Abstract
Remote deep-ocean sediment (DOS) ecosystems are among the least explored biomes on Earth. Genomic assessments of their biodiversity have failed to separate indigenous benthic organisms from sinking plankton. Here, we compare global-scale eukaryotic DNA metabarcoding datasets (18S-V9) from abyssal and lower bathyal surficial sediments and euphotic and aphotic ocean pelagic layers to distinguish plankton from benthic diversity in sediment material. Based on 1685 samples collected throughout the world ocean, we show that DOS diversity is at least threefold that in pelagic realms, with nearly two-thirds represented by abundant yet unknown eukaryotes. These benthic communities are spatially structured by ocean basins and particulate organic carbon (POC) flux from the upper ocean. Plankton DNA reaching the DOS originates from abundant species, with maximal deposition at high latitudes. Its seafloor DNA signature predicts variations in POC export from the surface and reveals previously overlooked taxa that may drive the biological carbon pump.
A global analysis of surface to seafloor eukaryotes distinguishes benthic diversity from sinking plankton in deep-sea sediments.
INTRODUCTION
Deep-ocean sediment (DOS) ecosystems cover more than half of Earth’s surface and remain one of the least explored ecosystems on the planet. This vast and heterogeneous environment provides habitats for diverse biological communities that support fundamental ecological processes and services, such as nutrient recycling for the healthy functioning of ocean ecosystems and carbon sequestration for the regulation of Earth’s climate over geological time scales (1). The DOS is exposed to growing anthropogenic pressures, notably from climate change (2, 3), deep-sea mining (4), oil and gas exploitation, and bottom trawling (5), making a scientifically informed protection of its biodiversity a matter of the highest importance (6–8).
For more than 50 years, a considerable effort has been devoted to understanding the diversity and biogeography of benthic organisms thriving in the DOS (9). However, the enormous extent of this habitat and its remote location under several kilometers of water means that only a minute proportion has ever been sampled. Most previous studies have focused on morphological analyses of the macro- and mega-fauna, which typically show high levels of a diversity and small-scale faunal patchiness (9–11), and have recently been proposed as biological indicators for deep-ocean monitoring and conservation (12). Less attention has been paid to the microbial and meiofaunal organisms that numerically dominate DOS communities (13, 14) but can hardly be identified using classical morphotaxonomic approaches. Studying planktonic organisms sinking to DOS is hampered by similar technical limitations related to great depths (resulting in poor spatial coverage of sinking plankton datasets) and limited morphological identification (but for shell-building taxa that can keep distinctive features once in the sediment). Their study is hence often approached indirectly, using sediment traps to capture the sinking flux of taxa over time that contribute most to the biologically driven carbon sequestration in the deep ocean before they reach the sediment (15–17).
The development of high-throughput environmental genomics has begun to fill these gaps in knowledge, revealing substantial unknown diversity among viruses (18) and prokaryotes (19–22) from DOS. Yet, the use of genomics to explore DOS eukaryotes has been limited and focused mostly on particular taxonomic groups (23–25) or geographic regions [(21, 26–28), but see (29)]. One major challenge in interpreting molecular data from DOS is to distinguish DNA reads that belong to indigenous benthic eukaryotes from those originating from pelagic organisms that sink through the water column and leave their DNA traces in the sediments (28, 30–32).
Here, we tackle these problems by comparing a newly generated, global-scale DNA metabarcoding dataset of total eukaryotic diversity from deep oceanic surficial sediments (418 samples collected during 15 oceanographic cruises from 2010 to 2016; table S1) to comparable published datasets from euphotic (1160 samples from the Tara Oceans expeditions) (33, 34) and aphotic (138 samples from the Tara Oceans and Malaspina expeditions) (35) zones across the world ocean. Together, these represent the first consistent molecular meta-dataset spanning the three main open-ocean realms (pelagic euphotic, pelagic aphotic, DOS) at a global scale across 447 sampling sites (Fig. 1A). We assembled ~2.42 billion DNA reads (table S2), produced by polymerase chain reaction (PCR) amplification of the V9 region of the 18S ribosomal RNA gene, and processed them using the DADA2 workflow to infer amplicon sequence variants (ASVs). On the basis of taxonomic annotations of ASVs using the SILVA and PR2 sequence databases and on the occurrence of a highly conserved DNA sequence motif across eukaryotes, we discarded prokaryotic, plastidic, and mitochondrial ASVs, as well as technical artifacts, allowing us to focus on eukaryotic diversity.
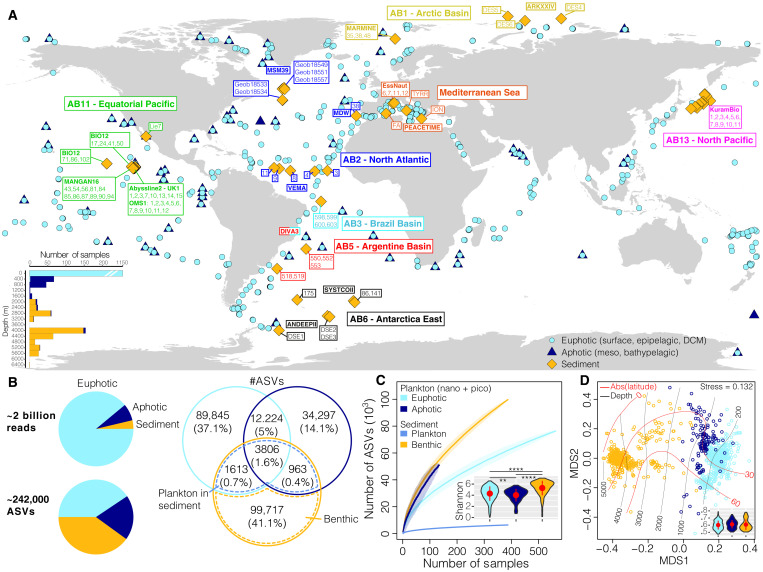
Fig. 1. Eukaryotic ribosomal DNA diversity in the pelagic euphotic and aphotic zones and in the deep-ocean surficial sediment.
(A) Location of the stations (n = 447) from which the samples (n = 1685) analyzed in this study were collected. The color of the sediment sampling station tags indicates approximate correspondence with abyssal provinces (42), and the names of deep-sea cruises are indicated in bold or within text boxes. The bottom left inset represents the depth distribution (in meters) of the samples. (B) Number of eukaryotic 18S V9 rDNA reads and of amplicon sequence variants (ASVs). A Venn diagram represents the distribution of ASV richness and their proportions within and across realms. The intersection of the pelagic and sediment datasets is used here to separate the indigenous benthic organisms from the sinking plankton (see Materials and Methods). (C) ASV accumulation curves as a function of sampling effort. For pelagic realms, we calculated the curves by focusing only on the nano- and picoplanktonic size fractions (see Materials and Methods). The inset displays the distribution of Shannon diversity for pelagic and benthic communities. The red dots and bars within violin plots represent means and SDs, and horizontal bars indicate significant differences (Wilcoxon tests, **P < 0.01 and ****P < 0.0001). (D) Nonmetric multidimensional scaling (NMDS) analysis of the Bray-Curtis dissimilarity matrix computed from the pelagic (only the nano- and pico-fractions) and sediment datasets. The red and black lines on the ordination represent, respectively, the absolute latitude and depth as fitted surfaces to the ordination. The inset represents the community dispersion within each realm (i.e., Bray-Curtis distances to the group centroid, higher values indicate more compositional variation). The red dots and bars within violin plots represent means and SDs, respectively, and no significant differences between realms were detected (Wilcoxon tests, P > 0.05).
RESULTS AND DISCUSSION
Eukaryotic diversity from the ocean surface to the DOS
We obtained a total of 242,465 eukaryotic ASVs represented by ~1.95 billion DNA reads (Fig. 1B). Only 3806 (1.6%) of these ASVs were detected in all three realms, while 6382 pelagic ASVs were detected in DOS. These ASVs were assumed to correspond to sinking pelagic organisms, mainly plankton, although 29 ASVs (representing 1.29% of the reads of these ASVs) could be ascribed to nekton (e.g., dead vertebrates), which also contribute to the downward flux of organic matter. From the metazoan fraction of sinking pelagic organisms, we curated benthic animals with known meroplanktonic larvae (224 ASVs; see Materials and Methods). The number of ASVs found exclusively in DOS, here assumed to correspond to indigenous deep-sea benthic organisms, was comparable to that found in the pelagic realms, although there were 25 times more pelagic DNA reads in our meta-dataset (Fig. 1B). To account for this variation in sequencing effort, we subsampled each aggregated dataset per realm 1000 times at identical sequencing depths (1 Mio reads) and analyzed the diversity of ASV, together with their distribution and abundances within and across the pelagic (euphotic and aphotic zones) and DOS (sinking pelagic and benthic organisms; fig. S1) realms. This indicated that, although nearly half of eukaryotic DNA reads represent sinking planktonic ASVs, the ASV richness in the DOS could be more than three times higher than in pelagic habitats, with more than 60% of ASVs being exclusively benthic.
The unique size fractionation of the pelagic samples from the Tara Oceans dataset has a strong effect on a diversity (richness and evenness) and b diversity (compositional variation) measures (fig. S2) (33). The samples from the micro- (20 to 180 μm) and meso- (180 to 2000 μm) plankton collected by 20- and 180-μm net tows concentrate mostly on copepods and collodarians that lower a diversity and inflate b diversity measures when compared to other pelagic samples. Since these taxa were also detected in the lower plankton size fractions, we confined our analyses to the richest nano- (3 to 20 μm) and pico- (0.2 to 5 μm) plankton fractions to compare a and b diversity patterns across Tara Oceans, Malaspina, and DOS samples.
Both the ASV accumulation curves as a function of sampling effort and Shannon diversity values confirmed that benthic eukaryotic diversity is much higher than that in the water column (Fig. 1C). The benthic accumulation curve is similar to that obtained for the pelagic aphotic zone, which may indicate that diversity in aphotic waters is also very high and largely undersampled [but see (36)]. Clustering of eukaryotic communities by their compositional similarity revealed a clear separation of the pelagic and DOS realms and changes along a gradient of absolute latitude (Fig. 1D and fig. S3). This was confirmed by permutational multivariate analysis of variance (PERMANOVA), both for type of realm (R2 = 0.144, P < 0.001) and for absolute latitude (R2 = 0.031, P < 0.001). The degree of eukaryotic community differentiation (b diversity dispersion) within each realm was similar (Wilcoxon test, P > 0.05; Fig. 1D). Our results therefore indicate that eukaryotic communities of the DOS are both more diverse and sharply different compared to those of pelagic realms.
