Abstract
Steller’s sea cow, an extinct sirenian and one of the largest Quaternary mammals, was described by Georg Steller in 1741 and eradicated by humans within 27 years. Here, we complement Steller’s descriptions with paleogenomic data from 12 individuals. We identified convergent evolution between Steller’s sea cow and cetaceans but not extant sirenians, suggesting a role of several genes in adaptation to cold aquatic (or marine) environments. Among these are inactivations of lipoxygenase genes, which in humans and mouse models cause ichthyosis, a skin disease characterized by a thick, hyperkeratotic epidermis that recapitulates Steller’s sea cows’ reportedly bark-like skin. We also found that Steller’s sea cows’ abundance was continuously declining for tens of thousands of years before their description, implying that environmental changes also contributed to their extinction.
Paleogenomes from Steller’s sea cows reveal functional changes responsible for their bark-like skin and adaptation to cold.
INTRODUCTION
During the Pleistocene, our planet was home to a broad diversity of megafaunal species, including mastodons, mammoths, giant camels, bears, and several large felids. Many of these disappeared suddenly during the Late Pleistocene and Early Holocene. Thus, our knowledge of these species is largely drawn from reconstructions of their ranges and phenotypes based on fossil remains. Several megafauna species, however, went extinct during the later Holocene, often on islands after human colonization. Some of these species survived long enough to be described by European explorers, who provided written details of behaviors and phenotypes that could not have been inferred from their scattered and fragmentary remains. Among the recently extinct megafaunal species is Steller’s sea cow, Hydrodamalis gigas, which was described by Georg Wilhelm Steller in 1741 and became extinct 27 years later, presumably as a result of human-driven habitat change (1) and overexploitation (2). The disappearance of Steller’s sea cow has been regarded as the first historical extinction of a marine mammal as a consequence of human actions (3) and is an iconic example of the catastrophic impact that human hunting can have on a species (2). At the time of Steller’s description, the Steller’s sea cows’ range had already been reduced to shallow waters around the uninhabited Commander Islands, Russia (Fig. 1A), where Steller reported a population of around 1000 individuals (2).
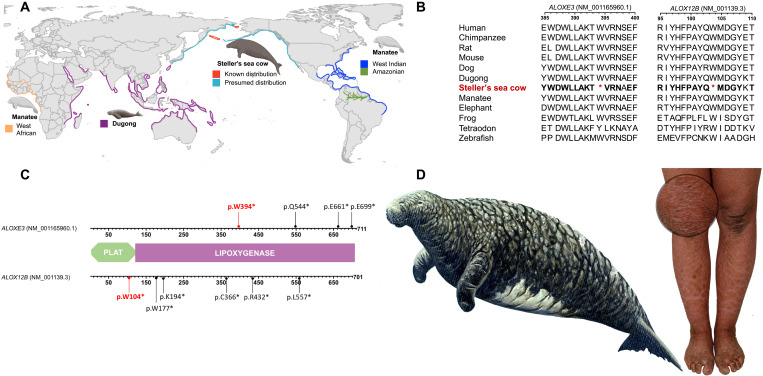
Fig. 1. Molecular basis for Steller’s sea cow’s skin phenotype.
(A) Sirenian distribution according to the International Union for Conservation of Nature Red List (51). All sequenced Steller’s sea cow individuals originate from the Commander Islands. (B) Translated multiple sequence alignment of the ALOXE3 and ALOX12B genes showing amino acid sequence conservation corresponding to the human proteins (bold in Steller’s sea cow) and the position of the premature stop codons. (C) Arachidonate lipoxygenases structure, which is composed of the PLAT (Polycystin-1, Lipoxygenase, and Alpha-Toxin) domain and the enzymatic LIPOXYGENASE core domain. Premature stop codons in Steller’s sea cow ALOXE3 and ALOX12B genes are depicted in red. Truncating variants described in human patients and located downstream from the Steller’s sea cow premature stop codons are depicted in black. (D) Left: Steller’s sea cow drawing according to Steller’s description from 1741 (image by R. Ellis). Right: Image of a patient with ichthyosis; detail depicts scaling and hyperkeratosis.
Adult Steller’s sea cows reached a length of approximately 10 m, weighed over 10 metric tons, and stored up to 10 cm of blubber in some areas of the body (4). This made them ideal resources for human hunters, who exploited them for meat, fat, and skin. Their skin was thick, hairless, and roughly textured, an attribute that led Steller to describe it as resembling “more the bark of an old oak tree, than the skin of an animal” (5). The rough, bark-like skin may have been an adaptation to their shallow water habitat, perhaps preventing abrasion on ice or rocks (6). For centuries, naturalists relied solely on phenotypes to describe species, lacking opportunity to explore the genetic underpinnings of those traits. Unlike for many other extinct species, the existence of a thorough phenotypic record for the Steller’s sea cow can facilitate the identification and understanding of the underlying molecular basis of their traits.
RESULTS AND DISCUSSION
Here, we generate paleogenomic data from Steller’s sea cow bones and explore the genetic underpinnings of their unique phenotype and population history. We extracted ancient DNA from fragmentary remains of 12 Steller’s sea cow individuals recovered from beaches of Bering Island, which we radiocarbon dated to 2205 to 1155 before the present (B.P.) (see section SM1, in the Supplementary Materials). We sequenced the genomes of two of the best preserved of these (SNMB N51667 from the Braunschweig Natural History Museum and SC16.JK045 from the collection of the Kamchatka Branch of the Pacific Geographical Institute, Russian Academy of Sciences Museum) to 15.86 and 15.63× coverage, respectively, and the remaining 10 to an average coverage of 2.78× (range 1.97 to 3.95×) (metrics for libraries and mapping quality are presented in table S1). Our analysis of mitochondrial genomes from the 12 Steller’s sea cows confirms that they are different individuals (section SM1). We mapped the recovered data to a new de novo–assembled draft genome of the dugong (Dugong dugon, originating from the Australian Museum; section SM2), which is the closest living relative of Steller’s sea cow (7, 8). To explore how genomic changes along the lineage to Steller’s sea cow might explain their unique phenotypes, we generated a comparative dataset consisting of 4877 orthologous genes. To this end, we annotated our new dugong genome and a previously published Florida manatee genome [Trichechus manatus latirostris (9); University of California, Santa Cruz (UCSC) GCA_000243295.1] using the MAKER pipeline (10) and annotations from human, mouse, and elephant genomes (11). Because pseudogenization can shape phenotypes (7, 12), we focused first on identification of inactivated genes and found two arachidonate lipoxygenases (ALOXE3 and ALOX12B) that have been inactivated via premature stop codons along the lineage to Steller’s sea cow (section SM3).
In humans, loss-of-function variants in the lipoxygenase genes ALOX12B and ALOXE3 are the second most common cause of autosomal recessive congenital ichthyosis, which is a disease characterized by hyperkeratotic, dry, thickened, scaling skin (13). The premature stop codons in Steller’s sea cow are located at position 104 in ALOX12B and position 394 in ALOXE3 (Fig. 1B) and are present in all our 12 sequenced individuals (section SM3), as well as in an additional individual recovered from the same site (14). In humans, inactivating variants with functional relevance in both genes have been described downstream of those present in Steller’s sea cow (15–17) (Fig. 1C) and are absent in healthy controls supporting the proposed functional relevance of the Steller’s sea cow’s pseudogenizations. Skin appearance as described by Steller in 1741 as resembling “the bark of an old oak tree” is phenotypically similar to skin appearance in ALOXE3- or ALOX12B-deficient humans (Fig. 1D) and the respective gene-deficient mouse models (13, 18).
