Abstract
In October 1998, the Centers for Disease Control and Prevention (CDC) assisted in an investigation of an outbreak of campylobacteriosis at a school in Salina, Kansas. Twenty-two isolates were submitted from the Kansas state public health laboratory to CDC, 9 associated with the outbreak and 13 epidemiologically unrelated sporadic isolates. Pulsed-field gel electrophoresis (PFGE) using SmaI and SalI was initially used to validate the epidemiologic data. We then tested the ability of other subtyping techniques to distinguish the outbreak-associated isolates from unrelated sporadic isolates. The methods employed were somatic O serotyping, PCR-restriction fragment length polymorphism (RFLP) analysis of flaA, DNA sequence analysis of 582 bp of flaA that included the short variable region (SVR), and sequencing of the entire flaA gene. PFGE was the most discriminatory technique, yielding 11 SmaI and 10 SalI restriction profiles. All outbreak isolates were indistinguishable by PFGE, somatic O serotyping, and sequencing of the 582-bp region of the flaA gene. fla typing by PCR-RFLP grouped one sporadic isolate with the outbreak strain. Analysis of the DNA sequence of a 582-bp segment of flaA produced strain groupings similar to that generated by PCR-RFLP but further differentiated two flaA PCR-RFLP types (with a 1-bp difference in the 582-bp region). Two sporadic strains were distinct by flaA PCR-RFLP but differed only by a single base substitution in the 582-bp region. The entire flaA gene was sequenced from strains differing by a single base pair in the 582-bp region, and the data revealed that additional discrimination may in some cases be obtained by sequencing outside the SVR. PFGE was superior to all other typing methods tested for strain discrimination; it was crucial for understanding the Kansas outbreak and, when SmaI was used, provided adequate discrimination between unrelated isolates.
The significance of campylobacters as important human pathogens is now well established. In the United States, Campylobacter jejuni is the most common cause of bacterial enteritis; an estimated 2.5 million cases of human Campylobacter infection occur each year (17). The rise in the number of Campylobacter infections is most likely the result of increased case ascertainment (34) and a growing awareness of the organism among the public, physicians, and the public health community. In 1998, 44% of laboratory-confirmed cases of bacterial gastroenteritis reported to the Centers for Disease Control and Prevention (CDC)-U.S. Department of Agriculture-Food and Drug Administration collaborating sites, food-borne disease active surveillance network (FoodNet) were caused by Campylobacter species (4).
Although outbreaks of Campylobacter infection occur (27), the majority of infections are sporadic. The control of Campylobacter infection will ultimately depend on a more thorough understanding of sources, transmission routes, and pathogen-host interactions (1). Our current understanding of the epidemiology of Campylobacter infection remains incomplete. To gain more insight, laboratory methods are needed that differentiate epidemiologically related isolates from unrelated isolates. Over the last 20 years, a large number of phenotypic and genotypic typing methods have been applied to Campylobacter isolates (25). Phenotypic techniques, such as biotyping (3, 16), serotyping (15, 28), and phage typing (7, 32), are useful for strain characterization and are still in widespread use, but they do have limitations. These include the considerable time and labor investment required for maintenance of reagents; cross-reactivity between antigens, notably in the somatic O (Penner) serotyping scheme; and the occurrence of nontypeable isolates (13, 25, 29). Genotyping offers greater capacity for differentiating strains and can be useful in making phylogenetic as well as epidemiologic inferences. Restriction endonuclease analysis, ribotyping, PCR-based methods, pulsed-field gel electrophoresis (PFGE), and, more recently, DNA sequencing-based typing of the flaA gene have all been used to subtype Campylobacter (38). However, there are questions regarding the stability of the Campylobacter genome (8, 37); at this time, no technique has been solely identified as the “gold standard” for typing Campylobacter.
