Keywords: bile acid metabolism, developmental programming, lipid metabolism, body mass index, microbiome
Abstract
Mice exposed in gestation to maternal high-fat/high-sucrose (HF/HS) diet develop altered bile acid (BA) homeostasis. We hypothesized that these reflect an altered microbiome and asked if microbiota transplanted from HF/HS offspring change hepatic BA and lipid metabolism to determine the directionality of effect. Female mice were fed HF/HS or chow (CON) for 6 wk and bred with lean males. 16S sequencing was performed to compare taxa in offspring. Cecal microbiome transplantation (CMT) was performed from HF/HS or CON offspring into antibiotic-treated mice fed chow or high fructose. BA, lipid metabolic, and gene expression analyses were performed in recipient mice. Gut microbiomes from HF/HS offspring segregated from CON offspring, with increased Firmicutes to Bacteriodetes ratios and Verrucomicrobial abundance. After CMT was performed, HF/HS-recipient mice had larger BA pools, increased intrahepatic muricholic acid, and decreased deoxycholic acid species. HF/HS-recipient mice exhibited downregulated hepatic Mrp2, increased hepatic Oatp1b2, and decreased ileal Asbt mRNA expression. HF/HS-recipient mice exhibited decreased cecal butyrate and increased hepatic expression of Il6. HF/HS-recipient mice had larger livers and increased intrahepatic triglyceride versus CON-recipient mice after fructose feeding, with increased hepatic mRNA expression of lipogenic genes including Srebf1, Fabp1, Mogat1, and Mogat2. CMT from HF/HS offspring increased BA pool and shifted the composition of the intrahepatic BA pool. CMT from HF/HS donor offspring increased fructose-induced liver triglyceride accumulation. These findings support a causal role for vertical transfer of an altered microbiome in hepatic BA and lipid metabolism in HF/HS offspring.
NEW & NOTEWORTHY We utilized a mouse model of maternal obesogenic diet exposure to evaluate the effect on offspring microbiome and bile acid homeostasis. We identified shifts in the offspring microbiome associated with changes in cecal bile acid levels. Transfer of the microbiome from maternal obesogenic diet-exposed offspring to microbiome-depleted mice altered bile acid homeostasis and increased fructose-induced hepatic steatosis.
INTRODUCTION
As obesity rates increase, so do the prevalence and impact of its complications, such as nonalcoholic fatty liver disease (NAFLD). The global prevalence of NAFLD (nearly 25%) makes this condition the most common chronic liver disease at all ages (1, 2). Individual lifestyle is an important factor in the development and progression of NAFLD, but genetic and environmental factors play important roles. Exposures beginning in utero can initiate the development of metabolic liver disease later in life. One exposure is maternal diet, a well-established predisposing factor for development of NAFLD in offspring (for review, see Ref. 3). Longitudinal analysis of several birth cohorts has identified maternal prepregnancy BMI ≥ 30 as an independent predictor of offspring NAFLD (4, 5). Animal models corroborate these findings and are being utilized to identify the mechanisms for this developmental programming of liver disease (6–8).
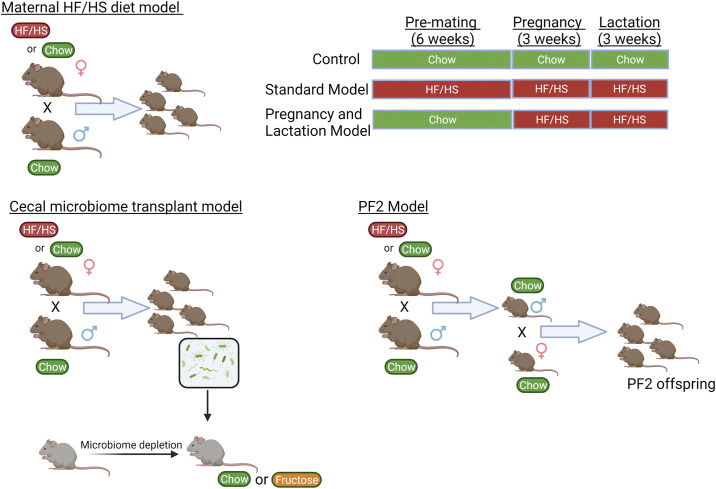
We have found that maternal high-fat/high-sucrose (HF/HS) diet changes bile acid (BA) homeostasis in mouse offspring (9), with increased BA pool size, increased expression and activity of hepatic Cyp7a1, and a shifted intrahepatic BA profile. However, the underlying mechanisms of transmission of altered BA homeostasis from dam to offspring is yet to be defined. One potential mode for transmission is through vertical transmission of an altered microbiome at birth. The role of resident gut bacteria in metabolizing bile acids has been well described (for review, see Ref. 10). In models of maternal obesogenic diet exposure (Fig. 1), the microbiome of offspring is altered (11, 12). There are also shifts in the microbiome of human offspring of mothers with BMI ≥ 30 (13).
Figure 1.
Diagram of mouse models. Schematic representation of the different mouse models used for the studies in this manuscript. HF/HS, high fat/high sucrose. Created with BioRender.com.
Although changes in bile acid metabolism and microbiome occur in models of maternal obesogenic diet exposure, a causal relationship for gut bacteria in regulating BA pool size and intrahepatic BA profile remains unproven. Here, we tested the hypothesis that vertical transmission of an altered microbiome after maternal HF/HS diet exposure affects BA homeostasis in the offspring. We also sought to define whether vertical transmission of an altered microbiome increases steatosis and alters expression of factors involved in hepatic lipid metabolism in offspring exposed to maternal obesogenic diet.
EXPERIMENTAL PROCEDURES
Mouse Breeding Scheme, Feeding Paradigm, and Metabolic Analysis
All procedures were approved by the Animal Studies Committee at Washington University School of Medicine and conformed to National Institutes of Health guidelines. Four-week-old female C57Bl/6J mice were fed either a high-fat/high-sugar (HF/HS) [Test Diet 58R3; 59% fat, 26% carbohydrates (17% sucrose) and 15% protein] or standard (CON) chow [Pico Lab Rodent diet 20; 13% fat, 62% carbohydrates (3.2% sucrose) and 25% protein] for 6 wk as we have reported previously (14). HF/HS- and CON-fed F0 female mice were mated with chow-fed male mice to produce HF/HS- and CON-exposed offspring. Tissues were collected at necropsy at 6–8 wk of age. The numbers of offspring and number of different litters represented are noted in each figure legend.
To evaluate maternal obesogenic diet during pregnancy and lactation only, we mated 10-wk-old lean female C57Bl/6J mice with lean males. If a vaginal plug was identified, the female mouse was separated and fed either HF/HS or standard chow (CON) throughout pregnancy and lactation.
To evaluate transmission across generations by male offspring, F1 male offspring from the CON or HF/HS were mated with lean female mice. Offspring from this breeding are denoted as PF2C if from the CON and PF2H if from the HF/HS.
For all breeding, potential dams were staged to identify the most likely period for successful mating. The sire was placed in the cage for only 24 h to limit cohousing effects.
Bile Acid Analysis
BA pool size was measured in offspring as previously reported (15). Briefly, after 4 h of fasting, gallbladder, liver, and intestines including luminal contents were collected, homogenized, and incubated in ethanol to extract BAs. Total BA content was determined enzymatically (Crystal Chem, Elk Grove Village, IL) and normalized to body weight. Individual intrahepatic BA species were measured using high-performance liquid chromatography (HPLC) and tandem mass spectrometry as we have previously described (15). Individual BA concentrations were normalized to tissue weight. The quantities of each BA type are presented as a percent of the total intrahepatic BA pool. Primary and secondary bile acids were combined to calculate a ratio.
Quantitative PCR
Hepatic and ileal total RNA were extracted and cDNA was prepared using an ABI high-capacity cDNA reverse transcription kit with 1 µg of total RNA. We applied real-time quantitative PCR to cDNAs from six animals per group, performed in duplicate on an Applied Biosystems 7500 Real-Time PCR System using SYBR Green PCR Master Mix (Applied Biosystems) and primer pairs (provided on request) designed by Primer Express software (Applied Biosystems). Relative mRNA abundance is expressed as fold change to maternal chow-offspring chow group after normalization to GAPDH.
