Abstract
From 1997 to 1999 seven isolates of Campylobacter-like organisms from five patients that were exhibiting symptoms of gastroenteritis, including fever, stomach malaise, and diarrhea, were investigated. The organisms were isolated from stool samples and found to exhibit a diverse colony morphology; hence multiple isolates were submitted from one of the patients. All isolates were found to be identical. The organisms were catalase, urease, alkaline phosphatase, and nitrate negative but oxidase and indoxyl acetate positive. They grew at 37°C but not at 42°C, and three of the isolates from two different patients were sensitive to nalidixic acid and cephalothin. Full 16S rRNA sequence analysis not only grouped these organisms within the Helicobacter genus but also differentiated them from previously identified Helicobacter species. The closest relative by phylogenetic analysis was Helicobacter sp. flexispira taxon 1. Electron microscopy showed that these isolates had one or two bipolar flagella; however, the periplasmic fibers, a characteristic of the known Helicobacter sp. flexispira taxa, were not observed. The present isolates also lacked a flagellar sheath, a trait shared with four other Helicobacter spp., H. canadensis, H. mesocricetorum, H. pullorum, and H. rodentium. On the basis of the unique phenotypic properties of these isolates and 16S rRNA sequence analysis, we propose the classification of a new Helicobacter species, Helicobacter winghamensis sp. nov.
Helicobacters have emerged as a burgeoning cause of enteric disease in humans. Members of this genus have gained recognition largely as a result of Helicobacter pylori (15), which colonizes the stomachs of humans and which has been associated with gastritis, peptic ulcer disease, and most recently with the development of adenocarcinoma and gastric mucosa-associated lymphoma (11, 17, 24, 27). Indeed, H. pylori constitutes a significant disease burden for the human population, and extensive investigations have been undertaken to better define the pathogenicity of this organism.
To date, the pathogenesis of many Helicobacter species which are isolated from the intestinal contents of humans and animals remains in doubt as often they are isolated in the absence of symptoms. Such is the case with H. pullorum, H. muridarum, and H. pametensis (2, 7, 19, 33). As a consequence, little has been done to elucidate the nature of the disease mechanisms of these intestinal helicobacters. The recent isolation and characterization of the novel H. canadensis and its demonstrated link to gastroenteritis have raised the profile of these intestinal helicobacters and have underscored their potential role in human disease (12).
In the past decade the taxonomy of Helicobacter has expanded dramatically with an average of two or three new species added to the group each year. Currently, the genus comprises 28 species isolated from mammalian and avian sources. Of these, 19 have been validated in accordance with the international rules of nomenclature, 7 have yet to be validated, and 2 are candidate species (10). Five of the 28 species (H. canadensis, H. canis, H. cinaedi, H. fennelliae, and H. pullorum) have been isolated from the intestinal contents of humans suffering from diarrhea, while H. bizzozeronii has been obtained from a single patient with gastritis (12, 33, 34, 37; K. Jalava, S. L. W. On, C. S. Harrington, L. P. Anderson, M.-L. Hänninen, and P. A. R. Vandamme, Abstr. 10th Int. Workshop Campylobacter, Helicobacter, Related Organisms, abstr. HD5, 1999). In addition, a Campylobacter-like organism with the provisional name “Flexispira rappini” was also isolated from the intestinal contents of humans with diarrhea (1). Members of the “Flexispira rappini” group, also named Helicobacter sp. flexispira (8), represent a collection of organisms with similar morphological and phenotypic properties. Based on 16S rRNA sequence analysis, these have been described as comprising 10 taxa within the Helicobacter genus (8).
In this study we describe seven Campylobacter-like isolates which, following phenotypic and genotypic analysis, were shown to belong to a new species of Helicobacter. All isolates came from the feces of humans with enteric tract symptoms, thus further underscoring the potential for other intestinal helicobacters to induce human disease. Phenotypically, these organisms were similar to H. canis but differed in their abilities to grow at 42°C and in their alkaline phosphatase activities. Following phylogenetic analysis using 16S rRNA sequencing, a close relationship to Helicobacter sp. flexispira taxon 1 was observed; however, major phenotypic and ultrastructural differences between these two were noted. Based on the unique characteristics of this group of isolates and in keeping with the recently described minimal standards for describing new species of Helicobacter (6) we propose a new Helicobacter species, H. winghamensis sp. nov.
MATERIALS AND METHODS
Bacterial isolates.
Over the 3-year period 1997 to 1999, seven isolates of Campylobacter-like organisms were submitted from the Provincial Laboratories of Public Health in Alberta, Ontario, and Manitoba to the National Laboratory for Enteric Pathogens for species identification and further characterization. All were clinical isolates from stools derived from two children and three adults all with symptoms of gastroenteritis. The bacterial strains used for comparison are outlined in Table 1.
TABLE 1.