In October 1998, CDC assisted the Kansas Department of Health and Environment in an investigation of an outbreak of campylobacteriosis at a school in Salina, Kansas, involving students, staff, and visitors. During the same period, additional persons with Campylobacter infection were identified in the surrounding community. It was unclear if they too were associated with the school outbreak, and PFGE was used to help determine that community cases were not linked to the outbreak (21). In this study, we sought to assess the abilities of additional subtyping techniques to correctly characterize this epidemiologically well-defined collection of Campylobacter isolates in terms of their abilities to distinguish between outbreak-associated isolates and non-outbreak-associated isolates causing sporadic infection in the Kansas community. In addition, a range of criteria, including the ease of use, cost, and rapidity of each method, was considered. The methods evaluated were somatic O serotyping, PCR-restriction fragment length polymorphism (RFLP) analysis of flaA, DNA sequence analysis of 585 bp of flaA that included the short variable region (SVR), sequencing of the entire flaA gene, and PFGE using SmaI and SalI. Our findings show that PFGE is the most discriminatory subtyping method for molecular epidemiologic studies of Campylobacter.
MATERIALS AND METHODS
We tested 22 isolates from stool specimens submitted by the Kansas Department of Health and Environment. They were nine outbreak-associated isolates from Saline County, five others from the same county, and eight epidemiologically unrelated isolates from other counties in the same state isolated during the same period. All isolates were cultivated at 37°C for 48 h on heart infusion agar with 5% (vol/vol) defibrinated rabbit blood (Becton Dickinson Biosciences, Franklin Lakes, N.J.) under microaerobic conditions. Isolates were speciated by standard procedures (2).
Somatic O serotyping was performed as described by Penner and Hennessy (28) using a panel of 24 antisera which represent common serotypes in the United States (26). Isolates that were nontypeable underwent passage on blood agar an additional eight times until an antigen was detected. Flagellin PCR-RFLP analysis was done as previously described (19). Sequencing of 582 bp of the flaA gene that included the SVR and the entire coding sequence was performed by the method of Meinersmann et al. (18).
Preparation of C. jejuni DNA, macrorestriction analysis using the restriction enzymes SmaI and SalI, and PFGE were performed previously (21). Electrophoresis was carried out for 22 h at 200 V and 14°C constant temperature in a CHEF-DRIII system (Bio-Rad, Richmond, Calif.) with pulse times ramped from 10 to 35 s for SmaI and 4 to 50 s for SalI. Simpson's index of diversity was calculated as described previously (9).
Nucleotide sequence accession numbers.
The sequences obtained in this study have been deposited in GenBank under accession numbers AF369577 to AF369587.
RESULTS
A summary of the typing results is given in Table 1.
TABLE 1.
Typing results for Kansas C. jejuni isolates
| Strain no. | Somatic O serotype | Flagellin gene type
|
PFGE type
|
|||
|---|---|---|---|---|---|---|
| PCR-RFLP (DdeI) | Sequencing
|
SmaI | SalI | |||
| SVR | Full | |||||
| Outbreak associated | ||||||
| D5475 | 19 | D1 | 1 | NTb | A | 1 |
| D5477 | 19 | D1 | 1 | 1 | A | 1 |
| D5479 | 19 | D1 | 1 | NT | A | 1 |
| D5482 | 19 | D1 | 1 | NT | A | 1 |
| D5483 | 19 | D1 | 1 | NT | A | 1 |
| D5484 | 19 | D1 | 1 | NT | A | 1 |
| D5486 | 19 | D1 | 1 | NT | A | 1 |
| D5476 | 19 | D1 | 1 | NT | A | 1 |
| D5480 | 19 | D1 | 1 | NT | A | 1 |
| Sporadic infection | ||||||
| D5478 | 4 complexa | D2 | 2 | 2 | B | 2 |
| D5487 | 4 complex | D2 | 2 | NT | B | 2 |
| D5498 | 4 complex | D2 | 2 | NT | B | 2 |
| D5493 | 4 complex | D2 | 2b | 2b | H | 7 |
| D5494 | 4 complex | D2 | 2 | NT | I | 8 |
| D5497 | 4 complex | D1 | 1b | 1b | K | 10 |
| D5492 | 4 complex | D6 | 5b | 4 | G | 5 |
| D5488 | 38, 29 | D4 | 4 | NT | C | 3 |
| D5489 | 38 | D4 | 4 | NT | C | 3 |
| D5490 | 8 | D5 | 5 | 3 | E | 4 |
| D5491 | 1, 8 | D5 | 5 | NT | F | 5 |
| D5495 | 2 | D7 | 6 | NT | J | 9 |
| D5481 | 5 | D3 | 3 | NT | D | 3 |
4 complex consists of strains expressing any combinations of antigens 4, 13, 16, 43, and 50.