On the basis of the presence of inactivating mutations in ALOX12B and ALOXE3 in our 12 sequenced individuals, we calculated the Bayesian credible interval for the population allele frequency to be 0.9 to 1 with a posterior probability of >0.95. This suggests that the mutations were either already fixed or in the process of reaching fixation in the population from which we sampled and may have provided an adaptive advantage to Steller’s sea cows.
To explore this possibility further, we scanned all available mammalian sequences including extant marine mammals. We found that both ALOX12B and ALOXE3 are also inactivated in extant cetaceans through gene loss (ALOX12B) and the accumulation of premature stop codons, deletions, or frameshifts (ALOXE3) but are active in pinnipeds (19, 20), sea otter, polar bear, and extant sirenians (Fig. 2, A and B, and section SM3). The absence of an ALOX12B ortholog in all extant cetaceans suggests that the gene loss happened in the lineage leading to their most recent common ancestor. Because the inactivation of ALOXE3 occurred via different changes in each cetacean lineage (Fig. 2B), this loss must have happened more recently through several independent events.
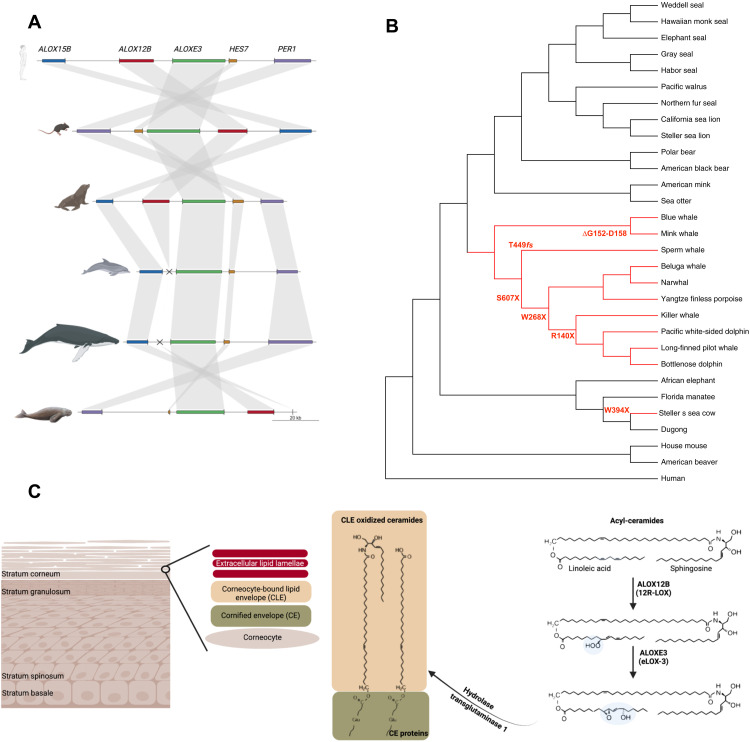
Fig. 2. Genomic locus and function of skin lipooxygenases.
(A) Synteny map of the genomic locus around ALOX12B and ALOXE3 in human, mouse, sea lion, dolphin, blue whale, and dugong. The ALOX12B locus is missing in cetaceans. (B) ALOXE3 inactivation occurred through different changes in cetaceans and the Steller’s sea cow; branches of species with ALOXE3 inactivation are depicted in red. (C) Structure of the epidermis. Acyl-ceramides are produced mainly in the stratum granulosum and partly in the stratum spinosum and stored in lamellar bodies as acyl-glucosylceramides. At the transition to stratum corneum, lamellar bodies release their contents into the extracellular space, where acyl-glucosylceramides are converted to acyl-ceramides. Released acyl-ceramides, fatty acids, and cholesterol form lipid lamellae in the stratum corneum. Some acyl-ceramides are oxidized in the presence of skin lipoxygenases, like enzymes encoded by ALOX12B and ALOXE3. The oxygenation is required to facilitate hydrolysis and subsequent covalent linkage of the ω-hydroxyceramides from the corneocyte-bound lipid envelope to proteins of the cornified envelope, which form the epidermis permeability barrier. ALOXE3’s accumulation of inactivating mutations in the absence of ALOX12B (B) confirms its downstream location in the biochemical pathway (C).
Understanding the constraints imposed by this ancient macroevolutionary innovation could provide insight into the mechanism of the human disease. The two encoded proteins are proposed to operate in the metabolic pathway involved in epidermal structural lipid formation (Fig. 2C) (13). From an evolutionary perspective, accumulation of different inactivating mutations in ALOXE3, probably as a result of genetic drift in the absence of ALOX12B, supports the proposed involvement of both genes in the same pathway, which becomes nonfunctional in the absence of ALOX12B.
Cetaceans have a high shedding rate of skin’s stratum corneum (13), which prevents the buildup of the ichthyotic leathery skin that occurs in the absence of skin lipoxygenases (13, 18). The exfoliation rate of cetaceans may be due to loss of the desmosome genes DSC1 and DSG4 (21), which form the strongest bonds in the outermost skin layer (22). We did not identify any inactivating mutations in these desmosome genes in Steller’s sea cow (section SM3), which is likely the reason why Steller’s sea cow, unlike cetaceans, developed the ichthyotic skin appearance, as suggested by Steller’s description.
Gene losses or pseudogenization may occur as an adaptation or because the gene function becomes obsolete and the gene sequence drifts. Previous studies have suggested that the loss of skin lipoxygenases accompanied by hair loss may be an adaptation to a fully aquatic environment (20). However, because we find that skin lipoxygenases are convergently inactivated in extant cetaceans and Steller’s sea cow, while the similarly fully aquatic extant sirenians retained active orthologs (Fig. 2, A and B, and section SM3), the marine environment is insufficient to explain this adaptation. A key difference between the environments of Steller’s sea cows and the two extant sirenian species is the temperature of the oceans in which the lineages live. We therefore hypothesize that the loss of lipoxygenase genes may have been adaptive in particular in a cold marine environment and may reflect a different adaptation strategy to retain heat compared to fur-bearing marine mammals.
In further support of convergent evolution in Steller’s sea cow and cetaceans regarding energy metabolism, we identified NPFFR2 as inactivated along both lineages but not in pinnipeds or extant sirenians (section SM3). NPFFR2 encodes for neuropeptide FF receptor-2, a G protein–coupled receptor that has been linked to thermogenesis. Usually, excess caloric intake results in increased fat accumulation and an elevated energy expenditure via diet-induced adaptive thermogenesis. Mice lacking Npfr2, however, develop obesity and fail to activate brown adipose tissue (BAT) thermogenesis [nonshivering thermogenesis (NST)] in response to increased caloric intake (23). Selection for large body size is predicted to reduce the importance of BAT for NST by enabling the development of size-dependent heat conservation measures, such as a decrease in relative surface area for heat loss (24). In addition to inactivation of NPFFR2, Steller’s sea cows and sequenced cetaceans also lack UCP1 (24), which is very important for NST in BAT. In contrast to pinnipeds, which express UCP1 and NPFFR2, UCP1-based NST may therefore not have played an important role in Steller’s sea cow’s adaptation to cold-water habitat, which is similar to cetaceans (24). While NPFFR2 absence does not fully explain the increased body size in cetaceans and Steller’s sea cows, it may contribute to their large size by helping to mitigate the cost of heat loss (25).
