Abstract
Two cyp51-related genes (cyp51A and cyp51B) encoding 14-α sterol demethylase-like enzymes were identified in the opportunistic human pathogen Aspergillus fumigatus. PCR amplification using degenerate oligonucleotides based on conserved areas of cytochrome P450 demethylases of other filamentous fungi and yeasts allowed the cloning and sequencing of two different homologue genes in A. fumigatus. Southern analysis confirmed that both genes hybridized to distinct genomic loci and that both are represented as single copies in the genome. Comparison of the deduced Cyp51A and Cyp51B proteins with the CYP51 proteins from Penicillium italicum, Aspergillus nidulans, Erysiphe graminis, Uncinula necator, Botrytis cinerea, Ustilago maydis, Cryptococcus neoformans, Candida albicans, Saccharomyces cerevisiae, Candida tropicalis, and Candida glabrata showed that the percentages of identity of the amino acid sequences (range, 40 to 70%) were high enough to consider Cyp51A and Cyp51B to be members of the fungal CYP51 family. Fragments from both genes were also cloned from other Aspergillus spp. (A. flavus, A. nidulans, and A. terreus). Phylogenetic analysis showed that, at least in the most pathogenic species of Aspergillus, there are two fungal CYP51 proteins. This is the first report of the existence of two homologue genes coding for 14-α sterol demethylase in the fungal kingdom. This finding could provide insights into the azole resistance mechanisms operating in fungi. The primers used here may be useful molecular tools for facilitating the cloning of novel 14-α sterol demethylase genes in other filamentous fungi.
Invasive aspergillosis is an increasingly common fungal infection and an important cause of morbidity and mortality in the immunocompromised host, especially in patients with acute leukemia, bone marrow or solid organ transplantation, or AIDS (23). Attributable mortality rates remain excessively high, despite treatment of affected patients with available antifungal agents, such as amphotericin B and itraconazole. About 85% of patients with invasive aspergillosis die (10). More than 100 species of Aspergillus have been described but only a few cause disease with any regularity (Aspergillus fumigatus, Aspergillus flavus, Aspergillus terreus, and Aspergillus niger). Of these, A. fumigatus is the most common fungus causing infection worldwide (23).
In spite of the fact that antifungal testing of filamentous fungi is in its infancy, the recent publication by the National Committee for Clinical Laboratory Standards (NCCLS) of a proposed standard has paved the way to understanding of the susceptibility of filamentous fungi to antifungal agents (26). Resistance to itraconazole, at least in Aspergillus spp., can now be readily detected by the NCCLS methodology and related methods (5, 6, 26). Resistant strains of A. fumigatus have been documented in the United Kingdom (11, 28), Sweden (4), and, more recently, in Spain (unpublished data). In addition, the high MICs obtained have been correlated with clinical outcomes and with animal models of fungal infection (12). In contrast, resistance to amphotericin B has not been detected in vitro, although clinical failures have been reported and seem to be related to the appearance of resistance (22). Recent failures in aspergillosis treatment, combined with improvements in performance and standardization of antifungal susceptibility testing, have drawn attention to the problem of antifungal resistance. Concomitantly, the use of antifungal drugs continues to expand as the immunocompromised population grows. The massive use of demethylation inhibitors (DMIs) in agriculture and the fact that fungi pathogenic for humans share ecological niches with phytopathogenic fungi also contribute to the emergence of resistance in molds. It is now clear that antifungal agents could, in the near future, create clinical and epidemiological situations similar to those found with antibiotic-resistant bacteria. Moreover, the emergence of strains resistant to azoles necessitates careful study of the molecular mechanisms implicated in this resistance, since most of the newer drugs are azole based. This is especially important with regard to voriconazole, a new triazole-derived agent which has shown potent and promising in vitro and in vivo activities (5, 6, 35).
The azoles inhibit the ergosterol biosynthesis pathway. Specifically, they inhibit the demethylation of precursor sterols at position 14 by sterol 14-α-demethylase (CYP51). This enzyme belongs to a superfamily of monooxygenases called cytochrome P450, members of which are involved in various biosynthetic functions (38). The chemical families that inhibit C-14 demethylation are the imidazoles and triazoles. Collectively, these compounds are called sterol DMIs, and they are widely used both clinically and agriculturally (37). The emergence of resistance to azoles in yeasts has accelerated studies of the mechanisms implicated in this resistance (20, 24, 32). However, the mechanisms of resistance to azoles in filamentous fungi are poorly understood. Although they have been studied in greater depth for some phytopathogenic fungi, the information about human-pathogenic fungi is very meager. In general, two classes of resistance mechanisms have been described up to now: altered affinity of CYP51 due to target site mutation (8, 9) and decreased accumulation of drugs due to enhanced energy-dependent drug efflux (7, 25, 33). Characterization of genes encoding CYP51 will contribute to better understanding of azole resistance mechanisms at the molecular level. The cyp51 genes from Saccharomyces cerevisiae, Candida tropicalis, Candida albicans, Candida glabrata, Cryptococcus neoformans, Ustilago maydis, Aspergillus nidulans, Botrytis cinerea, Penicillium italicum, Uncinula necator, and Erysiphe graminis have been cloned, and some of the proteins have been characterized to some extent (3, 8, 9, 14, 16, 19, 39). The availability of their sequences has facilitated the design of degenerate primers to amplify fragment homologues of fungal cyp51 in Aspergillus species. This work describes the identification, characterization, and phylogeny of two different cyp51 genes (cyp51A and cyp51B) from Aspergillus species, encoding proteins belonging to the same family of Cyp51 proteins. The implications of this new finding are discussed.
MATERIALS AND METHODS
Strains and plasmids.
A. fumigatus strain 237, which was used throughout this work, was originally cultured from open-lung biopsy material from a patient with invasive pulmonary aspergillosis at Hope Hospital, Manchester, United Kingdom, and was obtained as a gift from M. Keaney. A. terreus (CM-16) was a gift from D. W. Denning. A. nidulans (CM-1392) was initially cultured from a lung biopsy specimen, and A. flavus (CM-1155) was originally isolated from ear exudate; these two strains were obtained from our culture collection. The fungi were grown at 37°C in either GYEP (2% glucose, 0.3% yeast extract, 1% peptone) or Sabouraud (2% glucose, 1% mycologicalpeptone) medium. Escherichia coli JM109 was grown in Luria-Bertani (LB) medium (31), supplemented with ampicillin (100 μg/ml), for propagation of plasmids for DNA purification. For standard cloning and subcloning procedures vectors pGEM-3Z and pGEM-T (Promega, Madrid, Spain) were used.
Primer design and PCR conditions.