The taxonomic compositions of eukaryotic assemblages were clearly different in the pelagic euphotic, pelagic aphotic, and DOS realms (Fig. 2A; see fig. S4 for relative abundances and table S3 for details). While diversity in the euphotic zone is dominated taxonomically by Alveolata (30.8%), notably dinoflagellates (Dinophyceae, 19.4%), the aphotic zone is extremely rich in Diplonemea [46.5%; see (37–39)], mainly heterotrophic nanoflagellates in the family Eupelagonemidae (46.2%). The taxonomic composition of the deep, exclusively benthic, eukaryotic assemblage is very different, comprising various groups that do not occur or rarely occur in the water column (fig. S5 and table S3), e.g., Dactylopodida amoebae (6.5%), Chromadoria nematodes (5.3%), Monothalamid foraminifera (4.4%), and Oligohymenophorea ciliates (3.7%). Nearly two-thirds (60.1%) of the benthic eukaryotic ASVs (representing 47.8% of the reads) could not be taxonomically annotated using current reference taxonomic databases and a similarity cutoff of 85%, and many of them matched a reference sequence with less than 80% similarity (Fig. 2B). By comparison, the proportion of unassigned ASVs in the pelagic samples is 24.7% (2.6% of the reads) in the euphotic zone and 13.9% (4.1% of the reads) in the aphotic zone (Fig. 2C and table S3).
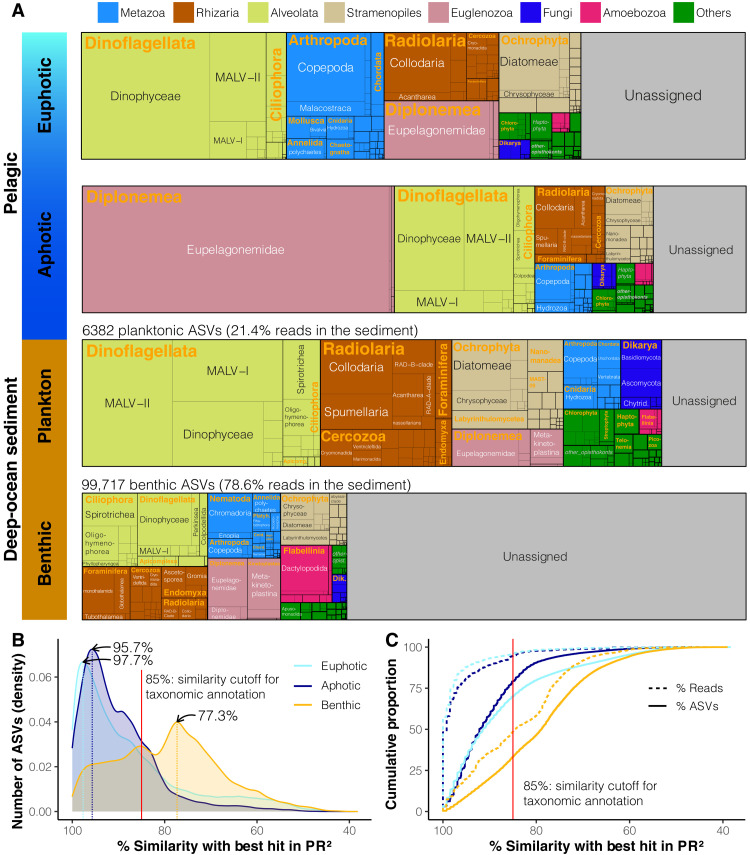
Fig. 2. Taxonomic composition of eukaryotes (ASV richness) in the pelagic euphotic, pelagic aphotic, and DOS (sinking plankton and benthic communities) realms.
(A) ASV richness of eukaryotic groups (see fig. S4 for relative abundances and table S3 for details). The number of ASVs and their relative abundance in the sediment are shown for ASVs of pelagic origin, as well as those derived from indigenous benthic taxa. (B) Number of ASVs (represented as density) as a function of their similarity with the best hit with a reference sequence in the PR2 database. The peaks in density for each realm are highlighted on the plot, and the corresponding similarity level with reference sequences is indicated. (C) Cumulative proportion of ASVs and read abundance as a function of their similarity with the best hit with a reference sequence in PR2. The red vertical lines indicate the similarity cutoff (85%) for taxonomic annotation used in this study.
To better characterize the taxonomic breadth of the unknown eukaryotic diversity in the ocean, we clustered all unassigned eukaryotic ASVs into operational taxonomic units (OTUs) at decreasing similarity thresholds (fig. S6). This revealed that more than 10,000 benthic OTUs are formed with a 90% similarity cutoff, well below the species/genus threshold levels (40). These results indicate that previously unknown high-rank eukaryotic groups with diverse and abundant sublineages likely make up most of the diversity thriving in DOS. A similar number of 90% cutoff OTUs is formed in the pelagic euphotic zone, but their relative abundance (2.6% of the reads) is much lower than for benthic diversity (47.8% of the reads). Many of these unassigned pelagic ASVs may thus correspond to rare unknown eukaryotes or rare intraspecific/intragenomic variants of known eukaryotes with unusually high polymorphism (41). Among the known taxa (39.9% of benthic ASVs and 52.2% of benthic reads), our data for selected typical deep benthic macrofaunal and meiofaunal groups show that some are relatively well represented in the current databases (e.g., polychaetes and nemerteans; fig. S7), while others remain poorly represented (e.g., foraminifera and nematodes).
Biogeography of deep-ocean benthic eukaryotes
Analysis of the strictly benthic eukaryotic diversity revealed global biogeographic patterns among DOS communities. The overall richness of benthic ASVs tends to decrease with increasing latitude (fig. S8). Benthic richness follows a bell-shaped trend with increasing export flux of particulate organic carbon (POC) from the surface and particularly with increasing POC reaching the seafloor (the latter explaining up to 10.7% of the variation in overall benthic richness). This pattern is not consistent across benthic groups, with nematodes, foraminifera, and molluscs being notably more diverse at higher latitudes and at sites with higher POC flux reaching the seafloor (fig. S8). The compositional structure of deep benthic communities is in broad agreement with abyssal biogeographic provinces (42) (PERMANOVA R2 = 0.136, P < 0.001) and somewhat structured along a gradient of absolute latitude at a global scale (R2 = 0.051, P < 0.001), although polar regions are separated on the ordination (Fig. 3A). We used a selection of environmental parameters (see Materials and Methods) in a stepwise model building for constrained ordination to explain the observed pattern. The model explained up to 15.1% of the benthic compositional variation, with seabed nitrate, POC export from the surface, and POC reaching the seafloor together explaining 11.2% (table S4), in line with previous findings on the role of POC export in shaping benthic prokaryotic communities (19).
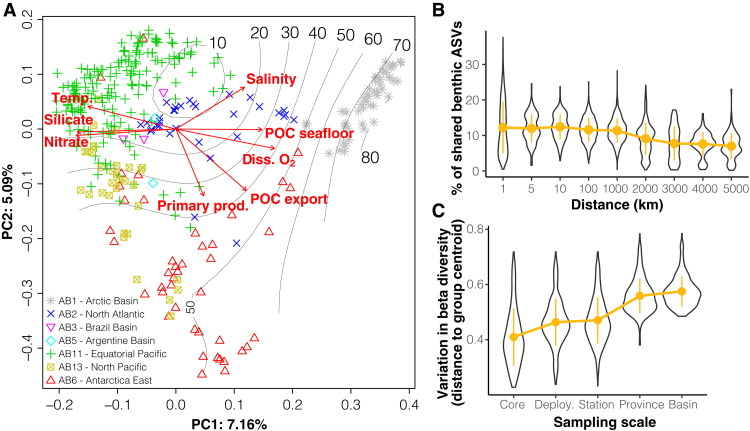
Fig. 3. Biogeography of the deep-ocean benthic eukaryotic communities.
(A) Principal coordinates analysis of the Bray-Curtis dissimilarity matrix computed from the normalized read counts of indigenous benthic ASVs using the cumulative sum scaling (CSS) method. The proportion of variance explained by the first two axes is indicated on the plot. The gray lines and numbers indicate the absolute latitude as a fitted smooth surface on the ordination, and the red arrows are fitted seafloor (temp.: temperature; diss. O2: dissolved oxygen; seabed salinity, silicate and nitrate concentrations, and POC reaching the seafloor) or surface water (primary prod.: primary productivity, POC export from the surface) environmental parameters to the ordination. Colors and symbols indicate location of sampling sites in the abyssal biogeographic provinces (AB1 to AB13) postulated by (42). (B) Proportion of shared benthic ASVs as a function of increasing distance between pairs of samples. The proportion of shared ASVs was computed only within the same oceanic basins. (C) Variation in b diversity, i.e., distribution of sample distances to group centroids, as a function of increasing spatial sampling scale. Higher values indicate higher compositional variation. Orange dots and bars represent means and standard deviations (SDs), respectively.
The average proportion of shared ASVs between pairs of samples as a function of their geographical distance was relatively stable up to 10 km, after which it decreased steadily (fig. 3B), possibly indicating dispersal limitation among benthic taxa or shifts in environmental drivers. We calculated key distance-decay parameters for selected benthic groups (table S5), i.e., the rate of decrease in their community similarity with increasing spatial separation. The initial similarities (i.e., similarity at 1 km distance that provides a measure of the local presence/absence patchiness) indicate that benthic macrofaunal groups (molluscs and polychaetes) tend to have a stronger local turnover than meiofaunal or protistan groups (nematodes, foraminifera, and amoebae). The former also generally have a steeper distance-decay, as indicated by the greater slope of linear models and lower halving distances, i.e., the distance after which the initial similarity is halved (table S5). These results indicate that dispersal limitation or environmental filtering (or a combination of both) may be stronger for macrofaunal benthic organisms than for meiofaunal or microbial eukaryotes, although macrofaunal taxa are usually thought not to be limited in their dispersion, owing to their common planktonic larval phases (43).
We lastly compared spatial structures and distance-decay parameters for whole benthic eukaryotic communities with those for water column communities (table S5). Overall, benthic communities are more spatially structured (mantel: r = 0.454, P < 0.001) than pelagic communities (euphotic: r = 0.147, P < 0.001; aphotic: r = 0.228, P < 0.001) and have lower initial similarity, steeper distance-decay, and smaller halving distances (table S5 and fig. S9), consistent with previous findings for benthic compared to pelagic communities of bacteria in the world ocean (44). This was also shown by fitting neutral community assembly models to each realm dataset, indicating that taxonomic groups of the pelagic euphotic zone tend to be more geographically widespread, while benthic groups tend to be less widespread than expected by neutral models (fig. S10). The importance of benthic community patchiness at local scales is reinforced by the variation of b diversity as a function of increasing sampling scale (Fig. 3C). The compositional variations within single sediment cores (8 to 10 cm in diameter) and between sediment cores collected from the same multicorer deployment (30 cm to 1 m apart) were comparable to that between deployments at a single station. This confirms the high degree of deep benthic community variation at local scales observed before in the case of selected groups of macro- and meiofauna by both morphological (45) and DNA-based (24) studies.
Eukaryotic plankton DNA signature on the DOS
Our study provides the first DNA-based insight into the qualitative and semiquantitative importance of the eukaryotic plankton diversity reaching the DOS at a global scale and thus driving the biological transfer of atmospheric carbon to the seafloor. The taxonomic composition of the 6382 planktonic eukaryotes in the DOS is roughly similar to that in the pelagic euphotic and aphotic zones (Fig. 2). In terms of relative abundance, however, planktonic DNA reads from sediment samples are mainly distributed among diatoms (15.4%) and various groups of rhizarians (26.9%) but include relatively few copepods (1.3%) and dinoflagellates (4.7%), which numerically dominate the plankton in upper ocean layers (fig. S4 and table S3). Plankton DNA on the DOS also include abundant ASVs assigned to the diplonemids (2.3%) and fungi (4.2%) that are common in the aphotic zone, supporting previous findings that the DOS accumulates DNA from organisms occurring throughout the entire water column (28).