Hepatic Triglyceride Measurement
Liver triglycerides (TG) were measured after 4 h of fasting at necropsy using a colorimetric biochemical assay (Diabetes Models Phenotyping Core, Diabetes Research Center at Washington University).
Sequencing for Gut Microbiome Analysis
Stool DNA was isolated from cecal contents using the PowerSoil DNA Isolation Kit (Qiagen) per the manufacturer’s instructions. The samples were lysed with beads and spin filtered to elute purified DNA, which was stored at −20°C until PCR amplification. A reengineered version of bTEFAP, a form of amplicon sequencing utilizing next generation sequencing, was used to evaluate the microbiota. This modern version of bTEFAP has been adapted to current next generation sequencing technologies. We used 16S rRNA primer pairs 515F GTGYCAGCMGCCGCGGTAA/806R GGACTACNVGGGTWTCTAAT, which were subjected to a single-step 30 cycle PCR was used for the HotStarTaq Plus Master Mix Kit (95°C (5 min), 30 cycles at 95°C (30 s), 53°C (40 s), 72°C (1 min), and 72°C (10 min). The amplification products from the different samples were mixed in equal concentrations, purified, and sequenced (Illumina NovaSeq). The Mr. DNA ribosomal and functional gene analysis pipeline was utilized to process the Q25 sequence data. To note, sequences with point errors, shorter than 150 bp, or containing ambiguous bases were removed. The quality of the sequences was assessed by dereplicating sequences with expected error >1.0. This process allows creation of a denoised sequence or Zero-radius Operational Taxonomic Unit (zOTU). The final sequence data were classified based on taxonomy using BLASTn. Statistical analysis was conducted using XLstat, NCSS 2007, “R,” and NCSS 2010. α and β diversity analyses were viewed through Qiime 2.
Microbiome Depletion and Cecal Microbiome Transplantation
Six-week-old male C57/Bl6 mice were purchased from Jackson laboratories and acclimated to our mouse facility for 1 wk before antibiotic treatment to deplete the resident gut bacterial microbiome. The ABX mix contained 300 mL of sterile water, to which vancomycin (150 mg), neomycin (300 mg), and artificial sweetener (equal) (2 g) were added. The antibiotic water was provided for 60 h followed by 12 h of water containing polyethylene glycol.
Cecal contents were collected from 6-wk-old female HF/HS and CON offspring and placed in reduced PBS with glycerol (adjusted to 16%) to create a slurry at 0.1 g/mL concentration, which was then stored (−80°C) (Repeated using male donor cecal contents in Supplemental Material; see https://doi.org/10.6084/m9.figshare.15044439.v1). The slurry mix was thawed on ice, homogenized with a glass bulb, passed through a 100-µm nylon cell strainer, centrifuged for (3,000 rpm, 30 s), and vortexed to create a homogeneous mix 30 min before oral gavage of the recipient mice. Microbiome-depleted mice were gavaged with stool slurry mix from CON or HF/HS offspring five times over 11 days.
Statistical Analysis
Unpaired Student’s t test and ANOVA were used when appropriate using GraphPad prism software. Data are presented as means (SE) with two-tailed P < 0.05 representing significance and P > 0.05 and < 0.10 representing a trend.
RESULTS
Altered Microbiome in Offspring Exposed to Maternal HF/HS Diet
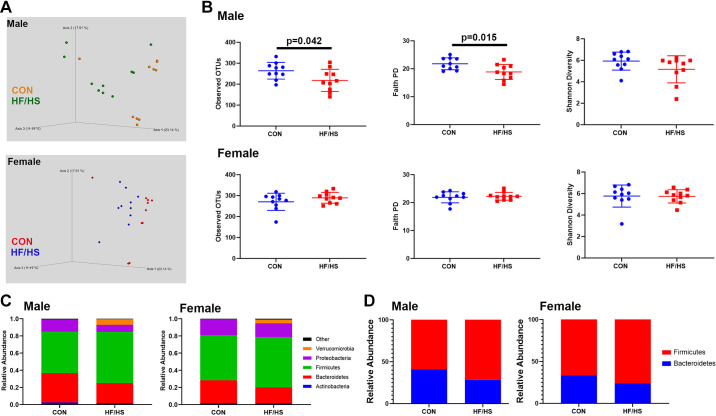
Male and female offspring exhibited separation of cecal bacterial communities between offspring exposed to maternal CON and HF/HS diet (Fig. 2A). This separation was also observed in principal component analysis (PCoA) of excreted stool in male offspring (Supplemental Fig. S1; all Supplemental figures are available at https://doi.org/10.6084/m9.figshare.15044439). Three measures of α-diversity (observed OTUs, Faith phylogenetic diversity (PD), and Shannon diversity) in male HF/HS offspring demonstrated decreased α-diversity by observed OTUs and Faith PD (Fig. 2B). At the family-phylum level, there was decreased abundance of Bacteroidetes and Proteobacteria in male and female offspring after exposure to maternal HF/HS diet (Fig. 2C). Conversely, Firmicutes and Verrucomicrobia increased (Fig. 2C). The Firmicutes to Bacteroidetes ratio was also increased in maternal HF/HS offspring (Fig. 2D).
Figure 2.
Shift in the cecal microbiome of high-fat/high-sucrose (HF/HS) male and female offspring. A: Bray–Curtis plots for β-diversity of cecal microbiome from HF/HS and chow (CON) offspring. B: measures of α-diversity (observed OTUs, Faith PD, and Shannon diversity) in cecal microbiome of HF/HS and CON offspring. C: relative abundance of each bacterial family in cecal microbiome of HF/HS and CON offspring. D: ratio of Firmicutes to Bacteroidetes in cecal microbiome of HF/HS and CON offspring. Quantitative data presented as means ± SE with n = 10 in each group and ≥5 separate litters represented in each group. Male and female offspring included in each section. P values as indicated on graph for t tests. OTU, operational taxonomic unit; PD, phylogenetic diversity.
Altered Cecal BA Profile and BA Metabolizing Bacteria in Offspring Exposed to Maternal HF/HS Diet
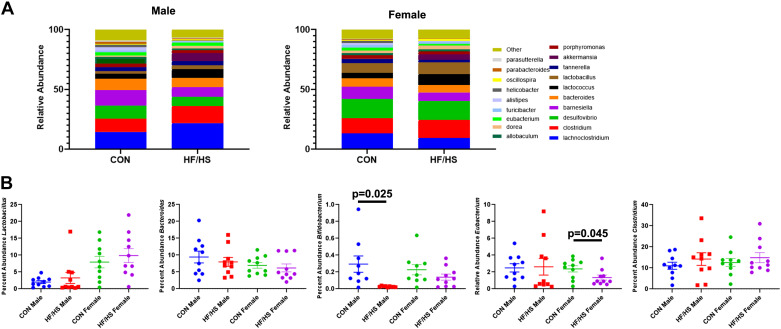
We identified shifted abundance of numerous genera of bacteria between HF/HS and CON offspring (Fig. 3A). We specifically compared the abundance of five genera known to metabolize BA’s between HF/HS and CON offspring(16). No significant differences were observed in Lactobacillus, Bacteroides, and Clostridium (Fig. 3B). There was a significant decrease in Bifidobacterium in males and Eubacterium in females (Fig. 3B).
Figure 3.
Abundance of bacterial genera involved in bile acid (BA) metabolism in offspring cecal contents. A: relative abundance of top 20 genera of bacteria present in cecal contents of high-fat/high-sucrose (HF/HS) and chow (CON) offspring. B: percent abundance of five specific genera of bacteria that can metabolize bile acids (Lactobacillus, Bacteroides, Bifidobacterium, Eubacterium, and Clostridium). Quantitative data presented as means ± SE with n = 10 in each group and ≥4 separate litters represented in each group. Male and female offspring included in each section. P values as indicated on graph for t tests.