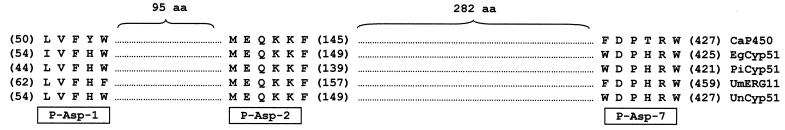
Two highly conserved regions of fungal 14-α sterol demethylase proteins were used to design degenerate primers Asp-1 and Asp-2 (Fig. 1). Primers Asp-1 and Asp-2 contained BamHI restriction sites at the 5′ ends to facilitate cloning of the PCR products. All the primers used in the present work were synthesized by Pharmacia (Madrid, Spain) (Table 1). The PCRs were carried out in a 100-μl volume, containing 10 mM (NH4)2SO4, 10 mM KCl, 20 mM Tris-Cl (pH 8.8), 2 mM MgSO4, 10 ng of bovine serum albumin, 0.1% Triton X-100, 250 μM each dATP, dGTP, dCTP, and dTTP (Perkin-Elmer Cetus, Madrid, Spain), 1 μM each primer, 2.5 U of Pfu DNA polymerase (Promega), and 50 ng of genomic DNA. Amplification was performed in a thermal cycler (Perkin-Elmer Cetus) for one cycle of 5 min at 94°C, 45 s at 58°C, and 2 min at 72°C, and then for 30 cycles of 30 s at 94°C, 45 s at 58°C, and 2 min at 72°C, followed by one final cycle similar to the previous one but with 10 min at 72°C. The PCR products were analyzed by electrophoresis on 0.8 or 1.3% agarose gels, depending on their sizes and were visualized by transillumination after staining with ethidium bromide.
FIG. 1.
Alignment of conserved CYP51 protein sequences used to design degenerate primers (Asp-1, Asp-2, and Asp-7). Numbers in parentheses refer to amino acid positions in the respective protein sequences. Shown are sequences from C. albicans (GenBank accession no. AAF00598.1), E. graminis (AAC97606.1), P. italicum (CAA89824.1), U. maydis (CAA88176.1), and U. necator (AAC49812.2).
TABLE 1.
Oligonucleotide primers used in this work
| Primera | 5′→3′ | Sequenceb (5′→3′) | Use | Species |
|---|---|---|---|---|
| Asp-1 | Sense | ATAGGATCCGTBGTBTTYCAYTGG | Universal | Aspergillus spp. |
| Asp-2 | Antisense | CGCGGATCCAAYTTYTTYTGYTCCAT | Universal | Aspergillus spp. |
| P-A1 | Sense | CTTCTTTGCGTGCAGAGA | Specific for cyp51A | A. fumigatus |
| P-A2 | Antisense | GGGGTCGTCAATGGACTA | Specific for cyp51A | A. fumigatus |
| P-A3 | Sense | TAGTCCATTGACGACCCC | Specific for cyp51A | A. fumigatus |
| P-B1 | Sense | CTTTTTCGACTGCCGCGC | Specific for cyp51B | A. fumigatus |
| P-B2 | Antisense | AGGCGTAGTGAGTGGAGA | Specific for cyp51B | A. fumigatus |
| P-B3 | Sense | TCTCCACTCACTACGCCT | Specific for cyp51B | A. fumigatus |
| Asp-7 | Antisense | CCARCGRTGNGGRTCCCA | Universal | A. fumigatus |
| P-A5 | Antisense | TCTCTGCACGCAAAGAAGAAC | cyp51A 5′ end | A. fumigatus |
| P-A6 | Sense | ACCCCTTACATGATTCCTCCCC | cyp51A 3′ end | A. fumigatus |
| P-A7 | Sense | TCATATGTTGCTCAGCGG | cyp51A 5′ end | A. fumigatus |
| P-B5 | Antisense | TTGCGCGGCAGTCGAAAAAGAAC | cyp51B 5′ end | A. fumigatus |
| P-B6 | Sense | CATGGCTGTGGATGGTACTTC | cyp51B 3′ end | A. fumigatus |
| P-B7 | Sense | ACTAGGGTCAGTTAGTCC | cyp51B 5′ end | A. fumigatus |
| P-B9 | Antisense | CCTCATAATATGCTAAGG | cyp51B 3′ end | A. fumigatus |
| T-7* | Sense | TAATACGACTCACTATAGGGCGA | 5′ and 3′ ends and sequencing | A. fumigatus |
| Sp-6* | Sense | ATTTAGGTGACACTATAGAATAC | 5′ and 3′ ends and sequencing | A. fumigatus |
Asterisks denote commercially available primers (Pharmacia).
DNA letter code: B = C or G or T; Y = C or T; R = G or A; N = A or G or C or T. Underlining indicates a BamHI restriction site.
Cloning and DNA sequencing.
The PCR products were purified by Spin Columns-200 (Clontech, Madrid, Spain), and cloned into the pGEM-T vector system (Promega). Insert DNAs of recombinant plasmids were sequenced by the BigDye terminator cycle sequencing ready reaction system (Perkin-Elmer Applied Biosystems, Madrid, Spain) according to the manufacturer's instructions. All the clones were sequenced on both strands. For each Aspergillus strain at least two inserts were analyzed. Primers T-7 and SP-6 (Pharmacia) were used for sequencing. Sequence analysis was performed on an ABI Prism 377 DNA sequencer (Perkin-Elmer) using the facilities available at the Sequencing Department at Instituto de Salud Carlos III, Majadahonda, Madrid, Spain.
RNA isolation and RT-PCR.
Mycelial mats were blot dried, frozen with liquid nitrogen, and then ground to a powder by using a mortar and pestle. RNA was isolated from the mycelial powder by using an RNAeasy plant mini-kit (Qiagen, Madrid, Spain) according to the manufacturer's instructions. Reverse transcription (RT) was carried out in a 20-μl reaction volume containing 10 mM Tris-HCl (pH 8.8), 50 mM KCl, 0.1% Triton X-100, 5 mM MgCl2, 1 mM each deoxynucleoside triphosphate, 0.5 μg of the specific primer [oligo(dT)15 primer], 20 U of rRNasin RNase inhibitor, and 15 U of avian myeloblastosis virus reverse transcriptase (Reverse Transcription System; Promega) on 2.5 μg of total A. fumigatus RNA. The reaction conditions were 1 h at 42°C. A tube containing all the reaction components except the avian myeloblastosis virus reverse transcriptase was always included as a negative control to check for the presence of contaminating DNA in the RNA sample. Ten microliters of cDNA product was then employed as target DNAs for amplification using the PCR protocol described above. Primers P-A1 and P-A2 were used to amplify the cDNA from the cyp51A gene, and primers P-B1 and P-B2 were used for the cyp51B gene (Table 1). The RT-PCR products were resolved by electrophoresis on 1.4% agarose gels and stained with ethidium bromide for photography.