NT, not tested.
Serotyping.
Seven different somatic O (heat-stable) serotypes were identified among the 22 C. jejuni isolates (Table 1). Seven of nine isolates associated with the outbreak were serotype O:19, and the remaining two were initially nontypeable by standard techniques. Testing of the nontypeable isolates after eight transfers resulted in detection of the O:19 antigen. The isolates associated with sporadic infection exhibited a range of serotypes; none were serotype O:19 (Table 1).
Flagellin gene typing.
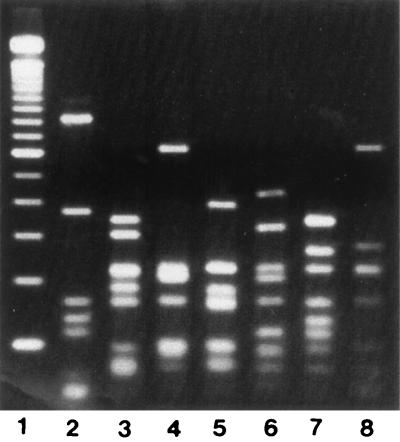
All isolates produced an flaA PCR amplicon of the expected 1.7-kb size. Amplicons were digested with DdeI, and flaA types were assigned based on the different fragment patterns. Restriction analysis by DdeI gave seven different flaA types among the 22 C. jejuni isolates (Fig. 1). All outbreak-associated isolates had an indistinguishable flaA type, designated D1 (Table 1). One sporadic strain (D5497; serotype O:4 complex) also had this flaA type. The second most common flaA type (D2) was seen in five of the seven O:4 complex isolates. The remaining strain with this serotype had a unique flaA type. Of the four remaining flaA types, two (D4 and D5) were each seen in two isolates, and two (D3 and D7) were unique.
FIG. 1.
DdeI flaA PCR-RFLP patterns of Kansas C. jejuni strains. Lanes: 1, 100-bp ladder marker; 2, D5475; 3, D5478; 4, D5481; 5, D5488; 6, D5490; 7, D5492; 8, D5495.
Flagellin gene sequencing.
A 582-bp segment including the SVR of flaA was sequenced and analyzed. A numerical designation was assigned to each unique sequence (Table 1). The 582-bp sequences from all nine of the outbreak-associated isolates were indistinguishable. Strain D5497, which had a flaA PCR-RFLP pattern indistinguishable from that of the outbreak strain, differed in its SVR sequence from the outbreak strain by a single base pair. flaA PCR-RFLP type D2 was also further differentiated by SVR sequencing; one strain (D5493) differed from the other four isolates with the same flaA type by a single base pair in the 582-bp region sequenced, which was outside the SVR. The SVR sequence of strain D5492 (flaA pattern D6) differed from the SVR sequence of strains D5490 and D5491 (flaA pattern D5) by a single base pair, though their RFLP patterns were related but different (Fig. 1, lane 6 [D6] versus lane 7 [D7]).
The entire coding region of flaA was sequenced from one isolate from each of the three groups of strains that differed by a single base pair in the 582-bp sequence. The outbreak strain and one sporadic strain, D5497, had indistinguishable flaA PCR-RFLP patterns and a 1-bp mismatch in the SVR. Two additional mismatches were found in the rest of flaA. Strains D5478 (as well as D5487, D5494, and D5498) and D5493 had identical flaA PCR-RFLP patterns and a 1-bp mismatch in the 582-bp sequence that was outside the SVR. There were no additional base substitutions in the rest of the gene in these strains. Strains D5490 (and D5491) and D5492 were distinct by flaA DdeI RFLP but differed by 1-bp in the SVR. The flaA sequences from these strains were 99.9% identical for the first approximately 800 bp of flaA (where the SVR is located) but more divergent at the 3′ end of the gene (94% identity).
