Abstract
Microbial cell factories (bacteria and fungi) are the leading producers of beneficial natural products such as lycopene, carotene, herbal medicine, and biodiesel etc. These microorganisms are considered efficient due to their effective bioprocessing strategy (monoculture- and consortial-based approach) under distinct processing conditions. Meanwhile, the advancement in genetic and process optimization techniques leads to enhanced biosynthesis of natural products that are known functional ingredients with numerous applications in the food, cosmetic and medical industries. Natural consortia and monoculture thrive in nature in a small proportion, such as wastewater, food products, and soils. In similitude to natural consortia, it is possible to engineer artificial microbial consortia and program their behaviours via synthetic biology tools. Therefore, this review summarizes the optimization of genetic and physicochemical parameters of the microbial system for improved production of natural products. Also, this review presents a brief history of natural consortium and describes the functional properties of monocultures. This review focuses on synthetic biology tools that enable new approaches to design synthetic consortia; and highlights the syntropic interactions that determine the performance and stability of synthetic consortia. In particular, the effect of processing conditions and advanced genetic techniques to improve the productibility of both monoculture and consortial based systems have been greatly emphasized. In this context, possible strategies are also discussed to give an insight into microbial engineering for improved production of natural products in the future. In summary, it is concluded that the coupling of genomic modifications with optimum physicochemical factors would be promising for producing a robust microbial cell factory that shall contribute to the increased production of natural products.
Keywords: Genetic and process optimization techniques, Monoculture, Natural products, Synthetic consortia, Synthetic biology tools
1. Background
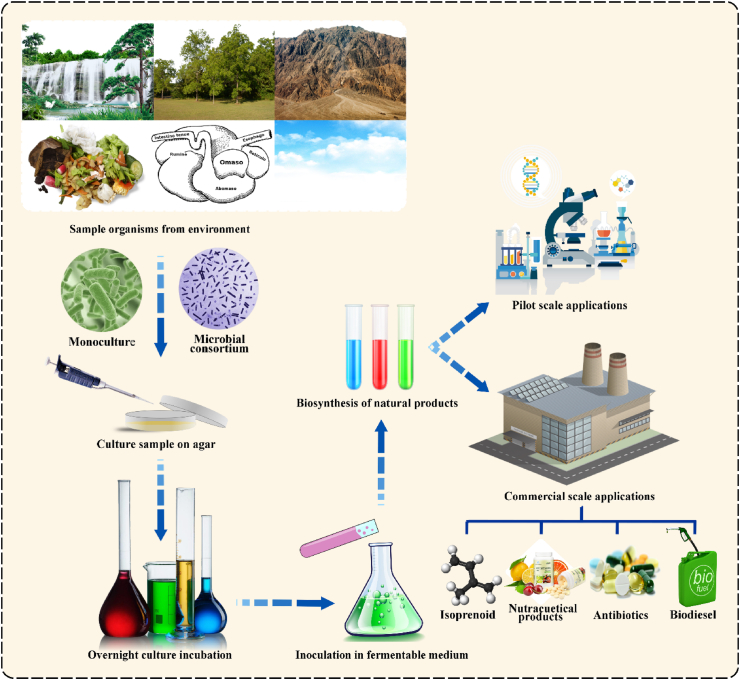
Green manufacturing industry, created from natural microorganisms, is a productive cell factory capable of valorizing polysaccharides (that are sourced from either plants or animals) into highly value-added natural products (NPs), such as flavonoid, carotenoid, and essential oil, etc. However, it is challenging to obtain a high yield due to involved stereochemical complexity [1]. Therefore, pretreatment methods before valorization have been investigated to overcome this bottleneck. Studies reveal that the optimum NPs production can be achieved by increasing microbial enzymes permeability that can be further enhanced by engineering microbial cells [2,3]. Generally, microbial-derived natural products exhibit lower water and land requirements and contribute as environmental benign (i.e., reduce global warming and pollution). Furthermore, these products are also cost-effective compared to products obtained from chemical plants. Normally, two major approaches, monoculture and consortial strategy, are adopted by microbial cells to produce diversified valuable products, as shown in Fig. 1 [4]. However, in the last decade, an increase in preference has been noticed for consortial systems because of higher production efficiency and fulfilment of more complicated processing tasks. Recently, a study on environmental biodiversity demonstrated the existence of 99% microorganisms in the form of consortia and highlighted their application areas such as wastewater treatment, bioremediation, composting, biomining, biofuel production and functional ingredients in the food industry [[5], [6], [7], [8]].
Fig. 1.
Schematic diagram shows the multi-step technique leading to biosynthesis of natural products via monoculture and consortium bioprocessing strategies.
Apart from the evolution of microbial platforms to transform nearly any carbon source into the desired product, a rather modest number of these cases have seen the successful transition to industrial-scale marketed products. Moreover, the quality and titer of obtained products in the laboratory setting are lower than that of commercial setup; hence, several techniques (i.e., genetic engineering and process optimization) have been considered to proceed with the scale-up. Recently, multiple reviews have been written on process engineering practices but are limited to particular product scale-up in microbial production systems [[9], [10], [11]]. This review extensively focuses on physical (i.e., temperature, pH, inoculum ratio, and media composition) and genetic parameters that allow a better scale-up microbial NPs production. While, the comparison between two production schemes, including monoculture and consortium, shows system limitations regarding optimum product yield [12]. Secondly, the present review comprehensively assesses the monoculture and consortial systems and their syntrophic interaction identified by advanced synthetic techniques.
2. Primary microbial systems for sustainable NPs development
It is challenging for the microbiologists to identify and isolate novel producing microorganisms to meet the demand for natural products. It has been found that less than 5% of fungi and 1% of bacterial species are currently known, indicating still a large number of NP's synthetic microbial species to be discovered [13]. These natural products, especially polyphenols, flavonoids and carotenoids are characterized by an adequate level of antimicrobial and antioxidant properties comparable to or even better than many synthetic antioxidants. Additionally, therapeutic drugs and functional foods formulated with natural products have also been developed because of the less toxicity, cost-effectiveness, and surplus availability [14]. In general, there are approximately 1 million types of natural products that are simply categorized into bioactive and inactive compounds. Of these natural products, biologically active compounds are around 25%, while 60% have plant origin and the rest are from microbial sources. Meanwhile, the terrestrial environment seems to be ideal for the obtainment of natural products; approximately 129 bioactive natural products were collected from marine microbes from 2000 to 2003 [15].
2.1. Bacteria and fungi: a robust and efficient microbial factory
Particularly, fungi and bacteria possess stronger adaptation ability for every ecological niche, making them the largest kingdom globally. There are approximately 1.5 million known species of fungi, and only 10% of them are known to microbiologists that mostly undergo some sort of mutualistic relationships, such as mycorrhiza, lichen, and bifidobacterium (gut microbe). While, the examples of symbiotic nitrogen-fixing bacteria include rhizobium and azospirillum, which are mainly associated with plants [16,17]. Because of the complex ecosystem in an animal's intestine, a symbiotic relationship between gut microbiota and the host has been developed in which Bacteroidetes and Firmicutes make up 99% of the total gut population. Intriguingly, the existence of bacteria on earth dates back to over 3 billion years ago, and eukaryotes even existed for over 1 billion years. In terms of economic and ecological roles, specific bacterial and fungal families, such as Lactobacillaceae, Enterobacteriaceae, Saccharomycetaceae, and Agaricaceae, etc., have been well-recognized and also effectively contribute towards environmental benefits.
However, since the discovery of penicillin, more than 23,000 natural products (i.e., antivirals, antimicrobials, anti-inflammatory and cytotoxic agents, etc.) have been isolated [15]. Of these products, 42% are made from fungi (Basidiomycota and Ascomycota) and 32% by filamentous bacteria (actinomycetes with an average mass range of 200–3,000 Da) [18]. More importantly, some ascomycetes, such as Aspergillus, Pencillium and Fusarium species, are identified as a major source of valuable compounds, from which 950, 900, and 350 bioactive compounds have been isolated, respectively [19]. The development of new antibiotics is facing an increasing trend, as the human's fatalities by pathogenic microbes are progressively increasing. Because of the high quality and functionality, commercial antibiotics have been developed from fungi and bacteria, with the production proportion of 50, 15, 20% by the actinomycetes, non-filamentous bacteria and filamentous fungi, respectively [15].
3. Growing market demand for microbial derived NPs
According to Business Communication Company (BCC), global market for microbial products shows an increasing trend, from $143.5 billion in 2014 to $306 billion in 2020 [20]. Moreover, the latest technologies for the manufacturing of microbial products exhibit a higher degree of technical and economic advantages relative to simple synthetic processing. These microbial-derived products include nutrition supplements like amino acids and vitamins, secondary metabolites, organic acids, enzymes, coloring agents, flavoring agents, and therapeutic products (i.e., drugs, and antibiotics) [21]. Approximately 75% of antibiotics production in actinomycetes is majorly due to the single genus Streptomyces, which alone had a market of $25 billion in 2001 [22]. The market for antifungal drugs is expected to reach approximately $4 billion in 2002 [23]. The isoprenoid production seems to fluctuate over the time, the overall trend has been rising with hundreds of new structures being reported each year; while, markets for isoprenoids, including polyketides and terpenoids are $17 and $12 billion, respectively.
4. Monoculture as primitive and simpler strategy for bio-manufacturing
In environmental diversity, a single productive cell possesses the strong capacity to endure harsh physiological changes, including nutrient deficiency and negative biological interactions, by adopting the dormant strategy. Compared with the dormant state, the single culture exhibits increased potential growth in non-dormant mode but is more susceptible to predation [24]. However, metabolic control effect and microbial growth behavior also regulate the cell behavior primarily because of interacting intra- and extracellular chemical and physical parameters and due to the genetically encoded characteristics. There are several methods to identify the growth and behavior of single cell culture (i.e., known as monoculture). Of these, molecular and spectroscopic methods exhibit better results than conventional techniques. Meanwhile, it is challenging for the small percentages of monocultures to grow under severe environmental conditions, as they require controlled laboratory conditions after isolation and purification from environmental samples (e.g. water, soil, air, and animal gut) [25]. Till now, several studies have demonstrated the effect of various processing conditions, such as medium composition, incubation temperature, pH, and inoculum ratio, etc. on monoculture's growth and behavior profiles. However, this review aims to motivate the potential of utilizing consortium and monoculture as natural product sources and functional additives in the food and non-food industries, with the highlight on the effect of physical and genetic parameters on their optimal growth and production in subsequent sections [26].
4.1. Monoculture as a bio-manufacturing system for NPs
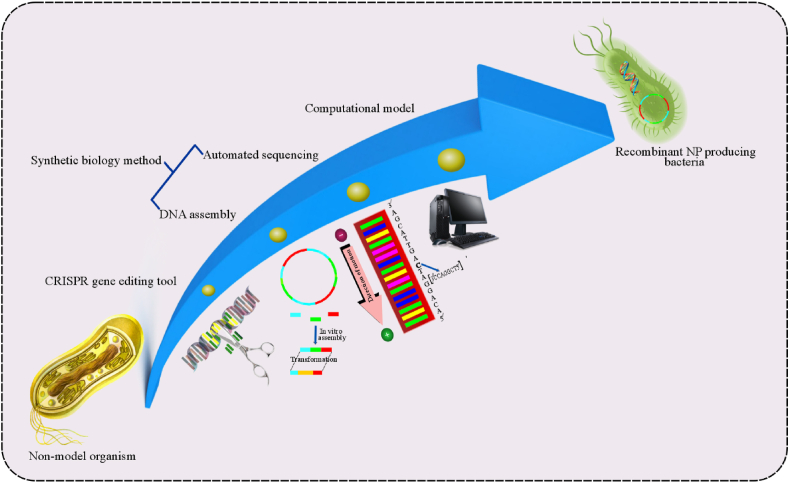
In industrial biotechnology, microbial strains isolated from nature are characterized by functional properties to execute the industrial NPs manufacturing process successfully. During process designing, the overall contribution of each microorganism also largely affects the plant and animal-derived natural products development. For instance, when comparison with non-model organisms, industrial natural products produced by model monocultures (i.e., E. coli and S. cerevisiae) are significantly higher, suggesting its simplified metabolic engineering and optimized bioprocess control [27]. Moreover, non-model organisms exhibit growth incompatibility at laboratory setting, but the recent advances in genome engineering technologies based on the combination of computational models, CRISPR/Cas tools, and synthetic biology methods, such as DNA assembly method are enabling their optimal growth in pre-defined laboratory model. Latter tools lead to rapid testing, construction, and characterization of genetic parts from different organisms [28]. With desired genetic characteristics, recombinant strains produce a higher level of natural products than nonrecombinant strains in an industrial setting, as displayed in Fig. 2 [29].
Fig. 2.
Illustration of combinational strategy including, CRISPR gene editing tool, synthetic biology method (DNA assembly and automated sequencing) and computational modeling in nonmodel organisms for rapid testing, and construction, and characterization of recombinants genetic parts for improved production of natural products.
Furthermore, hydrolysis of polysaccharides via enzymes significantly enhances NPs, such as isoprenes, aromatic compounds, peptides, etc. These products have attracted tremendous manufacturer interests due to on-demand production and higher yield as well as controlled and scalable production in fermentation facilities; see Fig. S1, Supporting Information [30]. In terms of the application of monoculture as functional food additives, they can be simply categorized into GRAS (Generally Regarded as Safe) microbes (i.e., S. cerevisiae, and C. glutamicum etc.) and non-conventional GRAS microbes (Y. lipolytica), which is further positively correlated with the development of therapeutical and nutraceutical products [[31], [32], [33]]. Besides the well-known monoculture drawbacks, multiple reports have also demonstrated certain benefits, such as cost-effectiveness, simpler cultivation procedure, and robust method when compared to mixed culture [[34], [35], [36]].
5. Bio-synthetic tool assisted shift of monoculture system to consortium one
Recently, comprehensive assessments of activity and complexity of microbial systems have gained much attention in several scientific studies, which might be due to its association with the enhanced NPs production via monoculture and/or consortial based production set-ups. During symbiotic evolution, the monocultures that thrive under different environmental conditions usually adopt the traits of communities. Interestingly, a functional consortium comes into being by the interaction of several groupings of microorganisms, which indicates its wide range of specificity and complexity in structure than that of monoculture [4]. Thus, the multi-organism design (i.e. raceway ponds) could be a competitive alternative to traditional monoculture strategy, saving time and exhibiting resilience to fluctuating environmental conditions. This transition from static to dynamic programme is thought to be a result of synthetic biology tools that enable genetic engineering and DNA assembly methods to control organisms within consortia [37,38]. These tools consist of exogenous molecules to control population behaviors, to develop communication systems between consortial members via intercellular signalling, and to construct codependent links of microorganisms by syntrophic interactions.