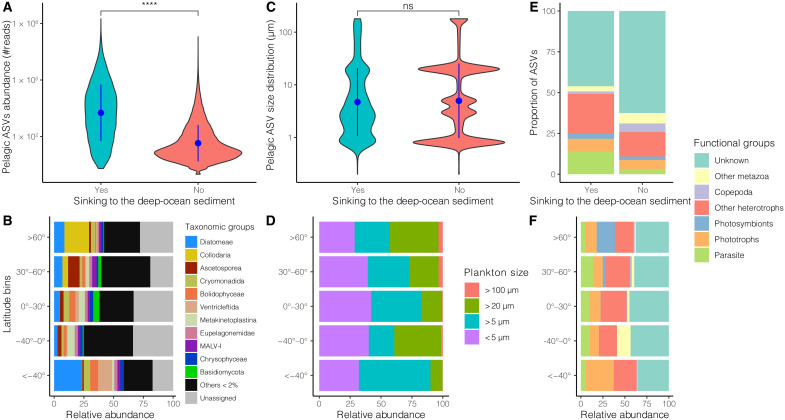
The pelagic ASVs reaching the DOS are generally among the most abundant planktonic eukaryotes in the water column (together representing 75.8% of the reads in the euphotic and 79.3% of the reads in the aphotic), although not all abundant pelagic ASVs are present in the DOS (Fig. 4A). We explored whether their occurrence in the DOS could be explained by their size distribution and by their trophic modes. We found no evidence that larger planktonic taxa are more likely to reach the DOS than smaller taxa (Fig. 4C), reinforcing the idea that most sinking plankton is transferred to the sediment through organic aggregates and not as individual organisms. However, the relative abundance of large planktonic taxa was higher in high-latitude sediments, especially in the Arctic (Fig. 4D), consistent with the trend of increasing sea-surface plankton size with increasing latitude and nutrient content (46, 47). Notably, the proportion of parasitic protists among sinking pelagic ASVs (13.7%) is greater than among nonsinking pelagic ASVs (2.9%; Fig. 4E), indicating that pelagic parasites are more likely to reach the DOS. Their relatively higher abundance in temperate and tropical latitude sediments suggests their ecological importance at these latitudes (Fig. 4F). Greater transfer of parasites to the DOS could reflect their ability to infect and kill larger hosts and/or the massive amounts of resistant and relatively dense propagules that they typically release after host infection and that could persist in sinking aggregates (48–51).
Fig. 4. Abundance and functional attributes of the sinking plankton compared to their nonsinking counterparts and the taxonomic and functional abundance breakdown of plankton DNA in DOSs.
Comparison of the DNA-based log-transformed read abundance (A), the inferred size distribution (C), and the functional breakdown (E) of pelagic ASV sinking to the DOS with their nonsinking counterparts in pelagic realms (euphotic and aphotic datasets have been combined). Results of Wilcoxon tests in (A) and (C) are indicated as follows: ****P < 0.0001; not significant (ns), P > 0.05. Breakdown of taxonomic (B), inferred size classes (D), and functional (F) relative abundances of plankton DNA in DOS as a function of latitudinal bins.
We aggregated our data for each entire realm to investigate whether the most abundant sinking pelagic ASVs in the water column are also the most abundant among sinking pelagic ASVs detected in the sediment, providing insight into their overall taphonomy (fig. S11). Similar abundance profiles would indicate that the structure of sinking plankton assemblages is overall preserved in surface sediment, whereas dissimilar profiles would indicate that sinking assemblages are consumed or repackaged during their downward transfer in a nonrandom manner. For instance, the abundance profiles of sinking pelagic ASVs of copepods, dinoflagellates, diatoms, acanthareans, eupelagonemids, hydrozoans, and spumellarians in both the euphotic and aphotic zones are similar to their abundance profiles in the DOS (fig. S11), suggesting that their structure in pelagic ecosystems is preserved on the seafloor. Their profiles in the DOS better mirrored those in the aphotic zone (higher R2 of linear models), suggesting that the transformation of sinking material occurs mostly in the upper oceanic layer. However, this was not the case for diatoms and collodarians, for which the abundance profiles in the euphotic and aphotic layer were similarly preserved in the DOS, likely because of their higher propensity to sink and form aggregates (52, 53). Although plankton DNA transfer to sediment has yet to be investigated [but see (54–56)], notably by accounting for physical processes (e.g., deep waters currents and vertical mixing) that interact with biological and ecological processes in the deposition of sinking material, our study reinforces the significance of the DOS as a DNA archive of upper-ocean biodiversity and ecology and a source of potential new proxies to document past environmental changes (57).
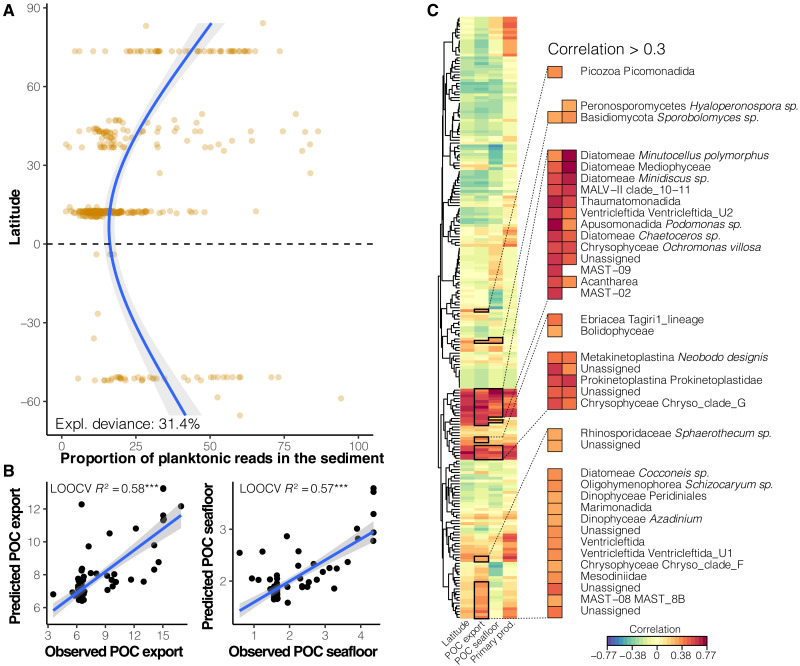
Last, we attempted to correlate our global DOS plankton biodiversity dataset to yearly average POC export from the surface and the fraction of it reaching the DOS, as estimated, respectively, by thorium-derived export measurements, modeled at a global scale (58), and by evaluating the efficiency of POC transfer through the water column based on sediment trap POC flux data, net primary production estimates, and sea surface temperatures (59). Overall, the pelagic ASVs detected in the DOS represent 21.4% of the DNA reads obtained from sediment samples. The proportion of DNA reads of pelagic origin in the sediment follows an increasing trend from low to high latitudes (Fig. 5A). This proportion also broadly approximates POC export from the surface (R2 = 0.23, P < 0.001), despite the higher remineralization rates at productive high latitudes (60). Furthermore, the composition of plankton DNA in the DOS can predict up to 58% of the variation in POC export from the surface and 57% of the POC reaching the seafloor using cross-validated random forest regressions (Fig. 5B). We used a multivariate regression method to identify the sinking pelagic ASVs that best explain the variation of POC export and POC reaching the seafloor (Fig. 5C). Not unexpectedly, diatoms and dinoflagellates (52, 61) were important contributors, but we also identified some previously overlooked taxa that are not usually considered to contribute to POC export, such as alveolate parasites (MALV-II), cercozoans, chrysophytes, and several unknown eukaryotes [see also (51)]. Our time-integrated data from the DOS therefore highlight previously underappreciated taxa that may be keystone drivers of the biological carbon pump.
Fig. 5. Plankton DNA proportion in the DOS as a function of latitude and planktonic taxa in sediment associated with POC export from the surface and its fraction reaching the seafloor.
(A) Proportion of DNA reads in DOSs representing sinking planktonic taxa as a function of latitude. The blue solid line represents a fitted generalized additive model (s = 3); the shades are displaying 95% confidence intervals. The explained variation of plankton DNA proportion in sediment is indicated on the plot. (B) Prediction of POC export and POC reaching the seafloor from the composition of plankton DNA in the DOS (POC export and POC seafloor are expressed in g Corg/m2 per year). Random forest regressions were used in a leave-one-out cross validation (LOOCV) approach at the station scale. Correlation coefficients of linear models are reported on the plots. Blue lines represent linear models (both with P < 0.001), and shades represent 95% confidence intervals. (C) Planktonic ASVs in sediment associated with the POC export and POC reaching the seafloor were identified with a sparse partial least square regression. Planktonic ASVs strongly associated (correlation > 0.3) with POC export and POC reaching the seafloor are detailed on the clustered heatmap (details in table S6).
Toward a holistic view of ocean biodiversity and ecosystem processes
Our global molecular meta-dataset from the ocean surface to the DOS provides the first unified vision of eukaryotic biodiversity patterns across the three dimensions of the world ocean (Fig. 1). It shows that the DOS is an extremely rich and unique realm with a strong connection to the water masses above that is reflected in the pelagic DNA signature (Figs. 1, 4, and 5 and fig. S11). Although focused on smaller-sized organisms (eukaryotic microbes and meiofauna), these DNA-based results are broadly consistent with morphological evidence from larger animals for high deep-sea benthic a diversities and small-scale patchiness (high local species turnover) (9–11).
The DOS appears to be much more diverse than oceanic waters (Fig. 1 and fig. S1) and is composed of communities of mostly unknown eukaryotes (Fig. 2) that display clear biogeographic patterns at global scales and considerable patchiness at local scales (Fig. 3). These patterns are likely driven by the flux of sinking organic aggregates and fecal pellets (Fig. 3, table S4, and fig. S8) (62, 63). Our data also show that the DNA-based plankton abundance profiles are broadly preserved in the DOS and that the transformation of sinking material appears to occur mostly between the euphotic and aphotic layers (fig. S11). The deposition of eukaryotic plankton is maximal at productive high latitudes (Fig. 5), and the plankton contribution to time-integrated sedimentary DNA broadly approximates the yearly average POC export from the surface. Moreover, the taxonomic composition of planktonic assemblages in the DOS is an even better predictor of the variation of POC export from the surface and the fraction of it reaching the seafloor (Fig. 5), indicating that biodiversity is key for ocean carbon export and burial. These DOS assemblages comprise not only taxa that are known to be important drivers of the biological carbon pump but also several taxonomic and functional groups that have been overlooked in what is arguably one of the most fundamental ecological processes of the world ocean.