DNA isolation and hybridization.
DNA was isolated from Aspergillus spp. using a rapid extraction procedure (36), digested with two different restriction enzymes (BamHI and EcoRV), and fractionated by electrophoresis through 0.8% agarose gels in TAE buffer (40 mM Tris-acetate, 1 mM EDTA). Southern analysis was performed as previously described (18). Probes for both cyp51A and cyp51B genes for each Aspergillus sp. were obtained by restriction digestion of the appropriate clones, fractionation in 0.7% low-melting-point agarose gels, and excision of the desired fragments for labeling. A random prime labeling system (ECL; Amersham Pharmacia Biotech, Madrid, Spain) was used to label DNA probes according to the manufacturer's instructions.
Obtaining of complete A. fumigatus cyp51A and cyp51B gene sequences.
Based on the information derived from the sequencing of the cloned PCR fragments, new specific primers named P-A3 and P-B3 were designed for each of the A. fumigatus genes cyp51A and cyp51B, respectively. A new degenerate primer (Asp-7), based on a conserved area downstream of the sequences used before, was used in combination with either P-A3 or P-B3 for PCR amplification of a single fragment over 1 kb of each of the genes (Table 1). The full sequences of the 1-kb fragments for both genes were obtained after cloning of the fragments as described above. To obtain the sequences of the regions flanking these 1-kb fragments (5′ and 3′ ends of the genes), a modification of the protocol described by Délye et al. (8) was used. Southern analysis (of different genomic restriction digestions) using each 1-kb gene fragment as a probe allowed the construction of restriction maps and the selection of fragments of appropriate sizes to ligate and PCR amplify. In brief, 200 ng of purified A. fumigatus genomic DNA was digested to completion with EcoRV, HindIII, or XhoI (Pharmacia) according to the manufacturer's instructions and ligated into the pGEM-3Z plasmid vector, which had already been digested with either SmaI, HindIII, or SalI, respectively, and treated with CIAP (calf intestinal alkaline phosphatase) (Promega). Ligation was performed overnight at 16°C using T4 DNA ligase (Promega) in a total volume of 10 μl. Four primer pairs (two pairs for each gene) were used to PCR amplify DNA fragments that included sequences located upstream and downstream of the sequence already amplified with primer sets P-A3–Asp-7 and P-B3–Asp-7; each primer pair was composed of one primer specifically directed to the sequence of the target gene and either the P-T7 or P-Sp6 primer directed to known sequences flanking the polylinker site of the pGEM-3Z plasmid vector (Table 1). Therefore, for the cyp51A gene, primer P-A5 in combination with primer P-T7 was used to amplify the 5′ end and primer P-A6 in combination with primer P-T7 was used to amplify the 3′ end. Likewise, for the cyp51B gene, primer P-B5 together with primer P-T7 was used to amplify the 5′ end and primer P-B6 in combination with primer P-Sp6 was used to amplify the 3′ end. (After several fruitless attempts at amplification of the 3′ end of the cyp51B gene, a cosmid clone containing the gene [provided by J. P. Latgé of the Pasteur Institute] was finally used to obtain the sequence of this part.) Primers were used at a final concentration of 0.1 μM each, while the rest of the components for the PCR mixtures were the same as described above. Amplifications were performed on 1:10 dilutions of the ligation mix in a total volume of 50 μl. The cycling program consisted of 30 cycles of 30 s of denaturation at 94°C, 45 s of annealing at 60°C, and 2 min of extension at 72°C, with an initial cycle of 5 min at 94°C. Amplified products were cloned into the pGEM-T vector system and sequenced as described before. Specific primers for both genes were designed to complete the sequence on both strands.
Computer analysis.
The amino acid sequences of putative 14-α sterol demethylase gene fragments were deduced from nucleotide sequences and analyzed using the MegAlign software package (Lasergene; DNAstar, Inc., Madison Wis.) run on a PC. The multiple amino acid alignments were carried out by CLUSTAL analysis (17), which first derives a dendrogram from a matrix of all pairwise sequence similarity scores and then aligns the most-similar sequences. The dendrograms produced by the CLUSTAL analysis were generated by the unweighted pair group method using arithmetic averages (UPGMA). The final phylogeny is produced by applying the neighbor-joining method (30) to the distance and alignment data. This occurs after the alignment step and is an independent calculation.
Nucleotide sequence accession numbers.
The full nucleotide sequences of the cyp51A and cyp51B genes from A. fumigatus determined in this work appear in GenBank under accession no. AF338659 and AF338660, respectively. The partial nucleotide sequences of the cyp51A and cyp51B genes from A. flavus, A. nidulans, and A. terreus appear in GenBank under accession no. AF343311 (cyp51A) and AF343312 (cyp51B) for A. flavus, AF343313 (cyp51A) and AF343314 (cyp51B) for A. nidulans, and AF343315 (cyp51A) and AF343316 (cyp51B) for A. terreus.
RESULTS
Isolation of A. fumigatus 14-α sterol demethylase gene fragments by PCR.
The primer set of Asp-1 and Asp-2 was used for priming PCR amplification of A. fumigatus genomic DNA (Fig. 1). Agarose gel analysis of the PCR product amplified by primers Asp-1 and Asp-2 revealed the presence of a band of approximately 350 bp. This fragment was purified, ligated into pGEM-T, and cloned. Partial DNA sequences were determined for several clones containing the 350-bp inserts. Analysis of 140 clones yielded 29 clones containing one sequence (designated cyp51A) and 111 clones containing another sequence (designated cyp51B). Both strands of at least three representatives of each of the two different fragments were then sequenced completely. The deduced 95-amino-acid sequence of A. fumigatus cyp51A is derived from 1 open reading frame (ORF) split by an intervening sequence of 71 bp between codons 29 (K) and 30 (Y). The deduced 95-amino acid sequence of A. fumigatus cyp51B is derived from 1 ORF split by an intervening sequence of 58 bp between codons 29 (K) and 30 (Y). Both intervening sequences have the intron splice junctions GT- and -AG at the 5′ and 3′ ends, respectively, and contain the internal consensus sequence for lariat formation described for filamentous fungi (15). A BLAST sequence similarity search was carried out in the SwissProt database of GenBank to identify which proteins the deduced amino acid sequences of the A. fumigatus gene fragments were most closely related to. The results showed that both Cyp51A and Cyp51B have high percentages of identity (40 to 70%) to the Cyp51 proteins from a variety of filamentous fungi and yeasts. These percentages of identity are high enough to consider both gene products to be members of the CYP51 family of cytochrome P450 (27).