PFGE analysis.
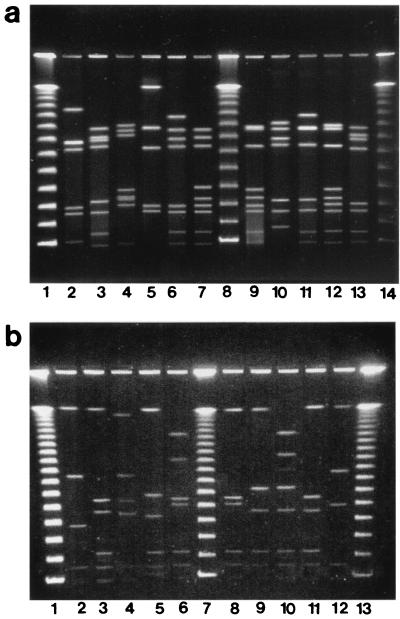
PFGE analysis of SmaI-digested DNA from all isolates yielded between 5 and 10 fragments ranging in size from approximately 40 to 480 kb (Fig. 2a). All outbreak-associated isolates had indistinguishable macrorestriction profiles (pattern A). Ten different macrorestriction profiles were seen among the remaining 13 isolates from sporadic infections; none of them was identical to pattern A. Two macrorestriction profiles were observed among multiple strains, with all isolates of a given profile having identical serotypes and flaA types. The remaining eight strains all had unique macrorestriction profiles. SalI digestion of DNA yielded up to eight fragments ranging in size from approximately 40 to 560 kb (Fig. 2b). The strains were grouped almost identically to those assigned using SmaI, except for two strains, D5491 and D5492. With SmaI, each strain had a unique profile, with the two profiles differing from each other by three bands; with SalI, they were indistinguishable.
FIG. 2.
(a) PFGE restriction profiles of SmaI-digested DNA of Kansas C. jejuni strains. Lanes: 2, D5482; 3, D5487; 4, D5488; 5, D5481; 6, D5490; 7, D5491; 9, D5492; 10, D5493; 11, D5494; 12, D5495; 13, D5497; 1, 8, and 14, 48.5-kb DNA ladder. (b) PFGE restriction profiles of SalI-digested DNA of Kansas C. jejuni strains. Lanes: 2, D5482; 3, D5487; 4, D5481; 5, D5488; 6, D5490; 8, D5491; 9, D5493; 10, D5494; 11, D5495; 12, D5497; 1, 7, and 13, 48.5-kb DNA ladder.
Discrimination potentials of the different typing methods.
A summary of the discrimination among the C. jejuni strains by the different techniques used is shown in Table 2. The numerical index of discrimination (9) ranged from 0.749 for serotyping up to 0.827 for PFGE using SmaI.
TABLE 2.
Discrimination indices for methods used to type C. jejuni
| Typing method | No. of types | Isolates in main type (%) | Discrimination index |
|---|---|---|---|
| Serotyping | 7 | 41 | 0.749 |
| PCR-RFLP of flaA gene | 7 | 45 | 0.753 |
| Sequencing of flaA gene | |||
| SVR | 9 | 41 | 0.804 |
| PFGE | |||
| SmaI | 11 | 41 | 0.827 |
| SalI | 10 | 41 | 0.823 |
DISCUSSION
We studied a collection of outbreak-associated and sporadic isolates from patients in Kansas who were all ill during the same time period in order to compare the relative usefulness of current subtyping techniques. The evaluation of the practical utility of these methods included both correct discrimination in this well-characterized event and the ability to perform the method rapidly and economically. Correct separation of outbreak cases from sporadic cases was achieved by several of the methods: PFGE, serotyping, and sequencing of the SVR. PFGE was the most discriminatory technique used in this study, with a numerical index of discrimination of 0.827 for SmaI and 0.823 for SalI.