Generally, biosynthesis of natural products via interactive approach is considered relatively difficult, mostly because of complex population dynamics, inter-modules metabolites transport, inappropriate precursors supply, different growth rate of both upstream and downstream modules, and long fermentation period [39]. Despite its limitations, recent researches have also demonstrated certain underlying consortium advantages that include increased substrates utilization spectra, lower metabolic stress through labor division approach, controlled intermediate products accumulation through changing strain–strain ratios, co-culture stability inside the fermentation reactors, reduced metabolite's production cost and by-product formation simultaneously [40,41].
5.1. Synthetic consortia as mimicking system from natural one
Since 1980, microbiologists aim to motivate the potential of utilizing community based activities (i.e., bioleaching, and degradation of pollutants) which are evolved from macro-to micro-sphere natural scenarios [42]. Recently, system biology tools have been introduced that determine the microbial community functioning and composition with the purpose to develop synthetic consortia [38,43]. The biological activities of microorganisms which are isolated from their natural communities have been widely reported. Meanwhile, in the laboratory, the unculturability of isolated strains might be due to niche mismatch, dormancy, antagonistic effects and obligate metabolic interactions. However, ‘unculturable’ indicates that present culturing techniques are not appropriate to grow a given microorganism in pure cultures under the laboratory setting [44]. Therefore, to resolve this problem microbial ecologists have adopted certain strategies such as selecting bacteria amenable to conventional culturing, and using culture-independent methods. In addition, many studies have shown that majority of these isolated strain prefer to grow as consortium rather than individual monocultures, and it is strongly indicating towards metabolic interactions in all possible combinations which might bring together strains that would not meet in their natural habitat [45,46].
In general, there are two major ways to procure microbial consortia involving either (i) a synthetic assembly obtained from scratch, or (ii) microbial community isolated from environmental samples [47]. Because of the high functional properties, natural consortia is commonly used to develop commercial food products, such as beverage and baked products, and also well-known for its role in some clinical studies [[48], [49], [50]]. Moreover, natural consortium synergism was commonly observed in herbivores’ gut and in bioleaching processing. It is mimicked by synthetic consortia, which leads to beneficial interaction for enhanced productivity of target natural products from cheap raw materials, such as lignocellulose wastes and animal manures; hence, no further acidic and enzymatic pretreatments are required [51,52].
6. Syntropic interaction in well-defined consortial system
Particularly, different members of microbial community that together respond well against industrial and environmental challenges in relative to monocultures, indicating the strong syntropic interaction among the diverse microorganisms like algae and bacteria [53,54]. It suggests that change in nutritional parameters affect the interaction pattern, which may be classified into two major types, either cooperative (symbiosis, mutualism, and commensalism) or non-cooperative interaction (parasitism, ammensalism and predation) [55]. Hence, cooperative interaction is practically important to achieve maximum beneficial outcomes in terms of valuable products quality and titer. In both cases, metabolic interactions exist along two main axes: (i) the investment by the involved partners (i.e. production cost of exchanged metabolite), and (ii) the degree of reciprocity (i.e. flow direction of metabolite). The microbial community is characterized by unique functional properties which emerge as a result of different interactions among inter-species. Moreover, shaping of function, structure, and dynamics of microbial communities have been aroused from inter-species interactions leading to the development of synthetic consortial system [56,57].
6.1. Spatially linked microbial consortia as a conceptual design to engineer consortia
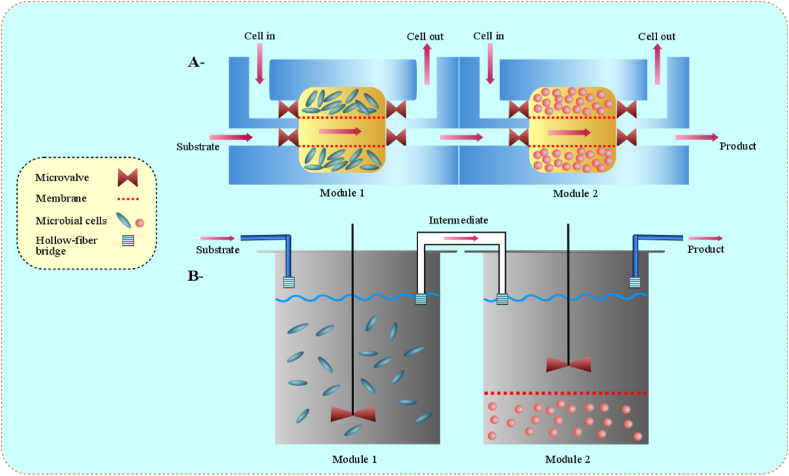
It has been found that an enhanced control over the system (consortium) is achieved by applying a compartmentalization strategy that provides ideal settings compatible with the consortial members. Additionally, cross-feeding interactions influence the growth of consortial members in spatially linked microbial consortia (SLMC); while, it has been affected by certain factors, for example, regulation of biosynthesis enzymes, allocation of limited resources (e.g. expression machinery, nutrients, and space), and biotic composition of bacterial community, as well as certain ecological parameters like chemical diffusivity and degree of spatial structuring, etc. The transfer rate of metabolite is higher through membrane transport than diffusion-based transport, which also bears some risks, such as consumption of nutrients by third party, degradation and loss of exchanged metabolites by fast diffusion [58]. At the laboratory scale, several synthetic structures (e.g. nanotubes, channels, and pili) facilitate the attachment of bacterial communities in order to exchange the metabolites. Furthermore, Dietz et al. [59] used dialysis membrane reactor to exchange metabolites on either side of a membrane at a commercial scale. On the other hand, Hollow-fibre bioreactor provides similar growth conditions to co-culture on each side of separate bioreactors, as demonstrated in Fig. 3.
Fig. 3.
Engineered laboratory (A) and commercial (B) scale approach to make consortium derived natural products under ideal growth conditions. At laboratory scale, microfluidic offers growing set of tools for manipulating the consortium growth under controlled environmental conditions, while hollow-fiber bioreactor is considered as commercial bioreactor, as it maintains the similar growth conditions for co-culture on each side of separate reactors.
Besides, spatial segregation coupled with in-line nutrient supplementation further improves the production of natural products. It has been considered an effective approach to retrieve consortial related drawbacks, such as growth inhibition by toxic intermediate compounds, inter-species dependency for nutrients, and lower product yield by downstream strains. Meanwhile, simple nutritional requirements are the key prerequisite characteristics for selecting upstream microorganisms limiting the metabolism interferences in downstream microorganisms. Moreover, the downstream microorganisms should possess some of the following characteristics: capable of co-habiting, matching growth rates, increasing the capability of upstream micro-organism to consume multiple feedstocks, nontoxic in nature, maintaining genetic integrity, and functioning as a bio producer [[60], [61], [62]].
7. Design and evaluation of consortial functioning by advanced synthetic technologies
Non-linearity in combination coupled with the immense complexity of microbial communities remains a major challenge to construct an efficient synthetic microbial consortium in which the role of each member is well-defined in the provision of community-derived fluxes under dynamic environments with varying physical and chemical conditions. The omics technology and genome-editing tools have gained much regard currently to achieve a rational design of synthetic microbial consortia. Omics tools arm researchers to have holistic views of growth dynamics and metabolic fluxes in defined consortia [63,64], but indicate a particular genetic pathway and interlinking between consortial members, leading to a consortium with the desired functional characteristics. Furthermore, the genetic-based treatments, such as transcriptional control, genome editing, and high-throughput mutagenesis significantly enhance the processing capabilities (i.e. enduring fluctuating reactor's environment, and producing optimum level of NPs) of synthetic microbial consortia, with more obvious improvement observed by CRISPR/Cas-based toolkits [[65], [66], [67]]. The aforementioned technologies such as omics and genetic engineering should be coupled to computer modelling to further contribute for a better understanding of microbial interactions [68,69]. Hence, comprehensive assessments of synthetic consortium structure, composition and functionality of consortial members are usually essential to promote their application in the industrial, medical, and environmental sectors [70,71].
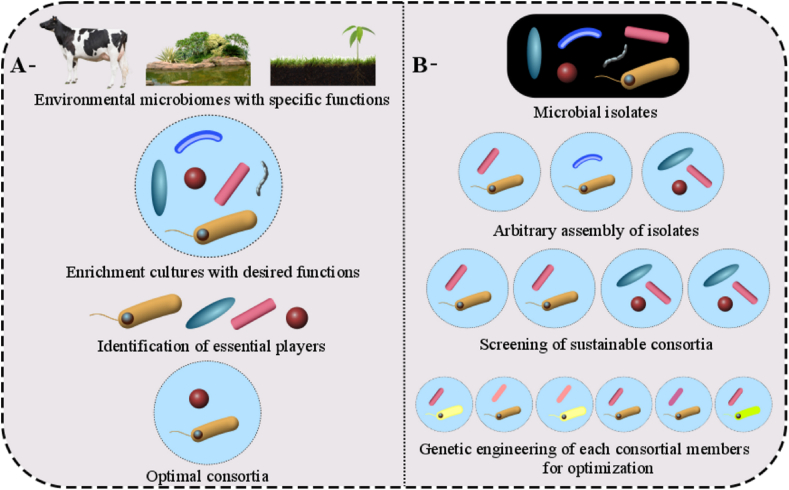
Modern technique, including omics approach (transcriptomics, metabolomics, and proteomics), is one of the most robust, precise and efficient techniques (coupled with a higher level of detection limit) to analyze monoculture, environmental microbial communities, and defined consortia to understand the underlying molecular mechanisms of microbial interactions; see Fig. S2, Supporting Information [72,73]. Interestingly, by applying current metatranscriptomics and metaproteomics technologies, the obtained informative data are employed to compare the temporal gene expression between consortia and monocultures, such as temporal proteomes when complemented with the metabolomic analysis demonstrated the positive interaction between K. vulgare and B. megaterium to produce 2-keto-gulonic acid. In general, there are two major strategies for the selection of consortium members, the top-down method that considers consortium members as the keystone players isolated from specific complex microbial community [74], and the bottom-up method considering consortium members having desired traits and selected from the pool of engineered microorganisms [40]. Meanwhile, comparing the top-down method which is based on multiple omics analysis [[75], [76], [77]], the bottom-up method is more preferable for constructing the stable artificial microbial consortia, probably due to the rational guidance of genetic engineering principles with comprehensive analysis on system's genetic elements, circuits, modules, and metabolic pathways, as shown in Fig. 4 [78,79].
Fig. 4.
Top-down (A) and bottom-up (B) approaches for synthetic consortia construction. In top down approach, the consortium members are isolated from complex microbial community, while in bottom up approach, consortium member are ascribed to specific traits and selected from pool of engineered microorganisms.
Several studies report that the process efficiency and ease to construct synthetic consortia via top-down approach is lower as compared to bottom-up strategy, which is mainly due to the unavailability of isolates, lack of genomic information, and suitable engineering tools for unconventional microorganisms. The bottom-up approach, which is facilitated by multiple synthetic biology tools, has thus been investigated for resolving this drawback to further lead in constructing robust and stable artificial consortia for the optimum production of NPs.
8. Optimum processing conditions for microbial bio-operational unit
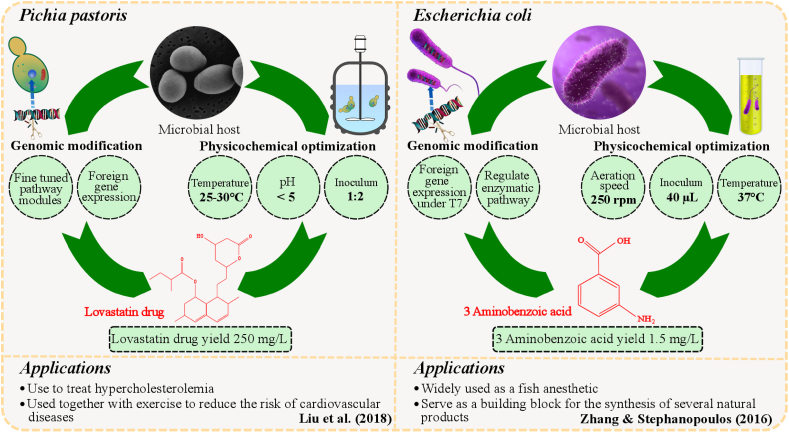
Particularly, the biosynthetic strain behavior, either in consortium or in monoculture, is dependent on several factors, such as genomic (mutagenesis, and genetic engineering), and physicochemical parameters including media composition, incubation temperature, pH, and inoculation ratio etc. Microbial cell factory overall represents a valuable source of NPs, and these compounds are capable of meeting the requirements of value-added foods and therapeutic drugs of the local population [[80], [81], [82]]. Interestingly, the higher level of microbial NPs has been obtained by compatible physicochemical conditions coupled with desired genetic modification, and both of these are specific for various bacterial or fungal species [83,84]. There are several tools for recombinant geneticists, including genetic recombination, targeted duplication, and deletion by genetic engineering and transposition mutagenesis, to achieve the desired production of natural products [85]. Recent additions to these techniques include transcriptome analysis, metabolic engineering and genome shuffling. Moreover, this review aims to motivate the potential of utilizing monoculture and consortium for the optimum microbial NPs (e.g., lovastatin, and 3 aminobenzoic acid, etc.) production at a commercial scale, with the highlights on its compatible physicochemical conditions and desired genetic modulations, as displayed in Fig. 5. These parameters will be discussed herein.
Fig. 5.
Multi-objective optimization of processing conditions and genomic engineering for enhanced production of microbial-derived natural products.
8.1. Effect of physical conditions on microbial derived-NPs production
In general, the microbial production of natural products and secondary metabolites is affected by external stimuli, i.e., the surrounding environment. Particularly, the microbial cell possesses the strong capacity to tolerate severe perturbation in environmental conditions. Considering the influence of environmental factors on microbial cells, it is possible to determine the most significant, as described in the following section.
