS, nNot significantot significant. (B) Combined probability of patients with autosomal recessive Alport syndrome developing ESKD on the basis of the number of alleles containing heterozygous mutations associated with urinary abnormalities. The solid line indicates patients with two such alleles (n=14), and the dashed-dotted line indicates those with one or no such allele (n=35). The median ages at ESKD onset were 15 and 45 years, respectively (P<0.001).
Discussion
This is our second report of a patient series of patients with ARAS; concurrently, it is the study performed that pays attention to the clinical information of individuals with heterozygous mutations.
In this study, in contrast to XLAS, there was no association between the presence of truncating mutations and the renal prognosis of the patients with ARAS, which is consistent with the report by Oka et al.(10) but contradictory to the results reported by Storey et al. (2), Savige et al. (8), and Lee et al. (9). A common feature among these previous reports is a bias toward identified variants that may be due to a “founder effect.” Because of this bias and the small number of patients included in these reports, it is likely that the characteristics of the individual variants affected the results. Although our report only included a small number of Japanese patients due to the rarity of the disease, it did not include hot spots in the Japanese population, and therefore, we believe that there was no extreme bias; however, patients with a family history may lead to an early genetic diagnosis, which may have introduced some bias. Moreover, as an unavoidable limitation of any genotype-phenotype investigation, this study does not provide evidence of whether the heterozygous mutations of interest caused the phenotype or were simply associated with it. Additionally, the relationship may become clear if protein prediction modeling can be carried out, instead of simply distinguishing from truncating and nontruncating.
Here, we compiled the first report on ARAS containing clinical information regarding heterozygous variant carriers such as patients’ parents or by including data extracted from a database of previously reported variants. Our study showed that patients with ARAS have more severe phenotypes when they carry certain heterozygous variants associated with urinary abnormalities, and it indicates the necessity of trio analysis combined with parent urinalysis for early diagnosis and improved patient management. It is evident that further cases of ARAS with genetic diagnosis and the clinical information regarding the presence of heterozygous variants are needed to confirm the prediction of the prognosis of ARAS.
It has been reported that heterozygous mutations in the COL4A3 and COL4A4 genes act as disease modifiers (18). Recently, common variants of UMOD, the causative gene of autosomal dominant nodular interstitial kidney disease, was found to be a susceptibility gene for CKD, hypertension, and renal calculi in the general population (24,25). In the same way, considering the broad spectrum of COL4A3 and COL4A4, these two genes may be disease susceptibility genes for certain renal diseases.
In conclusion, this study showed the clinical importance of noting the urinary findings of heterozygous mutation carriers, such as the parents of patients with ARAS. Notably, our results indicate that patients with ARAS tend to have a poorer prognosis when they carry more mutations associated with urinary abnormalities, even where these are present in the heterozygous state.
Disclosures
K. Iijima and K. Nozu have filed a patent application on the development of antisense nucleotides for exon skipping therapy in Alport syndrome. K. Nozu has received lecture fees from Novartis Pharmaceuticals and corporation and consulting fees from Kyowa Kirin Co., Ltd. All remaining authors have nothing to disclose.
Funding
This study was supported by Ministry of Education, Culture, Sports, Science and Technology of Japan Grants-in-Aid for Scientific Research 18K15712 (to T. Horinouchi), 26293203 (to K. Iijima), 17H04189 (to K. Iijima), and 19K08726 (to K. Nozu) and Japan Agency for Medical Research and Development grants 19ek0109231s0103 (to K. Iijima) and JP19ek0109231h0003 (to K. Nozu).
Supplemental Material
This article contains the following supplemental material online at http://kidney360.asnjournals.org/lookup/suppl/doi:10.34067/KID.0000372019/-/DCSupplemental.
Patient clinical and genetic information. Download Supplemental Table 1, PDF file, 223 KB (222.3KB, pdf)
Acknowledgments
We thank Dr. Natasha Beeton-Kempen from Edanz Group (www.edanzediting.com/ac) for editing a draft of this manuscript. Dr. Kazumoto Iijima reports personal fees from Boehringer Ingelheim; Chugai Pharmaceutical Co., Ltd.; Integrated Development Associates Co., Ltd.; JCR Pharmaceuticals Co. Ltd.; Kyowa Kirin Co. Ltd.; Ono Pharmaceutical Co., Ltd.; Takeda Pharmaceutical Company; and Zenyaku Kogyo Co., Ltd. and grants from Air Water Medical Inc.; Astellas Pharma Inc.; Daiichi Sankyo, Co., Ltd.; Eisai Co., Ltd.; Mochida Pharmaceutical Co., Ltd.; Otsuka Pharmaceutical Co., Ltd.; Shionogi & Co., Ltd.; and Zenyaku Kogyo Co., Ltd., all outside the submitted work.