Although serotyping is a practical and valid phenotypic method for epidemiologic typing of Campylobacter and has been useful in both clinical (12) and outbreak (23, 36) investigations, it can produce ambiguous results. This can be due to the occurrence of nontypeable strains, transient antigen expression, and cross-reactivity between certain antigens (25, 29). The method requires a panel of antisera that is costly to maintain; all these factors limit the use of this technique in surveillance studies. In practical terms, serotyping is laborious and requires at least 5 to 7 days to complete, considering the need to repeatedly subculture isolates before testing. A number of studies have reported that repeated subculturing resulted in nontypeable strains becoming typeable by serotyping (10, 23). In this study, initial serotyping results identified two of nine outbreak isolates (D5476 and D5480) as nontypeable in the panel of 24 antisera used, yet epidemiologic data supported the fact that these isolates were part of the school outbreak. Only after repeated subculturing (eight transfers) were the two nontypeable isolates identified as serotype O:19.
Somatic serotype O:19 has been reported as the cause of a number of Campylobacter outbreaks (11, 24, 31). The prevalence of serotype O:19 in sporadic cases of uncomplicated campylobacteriosis has been reported to be between 1 and 6% (20). Several studies suggest that Penner O:19 is overrepresented in Guillain-Barré syndrome-associated C. jejuni isolates (S. Fujimoto, N. Yuki, T. Itoh, and K. Amako, Letter, J. Infect. Dis. 165:183, 1992). No cases of Guillain-Barré syndrome were recognized in this outbreak.
Several subtyping methods based on flaA have been reported, including PCR-RFLP (19, 22) and flaA sequencing (18). They are generally simple, cost-effective, and relatively rapid (2 days). However, the use of a single genetic locus as an epidemiologic tool requires caution, especially when making inferences about clonal ancestry, since one gene may not be representative of the entire genome (38). Indeed, this has been reported previously for the flaA locus (8) and is also demonstrated in our study. One sporadic strain (D5497) had a flaA PCR-RFLP profile indistinguishable from that of the outbreak pattern (D1), yet it had a different serotype (O:4) and different PFGE profiles. The occurrence of strains with different serotypes having identical flaA types has been shown previously (22, 33). Thus, relying solely on flaA PCR-RFLP analysis can lead to misinterpretation of the data.
Sequence-based subtyping of the SVR, a 267-bp sequence located near the 5′ end of flaA that provides a level of discrimination similar to that detected in the entire flaA sequence (18), was more discriminatory than PCR-RFLP analysis (Table 2) and correctly differentiated the outbreak strain from the sporadic strains. Among the unrelated strains, sequencing of the SVR, as well as PFGE, further differentiated the O:4 complex strains. In our study, sequences outside the SVR provided additional discrimination not seen in the SVR. Although it is advantageous to sequence only a small region of the flaA gene, further evaluation may be necessary to clarify whether it is representive of the entire gene. Despite these limitations, molecular characterization of flaA via PCR-RFLP analysis or sequencing may be useful for rapid, preliminary characterization of strains when the aim is to establish an epidemiologic link in a well-defined setting. Furthermore, sequencing of flaA provides a precise measure of genetic variability, as it is based on the DNA sequence, not band matching. However, an initial investment in an automated DNA sequencer is necessary to carry out this method, making it less accessible for smaller laboratories. Recent advances in the development of high-throughput sequence capabilities, microarray technologies, and powerful bioinformatics tools means that sequence-based techniques are valuable and should be investigated further.
While PFGE is also somewhat labor-intensive, we regard it as the current gold standard because it examines polymorphisms throughout the genome and it has the highest discriminatory power of the typing methods tested. Recently CDC, in collaboration with state health departments and the Food and Drug Administration, established PulseNet, a computer network to rapidly analyze and compare PFGE patterns from different sources of several important food-borne pathogens (35). Until recently, one disadvantage of PFGE was the length of time required to perform the technique, which for Campylobacter was typically 3 to 4 days. The development of a rapid PFGE protocol (24 to 30 h) for Campylobacter (30) and the addition of this important enteric organism to the PulseNet system, which is currently under way, will make this a rapid and standardized technique more amenable to routine use. Together with traditional epidemiologic methods, this genotypic database should enable us to make more accurate and relevant epidemiologic conclusions.