8.1.1. Effect of temperature on microbial derived-NPs production
Many reports have shown that fermentation temperature can significantly affect microbial growth, specific enzyme activity, and enzyme folding [86]. Moreover, several temperature-related studies have also demonstrated its considerable impact on the production phases of NPs metabolism of microorganisms in monoculture and consortium and influence the interactions between various microbial groups [87,88]. In general, microorganisms are categorized into three major classes based on optimum growth temperature requirements: psychrophiles, mesophiles, and thermophiles. Psychrophiles, which grow as monoculture, appears to be an efficient source of novel metabolites, as they only perform under a certain temperature range of 0–15 °C. At the same time, these cold-adapted micro-organisms show enhanced adaptability against numerous stresses, like cold temperature stress, decreased fluidity of cell membrane, low nutrient availability, and transport protein efficiency [89]. Among the cold-adapted strains examined, the Penicillium griseofulvum exhibits increased fulvic acid, chanoclavine I, elymoclavine and mycelianamide production, all of which possess strong antimicrobial activity [90]. Additionally, Pseudogymnoascus sp. and Bacillus sp. are predicted to be among the most promising psychrophiles, based on their ability to produce higher concentrations of asteric acid derivatives and mixirins. These compounds are known to reduce fungal infection and proliferation of colon tumour cells, respectively [91,92].
More importantly, most monocultures prefer to grow and produce natural products at a moderate temperature range in between 20 and 45 °C. For example, Bacillus subtilis and Aspergillus sp. were cultured to produce optimum antifungal and insecticidal substances at a specific temperature, ranging from 25 °C to 37 °C as well as Aspergillus sp. also contributed in the production of aspochalasins antimalarial antibiotic [93]. A few findings have also been reported by Wiebe et al. 28, who demonstrated the microbial growth suppression by lack of substrate at minimum growth temperature (near 10 °C), which can be treated either by addition of substrates or rise in temperature [94]. Besides, Streptomyces thermoviolaceus, which grows as a thermophilic monoculture (from 40 °C to 55 °C) produced antibiotic granaticin and various extracellular proteins at a growth rate of 0.175 h-l at 45 °C [95].
A detailed study on the consortial growth behaviour during myeolchi-aekjeot (MA) fermentation demonstrates that the low temperature notably affects interaction among mesophilic bacterial communities and has adverse effects on microbial growth due to mycoplasma toxicity. Meanwhile, fermentation-derived amino acid production escalates many folds when the temperature is adjusted to 25–30 °C [96]. Anammox consortia, consisting of bacteria from phylum Chloroflexi, Chlorobi, and Proteobacteria, is a mesophilic consortium (20–25 °C), which is significantly linked to energy conservation strategy and capable of producing a higher level of amino acids [97]. Furthermore, the synthetic bacterial consortium consists of Streptomyces sp. and Methanosarcina sp. which have been recognized as a robust producer of actinorhodin (with an optimum growth between 25 °C and 37 °C) [98,99]. In shrimp fermentation, psychrophile community members exhibit a higher production of amino acids at 15 °C due to the growth inhibition of pathogenic strains (i.e., Vibrio, Aliivibrio, and Photobacterium sp.) after 105 days [100]. On the other hand, thermophilic methanogenic consortium TERIL63 was developed to produce a significant amount of volatile fatty acid (2037 mg/L), consisted of two strains, Methanothermobacter thermoautotrophicus and Thermoanaerobacter sp. It can grow in the wide range of 60–100 °C temperature in methogenic medium [101].
8.1.2. Effect of pH on microbial derived-NPs production
The pH level of the growth medium imposes selective pressure on microbial metabolism, growth, and secondary metabolites production. Interesting, both hydrogen and hydroxyl ions concentrations have an appreciable impact on microbial cell behaviour, or it may act indirectly by changing the degree of dissociation of valuable compounds in the growth medium. Hence, the pH variation largely impacts the enzymatic activity and normal nutrient solubility and influences the distribution of microorganisms in a diversified ecosystem [102,103]. In general, there are three major types of microorganisms, which are described as alkaliphiles, acidophiles, and neutrophiles on the basis of optimum pH requirement for growth. Importantly, microorganisms produce higher level of NPs at a wider range of pH (typically, most of the synthetic microbial cells prefer the pH range of 3–4 units) and pH variation remarkably affects the activity and composition of microbial community, indicating its significance more than temperature in certain industrial bioprocess (i.e. volatile fatty acid (VFA) production in bioreactor) [104].
For example, Bacteroides can adapt themselves over a wide range of pH rather than that of Streptococcus and Veillonella, which experience growth inhibition under acidic pH treatments [105]. Furthermore, in comparison with butyrogenic reactions (operated at acidic pH 5.5), propionate- and acetate-producing consortia, consisted of Veillonella, Escherichia, and Bacteroides, exhibited enhanced production of secondary matabolites at pH 6.5–6.9 (neutral pH) [106]. It has been found that basic mesophilic consortia, consisting of Bacteroidetes, Firmicutes, Actinobacteria, Tenericutes, Proteobacteria and Cyanobacteria, largely contributed in the production of longer chain VFAs at pH 9–10 and its yield was calculated up to 388 ± 28 mg VFA as HAceq g VS−1 added, where average composition comprises of 83% acetic acid, 6.3% propionic acid, 4.4% isovaleric acids, 3.6% isobutyric, and 2.7% n-butyric [104,107]. In addition, duckweed treatment via acidic mesophilic consortia (comprises of Bacteroidia, Gammaproteobacteria, and Clostridia) produces an appropriate level of volatile fatty acids; while, the optimization of processing conditions can further improve its yield. Later, genus Acidaminococcus, a mesophilic gram-negative cocci, is well determined for its amino acid production in acidic mesophilic fermenter [108].
In the case of monoculture, an increased selection has been noticed for both alkaliphile (i.e. B. aurantinus), and acidophile (i.e. A. terreus, and Streptomyces) to meet the demand of local population for antimicrobial products, such as aurantin, anti-influenza agent and secondary metabolites, etc., which are produced at pH value of 7.5, 6.4, and 5, respectively [109,110]. Aside from this, an appreciable level of granaticin antibiotic has been identified in the fermentation broth of S. therrnoviolaceus at pH 6.5–7.5 [111].
8.1.3. Effect of growth medium on microbial derived-NPs production
The prerequisite requirement for successful industrial fermentation is a good fermentation medium with a good nutritional profile, such as carbon, potassium, phosphorus, manganese, nitrogen, energy sources, etc. Also, some trace elements, including minimal salts, heavy metals and vitamins (growth factor), must meet the elemental needs for metabolite production and cell metabolism [112,113]. Thereafter, the high functional constituents in the medium serve as regulators which influence the quantity and type of metabolite produced during incubation. Moreover, it is important to determine the appropriate incubation period for the production of NPs because in a long incubation period, desired biologically active compounds convert into other less significant compounds. Cultivation medium can be simply categorized into complex media and minimal media. For instance, complex media exhibits higher cell density and microbial growth rates than minimal broth media, which has been widely reported for enhanced fatty acids, and sugars production [114].
As in monoculture, 2% of glucose supplementation appears as a superior substrate for higher antimicrobial metabolites production and cell growth in Streptomyces sp. RUPA-08PR and sucrose exhibit a similar pattern of result followed by xylose, mannose, and fructose, respectively [115]. However, it is not surprising to observe little antibiotic production in galactose and lactose supplemented medium due to its interference with microbial metabolism [116]. Particularly, the growth medium of B. subtilis is supplemented with glucose (40 g/L) and NH4NO3 (4.5 g/L), which positively contributes to enhanced surfactin production [117]. In the case of organic and inorganic nitrogen sources, yeast extract exhibited a higher yield of antimicrobial agents followed by peptone, NaNO3, beef extract, KNO3, etc. Meanwhile, 1% NaCl supplementation shows higher production of antimicrobial metabolites in various strains. Few studies have been reported that potato dextrose broth as an efficient growth medium, used to produce tropolone (antimalarial antibiotic) and cladospolide D (antifungal antibiotic) against Cordyceps sp. and Cladosporium sp., respectively [118].
Various cultivation media are identified in advanced microbial metabolomics studies, such as M9, minimal broth, universal 13C-labelled medium, nutrient broth, and Luria-Bertani broth [119]. It was found that too high substrate (phenanthrene, pyrene, and naphthalene) concentration cause toxicity to microbes, while low substrate concentration cannot meet the growth requirement of the microbial cell. Hence, selecting an ideal biosynthetic strain is necessary either by direct evolution or genetic engineering, allowing the microorganisms to co-utilize the multiple sugars in the cultivation medium [120,121]. A detailed assessment of consortia's co-utilization of various substrate mixtures demonstrates that engineered E. coli co-culture can co-utilize both xylose and glucose in a similar cultivation medium to produce flavonoid naringenin [122]. Moreover, in the fermenter, a periodic feeding of xylose, phosphate, and ammonium during the first 24 h, has been proved to optimize S. cerevisiae's growth in coculture to increase oxygenated taxane titer about threefold (16 mg/L in 90 h). Additionally, amino acids supplementation (i.e. Pro, Thr, Ile, Leu, and His) was added to the 2-Keto-d-gluconic acid producing consortium (G. oxydans-K. vulgare), which greatly improves 2-KGA (2-keto-l-gulonic acid) titer by 41.8% when compared to the original consortium (without amino acid supplementation) [123].
8.1.4. Effect of inoculation ratio on microbial derived-NPs production
Primarily, mixed synthetic communities are characterized by various kinds of species with varying growth rates in which the faster-growing species destabilizes other species through vigorous consumption of nutrients. Co-cultivation techniques have thus been investigated for resolving this drawback by optimizing the relative population ratio of microbial consortium either by varying the initial inoculum ratio or by inoculating a downstream strain during the culturing of upstream strain [[124], [125], [126]]. During fermentation, adjusting the initial inoculum ratio of upstream to downstream strains before strain cultivation could appreciably enhance population growth rate and specific performance of strains in the consortium. Moreover, population control strategies such as rational tuning of inoculation ratio among consortium partners should be applied to regulate overall production. For example, consortium with dominant ratios of upstream strains utilizes more substrates. However, it produces less final product rather than that of consortium with dominant downstream ratios, which exhibits decreased substrate utilization and enhanced final product production [124,127].
More importantly, the growth phase and inoculation timing are the important parameters that influence the consortium's performance and general structure. Generally, each growth phase (i.e., lag, log and stationary phase) associates with specific physiological changes which appear in the microbial cell [128]. A synthetic consortium, consisted of R. glutinis and C. vulgaris, displayed an enhanced lipid production (70.9%) at a ratio of 1:1 in their log phase. Besides, lipid producing consortia (Saccharomyces cerevisiae-Chlorella sp.) exhibits a higher level of metabolite production at ratio of 2:1 [129,130]. Additionally, a robust two member's consortium of Dinoroseobacter shibae strain and Thalassiosira pseudonana strain has been reported to provide higher metabolite yield when T. pseudonana is in exponential growth phase before the bacterial inoculation [131].
Meanwhile, it is challenging to achieve optimum production of β-carotene and flavonoids via synthetic consortia, such as Rhodotorula glutinis-Debaryomyces castellii (upstream:downstream) consortium and E. coli-E. coli consortium. Therefore, the adjustment of inoculum ratio at 1:1 and 8:2, respectively, have been investigated for enhanced production of β-carotene, and flavonoid [132,133]. It has been reported that the consortium's stability is negatively influenced by long fermentation period and large fermentation volume due to the incompatible growth requirements, metabolite dilution, and spatiotemporal dynamics inside the fermenter and thus, it is not surprising to exploit advanced genetic techniques to resolve these problems [134,135].
The inoculum density and age are now regarded as important parameters which influence the metabolite and biomass accumulation in monoculture as well as provide economic feasibility (in case of commercial fermentation) [136,137]. Bacillus subtilis SPB1, grown as a monoculture, provides an optimum lipopeptide yield (3.4 g/l) by adjusting second inoculum age and size to 4 h and 0.01, respectively, after a first inoculum age of 23 h [138]. In particular, glucose as a carbohydrate substrate increases the surfactin production in B. subtilis, which improves by optimum incubation time and inoculum size of 72 h and 1.5–2% (v/v), respectively. In B. braunii, maximum production of biomass and hydrocarbon up to 5.97 and 2.99 g m−2 day−1 can be achieved at controlled seed age and inoculum density of 14 days and 7.9–10.1 g m−2, respectively [139]. Finally, comprehensive assessments of physical requirements, including temperature, pH, medium composition, and inoculum density that are optimal for bacterial production, were conducted in Table 1.
Table 1.