Author Contributions
T. Horinouchi, K. Iijima, K. Nakanishi, and K. Nozu conceptualized the study; Y. Aoto, T. Horinouchi, K. Iijima, S. Ishiko, C. Nagano, K. Nozu, R. Rossanti, N. Sakakibara, and T. Yamamura were responsible for data curation; T. Horinouchi, C. Nagano, K. Nakanishi, K. Nozu, N. Sakakibara, and Y. Shima were responsible for investigation; T. Horinouchi, K. Iijima, K. Nakanishi, K. Nozu, Y. Shima, and T. Yamamura were responsible for methodology; K. Iijima, N. Morisada, K. Nakanishi, K. Nozu, Y. Shima, and T. Yamamura were responsible for validation; T. Horinouchi wrote the original draft; K. Iijima and K. Nozu were responsible for funding acquisition; K. Iijima and K. Nozu provided supervision; K. Nozu was responsible for project administration; K. Nozu was responsible for resources; and T. Horinouchi reviewed and edited the manuscript.
References
- 1.Kashtan CE: Alport syndrome and thin glomerular basement membrane disease. J Am Soc Nephrol 9: 1736–1750, 1998 [DOI] [PubMed] [Google Scholar]
- 2.Storey H, Savige J, Sivakumar V, Abbs S, Flinter FA: COL4A3/COL4A4 mutations and features in individuals with autosomal recessive Alport syndrome. J Am Soc Nephrol 24: 1945–1954, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gross O, Netzer KO, Lambrecht R, Seibold S, Weber M: Meta-analysis of genotype-phenotype correlation in X-linked Alport syndrome: Impact on clinical counselling. Nephrol Dial Transplant 17: 1218–1227, 2002 [DOI] [PubMed] [Google Scholar]
- 4.Jais JP, Knebelmann B, Giatras I, De Marchi M, Rizzoni G, Renieri A, Weber M, Gross O, Netzer KO, Flinter F, Pirson Y, Verellen C, Wieslander J, Persson U, Tryggvason K, Martin P, Hertz JM, Schröder C, Sanak M, Krejcova S, Carvalho MF, Saus J, Antignac C, Smeets H, Gubler MC: X-linked Alport syndrome: Natural history in 195 families and genotype- phenotype correlations in males. J Am Soc Nephrol 11: 649–657, 2000 [DOI] [PubMed] [Google Scholar]
- 5.Bekheirnia MR, Reed B, Gregory MC, McFann K, Shamshirsaz AA, Masoumi A, Schrier RW: Genotype-phenotype correlation in X-linked Alport syndrome. J Am Soc Nephrol 21: 876–883, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horinouchi T, Nozu K, Yamamura T, Minamikawa S, Omori T, Nakanishi K, Fujimura J, Ashida A, Kitamura M, Kawano M, Shimabukuro W, Kitabayashi C, Imafuku A, Tamagaki K, Kamei K, Okamoto K, Fujinaga S, Oka M, Igarashi T, Miyazono A, Sawanobori E, Fujimaru R, Nakanishi K, Shima Y, Matsuo M, Ye MJ, Nozu Y, Morisada N, Kaito H, Iijima K: Detection of splicing abnormalities and genotype-phenotype correlation in X-linked Alport syndrome. J Am Soc Nephrol 29: 2244–2254, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hashimura Y, Nozu K, Kaito H, Nakanishi K, Fu XJ, Ohtsubo H, Hashimoto F, Oka M, Ninchoji T, Ishimori S, Morisada N, Matsunoshita N, Kamiyoshi N, Yoshikawa N, Iijima K: Milder clinical aspects of X-linked Alport syndrome in men positive for the collagen IV α5 chain. Kidney Int 85: 1208–1213, 2014 [DOI] [PubMed] [Google Scholar]
- 8.Savige J, Storey H, Il Cheong H, Gyung Kang H, Park E, Hilbert P, Persikov A, Torres-Fernandez C, Ars E, Torra R, Hertz JM, Thomassen M, Shagam L, Wang D, Wang Y, Flinter F, Nagel M: X-linked and autosomal recessive Alport syndrome: Pathogenic variant features and further genotype-phenotype correlations. PLoS One 11: e0161802, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee JM, Nozu K, Choi DE, Kang HG, Ha IS, Cheong HI: Features of autosomal recessive Alport syndrome: A systematic review. J Clin Med 8: 178, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oka M, Nozu K, Kaito H, Fu XJ, Nakanishi K, Hashimura Y, Morisada N, Yan K, Matsuo M, Yoshikawa N, Vorechovsky I, Iijima K: Natural history of genetically proven autosomal recessive Alport syndrome. Pediatr Nephrol 29: 1535–1544, 2014 [DOI] [PubMed] [Google Scholar]
- 11.Heidet L, Arrondel C, Forestier L, Cohen-Solal L, Mollet G, Gutierrez B, Stavrou C, Gubler MC, Antignac C: Structure of the human type IV collagen gene COL4A3 and mutations in autosomal Alport syndrome. J Am Soc Nephrol 12: 97–106, 2001 [DOI] [PubMed] [Google Scholar]