Helicobacter strains used in this study
| Organism | Source no. | GenBank accession no. |
|---|---|---|
| H. winghamensis 1 | NLEP 97–1090c | AF246984 |
| H. winghamensis 2 | NLEP 98-2019 | AF246985 |
| H. winghamensis 3 | NLEP 98-2020 | AF246986 |
| H. winghamensis 4 | NLEP 98-2021 | AF246987 |
| H. winghamensis 5 | NLEP 99-4873 | AF246988 |
| H. winghamensis 6 | NLEP 97-1611 | AF363062 |
| H. winghamensis 7 | NLEP 98-0305 | AF363063 |
| H. aurati | MIT 97-5075c | AF 297868 |
| H. acinonychis | ATCC 51101 | M88148 |
| H. bilis | ATCC 51630 | U18766 |
| H. bizzozeronii | ATCC 700030 | Y09404 |
| H. bizzozeronii | ATCC 700031 | AF302107 |
| H. canadensis | ATCC 700968 | AF262037 |
| H. canis | ATCC 51401 | L13464 |
| H. cinaedi | CCUG 18818 | M88150 |
| H. cholecystus | Hkb-1 | U46129 |
| H. colifelis | N/Aa | AF142062 |
| H. felis | ATCC 49179 | M37642 |
| H. fennelliae | ATCC 35684 | M88154 |
| H. heilmannii | Str 1 | AF058768 |
| H. hepaticus | ATCC 51488 | AF302103b |
| H. mainz | ATCC 51800 | X81028 |
| H. mesocricetorum | MU97-1514 | AF072471 |
| H. muridarum | ATCC 49282 | AF302104b |
| H. mustelae | ATCC 43772 | M35048 |
| H. nemestrinae | ATCC 49396 | AF363064b |
| H. pametensis | ATCC 51478 | AF302105b |
| H. pullorum | ATCC 51801 | L36141 |
| H. pylori | ATCC 43504 | AF302106b |
| Helicobacter sp. flexispira | ATCC 43966 taxon 8 | M88138 |
| Helicobacter sp. flexispira | ATCC 43968 taxon 1 | U96300 |
| H. rodentium | ATCC 700285 | U96296 |
| H. salomonis | CCUG 37848 | Y09405 |
| H. suncus | Kaz-1 | AB006147 |
| H. trogontum | ATCC 700114 | U07574 |
| H. typhlonicus | Strain MU | AF061104 |
| H. westmeadii | Taxon 218 | U44756 |
N/A, not available (species unculturable).
GenBank resubmission.
Type strain.
Phenotypic characterization.
Cultures were recovered using Mueller-Hinton (MH) agar supplemented with 10% sheep blood and grown in a microaerobic atmosphere consisting of 3% O2, 7% H2, 7% CO2, and 83% N2 for 48 h. All phenotypic and biochemical tests requiring growth of the organisms employed the same microaerobic atmosphere. All media and reagents were obtained from Oxoid (Nepean, Ontario, Canada). Morphology was established using phase-contrast microscopy and Gram staining, while further traits were assessed using the biochemical tests for oxidase, catalase, indoxyl acetate hydrolysis (read at 15 min), alkaline phosphatase activity (read at 2 h), urease activity, and nitrate reduction (3, 5, 25, 28). Growth was assayed on MH agar containing 10% sheep blood at 25, 37, and 42°C, and results were read at 72 h. Growth tolerance studies were performed in modified brucella broth supplemented with 1% bile (23, 26). Disk diffusion assays, read at 48 h, using MH agar supplemented with 10% sheep blood were used to evaluate the susceptibility of organisms to nalidixic acid (30 μg) and cephalothin (30 μg) (Becton Dickinson, Cockeysville, Md.). A zone or no zone interpretation was used as the determining factor for resistance and susceptibility criteria as previously described for Campylobacter identification (18).
Electron microscopy.
The morphology of organisms including the flagellum arrangements and structure as well as the presence or absence of surface projections was investigated by negative-stain transmission electron microscopy (TEM) procedures (29, 30). Bacterial cells were suspended in modified brucella broth (23), added dropwise to carbon-coated 400 mesh TEM grids, drained, and negatively stained using 2% (wt/vol) phosphotungstic acid (Marivae, Halifax, Nova Scotia, Canada). Preparations were examined with a CM120 transmission electron microscope (Philips Electron Optics, Toronto, Ontario, Canada).
Genotypic characterization.
Preliminary genotypic analysis was achieved using the 16S rRNA PCR-restriction fragment length polymorphism (RFLP) procedure developed by Marshall et al. (20). In brief, chromosomal DNA was isolated from the organisms and subjected to a PCR procedure that amplified a 1-kb portion of the 16S rRNA gene. The resulting amplicon was then digested with endonucleases DdeI and BsrI (New England Biolabs, Mississauga, Ontario, Canada), and the resulting RFLP patterns were visualized after electrophoresis by ethidium bromide staining.
16S rRNA gene sequencing and analysis.