Effects of physico-chemical parameters on the production of natural products via monocultutre and consortial based system.
| Physico-chemical parameters | ||||||||
|---|---|---|---|---|---|---|---|---|
| Factor | Organism | Specifications | Production | Bioprocessing strategy | Reference | |||
| Temperature | ||||||||
| 33.7 °C | P. acidilactici | Observe bacterial growth at various temperature at a constant pH of 6.5 No growth was seen at 10 °C, 50 °C |
959.75 AU mL−1 Bacteriocin | Monoculture | [168] | |||
| 37 °C | A. senegalensis | Enzymes speed up metabolism Bacterial cells rapidly multiply |
57 mg mL− 1 Bacterial cellulose |
Monoculture | [103] | |||
| 37 °C | E. coli-E. coli | Heterologous enzyme folding and its activity in E. coli is dependent on temperature | 1.5 mg/L 3-amino-benzoic acid |
Consortium | [169] | |||
| 25–35 °C | Acinetobacter, Acetitomaculum,Bacillus | Mesophilic consortia operate well in acidogenic phase Microbial activity inhibited at 20 °C |
4403 mg/L Volatile fatty acid |
Consortium | [170] | |||
| pH | ||||||||
| 4.5 | A. senegalensis | Carried out oxidative reaction Aids nutrient solubility |
98 mg mL− 1 Bacterial cellulose |
Monoculture | [103] | |||
| 5.8–4.0 | P. pastoris | Medium pH effect the chemical structure of end-products | 60 mg/L &14.4 mg/L monacolin J, lovastatin | Monoculture | [171] | |||
| 6.0 | L. lactis-S. cerevisiae | pH control strategy without alkali supplementation was adapted | 150.3 mg/L Nisin |
Consortium | [172] | |||
| 6.0 | L. lactis-K. marxianus | pH controlled by NaOH supplementation In anaerobic cultivation, nisin production is higher as compared to aerobic conditions |
98 mg/L Nisin |
Consortium | [173] | |||
| Medium composition | ||||||||
| 41 g/L fructose + 38 g/L peptone with HS medium | K. intermedius | Optimization of culture condition via response surface methodology (RSM) Fulfill nutritional requirement |
3.906 g/L Bacterial cellulose |
Monoculture | [174] | |||
| WB + BHM-YEP media | B. altitudinis J208 | Optimization of culture condition through RSM based on central composite design (RSM-CCD) Cheap nutrient source Highly efficient and stable medium |
60.1 L-1 h-1 Xylanase 11343 L-1 h-1 Pectinase |
Monoculture | [175] | |||
| 55 g/L lactose + 15 g/L corn steep liquor + 5 g/L ammonium sulphate with MRS | Lactobacillus LMI8 | Optimization of culture condition through RSM Cost effectiveness Highly efficient source of medium |
52.37 g/L Lactic acid |
Monoculture | [176] | |||
| Glucose 20 g/L + yeast extract 2.5 g/L + KH2PO4 1 g/L + MgSO4 0.5 g/L + (NH4)2SO4 0.05 g/L + FeSO4 0.01 g/L | T. maxima, P. carneus | Optimization of culture condition via Plackett–Burman experimental design Maximum laccase production on the 3rd day in co-culture 13 Maximum MnP production on the 10th day in co-culture 15 |
12,382.5 U/mg protein Laccase activity 564.1 U/mg protein MnP activity |
Consortium | [177] | |||
| LB broth + antiboitics + sodium acetate + disodium malonate | E.coli-E.coli | Combination of modules I (Pc4CL2, VvSTS genes), IIc (S. coelicolor genes) and III (E.coli K-12 genes) generate piceatannol | 124 mg/L (piceatannol) | Consortium | [178] | |||
| LB broth + antiboitics + disodium malonate | E.coli-E.coli | Combination of module I (Pc4CL2, VvSTS genes) with module II (two different gene sets) to generate resv | 137 mg/L (resveratrol) | Consortium | [178] | |||
| Inoculum concentration/ratio | ||||||||
| 20% | A. senegalensis | Decrease competition between bacteria in consuming nutrients Promote bacterial growth |
395 g L− 1 Bacterial cellulose | Monoculture | [179] | |||
| 1:2 | P.pastoris-P. pastoris | Conversion capacity of intermediates was improved DML and ML totally converted to MJ |
93 mg/L monacolin J | Consortia | [171] | |||
| 20:1 | E. coli-E. coli | BW23 upstream module optimized for consolidated SAG production BW downstream module designed for the conversion of SA to SAG |
2500 mg/L salicylate 2-O-β-D-glucoside | Consortia | [180] | |||
| 1:1 | P.pastoris-P. pastoris | Ratio of P. p/FNsD_RFP_sAR should be optimized Downstream LV module could match upstream DML module |
24.6 mg/L lovastatin | Consortia | [171] | |||
Abbreviation: HS medium, Hestrin-Schramm medium; WB, wheat bran; BHM-YEP, Bushnell Haas Medium-Yeast extract and Peptone; MRS, De Man, Rogosa and Sharpe; LB broth, Luria-bertani; SA, salicylate; SAG, salicylate 2-O-β-D-glucoside; DML, dihydromonacolin L acid; ML, monacolin L acid; LV, lovastatin.
8.2. Effect of genetic modifications on microbial derived-NPs production
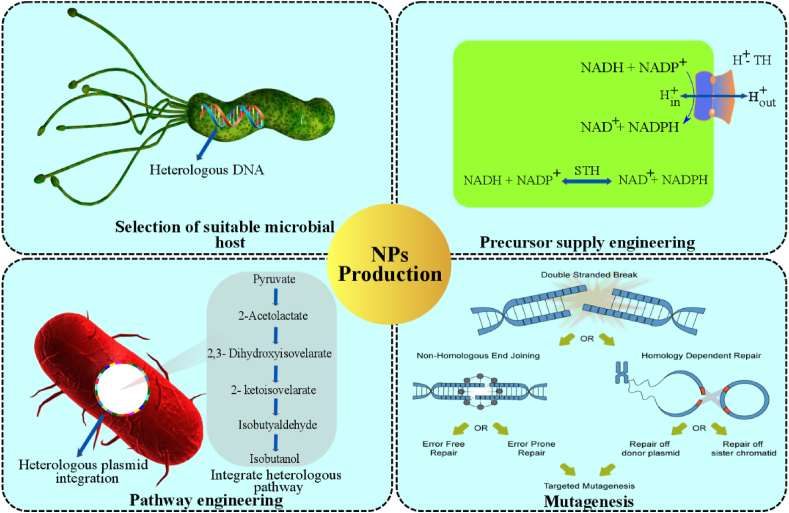
As a robust natural product producer, genetically modified microorganisms (GMOs) have gained growing popularity in food, cosmetics, pharmaceutical, and many other industries. These microorganisms have been developed by engineering the genome on inserting and deleting desirable genes or pathways which will be discussed herein and also shown in Fig. 6.
Fig. 6.
Overview of the various techniques, such as selection of suitable host, precursor engineering, pathway engineering and mutagenesis, employed in genetic engineering have led to the increased production of natural products.
8.2.1. Effect of heterologous host selection on microbial derived-NPs production
The optimum production of microbial-derived natural products, such as flavonoids, alkaloids, polyketides, and pharmaceutical products (i.e., antibiotic and antifungal drugs, etc.) have been widely reported via strain improvement, precursor supply engineering, pathway engineering, and mutagenesis. Interesting, deregulated organisms exhibit lower NPs production, probably due to the absence of appropriate biosynthetic enzymes. Thus genetic engineering (optimization of copy number and/or transcription frequency) is carried out in the suitable microbial host, including E. coli or S. cerevisiae, to resolve this low productivity issue [140]. However, an increased selection has been noticed for both Escherichia coli and Saccharomyces cerevisiae due to the availability of genetic manipulation tools, which are amenable to simple cultivation methods and production scale-up. Besides, other microorganisms, including Corynebacterium glutamicum and Streptomyces species have been extensively utilized as a heterologous host for NPs production [141,142].
Monoculture derived NPs have gained the growing popularity at commercial level because of its cost effectiveness and simpler cultivation protocols. Amorphadiene, an artemisinin precursor, has been produced by heterologous E. coli approximately 24 mg/L, by expressing S. cerevisiae's mevalonate pathway and synthetic amorphadiene synthase gene and its production will further improve up to 105 mg/L by optimization of processing conditions [143]. One additional avenue when selecting an appropriate host for NP production is to utilize co-culture (E. coli- S. cerevisiae) with components of a metabolic pathway split between distinct consortium's members to produce benzylisoquinoline alkaloids (BIAs) in which membrane-bound P450 enzymes expresses in upstream strain (S. cerevisiae) and (S)-reticuline biosynthesizes in downstream strain (E. coli), respectively [1,[144], [145], [146]]. Some examples of bioactive NPs derived from recombinant hosts have been shown in Table 2.
Table 2.
Effect of genetic parameters on the production of natural products via monocultutre and consortial based system.
| Genetic parameters | |||||
|---|---|---|---|---|---|
| Factor | Organism | Specifications | Production | Bioprocessing strategy | Reference |
| Host selection | E. coli | Plasmid having tryptophan synthase Induction with 3-indole acrylate |
180 g L −1 Tryptophan |
Monoculture | [181] |
| Host selection | E. coli | Overexpression of heterologous crt genes (P. ananatis) & ispDF in DXP pathway | 433 mg/L Astaxanthin |
Monoculture | [182] |
| Host selection | E. coli-E. coli | Modular nature of co-culture engineering to rapidly identify a particular E. coli strain that markedly improved the efficiency of 3AB biosynthesis | 48 mg/L 3-amino-benzoic acid (3AB) |
Consortium | [169] |
| Pathway engineering | E. coli | Integration of MVA and lycopene pathway Extra copies of the idi gene |
1.44 g/L Lycopene | Monoculture | [183] |
| Pathway engineering | E. coli | Combinatorial tuning of pathway enzymes (TAL, 4CL, CHS, CHI) Inducible promoter strengths Enhancement of intracellular tyrosine supply |
100 mg/L Naringenin | Monoculture | [184] |
| Pathway engineering | E. coli | Coupling of 3AB synthase, and PctV with the engineered shikimate pathway T7 promoter enhances the expression of pathway enzymes |
1.5 mg/L 3AB | Monoculture | [169] |
| Pathway engineering | Y. lipolytica | Introduction of β-carotene biosynthesis pathway Optimization of upstream MVA pathway Downregulation of squalene synthase Overproduction of astaxanthin synthesis |
10.4 mg/L Astaxanthin | Monoculture | [185] |
| Pathway engineering | P. pastoris | Lovastatin and monacolin J production improved by 55% and 71% in consortial approach Upstream and downstream modules accommodate in two fluorescent strains |
593.9 mg/L Monacolin J 250.8 mg/L Lovastatin |
Consortium | [171] |
| Pathway engineering | Three E. coli modules | Upstream strain produces pathway intermediate p-coumaric acid Midstream strain produces naringenin. Downstream strain converts naringenin into acacetin |
20.3 mg/L Acacetin | Consortium | [186] |
| Precursor engineering | S. clavuligerus | Supply of 10 mM methyl oleate to bacteria carrying (MCM) pathway Enhanced the conc. of methylmalonyl-CoA |
17.8 mg/L FK506 |
Monoculture | [187] |
| Precursor engineering | E. coli | MEP pathway (B. subtilis), GPPS2 (A. grandis), MVA pathway are coexpressed | 122.4 mg/L β-carotene | Monoculture | [188] |
| Precursor engineering | E. coli-E. coli | Co-culture enhance the availability of p-coumaric acid as a precursor for the production of sakuranetin | 29.7 mg/L Sakuranetin | Consortium | [189] |
| Precursor engineering | E. coli-S. cerevisiae |
E. coli excretes p-coumaric acid in mediumSaccharomyces cerevisiae expressing codon-optimized 4CL and STS for the conversion of p-coumaric acid into resveratrol |
28.5 mg/L Resveratrol | Consortium | [190] |
| Precursor engineering | E. coli-E. coli | Upstream E. coli BL21 engineered to produce caffeic acid by expressing HpaBC, RgTAL Downstream E.coli utilize caffeic acid to generate caffeoylmalic acid |
570.1 mg/L Caffeoylmalic acid |
Consortium | [191] |
| Mutagenesis | E. coli | Generation of GadB mutant (Glu89Gln/Δ452–466) upon pH shift | 4.8 g/L of GABA | Monoculture | [192] |
| Mutagenesis | S.viridochromogenes | Mutation in ribosome protein S12 (rps12) through gene shuffling and ribosome engineering | 1.4 g/L of Avilamycin | Monoculture | [193] |
| Mutagenesis | S. cerevisiae | 25 min of UV exposure Addition of 20 mM Zinc sulphate |
4.632 (v/v) Alcohol |
Monoculture | [194] |
| Mutagenesis | S. cerevisiae | Express CrtZ (A. aurantiacum) and CrtW (B. vesicularis) Subjected to ARTP mutagenesis |
217.9 mg/L Astaxanthin |
Monoculture | [195] |
| Mutagenesis | E. coli-E. coli | ALE and ARTP mutagenesis coupled with efflux pump in the acquisition of pinene tolerant strain E. coli YZFP | 166.5 mg/L α-Pinene |
Consortium | [196] |
Abbreviation: MCM, methylmalonyl-CoA mutase; GABA, gamma-aminobutyrate; ARTP, Atmospheric and room-temperature plasma, ALE, Adaptive laboratory evolution; GPPS2, geranyl diphosphate synthase; MEP, methylerythritol 4-phosphate pathway, MVA, mevalonate pathway; 4CL, 4-coumarate-CoA ligase; STS, resveratrol synthase; TAL, tyrosine ammonia lyase; CHS, chalcone synthase; CHI, chalcone isomerase; DXP, 1-deoxy-d-xylulose-5-phosphate pathway; crt, carotenogenic gene.
8.2.2. Effect of precursor engineering on microbial derived-NPs production
After the host selection, strains exposure to the suitable genetic engineering techniques, such as overexpression of metabolic genes, gene deletion and replacement of existing enzymes with more active homologues have been proved to enhance the supply of biosynthetic precursors. This suggests the potent role of precursor supply engineering which can be achieved by manipulating either the enzymes (i.e., phosphoenolpyruvate carboxylase in E. coli and, Glc-6-phosphate dehydrogenase in S. cerevisiae) or pathway (i.e., NADPH and Malonyl-CoA generating pathway) participated in precursor supply [147,148]. As an example of precursors supply in monoculture, the addition of propionyl-CoA carboxylase coupled with propionate supplementation could further improve rapamycin and methylmalonyl-CoA titers upto 23.6 mg/L in the UV irridiated mutant S. rapamycinicus strain which is 3.2-fold higher than that of parental strain (7 mg/L) [149].
Aside from the overexpression of endogenous genes, an appreciable level of biosynthetic precursors can be achieved by the deletion of undesired side or competing pathways in the host. For example, the double deletion of ATF1 and OYE2 enzymes in strictosidine producing strain resulted in reduction of geraniol synthesis which could enhance the strictosidine production by 6-folds [150]. Meanwhile, in recent study, the two members consortium (E. coli-S. cerevisiae) has been investigated for resolving the problem of lower precursor supply during the taxanes production by carrying out oxygenation reaction in S. cerevisiae and enhancing taxadiene production in E. coli, leading to increased production of oxygenated taxanes (33 mg/L) [151,152]. Many reports have shown that precursor supply techniques have positive effects on optimum NPs production, shown in Table 2.
8.2.3. Effect of pathway engineering on microbial derived-NPs production
Metabolic pathway engineering comprises gene knockout, gene overexpression, and introduction of the functional genes capable of enhancing NP production [153]. Many studies have shown that deletion of genes can improve the yields of natural products, probably due to the elimination of competing pathways that involve unnecessary consumption of cellular resources. In the case of monoculture, the streptomycin gene cluster is introduced into the genome-minimized S. avermitilis (83% of its original size) and result in the maximum level of streptomycin titer when compared to parental strain carrying a similar heterologous gene cluster [154]. The overexpression of protein disulfide isomerase and actinorhodin gene cluster (4–12 tandem copies) greatly improves the production of Na-ASP1 protein and actinorhodin in P. pastoris and S. coelicolor, respectively [155,156]. Furthermore, compared with monoculture, the co-cultivation system possesses more flexibility and stability, leading to the optimum conversion of substrate to natural products [151].