Because evolution does not only proceed by functionally inactivating genes via obvious pseudogenization (nonsense and frame-shifting mutations), we next searched for intact genes evolving at significantly different evolutionary rates in Steller’s sea cow compared to extant sirenians (section SM3). Of the 197 genes evolving significantly faster and 41 evolving significantly slower in Steller’s sea cow, 20 have been shown to modulate body weight and energy metabolism (section SM3). This significant enrichment in genes involved with energy homeostasis and weight regulation (hypergeometric test P value = 0.006) is accompanied by an apparent lack of signal in the gene ontology category related to BAT (GO:0050873; 16 annotated genes in our orthologous set; section SM3), supporting the suggestion that BAT-involving NST indeed did not play a role in thermoregulation in Steller’s sea cows. In addition, several genes with significantly different evolutionary rate on the Steller’s sea cow lineage (ACP6, ACSF3, ACSL5, EHHADH, IVD, PLA2G2A, and PLA2G4F), two of which (ACSL5 and PLA2G4F) are faster evolving in cetaceans, are components of the lipid-degrading metabolic pathways. Some of these are involved in thermogenic futile cycles based on adenosine 5′-triphosphate (ATP)–demanding lipolysis/re-esterification (26), which generate heat by ATP hydrolysis and thus contribute to NST (section SM3) (26, 27).
A shift in the energetic balance toward fat accumulation renders an increased capacity for fasting, which may have been key to both survival and long-distance movements of Steller’s sea cows. Steller’s sea cows lacked dentition (4, 7), and their diet depended almost exclusively on kelp (28). Dragon kelp (Eularia fistulosa), the predominant surface canopy kelp species in the western North Pacific, is present only from about May through September (1), such that animals would have entered a fasting state for many months each year. As the Pleistocene fossil record shows that Steller’s sea cows were widespread in shallow Pacific waters from present-day Japan to the Mexican Baja Peninsula (Fig. 1A), fasting may have been critical to dispersing between oceanic islands.
The only published information on historical Steller’s sea cow population size is Steller’s first-hand report from Bering Island during the winter of 1741–1742. That account indicates that Steller’s sea cows numbered in the general range of a thousand animals (2). If the pre-Holocene population density of Steller’s sea cows was roughly similar in other kelp forest habitats across the Pacific rim, then this translates to a potential peak abundance of roughly 200,000 animals across North America, Kamchatka, and the Kurile Islands (section SM4).
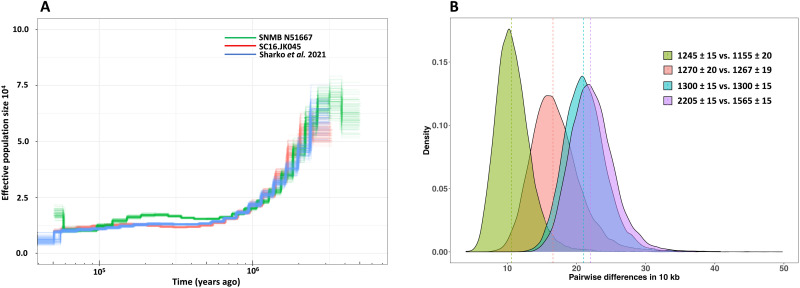
We used the pairwise sequentially Markovian coalescent (PSMC) (29) method to estimate effective population size over time. Our results suggest that Steller’s sea cow populations have been declining for at least the last 500,000 years (Fig. 3A and section SM4), perhaps stabilizing during the warm Marine Isotope Stage 5 Interglacial, and then continued along a slow trajectory of decline until the approach loses power to detect demographic change some 10,000 to 30,000 years ago. The demographic trajectories inferred from our two individuals are concordant with that from another undated individual from the Commander Islands (Fig. 3A) (14). While there is little doubt that the Steller’s sea cow’s final demise was caused either directly (6) or indirectly (1) by humans, this genetic evidence of long-term decline suggests that the species’ demographic trajectory was additionally influenced by environmental factors. We note, however, that all available samples to date come from a single location. Although Steller’s sea cows were probably capable of long-distance inter-island dispersal, undetected population structure, in addition to the small number of available high coverage genomes (N = 3), could confound this reconstructed demographic history.
Fig. 3. Steller’s sea cow’s population history.
(A) Inferred effective population size trajectories for three Steller’s sea cow individuals. All three trajectories show a consistent decrease in population size from at least 500,000 years ago. (B) Pairwise differences between individuals are estimated as the average number of differences per 10 kbp in nonoverlapping 50-kbp regions in 10,000 randomly chosen blocks. The amount of divergence between individuals declines over time (section SM4). The vertical dashed lines correspond to the median of the number of pairwise differences per 10 kbp. Numbers on the graph represent the 14C uncalibrated date (years B.P.).
To gauge the level of genetic diversity within the Steller’s sea cow population in the nearer term past, we performed pairwise comparisons between our individuals, grouped according to radiocarbon date (Fig. 3B and section SM4). We find that Steller’s sea cow diversity was already declining some 900 to 800 years before their discovery by Steller (Fig. 3B). Our ~15×-coverage individuals, dated to approximately 1250 B.P., show ∼17 differences in 10,000 sites, a level similar to extant brown bears (30). This relatively high level of genetic diversity, despite the small long-term effective population size, may have been maintained by inter-island dispersal. These results show that Steller’s sea cow populations had most probably been small for many thousands of years before their observation by Steller on Commander Island.
Our findings exemplify the power of paleogenomics to reveal unique aspects of species’ historical ecology and evolution, here addressing the phenotypic traits and population decline of Steller’s sea cow, an iconic member of the now-extinct Pleistocene megafauna. We find that Steller’s sea cows’ population decline began during the Middle Pleistocene and continued until their extinction. We also explored the genetic basis of Steller’s sea cow’s skin phenotype, exceptionally large size, and reportedly thick blubber. Here, we found evidence for convergent evolution with cetaceans in regard to inactivations of genes that are active in the extant sirenians. The thorough phenotypic record provided by Steller studied within a paleogenomics framework revealed the molecular basis of Steller’s sea cows’ peculiar skin appearance almost three centuries after their description.
MATERIALS AND METHODS
For detailed methods, please refer to the Supplementary Materials.
Sampling, DNA extraction, library preparation, and sequencing
We sampled bone fragments from 25 H. gigas individuals that were collected along the beaches of Commander Island, Russia and one sample (SNMB N51667) that is part of the collection at the Braunschweig Natural History Museum (section SM1). Of these, 12 samples contained sufficient endogenous DNA to allow sequencing of the nuclear genome (section SM1). For each specimen excluding SNMB N51667, we removed a thin layer of surface material and then powdered the remaining sample using a Mixer Mill MM400 (Restch). We divided the resulting powder and used ~30 mg for ancient DNA extraction and the remainder for radiocarbon dating, which was performed at the W. M. Keck Accelerator Mass Spectrometry Center at University of California, Irvine using ultrafiltration and following Shammas et al. (31) (section SM1).
With the help of a computed tomography (CT) scan (IZW Berlin; section SM1), we identified the densest regions of the petrous bone that was then drilled to obtain bone powder from SNMB N51667. This sample was radiocarbon dated at the Klaus-Tschira-AMS facility in the Curt-Engelhorn-Centre of Archaeometry in Mannheim (laboratory number GMP391).