Gene expression.
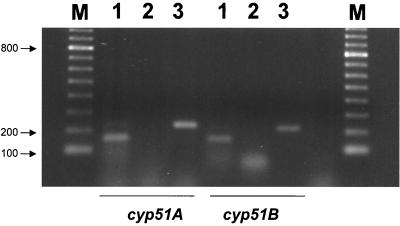
RT-PCR amplification was used to show that both genes are expressed during hyphal growth in submerged culture (Fig. 2). RT-dependent products of the expected sizes were amplified for each of the cyp51 genes, confirming the presence of one intron in each of the two genes analyzed. In the case of cyp51A, one band that corresponded to the fragment of 159 bases amplified by the specific primer set P-A1 and P-A2 was obtained from the cDNA of A. fumigatus. For cyp51B, one band of a similar size was obtained with the specific primer set P-B1 and P-B2 from the cDNA, corresponding to the expected size of 153 bases. Positive controls using A. fumigatus genomic DNA as the target were included for both sets of primers, and the expected bands of 230 and 210 bp were obtained, respectively, with primer sets P-A1–P-A2 and P-B1–P-B2.
FIG. 2.
Detection of A. fumigatus cyp51A and cyp51B gene transcripts. Shown is an ethidium bromide-stained agarose gel of RT-PCR products from cyp51 genes. Lanes 1, RT-PCR products using A. fumigatus total RNA as the target; lanes 2, negative controls using RNA as the target but without reverse transcriptase; lanes 3, positive controls using A. fumigatus genomic DNA as the target; lanes M, 100-bp molecular marker. Sizes of the bands (in base pairs) are given.
Cloning of the putative cyp51A and cyp51B 14-α sterol demethylase genes from A. fumigatus.
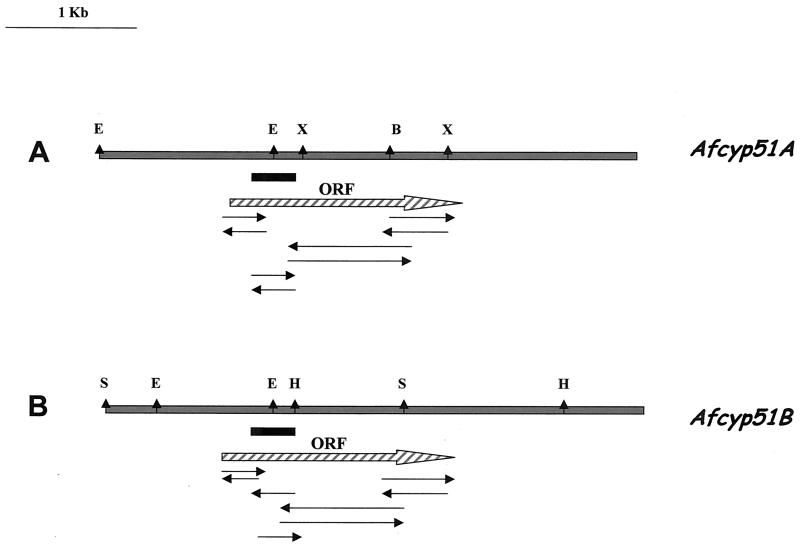
Restriction maps and the full genomic sequence for both genes were obtained as described in Materials and Methods (Fig. 3). Therefore, the 2,048-bp sequence of A. fumigatus cyp51A can be deduced as an ORF encoding a protein of 515 amino acids that identifies a putative 14-α sterol demethylase gene. The ORF is interrupted once by an intron, based on the presence of matching consensus splice junctions (15). The 2,239-bp cyp51B sequence can be deduced as an ORF encoding a protein of 524 amino acids that identifies another putative 14-α sterol demethylase gene. The ORF is interrupted by three introns, based on the presence of matching consensus splice junctions, that are located at the same position in the deduced protein as those of P. italicum and the first two introns of E. graminis and U. necator (7, 8, 39).
FIG. 3.
Restriction maps of A. fumigatus genomic DNA containing the cyp51A (A) and cyp51B (B) genes. ORFs are represented by hatched arrows. Solid boxes represent probes used for Southern analysis. Arrows below the maps indicate the sequence strategy. Restriction enzymes are abbreviated as follows: X, XhoI; H, HindIII; E, EcoRV; B, BamHI; S, SalI.
Sequence analysis: alignments and similarity.
The deduced 515-amino-acid protein encoded by the 1,545-bp coding sequence of the 1,619-bp cyp51A gene was compared to the known complete amino acid sequences of fungal CYP51s. The strongest homologies were shown with A. nidulans Cyp51 (71.8%), P. italicum CYP51 (63.3%), and the other A. fumigatus (Cyp51B) deduced protein (59.4%). In addition, percentages of identity with the other proteins compared were very high (U. necator, 58.1%; E. graminis, 56%; U. maydis, 43%; C. neoformans, 43%; C. albicans, 44%; C. tropicalis, 45%; S. cerevisiae, 45%; and C. glabrata, 43%). The deduced 524-amino-acid protein encoded by the 1,575-bp coding sequence of the 1,731-bp A. fumigatus cyp51B gene was compared to the same CYP51 sequences. The strongest homologies were shown with U. necator (61.6%) and E. graminis (60.2%), apart from the identity with its homologous Cyp51A (59.4%). Homologies with the rest of the compared CYP51 proteins from fungi were as follows: P. italicum, 59%; A. nidulans, 57%; U. maydis, 44%; C. albicans, 44%; C. tropicalis, 44%; S. cerevisiae, 46%; and C. glabrata, 46%. Alignments between the amino acid sequences derived from Cyp51A and Cyp51B and the corresponding predicted amino acid sequences of P. italicum, U. necator, U. maydis, E. graminis, C. neoformans, C. tropicalis, S. cerevisiae, C. glabrata, and C. albicans were also made (data not shown but available on request). The alignment of the 11 full amino acid sequences showed some conserved domains present in all cytochrome P450 enzymes, such as the areas related to substrate recognition or the heme-binding motif at the C terminus, and others specific to CYP51 proteins (38, 39). Each of these domains is present at the expected position in both the A. fumigatus Cyp51A and Cyp51B deduced proteins. The region known as HR-1 is located at the N terminus of the protein and corresponds to amino acids 110 to 133 in Cyp51A and amino acids 115 to 140 in Cyp51B. The function of this domain (conserved among CYP51s) is not known, but it is presumably involved in substrate recognition (9, 38, 39). The HR-2 region is located at the C terminus of the protein and corresponds to amino acids 447 to 461 in Cyp51A and amino acids 456 to 468 in Cyp51B. This motif, known as the heme-binding motif, has a conserved cysteine residue that is responsible for binding to the fifth ligand of the heme iron (38) and that is present in all CYP51, proteins, including Cyp51A and Cyp51B from A. fumigatus. Other domains important for substrate specificity and/or recognition are the central helix and the CR-4 motif, which are also highly conserved among, CYP51s and are present in both genes from A. fumigatus (20, 24, 39).