As in the flavonoid synthesis pathway, six genes were split between the two modules, each carrying three genes, as per the co-factor requirement, i.e. NADPH and malonyl-CoA. This technique leads to enhanced flavan-3-ol production upto 40.7 mg/L, which is 970-fold higher than mono-cultivation system [37,157]. Meanwhile, the stability of engineered synthetic consortium (K. vulgare-G. oxydans) seems to be enhanced by knocking out specific metabolic genes and make it an ideal consortium for optimum production of 2-KGA (reached 89.7% within 36 h) against d-sorbitol [123]. Additionally, stable consortium and monoculture can be used to prepare amino acid, purine, antibiotics and fatty acid, as demonstrated in Table 2.
8.2.4. Effect of mutagenesis on microbial derived-NPs production
Mutation strategies have been utilized to procure overproducing cell lines that are resistant to environmental stress and toxic inhibitor [158]. Mutagenesis can be simply categorized into physical mutagenesis (e.g., by X-rays or ultraviolet light) and chemical mutagenesis (e.g., by ethyl methanesulfonate, nitrous acid, p-fluorophenylalanine or N-methyl-N′-nitro-N-nitrosoguanidine). Of these, p-fluorophenylalanine was employed to select overproducing cell lines with respect to phenolics [159]. As in fermentation industry, introduction of mutant strains of Brevibacterium, Serratia and Corynebacterium have been considered to improve the production of several natural products, such as antibiotics, amino acids, and nucleotides. Moreover, certain genetic techniques like protoplast fusion, and genomic shuffling are used extensively for strain improvement at industrial scale [[160], [161], [162], [163]]. Genome shuffling introduces a point mutations at a very low controlled rate which has been successfully improved the NPs titers in several strains like 130-fold increase in production of epothilone (104 mg/L) was observed in mutant strain of S. cellulosum GSUV3-205 in comparison with parental strain S. cellulosum So0157-2 (0.8 mg/L) [164].
Serratia marcescens, which grows as a monoculture, undergoes control mutation to achieve optimum production of threonine (25 g/l) and its titer was further improved up to 63 g/l by recombinant DNA technology [165,166]. Recently, a direct evolution technique is applied to microbial consortia to generate mutant strains of individual consortium members to construct productive consortia. For example, in mutated anammox consortium, the candidate genes in dominant organism Candidatus Kuenenia stuttgartiensis has been reported to efficiently perform certain tasks, such as the hydrazine metabolism and ladderane synthesis [167]. In addition, optimum productions of NPs were obtained via mutant strains have been demonstrated in Table 2.
9. Concluding remarks and future perspectives
This review emphasizes the significance of monoculture- and consortium-based bioprocessing approaches for the development of natural products that have gained popularity in many food and non-food industries because of their high yields, ease of optimization, economic feasibility, robust growth on inexpensive media, and stability. More importantly, these microbial-derived natural products are currently grabbing attention due to the rapid development of controllable consortia and monoculture through advanced genetic and process optimization techniques.
In particular, we briefly foreground some of the practical implementation of microbial consortium and a monoculture and some of the directions for future research that offer the potential of enhanced production yields and reduced processing costs. Potential directions include (i) development of inexpensive gene-chip assay and high-throughput screening tools that cause directed evolution in multiple communities (ii) in-depth understanding of microbial interactions (in case of a synthetic consortium), and metabolic networks to develop rational metabolic engineering design (iii) Advancement in omics approaches (such as transcriptomics and metabolomics) appears to be an effective way to quantify biochemical changes and metabolic mechanisms, as well as advances in metagenomics positively contributes to enhanced understanding of immensely diverse microbial sources, such as rivers, lakes, sub-seafloor sites, and ice cores. Ultimately, the scientific community will utilize advanced biotechnological data and computational models to achieve optimum natural products production via stable monoculture and consortium.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgements
This review was supported by China Postdoctoral Science Fund (2020M671026).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.synbio.2021.12.012.
Contributor Information
Ali Mohsin, Email: alimohsin@ecust.edu.cn.
Meijin Guo, Email: guo_mj@ecust.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Cravens A., Payne J., Smolke C.D. Synthetic biology strategies for microbial biosynthesis of plant natural products. Nat Commun. 2019;10(1):2142. doi: 10.1038/s41467-019-09848-w/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chae T.U., Choi S.Y., Kim J.W., Ko Y.S., Lee S.Y. Recent advances in systems metabolic engineering tools and strategies. Curr Opin Biotechnol. 2017;47:67–82. doi: 10.1016/j.copbio.2017.06.007. [DOI] [PubMed] [Google Scholar]
- 3.Chubukov V., Aindrila M., Christopher J.P., Keasling J.D., Martín H.G. Synthetic and systems biology for microbial production of commodity chemicals. NPJ Syst Biol Appl. 2016;2:16009. doi: 10.1038/npjsba.2016.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang Y., Wu Ruofan, Zhou Jie, et al. Recent advances of biofuels and biochemicals production from sustainable resources using co-cultivation systems. Biotechnol Biofuels. 2019;12(1):155. doi: 10.1186/s13068-019-1495-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiang L.-L., Zhou J.-J., Quan C.S., Xiu Z.L. Advances in industrial microbiome based on microbial consortium for biorefinery. Bioresour Bioprocess. 2017;4(1):11. doi: 10.1186/s40643-017-0141-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zuroff T.R., Curtis W.R. Developing symbiotic consortia for lignocellulosic biofuel production. Appl Microbiol Biotechnol. 2012;93(4):1423–1435. doi: 10.1007/s00253-011-3762-9. [DOI] [PubMed] [Google Scholar]
- 7.He Y., Xie K., Xu P., Huang X., Gu W., Zhang F. Evolution of microbial community diversity and enzymatic activity during composting. Res Microbiol. 2013;164(2):189–198. doi: 10.1016/j.resmic.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 8.Brune K., Bayer T. Engineering microbial consortia to enhance biomining and bioremediation. Front Microbiol. 2012;3(203) doi: 10.3389/fmicb.2012.00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rugbjerg P., Petersen N.M., Porse A., Sarup-Lytzen K., Morten A. Diverse genetic error modes constrain large-scale bio-based production. Nat Commun. 2018;9(1):787. doi: 10.1038/s41467-018-03232-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmidt F.R. Optimization and scale up of industrial fermentation processes. Appl Microbiol Biotechnol. 2005;68(4):425–435. doi: 10.1007/s00253-005-0003-0. [DOI] [PubMed] [Google Scholar]
- 11.Young T.B. Fermentation scaleup: industrial experience with a total environmental approach. Ann N Y Acad Sci. 1979;326(1):165–180. [Google Scholar]
- 12.Zhang J., Zeng G., Chen Y. Effects of physico-chemical parameters on the bacterial and fungal communities during agricultural waste composting. Bioresour Technol. 2011;102(3):2950–2956. doi: 10.1016/j.biortech.2010.11.089. [DOI] [PubMed] [Google Scholar]
- 13.Pham J.V., Mariamawit N., Yilma A., et al. A review of the microbial production of bioactive natural products and biologics. Front Microbiol. 2019;10(1404) doi: 10.3389/fmicb.2019.01404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Silva B., Barreira J., Oliveira M. Natural phytochemicals and probiotics as bioactive ingredients for functional foods: extraction, biochemistry and protected-delivery technologies. Trends Food Sci Technol. 2016;50 doi: 10.1016/j.tifs.2015.12.007. [DOI] [Google Scholar]
- 15.Demain A.L. Importance of microbial natural products and the need to revitalize their discovery. J Ind Microbiol Biotechnol. 2014;41(2):185–201. doi: 10.1007/s10295-013-1325-z. [DOI] [PubMed] [Google Scholar]
- 16.Püschel D., Martina J., Alena V., Hana G., Miroslav V., Jan J. Arbuscular mycorrhiza stimulates biological nitrogen fixation in two medicago spp. through improved phosphorus acquisition. Front Plant Sci. 2017;8(390) doi: 10.3389/fpls.2017.00390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stajich J.E., Wilke Sarah K. Dag Ahrén, Chun Hang Au, Bruce W. Birren, Mark Borodovsky et al., Insights into evolution of multicellular fungi from the assembled chromosomes of the mushroom Coprinopsis cinerea (Coprinus cinereus) Proc Natl Acad Sci USA. 2010;107(26):11889–11894. doi: 10.1073/pnas.1003391107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bills G.F., Gloer J.B. Biologically active secondary metabolites from the fungi, in the fungal kingdom. Microbiol Spectr. 2017:1087–1119. doi: 10.1128/microbiolspec.FUNK-0009-2016. [DOI] [PubMed] [Google Scholar]
- 19.Cadamuro R.D., Isabela M.A., da Silveira B., Izabella T.S., et al. Bioactive compounds from mangrove endophytic fungus and their uses for microorganism control. J Fungi (Basel) 2021;7(6):455. doi: 10.3390/jof7060455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singh R., Kumar M., Mittal A., Kumar P. Microbial metabolites in nutrition, healthcare and agriculture. J Biotechnol. 2017;7(1):15. doi: 10.1007/s13205-016-0586-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Demain A. REVIEWS: the business of biotechnology. Ind Biotechnol. 2007;3:269–283. doi: 10.1089/ind.2007.3.269. [DOI] [Google Scholar]
- 22.Hranueli D., John C., Bojan B., Pavle G., Long P.F. Plasticity of the streptomyces genome-evolution and engineering of new antibiotics. Curr Med Chem. 2005;12(14):1697–1704. doi: 10.2174/0929867054367176. [DOI] [PubMed] [Google Scholar]
- 23.Chen L., Yue Q., Li Y., Niu X. Engineering of glarea lozoyensis for exclusive production of the pneumocandin B precursor of the antifungal drug caspofungin acetate. Appl Environ Microbiol. 2015;81(5):1550–1558. doi: 10.1128/AEM.03256-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gray D.A., Dugar G., Gamba P. Extreme slow growth as alternative strategy to survive deep starvation in bacteria. Nat Commun. 2019;10(1):890. doi: 10.1038/s41467-019-08719-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garcia S. Mixed cultures as model communities: hunting for ubiquitous microorganisms, their partners, and interactions. Aquat Microb Ecol. 2016;77 doi: 10.3354/ame01789. [DOI] [Google Scholar]
- 26.D'Souza G., Shraddha S., Daniel P., Yousif G., Waschina S. Ecology and evolution of metabolic cross-feeding interactions in bacteria. Nat Prod Rep. 2018;35(5):455–488. doi: 10.1039/C8NP00009C. [DOI] [PubMed] [Google Scholar]
- 27.Hanly T., Urello M., Henson M. Dynamic flux balance modeling of S. cerevisiae and E. coli co-cultures for efficient consumption of glucose/xylose mixtures. Appl Microbiol Biotechnol. 2012;93:2529–2541. doi: 10.1007/s00253-011-3628-1. [DOI] [PubMed] [Google Scholar]
- 28.Smanski M.J., Zhou H., Claesen J., et al. Synthetic biology to access and expand nature's chemical diversity. Nat Rev Microbiol. 2016;14(3):135–149. doi: 10.1038/nrmicro.2015.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nai C., Meyer V. From axenic to mixed cultures: technological advances accelerating a paradigm shift in microbiology. Trends Microbiol. 2018;26(6):538–554. doi: 10.1016/j.tim.2017.11.004. [DOI] [PubMed] [Google Scholar]
- 30.Ruiz B., Chávez A., Forero A., et al. Production of microbial secondary metabolites: regulation by the carbon source. Crit Rev Microbiol. 2010;36(2):146–167. doi: 10.3109/10408410903489576. [DOI] [PubMed] [Google Scholar]
- 31.Becker J., Rohles C.M., Wittmann C. Metabolically engineered Corynebacterium glutamicum for bio-based production of chemicals, fuels, materials, and healthcare products. Metab Eng. 2018;50:122–141. doi: 10.1016/j.ymben.2018.07.008. [DOI] [PubMed] [Google Scholar]
- 32.Pontrelli S., Tsan-Yu C., Ethan I.L., Chang P., Liao J.C. Escherichia coli as a host for metabolic engineering. Metab Eng. 2018;50:16–46. doi: 10.1016/j.ymben.2018.04.008. [DOI] [PubMed] [Google Scholar]
- 33.Darvishi F., Ariana M., Marella E.R., Borodina I. Advances in synthetic biology of oleaginous yeast Yarrowia lipolytica for producing non-native chemicals. Appl Microbiol Biotechnol. 2018;102(14):5925–5938. doi: 10.1007/s00253-018-9099-x. [DOI] [PubMed] [Google Scholar]
- 34.Novoveská L., Frank D.T., Wulfer T.A., William J.H. Stabilizing continuous mixed cultures of microalgae. Algal Res. 2016;13:126–133. doi: 10.1016/j.algal.2015.11.021. [DOI] [Google Scholar]
- 35.Garcia S.L., Katherine D.M., Hans-Peter G., Falk W. Successful enrichment of the ubiquitous freshwater acI Actinobacteria. Environ Microbiol Rep. 2014;6(1):21–27. doi: 10.1111/1758-2229.12104. [DOI] [PubMed] [Google Scholar]
- 36.Eilers H., Pernthaler F., Glöckner O., Amann R. Culturability and in situ abundance of pelagic bacteria from the North sea. Appl Environ Microbiol. 2000;66(7):3044–3051. doi: 10.1128/AEM.66.7.3044-3051.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johns N.I., Tomasz B., Antonio L.G., Wang H.H. Principles for designing synthetic microbial communities. Curr Opin Microbiol. 2016;31:146–153. doi: 10.1016/j.mib.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zengler K., Palsson B.O. A road map for the development of community systems (CoSy) biology. Nat Rev Microbiol. 2012;10(5):366–372. doi: 10.1038/nrmicro2763. [DOI] [PubMed] [Google Scholar]