For 16S rRNA gene sequencing, chromosomal DNA was first extracted from the isolates using either DNAzol (Molecular Research Center, Inc., Cincinnati, Ohio) or Integrated Separation Systems automated DNA extractor Autogen 540 according to the manufacturer's specifications (Enprotech, Natick, Mass.). Approximately 1.5 kb of DNA from each isolate was amplified using PCR for the16S rRNA gene with primers pA and pHr from Edwards et al. (9). The PCR products were subjected to electrophoresis in low-melting-point agarose (Eclipse Molecular Biologicals, Missisauga, Ontario, Canada), excised, and purified using the Wizard PCR Preps DNA purification system (Promega, Madison, Wis.). The resulting PCR product was sequenced in six fragments using pA, pC, pDr, pE, pFr, and pHr primers also described by Edwards et al. (9). The sequenced fragments were assembled using the software program Sequencher 3.0 (Gene Codes Corp., Ann Arbor, Mich.). The complete sequences from each isolate were then compared to the GenBank database through the National Center for Biotechnology Information (National Institutes of Heath, Bethesda, Md.). The sequences were subsequently aligned with other Helicobacter sequences using the ClustalW method incorporated in the MegAlign software of Lasergene (DNASTAR Inc., Madison, Wis.). Finally, phylogenetic analysis was done by first converting the files using ForCon software (Department of Biochemistry, University of Antwerp, Antwerp, Belgium) and drawing phylogenetic relationships using an unrooted neighbor-joining tree, generated from a distance matrix calculated with the Kimura two-parameter model of nucleotide substitution using MEGA 1.01 software (Institute of Molecular Evolutionary Genetics, Pennsylvania State University, University Park, Pa.).
Nucleotide sequence accession numbers.
The 16S rRNA gene for the reference strain was submitted to GenBank under the accession number AF246984. Accession numbers for the other H. winghamensis isolates are as follows: AF246985, AF246986, AF246987, AF246988, AF363062, and AF363063.
RESULTS
Phenotypic characterization and electron microscopy.
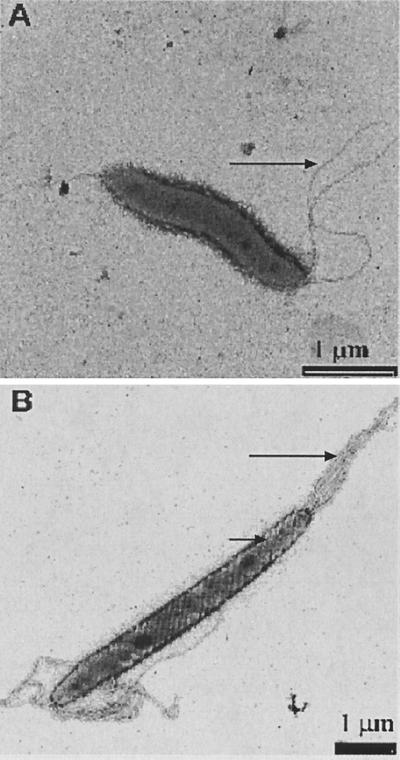
A summary of the phenotypic profile and the ultrastructural observations of the seven isolates together with other intestinal Helicobacter species is presented in Table 2. All of the clinical isolates possessed similar biochemical properties with the exception that three showed susceptibility to the antibiotics nalidixic acid and cephalothin. Like many other intestinal helicobacters the seven isolates were urease negative. They were oxidase and indoxyl acetate positive and tolerated 1% bile, but, most notably, they were uniformly negative for catalase. H. canis and Helicobacter sp. flexispira taxon 7 and taxon 8 are the only other Helicobacter spp. to share the unusual negative catalase reaction; however, they differ from the presently described isolates by a number of phenotypic traits. Both H. canis and the two Helicobacter sp. flexispira taxa grow at 42°C, whereas the present isolates do not. Furthermore, H. canis exhibits alkaline phosphatase activity and Helicobacter sp. flexispira taxon 7 and taxon 8 are urease positive and lack the ability to hydrolyze indoxyl acetate, properties that were not found with our group of isolates. H. cholecystus and H. pametensis, which were differentiated from each other by their antibiotic sensitivity profiles, differed from our isolates to the greatest degree, with five divergent reactions: catalase, nitrate, indoxyl acetate, alkaline phosphatase, and growth at 42°C. Standard microscopic observation indicated that these organisms, in keeping with most helicobacters, were motile, gram-negative, non-spore-forming, slightly curved bacilli. By TEM these organisms were approximately 2 μm in length and 0.3 to 0.6 μm in diameter with one or two unsheathed bipolar flagella (Fig. 1A). The absence of a flagellar sheath, uncharacteristic of most helicobacters, is shared by four other Helicobacter species, H. canadensis, H. mesocricetorum, H. pullorum, and H. rodentium (12, 31–33). However, these species differed by several significant traits from this group of isolates, including catalase activity, nitrate reduction, indoxyl acetate hydrolysis, alkaline phosphatase activity, and growth at 42°C. Periplasmic fibers, a structure found by TEM on H. muridarum and all Helicobacter sp. flexispira taxa (Fig. 1B), were not found on these isolates.