Together, our results highlight the DOS as one of Earth’s richest modern ecosystems and fossil archives. They underline the need for concerted international efforts to further understand DOS biodiversity and its ecological role in planetary biogeochemical cycles. Our study, together with recent evidence that plankton DNA signal can be preserved in subseafloor sediments (54–57), paves the way for using sedimentary planktonic DNA to complement the microfossil-based proxies currently used to reconstruct ancient oceans, including their biological carbon sequestration processes. We hope it will also provide the basis for a more informed and effective stewardship strategy for protecting unique and relatively pristine deep-ocean ecosystems as the exploitation of seabed resources gathers pace.
MATERIALS AND METHODS
DOS sample collection
DOSs have been collected during two main projects (deep_sea and eDNAbyss). For the deep_sea project, sediment samples were collected at abyssal depths during eight expeditions to the Arctic, Atlantic, Southern, and Pacific Oceans (table S1). We used disposable sterile spoons to subsample the top surface sediment centimeter (c.a. 2 g from 0 to 1 cm) following a nested sampling design: up to three pseudo-replicates per core, up to two cores per deployment (multicorer), and up to three deployments per station (detailed list in table S7). Sediment samples were placed in sterile falcon tubes with (VEMA, SYSTCOII, KuramBio I, MANGAN’16, ABYSSLINE) and without (MSM39, DIVA3, and BIONOD) 6 ml of Lifeguard Preservation Solution (QIAGEN) before being frozen onboard at −20°C. The samples were shipped within −20°C containers to the University of Geneva (Switzerland). Upon arrival, sediment samples were stored at −80°C until extraction of nucleic acids.
For the eDNAbyss project, lower bathyal and abyssal sediments were collected in the Mediterranean Sea, North Atlantic, and Arctic Oceans during five cruises (Arctic: MarMine; North Atlantic: MEDWAVES; Mediterranean: PEACETIME, CANHROV, and ESSNAUT; table S1). For each station, triplicate cores (10 cm in diameter) were collected with a multicorer or with a remotely operated vehicle. Surface sediment (0 to 1 cm) was collected using metallic spatulas previously sterilized with bleach or DNA Exitus, rinsed with ethanol 96° and then nanopure water, and transferred into sterile zip-locked bags, homogenized by mixing and flattened to be stored at −80°C until DNA extraction. When possible, other layers (1 to 3, 3 to 5, 5 to 10, and 10 to 15 cm) were also collected from sediment cores. An empty zip lock bag from the stock used served as a blank sampling and extraction control in several stations along each cruise.
Nucleic acid extractions, PCR amplification, and illumina sequencing
For each of the 320 surface sediment samples collected in abyssal plains of the deep_sea project, we extracted the total RNA and DNA contents of c.a. 2 g of material as in (64), and we generated cDNA from deoxyribonuclease-treated RNA as in (65). We controlled that no carried-over DNA molecules remained in the RNA extracts based on the absence of PCR products after 60 cycles. We amplified by PCR the V9 hypervariable region of the ribosomal 18S gene with the following primer pair: the forward 1389F (5′-TTGTACACACCGCCC-3′) and the reverse 1510R (5′-CCTTCYGCAGGTTCACCTAC-3′) as designed in (66). Tag-encoded versions of the primers (a unique 8-nt sequence was added in the 5′ end of each primer) were used to multiplex up to 40 samples per sequencing library. Each sample was amplified in duplicate PCR reactions, and each PCR was performed in a total volume of 25 μl as follows: 19.4 μl of H2O, 2.5 μl buffer (FastStart, Roche), 0.5 μl of bovine serum albumin (20 mg/ml; Invitrogen Ultrapure), 0.5 μl of 10 mM dNTPs (deoxyribonucleotide triphosphate) (Roche), 0.1 μl of FastStart DNA Polymerase (5 U/μl; FastStart, Roche), 0.5 μl of forward and reverse primers at 10 mM, and lastly, 1 μl of DNA or RNA template (or 1.5 μl for some samples that did not amplify with 1 μl). All DNA and RNA samples were measured using the double-stranded DNA (dsDNA) High-Sensitivity Assay Kit and the RNA High-Sensitivity Assay Kit on the Qubit 4 fluorometer (Thermo Fisher Scientific) and diluted at 7 ng/μl prior PCR amplification. The PCR reaction conditions were as follows: predenaturation step at 94°C for 3 min, followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 57°C for 1 min, extension at 72°C for 1.5 min, and a final extension at 72°C for 2 min. A PCR-negative control for each unique combination of tag-encoded primers was verified by agarose gel electrophoresis. The two PCR replicates for each sample were combined and quantified using high-resolution capillary electrophoresis (QIAxcel System, QIAGEN). The PCR products were pooled in equimolar concentration within each multiplexed library. Each pool of PCR products was purified using a High Pure PCR Product Purification kit (Roche), following the manufacturer’s instructions. The sequencing libraries were prepared using the TruSeq DNA PCR-Free Library Preparation Kit (Illumina), following the manufacturer’s instructions. The libraries were quantified by quantitative PCR (qPCR) using the Kapa Library Quantification Kit for Illumina Platforms (Kapa Biosystems) and sequenced on a MiSeq instrument (Illumina) using paired-end sequencing for 300 cycles with kit v2.
Within the project eDNAbyss, DNA extractions were performed on about 10 g of sediment using the PowerMax Soil DNA Isolation Kit (QIAGEN, Hilden, Germany), following the manufacturer protocol, except for the last step where incubation of the elution buffer was prolonged 10 min on the spin filter membrane to increase the DNA yield. The first solution of the kit was poured into empty field control ziplock bags, before being extracted along with sediment samples, following the exact same protocol. All DNA extracts were then stored at −80°C (and transported to Genoscope on dry ice) until PCR amplifications. The V9 hypervariable region of the 18S ribosomal RNA (rRNA) gene was amplified by PCR using the same primer pair (1389F and 1510R). Each sample was amplified in triplicates, and each PCR reaction was performed in a total volume of 25 μl with the Phusion High-Fidelity PCR Master Mix with GC buffer (Thermo Fisher Scientific), 0.4 μM final concentration of each primer, 3% of dimethyl sulfoxide, 1× Phusion Master Mix, and 2.5 ng of template DNA (less for few extracts with very low DNA concentration). The PCR reaction conditions were as follows: predenaturation step at 98°C for 30 s, followed by 25 cycles of denaturation at 98°C for 10 s, annealing at 57°C for 30 s, extension at 72°C for 30 s, and a final extension at 72°C for 10 min. PCR products were purified using 1.8× AMPure XP beads cleanup (Beckmann Coulter Genomics). Aliquots of purified amplicons were then run on an Agilent Bioanalyzer using the DNA High Sensitivity LabChip kit to check their lengths and quantified with a Qubit Fluorometer to check their quality and concentration. Amplicons generated were then used for preparation of sequencing libraries. Amplicons (100 ng) were directly end-repaired, A-tailed, and ligated to Illumina adapters on a Biomek FX Laboratory Automation Workstation. Library amplification was then performed using a Kapa Hifi HotStart NGS library Amplification kit with the same cycling conditions applied for previous steps and cleaned up by AMPure XP purification (1 to 1 volume). All libraries were then quantified first by Quant-it dsDNA HS (high-sensitivity) assay using a Fluoroskan Ascent instrument (Thermo Fisher Scientific) and then by qPCR with the KAPA Library Quantification Kit for Illumina Libraries (Kapa Biosystems) on an MXPro instrument (Agilent Technologies). Library profiles were checked using high-throughput microfluidic capillary electrophoresis system (LabChip GX, PerkinElmer, Waltham, MA). Libraries were then normalized to 10 nM by addition of 10 mM tris-Cl (pH 8.5) and applied to cluster generation according to the Illumina Cbot User Guide (part no. 15006165). PhiX DNA spike-in was adapted for some libraries (20% instead of 1%) to minimize the loss of data due to low nucleotide diversity at the beginning of the sequencing run. Libraries were sequenced on HiSeq4000 or HiSeq2500 instruments (Illumina) on a paired-end mode. The raw sediment sequencing data have been deposited to the European Nucleotide Archive (ENA) under project accessions PRJEB33873 (eDNAbyss) and PRJEB48517 (deep_sea).
Public 18S-V9 rDNA sequencing datasets
We gathered published datasets (table S2) targeting the V9 hypervariable region of the eukaryotic 18S rRNA gene and using the same PCR primers pair (1389F and 1510R) used here for the DOS samples. These datasets were produced by studies sampling the euphotic/aphotic zones (33–35, 67–69) and the DOS (30, 67).
Environmental variables
Although some environmental variables and sediment descriptors were collected during the oceanic expeditions from which we collected sediment samples, their heterogeneity led us to extract more homogeneous environmental layers from the Global Marine Environment Datasets (http://gmed.auckland.ac.nz) to standardize our concatenated dataset across multiple studies. These variables included the surface calcite (calcite, in mole per cubic meter), surface nitrate (nitrate, in micromole per liter), surface silicate (silicate, in micromole per liter), surface phosphate (phosphate, in micromole per liter), average photosynthetically active radiation (PAR_mean, in Einstein per square meter per day), surface pH, average sea surface temperature (sst_mean, in celcius), variation in sea surface temperature (sst_range, in celsius), average surface currents strength (srf_current, in meter per second), primary production (primprod, in mgC·m2/day/cell), average stock of particulate inorganic carbon (PIC_mean, in mole per cubic meter), average stock of POC (POC_mean, in mole per cubic meter), total suspended matter (tsm_mean, in grams per cubic meter), seabed slope (slope, degree), seabed nitrate (sb_nitrate, in micromole per liter), seabed silicate (sb_silicate, in micromole per liter), seabed-dissolved oxygen (sb_o2dissolve, in milliliter per liter), seabed-utilized oxygen (sb_o2utilized, in milliliter per liter), seabed temperature (sb_temp, in celsius), seabed salinity [sb_salinity, practical salinity scale (PSS)], and average temperature in the water column (wat_col_temp, in celsius). We also extracted the estimated POC export at 100 m depth below the surface (POC_export, g Corg/m2 per year) (58) and the POC fraction reaching the seafloor (POC_seafloor, g Corg/m2 per year) (59, 70). The values of each environmental variable for each sample analyzed in this study were extracted from the environmental layers with their Global Positioning System (GPS) coordinates (table S7).