- 39.Roell G.W., Rhiannon R., Carr, Stephen S.F., Yinjie J.T. Engineering microbial consortia by division of labor. Microb Cell Factories. 2019;18(1):35. doi: 10.1186/s12934-019-1083-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Che S., Men Y. Synthetic microbial consortia for biosynthesis and biodegradation: promises and challenges. J Ind Microbiol Biotechnol. 2019;46(9–10):1343–1358. doi: 10.1007/s10295-019-02211-4. [DOI] [PubMed] [Google Scholar]
- 41.Jiang Y., Wu R., Zhou J., et al. Recent advances of biofuels and biochemicals production from sustainable resources using co-cultivation systems. Biotechnol Biofuels. 2019;12 doi: 10.1186/s13068-019-1495-7. 155-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.D'Souza G., Shraddha S., Daniel P. Ecology and evolution of metabolic cross-feeding interactions in bacteria. Nat Prod Rep. 2018;35(5):455–488. doi: 10.1039/C8NP00009C. [DOI] [PubMed] [Google Scholar]
- 43.Röling W.F., Ferrer M., Golyshin P.N. Systems approaches to microbial communities and their functioning. Curr Opin Biotechnol. 2010;21(4):532–538. doi: 10.1016/j.copbio.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 44.Stewart E.J. Growing unculturable bacteria. J Bacteriol. 2012;194(16):4151–4160. doi: 10.1128/JB.00345-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Madsen J.S., Henriette L.R., Jakob R. Coexistence facilitates interspecific biofilm formation in complex microbial communities. Environ Microbiol. 2016;18(8):2565–2574. doi: 10.1111/1462-2920.13335. [DOI] [PubMed] [Google Scholar]
- 46.Rosenthal A.Z., Eric G.M., Avigdor E., Jared R.L. RNA-seq reveals cooperative metabolic interactions between two termite-gut spirochete species in co-culture. ISME J. 2011;5(7):1133–1142. doi: 10.1038/ismej.2011.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kang D., Samuel J., Jakob H., Wei S., et al. Construction of simplified microbial consortia to degrade recalcitrant materials based on enrichment and dilution-to-extinction cultures. Front Microbiol. 2020;10(3010) doi: 10.3389/fmicb.2019.03010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kleerebezem R., van Loosdrecht M.C.M. Mixed culture biotechnology for bioenergy production. Curr Opin Biotechnol. 2007;18(3):207–212. doi: 10.1016/j.copbio.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 49.Bader J., Mast-Gerlach E., Popović M.K., Bajpai R., Stahl U. Relevance of microbial coculture fermentations in biotechnology. J Appl Microbiol. 2010;109(2):371–387. doi: 10.1111/j.1365-2672.2009.04659.x. [DOI] [PubMed] [Google Scholar]
- 50.Sabra W., Dietz D., Tjahjasari D. An-Ping Z. Biosystems analysis and engineering of microbial consortia for industrial biotechnology. Eng Life Sci. 2010;10:407–421. doi: 10.1002/elsc.201000111. [DOI] [Google Scholar]
- 51.Diender M., Stams A.J., Sousa D.Z. Production of medium-chain fatty acids and higher alcohols by a synthetic co-culture grown on carbon monoxide or syngas. Biotechnol Biofuels. 2016;9:82. doi: 10.1186/s13068-016-0495-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bhatia S.K., Jeong-Jun Y., Hyun-Joong K., Ju Won H., Yoon Gi H. Engineering of artificial microbial consortia of Ralstonia eutropha and Bacillus subtilis for poly(3-hydroxybutyrate-co-3-hydroxyvalerate) copolymer production from sugarcane sugar without precursor feeding. Bioresour Technol. 2018;257:92–101. doi: 10.1016/j.biortech.2018.02.056. [DOI] [PubMed] [Google Scholar]
- 53.Tsoi R., Zhang C., Bewick S., Karig D., You L. Metabolic division of labor in microbial systems. Proc Natl Acad Sci U S A. 2018;115(10):2526–2531. doi: 10.1073/pnas.1716888115/-/DCSupplemental. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stenuit B., Agathos S.N. Deciphering microbial community robustness through synthetic ecology and molecular systems synecology. Curr Opin Biotechnol. 2015;33:305–317. doi: 10.1016/j.copbio.2015.03.012. [DOI] [PubMed] [Google Scholar]
- 55.Loccoz M., Mavingui P., Combes C., Normand P., Steinberg C.Y. Microorganisms and biotic interactions. Environmental microbiology: fundamentals and applications. Microb Ecol. 2014:395–444. doi: 10.1007/978-94-017-9118-2_11. [DOI] [Google Scholar]
- 56.Haruta S., Kato S., Yamamoto K., Igarashi Y. Intertwined interspecies relationships: approaches to untangle the microbial network. Environ Microbiol. 2009;11:2963–2969. doi: 10.1111/j.1462-2920.2009.01956.x. [DOI] [PubMed] [Google Scholar]
- 57.Little A.E., Robinson C.J., Peterson S.B., Raffa K.F., Handelsman J. Rules of engagement: interspecies interactions that regulate microbial communities. Annu Rev Microbiol. 2008;62:375–401. doi: 10.1146/annurev.micro.030608.101423. [DOI] [PubMed] [Google Scholar]
- 58.Zhou K., Ruiyang Z., Gregory S., Heng-Phon T. Metabolite profiling identified methylerythritol cyclodiphosphate efflux as a limiting step in microbial isoprenoid production. PLoS One. 2012;7 doi: 10.1371/journal.pone.0047513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dietz D., Sabra W., Zeng A.-P. Co-cultivation of Lactobacillus zeaeand Veillonella criceti for the production of propionic acid. Amb Express. 2013;3(1) doi: 10.1186/2191-0855-3-29. 29-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Minty J.J., Marc E.S., Scott A.S., hang-Hoon B. Design and characterization of synthetic fungal-bacterial consortia for direct production of isobutanol from cellulosic biomass. Proc Natl Acad Sci U S A. 2013;110(36):14592–14597. doi: 10.1073/pnas.1218447110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schmidtke A., Gaedke U., Weithoff G. A mechanistic basis for underyielding in phytoplankton communities. Ecology. 2010;91(1):212–221. doi: 10.1890/08-2370.1. [DOI] [PubMed] [Google Scholar]
- 62.Watanabe K. Microorganisms relevant to bioremediation. Curr Opin Biotechnol. 2001;12(3):237–241. doi: 10.1016/S0958-1669(00)00205-6. [DOI] [PubMed] [Google Scholar]
- 63.Naseri G., Koffas M.A.G. Application of combinatorial optimization strategies in synthetic biology. Nat Commun. 2020;11(1):2446. doi: 10.1038/s41467-020-16175-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xu X., Liu Y., Guocheng D., Ledesma-Amaro R., Liu L. Microbial chassis development for natural product biosynthesis. Trends Biotechnol. 2020;38 doi: 10.1016/j.tibtech.2020.01.002. [DOI] [PubMed] [Google Scholar]
- 65.Che S., Men Y. Synthetic microbial consortia for biosynthesis and biodegradation: promises and challenges. J Ind Microbiol Biotechnol. 2019;46 doi: 10.1007/s10295-019-02211-4. [DOI] [PubMed] [Google Scholar]
- 66.Cobb R.E., Wang Y., Zhao H. High-efficiency multiplex genome editing of Streptomyces species using an engineered CRISPR/Cas system. ACS Synth Biol. 2015;4(6):723–728. doi: 10.1021/sb500351f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wen Z., Minton N., Zhang Y., Li Q., et al. Enhanced solvent production by metabolic engineering of a twin-clostridial consortium. Metab Eng. 2017;39:38–48. doi: 10.1016/j.ymben.2016.10.013. [DOI] [PubMed] [Google Scholar]
- 68.Schofield M.M., Sherman D.H. Meta-omic characterization of prokaryotic gene clusters for natural product biosynthesis. Curr Opin Biotechnol. 2013;24(6):1151–1158. doi: 10.1016/j.copbio.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.van Baarlen P., Kleerebezem M., Wells J.M. Omics approaches to study host-microbiota interactions. Curr Opin Microbiol. 2013;16(3):270–277. doi: 10.1016/j.mib.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 70.El-Ali J., Sorger P.K., Jensen K.F. Cells on chips. Nature. 2006;442(7101):403–411. doi: 10.1038/nature05063. [DOI] [PubMed] [Google Scholar]
- 71.Goers L., Freemont P., Polizzi K.M. Co-culture systems and technologies: taking synthetic biology to the next level. J R Soc Interface. 2014;11(96) doi: 10.1098/rsif.2014.0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Segal J.P., Benjamin H.M., Mohammed N.Q., Acharjee A. The application of omics techniques to understand the role of the gut microbiota in inflammatory bowel disease. Therap Adv Gastroenterol. 2019;12:17–24. doi: 10.1177/1756284818822250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chandran H., Meena M., Sharma K. Microbial biodiversity and bioremediation assessment through omics approaches. Front Environ Chem. 2020;1(9) doi: 10.3389/fenvc.2020.570326. [DOI] [Google Scholar]
- 74.Ma Q., Zhou J., Zhang W., Meng X., Sun J., Ying-Jin Y. Integrated proteomic and metabolomic analysis of an artificial microbial community for two-step production of vitamin C. PLoS One. 2011;6(10) doi: 10.1371/journal.pone.0026108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ma Q., Wang J., Lu S., Lv Y., Yuan Y. Quantitative proteomic profiling reveals photosynthesis responsible for inoculum size dependent variation in Chlorella sorokiniana. Biotechnol Bioeng. 2013;110(3):773–784. doi: 10.1002/bit.24762. [DOI] [PubMed] [Google Scholar]
- 76.Wang D.-Z., Xie Z.-X., Zhang S.-F. Marine metaproteomics: current status and future directions. J Proteomics. 2014;97:27–35. doi: 10.1016/j.jprot.2013.08.024. [DOI] [PubMed] [Google Scholar]
- 77.Li B.-Z., Yuan Y.-J. Transcriptome shifts in response to furfural and acetic acid in Saccharomyces cerevisiae. Appl Microbiol Biotechnol. 2010;86(6):1915–1924. doi: 10.1007/s00253-010-2518-2. [DOI] [PubMed] [Google Scholar]
- 78.Lim W.A. Designing customized cell signalling circuits. Nat Rev Mol Cell Biol. 2010;11(6):393–403. doi: 10.1038/nrm2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Slusarczyk A.L., Lin A., Weiss R. Foundations for the design and implementation of synthetic genetic circuits. Nat Rev Genet. 2012;13(6):406–420. doi: 10.1038/nrg3227. [DOI] [PubMed] [Google Scholar]
- 80.Tang J.C., Shibata A., Zhou Q., Katayama A. Effect of temperature on reaction rate and microbial community in composting of cattle manure with rice straw. J Biosci Bioeng. 2007;104(4):321–328. doi: 10.1263/jbb.104.321. [DOI] [PubMed] [Google Scholar]
- 81.Tang L., Zeng G., Shen G., et al. Sensitive detection of lip genes by electrochemical DNA sensor and its application in polymerase chain reaction amplicons from Phanerochaete chrysosporium. Biosens Bioelectron. 2009;24(5):1474–1479. doi: 10.1016/j.bios.2008.09.031. [DOI] [PubMed] [Google Scholar]
- 82.Cahyani V., Matsuya K., Asakawa S., Kimura M. Succession and phylogenetic composition of bacterial communities responsible for the composting process of rice straw estimated by PCR-DGGE Analysis. Soil Sci Plant Nutr. 2003;49:619–630. doi: 10.1080/00380768.2003.10410052. [DOI] [Google Scholar]
- 83.Klamer M., Bååth M. Microbial community dynamics during composting of straw material studied using phospholipid fatty acid analysis. FEMS Microbiol Ecol. 1998;27(1):9–20. doi: 10.1016/S0168-6496(98)00051-8. [DOI] [Google Scholar]
- 84.Herrmann R.F., Shann J.F. Microbial community changes during the composting of municipal solid waste. Microb Ecol. 1997;33(1):78–85. doi: 10.1007/s002489900010. [DOI] [PubMed] [Google Scholar]
- 85.Khan S., Wajid Ullah M., Siddique R. Role of recombinant DNA technology to improve life. Int J Genomics. 2016;2016 doi: 10.1155/2016/2405954. 2405954-2405954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bhatia S. Introduction to pharmaceutical biotechnology. second ed. IOP Publisher; 2018. Introduction to enzymes and their applications. [Google Scholar]
- 87.Rademacher A., Christine N., Mandy S., Michael K. Temperature increases from 55 to 75 °C in a two-phase biogas reactor result in fundamental alterations within the bacterial and archaeal community structure. Appl Microbiol Biotechnol. 2012;96(2):565–576. doi: 10.1007/s00253-012-4348-x. [DOI] [PubMed] [Google Scholar]
- 88.Xiong H., Chen J., Wang H., Shi H. Influences of volatile solid concentration, temperature and solid retention time for the hydrolysis of waste activated sludge to recover volatile fatty acids. Bioresour Technol. 2012;119:285–292. doi: 10.1016/j.biortech.2012.05.126. [DOI] [PubMed] [Google Scholar]
- 89.D'Amico S., Collins T., Jean-Claude M., et al. Psychrophilic microorganisms: challenges for life. EMBO Rep. 2006;7(4):385–389. doi: 10.1038/sj.embor.7400662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yogabaanu U., Jean-Frederic F.W., Peter C., Rizman-Idid M., Alias S.A. Antimicrobial properties and the influence of temperature on secondary metabolite production in cold environment soil fungi. Polar Sci. 2017;14:60–67. doi: 10.1016/j.polar.2017.09.005. [DOI] [Google Scholar]
- 91.Li Y., Guo F., Guan Y., et al. Novel anthraquinone compounds inhibit colon cancer cell proliferation via the reactive oxygen species/JNK pathway. Molecules. 2020;25(7):1672. doi: 10.3390/molecules25071672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang H., Ming-Hua H., Hu-Pei Y., Yao X.S. Three new cytotoxic cyclic acylpeptides from marine Bacillus sp. Chem Pharm Bull (Tokyo) 2004;52:1029–1030. doi: 10.1248/cpb.52.1029. [DOI] [PubMed] [Google Scholar]
- 93.Oakley K.L., Moore C.B., Denning D.W. In vitro activity of the echinocandin antifungal agent LY303,366 in comparison with itraconazole and amphotericin B against Aspergillus spp. Antimicrob Agents Chemother. 1998;42(10):2726–2730. doi: 10.1128/AAC.42.10.2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wiebe W.J., Sheldon W.M., Pomeroy L.R. Evidence for an enhanced substrate requirement by marine mesophilic bacterial isolates at minimal growth temperatures. Microb Ecol. 1993;25(2):151–159. doi: 10.1007/BF00177192. [DOI] [PubMed] [Google Scholar]