TABLE 2.
Phenotypic characteristics of H. winghamensis and other intestinal Helicobacter spp.
| Organism | Resulta of test for:
|
Flagellac (type, no.) | Presence of
|
Reference | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Oxidase | Catalase | Urease | Nitrate | Indoxyl acetate | Alkaline phosphatase | Growth at 42°C | Tolerance of 1% bile | Susceptibility tob:
|
Flagellar sheath | Periplasmic fibers | ||||
| Na | Ceph | |||||||||||||
| H. winghamensis | + (7/7) | − (7/7) | − (7/7) | − (7/7) | + (7/7) | − (7/7) | − (7/7) | + (7/7) | R (4/7) | R (4/7) | BP, 1–2 | − | − | |
| H. bilis | + | + | + | + | − | − | + | NDf | R | R | BP, 3–14 | + | + | 13 |
| H. bizzozeronii | + | + | + | + | + | + | + | − | R | S | BP, 10–20 | + | − | 16 |
| H. canadensis | + | + | − | +/−e | + | − | + | + | R | R | BP, 1–2 | − | − | 12 |
| H. canis | + | − | − | − | + | + | + | + | S | S | BP, 2 | + | − | 34 |
| H. cinaedi | + | + | − | + | − | − | − | − | S | S | MP, BP, 1–2 | + | − | 37 |
| H. fennelliae | + | + | − | − | + | + | − | − | S | S | MP, BP, 1–2 | + | − | 37 |
| H. hepaticus | + | + | + | + | + | − | − | ND | R | R | BP, 2 | + | − | 14 |
| H. mesocricetorum | + | + | − | + | ND | + | + | ND | R | S | BP, 2 | − | − | 32 |
| H. muridarum | + | + | + | − | − | + | − | ND | R | R | BP, 10–14 | + | + | 19 |
| H. pametensis | + | + | − | + | − | + | + | + | S | S | BP, 2 | + | − | 7 |
| H. pullorum | + | + | − | + | − | − | + | + | S | R | MP, 1 | − | − | 33 |
| H. pylori | + | + | + | − | − | + | − | − | R | S | BP, 4–8 | + | − | 15 |
| Helicobacter sp. flexispira taxon 1 | + | + | + | − | + | − | + | ND | R | R | BP, 10–20 | + | + | 8 |
| Helicobacter sp. flexispira taxon 8 | + | − | + | − | + | − | + | ND | R | R | BP, 10–20 | + | + | 8 |
| H. rodentium | + | + | − | + | − | − | + | ND | R | R | BP, 2 | − | − | 31 |
| H. trogontum | + | + | + | + | − | − | + | ND | R | R | BP, 5–7 | + | + | 22 |
+, positive reaction; −, negative reaction; S, sensitive; R, resistant. Numbers in parentheses, number of isolates exhibiting the indicated reaction per number of isolates tested.
Na, nalidixic acid; Ceph, cephalothin.
MP, monopolar, BP, bipolar.
+, presence; −, absence.
+/−, variable trait among identified isolates.
ND, not determined.
FIG. 1.
(A) Electron micrograph of H. winghamensis. Note unsheathed flagella (arrow) and a lack of periplasmic fibers on the surface of the organism. (B) Electron micrograph of Helicobacter sp. flexispira taxon 1 showing sheathed flagella (long arrow) and periplasmic fibers (short arrow).
Genotypic characterization.
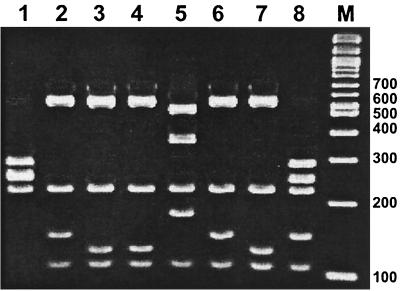
Preliminary genotypic analysis by 16S rRNA PCR-RFLP showed that these isolates had restriction patterns that were identical but that they were different from those described for a range of Campylobacter, Arcobacter, and Helicobacter species by Marshall et al. (20). The present isolates also differed in their 16S rRNA patterns from the other intestinal helicobacters (H. bizzozeronii, H. canadensis, and Helicobacter sp. flexispira taxon 8). The common Helicobacter pattern H2 (750 and 230 bp) was produced by restriction with enzyme DdeI; however, following digestion with BsrI, a distinctive, species-specific pattern comprising bands of 290, 250, 220, 150, and 110 bp resulted (Fig. 2).
FIG. 2.