- 12.Wang Y, Sivakumar V, Mohammad M, Colville D, Storey H, Flinter F, Dagher H, Savige J: Clinical and genetic features in autosomal recessive and X-linked Alport syndrome. Pediatr Nephrol 29: 391–396, 2014 [DOI] [PubMed] [Google Scholar]
- 13.Nozu K, Nakanishi K, Abe Y, Udagawa T, Okada S, Okamoto T, Kaito H, Kanemoto K, Kobayashi A, Tanaka E, Tanaka K, Hama T, Fujimaru R, Miwa S, Yamamura T, Yamamura N, Horinouchi T, Minamikawa S, Nagata M, Iijima K: A review of clinical characteristics and genetic backgrounds in Alport syndrome. Clin Exp Nephrol 23: 158–168, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Savige J, Gregory M, Gross O, Kashtan C, Ding J, Flinter F: Expert guidelines for the management of Alport syndrome and thin basement membrane nephropathy. J Am Soc Nephrol 24: 364–375, 2013 [DOI] [PubMed] [Google Scholar]
- 15.Savige J: Should we diagnose autosomal dominant Alport syndrome when there is a pathogenic heterozygous COL4A3 or COL4A4 variant? Kidney Int Rep 3: 1239–1241, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Strasser K, Hoefele J, Bergmann C, Büscher AK, Büscher R, Hoyer PF, Weber S: COL4A5-associated X-linked Alport syndrome in a female patient with early inner ear deafness due to a mutation in MYH9. Nephrol Dial Transplant 27: 4236–4240, 2012 [DOI] [PubMed] [Google Scholar]
- 17.Tonna S, Wang YY, Wilson D, Rigby L, Tabone T, Cotton R, Savige J: The R229Q mutation in NPHS2 may predispose to proteinuria in thin-basement-membrane nephropathy. Pediatr Nephrol 23: 2201–2207, 2008 [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y, Ding J, Zhang H, Yao Y, Xiao H, Wang S, Wang F: Effect of heterozygous pathogenic COL4A3 or COL4A4 variants on patients with X-linked Alport syndrome. Mol Genet Genomic Med 7: e647, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uemura O, Nagai T, Ishikura K, Ito S, Hataya H, Gotoh Y, Fujita N, Akioka Y, Kaneko T, Honda M: Creatinine-based equation to estimate the glomerular filtration rate in Japanese children and adolescents with chronic kidney disease. Clin Exp Nephrol 18: 626–633, 2014 [DOI] [PubMed] [Google Scholar]
- 20.Matsuo S, Imai E, Horio M, Yasuda Y, Tomita K, Nitta K, Yamagata K, Tomino Y, Yokoyama H, Hishida A; Collaborators developing the Japanese equation for estimated GFR : Revised equations for estimated GFR from serum creatinine in Japan. Am J Kidney Dis 53: 982–992, 2009 [DOI] [PubMed] [Google Scholar]
- 21.Yamamura T, Nozu K, Fu XJ, Nozu Y, Ye MJ, Shono A, Yamanouchi S, Minamikawa S, Morisada N, Nakanishi K, Shima Y, Yoshikawa N, Ninchoji T, Morioka I, Kaito H, Iijima K: Natural history and genotype-phenotype correlation in female X-linked Alport syndrome. Kidney Int Rep 2: 850–855, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kamiyoshi N, Nozu K, Fu XJ, Morisada N, Nozu Y, Ye MJ, Imafuku A, Miura K, Yamamura T, Minamikawa S, Shono A, Ninchoji T, Morioka I, Nakanishi K, Yoshikawa N, Kaito H, Iijima K: Genetic, clinical, and pathologic backgrounds of patients with autosomal dominant Alport syndrome. Clin J Am Soc Nephrol 11: 1441–1449, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Longo I, Porcedda P, Mari F, Giachino D, Meloni I, Deplano C, Brusco A, Bosio M, Massella L, Lavoratti G, Roccatello D, Frascá G, Mazzucco G, Muda AO, Conti M, Fasciolo F, Arrondel C, Heidet L, Renieri A, De Marchi M: COL4A3/COL4A4 mutations: From familial hematuria to autosomal-dominant or recessive Alport syndrome. Kidney Int 61: 1947–1956, 2002 [DOI] [PubMed] [Google Scholar]
- 24.Devuyst O, Olinger E, Rampoldi L: Uromodulin: From physiology to rare and complex kidney disorders. Nat Rev Nephrol 13: 525–544, 2017 [DOI] [PubMed] [Google Scholar]
- 25.Baek JI, Choi SJ, Park SH, Choi JY, Kim CD, Kim YL, Kim UK: Identification of novel variants in the COL4A4 gene in Korean patients with thin basement membrane nephropathy. Indian J Med Res 129: 525–533, 2009 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Patient clinical and genetic information. Download Supplemental Table 1, PDF file, 223 KB (222.3KB, pdf)