Raw sequencing data processing
For the deep_sea dataset, the sequencing libraries were demultiplexed using Double Tag Demultiplexer (DTD) software (https://github.com/yoann-dufresne/DoubleTagDemultiplexer) to screen the R1 and R2 files of each library and retrieve unique tag-encoded primer combinations associated to each sample (allowing no mismatches). We thus produced pairs of fastq files for each sample. For all other illumina datasets (see table S2), we obtained at least one pair of fastq files (paired-end) per sample (some samples were sequenced several times to obtain enough reads that were subsequently merged before statistical analysis). For the datasets produced with the 454 sequencing technology, we obtained one fastq file per sample. We used two R scripts (for paired-end illumina datasets and for 454 dataset, see the “rds_pipeline_illumina.R” and “rds_pipeline_454.R” scripts) to process all the fastq files by batch of 10 samples per job on a High-Performance Computing cluster (Baobab, University of Geneva). The R scripts implemented the key steps of the DADA2 workflow (71) and additional quality filtering steps (see below). The scripts performed the quality filtering with the filterAndTrim function of the DADA2 v1.12.1 R package with default settings, the trimming of primers using the cutadapt v2.4 software (72), the filtering of any read that still contain traces of primers (fastqFilterPrimersMatchs function in the fastqUtils.R script), the filtering of any read below 20 bp (fastqFilterWidth function in the fastqUtils.R script), the training of errors models using the learnErrors function of DADA2 with default settings, the inference of ASVs using the dada function with default settings (but for the 454 data, for which we used the HOMOPOLYMER_GAP_PENALTY = −1, BAND_SIZE = 32 options, as recommended by the DADA2 package developing team), and the merging for the overlapping paired-end reads using the mergePairs function with the option “trimOverhang.” Last, we exported the output of the DADA2 workflow, i.e., the “.rds” files that contain all the ASV sequences and their counts for each sample. We also collected summary statistics on each processing step, the trained errors models, and processing time for each sample.
Combining the datasets into a single ASV-to-sample table, taxonomic and functional annotations, matrix curation
We reimported the rds files into R to build an ASV-to-sample table. We first produced an ASV table per dataset, filtered chimeric ASVs with the option “consensus” within each dataset, and aggregated the replicated libraries per biological sample for the deep_sea, tara, and tara_polar datasets. We also aggregated the reads obtained from DNA and RNA libraries generated for each deep_sea sediment samples, since a comparative analysis revealed that the diversity and biogeographic patterns of eukaryotic communities are mostly similar between DNA and RNA [fig. S12; in line with (29, 73)]. We filtered the ASVs detected in the negative controls of the eDNAbyss dataset across the eDNAbyss ASV table. Last, we concatenated each dataset-based ASV-to-sample table into a single one and exported all the ASV sequences into a fasta file for taxonomic annotations. We used the “assignment-fasta-vsearch” module of the SLIM v0.6 software (74) that wraps the vsearch v2.2.2 software (75). The ASVs were compared to a custom version of PR2 (33) that focus on the 18S V9 region and that include functional annotations (available at https://doi.org/10.5281/zenodo.3768950) and with the SILVA v138 database (76). Taxonomic annotations were the consensus among up to three candidate reference sequences that are above 85% similarity with the query or directly assigned to the reference sequence if the query had a similarity of at least 99%. We also performed another search without restricting a minimum similarity threshold to match an entry reference sequence in the custom V9 version of PR2, to identify non-18S V9 sequences. We focused our analysis on the eukaryotic diversity by discarding any prokaryotic, plastidic ASVs, or any other artifactual ASV. We used taxonomic annotations obtained with the SILVA database to discard prokaryotic ASVs and the annotations obtained with PR2 to discard organelle-derived ASVs. All ASVs that only loosely match any V9 reference sequence (i.e., <20% similarity) were considered as non-18S V9 sequence and were discarded. We also filtered ASVs that did not contain the “GTCG” motif in the first four nucleotides in the 5′ end. This motif is widely conserved across eukaryotes, whereas prokaryotes have a “GTCA” motif highly conserved in those positions. We lastly used the length distribution of eukaryotic and prokaryotic ASVs to discard any possible prokaryotic unassigned ASVs (filter set at 116 bp; fig. S13). For downstream taxonomic analyses, we used PR2-based annotations using the 85% minimum similarity threshold. We also inferred the trophic mode of pelagic ASVs (phototrophic/photosymbiotic/parasitic/heterotrophic protists and zooplankton/other metazoan) based on the matching candidate reference sequences in PR2 (up to three candidates per queried ASV). We ascribed our ASVs to functional groups only if the functional attributes across candidates above 95% similarity were unambiguous, i.e., all the candidates for taxonomic assignment share similar functional attributes. We also used the different size fractions of the Tara Oceans samples to infer the size of pelagic ASVs, by using a weighted average of relative abundances across the size fractions of plankton samples (using the lowest mesh size, e.g., from the 20- to 180-μm size fraction, we used 20 μm in our calculation for this size fraction).
Classifying eukaryotic ASVs into pelagic or benthic taxa
We considered the ASVs being detected in pelagic samples as planktonic (or nektic), the ASVs detected exclusively in sediment samples as benthic, and the ASVs detected in both pelagic and sediment datasets as sinking plankton [although 29 vertebrates ASVs, comprising most of the nekton, represented ~1.29% of the sequences detected in both pelagic and sediment samples (table S3), we hereafter refer only to sinking plankton]. However, because multiple benthic groups have meroplanktonic larvae and hence could be detected in pelagic samples, we manually curated the ASVs assigned to metazoans within the sinking plankton fraction, based on their known lifestyles. This was, for instance, the case for some polychaetes, molluscs, echinoderms, or harpacticoid copepods that were “forced” into benthic diversity but not for pteropods that were left in the sinking plankton. Of the 546 metazoan ASVs in the sinking plankton fraction, 224 were curated as benthic (see table S8 for the details of this manual curation).
Eukaryotic community diversity and structural analysis
For a and b diversity analyses, we used functions of the vegan R package v2.5-3 (77), unless specified differently. Because the size fractionation of the pelagic samples from the Tara Oceans datasets has a strong effect on a and b diversity measures, we compared eukaryotic diversity patterns across pelagic and benthic realms by considering only the richest nano- (3 to 20 μm) and pico- (0.2 to 5 μm) size fractions of pelagic samples. The eukaryotic ASV accumulation curves as a function of sampling effort were computed with the specaccum function with the “random” method. We calculated the Shannon diversity for each sample and compared the distribution of sample diversity across both pelagic euphotic and aphotic with the strictly benthic diversity using the stat_compare_means function of the ggpubr R package v0.2.5 (78) with default settings. For b diversity analysis, we removed samples with less than 1000 reads and discarded ASVs represented by less than 100 reads throughout the dataset. We then normalized the ASV-to-sample matrix with the cumulative sum scaling (CSS) method (79) and computed a Bray-Curtis dissimilarity matrix between pairs of samples. The dissimilarity matrix was used to perform a nonmetric multidimensional scaling (NMDS) ordination on two axes. Sampling depth and absolute latitude variables were fit to the NMDS as smooth surfaces using the ordisurf function. The dissimilarity matrix was also used as input of the adonis function for PERMANOVA models testing for differences between eukaryotic compositional structure between realms (pelagic euphotic, pelagic aphotic, and sediment) and along a gradient of absolute latitude (nested in type of realm and restricting permutations within type of realm with the “strata” option), using 999 permutations. Last, we measured the b diversity dispersion within each realm using the betadisper function and compared the distances distribution to group centroids between realms using the stat_compare_means function of the ggpubr R package.
For a diversity and b diversity analyses of the deep-ocean benthic communities, we focused on oceanic samples only, i.e., we did not consider the samples from the Mediterranean Sea nor the ones from the Gulf of California, to avoid potential effects from coastal ecosystems. We calculated the normalized ASV richness per sample for the overall benthic communities and for selected benthic groups (nematodes, foraminifera, platyhelminths, polychaetes, molluscs, and ciliates) by rarefying each benthic sample at the lowest remaining sequencing depth (after removing planktonic ASVs and after focusing on a given benthic taxonomic group). We used generalized additive models (GAMs) to investigate the possible nonlinear variation of richness and Shannon diversity along gradients of latitude, primary production, and POC export from the surface and reaching the seafloor using the gam function of the mgcv R package (https://cran.r-project.org/web/packages/mgcv/) and the smoothing parameter set to 3. For b diversity analyses, we used a similar approach than detailed above (CSS-normalized and Bray-Curtis dissimilarity matrix), although here, we did not filter rare ASVs. We used the pcoa function of the ape R package (80) to perform a principal coordinate analysis of the Bray-Curtis dissimilarity matrix and calculate the structural variation explained by the first two axis of the ordination. We used the ordisurf and envfit functions to respectively fit the absolute latitude and a selection of environmental variables (seabed variables: salinity, temperature, silicate, nitrate, dissolved oxygen, POC reaching the seafloor, and pelagic variables that connect the surface to the DOS, namely, the primary productivity and the POC export from the surface) to the ordination. PERMANOVA models were used to test for differences in benthic composition between abyssal postulated biogeographic provinces (42) and along a gradient of absolute latitude using 999 permutations. Then, we used the selected environmental variables in a stepwise model building for constrained ordination (distance-based redundancy analysis) using the ordi2step function in a forward direction and using 999 permutations, to explain the observed benthic community structure. We calculated the proportion of shared ASVs between pairs of benthic samples to investigate the decrease of shared ASV proportion as a function of increasing spatial distance (calculated from GPS coordinates, see the “companionFunctions.R” script). We also calculated key distance-decay parameters as in (81), i.e., the initial similarities (Sørensen similarities between pairs of samples distant to each other by less than a kilometer), the slope of distance-decay relationship (here in a log-linear regression form), and the halving distances, i.e., the spatial distance after which the initial similarities are halved. We calculated these parameters on an average Sørensen dissimilarity matrix calculated over 10 rarefaction draws at the minimum sequencing depth possible (the sequencing depth of the sample with the lowest number of reads) and by considering the full benthic community or by focusing on selected benthic groups only, e.g., polychaetes, molluscs, or platyhelminths (macrofaunal size classes); nematodes or foraminifera (meiofauna); and amoebae or ciliates (microbes). We used the mantel function to test for correlation between spatial distance and community dissimilarities using 999 permutations. We used the betadisper function to calculate the b dispersion of benthic communities at increasing sampling spatial scale (between replicates samples of a sediment core, between cores of the same deployment, between deployments at a given station, and within a given abyssal basin). Last, we aggregated all samples at the station scale and fitted neutral community assembly models as in (82) to investigate whether the distribution of ASVs within the pelagic and benthic realms are less or more geographically widespread than expected by neutral models.
We compared the inferred functional attributes (size and trophic mode) of sinking pelagic ASVs with their nonsinking counterparts to explore whether these traits could explain their transfer to the DOS. We also explored the variation of functional groups and size classes of the sinking planktonic communities in the sediment along the gradient of latitude. We investigated the spatial pattern of planktonic abundance on the seafloor by fitting a GAM on the proportion of planktonic DNA reads in the sediment as function of latitude (with smoothing parameter set to 3). Then, we aggregated the planktonic DNA reads of all sediment samples at the station scale and used random forest models to predict the POC export from the surface and the POC reaching the seafloor in a leave-one-out cross-validation approach. We used the ranger function of the ranger R package (83) in a regression mode, growing 300 trees and setting the “mtry” parameter at one-third of the total number of features (number of sinking pelagic ASVs). Linear models were used to measure the performance of predictive models. Last, we used a sparse partial least square regression [mixOmics R package (84)] to identify the pelagic ASVs detected in the sediment that are best correlated with the variation of POC export and POC reaching the seafloor and with primary productivity and latitudes. We then focused on the pelagic ASVs that were reported with a correlation coefficient above 0.3 with POC export and POC reaching the seafloor and presented them in a clustered heatmap.