- 95.James P.D., Edwards C., Dawson M. The effects of temperature, pH and growth rate on secondary metabolism in Streptomyces thermoviolaceus grown in a chemostat. J Gen Microbiol. 1991;137(7):1715–1720. doi: 10.1099/00221287-137-7-1715. [DOI] [PubMed] [Google Scholar]
- 96.Hanke A., Jasmine B., Theresa H., Halina E.T., Christine E.S., Marc S. Selective pressure of temperature on competition and cross-feeding within denitrifying and fermentative microbial communities. Front Microbiol. 2015;6:1461. doi: 10.3389/fmicb.2015.01461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lawson C.E., Wu S., Bhattacharjee A.S. Metabolic network analysis reveals microbial community interactions in anammox granules. Nat Commun. 2017;8(1):15416. doi: 10.1038/ncomms15416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chen W., Qin Z. Development of a gene cloning system in a fast-growing and moderately thermophilic Streptomyces species and heterologous expression of Streptomyces antibiotic biosynthetic gene clusters. BMC Microbiol. 2011;11:243. doi: 10.1186/1471-2180-11-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gunnigle E., McCay P., Matthew F., Catherine H.B., Florence A., Vincent F. A functional approach to uncover the low-temperature adaptation strategies of the archaeon Methanosarcina barkeri. Appl Environ Microbiol. 2013;79(14):4210–4219. doi: 10.1128/AEM.03787-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lee S.H., Jung J.Y., Jeon C.O. Effects of temperature on microbial succession and metabolite change during saeu-jeot fermentation. Food Microbiol. 2014;38:16–25. doi: 10.1016/j.fm.2013.08.004. [DOI] [PubMed] [Google Scholar]
- 101.Rathi R., Meeta L., Manoj S., Vipin K., Kumar S., Banwari L. Stimulation of an indigenous thermophillic anaerobic bacterial consortium for enhanced oil recovery. RSC Adv. 2015;5(107):88115–88124. doi: 10.1039/C5RA10489K. [DOI] [Google Scholar]
- 102.Rizk M., Abdel-Rahman T., Metwally H. Factors affecting growth and antifungal activity of some Streptomyces species against Candida albicans. J Food Agric Environ. 2007;5 https://www.researchgate.net/publication/268350948 [Google Scholar]
- 103.Aswini K., Gopal N.O., Uthandi S. Optimized culture conditions for bacterial cellulose production by Acetobacter senegalensis MA1. BMC Biotechnol. 2020;20(1):46. doi: 10.1186/s12896-020-00639-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Calicioglu O., Michael J.S., Tom L.R., Rachel A.B. Effect of pH and temperature on microbial community structure and carboxylic acid yield during the acidogenic digestion of duckweed. Biotechnol Biofuels. 2018;11(1):275. doi: 10.1186/s13068-018-1278-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ilhan Z.E., Andrew K.M., Dae-Wook K., Bruce E.R., Krajmalnik-Brown R. pH-Mediated microbial and metabolic interactions in fecal enrichment cultures. mSphere. 2017;2(3):e00047–17. doi: 10.1128/mSphere.00047-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kolenbrander P. The genus Veillonella. J Bacteriol. 2006:1022–1040. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC276976 [Google Scholar]
- 107.Yu G.-H., Pin-Jing H., Li-Ming S., Pei-Pei H. Toward understanding the mechanism of improving the production of volatile fatty acids from activated sludge at pH 10.0. Water Res. 2008;42(18):4637–4644. doi: 10.1016/j.watres.2008.08.018. [DOI] [PubMed] [Google Scholar]
- 108.Shetty S.A., Nachiket P.M., Vikram L., Dilip R., Yogesh S.S. Comparative genome analysis of Megasphaera sp. reveals niche specialization and its potential role in the human gut. PLoS One. 2013;8(11) doi: 10.1371/journal.pone.0079353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Nishihara Y., Tsujii E., Yamagishi Y., et al. A new anti-influenza agent isolated from Apsergillus terreus No 13830. I. Taxomony, fermentation, isolation, physico- chemical properties and biological activities. J Antibiot. 2001;54(2):136–143. doi: 10.7164/antibiotics.54.136. FR198248. [DOI] [PubMed] [Google Scholar]
- 110.Rajan B.M., Kannabiran K. Extraction and identification of antibacterial secondary metabolites from marine streptomyces sp. VITBRK2. Int J Mol Cell Med. 2014;3(3):130–137. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4170486 [PMC free article] [PubMed] [Google Scholar]
- 111.James P.D.A., Edwards C. The effects of cultural conditions on growth and secondary metabolism in Streptomyces thermoviolaceus. FEMS Microbiol Lett. 1988;52(1–2):1–5. doi: 10.1111/j.1574-6968.1988.tb02562.x. [DOI] [Google Scholar]
- 112.Rizk M., Abdel-rahman T., Metwally H. Factors affecting growth and antifungal activity of some Streptomyces species against Candida albicans. J Food Agric Environ. 2007;5:446–449. https://www.researchgate.net/publication/268350948 [Google Scholar]
- 113.El-Tayeb O., Hussein M., El-Sedawyet H.F. 2004. Full Length Research Paper - optimization of industrial production of rifamycin B by Amycolatopsis mediterranei. II. The role of gene amplification and physiological factors in productivity in shake flasks. [DOI] [Google Scholar]
- 114.Kim J., Kim K.H. Effects of minimal media vs. complex media on the metabolite profiles of Escherichia coli and Saccharomyces cerevisiae. Process Biochem. 2017;57:64–71. doi: 10.1016/j.procbio.2017.04.003. [DOI] [Google Scholar]
- 115.Ripa F.A., Nikkon F., Zaman S., Khondkar P. Optimal conditions for antimicrobial metabolites production from a new streptomyces sp. RUPA-08PR isolated from Bangladeshi soil. Mycobiology. 2009;37(3):211–214. doi: 10.4489/MYCO.2009.37.3.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Demain A.L., Fang A. Emerging concepts of secondary metabolism in actinomycetes. Actinomycetologica. 1995;9(2):98–117. doi: 10.3209/saj.9_98. [DOI] [Google Scholar]
- 117.Ghribi D., Ellouze-Chaabouni S. Enhancement of Bacillus subtilis lipopeptide biosurfactants production through optimization of medium composition and adequate control of aeration. Biotechnol Res Int. 2011;2011:653654. doi: 10.4061/2011/653654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Jain P., Pundir R. Effect of fermentation medium, pH and temperature variation on antibacterial soil fungal metabolite production. J Agric Technol. 2011;7:247–269. https://www.researchgate.net/publication/284976774 [Google Scholar]
- 119.Marcinowska R., Johan T., Wolf-Watz H., Mortiz T., Surowiec I. Optimization of a sample preparation method for the metabolomic analysis of clinically relevant bacteria. J Microbiol Methods. 2011;87:24–31. doi: 10.1016/j.mimet.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 120.Ishii K., Takii S. Comparison of microbial communities in four different composting processes as evaluated by denaturing gradient gel electrophoresis analysis. J Appl Microbiol. 2003;95(1):109–119. doi: 10.1046/j.1365-2672.2003.01949.x. [DOI] [PubMed] [Google Scholar]
- 121.Maeda K., Morioka R., Osada T. Effect of covering composting piles with mature compost on ammonia emission and microbial community structure of composting process. J Environ Qual. 2009;38(2):598–606. doi: 10.2134/jeq2008.0083. [DOI] [PubMed] [Google Scholar]
- 122.Lou H., Hu L., Lu H., Wei T., Chen Q. Metabolic engineering of microbial cell factories for biosynthesis of flavonoids: a review. Molecules. 2021;26(15):4522. doi: 10.3390/molecules26154522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wang E.-X. Reorganization of a synthetic microbial consortium for one-step vitamin C fermentation. Microb Cell Factories. 2016;15 doi: 10.1186/s12934-016-0418-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Jones J.A., Koffas M.A. Methods in enzymology. fourth ed. Academic Press; 2016. Chapter eight - optimizing metabolic pathways for the improved production of natural products. [DOI] [PubMed] [Google Scholar]
- 125.Liu Y., Tu X., Xu Q., et al. Engineered monoculture and co-culture of methylotrophic yeast for de novo production of monacolin J and lovastatin from methanol. Metab Eng. 2018;45:189–199. doi: 10.1016/j.ymben.2017.12.009. [DOI] [PubMed] [Google Scholar]
- 126.Saini M., Chen M.H., Chung-Jen C., Yun-Peng C. Potential production platform of n-butanol in Escherichia coli. Metab Eng. 2015;27:76–82. doi: 10.1016/j.ymben.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 127.Li Z., Wan J., Zhan H., Cheng X., Wei C., Huang K. Particle size distribution on Forchheimer flow and transition of flow regimes in porous media. J Hydrol. 2019;574:1–11. doi: 10.1016/j.jhydrol.2019.04.026. [DOI] [Google Scholar]
- 128.Pepper I.L., Charles G.T. third ed. Academic press; 2011. Environmental microbiology. [Google Scholar]
- 129.Zhang Z., Hairui J., Guiping G., Xu Z., Tianwei T. Synergistic effects of oleaginous yeast Rhodotorula glutinis and microalga Chlorella vulgaris for enhancement of biomass and lipid yields. Bioresour Technol. 2014;164:93–99. doi: 10.1016/j.biortech.2014.04.039. [DOI] [PubMed] [Google Scholar]
- 130.Ashtiani F.-R., Hasan J., Mahdi R., Mahsa S., Abdeltif A. Effect of mixed culture of yeast and microalgae on acetyl-CoA carboxylase and Glycerol-3-phosphate acyltransferase expression. J Biosci Bioeng. 2021;131(4):364–372. doi: 10.1016/j.jbiosc.2020.11.006. [DOI] [PubMed] [Google Scholar]
- 131.Paul C., Mausz M.A., Pohnert G. A co-culturing/metabolomics approach to investigate chemically mediated interactions of planktonic organisms reveals influence of bacteria on diatom metabolism. Metabolomics. 2013;9(2):349–359. doi: 10.1007/s11306-012-0453-1. [DOI] [Google Scholar]
- 132.Jones J.A., Victoria R.V., Andrew L.S., et al. Experimental and computational optimization of an Escherichia coli co-culture for the efficient production of flavonoids. Metab Eng. 2016;35:55–63. doi: 10.1016/j.ymben.2016.01.006. [DOI] [PubMed] [Google Scholar]
- 133.Buzzini P. Batch and fed-batch carotenoid production by Rhodotorula glutinis-Debaryomyces castellii co-cultures in corn syrup. J Appl Microbiol. 2001;90(5):843–847. doi: 10.1046/j.1365-2672.2001.01319.x. [DOI] [PubMed] [Google Scholar]
- 134.Harcombe W.R., William J.R., Ilija D., et al. Metabolic resource allocation in individual microbes determines ecosystem interactions and spatial dynamics. Cell Rep. 2014;7(4):1104–1115. doi: 10.1016/j.celrep.2014.03.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Shou W., Ram S., Vilar J.M. Synthetic cooperation in engineered yeast populations. Proc Natl Acad Sci U S A. 2007;104(6):1877–1882. doi: 10.1073/pnas.0610575104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lee C.W., Shuler M.L. The effect of inoculum density and conditioned medium on the production of ajmalicine and catharanthine from immobilized Catharanthus roseus cells. Biotechnol Bioeng. 2000;67(1):61–71. doi: 10.1002/(sici)1097-0290(20000105)67:1<61::aid-bit7>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 137.Matsubara K., Kitani S., Yoshioka T., Morimoto T., Fujita Y., Yamada Y. High density culture of Coptis japonica cells increases berberine production. J Chem Technol Biotechnol. 1989;46(1):61–69. doi: 10.1002/jctb.280460107. [DOI] [Google Scholar]
- 138.Mnif I., Ellouze-Chaabouni S., Ghribi D. Optimization of inocula conditions for enhanced biosurfactant production by Bacillus subtilis SPB1, in submerged culture, using Box–Behnken design. Probiotic Antimicrob Proteins. 2012;5 doi: 10.1007/s12602-012-9113-z. [DOI] [PubMed] [Google Scholar]
- 139.Cheng P., Wang Y., Osei-Wusu D., Liu T., Liu D. Effects of seed age, inoculum density, and culture conditions on growth and hydrocarbon accumulation of Botryococcus braunii SAG807-1 with attached culture. Bioresour Bioprocess. 2018;5(1):15. doi: 10.1186/s40643-018-0198-4. [DOI] [Google Scholar]
- 140.Mahalik S., Sharma A.K., Mukherjee K.J. Genome engineering for improved recombinant protein expression in Escherichia coli. Microb Cell Factories. 2014;13(1):177. doi: 10.1186/s12934-014-0177-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wendisch V.F., Jorge M.P., Pérez-García F., Sgobba E. Updates on industrial production of amino acids using Corynebacterium glutamicum. World J Microbiol Biotechnol. 2016;32(6):105. doi: 10.1007/s11274-016-2060-1. [DOI] [PubMed] [Google Scholar]
- 142.Demain A.L. Pharmaceutically active secondary metabolites of microorganisms. Appl Microbiol Biotechnol. 1999;52(4):455–463. doi: 10.1007/s002530051546. [DOI] [PubMed] [Google Scholar]
- 143.Chang M.C., Rachel A.E., William T., Dae-Kyun R., Jay D.K. Engineering Escherichia coli for production of functionalized terpenoids using plant P450s. Nat Chem Biol. 2007;3(5):274–277. doi: 10.1038/nchembio875. [DOI] [PubMed] [Google Scholar]
- 144.Fang Z., Jones J.A., Zhou J., Koffas A.G. Engineering Escherichia coli co‐cultures for production of curcuminoids from glucose. Biotechnol J. 2018;13(5):1700576. doi: 10.1002/biot.201700576. [DOI] [PubMed] [Google Scholar]