Laboratory work for SNMB N51667 was performed in dedicated ancient DNA facilities at the University of Potsdam and included the parallel processing of blanks to detect possible contamination of laboratory equipment and reagents. DNA was extracted from 49.5 mg of bone powder following the protocol of Dabney and colleagues (32). The DNA extract was converted into a single-stranded, double-indexed Illumina library and treated with uracil-DNA glycosylase and endonuclease VIII before library preparation, effectively reducing DNA damage (33). The optimal cycle number for indexing was established by quantitative polymerase chain reaction [qPCR; Thermo Fisher Scientific PikoReal Real-Time PCR System (33)]. The library was sequenced at the SciLifeLab facilities on an Illumina HiSeq 2500 using 151-cycle paired-end runs.
The remaining 11 samples were processed at the Paleogenomics Laboratory at the University of California, Santa Cruz in a clean laboratory facility dedicated to ancient DNA research. These samples were not treated with uracil-DNA glycosylase and endonuclease VIII before library preparation. We pretreated each powdered sample with a 0.5% sodium hypochlorite solution to remove surface contaminants (34), extracted DNA as described in (32), and eluted in 50 μl of 10 mM tris-HCl, 1 mM EDTA, and 0.05% Tween 20. We prepared dual-indexed, single-stranded DNA libraries (35), which we pooled and sequenced at the SciLifeLab facilities on an Illumina HiSeq 2500 using 151-cycle paired-end runs adapted to double-indexed libraries. SC16.JK045 was sequenced at the University of California, San Francisco Center for Advanced Technology on an Illumina NovaSeq 6000 S4 2x100 lane.
To generate a de novo assembly of D. dugon, high–molecular weight DNA was extracted by staff at the Australian Museum from a fresh kidney sample (specimen M39374.001) using the Bioline Isolate ll Genomic DNA Kit (Bioline). DNA was shipped to the Swedish Museum of Natural History under the Convention on International Trade in Endangered Species (CITES) regulation of shipments between registered scientific institutions entitled to the exemption (Article VII, paragraph 6, of the Convention). The DNA was subsequently converted into a linked-read library using the Chromium Genome v2 kit (10x Genomics) and sequenced at the National Genomics Infrastructure (SciLifeLab) in Stockholm using two lanes on an Illumina HiSeqX instrument [2 × 150 base pairs (bp)] to reach approximately 60× genomic coverage.
Steller's sea cow sample collection and experiments are in accordance with ethics committee regulations from the University of Postdam and the University of California, Santa Cruz. The dugong tissue sample was transferred from the Australian Museum to the Swedish Museum of Natural History in accordance with the CITES Convention exemption provided by Article VII, paragraph 6.
De novo genome assembly, genome annotation, and orthology assignment
We assembled the dugong genome initially using Supernova (version 2.1.1) (36), followed by another round of scaffolding using the ARKS+LINKS pipeline (versions 1.0.2 and 1.8.6) (37) with the 10X linked reads. Parameters used for ARKS+LINKS were as follows: -c 5, -k 30, -j 0.5, -z 500, -e 30000, -m 50-10000, -l 5, and -a 0.3.
We evaluated assembly quality using BUSCO (version 3.0.2) (38) with both eukaryotic and mammalian ortholog datasets. We used QUAST (version 4.5.4) (39) for contiguity statistics.
We annotated the dugong (D. dugon) and the manatee [T. manatus latirostris (9); UCSC GCA_000243295.1] genomes using the MAKER pipeline (10) and RNA and protein sequences of three high-quality genomes (human, GRCh38.p13; mouse, GRCm38.p6; and elephant, Loxafr3.0) (11), as previously described (40). Triplet orthologs between human, mouse, and elephant were downloaded from Ensembl 98 (11). Manatee and dugong genes were considered orthologs to a triplet if the ortholog assignment from MAKER agreed with the orthologous gene assigned in each triplet orthologous pair.
Data processing, selection analysis, and demographic history reconstruction
We used the Illumina Bustard software for base calling, deML for demultiplexing (41), and leeHom (42) for adapter trimming and merging of overlapping paired-end reads. We merged sequences from a given library using SAMtools (43) and aligned reads from our individuals, as well as the publicly available data from Sharko et al. (14) to the dugong genome using Burrows-Wheeler Aligner (BWA) (44) with parameters adjusted for ancient DNA (45). We removed PCR duplicates using bam-rmdup (https://doi.org/10.5281/zenodo.4616582) and filtered for DNA fragments of length ≥ 32 bases with a mapping quality of 30 or higher.
We used the ancient DNA damage-aware genotyper snpAD (46) on alignments to the dugong genome to call genotypes on scaffolds >100 kbp. We calculated pairwise distances of all samples based on mitochondrial positions with mapping quality (MQ) ≥ 30, genotype quality (GQ) ≥ 20, and coverage ≥10 identified using SAMtools mpileup (43) to ensure that the 12 sequenced specimens originated from different individuals.
Using the snpAD variant calling on the Steller’s sea cow SNMB N51667, which was treated with uracil-DNA glycosylase and endonuclease VIII and had the lowest ancient DNA damage (section SM1), and the dugong gene predictions, we lifted the corresponding coordinates and inferred the Steller’s sea cow sequence by replacing heterozygous/homozygous alternative positions with the alternative allele. For positions where Steller’s sea cow displayed two alternative alleles, we randomly picked one to reconstruct the sequence. To this end, we considered positions with coverage ≥5× and GQ ≥ 20, while all others were deemed as unresolved and replaced by “N”s. Codon-based alignments were obtained using MACSE version 2 (47), followed by exclusion of regions containing gaps in any of the species.
We used CODEML (48) to scan for differently evolving genes under a branch model (model = 2 versus model = 0 compared via likelihood ratio test) using the set of orthologs as defined above in the five species, and the inferred Steller’s sea cow sequence. Significance of differently evolving genes was established using a chi-square test with one degree of freedom.
We estimated the allele frequency of stop codons in ALOXE3 and ALOX12B using the noninformative Jeffreys’ prior for a binomial distribution incorporated in the jeffreysci function from the ratesci package in R (49).
For pairwise differences between individuals, we used Consensify (50) to generate a consensus pseudohaploid genome sequence. PSMC (29) was run using the parameters -t 40 and -p 4+25*2+4+6. We estimated the mutation rate using polymorphic sites between Steller’s sea cow and dugong or manatee and the divergence time for each pair of species, respectively (7). Prehistorical population estimates for regions beyond Bering Island were obtained by extrapolating the number of sea cows per square kilometer of habitat at Bering Island to the square kilometer of habitat (x) in any region of interest, i.e., as (x km2 / 429 km2) × 1000.
Acknowledgments
We are grateful to our patient who agreed with the publication of the photo to exemplify skin changes caused by ichthyosis. We acknowledge support from Science for Life Laboratory, the Knut and Alice Wallenberg Foundation, the National Genomics Infrastructure funded by the Swedish Research Council, and Uppsala Multidisciplinary Center for Advanced Computational Science for assistance with massively parallel sequencing and access to the UPPMAX computational infrastructure. We express gratitude to A. King and A. Divljan at the Australian Museum who conducted sampling and extracted DNA from the dugong specimen analyzed here and K. L. Laidre, who contributed to the estimation of Steller’s sea cow population size.