Acknowledgments
We thank the captains and crews of all 15 expeditions on RV Polarstern (SYSTCOII, ARK XXIV, and ANDEEP II), RV Sonne (KuramBio I, Vema-TRANSIT), RV Maria S. Merian (MSM39), RV Meteor (DIVA3), RV Kilo Moana (MANGAN-16), RV Thompson (ABYSSLINE2), RV L'Atalante (BIONOD), F. Pradillon (ESSNAUT), RV “Pourquoi pas?,” C. Guieu and C. Tamburini (PEACETIME), RV Sarmiento de Gamboa (MEDWAVES), RV L’Europe and M.-C. Fabri (CANHROV), RV Polar King and E. Ramirez-Llodra (MarMine), the Carbon Group at Bjerknes Centre for Climate Research and the Tara Ocean Foundation and its partners. Samples from the KM16 license area were made available for the investigations by C. Rühlemann and A. Vink from the Federal Institute for Geosciences and Raw Materials (BGR) in Hanover. The computations were performed at the University of Geneva on the Baobab cluster. This article is contribution 125 of Tara Oceans.
Funding: We further thank the following sponsors for support: the Swiss National Science Foundation (grants 31003A_159709, 31003A_1791259, and P2GEP3_171829), the Swiss Network for International Studies award (20170024), the European Research Council (grant 818449 AGENSI), CNRS (in particular, FR022), Sorbonne University, the French Government “Investissement d’Avenir” program OCEANOMICS (ANR-11-BTBR-0008), France Génomique, the Genoscope-CEA (ANR-10-INBS-09) for the project eDNAbyss (AP2016-228), Ifremer for the project Merlin “Pourquoi pas les Abysses,” the German Research Foundation (grants BR1121/20-1 and BR1121/41-1 and the Center/Cluster of Excellence “The Ocean Floor—Earth’s Uncharted Interface”), the German Ministry for Science and Education (grants 03G0223A and 03G0227A), EU JPIO-Oceans project MinigImpact-2 (German BMBF contract 03F0812E), Spanish Ministry of Economy and Competitiveness (CTM2016-75083-R), European Union’s Horizon 2020 Research and Innovation Program (grant 678760 ATLAS), and the Gordon and Betty Moore Foundation (grant GBMF5257 UniEuk). Samples from the UK-1 and OMS license areas in the Clarion-Clipperton Zone were collected as part of the ABYSSLINE project, funded by UK Seabed Resources Development Ltd. (contract SRDL SRD100100).
Author contributions: T.C., C.d.V., and J.Pa. conceived the study. S.A.-H., A.B., M.-A.C.-B., P.M.A., R.Mo., C.O., and C.R.S. organized deep-sea expeditions and provided sediment material or raw sediment sequencing data. I.B.A., F.L., T.C., S.A.-H., J.Po., and P.W. prepared sediment samples and produced data. T.C. performed the data analysis. I.B.A., N.H., F.L., C.B., L.G., F.L., P.M.A., R.Ma., C.R.S., C.d.V., A.J.G., and J.Pa. contributed to improve the data analysis. T.C., A.J.G., C.d.V., and J.Pa. led the writing of the manuscript. All authors contributed to the writing of the manuscript and approved the final version.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: The raw sediment sequencing data have been deposited to the ENA under project accessions PRJEB48517 (deep_sea) and PRJEB33873 (eDNAbyss). Accession numbers of additional raw sequencing data of pelagic and sediment samples analyzed in this study can be found in respective publications listed in table S2. All the metadata and R code to process the raw sequencing data and to reproduce the results and figures are available through a GitHub repository: https://github.com/trtcrd/DOS_V9.
Supplementary Materials
This PDF file includes:
Figs. S1 to S13
Tables S1 and S4
Legends for tables S2, S3, S5 to S8
References
Other Supplementary Material for this manuscript includes the following:
Tables S2, S3, S5 to S8
REFERENCES AND NOTES
- 1.Thurber A. R., Sweetman A. K., Narayanaswamy B. E., Jones D. O. B., Ingels J., Hansman R. L., Ecosystem function and services provided by the deep sea. Biogeosciences 11, 3941–3963 (2014). [Google Scholar]
- 2.Levin L. A., Le Bris N., The deep ocean under climate change. Science 350, 766–768 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Breitburg D., Levin L. A., Oschlies A., Grégoire M., Chavez F. P., Conley D. J., Garçon V., Gilbert D., Gutiérrez D., Isensee K., Jacinto G. S., Limburg K. E., Montes I., Naqvi S. W. A., Pitcher G. C., Rabalais N. N., Roman M. R., Rose K. A., Seibel B. A., Telszewski M., Yasuhara M., Zhang J., Declining oxygen in the global ocean and coastal waters. Science 359, eaam7240 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Smith C. R., Tunnicliffe V., Colaço A., Drazen J. C., Gollner S., Levin L. A., Mestre N. C., Metaxas A., Molodtsova T. N., Morato T., Sweetman A. K., Washburn T., Amon D. J., Deep-sea misconceptions cause underestimation of seabed-mining impacts. Trends Ecol. Evol. 35, 853–857 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Pusceddu A., Bianchelli S., Martín J., Puig P., Palanques A., Masqué P., Danovaro R., Chronic and intensive bottom trawling impairs deep-sea biodiversity and ecosystem functioning. Proc. Natl. Acad. Sci. U.S.A. 111, 8861–8866 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barbier E. B., Moreno-Mateos D., Rogers A. D., Aronson J., Pendleton L., Danovaro R., Henry L. A., Morato T., Ardron J., Van Dover C. L., Protect the deep sea. Nature 505, 475–477 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Mengerink K. J., Van Dover C. L., Ardron J., Baker M., Escobar-Briones E., Gjerde K., Koslow J. A., Ramirez-Llodra E., Lara-Lopez A., Squires D., Sutton T., Sweetman A. K., Levin L. A., A call for deep-ocean stewardship. Science 344, 696–698 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Levin L. A., Amon D. J., Lily H., Challenges to the sustainability of deep-seabed mining. Nat. Sustain. 3, 784–794 (2020). [Google Scholar]
- 9.M. A. Rex, R. J. Etter, Deep-Sea Biodiversity (Harvard Univ. Press, 2010). [Google Scholar]
- 10.Grassle J. F., Morse-Porteous L. S., Macrofaunal colonization of disturbed deep-sea environments and the structure of deep-sea benthic communities. Deep Sea Res. Part A 34, 1911–1915 (1987). [Google Scholar]
- 11.McClain C. R., Nekola J. C., Kuhnz L., Barry J. P., Local-scale faunal turnover on the deep Pacific seafloor. Mar. Ecol. Prog. Ser. 422, 193–200 (2011). [Google Scholar]
- 12.Danovaro R., Fanelli E., Aguzzi J., Billett D., Carugati L., Corinaldesi C., Dell’Anno A., Gjerde K., Jamieson A. J., Kark S., McClain C., Levin L., Levin N., Ramirez-Llodra E., Ruhl H., Smith C. R., Snelgrove P. V. R., Thomsen L., Van Dover C. L., Yasuhara M., Ecological variables for developing a global deep-ocean monitoring and conservation strategy. Nat. Ecol. Evol. 4, 181–192 (2020). [DOI] [PubMed] [Google Scholar]
- 13.Gooday A. J., Schoenle A., Dolan J. R., Arndt H., Protist diversity and function in the dark ocean – Challenging the paradigms of deep-sea ecology with special emphasis on foraminiferans and naked protists. Eur. J. Protistol. 75, 125721 (2020). [DOI] [PubMed] [Google Scholar]
- 14.Ingels J., Vanreusel A., Pape E., Pasotti F., Macheriotou L., Arbizu P. M., Sørensen M. V., Edgcomb V. P., Sharma J., Sánchez N., Homoky W. B., Woulds C., Leduc D., Gooday A. J., Pawlowski J., Dolan J. R., Schratzberger M., Gollner S., Schoenle A., Arndt H., Zeppilli D., Ecological variables for deep-ocean monitoring must include microbiota and meiofauna for effective conservation. Nat. Ecol. Evol. 5, 27–29 (2021). [DOI] [PubMed] [Google Scholar]
- 15.Billett D. S. M., Lampitt R. S., Rice A. L., Mantoura R. F. C., Seasonal sedimentation of phytoplankton to the deep-sea benthos. Nature 302, 520–522 (1983). [Google Scholar]
- 16.Thiel H., Pfannkuche O., Schriever G., Lochte K., Gooday A. J., Hemleben C., Mantoura R. F. G., Turley C. M., Patching J. W., Riemann F., Phytodetritus on the deep-sea floor in a central oceanic region of the northeast Atlantic. Biol. Oceanogr. 6, 203–239 (1988). [Google Scholar]
- 17.Silver M. W., Gowing M. M., The “particle” flux: Origins and biological components. Prog. Oceanogr. 26, 75–113 (1991). [Google Scholar]
- 18.Zheng X., Liu W., Dai X., Zhu Y., Wang J., Zhu Y., Zheng H., Huang Y., Dong Z., Du W., Zhao F., Huang L., Extraordinary diversity of viruses in deep-sea sediments as revealed by metagenomics without prior virion separation. Environ. Microbiol. 23, 728–743 (2020). [DOI] [PubMed] [Google Scholar]
- 19.Danovaro R., Molari M., Corinaldesi C., Dell’Anno A., Macroecological drivers of archaea and bacteria in benthic deep-sea ecosystems. Sci. Adv. 2, e1500961 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bienhold C., Zinger L., Boetius A., Ramette A., Diversity and biogeography of bathyal and abyssal seafloor bacteria. PLOS ONE 11, e0148016 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shulse C. N., Maillot B., Smith C. R., Church M. J., Polymetallic nodules, sediments, and deep waters in the equatorial North Pacific exhibit highly diverse and distinct bacterial, archaeal, and microeukaryotic communities. Microbiology 6, e00428 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoshino T., Doi H., Uramoto G. I., Wörmer L., Adhikari R. R., Xiao N., Morono Y., D’Hondt S., Hinrichs K. U., Inagaki F., Global diversity of microbial communities in marine sediment. Proc. Natl. Acad. Sci. U.S.A. 117, 27587–27597 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lecroq B., Lejzerowicz F., Bachar D., Christen R., Esling P., Baerlocher L., Osterås M., Farinelli L., Pawlowski J., Ultra-deep sequencing of foraminiferal microbarcodes unveils hidden richness of early monothalamous lineages in deep-sea sediments. Proc. Natl. Acad. Sci. U.S.A. 108, 13177–13182 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lejzerowicz F., Esling P., Pawlowski J., Patchiness of deep-sea benthic Foraminifera across the Southern Ocean: Insights from high-throughput DNA sequencing. Deep Sea Res. II Top. Stud. Oceanogr. 108, 17–26 (2014). [Google Scholar]