- 145.Minami H., Ju-Sung K., Nobuhiro I., Tomoya T., et al. Microbial production of plant benzylisoquinoline alkaloids. Proc Natl Acad Sci U S A. 2008;105(21):7393–7398. doi: 10.1073/pnas.0802981105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Camacho-Zaragoza J.M., Hernández-Chávez G., Moreno-Avitia F., et al. Engineering of a microbial coculture of Escherichia coli strains for the biosynthesis of resveratrol. Microb Cell Factories. 2016;15(1):1–11. doi: 10.1186/s12934-016-0562-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Gold N.D., Christopher M.G., Francois-Xavier L., et al. Metabolic engineering of a tyrosine-overproducing yeast platform using targeted metabolomics. Microb Cell Factories. 2015;14:73. doi: 10.1186/s12934-015-0252-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Chemler J.A., Zachary L.F., Kyle P.M., Mattheos A., Koffas G. Improving NADPH availability for natural product biosynthesis in Escherichia coli by metabolic engineering. Metab Eng. 2010;12(2):96–104. doi: 10.1016/j.ymben.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 149.Jung W.S., Young J.Y., Je W.P., et al. A combined approach of classical mutagenesis and rational metabolic engineering improves rapamycin biosynthesis and provides insights into methylmalonyl-CoA precursor supply pathway in Streptomyces hygroscopicus ATCC 29253. Appl Microbiol Biotechnol. 2011;91(5):1389–1397. doi: 10.1007/s00253-011-3348-6. [DOI] [PubMed] [Google Scholar]
- 150.Brown S., Clastre M., Courdavault V., Connoret S. De novo production of the plant- derived alkaloid strictosidine in yeast. Proc Natl Acad Sci U S A. 2015;112(11):3205–3210. doi: 10.1073/pnas.1423555112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Zhou K., Qiao K., Edgar S., Stephanopoulos G. Distributing a metabolic pathway among a microbial consortium enhances production of natural products. Nat Biotechnol. 2015;33(4):377–383. doi: 10.1038/nbt.3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Wu G., Yan Q., Jones J.A., Tang Y.J., Fong S.S. Metabolic burden: cornerstones in synthetic biology and metabolic engineering applications. Trends Biotechnol. 2016;34(8):652–664. doi: 10.1016/j.tibtech.2016.02.010. [DOI] [PubMed] [Google Scholar]
- 153.Pickens L.B., Tang Y., Chooi Y.-H. Metabolic engineering for the production of natural products. Annu Rev Chem Biomol Eng. 2011;2:211–236. doi: 10.1146/annurev-chembioeng-061010-114209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Komatsu M., Uchiyama T., Ōmura S., Cane D.E., Ikeda H. Genome-minimized Streptomyces host for the heterologous expression of secondary metabolism. Proc Natl Acad Sci U S A. 2010;107(6):2646–2651. doi: 10.1073/pnas.0914833107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Murakami T., Burian J., Thompson C. A system for the targeted amplification of bacterial gene clusters multiplies antibiotic yield in Streptomyces coelicolor. Proc Natl Acad Sci U S A. 2011;108(38):16020–16025. doi: 10.1073/pnas.1108124108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Inan M., Dinesh A., Jayanta S., Michael M.M. Enhancement of protein secretion in Pichia pastoris by overexpression of protein disulfide isomerase. Biotechnol Bioeng. 2006;93(4):771–778. doi: 10.1002/bit.20762. [DOI] [PubMed] [Google Scholar]
- 157.Chemler J.A., Lock L.T., Koffas A.G., Tzanakakiset E.S. Standardized biosynthesis of flavan-3-ols with effects on pancreatic beta-cell insulin secretion. Appl Microbiol Biotechnol. 2007;77(4):797–807. doi: 10.1007/s00253-007-1227-y. [DOI] [PubMed] [Google Scholar]
- 158.Rao S.R., Ravishankar G. Biotransformation of isoeugenol to vanilla flavour metabolites and capsaicin in suspended and immobilized cell cultures of Capsicum frutescens: study of the influence of β-cyclodextrin and fungal elicitor. Process Biochem. 1999;35(3–4):341–348. doi: 10.1007/102-008-103. [DOI] [Google Scholar]
- 159.Smetanska I. Production of secondary metabolites using plant cell cultures. Adv Biochem Eng Biotechnol. 2008;111:187–228. doi: 10.1007/10-2008-103. [DOI] [PubMed] [Google Scholar]
- 160.Zhou M., Jing X., Xie P., Chen W., Wang T., Xia H., Qin Z. Sequential deletion of all the polyketide synthase and nonribosomal peptide synthetase biosynthetic gene clusters and a 900-kb subtelomeric sequence of the linear chromosome of Streptomyces coelicolor. FEMS Microbiol Lett. 2012;333(2):169–179. doi: 10.1111/j.1574-6968.2012.02609.x. [DOI] [PubMed] [Google Scholar]
- 161.Baltz R.H. Genomics and the ancient origins of the daptomycin biosynthetic gene cluster. J Antibiot (Tokyo) 2010;63(8):506–511. doi: 10.1038/ja.2010.82. [DOI] [PubMed] [Google Scholar]
- 162.Pullan S.T., Chandra G., Bibb M.J., Merrick M. Genome-wide analysis of the role of GlnR in Streptomyces venezuelae provides new insights into global nitrogen regulation in actinomycetes. BMC Genom. 2011;12:175. doi: 10.1186/1471-2164-12-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Lopatniuk M., Ostash B., Luzhetskyy A., Walker S., Fedorenko V. Generation and study of the strains of streptomycetes - heterologous hosts for production of moenomycin. Russ J Genet. 2014;50(4):360–365. doi: 10.1134/S1022795414040085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Gong G., Sun X., Xin-li L., et al. Mutation and a high-throughput screening method for improving the production of Epothilones of Sorangium. J Ind Microbiol Biotechnol. 2007;34(9):615–623. doi: 10.1007/s10295-007-0236-2. [DOI] [PubMed] [Google Scholar]
- 165.Sugita T., Komatsubara S. Construction of a threonine-hyperproducing strain of Serratia marcescens by amplifying the phosphoenolpyruvate carboxylase gene. Appl Microbiol Biotechnol. 1989;30(3):290–293. doi: 10.4161/bbug.1.2.10484. [DOI] [Google Scholar]
- 166.Komatsubara S., Kisumi M., Chibata I. Transductional construction of a threonine-producing strain of Serratia marcescens. Appl Environ Microbiol. 1979;38(6):1045–1051. doi: 10.1128/aem.38.6.1045-1051.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Ding C., Adrian L. Comparative genomics in “Candidatus Kuenenia stuttgartiensis” reveal high genomic plasticity in the overall genome structure, CRISPR loci and surface proteins. BMC Genom. 2020;21(1):851. doi: 10.1186/s12864-020-07242-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Zhang J., Zhang Y., Shan-na L., Han Y., Zhi-jiang Z. Modelling growth and bacteriocin production by Pediococcus acidilactici PA003 as a function of temperature and pH value. Appl Biochem Biotechnol. 2012;166(6):1388–1400. doi: 10.1007/s12010-011-9532-4. [DOI] [PubMed] [Google Scholar]
- 169.Zhang H., Stephanopoulos G. Co-culture engineering for microbial biosynthesis of 3-amino-benzoic acid in Escherichia coli. Biotechnol J. 2016;11(7):981–987. doi: 10.1002/biot.201600013. [DOI] [PubMed] [Google Scholar]
- 170.Wang S., Fang M., Weiwei M., Wang P., Zhao G., Lu X. Influence of temperature on biogas production efficiency and microbial community in a two-phase Anaerobic digestion system. Water. 2019;11:133. doi: 10.3390/w11010133. [DOI] [Google Scholar]
- 171.Liu Y., Tu X., Xu Q. Engineered monoculture and co-culture of methylotrophic yeast for de novo production of monacolin J and lovastatin from methanol. Metab Eng. 2018;45:189–199. doi: 10.1016/j.ymben.2017.12.009. [DOI] [PubMed] [Google Scholar]
- 172.Liu C., Hu B., Liu Y., Chen S. Stimulation of Nisin production from whey by a mixed culture of Lactococcus lactis and Saccharomyces cerevisiae. Appl Biochem Biotechnol. 2006;131(1):751–761. doi: 10.1385/ABAB:131:1:751. [DOI] [PubMed] [Google Scholar]
- 173.Shimizu H., Mizuguchi T., Tanaka E., Shioya S. Nisin production by a mixed-culture system consisting of Lactococcus lactis and Kluyveromyces marxianus. Appl Environ Microbiol. 1999;65(7):3134–3141. doi: 10.1128/aem.65.7.3134-3141.1999. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC91467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Santoso S., Chih-Chan C., Shin-Ping L., et al. Enhanced production of bacterial cellulose by Komactobacter intermedius using statistical modeling. Cellulose. 2020;27:1–13. doi: 10.1007/s10570-019-02961-5. [DOI] [Google Scholar]
- 175.Thite V.S., Nerurkar A.S., Baxi N.N. Optimization of concurrent production of xylanolytic and pectinolytic enzymes by Bacillus safensis M35 and Bacillus altitudinis J208 using agro-industrial biomass through Response Surface Methodology. Sci Rep. 2020;10(1):3824. doi: 10.1038/s41598-020-60760-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Lima C., Fontes L., Contiero J. The use of response surface methodology in optimization of lactic acid production: focus on medium supplementation, temperature and pH control. Food Technol Biotechnol. 2010;48 https://www.researchgate.net/publication/268337823 [Google Scholar]
- 177.Chan-Cupul W., Heredia G., Carrera D.M., Vazquez R.R. Enhancement of ligninolytic enzyme activities in a Trametes maxima–Paecilomyces carneus co-culture: key factors revealed after screening using a Plackett– Burman experimental design. Electron J Biotechnol. 2014;17:114–121. doi: 10.1016/j.ejbt.2014.04.007. [DOI] [Google Scholar]
- 178.Shrestha A., Pandey R.P., Pokhrel A.R. Modular pathway engineering for resveratrol and piceatannol production in engineered Escherichia coli. Appl Microbiol Biotechnol. 2018;102(22):9691–9706. doi: 10.1007/s00253-018-9323-8. [DOI] [PubMed] [Google Scholar]
- 179.Rangaswamy B.E., Vanitha K.P., Hungund B.S. Microbial cellulose production from bacteria isolated from rotten fruit. Int J Polym Sci. 2015;2015:280784. doi: 10.1155/2015/280784. [DOI] [Google Scholar]
- 180.Ahmadi M.K., Fang L., Moscatello N., Pfeifer B.A.E. Coli metabolic engineering for gram scale production of a plant-based anti-inflammatory agent. Metab Eng. 2016;38:382–388. doi: 10.1016/j.ymben.2016.10.001. [DOI] [PubMed] [Google Scholar]
- 181.Yukawa H., Kurusu Y., Shimazu M., Yamagata H., Terasawa M. Stabilization of an E. coli plasmid by a mini-F fragment of DNA. J Ind Microbiol Biotechnol. 1988;2(6):323–328. doi: 10.1007/BF01569570. [DOI] [Google Scholar]
- 182.Park S.Y., Kim W.J., Lee M.H., Lee S.Y. Metabolic engineering of Escherichia coli for high- level astaxanthin production with high productivity. Metab Eng. 2018;49:105–115. doi: 10.1016/j.ymben.2018.08.002. [DOI] [PubMed] [Google Scholar]
- 183.Zhu F., Zhong X., Hu M., Deng Z., Liu T. Targeted engineering and scale up of lycopene overproduction in Escherichia coli. Process Biochem. 2015;50(3):341–346. doi: 10.1016/j.procbio.2014.12.008. [DOI] [Google Scholar]
- 184.Wu J., Zhou T., Du G., Zhou J., Chen J. Modular optimization of heterologous pathways for de novo synthesis of (2S)-naringenin in Escherichia coli. [DOI] [PMC free article] [PubMed]
- 185.Kildegaard K.R., Adiego-Pérez B., Doménech B. Engineering of Yarrowia lipolytica for production of astaxanthin. Synth Syst Biotechnol. 2017;2(4):287–294. doi: 10.1016/j.synbio.2017.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Wang X., Shao A., Li Z., Policarpio L., Zhang H. Constructing E. coli Co-cultures for de novo biosynthesis of natural product acacetin. Biotechnol J. 2020;15(9):2000131. doi: 10.1002/biot.202000131. [DOI] [PubMed] [Google Scholar]
- 187.Mo S., Young J.Y., Yeon H.B., Sung-Kwon L., Eunji K., Joo-Won S., Yeo J.Y. Roles of fkbN in positive regulation and tcs7 in negative regulation of FK506 biosynthesis in Streptomyces sp. strain KCTC 11604BP. Appl Environ Microbiol. 2012;78(7):2249–2255. doi: 10.1128/AEM.06766-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Yang J., Guo L. Biosynthesis of β-carotene in engineered E. coli using the MEP and MVA pathways. Microb Cell Factories. 2014;13(1):160. doi: 10.1186/s12934-014-0160-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Wang L., Zhenghong L., Lizelle P., Mattheos A.G., Zhang H. De novo biosynthesis of complex natural product sakuranetin using modular co-culture engineering. Appl Microbiol Biotechnol. 2020;104(11):4849–4861. doi: 10.1007/s00253-020-10576-1. [DOI] [PubMed] [Google Scholar]
- 190.Yuan S.-F., Xiunan Y., Trevor G.J., Hal S.A. De novo resveratrol production through modular engineering of an Escherichia coli–Saccharomyces cerevisiae co-culture. Microb Cell Factories. 2020;19(1):143. doi: 10.1186/s12934-020-01401-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Li T., Wei Z., Huiping B., Zhuang Y., Zhang T., Liu T. Production of caffeoylmalic acid from glucose in engineered Escherichia coli. Biotechnol Lett. 2018;40(7):1057–1065. doi: 10.1007/s10529-018-2580-x. [DOI] [PubMed] [Google Scholar]
- 192.Soma Y., Fujiwara Y., Nakagawa T., Tsuruno K., Hanai T. Reconstruction of a metabolic regulatory network in Escherichia coli for purposeful switching from cell growth mode to production mode in direct GABA fermentation from glucose. Metab Eng. 2017;43:54–63. doi: 10.1016/j.ymben.2017.08.002. [DOI] [PubMed] [Google Scholar]
- 193.Lv X.-A., Ying-Yan J., Yu-Dong L., Zhang H., Xin-Le L. Genome shuffling of Streptomyces viridochromogenes for improved production of avilamycin. Appl Microbiol Biotechnol. 2013;97(2):641–648. doi: 10.1007/s00253-012-4322-7. [DOI] [PubMed] [Google Scholar]
- 194.Taloria D., Sujata S.S., Pututunda D.C. Increase in bioethanol production by random UV mutagenesis of S.cerevisiae and by addition of zinc ions in the alcohol production media. APCBEE Procedia. 2012;2:43–49. doi: 10.1016/j.apcbee.2012.06.009. [DOI] [Google Scholar]
- 195.Jin J., Wang Y., Yao M., Gu X., et al. Astaxanthin overproduction in yeast by strain engineering and new gene target uncovering. Biotechnol Biofuels. 2018;11(1):230. doi: 10.1186/s13068-018-1227-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Niu F.-X., He Xin, Ya-Qin W., Jian-Zhong L. Enhancing production of pinene in Escherichia coli by using a combination of tolerance, evolution, and modular Co-culture engineering. Front Microbiol. 2018;9(1623) doi: 10.3389/fmicb.2018.01623. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.