16S rRNA PCR-RFLP patterns for intestinal helicobacters generated using restriction enzyme BsrI. Lane 1, H. bizzozeronii; lane 2, H. canadensis; lane 3, H. canis; lane 4, H. cinaedi; lane 5, H. fennelliae; lane 6, H. pullorum; lane 7, Helicobacter sp. flexispira taxon 8; lane 8, H. winghamensis; lane M, 100-bp molecular weight marker (New England Biolabs)
16S rRNA sequence analysis.
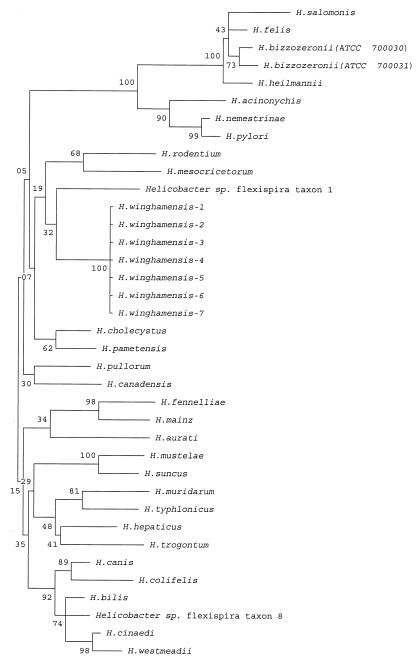
The phylogenetic relationship of these isolates and of other Helicobacter species based on full 16S rRNA sequence data and incorporating the Kimura two-parameter model of nucleotide substitution is shown in Fig. 3. Close analysis of the GenBank sequences for some of the reference strains revealed several gaps and “N” nucleotide designations. The 16S rRNA genes for these reference strains were resequenced and corrected. The H. winghamensis organisms formed a distinct group with a bootstrap value of 100 and cluster most closely to Helicobacter sp. flexispira taxon 1. By using the MegAlign software, the phylogenetic tree was augmented to give a divergence matrix created from the 16S rRNA sequences. A 2.7% divergence compared to Helicobacter sp. flexispira taxon 1 and H. cholecystus was observed, a figure considerably lower than those for any of the other Helicobacter 16S rRNA gene sequence comparisons. This supported the phylogenetic relationship between these organisms and the H. winghamensis isolates as can be seen in Fig. 3. The next most highly divergent species was H. rodentium at 2.8%. This species also shared with H. winghamensis the presence of unsheathed flagella, and this, together with the 16S rRNA similarity, suggested a phylogenetic relationship between these two species.
FIG. 3.
Phylogenetic dendrogram for Helicobacter. Numbers at nodes represent bootstrap support values (percentage of 1,000 resampled data sets that supported the node in the recalculated tree). Scale: 1.6 cm, approximate genetic distance of 0.01 base substitutions per nucleotide pair.
DISCUSSION
An increasing number of novel Helicobacter and Helicobacter-like organisms have been isolated from the stools of humans with gastrointestinal symptoms. Agents such as H. bizzozeronii, H. canadensis, H. canis, H. pullorum, H. cinaedi, H. fennelliae, Helicobacter sp. flexispira taxon 8, and now H. winghamensis (1, 12, 33, 34, 37; K. Jalava et al., Abstr. 10th Int. Workshop Campylobacter, Helicobacter, Related Organisms) are often identified as Campylobacter species that prove difficult to fully characterize in a routine clinical microbiology laboratory setting. In such situations, unconventional or nonstandard phenotypic markers or complex genotypic identifiers are required, tests for which are often performed in reference facilities. That this group has expanded rapidly in the past few years strongly suggests that the etiology of diarrhea induced by these Campylobacter-like organisms is far from clear.
Campylobacteriosis is the most common bacterial human enteric disease in Canada (D. L. Woodward, Y. D. Yaschuck, L. J. Price, A. Moterassed, J. G. Moses, W. M. Johnson, and F. G. Rodgers, Abstr. 10th Int. Workshop Campylobacter, Helicobacter, Related Organisms, abstr. CD9, 1999). Despite this, the identification of Campylobacter as the causative agent of disease is often determined using a limited phenotypic analysis based on Gram stain, size and shape, microaerobic growth, and catalase, oxidase, and hippuricase activity. This has resulted in erroneous reports of Campylobacter coli and Campylobacter lari isolates from the stools of patients with gastroenteritis that eventually proved to be H. pullorum; indeed, these groups of organisms have proved difficult to differentiate (2, 21). As a result of these problems H. pullorum is considered underreported as a human enteric pathogen (4, 21, 35). Like H. pullorum, the H. winghamensis isolates studied in this investigation are similar to Campylobacter in that they are gram negative and have common properties of morphology, oxidase activity, and microaerobic growth. Hence, they too are almost certainly underrepresented in the spectrum of disease agents causing human gastroenteritis. That the five isolates included in the present study were from unrelated individuals and from different geographic locations in Canada supports the potential for underreporting and suggests that H. winghamensis might play a more prominent role in gastroenteritis.