Isolation of A. flavus, A. terreus, and A. nidulans 14-α sterol demethylases gene fragments by PCR.
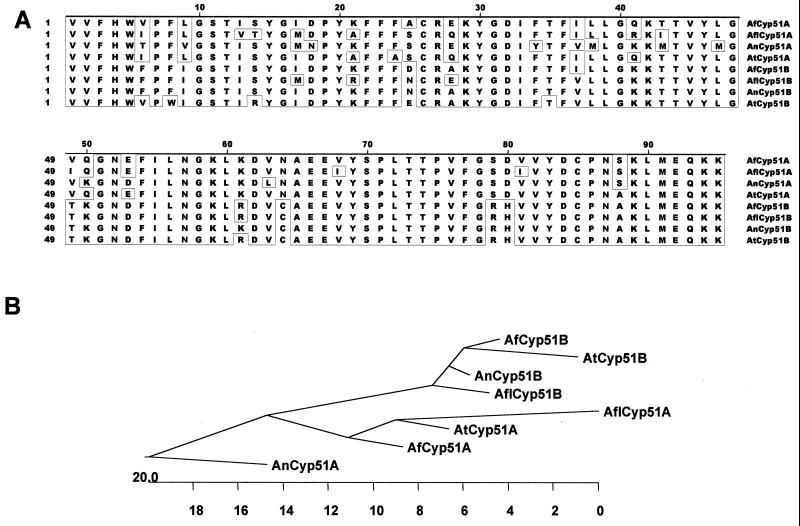
The oligonucleotide primer set Asp-1–Asp-2 (Table 1) was used for priming PCR amplification of A. terreus, A. flavus, and A. nidulans genomic DNAs. Agarose gel analysis of PCR products amplified by primers Asp-1 and Asp-2 revealed the presence of a single band of 350 bp in each of the Aspergillus species tested. These fragments were purified and ligated into pGEM-T. Partial DNA sequences were determined in several clones of each species containing the approximately 350-bp inserts. For every species of Aspergillus tested, two different sequences were found: A. flavus cyp51A and cyp51B, A. terreus cyp51A and cyp51B, and A. nidulans cyp51A and cyp51B. Both strands of two representatives of each of the different fragments were then sequenced completely for each species analyzed. The deduced 95-amino-acid sequences of all cyp51A and cyp51B genes are derived from one ORF split by an intervening sequence (variable in size but not in location, as in A. fumigatus) between codons 29 (K) and 30 (Y). All the intervening sequences have the intron splice junctions GT- and -AG at the 5′ and 3′ ends, respectively, and contain the internal consensus sequence for lariat formation described for filamentous fungi (15). The sizes for the putative introns were different for each gene and Aspergillus species analyzed (for A. flavus cyp51A, 67 bp; for A. flavus cyp51B, 54 bp; for A. nidulans cyp51A, 49 bp; for A. nidulans cyp51B, 61 bp; for A. terreus cyp51A, 69 bp; and for A. terreus cyp51B, 61 bp). Figure 4A shows the alignment of the derived amino acid sequences of the fragments obtained for all the other Aspergillus species tested with the corresponding regions of the proteins from A. fumigatus.
FIG. 4.
(A) Alignment of the predicted amino acid sequences of the cyp51A and cyp51B gene fragments from A. fumigatus, A. flavus, A. nidulans, and A. terreus. Amino acid residues identical in the eight gene fragments are boxed. (B) Phylogenetic tree obtained by CLUSTAL analysis. The length of each branch represents the distance between sequence pairs. Numbers at the bottom of the tree are numbers of substitution events.
Genomic organization.
A. fumigatus, A. terreus, A. flavus, and A. nidulans genomic DNAs digested with either BamHI or EcoRV and hybridized with the corresponding probe (one for each of the eight genes, i.e., the cyp51A and cyp51B genes of each of these four species), resulted in hybridization to single DNA fragments, or to two DNA fragments where there was a corresponding restriction site present in the probe, as in the case of the EcoRV digestions probed with the A. fumigatus cyp51A and cyp51B probes, the A. flavus cyp51B probe, and the A. terreus cyp51B probe (data not shown). These results show that each gene is present as a single copy in the genome of each of the Aspergillus species tested.
Phylogenetic analysis.
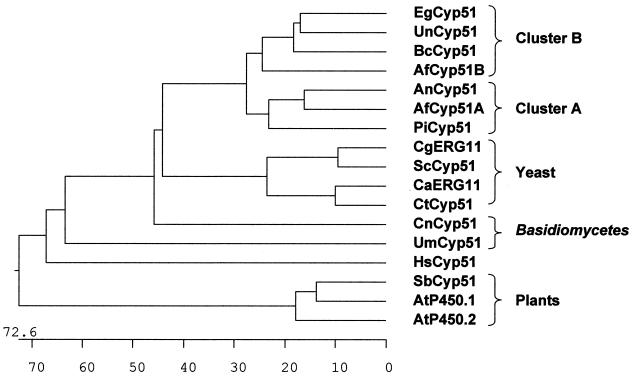
Phylogenetic trees were obtained by CLUSTAL analysis. First, we analyzed the comparison of all the amino acid sequences derived from Aspergillus sp. cyp51A and cyp51B gene fragments. Two clusters were clearly generated, one for the Cyp51B-derived protein fragments and another for the Cyp51A-derived protein fragments. Moreover, proteins in the Cyp51B cluster were closer to each other than were the Cyp51A protein fragments (Fig. 4B). The phylogenetic tree derived from the comparison of all the full known CYP51 protein sequences is shown in Fig. 5. We have included the Cyp51 sequences from two plants (Arabidopsis thaliana and Sorghum bicolor) and Homo sapiens (1, 29, 34). The main reason for including these sequences (although the percentages of similarity to these Cyp51s are not as high as those to the sequences of other fungi), is to point out the existence of two homologous Cyp51 proteins in A. thaliana (27, 29). Two sharply defined clusters can be detected, one for all the yeast sequences and another for those of filamentous fungi. Within the latter, two subclusters can be observed: one comprising A. fumigatus Cyp51A together with P. italicum Cyp51 and A. nidulans Cyp51 and the other comprising A. fumigatus Cyp51B, U. necator Cyp51, B. cinerea Cyp51, and E. graminis Cyp51. The Basidiomycetes (C. neoformans Cyp51 and U. maydis Cyp51) presented two branches that are quite distant from both yeasts and filamentous fungi.