Interestingly, analysis of our study data revealed two small clusters of related isolates among the sporadic cases, one containing three and the other two isolates. This suggests that the Campylobacter isolates in both cases came from a common source, although we have no epidemiologic evidence to support this. Epidemiologic and microbiological analysis of a larger panel of sporadic infections may be helpful in determining whether these related isolates may have had a common source.
The observations described in this study provide an insight into the usefulness of some of the currently available subtyping methods. Two additional high-resolution genotyping techniques have recently been described that may have utility for the subtyping of Campylobacter: amplified fragment length polymorphism (AFLP) analysis (5) and multilocus sequence typing (MLST) (6). AFLP has the advantage of whole-genome analysis, as does PFGE, and provides automated data acquisition and analysis. MLST, which involves comparative DNA sequencing of several genetic loci, provides precise information regarding strain relationships and simplifies interlaboratory comparisons, both of which have proven difficult with PFGE. Further investigation into the utility of these methods is needed, and to address this, we have initiated a project to assess the application of DNA sequence-based subtyping methods for national and global epidemiologic studies of Campylobacter.
REFERENCES
- 1.Advisory Committee on the Microbiological Safety of Food. Interim report on Campylobacter. London, United Kingdom: Her Majesty's Stationery Office; 1993. [Google Scholar]
- 2.Barrett T J, Patton C M, Morris G K. Differentiation of Campylobacter species using phenotypic characterization. Lab Med. 1988;19:96–102. [Google Scholar]
- 3.Bolton F J, Wareing D R A, Skirrow M B, Hutchinson D N. Identification and biotyping of campylobacters. In: Board R G, Jones D, Skinner F A, editors. Identification methods in applied and environmental microbiology. SAB Technical Series 29. London, United Kingdom: Academic Press; 1992. pp. 151–161. [Google Scholar]
- 4.Centers for Disease Control and Prevention. Incidence of foodborne illnesses: preliminary data from the Foodborne Diseases Active Surveillance Network (Foodnet)–United States. Morb Mortal Wkly Rep. 1999;48:189–194. [PubMed] [Google Scholar]
- 5.de Boer P, Duim B, Rigter A, van Der Plas J, Jacobs-Reitsma W F, Wagenaar J A. Computer-assisted analysis and epidemiological value of genotyping methods for Campylobacter jejuni and Campylobacter coli. J Clin Microbiol. 2000;38:1940–1946. doi: 10.1128/jcm.38.5.1940-1946.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dingle K E, Colles F M, Wareing D R A, Ure R, Fox A J, Bolton F E, Bootsma H J, Willems R J L, Urwin R, Maiden M C J. Multilocus sequence typing system for Campylobacter jejuni. J Clin Microbiol. 2001;39:14–23. doi: 10.1128/JCM.39.1.14-23.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grajewski B A, Kusek J W, Gelfand H M. Development of a bacteriophage typing system for Campylobacter jejuni and Campylobacter coli. J Clin Microbiol. 1985;22:13–18. doi: 10.1128/jcm.22.1.13-18.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harrington C S, Thomson-Carter F M, Carter P E. Evidence for recombination in the flagellin locus of Campylobacter jejuni: implications for the flagellin gene typing scheme. J Clin Microbiol. 1997;35:2386–2392. doi: 10.1128/jcm.35.9.2386-2392.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hunter P R, Gaston M A. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J Clin Microbiol. 1988;26:2465–2466. doi: 10.1128/jcm.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacobs-Reitsma W F, Maas H M E, Jansen W H. Penner serotyping of Campylobacter isolates from poultry with absorbed pooled antisera. J Appl Bacteriol. 1995;79:286–291. doi: 10.1111/j.1365-2672.1995.tb03139.x. [DOI] [PubMed] [Google Scholar]