Funding: This work was supported by Clinician Scientist Programm, Medizinische Fakultät der Universität Leipzig (D.L.D.); FORMAS grant 2018-01640 (L.D.); German Research Foundation grants HO 3492/15-1 and SCHO 624/13-1 (M.H., T.S., D.L.D., and J.K.); German Research Foundation grants CRC1052 Projektnummer 209933838 (D.L.D. and T.S.); and Revive & Restore grant 03012019-5571 (B.S.).
Author contributions: M.C.-J. and S.B. extracted DNA, prepared libraries, and conducted laboratory work. L.D. supervised sequencing and dugong genome assembly. R.-A.O. assembled and evaluated the dugong genome. A.V., D.L.D., and M.C.-J. processed and mapped sequence data and performed tests on DNA authenticity. A.V. with support from D.L.D. annotated the dugong and manatee genomes, established orthologous genes, identified inactivated genes, and performed selection analyses. A.V. with the input from M.-S.W. performed demographic analyses. C.-C.L. contributed to GO analyses. S.R. and M.C.-J. performed phylogenetic analyses. T.S. manually curated data and inquired the functional relevance of candidate genes. D.L.D., J.R.L., and S.G. contributed to assessment of mutations in patients with ichthyosis. A.B. and U.J. supplied the Steller’s sea cow samples. T.B.H. and G.F. performed the CT scan and contributed to bone powder sampling. M.C.-J. prepared samples for radiocarbon dating. J.R.S. and W.R. performed radiocarbon dating. J.A.E. estimated the Steller’s sea cow population size and evaluated their demographic history. J.K. provided advice on the bioinformatic analyses and helped interpret the results. Conceptualization: D.L.D., T.S., M.H., B.S., and J.K. Funding acquisition: D.L.D., T.S., M.H., J.K., and B.S. Supervision: T.S., M.H., and B.S. Writing (original draft): D.L.D., A.V., J.A.E., T.S., M.H., and B.S. Writing (review and editing): All authors contributed to reviewing and approved the final draft.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: The workflow and scripts used for all analyses have been deposited under Data Dryad. Sequences generated within this study and the de novo D. dugon assembly and annotations have been deposited in the European Nucleotide Archive under the following study accession numbers: PRJEB43951 for Steller’s sea cow reads and PRJEB43952 for the dugong assembly. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Sections SM1 to SM4
Tables S1 to S3 and S5 to S12
Figs. S1 to S6
References
Other Supplementary Material for this manuscript includes the following:
Table S4
REFERENCES AND NOTES
- 1.Estes J. A., Burdin A., Doak D. F., Sea otters, kelp forests, and the extinction of Steller’s sea cow. Proc. Natl. Acad. Sci. U.S.A. 113, 880–885 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turvey S. T., Risley C. L., Modelling the extinction of Steller’s sea cow. Biol. Lett. 2, 94–97 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.J. Davis, Steller’s sea cow: The first historical extinction of marine mammal at human hands | Natural History Museum; www.nhm.ac.uk/discover/stellers-sea-cow-first-historical-extinction-of-marine-mammal-at-human-hands.html.
- 4.Domning D. P., An ecological model for late Tertiary sirenian evolution in the North Pacific Ocean. Syst. Zool. 25, 352–362 (1976). [Google Scholar]
- 5.Forsten A., Youngman P. M., Hydrodamalis gigas. Mamm. Species. 165, 1 (1982). [Google Scholar]
- 6.H. Marsh, T. J. O’Shea, J. E. Reynolds, J. E. Reynolds III, Ecology and Conservation of the sirenia: Dugongs and Manatees (Cambridge Univ. Press, 2012), vol. 18. [Google Scholar]
- 7.Springer M. S., Signore A. V., Paijmans J. L., Vélez-Juarbe J., Domning D. P., Bauer C. E., He K., Crerar L., Campos P. F., Murphy W. J., Meredith R. W., Gatesy J., Willerslev E., MacPhee R. D., Hofreiter M., Campbell K. L., Interordinal gene capture, the phylogenetic position of Steller’s sea cow based on molecular and morphological data, and the macroevolutionary history of sirenia. Mol. Phylogenet. Evol. 91, 178–193 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Sharko F. S., Rastorguev S. M., Boulygina E. S., Tsygankova S. V., Ibragimova A. S., Tikhonov A. N., Nedoluzhko A. V., Molecular phylogeny of the extinct Steller’s sea cow and other sirenia species based on their complete mitochondrial genomes. Genomics 111, 1543–1546 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Foote A. D., Liu Y., Thomas G. W., Vinař T., Alföldi J., Deng J., Dugan S., van Elk C. E., Hunter M. E., Joshi V., Khan Z., Kovar C., Lee S. L., Lindblad-Toh K., Mancia A., Nielsen R., Qin X., Qu J., Raney B. J., Vijay N., Wolf J. B., Hahn M. W., Muzny D. M., Worley K. C., Gilbert M. T., Gibbs R. A., Convergent evolution of the genomes of marine mammals. Nat. Genet. 47, 272–275 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cantarel B. L., Korf I., Robb S. M., Parra G., Ross E., Moore B., Holt C., Sánchez Alvarado A., Yandell M., MAKER: An easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 18, 188–196 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cunningham F., Achuthan P., Akanni W., Allen J., Amode M. R., Armean I. M., Bennett R., Bhai J., Billis K., Boddu S., Cummins C., Davidson C., Dodiya K. J., Gall A., Girón C. G., Gil L., Grego T., Haggerty L., Haskell E., Hourlier T., Izuogu O. G., Janacek S. H., Juettemann T., Kay M., Laird M. R., Lavidas I., Liu Z., Loveland J. E., Marugán J. C., Maurel T., McMahon A. C., Moore B., Morales J., Mudge J. M., Nuhn M., Ogeh D., Parker A., Parton A., Patricio M., Abdul Salam A. I., Schmitt B. M., Schuilenburg H., Sheppard D., Sparrow H., Stapleton E., Szuba M., Taylor K., Threadgold G., Thormann A., Vullo A., Walts B., Winterbottom A., Zadissa A., Chakiachvili M., Frankish A., Hunt S. E., Kostadima M., Langridge N., Martin F. J., Muffato M., Perry E., Ruffier M., Staines D. M., Trevanion S. J., Aken B. L., Yates A. D., Zerbino D. R., Flicek P., Ensembl 2019. Nucleic Acids Res. 47, D745–D751 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McGowen M. R., Gatesy J., Wildman D. E., Molecular evolution tracks macroevolutionary transitions in Cetacea. Trends Ecol. Evol. 29, 336–346 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Krieg P., Rosenberger S., de Juanes S., Latzko S., Hou J., Dick A., Kloz U., van der Hoeven F., Hausser I., Esposito I., Rauh M., Schneider H., Aloxe3 knockout mice reveal a function of epidermal lipoxygenase-3 as hepoxilin synthase and its pivotal role in barrier formation. J. Invest. Dermatol. 133, 172–180 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Sharko F. S., Boulygina E. S., Tsygankova S. V., Slobodova N. V., Alekseev D. A., Krasivskaya A. A., Rastorguev S. M., Tikhonov A. N., Nedoluzhko A. V., Steller’s sea cow genome suggests this species began going extinct before the arrival of Paleolithic humans. Nat. Commun. 12, 2215 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simpson J. K., Martinez-Queipo M., Onoufriadis A., Tso S., Glass E., Liu L., Higashino T., Scott W., Tierney C., Simpson M. A., Desomchoke R., Youssefian L., SaeIdian A. H., Vahidnezhad H., Bisquera A., Ravenscroft J., Moss C., O’Toole E. A., Burrows N., Leech S., Jones E. A., Lim D., Ilchyshyn A., Goldstraw N., Cork M. J., Darne S., Uitto J., Martinez A. E., Mellerio J. E., McGrath J. A., Genotype-phenotype correlation in a large English cohort of patients with autosomal recessive ichthyosis. Br. J. Dermatol. 182, 729–737 (2020). [DOI] [PubMed] [Google Scholar]