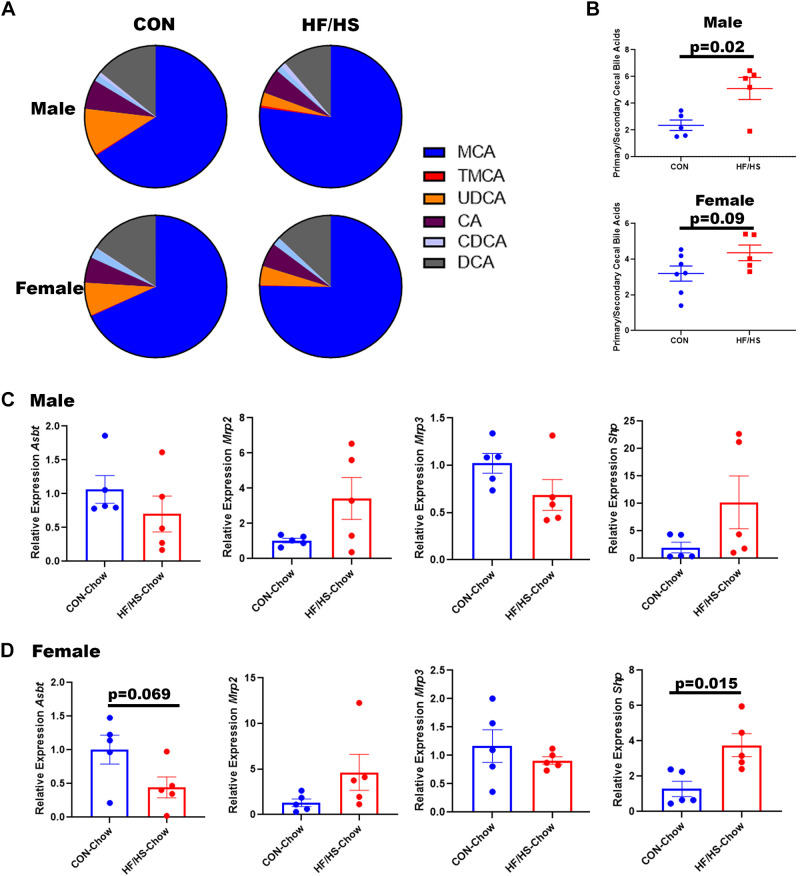
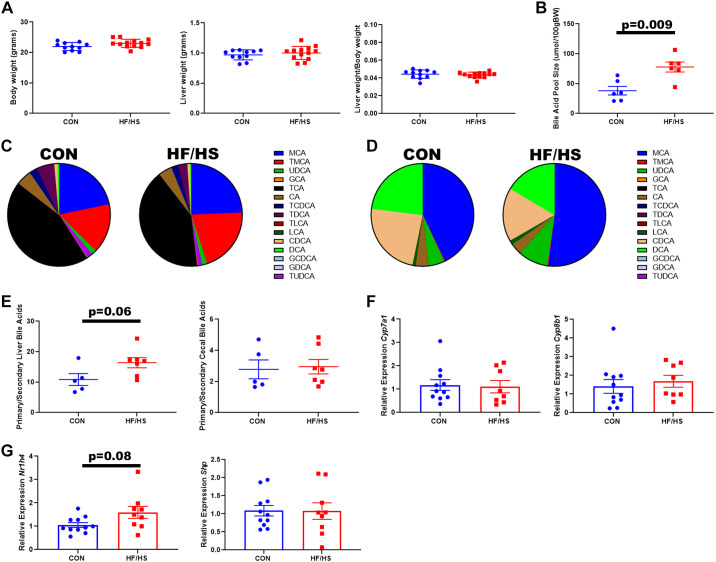
If the microbiome is playing a role in changing BA homeostasis, we would expect a shift in BA concentrations in stool. Consistent with previous reports of intrahepatic BA profile changes (9), cecal BA from HF/HS offspring showed increased muricholic acid (MCA) and decreased deoxycholic acid (DCA) (Fig. 4A). There was also decreased abundance of urso-DCA (UDCA) species. These changes were similar in male and female offspring. Consistent with these specific shifts in BA abundance, the ratio of primary to secondary bile acids was significantly increased in male HF/HS offspring (Fig. 4B). A trend toward this increase was observed in female HF/HS offspring (Fig. 4B). Over 99% of cecal BAs were unconjugated in each group. We measured expression of bile acid transporters in the ileum of HF/HS and CON offspring. Male offspring did not show any differences in Asbt, Mrp2, or Mrp3 (Fig. 4C). Female offspring exhibited a trend toward a decrease in Asbt expression (Fig. 4D). Bile acid-mediated activation of FXR represses Asbt expression via an increase in Shp (17). Ileum expression of Shp was increased in female HF/HS offspring, but not in males (Fig. 4, C and D).
Figure 4.
Shift in cecal bile acid concentrations in high-fat/high-sucrose (HF/HS) offspring. A: relative abundance of individual bile acids present in cecal contents of HF/HS and CON offspring. B: ratio of primary to secondary bile acids present in cecal contents of HF/HS and chow (CON) offspring. C: relative expression of Asbt, Mrp2, Mrp3, and Shp in ileum of male HF/HS and CON offspring. D: relative expression of Asbt, Mrp2, Mrp3, and Shp in ileum of female HF/HS and CON offspring. Quantitative data presented as means ± SE with n ≥ 5 in each group and ≥5 separate litters represented in each group. Male and female offspring included in each section unless noted. P values as indicated on graph for t tests. CA, cholic acid; DCA, deoxycholic acid; MCA, muricholic acid; TMCA, tauro-muricholic acid; UDCA, urso-deoxycholic acid. CDCA, chenodeoxycholic acid.
Cecal Microbiome Transplantation from HF/HS Donor Alters BA Homeostasis in Recipient Mice
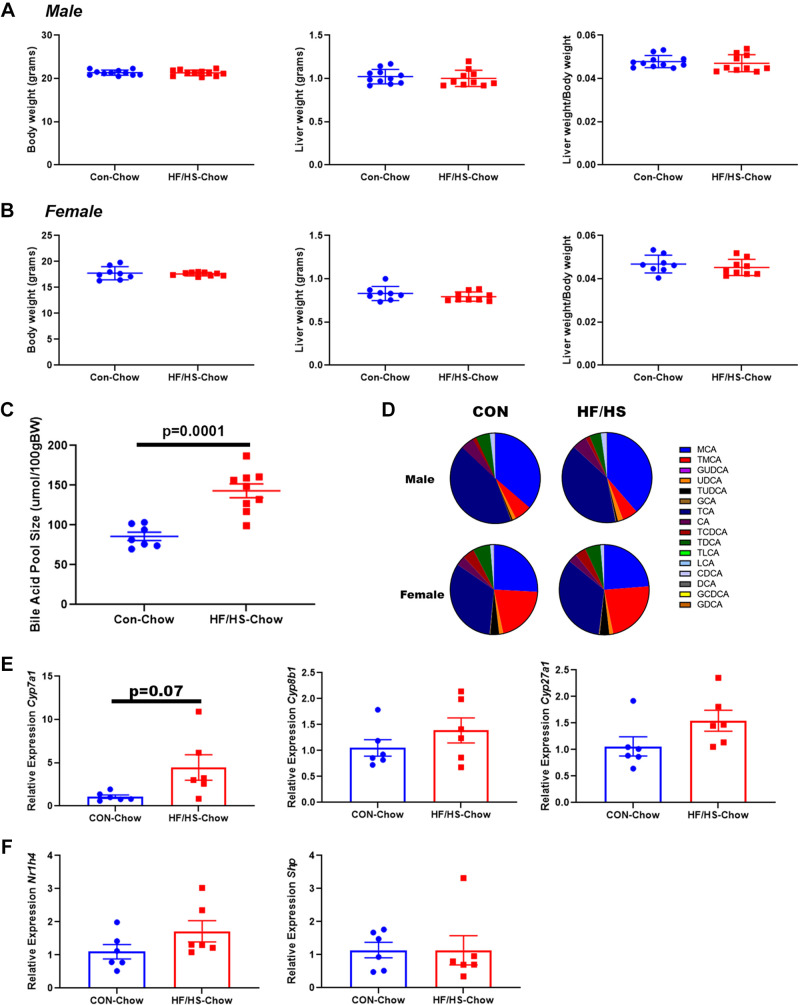
To determine if changes in the microbiome causally relate to changes in BA homeostasis in the offspring, we performed cecal microbiome transplantation (CMT) from HF/HS and CON offspring to mice whose microbiome was depleted by antibiotic treatment. After CMT, HF/HS and CON recipients exhibited similar body and liver weights (Fig. 5A). BA pool size was significantly greater in HF/HS recipients (Fig. 5B). Intrahepatic BA profiles were shifted in HF/HS recipient mice with increased abundance of MCA and tauro-MCA (TMCA), whereas tauro-DCA (TDCA) abundance decreased (Fig. 5C). The cecal BA profile of HF/HS recipient mice showed increased abundance of MCA and decreased abundance of DCA (Fig. 5D). We observed a trend toward an increased primary to secondary BAs in livers of HF/HS recipients but not in cecal BAs (Fig. 5E). Expression of two primary BA synthetic genes, Cyp7a1 and Cyp8b1, was not affected in HF/HS recipient livers (Fig. 5F). A trend toward an increase in Nr1h4 expression was observed; however, there was no associated difference in Shp expression (Fig. 5G).