- 25.Sinniger F., Pawlowski J., Harii S., Gooday A. J., Yamamoto H., Chevaldonné P., Cedhagen T., Carvalho G., Creer S., Worldwide analysis of sedimentary DNA reveals major gaps in taxonomic knowledge of deep-sea benthos. Front. Mar. Sci. 3, 1–14 (2016). [Google Scholar]
- 26.Danovaro R., Company J. B., Corinaldesi C., D’Onghia G., Galil B., Gambi C., Gooday A. J., Lampadariou N., Luna G. M., Morigi C., Olu K., Polymenakou P., Ramirez-Llodra E., Sabbatini A., Sardá F., Sibuet M., Tselepides A., Deep-sea biodiversity in the Mediterranean Sea: The known, the unknown, and the unknowable. PLOS ONE 5, e11832 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fonseca V. G., Sinniger F., Gaspar J. M., Quince C., Creer S., Power D. M., Peck L. S., Clark M. S., Revealing higher than expected meiofaunal diversity in Antarctic sediments: A metabarcoding approach. Sci. Rep. 7, 6094 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laroche O., Kersten O., Smith C. R., Goetze E., From sea surface to seafloor: A benthic allochthonous eDNA survey for the abyssal ocean. Front. Mar. Sci. 7, 1–16 (2020).32802822 [Google Scholar]
- 29.Lejzerowicz F., Gooday A. J., Angeles I. B., Cordier T., Morard R., Apothéloz-perret-gentil L., Lins L., Eukaryotic biodiversity and spatial patterns in the Clarion-Clipperton Zone and other abyssal regions: Insights from sediment DNA and RNA metabarcoding. Front. Mar. Sci. 8, 1–23 (2021).35685121 [Google Scholar]
- 30.Pawlowski J., Christen R., Lecroq B., Bachar D., Shahbazkia H. R., Amaral-Zettler L., Guillou L., Eukaryotic richness in the abyss: Insights from pyrotag sequencing. PLOS ONE 6, e18169 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lindh M. V., Maillot B. M., Shulse C. N., Gooday A. J., Amon D. J., Smith C. R., Church M. J., From the surface to the deep-sea: Bacterial distributions across polymetallic nodule fields in the Clarion-Clipperton Zone of the Pacific Ocean. Front. Microbiol. 8, 1696 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morard R., Lejzerowicz F., Darling K. F., Lecroq-Bennet B., Pedersen M. W., Orlando L., Pawlowski J., Mulitza S., De Vargas C., Kucera M., Planktonic foraminifera-derived environmental DNA extracted from abyssal sediments preserves patterns of plankton macroecology. Biogeosciences 14, 2741–2754 (2017). [Google Scholar]
- 33.de Vargas C., Audic S., Henry N., Decelle J., Mahe F., Logares R., Lara E., Berney C., Le Bescot N., Probert I., Carmichael M., Poulain J., Romac S., Colin S., Aury J.-M., Bittner L., Chaffron S., Dunthorn M., Engelen S., Flegontova O., Guidi L., Horak A., Jaillon O., Lima-Mendez G., Luke J., Malviya S., Morard R., Mulot M., Scalco E., Siano R., Vincent F., Zingone A., Dimier C., Picheral M., Searson S., Kandels-Lewis S., Acinas S. G., Bork P., Bowler C., Gorsky G., Grimsley N., Hingamp P., Iudicone D., Not F., Ogata H., Pesant S., Raes J., Sieracki M. E., Speich S., Stemmann L., Sunagawa S., Weissenbach J., Wincker P., Karsenti E., Boss E., Follows M., Karp-Boss L., Krzic U., Reynaud E. G., Sardet C., Sullivan M. B., Velayoudon D., Eukaryotic plankton diversity in the sunlit ocean. Science 348, 1261605 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Ibarbalz F. M., Henry N., Brandão M. C., Martini S., Busseni G., Byrne H., Coelho L. P., Endo H., Gasol J. M., Gregory A. C., Mahé F., Rigonato J., Royo-Llonch M., Salazar G., Sanz-Sáez I., Scalco E., Soviadan D., Zayed A. A., Zingone A., Labadie K., Ferland J., Marec C., Kandels S., Picheral M., Dimier C., Poulain J., Pisarev S., Carmichael M., Pesant S., Babin M., Boss E., Iudicone D., Jaillon O., Acinas S. G., Ogata H., Pelletier E., Stemmann L., Sullivan M. B., Sunagawa S., Bopp L., de Vargas C., Karp-Boss L., Wincker P., Lombard F., Bowler C., Zinger L., Acinas S. G., Babin M., Bork P., Boss E., Bowler C., Cochrane G., de Vargas C., Follows M., Gorsky G., Grimsley N., Guidi L., Hingamp P., Iudicone D., Jaillon O., Kandels S., Karp-Boss L., Karsenti E., Not F., Ogata H., Pesant S., Poulton N., Raes J., Sardet C., Speich S., Stemmann L., Sullivan M. B., Sunagawa S., Wincker P., Lombard F., Bowler C., Zinger L., Global trends in marine plankton diversity across kingdoms of life. Cell 179, 1084–1097.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Obiol A., Giner C. R., Sánchez P., Duarte C. M., Acinas S. G., Massana R., A metagenomic assessment of microbial eukaryotic diversity in the global ocean. Mol. Ecol. Resour. 718–731 (2020). [DOI] [PubMed] [Google Scholar]
- 36.Pernice M. C., Giner C. R., Logares R., Perera-Bel J., Acinas S. G., Duarte C. M., Gasol J. M., Massana R., Large variability of bathypelagic microbial eukaryotic communities across the world’s oceans. ISME J. 10, 945–958 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Flegontova O., Flegontov P., Malviya S., Audic S., Wincker P., de Vargas C., Bowler C., Lukeš J., Horák A., Extreme diversity of diplonemid eukaryotes in the Ocean. Curr. Biol. 26, 3060–3065 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Morgan-Smith D., Clouse M. A., Herndl G. J., Bochdansky A. B., Diversity and distribution of microbial eukaryotes in the deep tropical and subtropical North Atlantic Ocean. Deep. Res. I Oceanogr. Res. Pap. 78, 58–69 (2013). [Google Scholar]
- 39.Lara E., Moreira D., Vereshchaka A., López-García P., Pan-oceanic distribution of new highly diverse clades of deep-sea diplonemids. Environ. Microbiol. 11, 47–55 (2009). [DOI] [PubMed] [Google Scholar]
- 40.Forster D., Lentendu G., Filker S., Dubois E., Wilding T. A., Stoeck T., Improving eDNA-based protist diversity assessments using networks of amplicon sequence variants. Environ. Microbiol. 21, 4109–4124 (2019). [DOI] [PubMed] [Google Scholar]
- 41.Zhao F., Filker S., Xu K., Li J., Zhou T., Huang P., Effects of intragenomic polymorphism in the SSU rRNA gene on estimating marine microeukaryotic diversity: A test for ciliates using single-cell high-throughput DNA sequencing. Limnol. Oceanogr. Methods 17, 533–543 (2019). [Google Scholar]
- 42.Watling L., Guinotte J., Clark M. R., Smith C. R., A proposed biogeography of the deep ocean floor. Prog. Oceanogr. 111, 91–112 (2013). [Google Scholar]
- 43.McClain C. R., Rex M. A., Toward a conceptual understanding of β-diversity in the deep-sea benthos. Annu. Rev. Ecol. Evol. Syst. 46, 623–642 (2015). [Google Scholar]
- 44.Zinger L., Amaral-Zettler L. A., Fuhrman J. A., Horner-Devine M. C., Huse S. M., Welch D. B. M., Martiny J. B. H., Sogin M., Boetius A., Ramette A., Global patterns of bacterial beta-diversity in seafloor and seawater ecosystems. PLOS ONE 6, e24570 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zeppilli D., Pusceddu A., Trincardi F., Danovaro R., Seafloor heterogeneity influences the biodiversity-ecosystem functioning relationships in the deep sea. Sci. Rep. 6, 26352 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Acevedo-Trejos E., Marañón E., Merico A., Phytoplankton size diversity and ecosystem function relationships across oceanic regions. Proc. R. Soc. B 285, 20180621 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barton A. D., Pershing A. J., Litchman E., Record N. R., Edwards K. F., Finkel Z. V., Kiørboe T., Ward B. A., The biogeography of marine plankton traits. Ecol. Lett. 16, 522–534 (2013). [DOI] [PubMed] [Google Scholar]
- 48.Boeuf D., Edwards B. R., Eppley J. M., Hu S. K., Poff K. E., Romano A. E., Caron D. A., Karl D. M., DeLong E. F., Biological composition and microbial dynamics of sinking particulate organic matter at abyssal depths in the oligotrophic open ocean. Proc. Natl. Acad. Sci. U.S.A. 116, 11824–11832 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Poff K. E., Leu A. O., Eppley J. M., Karl D. M., DeLong E. F., Microbial dynamics of elevated carbon flux in the open ocean’s abyss. Proc. Natl. Acad. Sci. U.S.A. 118, e2018269118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Preston C. M., Durkin C. A., Yamahara K. M., DNA metabarcoding reveals organisms contributing to particulate matter flux to abyssal depths in the North East Pacific ocean. Deep. Res. II Top. Stud. Oceanogr. 173, 104708 (2020). [Google Scholar]
- 51.Guidi L., Chaffron S., Bittner L., Eveillard D., Larhlimi A., Roux S., Darzi Y., Audic S., Berline L., Brum J. R., Coelho L. P., Espinoza J. C. I., Malviya S., Sunagawa S., Dimier C., Kandels-Lewis S., Picheral M., Poulain J., Searson S., Stemmann L., Not F., Hingamp P., Speich S., Follows M., Karp-Boss L., Boss E., Ogata H., Pesant S., Weissenbach J., Wincker P., Acinas S. G., Bork P., De Vargas C., Iudicone D., Sullivan M. B., Raes J., Karsenti E., Bowler C., Gorsky G., Plankton networks driving carbon export in the oligotrophic ocean. Nature 532, 465–470 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Agusti S., González-Gordillo J. I., Vaqué D., Estrada M., Cerezo M. I., Salazar G., Gasol J. M., Duarte C. M., Ubiquitous healthy diatoms in the deep sea confirm deep carbon injection by the biological pump. Nat. Commun. 6, 7608 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Biard T., Bigeard E., Audic S., Poulain J., Gutierrez-Rodriguez A., Pesant S., Stemmann L., Not F., Biogeography and diversity of Collodaria (Radiolaria) in the global ocean. ISME J. 11, 1331–1344 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barrenechea Angeles I., Lejzerowicz F., Cordier T., Scheplitz J., Mulitza S., Kucera M., Ariztegui D., Pawlowski J., Morard R., Planktonic foraminifera eDNA signature deposited on the seafloor remains preserved after burial in marine sediments. Sci. Rep. 20351, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Armbrecht L., Hallegraeff G., Bolch C. J. S., Woodward C., Cooper A., Hybridisation capture allows DNA damage analysis of ancient marine eukaryotes. Sci. Rep. 11, 3220 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kirkpatrick J. B., Walsh E. A., D’Hondt S., Fossil DNA persistence and decay in marine sediment over hundred-thousand-year to million-year time scales. Geology 44, 615–618 (2016). [Google Scholar]