- 16.Bučková H., Nosková H., Borská R., Réblová K., Pinková B., Zapletalová E., Kopečková L., Horký O., Němečková J., Gaillyová R., Nagy Z., Veselý K., Hermanová M., Stehlíková K., Fajkusová L., Autosomal recessive congenital ichthyoses in the Czech Republic. Br. J. Dermatol. 174, 405–407 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Borská R., Pinková B., Réblová K., Bučková H., Kopečková L., Němečková J., Puchmajerová A., Malíková M., Hermanová M., Fajkusová L., Inherited ichthyoses: Molecular causes of the disease in Czech patients. Orphanet J. Rare Dis. 14, 92 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Juanes S., Epp N., Latzko S., Neumann M., Fürstenberger G., Hausser I., Stark H. J., Krieg P., Development of an ichthyosiform phenotype in Alox12b-deficient mouse skin transplants. J. Invest. Dermatol. 129, 1429–1436 (2009). [DOI] [PubMed] [Google Scholar]
- 19.Springer M. S., Guerrero-Juarez C. F., Huelsmann M., Collin M. A., Danil K., McGowen M. R., Oh J. W., Ramos R., Hiller M., Plikus M. V., Gatesy J., Genomic and anatomical comparisons of skin support independent adaptation to life in water by cetaceans and hippos. Curr. Biol. 31, 2124–2139.e3 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reisch F., Kakularam K. R., Stehling S., Heydeck D., Kuhn H., Eicosanoid biosynthesis in marine mammals. FEBS J. 288, 1387–1406 (2021). [DOI] [PubMed] [Google Scholar]
- 21.Sharma V., Hecker N., Roscito J. G., Foerster L., Langer B. E., Hiller M., A genomics approach reveals insights into the importance of gene losses for mammalian adaptations. Nat. Commun. 9, 1215 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harrison O. J., Brasch J., Lasso G., Katsamba P. S., Ahlsen G., Honig B., Shapiro L., Structural basis of adhesive binding by desmocollins and desmogleins. Proc. Natl. Acad. Sci. U.S.A. 113, 7160–7165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang L., Ip C. K., Lee I. C. J., Qi Y., Reed F., Karl T., Low J. K., Enriquez R. F., Lee N. J., Baldock P. A., Herzog H., Diet-induced adaptive thermogenesis requires neuropeptide FF receptor-2 signalling. Nat. Commun. 9, 4722 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gaudry M. J., Jastroch M., Treberg J. R., Hofreiter M., Paijmans J. L. A., Starrett J., Wales N., Signore A. V., Springer M. S., Campbell K. L., Inactivation of thermogenic UCP1 as a historical contingency in multiple placental mammal clades. Sci. Adv. 3, e1602878 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gearty W., McClain C. R., Payne J. L., Energetic tradeoffs control the size distribution of aquatic mammals. Proc. Natl. Acad. Sci. U.S.A. 115, 4194–4199 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roesler A., Kazak L., UCP1-independent thermogenesis. Biochem. J. 477, 709–725 (2020). [DOI] [PubMed] [Google Scholar]
- 27.Ikeda K., Yamada T., UCP1 dependent and independent thermogenesis in brown and beige adipocytes. Front. Endocrinol. 11, 498 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.G. W. Steller, O. W. Frost, Journal of a Voyage with Bering, 1741–1742 (Stanford Univ. Press, 1993). [Google Scholar]
- 29.Li H., Durbin R., Inference of human population history from individual whole-genome sequences. Nature 475, 493–496 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cahill J. A., Green R. E., Fulton T. L., Stiller M., Jay F., Ovsyanikov N., Salamzade R., John J. S., Stirling I., Slatkin M., Shapiro B., Genomic evidence for island population conversion resolves conflicting theories of polar bear evolution. PLOS Genet. 9, e1003345 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shammas N., Walker B., Martinez De La Torre H., Bertrand C., Southon J., Effect of ultrafilter pretreatment, acid strength and decalcification duration on archaeological bone collagen yield. Nucl. Instrum. Methods Phys. Res. B 456, 283–286 (2019). [Google Scholar]
- 32.Dabney J., Knapp M., Glocke I., Gansauge M. T., Weihmann A., Nickel B., Valdiosera C., García N., Pääbo S., Arsuaga J. L., Meyer M., Complete mitochondrial genome sequence of a Middle Pleistocene cave bear reconstructed from ultrashort DNA fragments. Proc. Natl. Acad. Sci. U.S.A. 110, 15758–15763 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gansauge M. T., Meyer M., Single-stranded DNA library preparation for the sequencing of ancient or damaged DNA. Nat. Protoc. 8, 737–748 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Korlević P., Gerber T., Gansauge M. T., Hajdinjak M., Nagel S., Aximu-Petri A., Meyer M., Reducing microbial and human contamination in DNA extractions from ancient bones and teeth. Biotechniques 59, 87–93 (2015). [DOI] [PubMed] [Google Scholar]
- 35.Kapp J. D., Green R. E., Shapiro B., A fast and efficient single-stranded genomic library preparation method optimized for ancient DNA. J. Hered. 112, 241–249 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weisenfeld N. I., Kumar V., Shah P., Church D. M., Jaffe D. B., Direct determination of diploid genome sequences. Genome Res. 27, 757–767 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coombe L., Zhang J., Vandervalk B. P., Chu J., Jackman S. D., Birol I., Warren R. L., ARKS: Chromosome-scale scaffolding of human genome drafts with linked read kmers. BMC Bioinformatics 19, 234 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simão F. A., Waterhouse R. M., Ioannidis P., Kriventseva E. V., Zdobnov E. M., BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Mikheenko A., Valin G., Prjibelski A., Saveliev V., Gurevich A., Icarus: Visualizer for de novo assembly evaluation. Bioinformatics 32, 3321–3323 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Le Duc D., Renaud G., Krishnan A., Almén M. S., Huynen L., Prohaska S. J., Ongyerth M., Bitarello B. D., Schiöth H. B., Hofreiter M., Stadler P. F., Prüfer K., Lambert D., Kelso J., Schöneberg T., Kiwi genome provides insights into evolution of a nocturnal lifestyle. Genome Biol. 16, 147 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Renaud G., Stenzel U., Maricic T., Wiebe V., Kelso J., deML: Robust demultiplexing of Illumina sequences using a likelihood-based approach. Bioinformatics 31, 770–772 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Renaud G., Stenzel U., Kelso J., leeHom: Adaptor trimming and merging for Illumina sequencing reads. Nucleic Acids Res. 42, e141 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.; 1000 Genome Project Data Processing Subgroup , The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li H., Durbin R., Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meyer M., Kircher M., Gansauge M. T., Li H., Racimo F., Mallick S., Schraiber J. G., Jay F., Prüfer K., de Filippo C., Sudmant P. H., Alkan C., Fu Q., Do R., Rohland N., Tandon A., Siebauer M., Green R. E., Bryc K., Briggs A. W., Stenzel U., Dabney J., Shendure J., Kitzman J., Hammer M. F., Shunkov M. V., Derevianko A. P., Patterson N., Andrés A. M., Eichler E. E., Slatkin M., Reich D., Kelso J., Pääbo S., A high-coverage genome sequence from an archaic Denisovan individual. Science 338, 222–226 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prüfer K., snpAD: An ancient DNA genotype caller. Bioinformatics 34, 4165–4171 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ranwez V., Douzery E. J. P., Cambon C., Chantret N., Delsuc F., MACSE v2: Toolkit for the alignment of coding sequences accounting for frameshifts and stop codons. Mol. Biol. Evol. 35, 2582–2584 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang Z., PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591 (2007). [DOI] [PubMed] [Google Scholar]
- 49.P. J. Laud, ratesci: Confidence Intervals for Comparisons of Binomial or Poisson Rates. R Packag. version 0.3–0 (2018); https//CRAN.R-project.org/package= ratesci.