H. winghamensis isolates atypically produce a negative catalase reaction, which sets them apart from most Helicobacter species except H. canis and Helicobacter sp. flexispira taxon 7 and taxon 8; however, these organisms may be differentiated by a number of phenotypic traits. Although a negative catalase reaction is not uncommon among campylobacters, they have the common property of nitrate reduction. The negative nitrate reaction for H. winghamensis should facilitate the separation of this newly described species from all other Campylobacter species, with the exception of Campylobacter jejuni subspecies doylei. Overall these organisms are relatively biochemically distinct and may be easily identified from related groups of bacteria by applying a more extensive range of biochemical tests including those for nitrate reduction, alkaline phosphatase activity, and indoxyl acetate hydrolysis. The presence of a species-specific 16S rRNA PCR-RFLP pattern (20) also contributes to the accurate identification of these agents. 16S rRNA similarities and differences form the basis for most phylogenetic dendrograms to define bacterial species. It is possible that 16S rRNA similarities may be an indication of a common origin among prokaryotic organisms. The phylogenetic similarity of these isolates to Helicobacter sp. flexispira taxon 1 and the low 16S rRNA sequence divergence from H. cholecystus and H. rodentium, isolated from murine sources, may be indicative of a common origin for these organisms.
The unique phenotypic and genotypic characteristics of these organisms should facilitate the detection of this newly proposed species. These identification traits will provide valuable laboratory-based epidemiological markers to better understand the role that these helicobacters play in gastroenteritis and will facilitate investigations of the virulence mechanisms they employ to induce human disease.
Description of H. winghamensis sp. nov.
H. winghamensis is a gram-negative, slightly curved to spiral, non-spore-forming bacillus. The organism is approximately 2 μm in length by 0.3 to 0.6 μm in width, and it is motile by one or two bipolar, unsheathed flagella. Cultures grow on solid agar media supplemented with 10% sheep blood and exhibit a seemingly diverse colonial morphology of nonspreading and spreading colonies. The organisms grow in a microaerobic atmosphere at 37°C but fail to grow at 42°C or in aerobic or anaerobic atmospheres. This Helicobacter is oxidase and indoxyl acetate positive, tolerates 1% bile, and is alkaline phosphatase, catalase, and urease negative. It does not reduce nitrate. All isolates induced similar symptoms of gastroenteritis in humans, and these included general stomach malaise, vomiting, diarrhea, cramping, and mild fever. Currently the recorded host range for this organism does not extend beyond humans. The first isolate fitting the characterized description of this species was from Wingham, Ontario, Canada; thus, we propose the name Helicobacter winghamensis. The type strain is NLEP 97–1090 and has GenBank accession no. AF246984.
ACKNOWLEDGMENTS
We thank the Provincial Laboratories of Public Health in Alberta, Manitoba, and Ontario for submitting these Campylobacter-like organisms to the NLEP for investigation. Thanks are also extended to Lawrence Price, Ali Moterassed, Jason Moses, and Yvonne Yaschuk for their technical assistance.
REFERENCES
- 1.Archer J R, Romero S, Ritche A E, Hamacher M E, Stener B M, Bryner J H, Schell R F. Characterization of an unclassified microaerophilic bacterium associated with gastroenteritis. J Clin Microbiol. 1988;26:101–105. doi: 10.1128/jcm.26.1.101-105.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atabay H I, Corry J E L, On S L W. Identification of unusual Campylobacter-like isolates from poultry products as Helicobacter pullorum. J Appl Microbiol. 1998;84:1017–1024. doi: 10.1046/j.1365-2672.1998.00438.x. [DOI] [PubMed] [Google Scholar]
- 3.Barret T J, Patton C M, Morris G K. Differentiation of Campylobacter species using phenotypic characterization. Lab Med. 1988;19:96–102. [Google Scholar]
- 4.Burnens A P, Stanley J, Morgenstern R, Nicolet J. Gastroenteritis associated with Helicobacter pullorum. Lancet. 1994;344:1569–1570. doi: 10.1016/s0140-6736(94)90376-x. [DOI] [PubMed] [Google Scholar]