Figure 5.
Recipients of cecal microbiome transplant from high-fat/high-sucrose (HF/HS) offspring develop altered bile acid (BA) homeostasis. A: body weight, liver weight, and liver weight-to-body weight ratio of HF/HS and chow (CON) recipient mice. B: BA pool size in HF/HS and CON recipient mice. C: abundance of individual BAs in liver tissue from HF/HS and CON recipient mice. D: abundance of individual BAs in cecal contents from HF/HS and CON recipient mice. E: ratio of primary to secondary BAs in liver and cecal contents of HF/HS and CON recipient mice. F: relative expression of Cyp7a1 and Cyp8b1 in liver of HF/HS and CON recipient mice. G: relative expression of Nr1h4 and Shp in liver of HF/HS and CON recipient mice. Quantitative data presented as means ± SE with n ≥ 5 in each group with representation from ≥5 litters. All data from male mice receiving cecal microbiome transplantation (CMT) from female donors. P values as indicated on graph for t tests.
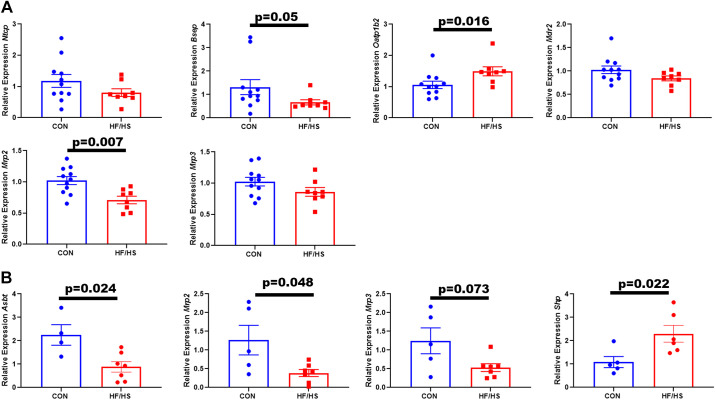
CMT from HF/HS Lineage Donor Altered Expression of Bile Acid Transport Genes in Liver and Ileum
The expression of apical transporters Bsep and Mrp2 significantly decreased in livers of HF/HS recipient mice after CMT, whereas the expression of the basolateral transporter Oatp1b2 significantly increased (Fig. 6A). Hepatic expression of Ntcp, Mdr2, and Mrp3 remained constant. Ileal Asbt, Mrp2, and Mrp3 expression decreased in HF/HS recipients (Fig. 6B). Ileal Shp expression was increased (Fig. 6B).
Figure 6.
Altered expression of bile acid (BA) transport genes in recipients of cecal microbiome from high-fat/high-sucrose (HF/HS) offspring. A: relative expression of hepatic BA transporters in HF/HS and chow (CON) recipient mice. B: relative expression of ileal BA transporters and Shp in HF/HS and CON recipient mice. Quantitative data presented as means ± SE with n ≥ 4 in each group with representation from ≥4 litters. All data from male mice receiving cecal microbiome transplantation (CMT) from female donors. P values as indicated on graph for t tests.
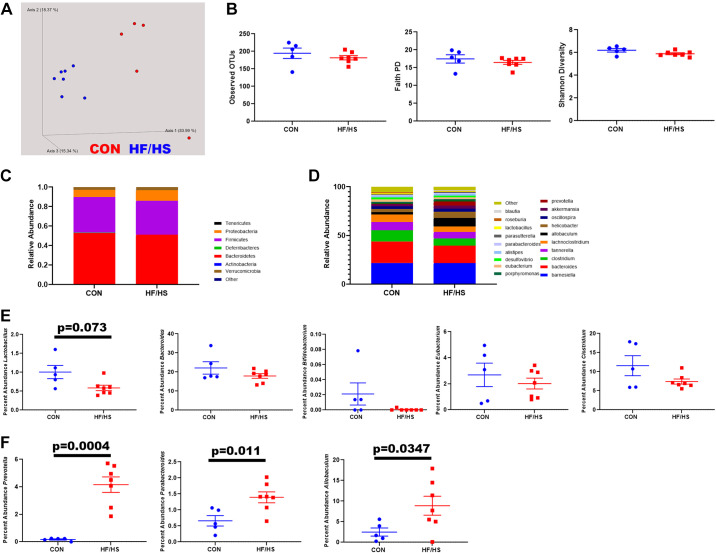
Gut Microbiome Altered in HF/HS CMT Recipients Compared with CON
β-Diversity of gut microbial communities differed between HF/HS and CON recipients. In contrast, three measures of α-diversity remained unchanged between groups (Fig. 7B). Analysis at the phylum level only showed minor differences in the abundance of specific bacterial phyla with more differences present at the genus level (Fig. 7, C and D). Of five different genera known to be involved in BA metabolism, only one (Lactobacillus) showed a trend toward decreased abundance (Fig. 7E), whereas Prevotella, Parabacteroides, and Allobaculum significantly increased (Fig. 7F).
Figure 7.
Shifts in gut microbiome in high-fat/high-sucrose (HF/HS) recipient mice compared with chow (CON) recipient mice. A: Bray–Curtis plot for β-diversity of cecal contents from HF/HS and CON recipient mice. B: measures of α-diversity (observed OTUs, Faith PD, and Shannon diversity) in cecal microbiome of HF/HS and CON recipient mice. C: relative abundance of each bacterial family in cecal microbiome of HF/HS and CON offspring. D: relative abundance of top 20 genera of bacteria present in cecal contents of HF/HS and CON offspring. E: percent abundance of five specific genera of bacteria that can metabolize bile acids (Lactobacillus, Bacteroides, Bifidobacterium, Eubacterium, and Clostridium). F: percent abundance of additional genera of bacteria that are significantly increased in cecal microbiome of HF/HS recipient mice. Five separate litters represented in each group with triglyceride and cholesterol concentrations, ≥6 separate litters represented for all other quantitative data. All data from male mice receiving cecal microbiome transplantation (CMT) from female donors. P values as indicated on graph for t tests. OTU, operational taxonomic unit; PD, phylogenetic diversity.
Changes in Short-Chain Fatty Acid Concentrations in HF/HS Recipient Mice
Short-chain fatty acids (SCFAs) of bacterial origin can also affect hepatic physiology. We measured concentrations of SCFAs in cecal contents and serum from CMT recipient mice. Cecal butyrate was significantly decreased in HF/HS recipient mice whereas cecal acetate and propionate were unchanged (Supplemental Fig. S2A), with no change in serum SCFA concentrations (Supplemental Fig. S2B). Hepatic gene expression of Il6 was increased in HF/HS recipient mice (Supplemental Fig. S2C).