- 11.Jones D M, Sutcliffe E M, Abbott J D. Serotyping of Campylobacter species by combined use of two methods. Eur J Clin Microbiol. 1985;4:562–565. doi: 10.1007/BF02013395. [DOI] [PubMed] [Google Scholar]
- 12.Karmali M A, Penner J L, Fleming P C, Williams A, Hennessy J N. The serotype and biotype distribution of clinical isolates of Campylobacter jejuni and Campylobacter coli over a three-year period. J Infect Dis. 1983;147:243–246. doi: 10.1093/infdis/147.2.243. [DOI] [PubMed] [Google Scholar]
- 13.Khakhria R, Lior H. Extended phage-typing scheme for Campylobacter jejuni and Campylobacter coli. Epidemiol Infect. 1992;108:403–414. doi: 10.1017/s0950268800049918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuroki S, Saida T, Nukina M, Haruta T, Yoshioka M, Kobayashi Y, Nakanishi H. Campylobacter jejuni strains from patients with Guillain-Barré syndrome belong mostly to Penner serogroup 19 and contain beta-N-acetylglucosamine residues. Ann Neurol. 1993;33:570–576. doi: 10.1002/ana.410330304. [DOI] [PubMed] [Google Scholar]
- 15.Lior H, Woodward D L, Edgar J A, Laroche L J, Gill P. Serotyping of Campylobacter jejuni by slide agglutination based on heat-labile antigenic factors. J Clin Microbiol. 1982;15:761–768. doi: 10.1128/jcm.15.5.761-768.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lior H. New extended biotyping scheme for Campylobacter jejuni, Campylobacter coli, and Campylobacter laridis. J Clin Microbiol. 1984;20:636–640. doi: 10.1128/jcm.20.4.636-640.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mead P S, Slutsker L, Dietz V, McCaig L F, Bresee J S, Shapiro C, Griffin P M, Tauxe R V. Food-related illness and death in the United States. Emerg Infect Dis. 1999;5:607–625. doi: 10.3201/eid0505.990502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meinersmann R J, Helsel L O, Fields P I, Hiett K L. Discrimination of Campylobacter jejuni isolates by fla gene sequencing. J Clin Microbiol. 1997;35:2810–2814. doi: 10.1128/jcm.35.11.2810-2814.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nachamkin I, Ung H, Patton C M. Analysis of HL and O serotypes of Campylobacter strains by the flagellin gene typing system. J Clin Microbiol. 1996;34:277–281. doi: 10.1128/jcm.34.2.277-281.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nachamkin I, Allos B M, Ho T. Campylobacter species and Guillain-Barré syndrome. Clin Microbiol Rev. 1998;11:555–567. doi: 10.1128/cmr.11.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olsen S J, Hansen G, Bartlett L, Fitzgerald C, Sonder A, Manjrekar R S, Riggs T, Kim J, Flahart R, Pezzino G, Swerdlow D L. An outbreak of Campylobacter jejuni infections associated with food handler contamination: the use of pulsed-field gel electrophoresis. J Infect Dis. 2001;183:164–167. doi: 10.1086/317657. [DOI] [PubMed] [Google Scholar]
- 22.Owen R J, Fitzgerald C, Sutherland K, Borman P. Flagellin gene polymorphism analysis of Campylobacter jejuni infecting man and other hosts and comparison with biotyping and somatic antigen serotyping. Epidemiol Infect. 1994;113:221–234. doi: 10.1017/s0950268800051657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patton C M, Barrett T J, Morris G K. Comparison of the Penner and Lior method for serotyping Campylobacter spp. J Clin Microbiol. 1985;22:558–565. doi: 10.1128/jcm.22.4.558-565.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patton C M, Wachsmuth I K, Evans G M, Kiehlbauch J A, Plikaytis B D, Troup N, Tompkins L, Lior H. Evaluation of 10 methods to distinguish epidemic-associated Campylobacter strains. J Clin Microbiol. 1991;29:680–688. doi: 10.1128/jcm.29.4.680-688.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patton C M, Wachsmuth I K. Typing schemes: are current methods useful? In: Nachamkin I, Blaser M J, Tompkins L S, editors. Campylobacter jejuni: current status and future trends. Washington, D.C.: American Society for Microbiology; 1992. pp. 110–128. [Google Scholar]