- 5.Cook G T. A plate test for nitrate reduction. J Clin Pathol. 1950;3:359–362. doi: 10.1136/jcp.3.4.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dewhirst F E, Fox J G, On S L W. Recommended minimal standards for describing new species of the genus Helicobacter. Int J Syst Evol Microbiol. 2000;50:2231–2237. doi: 10.1099/00207713-50-6-2231. [DOI] [PubMed] [Google Scholar]
- 7.Dewhirst F E, Seymour C, Fraser G J, Paster B J, Fox J G. Phylogeny of Helicobacter isolated from birds and swine feces and description of Helicobacter pametensis sp. nov. Int J Syst Bacteriol. 1994;44:553–560. doi: 10.1099/00207713-44-3-553. [DOI] [PubMed] [Google Scholar]
- 8.Dewhirst F E, Fox J G, Mendes E N, Paster B J, Gates C E, Kirkbride C A, Eaton K A. “Flexispira rappini” strains represent at least 10 Helicobacter taxa. Int J Syst Evol Microbiol. 2000;50:1781–1787. doi: 10.1099/00207713-50-5-1781. [DOI] [PubMed] [Google Scholar]
- 9.Edwards U, Rogall T, Blöcker H, Emde M, Böttger E C. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 1989;17:7843–7853. doi: 10.1093/nar/17.19.7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Euzeby J P. List of bacterial names with standing in nomenclature: a folder available on the Internet (URL: http://www.bacterio.cict.fr/) Int J Syst Bacteriol. 1997;47:590–592. doi: 10.1099/00207713-47-2-590. [DOI] [PubMed] [Google Scholar]
- 11.Forman D, Newal D G, Fullerton F, Yarnell J W, Stacey A R, Wald N, Sitas F. Association between infection with Helicobacter pylori and risk of gastric cancer: evidence from a prospective investigation. Br Med J. 1991;302:1302–1305. doi: 10.1136/bmj.302.6788.1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fox J G, Chien C C, Dewhirst F E, Paster B J, Shen Z, Melito P L, Woodward D L, Rodgers F G. Helicobacter canadensis sp. nov. isolated from humans with diarrhea as an example of an emerging pathogen. J Clin Microbiol. 2000;38:2546–2549. doi: 10.1128/jcm.38.7.2546-2549.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fox J G, Yan L L, Dewhirst F E, Paster B J, Shames B, Murphy J C, Hayward A, Belcher T C, Mendes E N. Helicobacter bilis sp. nov., a novel Helicobacter sp. isolated from bile, livers, and intestines of aged, inbred mice. J Clin Microbiol. 1995;33:445–454. doi: 10.1128/jcm.33.2.445-454.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fox J G, Dewhirst F E, Tully J G, Paster B J, Yan L, Taylor N S, Collins M J, Jr, Gorelick P L, Ward J M. Helicobacter hepaticus sp. nov., a microaerophilic bacterium isolated from livers and intestinal mucosal scrapings of mice. J Clin Microbiol. 1994;32:1238–1245. doi: 10.1128/jcm.32.5.1238-1245.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodwin C S, Armstrong J A, Chilvers T, Peters M, Collins M D, Sly L, McConnell W, Harper W E S. Transfer of Campylobacter pylori and Campylobacter mustelae to Helicobacter gen. nov. as Helicobacter pylori comb. nov. and Helicobacter mustelae comb. nov., respectively. Int J Syst Bacteriol. 1989;39:397–405. [Google Scholar]
- 16.Hänninen M L, Happonen J, Saari S, Jalava K. Culture and characteristics of Helicobacter bizzozeronii, a new canine gastric Helicobacter sp. Int J Syst Bacteriol. 1996;46:160–166. doi: 10.1099/00207713-46-1-160. [DOI] [PubMed] [Google Scholar]
- 17.Itoh T, Yanagawa Y, Shingaki M, Takahashi M, Kai A, Ohashi M, Hamana G. Isolation of Campylobacter pyloridis from human gastric mucosa and characterization of the isolates. Microbiol Immunol. 1987;31:603–614. doi: 10.1111/j.1348-0421.1987.tb03121.x. [DOI] [PubMed] [Google Scholar]
- 18.Karmali M A, De Grandis S, Fleming P C. Antimicrobial susceptibility of Campylobacter fetus subsp. fetus to eight cephalosporins with special reference to species differentiation. Antimicrob Agents Chemother. 1980;18:948–951. doi: 10.1128/aac.18.6.948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee A, Phillips M W, O'Rourke J L, Paster B J, Dewhirst F E, Fraser G J, Fox J G, Sly L I, Romaniuk P J, Trust T J. Helicobacter muridarum sp. nov., a microaerophilic helical bacterium with a novel ultrastructure isolated from the intestinal mucosa of rodents. Int J Syst Bacteriol. 1992;42:27–36. doi: 10.1099/00207713-42-1-27. [DOI] [PubMed] [Google Scholar]
- 20.Marshall S M, Melito P L, Woodward D L, Rodgers F G, Johnson W M, Mulvey M R. Rapid identification of Campylobacter, Arcobacter, and Helicobacter isolates by PCR-restriction fragment length polymorphism analysis of the 16S rRNA gene. J Clin Microbiol. 1999;37:4158–4160. doi: 10.1128/jcm.37.12.4158-4160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Melito P L, Woodward D L, Bernard K A, Price L, Khakhria R, Johnson W M, Rodgers F G. Differentiation of clinical Helicobacter pullorum isolates from related Helicobacter and Campylobacter species. Helicobacter. 2000;5:142–147. doi: 10.1046/j.1523-5378.2000.00022.x. [DOI] [PubMed] [Google Scholar]