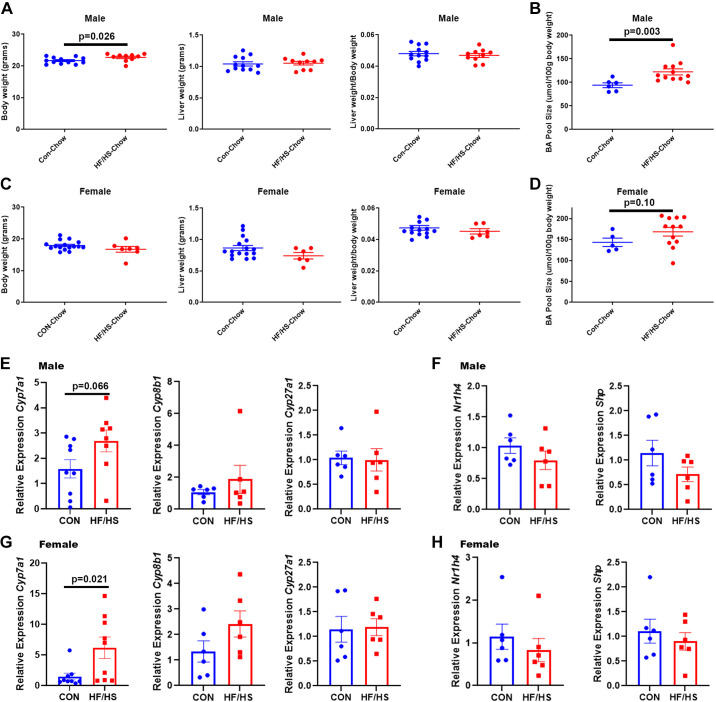
Maternal HF/HS Exposures Restricted to Pregnancy and Weaning Alters BA Homeostasis in Offspring
To determine whether exposure to maternal HF/HS diet restricted to gestation affects BA homeostasis, F0 female mice were placed on HF/HS diet the day after they were paired with a male and continued on the diet to the day of weaning. The weight of male offspring increased after maternal HF/HS diet exposure during pregnancy and lactation (Fig. 8, A and C). Liver weights in male and female offspring did not change (Fig. 8, A and C), but BA pool size increased in both sexes after maternal HF/HS exposure (Fig. 8, B and D).
Figure 8.
Altered bile acid (BA) homeostasis in offspring exposed to maternal obesogenic diet during pregnancy and lactation only. A: body weight, liver weight, and liver weight-to-body weight ratio of male offspring from chow (CON) or high fat/high sucrose (HF/HS) (P&L only). B: BA pool size in male offspring from CON or HF/HS (P&L only). C: body weight, liver weight, and liver weight-to-body weight ratio of female offspring from CON or HF/HS (P&L only). D: BA pool size in female offspring from CON or HF/HS (P&L only). E: relative expression of bile acid metabolism genes in male offspring from CON or HF/HS (P&L only). F: relative expression of Nr1h4 and Shp in male offspring from CON or HF/HS (P&L only). G: relative expression of bile acid metabolism genes in female offspring from CON or HF/HS (P&L only). H: Relative expression of Nr1h4 and Shp in female offspring from CON or HF/HS (P&L only). Quantitative data presented as means ± SE with n ≥ 5 in each group with ≥5 separate litters represented in each group. Male and female offspring in each section. P values as noted on each graph for t tests. P&L, pregnancy and lactation.
A significant increase in Cyp7a1 expression was observed in female offspring with a trend toward an increase (P = 0.066) in male offspring (Fig. 8, E and G). Neither male nor female offspring exhibited differences in expression of Cyp8b, and Cyp27a1 (Fig. 8, E and G), or of the BA nuclear receptor FXR or transcriptional repressor Shp (Fig. 8, F and H). We observed a trend toward an increase in hepatic Bsep expression and a decrease in Ntcp expression in male offspring exposed to maternal HF/HS diet between conception and weaning (Supplemental Fig. S3A), but no difference in female offspring was observed (Supplemental Fig. S3B). Likewise, we found no differences in mRNA expression of Oatp1b, Mrp2, and Mrp3 in offspring (Supplemental Fig. S3, A and B).
Intergenerational Transmission of Altered BA Homeostasis after Maternal HF/HS Exposure
We previously reported that F2 offspring exhibited increased BA pool sizes if bred from F1 female offspring from an F0 female that had been fed HF/HS diet (9). To evaluate whether an altered BA homeostasis phenotype is also passed by male F1 offspring from an F0 female fed HF/HS diet, we bred F1 generation offspring with lean females and evaluated BA homeostasis in the subsequent F2 offspring (termed PF2). No difference in body weight, liver weight, or liver weight-to-body weight ratio was observed in male or female offspring (Fig. 9, A and B). PF2H offspring exhibited increased BA pool size (Fig. 9C), without a difference in intrahepatic BA profile (Fig. 9D). Expression of Cyp7a1 mRNA exhibited a trend toward an increase (P = 0.07) in PF2H offspring liver, but other BA metabolism genes were unchanged (Fig. 9, E and F).
Figure 9.
Altered bile acid (BA) homeostasis in second generation offspring of male exposed to maternal obesogenic diet. A: body weight, liver weight, and liver weight/body weight in male offspring. B: body weight, liver weight, and liver weight-to-body weight ratio in female offspring. C: BA pool size of PF2 high-fat/high-sucrose (HF/HS) and chow (CON) male offspring. D: abundance of individual BAs in liver of PF2 HF/HS and CON male and female offspring. E: relative expression of bile acid metabolism genes in PF2 HF/HS and CON male offspring liver. F: relative expression of Nr1h4 and Shp in PF2 HF/HS and CON male offspring liver. Quantitative data presented as means ± SE with n ≥ 6 in each group and ≥6 separate litters represented in each group. P values as noted on each graph for t tests.
Microbiome from HF/HS Offspring is Sufficient to Enhance Fructose-Induced Hepatic Steatosis
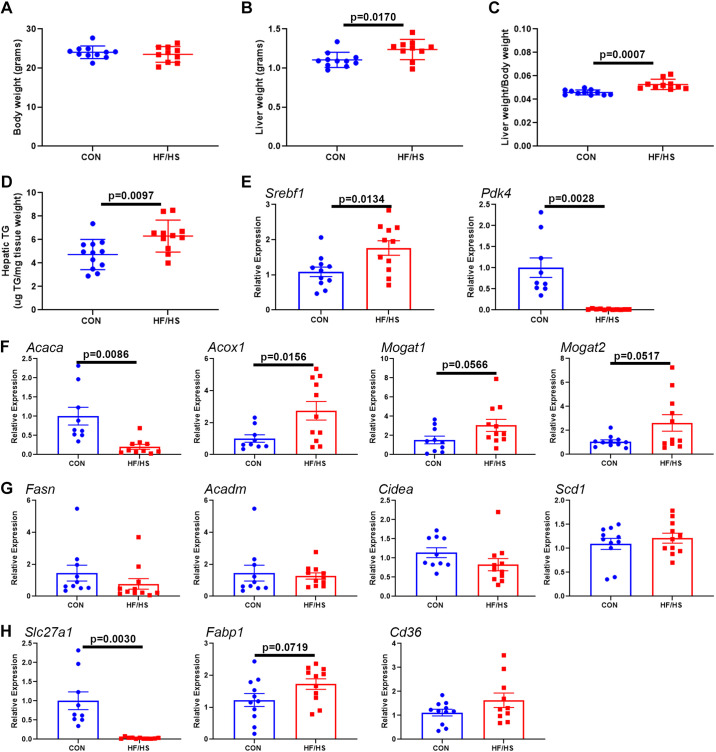
We next asked if the microbiome alteration due to maternal obesogenic diet exposure alters hepatic lipid metabolism, we performed CMT from HF/HS and CON offspring to antibiotic-treated mice followed by feeding a high-fructose diet for 7 days. No difference in body weight was observed (Fig. 10A), but liver weight and liver weight-to-body weight ratio were increased in HF/HS recipients (Fig. 10, B and C) along with increased hepatic TG (Fig. 10D). Gene expression analysis identified an increase in Srebf1 expression and a decrease in Pdk4 expression (Fig. 10E). Analysis of genes involved in lipid metabolism identified a significant decrease in Acaca and significant increase in Acox1 (Fig. 10F), and trend toward increased Mogat1 and Mogat2. No change in expression was observed in several other lipid metabolism genes (Fig. 10G). Genes involved in lipid transport were also affected with a decrease in Slc27a1 and a trend toward an increase in Fabp1 (P = 0.0719) (Fig. 10H).
Figure 10.