- 50.Barlow A., Hartmann S., Gonzalez J., Hofreiter M., Paijmans J. L. A., Consensify: A method for generating pseudohaploid genome sequences from palaeogenomic datasets with reduced error rates. Genes 11, 50 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.IUCN, The IUCN Red List of Threatened Species. Version 2020–3 (2020).
- 52.Korf I., Gene finding in novel genomes. BMC Bioinformatics 5, 59 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.A. F. Smit, R. Hubley, P. Green, Repeat masker open 4.0; www.repeatmasker.org/.
- 54.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- 55.Prüfer K., Muetzel B., Do H. H., Weiss G., Khaitovich P., Rahm E., Pääbo S., Lachmann M., Enard W., FUNC: A package for detecting significant associations between gene sets and ontological annotations. BMC Bioinformatics 8, 41 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Drost H.-G., Paszkowski J., Biomartr: Genomic data retrieval with R. Bioinformatics 33, 1216–1217 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cheng R., Liang J., Li Y., Zhang J., Ni C., Yu H., Kong X., Li M., Yao Z., Next-generation sequencing through multi-gene panel testing for diagnosis of hereditary ichthyosis in Chinese. Clin. Genet. 97, 770–778 (2020). [DOI] [PubMed] [Google Scholar]
- 58.Israeli S., Goldberg I., Fuchs-Telem D., Bergman R., Indelman M., Bitterman-Deutsch O., Harel A., Mashiach Y., Sarig O., Sprecher E., Non-syndromic autosomal recessive congenital ichthyosis in the Israeli population. Clin. Exp. Dermatol. 38, 911–916 (2013). [DOI] [PubMed] [Google Scholar]
- 59.Eckl K. M., Krieg P., Küster W., Traupe H., André F., Wittstruck N., Fürstenberger G., Hennies H. C., Mutation spectrum and functional analysis of epidermis-type lipoxygenases in patients with autosomal recessive congenital ichthyosis. Hum. Mutat. 26, 351–361 (2005). [DOI] [PubMed] [Google Scholar]
- 60.Espregueira Themudo G., Alves L. Q., Machado A. M., Lopes-Marques M., da Fonseca R. R., Fonseca M., Ruivo R., Castro L. F. C., Losing Genes: The evolutionary remodeling of Cetacea skin. Front. Mar. Sci. 7, 592375 (2020). [Google Scholar]
- 61.Hicks B. D., Aubin D. J. S., Geraci J. R., Brown W. R., Epidermal growth in the bottlenose dolphin, Tursiops truncatus. J. Invest. Dermatol. 85, 60–63 (1985). [DOI] [PubMed] [Google Scholar]
- 62.H.-G. Drost, Genomic Data Retrieval, doi: 10.1186/1471-2164-10-22. [DOI]
- 63.Smedley D., Haider S., Ballester B., Holland R., London D., Thorisson G., Kasprzyk A., BioMart—Biological queries made easy. BMC Genomics 10, 22 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hessel S., Eichinger A., Isken A., Amengual J., Hunzelmann S., Hoeller U., Elste V., Hunziker W., Goralczyk R., Oberhauser V., Von Lintig J., Wyss A., CMO1 deficiency abolishes vitamin A production from β-carotene and alters lipid metabolism in mice. J. Biol. Chem. 282, 33553–33561 (2007). [DOI] [PubMed] [Google Scholar]
- 65.Lanaspa M. A., Epperson L. E., Li N., Cicerchi C., Garcia G. E., Roncal-Jimenez C. A., Trostel J., Jain S., Mant C. T., Rivard C. J., Ishimoto T., Shimada M., Sanchez-Lozada L. G., Nakagawa T., Jani A., Stenvinkel P., Martin S. L., Johnson R. J., Opposing activity changes in AMP deaminase and AMP-activated protein kinase in the hibernating ground squirrel. PLOS ONE 10, e0123509 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Böttcher Y., Unbehauen H., Klöting N., Ruschke K., Körner A., Schleinitz D., Tönjes A., Enigk B., Wolf S., Dietrich K., Koriath M., Scholz G. H., Tseng Y. H., Dietrich A., Schön M. R., Kiess W., Stumvoll M., Blüher M., Kovacs P., Adipose tissue expression and genetic variants of the bone morphogenetic protein receptor 1A gene (BMPR1A) are associated with human obesity. Diabetes 58, 2119–2128 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Petersen P. S., Jin C., Madsen A. N., Rasmussen M., Kuhre R., Egerod K. L., Nielsen L. B., Schwartz T. W., Holst B., Deficiency of the GPR39 receptor is associated with obesity and altered adipocyte metabolism. FASEB J. 25, 3803–3814 (2011). [DOI] [PubMed] [Google Scholar]
- 68.Moechars D., Depoortere I., Moreaux B., de Smet B., Goris I., Hoskens L., Daneels G., Kass S., Ver Donck L., Peeters T., Coulie B., Altered gastrointestinal and metabolic function in the GPR39-obestatin receptor-knockout mouse. Gastroenterology 131, 1131–1141 (2006). [DOI] [PubMed] [Google Scholar]
- 69.Akbari P., Gilani A., Sosina O., Kosmicki J. A., Khrimian L., Fang Y. Y., Persaud T., Garcia V., Sun D., Li A., Mbatchou J., Locke A. E., Benner C., Verweij N., Lin N., Hossain S., Agostinucci K., Pascale J. V., Dirice E., Dunn M., Kraus W. E., Shah S. H., Chen Y. D. I., Rotter J. I., Rader D. J., Melander O., Still C. D., Mirshahi T., Carey D. J., Berumen-Campos J., Kuri-Morales P., Alegre-Díaz J., Torres J. M., Emberson J. R., Collins R., Balasubramanian S., Hawes A., Jones M., Zambrowicz B., Murphy A. J., Paulding C., Coppola G., Overton J. D., Reid J. G., Shuldiner A. R., Cantor M., Kang H. M., Abecasis G. R., Karalis K., Economides A. N., Marchini J., Yancopoulos G. D., Sleeman M. W., Altarejos J., Gatta G. D., Tapia-Conyer R., Schwartzman M. L., Baras A., Ferreira M. A. R., Lotta L. A., Sequencing of 640,000 exomes identifies GPR75 variants associated with protection from obesity. Science 373, eabf8683 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Carbon S., Douglass E., Dunn N., Good B., Harris N. L., Lewis S. E., Mungall C. J., Basu S., Chisholm R. L., Dodson R. J., Hartline E., Fey P., Thomas P. D., Albou L. P., Ebert D., Kesling M. J., Mi H., Muruganujan A., Huang X., Poudel S., Mushayahama T., Hu J. C., LaBonte S. A., Siegele D. A., Antonazzo G., Attrill H., Brown N. H., Fexova S., Garapati P., Jones T. E. M., Marygold S. J., Millburn G. H., Rey A. J., Trovisco