- 26.Patton C M, Nicholson M A, Ostroff S M, Ries A A, Wachsmuth I K, Tauxe R V. Common somatic O and heat-labile serotypes among Campylobacter strains from sporadic infections in the United States. J Clin Microbiol. 1993;31:1525–1530. doi: 10.1128/jcm.31.6.1525-1530.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pebody R G, Ryan M J, Wall P G. Outbreaks of Campylobacter infection: rare events for a common pathogen. Commun Dis Rep Rev. 1997;3:33–37. [PubMed] [Google Scholar]
- 28.Penner J L, Hennessy J N. Passive hemagglutination technique for serotyping Campylobacter fetus subsp. jejuni on the basis of soluble heat-stable antigens. J Clin Microbiol. 1980;12:732–737. doi: 10.1128/jcm.12.6.732-737.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Penner J L, Hennessy J N, Congi R V. Serotyping of Campylobacter jejuni and Campylobacter coli on the basis of thermostable antigens. Eur J Clin Microbiol. 1983;2:378–383. doi: 10.1007/BF02019474. [DOI] [PubMed] [Google Scholar]
- 30.Ribot E M, Fitzgerald C, Kubota K, Barrett T J, Swaminathan B. Rapid pulsed-field gel electrophoresis protocol for subtyping of Campylobacter jejuni. J Clin Microbiol. 2001;39:1889–1894. doi: 10.1128/JCM.39.5.1889-1894.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sacks J J, Lieb S, Baldy L M, Berta S, Patton C M, White M C, Bigler W J, Witte J J. Epidemic campylobacteriosis associated with a community water supply. Am J Public Health. 1986;76:424–428. doi: 10.2105/ajph.76.4.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salama S M, Bolton F J, Hutchinson D N. Application of a new phagetyping scheme to campylobacters isolated during outbreaks. Epidemiol Infect. 1990;104:405–411. doi: 10.1017/s0950268800047427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Santesteban E, Gibson J, Owen R J. Flagellin gene profiling of Campylobacter jejuni heat-stable serotype 1 and 4 complex. Res Microbiol. 1996;147:641–649. doi: 10.1016/0923-2508(96)84021-6. [DOI] [PubMed] [Google Scholar]
- 34.Skirrow M B, Blaser M J. Clinical and epidemiologic considerations. In: Nachamkin I, Blaser M J, Tompkins L S, editors. Campylobacter jejuni: current status and future trends. Washington, D.C.: American Society for Microbiology; 1992. pp. 3–8. [Google Scholar]
- 35.Swaminathan B, Barrett T J . the CDC Pulsenet Task Force. A national molecular subtyping network for food-borne bacterial disease surveillance in the United States. In: Nachamkin I, Blaser M J, editors. Campylobacter. 2nd ed. Washington, D.C.: American Society for Microbiology; 2000. pp. 529–535. [Google Scholar]
- 36.Vogt R L, Little A A, Patton C M, Barrett T J, Orciari L A. Serotyping and serology studies of campylobacteriosis associated with consumption of raw milk. J Clin Microbiol. 1984;20:998–1000. doi: 10.1128/jcm.20.5.998-1000.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wassenaar T M, Geilhausen B, Newell D G. Evidence of genomic instability in Campylobacter jejuni isolated from poultry. Appl Environ Microbiol. 1998;64:1816–1821. doi: 10.1128/aem.64.5.1816-1821.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wassenaar T M, Newell D G. Genotyping of Campylobacter spp. Appl Environ Microbiol. 2000;66:1–9. doi: 10.1128/aem.66.1.1-9.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]