FIG. 5.
Phylogenetic tree derived by CLUSTAL analysis after the alignment of the full amino acid sequences of Cyp51A and Cyp51B from A. fumigatus with CYP51s from C. albicans (CaErg11; GenBank accession no. AAF00598.1), C. glabrata (CgERG11; GenBank accession no. AAB02329.1), C. tropicalis (CtCyp51; GenBank accession no. AAA53284.1), S. cerevisiae (ScCyp51; GenBank accession no. AAA34546.1), P. italicum (PiCyp51; GenBank accession no. CAA89824.1), A. nidulans (AnCyp51; GenBank accession no. AAF79204.1), E. graminis (EgCyp51; GenBank accession no. AAC97606.1), U. necator (UnCyp51; GenBank accession no. AAC49812.2), B. cinerea (BcCyp51; GenBank accession no. AAF85983.1), U. maydis (UmCyp51; GenBank accession no. CAA88176.1), C. neoformans (CnCyp51; GenBank accession no. AAF35366.1), A. thaliana (AtP450.1 [GenBank accession no. AAD.30262.1] and AtP450.2 [GenBank accession no. AAB86510.1]), S. bicolor (SbCyp51; GenBank accession no. AAC49659.1), and H. sapiens (HsCyp51; GenBank accession no. XP004663.1). The length of each branch represents the distance between sequence pairs. Numbers at the bottom of the tree are numbers of substitution events.
DISCUSSION
All fungal cytochrome P450 genes isolated are categorized in CYP gene families 51 to 66 (27). One of the best characterized is the CYP51 gene family, encoding eburicol or lanosterol 14-α demethylase (EDM, CYP51, ERG11, or P45014DM). Sterol 14-α demethylase is the rate-limiting enzyme in the ergosterol biosynthesis pathway, and this fact makes cyp51 genes attractive as targets for azole agents (37). This interest has resulted in the isolation of cyp51 genes from many different yeasts and some filamentous fungi (3, 8, 9, 14, 16, 19, 39). The availability of some cyp51 genes and their derived proteins from different yeasts and fungi has been used, in this work, to design degenerate primers directed to conserved areas of this enzyme. Surprisingly, we have found two different, but closely related, cyp51 genes in A. fumigatus (cyp51A and cyp51B) as well as in A. terreus, A. flavus, and A. nidulans. This is the first report of the existence of two genes in the CYP51 fungal family. The percentages of identity found between CYP51s from other fungi and those from A. fumigatus are high enough to consider both genes to be members of the cytochrome P450 cyp51 gene family (27). In A. thaliana, the P450 cytochrome genes fall into 41 families, and none of them is a clear homologue to fungal or animal P450s except the cyp51 genes, which have been described as encoding the first eukaryotic P450 and probably the precursor of them all (27). The existence of two cyp51 genes in A. thaliana corroborates our findings (27, 29). These two genes are thought to encode obtusifoliol 14-α demethylases (the plant equivalents of fungal lanosterol and eburicol 14-α demethylases), and they are also highly related to each other (72% similarity). These data suggest that it could be possible to find cyp51A and cyp51B homologues in other filamentous fungi and yeasts. Therefore, the availability of the sequences of cyp51 genes from Aspergillus spp. may lead to the isolation of homologous genes in other filamentous fungi. This is of great importance, since basic knowledge of the cyp51 gene family and the enzymes they encode may facilitate additional studies, resulting in fungal inhibitors with increased activity or a broader spectrum. The products of these genes might perform different functions in the cell, might perform similar functions but have different localizations, or might have different substrate affinities. It has been pointed out that gene families arose through a process of duplication of an ancestral gene followed by functional and structural specialization (divergence) of both copies. Because a duplicated gene is likely to be lost unless it acquires a novel and important use, Cyp51A and Cyp51B probably have different functions (13). Therefore, the next step in the study of both A. fumigatus cyp51 genes will be the functional analysis of cyp51A and cyp51B by the construction of cyp51A and cyp51B single-mutant strains and, if possible, of a cyp51A cyp51B double-knockout mutant strain. Although disruption of the cyp51 gene in S. cerevisiae has been described as lethal (2), in A. fumigatus the existence of two cyp51 genes could facilitate the analysis of the single disruptants, as one enzyme may compensate for the lack of the other. These experiments are currently under way in our laboratory.
Once the function of each of these proteins has been elucidated, it might have some implications for the study of resistance mechanisms of fungi against azole derivatives. To date, two different fungal Cyp51 mechanisms have been related to azole resistance: alteration of the primary target (14-α sterol demethylase) by mutations and/or overexpression of the cyp51 gene. Several lines of evidence have shown that both mechanisms seem to be present in yeasts (20, 21, 24, 32). Numerous works have been published reporting azole resistance in C. albicans due to single point mutations which seem to have a cooperative effect (20, 24, 32). On the other hand, single point mutations have been shown to be directly related to fungal DMI resistance in filamentous fungi. This fact has been proven for E. graminis and resistance to benzimidazole (8) and for U. necator and resistance to triadimenol (9). In addition, overexpression of the cyp51 gene has been related to resistance of P. italicum to fenarimol (39). It seems obvious that the existence of two cyp51 genes in Aspergillus spp. could probably explain the intrinsic resistance of these fungi to some azole derivatives. The attribution of resistance to Cyp51 mutations will depend on the full analysis of both proteins, at least in Aspergillus spp.
In the phylogenetic tree derived from the comparison of all known full CYP51 protein sequences, two clear clusters can be detected: one for all the yeast sequences and another for those of filamentous fungi (Fig. 5). Within the latter, two subclusters can be observed: one branch for A. fumigatus Cyp51A together with P. italicum Cyp51 and A. nidulans Cyp51 and another branch with the proteins A. fumigatus Cyp51B, U. necator Cyp51, E. graminis Cyp51, and B. cinerea Cyp51. The Basidiomycetes (C. neoformans Cyp51 and U. maydis Cyp51) presented similar values compared with both yeasts and filamentous fungi. We have also included Cyp51s from two plants (A. thaliana and S. bicolor) and H. sapiens. Although the percentages of similarity are quite low for true comparison, the structures of the proteins are quite similar (27). The existence of two cyp51 genes in A. thaliana encourages the search for cyp51A and cyp51B homologues or orthologues, at least in filamentous fungi. Moreover, the fact that Cyp51B is closer to U. necator Cyp51, B. cinerea Cyp51, and E. graminis Cyp51 may indicate that other genes coding for Cyp51A homologues could be present, at least in this three species. Likewise, Cyp51B homologues could be present in P. italicum and A. nidulans. The finding of two genes in other fungi together with the functional analysis could help to clarify the mechanisms of resistance of filamentous fungi to azole agents. When more protein sequences from different fungi are obtained, a clearer picture can be drawn to establish if the apparent division found here could allow for division of the CYP51 family into functional subgroups.