Transfer of microbiome from high-fat/high-sucrose (HF/HS) offspring increases fructose-induced hepatic triglyceride (TG) accumulation and alters hepatic lipid metabolism. A: body weight of HF/HS and chow (CON) recipient mice after fructose diet feeding. B: liver weight of HF/HS and CON recipient mice after fructose diet feeding. C: liver weight-to-body weight ratio of HF/HS and CON recipient mice after fructose diet feeding. D: hepatic triglyceride concentration in HF/HS and CON recipient mice after fructose diet feeding. E: relative expression of transcription factors that control lipid metabolism in liver of HF/HS and CON recipient mice after fructose diet feeding. F: relative expression of lipid metabolism genes with significant or trend toward a change in liver of HF/HS and CON recipient mice after fructose diet feeding. G: relative expression of lipid metabolism genes that are unchanged in liver of HF/HS and CON recipient mice after fructose diet feeding. H: relative expression of lipid transport genes in liver of HF/HS and CON recipient mice after fructose diet feeding. Quantitative data presented as means ± SE with n ≥ 6 in each group and ≥6 separate litters represented in each group. All data from male mice that received cecal microbiome transplantation (CMT) from female donors. P values as noted on each graph for t tests.
DISCUSSION
Developmental programming of NAFLD due to maternal obesogenic diet has been described in multiple models with an altered microbiome emerging as a candidate mode of intergenerational transmission (6–9, 13, 18, 19). We and others have reported changes in BA homeostasis after maternal obesogenic diet exposure. Here, we demonstrate 1) increased bile acid pool size and a shift in the intrahepatic BA profile after CMT from HF/HS offspring to antibiotic-treated mice, 2) altered expression of several hepatic and intestinal BA transporters following CMT for HF/HS recipient mice, 3) decreased fecal butyrate concentrations in HF/HS recipient mice, 4) altered BA homeostasis in offspring exposed to time-limited maternal obesogenic diet (pregnancy and lactation only), and 5) enhanced fructose-induced hepatic steatosis in HF/HS recipient mice. Several of these findings warrant discussion.
Maternal Obesogenic Diet Induced Shifts in the Offspring Microbiome Regulate BA Homeostasis
We initially hypothesized that an altered microbiome in HF/HS offspring would be sufficient to induce the observed changes in BA homeostasis. Indeed, we found that following CMT from HF/HS offspring to microbiome-depleted mice similarly increases BA pool size and shifts the intrahepatic BA profile of the recipient mice. Although these changes are consistent with our prior work (9), we did not observe a similar increase in Cyp7a1 following CMT from HF/HS offspring. It is worth noting that Soderborg et al. (13) observed increased Cyp7a1 expression in their human fecal microbiome transplantation transplant model. One possible explanation is that expression of some of these genes may be under epigenetic regulation rather being specifically related to microbiome changes or to concentrations of specific BAs in our model. It is likely that the phenotypes observed after maternal diet-driven developmental programming will combine microbiome and epigenetic drivers. Another possibility is that differences in the type of maternal obesogenic diets administered induces variability in offspring microbiomes and BA metabolism. Changes in the offspring microbiomes might also reflect sex (20). Although no difference in Cyp7a1 was observed after CMT, we did find differences in multiple ileal and hepatic genes involved in BA transport. The decrease in all three ileal BA transporters could reflect a compensatory response to the increased BA pool size in an attempt to limit BA reuptake. We also observed increased hepatic Oatp1b2 (BA uptake into the hepatocyte) expression and decreased Bsep and Mrp2 (BA export from hepatocytes) expression. These findings would support overall BA retention within the hepatocyte. It is unclear if this reflects a response to the increased BA pool size or if this represents primary mediation of the changes in BA homeostasis secondary to microbiome changes. It is important to note that a scenario of increased BA retention within the hepatocyte could be cytotoxic or alter cell signaling (21, 22). Taken together, these studies implicate alterations in the microbiome as one determinant of altered BA homeostasis in offspring exposed to maternal high-fat diet (HFD).
Exposure during Pregnancy and Lactation Alone is Sufficient to Change BA Homeostasis
We (9) and others (13, 20) have shown that maternal diet affects bile acid metabolism when feeding starts before pregnancy. We now show that maternal HF/HS diet exposure during and immediately after pregnancy is sufficient to alter offspring BA homeostasis. We observed increased BA pool sizes in male and female offspring with an associated increase in Cyp7a1. These effects could be microbiome mediated as there is likely an effect on the microbiome after this shorter-term HF/HS diet exposure. Indeed, HFD exposure during gestation and lactation is sufficient to shift the microbiomes of Japanese macaques (23). Microbiome alterations are also present in mice offspring after maternal HFD exposure during gestation and lactation only with associated impairments in the gut barrier (24). Exposure to HFD perinatally exacerbates HFD-induced hepatic stetatosis in the offspring (25). It is clear that exposure during this key period is sufficient to drive changes in the offspring, so it will be important to differentiate and quantify the effect of prenatal exposure versus exposure during gestation/lactation alone, as transition to standard chow 1 wk before conception worsens steatosis (26).
Changes in the Microbiome Driven by Maternal HF/HS Exposure Alter SCFA Metabolism in Offspring
Butyrate protects from hepatic steatosis and maintains the intestinal barrier (27, 28). We did not observe changes in circulating SCFAs leaving unanswered the impact on systemic SCFA metabolism, but the changes in SCFA concentrations in portal blood might be more informative of SCFA delivery to the liver, and form a basis for future study.
Phenotypes May Be Affected by Timing of Microbiome Changes
It is important to consider the timing of microbiome changes and how that may affect phenotypes in the offspring. In the current study, we collected cecal contents from 6-wk-old mice and transplanted into adult mice. However, recent work has identified a key role for the early microbiome in programming the responsiveness of the immune system to future inflammatory insults (29), specifically a weaning reaction that occurs between 2 and 4 wk and is dependent on the early microbiome. In the absence of the microbiome, the weaning reaction, defined by temporary increases in colonic TNF-α and IFN-γ, did not occur and subsequently led to worse inflammatory response in the adult. It is not yet defined how maternal HFD may impact this weaning reaction, but there is increased intestinal inflammation and gut permeability in weaning age offspring after such exposure (30). The consequences of maternal diet-induced changes in the offspring will need to be more closely studied in the perinatal period given the programming impact of the early microbiome.
Intergenerational Transmission of Altered Bile Acid Homeostasis
Developmental programming studies can evaluate whether phenotypes are passed in an intergenerational (to F2) or transgenerational (to F3) manner. We previously reported that F2 offspring after F0 maternal HF/HS exposure have increased BA pool size but no difference in intrahepatic BA profile was observed (9). F2 offspring in these studies came from F1 females exposed to maternal HF/HS. In this report, we show that F2 offspring from F1 males exposed to maternal HF/HS also have increased BA pool size. This shows that this phenotype can be passed through male offspring and also that changes in BA pool size may also be driven by factors other than the microbiome as the F1 males are not contributing directly to the offsprings microbiome. In future, more mechanistic studies will be necessary to evalaute the factors that are driving the nonmicrobiome-mediated changes in offspring BA homeostasis after maternal HF/HS diet exposure.
Potential Sex Differences in Response to Maternal Obesogenic Diet
Previous studies have identified sex differences in response to maternal obesogenic diet and the development of offspring NAFLD (31–33). Although some commonalities exist beteween male and female offspring in BA homeostasis in our studies, there are also differences that merit consideration. Male offspring exposed to maternal obesogenic diet exhibited an increase in the ratio of primary to secondary BAs in cecal contents; however, only a trend was observed in females. This could suggest variability in how the microbiome is modifying BAs between male and female offspring, which could be a direct result of differences in the microbiome makeup. We observed similarites in the microbiome between sexes at the phylum level; however, analysis at the genus level revealed important differences, most notably a decrease in Bifidobacterium in males and a decrese in Eubacterium in females. Further studies will be necessary to decipher how these specific genus level differences affect offspring bile acid homeostasis through more reductionist approaches. Another sex difference we observed is that the expression of the ileal bile acid transporter Asbt is decreased in female HF/HS offspring as the BA pool increases, but those changes were not observed in male HF/HS offspring. We speculate that Asbt is decreased in response to elevated BA levels, possibly as a compensatory adaptation to decrease reabsorption and normalize the total BA pool. Since we do not observe this same adaptation in male offspring, that difference may in part explain why we observe more sigificant alterations in BA homeostasis in male HF/HS offspring. This speculation, however, will be a focus of future work to delineate sex differences in phenotypes driven by maternal obesogenic diet exposure.