- 57.De Schepper S., Ray J. L., Skaar K. S., Sadatzki H., Ijaz U. Z., Stein R., Larsen A., The potential of sedimentary ancient DNA for reconstructing past sea ice evolution. ISME J. 13, 2566–2577 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Henson S. A., Sanders R., Madsen E., Morris P. J., Le Moigne F., Quartly G. D., A reduced estimate of the strength of the ocean’s biological carbon pump. Geophys. Res. Lett. 38, L04606 (2011). [Google Scholar]
- 59.Lutz M. J., Caldeira K., Dunbar R. B., Behrenfeld M. J., Seasonal rhythms of net primary production and particulate organic carbon flux to depth describe the efficiency of biological pump in the global ocean. J. Geophys. Res. 112, C10011 (2007). [Google Scholar]
- 60.Guidi L., Legendre L., Reygondeau G., Uitz J., Stemmann L., Henson S. A., A new look at ocean carbon remineralization for estimating deepwater sequestration. Global Biogeochem. Cycles 29, 1044–1059 (2015). [Google Scholar]
- 61.Billett D. S. M., Lampitt R. S., Rice A. L., Mantoura R. C. F., Seasonal sedimentation of phytoplankton to the deep-sea benthos, 520–522. Nature 302, 520–522 (1983). [Google Scholar]
- 62.Steinberg D. K., Landry M. R., Zooplankton and the Ocean Carbon Cycle. Ann. Rev. Mar. Sci. 9, 413–444 (2017). [DOI] [PubMed] [Google Scholar]
- 63.Smith C. R., De Leo F. C., Bernardino A. F., Sweetman A. K., Arbizu P. M., Abyssal food limitation, ecosystem structure and climate change. Trends Ecol. Evol. 23, 518–528 (2008). [DOI] [PubMed] [Google Scholar]
- 64.Lejzerowicz F., Esling P., Pillet L., Wilding T. A., Black K. D., Pawlowski J., High-throughput sequencing and morphology perform equally well for benthic monitoring of marine ecosystems. Sci. Rep. 5, 13932 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pawlowski J., Esling P., Lejzerowicz F., Cedhagen T., Wilding T. A., Environmental monitoring through protist next-generation sequencing metabarcoding: Assessing the impact of fish farming on benthic foraminifera communities. Mol. Ecol. Resour. 14, 1129–1140 (2014). [DOI] [PubMed] [Google Scholar]
- 66.Amaral-Zettler L. A., McCliment E. A., Ducklow H. W., Huse S. M., A method for studying protistan diversity using massively parallel sequencing of V9 hypervariable regions of small-subunit ribosomal RNA Genes. PLOS ONE 4, e6372 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lie A. A. Y., Liu Z., Hu S. K., Jones A. C., Kim D. Y., Countway P. D., Amaral-Zettler L. A., Cary S. C., Sherr E. B., Sherr B. F., Gast R. J., Caron D. A., Investigating microbial eukaryotic diversity from a global census: Insights from a comparison of pyrotag and full-length sequences of 18S rRNA genes. Appl. Environ. Microbiol. 80, 4363–4373 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xu D., Li R., Hu C., Sun P., Jiao N., Warren A., Microbial eukaryote diversity and activity in the water column of the South China sea based on DNA and RNA high throughput sequencing. Front. Microbiol. 8, 1121 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.de Vargas C., Pollina T., Romac S., Le Bescot N., Henry N., Berger C., Colin S., Haëntjens N., Carmichael M., Le Guen D., Decelle J., Mahé F., Malpot E., Beaumont C., Hardy M., Guiffant D., Probert I., Gruber D. F., Allen A., Gorsky G., Follows M., Cael B. B., Pochon X., Troublé R., Lombard F., Boss E., Prakash M., Plankton Planet: ‘Seatizen’ oceanography to assess open ocean life at the planetary scale. bioRxiv , 2020.08.31.263442 (2020). [Google Scholar]
- 70.Sweetman A. K., Thurber A. R., Smith C. R., Levin L. A., Mora C., Wei C.-L., Gooday A. J., Jones D. O. B., Rex M., Yasuhara M., Ingels J., Ruhl H. A., Frieder C. A., Danovaro R., Würzberg L., Baco A., Grupe B. M., Pasulka A., Meyer K. S., Dunlop K. M., Henry L.-A., Roberts J. M., Major impacts of climate change on deep-sea benthic ecosystems. Elementa 5, 4 (2017). [Google Scholar]
- 71.Callahan B. J., McMurdie P. J., Rosen M. J., Han A. W., Johnson A. J. A., Holmes S. P., DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Martin M., Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10 (2011). [Google Scholar]
- 73.Guardiola M., Wangensteen O. S., Taberlet P., Coissac E., Uriz M. J., Turon X., Spatio-temporal monitoring of deep-sea communities using metabarcoding of sediment DNA and RNA. PeerJ. 4, e2807 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dufresne Y., Lejzerowicz F., Perret-Gentil L. A., Pawlowski J., Cordier T., SLIM: A flexible web application for the reproducible processing of environmental DNA metabarcoding data. BMC Bioinformatics 20, 88 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rognes T., Flouri T., Nichols B., Quince C., Mahé F., VSEARCH: A versatile open source tool for metagenomics. PeerJ. 4, e2584 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., Peplies J., Glöckner F. O., The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.J. Oksanen, F. G. Blanchet, M. Friendly, R. Kindt, P. Legendre, D. McGlinn, P. R. Minchin, R. B. O’Hara, G. L. Simpson, P. Solymos, M. H. H. Stevens, E. Szoecs, H. Wagner, vegan: Community Ecology Package. R package version 2.5–3 (2018), doi: 10.4135/9781412971874.n145. [DOI]
- 78.A. Kassambara, ggpubr: “ggplot2” Based Publication Ready Plots (2020).
- 79.Paulson J. N., Stine O. C., Bravo H. C., Pop M., Differential abundance analysis for microbial marker-gene surveys. Nat. Methods 10, 1200–1202 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Paradis E., Schliep K., ape 5.0: An environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 35, 526–528 (2019). [DOI] [PubMed] [Google Scholar]
- 81.Soininen J., McDonald R., Hillebrand H., The distance decay of similarity in ecological communities. Ecography 30, 3–12 (2007). [Google Scholar]
- 82.Sloan W. T., Woodcock S., Lunn M., Head I. M., Curtis T. P., Modeling taxa-abundance distributions in microbial communities using environmental sequence data. Microb. Ecol. 53, 443–455 (2007). [DOI] [PubMed] [Google Scholar]
- 83.Wright M. N., Ziegler A., Ranger: A fast implementation of random forests for high dimensional data in C++ and R. J. Stat. Softw. 77, 10.18637/jss.v077.i01, (2017). [Google Scholar]
- 84.Rohart F., Gautier B., Singh A., Lê Cao K. A., mixOmics: An R package for ‘omics feature selection and multiple data integration. PLOS Comput. Biol. 13, e1005752 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.S. Mulitza, F. Bergmann, L. Brück, C. M. Chiessi, A. Govin, M. Klann, H. Kuhnert, B. Lübben, L. Max, V. Meyer, R. Morard, V. Müller, G. Patton, A. Paul, A. Poirier, P. Riesen, T. Schade, U. Stöber, G. S. Völker, R. Völpel, T. von Dobeneck, Cruise No. MSM39 - June 07-June 25, 2014 - St. John’s (Canada) - St. John’s (Canada). In MARIA S. MERIAN-Berichte (2015); 10.2312/cr_msm39. [DOI]
- 86.C. W. Devey, Ed., RV SONNE Fahrtbericht / Cruise Report SO237 Vema-TRANSIT : bathymetry of the Vema-Fracture-Zone and Puerto Rico TRench and Abyssal AtlaNtic BiodiverSITy Study, Las Palmas (Spain) - Santo Domingo (Dom. Rep.) 14.12.14–26.01.15 (2015); 10.3289/GEOMAR_REP_NS_23_2015. [DOI]
- 87.D. Wolf-Gladrow, The expedition of the research vessel “Polarstern” to the Antarctic in 2012 (ANT-XXVIII/3). Berichte zur Polar- und Meeresforsch. = Reports polar Mar. Res. 661 (2013); http://hdl.handle.net/10013/epic.41332.
- 88.A. Brandt, M. Malyutina, I. Alalykina, B. N., C. A., N. Elsner, F. V., O. Golovan, G. Kamenev, V. Kharlamenko, A. Lavrenteva, F. Lejzerowicz, A. Maiorova, M. I., D. Miljutin, K. Minin, J. Packmor, K. Richter, T. Riehl, L. Schwabe, The German-Russian deep-sea expedition KuramBio (Kurile Kamchatka Biodiversity Study) to the Kurile Kamchatka Trench and abyssal plain on board of the R/V Sonne, 223rd Expedition (July 21th – September 7th 2012). Cruise report. (2012); 10.13140/RG.2.1.2473.6401. [DOI]
- 89.L. Menot, BIONOD cruise, RV L’Atalante (2012); 10.17600/12010030. [DOI]
- 90.M. Ludvigsen, K. Aasly, S. L. Ellefmo, A. Hilário, E. Ramirez-Llodra, F. X. Søreide, I. Falcon-Suarez, C. J. Juliani, A. Kieswetter, A. Lim, M. Christian, S. M. Nornes, H. Reimers, E. Paulsen, Ø. Sture, MarMine cruise report - Arctic Mid-Ocean Ridge 15.08.2016–05.09.2016 (2016); http://hdl.handle.net/11250/2427715.
- 91.C. Orejas, A. Addamo, M. Alvarez, A. Aparicio, D. Alcoverro, S. Arnaud-Haond, M. Bilan, J. Boavida, V. Cainzos, R. Calderon, P. Cambeiro, M. Castano, A. Fox, M. Gallardo, A. Gori, C. Guitierrez, L.-A. Henry, M. Hermida, J. A. Jimenez, J. L. Lopez-Jurado, P. Lozano, A. Mateo-Ramirez, G. Mateu, J. L. Matoso, C. Mendez, A. Morillas, J. Movilla, A. Olariaga, M. Paredes, V. Pelayo, S. Pineiro, M. Rakka, T. Ramirez, M. Ramos, J. Reis, J. Rivera, A. Romero, J. L. Rueda, T. Salvador, I. Sampaio, H. Sanchez, R. Santiago, A. Serrano, G. Taranto, J. Urra, P. Velez-Belchi, N. Viladrich, M. Zein, Cruise Summary Report - MEDWAVES survey (MEDiterranean out flow WAter and Vulnerable EcosystemS). (2017); 10.5281/zenodo.556516. [DOI]
- 92.C. Guieu, K. Desboeufs, PEACETIME cruise, RV Pourquoi pas ? (2017); 10.17600/17000300. [DOI]
- 93.M.-A. Cambon-Bonavita, ESSNAUT 2017 cruise, RV Pourquoi pas ? (2017); 10.17600/17009100. [DOI]
- 94.E. Raugel, CANHROV cruise, RV L’Europe, (2016); 10.17600/16012300. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S13
Tables S1 and S4
Legends for tables S2, S3, S5 to S8
References
Tables S2, S3, S5 to S8