V., Santos G. D., Emmert D. B., Falls K., Zhou P., Goodman J. L., Strelets V. B., Thurmond J., Courtot M., Osumi D. S., Parkinson H., Roncaglia P., Acencio M. L., Kuiper M., Lreid A., Logie C., Lovering R. C., Huntley R. P., Denny P., Campbell N. H., Kramarz B., Acquaah V., Ahmad S. H., Chen H., Rawson J. H., Chibucos M. C., Giglio M., Nadendla S., Tauber R., Duesbury M. J., Del N. T., Meldal B. H. M., Perfetto L., Porras P., Orchard S., Shrivastava A., Xie Z., Chang H. Y., Finn R. D., Mitchell A. L., Rawlings N. D., Richardson L., Sangrador-Vegas A., Blake J. A., Christie K. R., Dolan M. E., Drabkin H. J., Hill D. P., Ni L., Sitnikov D., Harris M. A., Oliver S. G., Rutherford K., Wood V., Hayles J., Bahler J., Lock A., Bolton E. R., De Pons J., Dwinell M., Hayman G. T., Laulederkind S. J. F., Shimoyama M., Tutaj M., Wang S. J., D’Eustachio P., Matthews L., Balhoff J. P., Aleksander S. A., Binkley G., Dunn B. L., Cherry J. M., Engel S. R., Gondwe F., Karra K., MacPherson K. A., Miyasato S. R., Nash R. S., Ng P. C., Sheppard T. K., Vp A. S., Simison M., Skrzypek M. S., Weng S., Wong E. D., Feuermann M., Gaudet P., Bakker E., Berardini T. Z., Reiser L., Subramaniam S., Huala E., Arighi C., Auchincloss A., Axelsen K., Argoud G. P., Bateman A., Bely B., Blatter M. C., Boutet E., Breuza L., Bridge A., Britto R., Bye-A-Jee H., Casals-Casas C., Coudert E., Estreicher A., Famiglietti L., Garmiri P., Georghiou G., Gos A., Gruaz-Gumowski N., Hatton-Ellis E., Hinz U., Hulo C., Ignatchenko A., Jungo F., Keller G., Laiho K., Lemercier P., Lieberherr D., Lussi Y., Mac-Dougall A., Magrane M., Martin M. J., Masson P., Natale D. A., Hyka N. N., Pedruzzi I., Pichler K., Poux S., Rivoire C., Rodriguez-Lopez M., Sawford T., Speretta E., Shypitsyna A., Stutz A., Sundaram S., Tognolli M., Tyagi N., Warner K., Zaru R., Wu C., Chan J., Cho J., Gao S., Grove C., Harrison M. C., Howe K., Lee R., Mendel J., Muller H. M., Raciti D., Van Auken K., Berriman M., Stein L., Sternberg P. W., Howe D., Toro S., Westerfield M., The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 47, D330–D338 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., Davis A. P., Dolinski K., Dwight S. S., Eppig J. T., Harris M. A., Hill D. P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J. C., Richardson J. E., Ringwald M., Rubin G. M., Sherlock G., Gene ontology: Tool for the unification of biology. Nat. Genet. 25, 25–29 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Watkins P. A., Maiguel D., Jia Z., Pevsner J., Evidence for 26 distinct acyl-coenzyme A synthetase genes in the human genome. J. Lipid Res. 48, 2736–2750 (2007). [DOI] [PubMed] [Google Scholar]
- 73.Yudin N. S., Larkin D. M., Ignatieva E. V., A compendium and functional characterization of mammalian genes involved in adaptation to Arctic or Antarctic environments. BMC Genet. 18, 111 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lynch V. J., Bedoya-Reina O. C., Ratan A., Sulak M., Drautz-Moses D. I., Perry G. H., Miller W., Schuster S. C., Elephantid genomes reveal the molecular bases of woolly mammoth adaptations to the Arctic. Cell Rep. 12, 217–228 (2015). [DOI] [PubMed] [Google Scholar]
- 75.Liu S., Lorenzen E. D., Fumagalli M., Li B., Harris K., Xiong Z., Zhou L., Korneliussen T. S., Somel M., Babbitt C., Wray G., Li J., He W., Wang Z., Fu W., Xiang X., Morgan C. C., Doherty A., O’Connell M. J., McInerney J. O., Born E. W., Dalén L., Dietz R., Orlando L., Sonne C., Zhang G., Nielsen R., Willerslev E., Wang J., Population genomics reveal recent speciation and rapid evolutionary adaptation in polar bears. Cell 157, 785–794 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fath M. A., Mullins R. F., Searby C., Nishimura D. Y., Wei J., Rahmouni K., Davis R. E., Tayeh M. K., Andrews M., Yang B., Sigmund C. D., Stone E. M., Sheffield V. C., Mkks-null mice have a phenotype resembling Bardet-Biedl syndrome. Hum. Mol. Genet. 14, 1109–1118 (2005). [DOI] [PubMed] [Google Scholar]
- 77.Min S. Y., Desai A., Yang Z., Sharma A., DeSouza T., Genga R. M. J., Kucukural A., Lifshitz L. M., Nielsen S., Scheele C., Maehr R., Garber M., Corvera S., Diverse repertoire of human adipocyte subtypes develops from transcriptionally distinct mesenchymal progenitor cells. Proc. Natl. Acad. Sci. U.S.A. 116, 17970–17979 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kong X., Yao T., Zhou P., Kazak L., Tenen D., Lyubetskaya A., Dawes B. A., Tsai L., Kahn B. B., Spiegelman B. M., Liu T., Rosen E. D., Brown adipose tissue controls skeletal muscle function via the secretion of myostatin. Cell Metab. 28, 631–643.e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kong X., Banks A., Liu T., Kazak L., Rao R. R., Cohen P., Wang X., Yu S., Lo J. C., Tseng Y. H., Cypess A. M., Xue R., Kleiner S., Kang S., Spiegelman B. M., Rosen E. D., IRF4 is a key thermogenic transcriptional partner of PGC-1α. Cell 158, 69–83 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Palkopoulou E., Mallick S., Skoglund P., Enk J., Rohland N., Li H., Omrak A., Vartanyan S., Poinar H., Götherström A., Reich D., Dalén L., Complete genomes reveal signatures of demographic and genetic declines in the woolly mammoth. Curr. Biol. 25, 1395–1400 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Behr B., Rath D., Hildebrandt T. B., Goeritz F., Blottner S., Portas T. J., Bryant B. R., Sieg B., Knieriem A., De Graaf S., Maxwell W. M. C., Hermes R., Germany/Australia index of sperm sex sortability in elephants and rhinoceros. Reprod. Domest. Anim. 44, 273–277 (2009). [DOI] [PubMed] [Google Scholar]
- 82.H. Wickham, ggplot2: Elegant Graphics for Data Analysis (Springer New York, 2016). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sections SM1 to SM4
Tables S1 to S3 and S5 to S12
Figs. S1 to S6
References
Table S4