There are additional limitations we want to highlight in relation to the current studies. The molecular analysis presented here centers around changes in gene expression by quantitative PCR. These changes may not reflect what is happening at the level of protein function or localization within the cell. In particular, FXR and Shp control gene expression in the nucleus and their cell-specific localization and function will be important to evaluate in future studies. Another limitation is that some but not all analyses were completed in both sexes in the offpsring. Although there is clear overlap in some of the changes in BA homeostasis and microbiome shifts, there appear to be sex differences that warrant further evaluation. Most of our analysis in the CMT studies utilized female donor cecal contents, and it is possible that there may be sex differences reflecting the source of donor microbiome. Nevertheless, we observed a similar increase in BA pool size when utilizing donor cecal contents from male offspring (Supplemental Fig. S4). Likewise, CMT studies were only performed in male recipients and it is possible the response to CMT in female recipients may be different. There are also inherent limitations in relation to performance of CMT studies, including the observation that antibiotic treatment followed by microbiome transplantation does not result in an exact engraftment of the same bacterial abundances (34). Although we observed similarities in the general directionality of abundances in donor and recipient abundances at the genus level, none of the differences achieve statistical significance in both groups. An alternative approach to increase efficiency of engraftment is to perform similar studies in gnotobiotic mice. However, gnotobiotic mice exhibit developmental defects due to the importance of the early microbiome, which could affect the outcomes of analyses performed here (for review, see Ref. 35). Given these limitations, the current data do not definitively show that certain specific changes in the offspring microbiome cause the observed changes in BA homeostasis and thus can only be attributed to global changes in the microbiome following maternal obesogenic diet exposure.
In summary, alteration of the offspring microbiome by maternal HF/HS diet exposure is sufficient to change BA homeostasis in offspring, increasing the bile acid pool size and shifting the intrahepatic BA pool. Furthermore, transferring the microbiome after maternal obesogenic diet exposure increases fructose-induced steatosis. Given the clear effect of maternal diet-driven microbiome alteration on offspring, interventions that reverse these changes could provide a pathway for disease prevention. Future experiments will need to further define the role of specific bacteria that are affected in the offspring microbiome. We anticipate that that future studies, including epigenetic effects of the microbiome, will guide the path for reversing developmental programming of NAFLD.
SUPPLEMENTAL DATA
Supplemental Figs. S1–S4: https://doi.org/10.6084/m9.figshare.15044439;
Supplemental Material: https://doi.org/10.6084/m9.figshare.15044439.v1.
GRANTS
This study was supported by National Institutes of Health (NIH) Grants DK-122018 (to M. D. Thompson); AGA Research Scholar Award (to M. D. Thompson); NIH Washington University Digestive Diseases Research Core Center Grant No. NIDDK P30 DK052574 (to M. D. Thompson, P. I. Tarr and N. O. Davidson); NIH Grant P30 DK056341 (Nutrition Obesity Research Center) (to M. D. Thompson); and by the Washington University Diabetes Research Center, Grant No. P30 DK020579 (to M. D. Thompson). N. O. Davidson was also supported by NIH Grants DK-119437, HL-151328, and DK-128169.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
M.D.T., P.I.T., and N.O.D. conceived and designed research; M.D.T., J.K., A.F., H.H., O.Ö., J.C., and Y.X. performed experiments; M.D.T., J.K., A.F., H.H., O.Ö., and J.C. analyzed data; M.D.T., J.K., and N.O.D. interpreted results of experiments; M.D.T., J.K., A.F., and O.Ö. prepared figures; M.D.T. drafted manuscript; M.D.T., J.K., H.H., O.Ö., P.I.T., and N.O.D. edited and revised manuscript; M.D.T., J.K., H.H., O.Ö., P.I.T., and N.O.D. approved final version of manuscript.
ACKNOWLEDGMENTS
The authors acknowledge the expert assistance of David Scherrer and Xuntian Jiang in the Metabolomics Core for performance of BA composition and SCFA analysis and Sangeeta Adak for completing triglyceride measurements.
REFERENCES
- 1.Younossi ZM, Koenig AB, Abdelatif D, Fazel Y, Henry L, Wymer M. Global epidemiology of nonalcoholic fatty liver disease-meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology 64: 73–84, 2016. doi: 10.1002/hep.28431. [DOI] [PubMed] [Google Scholar]
- 2.Younossi Z, Anstee QM, Marietti M, Hardy T, Henry L, Eslam M, George J, Bugianesi E. Global burden of NAFLD and NASH: trends, predictions, risk factors and prevention. Nat Rev Gastroenterol Hepatol 15: 11–20, 2018. doi: 10.1038/nrgastro.2017.109. [DOI] [PubMed] [Google Scholar]
- 3.Thompson MD. Developmental programming of NAFLD by parental obesity. Hepatol Commun 4: 1392–1403, 2020. doi: 10.1002/hep4.1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ayonrinde OT, Oddy WH, Adams LA, Mori TA, Beilin LJ, de Klerk N, Olynyk JK. Infant nutrition and maternal obesity influence the risk of non-alcoholic fatty liver disease in adolescents. J Hepatol 67: 568–576, 2017. doi: 10.1016/j.jhep.2017.03.029. [DOI] [PubMed] [Google Scholar]
- 5.Bellatorre A, Scherzinger A, Stamm E, Martinez M, Ringham B, Dabelea D. Fetal overnutrition and adolescent hepatic fat fraction: the exploring perinatal outcomes in children study. J Pediatr 192: 165–170.e1, 2018. doi: 10.1016/j.jpeds.2017.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruce KD, Cagampang FR, Argenton M, Zhang J, Ethirajan PL, Burdge GC, Bateman AC, Clough GF, Poston L, Hanson MA, McConnell JM, Byrne CD. Maternal high-fat feeding primes steatohepatitis in adult mice offspring, involving mitochondrial dysfunction and altered lipogenesis gene expression. Hepatology 50: 1796–1808, 2009. doi: 10.1002/hep.23205. [DOI] [PubMed] [Google Scholar]
- 7.Mouralidarane A, Soeda J, Visconti-Pugmire C, Samuelsson AM, Pombo J, Maragkoudaki X, Butt A, Saraswati R, Novelli M, Fusai G, Poston L, Taylor PD, Oben JA. Maternal obesity programs offspring nonalcoholic fatty liver disease by innate immune dysfunction in mice. Hepatology 58: 128–138, 2013. doi: 10.1002/hep.26248. [DOI] [PubMed] [Google Scholar]