In summary, we have reported for the first time the presence of two different genes encoding 14-α sterol demethylases in fungi. Homologues of these genes could be present in other filamentous fungi and yeasts, and this fact could change what is already known about azole resistance mechanisms. Functional analysis and further studies are warranted.
ACKNOWLEDGMENTS
This work was supported in part by grant 1078/99 from Instituto de Salud Carlos III. T.M.D.-G. is a fellow of the Instituto de Salud Carlos III.
We thank J. P. Latgé for valuable suggestions and critical reading of the manuscript. We also thank, M. Jose Buitrago for helping with RNA work at Unite des Aspergillus, Instituto Pasteur, Paris, France.
REFERENCES
- 1.Bak S, Kahn R A, Olsen C E, Halkier B A. Cloning and expression in Escherichia coli of the obtusifoliol 14-α-demethylase of Sorghum bicolor(L.) Moench, a cytochrome P450 orthologous to the sterol 14-α-demethylases (CYP51) from fungi and mammals. Plant J. 1997;11:191–201. doi: 10.1046/j.1365-313x.1997.11020191.x. [DOI] [PubMed] [Google Scholar]
- 2.Bard M, Lees N D, Turi T, Craft D, Cofrin L, Barbuch R, Koegel C, Loper J C. Sterol synthesis and viability of erg11 (cytochrome P450 lanosterol demethylase) mutations in Saccharomyces cerevisiae and Candida albicans. Lipids. 1993;28:963–967. doi: 10.1007/BF02537115. [DOI] [PubMed] [Google Scholar]
- 3.Chen C, Kalb V F, Turi T G, Loper J C. Primary structure of the cytochrome P450 lanosterol 14-α-demethylase gene from Candida tropicalis. DNA. 1988;7:617–626. doi: 10.1089/dna.1988.7.617. [DOI] [PubMed] [Google Scholar]
- 4.Chryssanthou E. In vitro susceptibility of respiratory isolates of Aspergillusspecies to itraconazole and amphotericin B. Acquired resistance to itraconazole. Scand J Infect Dis. 1997;29:509–512. doi: 10.3109/00365549709011864. [DOI] [PubMed] [Google Scholar]
- 5.Cuenca-Estrella M, Rodriguez-Tudela J L, Mellado E, Martinez-Suarez J V, Monzon A. Comparison of the in vitro activity of voriconazole (UK-109,496), itraconazole and amphotericin B against clinical isolates of Aspergillus fumigatus. J Antimicrob Chemother. 1998;42:531–533. doi: 10.1093/jac/42.4.531. [DOI] [PubMed] [Google Scholar]
- 6.Cuenca-Estrella M, Ruiz-Diez B, Martinez-Suarez J V, Monzon A, Rodriguez-Tudela J L. Comparative in vitro activity of voriconazole (UK-109,496) and six other antifungal agents against clinical isolates of Scedosporium prolificans and Scedosporium apiospermum. J Antimicrob Chemother. 1999;43:49–151. doi: 10.1093/jac/43.1.149. [DOI] [PubMed] [Google Scholar]
- 7.Del Sorbo G, Andrade A C, van Nistelrooy J G M, van Kan J A L, Balzi E, de Waard M A. Multidrug resistance in Aspergillus nidulansinvolves novel ATP-binding cassette transporters. Mol Gen Genet. 1997;254:417–426. doi: 10.1007/s004380050434. [DOI] [PubMed] [Google Scholar]
- 8.Délye C, Bousset L, Corio-Costet M F. PCR cloning and detection of point mutations in the eburicol 14-α-demethylase (CYP51) gene from Erysiphe graminis f. sp. hordei, a “recalcitrant” fungus. Curr Genet. 1998;34:399–403. doi: 10.1007/s002940050413. [DOI] [PubMed] [Google Scholar]
- 9.Délye C, Laigret F, Corio-Costet M F. A mutation in the 14-α-demethylase gene of Uncinula necatorthat correlates with resistance to a sterol biosynthesis inhibitor. Appl Environ Microbiol. 1997;63:2966–2970. doi: 10.1128/aem.63.8.2966-2970.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Denning D W. Invasive aspergillosis. Clin Infect Dis. 1998;26:781–805. doi: 10.1086/513943. [DOI] [PubMed] [Google Scholar]
- 11.Denning D W, Venkateswarlu K, Oakley K L, Anderson M J, Manning N J, Stevens D A, Warnock D W, Kelly S L. Itraconazole resistance in Aspergillus fumigatus. Antimicrob Agents Chemother. 1997;41:1364–1368. doi: 10.1128/aac.41.6.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Denning D W, Radford S A, Oakley K L, Hall L, Johnson E M, Warnock D W. Correlation between in-vitro susceptibility testing to itraconazole and in-vivo outcome of Aspergillus fumigatusinfection. J Antimicrob Chemother. 1997;40:401–414. doi: 10.1093/jac/40.3.401. [DOI] [PubMed] [Google Scholar]
- 13.Fryxell K J. The coevolution of gene family trees. Trends Genet. 1996;2:364–369. doi: 10.1016/s0168-9525(96)80020-5. [DOI] [PubMed] [Google Scholar]
- 14.Geber A, Hitchcock C A, Swartz J E, Pullen F S, Marsden K E, Kwon-Chung K J, Bennett J E. Deletion of the Candida glabrataERG3 and ERG11 genes: effect on cell viability, cell growth, sterol composition, and antifungal susceptibility. Antimicrob Agents Chemother. 1995;39:2708–2717. doi: 10.1128/aac.39.12.2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gurr S J, Unkles S E, Kinghorn J R. The structure and organization of nuclear genes of filamentous fungi. In: Kinghorn J R, editor. Gene structure in eukaryotic microbes. Oxford, United Kingdom: IRL Press; 1987. pp. 93–139. [Google Scholar]
- 16.Hargreaves J A, Keon J P R. Isolation of an Ustilago maydisERG11 gene and its expression in a mutant deficient in sterol-14-α-demethylase activity. FEMS Microbiol Lett. 1996;139:203–207. doi: 10.1111/j.1574-6968.1996.tb08203.x. [DOI] [PubMed] [Google Scholar]