- 22.Mendes E N, Queiruz D M M, Dewhirst F E, Paster B J, Moura S B, Fox J G. Helicobacter trogontum sp. nov., isolated from the rat intestine. Int J Syst Bacteriol. 1996;46:916–921. doi: 10.1099/00207713-46-4-916. [DOI] [PubMed] [Google Scholar]
- 23.Ng L K, Stiles M E, Taylor D E. Comparison of basal media for culturing Campylobacter jejuni and Campylobacter coli. J Clin Microbiol. 1985;21:226–230. doi: 10.1128/jcm.21.2.226-230.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nomura A, Stemmerman G N, Chyou P H, Kato I, Perez-Perez G I, Blaser M J. Helicobacter pylori infection and gastric carcinoma among Japanese Americans in Hawaii. N Engl J Med. 1991;325:1132–1136. doi: 10.1056/NEJM199110173251604. [DOI] [PubMed] [Google Scholar]
- 25.On S L W. Identification methods for Campylobacters, Helicobacters, and related organisms. Clin Microbiol Rev. 1996;9:405–422. doi: 10.1128/cmr.9.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.On S L W, Holmes B. Reproducibility of tolerance tests that are useful in the identification of Campylobacteria. J Clin Microbiol. 1991;29:1785–1788. doi: 10.1128/jcm.29.9.1785-1788.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parsonnet J, Friedman G D, Vandersteen D P, Chang Y, Vogelman J H, Orentreich N, Sibley R K. Helicobacter pylori infection and the risk of gastric carcinoma. N Engl J Med. 1991;325:1127–1131. doi: 10.1056/NEJM199110173251603. [DOI] [PubMed] [Google Scholar]
- 28.Popovic-Uroic T, Patton C M, Nicholson M A, Kiehlbauch J A. Evaluation of indoxyl acetate hydrolysis test for rapid differentiation of Campylobacter, Helicobacter, and Wolinella species. J Clin Microbiol. 1990;28:2335–2339. doi: 10.1128/jcm.28.10.2335-2339.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodgers F G. Ultrastructure of Legionella pneumophila. J Clin Pathol. 1979;32:1195–1202. doi: 10.1136/jcp.32.12.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodgers F G, Greaves P W, Macrae A D, Lewis M J. Electron microscopic evidence of flagella and pili on Legionella pneumophila. J Clin Pathol. 1980;33:1184–1188. doi: 10.1136/jcp.33.12.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen Z, Fox J G, Dewhirst F E, Paster B J, Foltz C J, Yan L, Shames B, Perry L. Helicobacter rodentium sp. nov., a urease-negative Helicobacter species isolated from laboratory mice. Int J Syst Bacteriol. 1997;47:627–634. doi: 10.1099/00207713-47-3-627. [DOI] [PubMed] [Google Scholar]
- 32.Simmons J H, Riley L K, Besch-Williford C L, Franklin C L. Helicobacter mesocricetorum sp. nov., a novel Helicobacter isolated from the feces of Syrian hamsters. J Clin Microbiol. 2000;38:1811–1817. doi: 10.1128/jcm.38.5.1811-1817.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stanley J, Linton D, Burnens A P, Dewhirst F E, On S L W, Porter A, Owen R J, Costas M. Helicobacter pullorum sp. nov.—genotype and phenotype of a new species isolated from poultry and from human patients with gastroenteritis. Microbiology. 1994;140:3441–3449. doi: 10.1099/13500872-140-12-3441. [DOI] [PubMed] [Google Scholar]
- 34.Stanley J, Linton D, Burnens A P, Dewhirst F E, Owen R J, Porter A, On S L W, Costas M. Helicobacter canis sp. nov., a new species from dogs: an integrated study of phenotype and genotype. J Gen Microbiol. 1993;139:2495–2504. doi: 10.1099/00221287-139-10-2495. [DOI] [PubMed] [Google Scholar]
- 35.Steinbrueckner B, Haerter G, Pelz K, Burnens A, Kist M. Discrimination of Helicobacter pullorum and Campylobacter lari by analysis of whole cell fatty acid extracts. FEMS Microbiol Lett. 1998;168:209–212. doi: 10.1111/j.1574-6968.1998.tb13275.x. [DOI] [PubMed] [Google Scholar]
- 36.Steinbrueckner B, Haerter G, Pelz K, Weiner S, Rump J, Deissler W, Bereswill S, Kist M. Isolation of Helicobacter pullorum from patients with enteritis. Scand J Infect Dis. 1997;29:315–318. doi: 10.3109/00365549709019053. [DOI] [PubMed] [Google Scholar]
- 37.Totten P A, Fennell C L, Tenover F C, Wezenberg J M, Perine P L, Stamm W E, Holmes K K. Campylobacter cinaedi (sp. nov.) and Campylobacter fennelliae (sp. nov.): two new Campylobacter species associated with enteric disease in homosexual men. J Infect Dis. 1985;151:131–139. doi: 10.1093/infdis/151.1.131. [DOI] [PubMed] [Google Scholar]