- 8.Gutierrez Sanchez LH, Tomita K, Guo Q, Furuta K, Alhuwaish H, Hirsova P, Baheti S, Alver B, Hlady R, Robertson KD, Ibrahim SH. Perinatal nutritional reprogramming of the epigenome promotes subsequent development of nonalcoholic steatohepatitis. Hepatol Commun 2: 1493–1512, 2018. doi: 10.1002/hep4.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thompson MD, Derse A, Ferey J, Reid M, Xie Y, Christ M, Chatterjee D, Nguyen C, Harasymowicz N, Guilak F, Moley KH, Davidson NO. Transgenerational impact of maternal obesogenic diet on offspring bile acid homeostasis and nonalcoholic fatty liver disease. Am J Physiol Endocrinol Physiol 316: E674–E686, 2019. doi: 10.1152/ajpendo.00474.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guzior DV, Quinn RA. Review: microbial transformations of human bile acids. Microbiome 9: 140, 2021. doi: 10.1186/s40168-021-01101-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wankhade UD, Zhong Y, Kang P, Alfaro M, Chintapalli SV, Thakali KM, Shankar K. Enhanced offspring predisposition to steatohepatitis with maternal high-fat diet is associated with epigenetic and microbiome alterations. PLoS One 12: e0175675, 2017. doi: 10.1371/journal.pone.0175675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma J, Prince AL, Bader D, Hu M, Ganu R, Baquero K, Blundell P, Alan Harris R, Frias AE, Grove KL, Aagaard KM. High-fat maternal diet during pregnancy persistently alters the offspring microbiome in a primate model. Nat Commun 5: 3889, 2014. doi: 10.1038/ncomms4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Soderborg TK, Clark SE, Mulligan CE, Janssen RC, Babcock L, Ir D, Young B, Krebs N, Lemas DJ, Johnson LK, Weir T, Lenz LL, Frank DN, Hernandez TL, Kuhn KA, D'Alessandro A, Barbour LA, El Kasmi KC, Friedman JE. The gut microbiota in infants of obese mothers increases inflammation and susceptibility to NAFLD. Nat Commun 9: 4462, 2018. doi: 10.1038/s41467-018-06929-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saben JL, Boudoures AL, Asghar Z, Thompson A, Drury A, Zhang W, Chi M, Cusumano A, Scheaffer S, Moley KH. Maternal metabolic syndrome programs mitochondrial dysfunction via germline changes across three generations. Cell Rep 16: 1–8, 2016. doi: 10.1016/j.celrep.2016.05.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xie Y, Cifarelli V, Pietka T, Newberry EP, Kennedy SM, Khalifeh-Soltani A, Clugston R, Atabai K, Abumrad NA, Davidson NO. Cd36 knockout mice are protected against lithogenic diet-induced gallstones. J Lipid Res 58: 1692–1701, 2017. doi: 10.1194/jlr.M077479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, Pettersson S. Host-gut microbiota metabolic interactions. Science 336: 1262–1267, 2012. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 17.Li H, Chen F, Shang Q, Pan L, Shneider BL, Chiang JY, Forman BM, Ananthanarayanan M, Tint GS, Salen G, Xu G. FXR-activating ligands inhibit rabbit ASBT expression via FXR-SHP-FTF cascade. Am J Physiol Gastrointest Liver Physiol 288: G60–G66, 2005. doi: 10.1152/ajpgi.00170.2004. [DOI] [PubMed] [Google Scholar]
- 18.Thompson MD, Cismowski MJ, Trask AJ, Lallier SW, Graf AE, Rogers LK, Lucchesi PA, Brigstock DR. Enhanced steatosis and fibrosis in liver of adult offspring exposed to maternal high-fat diet. Gene Expr 17: 47–59, 2016. doi: 10.3727/105221616X692135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thorn SR, Baquero KC, Newsom SA, El Kasmi KC, Bergman BC, Shulman GI, Grove KL, Friedman JE. Early life exposure to maternal insulin resistance has persistent effects on hepatic NAFLD in juvenile nonhuman primates. Diabetes 63: 2702–2713, 2014. doi: 10.2337/db14-0276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wankhade UD, Zhong Y, Kang P, Alfaro M, Chintapalli SV, Piccolo BD, Mercer KE, Andres A, Thakali KM, Shankar K. Maternal high-fat diet programs offspring liver steatosis in a sexually dimorphic manner in association with changes in gut microbial ecology in mice. Sci Rep 8: 16502, 2018. doi: 10.1038/s41598-018-34453-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allen K, Jaeschke H, Copple BL. Bile acids induce inflammatory genes in hepatocytes: a novel mechanism of inflammation during obstructive cholestasis. Am J Pathol 178: 175–186, 2011. doi: 10.1016/j.ajpath.2010.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perez MJ, Briz O. Bile-acid-induced cell injury and protection. World J Gastroenterol 15: 1677–1689, 2009. doi: 10.3748/wjg.15.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prince AL, Pace RM, Dean T, Takahashi D, Kievit P, Friedman JE, Aagaard KM. The development and ecology of the Japanese macaque gut microbiome from weaning to early adolescence in association with diet. Am J Primatol 81: e22980, 2019. doi: 10.1002/ajp.22980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xie R, Sun Y, Wu J, Huang S, Jin G, Guo Z, Zhang Y, Liu T, Liu X, Cao X, Wang B, Cao H. Maternal high fat diet alters gut microbiota of offspring and exacerbates DSS-induced colitis in adulthood. Front Immunol 9: 2608, 2018. doi: 10.3389/fimmu.2018.02608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kruse M, Seki Y, Vuguin PM, Du XQ, Fiallo A, Glenn AS, Singer S, Breuhahn K, Katz EB, Charron MJ. High-fat intake during pregnancy and lactation exacerbates high-fat diet-induced complications in male offspring in mice. Endocrinology 154: 3565–3576, 2013. doi: 10.1210/en.2012-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou Y, Peng H, Xu H, Li J, Golovko M, Cheng H, Lynch EC, Liu L, McCauley N, Kennedy L, Alpini G, Zhang KK, Xie L. Maternal diet intervention before pregnancy primes offspring lipid metabolism in liver. Lab Invest 100: 553–569, 2020. doi: 10.1038/s41374-019-0344-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao ZH, Wang ZX, Zhou D, Han Y, Ma F, Hu Z, Xin FZ, Liu XL, Ren TY, Zhang F, Xue Y, Cui A, Liu Z, Bai J, Liu Y, Cai G, Su W, Dai X, Shen F, Pan Q, Li Y, Fan JG. Sodium butyrate supplementation inhibits hepatic steatosis by stimulating liver kinase B1 and insulin-induced gene. Cell Mol Gastroenterol Hepatol 12: 857–871, 2021. doi: 10.1016/j.jcmgh.2021.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou D, Pan Q, Xin FZ, Zhang RN, He CX, Chen GY, Liu C, Chen YW, Fan JG. Sodium butyrate attenuates high-fat diet-induced steatohepatitis in mice by improving gut microbiota and gastrointestinal barrier. World J Gastroenterol 23: 60–75, 2017. doi: 10.3748/wjg.v23.i1.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Al Nabhani Z, Dulauroy S, Marques R, Cousu C, Al Bounny S, Dejardin F, Sparwasser T, Berard M, Cerf-Bensussan N, Eberl G. A weaning reaction to microbiota is required for resistance to immunopathologies in the adult. Immunity 50: 1276–1288.e5, 2019. doi: 10.1016/j.immuni.2019.02.014. [DOI] [PubMed] [Google Scholar]
- 30.Al Nabhani Z, Dulauroy S, Lécuyer E, Polomack B, Campagne P, Berard M, Eberl G. Excess calorie intake early in life increases susceptibility to colitis in adulthood. Nat Metab 1: 1101–1109, 2019. [Erratum in Nat Metab 1: 1169, 2019]. doi: 10.1038/s42255-019-0129-5. [DOI] [PubMed] [Google Scholar]
- 31.Dahlhoff M, Pfister S, Blutke A, Rozman J, Klingenspor M, Deutsch MJ, Rathkolb B, Fink B, Gimpfl M, Hrabě de Angelis M, Roscher AA, Wolf E, Ensenauer R. Peri-conceptional obesogenic exposure induces sex-specific programming of disease susceptibilities in adult mouse offspring. Biochim Biophys Acta 1842: 304–317, 2014. doi: 10.1016/j.bbadis.2013.11.021. [DOI] [PubMed] [Google Scholar]
- 32.Lomas-Soria C, Reyes-Castro LA, Rodríguez-González GL, Ibáñez CA, Bautista CJ, Cox LA, Nathanielsz PW, Zambrano E. Maternal obesity has sex-dependent effects on insulin, glucose and lipid metabolism and the liver transcriptome in young adult rat offspring. J Physiol 596: 4611–4628, 2018. doi: 10.1113/JP276372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ayonrinde OT, Adams LA, Mori TA, Beilin LJ, de Klerk N, Pennell CE, White S, Olynyk JK. Sex differences between parental pregnancy characteristics and nonalcoholic fatty liver disease in adolescents. Hepatology 67: 108–122, 2018. doi: 10.1002/hep.29347. [DOI] [PubMed] [Google Scholar]
- 34.Ericsson AC, Personett AR, Turner G, Dorfmeyer RA, Franklin CL. Variable colonization after reciprocal fecal microbiota transfer between mice with low and high richness microbiota. Front Microbiol 8: 196, 2017. doi: 10.3389/fmicb.2017.00196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kennedy EA, King KY, Baldridge MT. Mouse microbiota models: comparing germ-free mice and antibiotics treatment as tools for modifying gut bacteria. Front Physiol 9: 1534, 2018. doi: 10.3389/fphys.2018.01534. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figs. S1–S4: https://doi.org/10.6084/m9.figshare.15044439;
Supplemental Material: https://doi.org/10.6084/m9.figshare.15044439.v1.