- 17.Higgins D G, Sharp P M. CLUSTAL: a package for performing multiple sequence alignments on a microcomputer. Gene. 1998;73:237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- 18.Holden D W, Kronstad J W, Leong S A. Mutation in a heat regulated hsp70 gene of Ustilago maydis. EMBO J. 1989;8:1927–1934. doi: 10.1002/j.1460-2075.1989.tb03596.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishida N, Aoyama Y, Hata R, Oyama Y, Imajo S, Ishiguro M, Oshima T, Nakazato H, Noguchi T, Maitra U S, Mohan V P, Sprinson D B, Yoshida Y. A single amino acid substitution converts cytochrome P450–14DM to an inactive form, cytochrome P450-SG1: complete primary structures deduced from cloned DNAs. Biochem Biophys Res Commun. 1988;15:317–323. doi: 10.1016/s0006-291x(88)81087-8. [DOI] [PubMed] [Google Scholar]
- 20.Joseph-Horne T, Hollomon D W. Molecular mechanisms of azole resistance in fungi. FEMS Microbiol Lett. 1997;149:141–149. doi: 10.1111/j.1574-6968.1997.tb10321.x. [DOI] [PubMed] [Google Scholar]
- 21.Kontoyiannis D P, Sagar N, Hirschi K D. Overexpression of Erg11p by the regulatable GAL1 promoter confers fluconazole resistance in Saccharomyces cerevisiae. Antimicrob Agents Chemother. 1999;43:2798–2800. doi: 10.1128/aac.43.11.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lass-Florl C, Kofler G, Kropshofer G, Hermans J, Kreczy A, Dierich M P, Niederwieser D. In vitro testing of susceptibility to amphotericin B is a reliable predictor of clinical outcome in invasive aspergillosis. J Antimicrob Chemother. 1998;42:497–502. doi: 10.1093/jac/42.4.497. [DOI] [PubMed] [Google Scholar]
- 23.Latgé J-P. Aspergillus fumigatus and aspergillosis. Clin Microbiol Rev. 1999;12:310–350. doi: 10.1128/cmr.12.2.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marichal P, Koymas L, Willemsens S, Bellens D, Verhasselt P, Luyten W, Borgers M, Ramaekers F C S, Odds F C, Vanden Bossche H. Contribution of mutations in the cytochrome P450 14-α-demethylase (Erg11p, Cyp51p) to azole resistance in Candida albicans. Microbiology. 1999;145:2701–2713. doi: 10.1099/00221287-145-10-2701. [DOI] [PubMed] [Google Scholar]
- 25.Nakaune R, Adachi D, Nawata O, Tomiyama M, Akutsu K, Hibi T. A novel ATP-binding cassette transporter involved in multidrug resistance in the phytopathogenic fungus Penicillium digitatum. Appl Environ Microbiol. 1998;64:3983–3988. doi: 10.1128/aem.64.10.3983-3988.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.National Committee for Clinical Laboratory Standards. Reference method for broth dilution antifungal susceptibility. Testing of filamentous fungi: proposed standard. Document M38-P. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1998. [Google Scholar]
- 27.Nelson D R. Cytochrome P450 and the individuality of species. Arch Biochem Biophys. 1999;369:1–10. doi: 10.1006/abbi.1999.1352. [DOI] [PubMed] [Google Scholar]
- 28.Oakley K L, Morrissey G, Denning W. Efficacy of SCH-56592 in a temporarily neutropenic murine model of invasive aspergillosis with an itraconazole-susceptible and an itraconazole-resistant isolate of Aspergillus fumigatus. Antimicrob Agents Chemother. 1997;41:1504–1507. doi: 10.1128/aac.41.7.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paquette S M, Bak S, Feyereisen R. Intron-exon organization and phylogeny in a large superfamily, the paralogous cytochrome P450 genes of Arabidopsis thaliana. DNA Cell Biol. 2000;19:307–317. doi: 10.1089/10445490050021221. [DOI] [PubMed] [Google Scholar]
- 30.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 32.Sanglard D, Ischer F, Koymans K, Bille J. Amino acid substitutions in the cytochrome P450 14-α-demethylase (CYP51A1) from azole-resistant Candida albicansclinical isolates contribute to resistance to azole antifungal agents. Antimicrob Agents Chemother. 1998;42:241–253. doi: 10.1128/aac.42.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stehmann C, de Waard M A. Accumulation of tebuconazole by isolates of Botrytis cinereadiffering in sensitivity to sterol demethylation inhibiting fungicides. Pestic Sci. 1995;45:311–318. [Google Scholar]
- 34.Stromstedt M, Rozman D, Waterman M R. The ubiquitously expressed human CYP51 encodes lanosterol 14-α-demethylase, a cytochrome P450 whose expression is regulated by oxysterols. Arch Biochem Biophys. 1996;329:73–81. doi: 10.1006/abbi.1996.0193. [DOI] [PubMed] [Google Scholar]
- 35.Sutton D A, Sanche S E, Revankar S G, Fothergill A W, Rinaldi M G. In vitro amphotericin B resistance in clinical isolates of Aspergillus terreus, with a head-to-head comparison to voriconazole. J Clin Microbiol. 1999;37:2343–2345. doi: 10.1128/jcm.37.7.2343-2345.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang C M, Cohen J, Holden D W. An Aspergillus fumigatusalkaline protease mutant constructed by gene disruption is deficient in extracellular elastase activity. Mol Microbiol. 1992;6:1663–1671. doi: 10.1111/j.1365-2958.1992.tb00891.x. [DOI] [PubMed] [Google Scholar]
- 37.Vanden Bossche H, Koymans L, Moereels H. P450 inhibitors of use in medical treatment: focus on mechanism of action. Pharmacol Ther. 1995;67:79–100. doi: 10.1016/0163-7258(95)00011-5. [DOI] [PubMed] [Google Scholar]
- 38.van den Brink H J M, van Gorcom R F M, van den Hondel C A, Punt P J. Cytochrome P450 enzyme systems in fungi. Fungal Genet Biol. 1998;23:1–17. doi: 10.1006/fgbi.1997.1021. [DOI] [PubMed] [Google Scholar]
- 39.van Nistelrooy J G M, van den Brink J M, van Kan J A L, van Gorcom R F M, de Waard M A. Isolation and molecular characterization of the gene encoding eburicol 14-α-demethylase (CYP51) from Penicillium italicum. Mol Gen Genet. 1996;250:725–733. doi: 10.1007/BF02172984. [DOI] [PubMed] [Google Scholar]