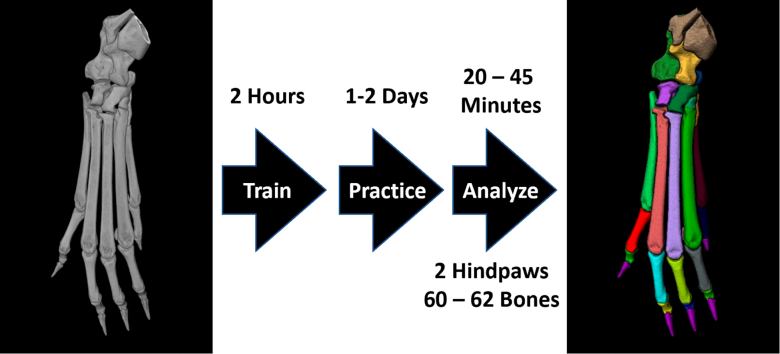
Abstract
Introduction
Micro-computed tomography (μCT) is a valuable imaging modality for longitudinal quantification of bone volumes to identify disease or treatment effects for a broad range of conditions that affect bone health. Complex structures, such as the hindpaw with up to 31 distinct bones in mice, have considerable analytic potential, but quantification is often limited to a single bone volume metric due to the intensive effort of manual segmentation. Herein, we introduce a high-throughput, user-friendly, and semi-automated method for segmentation of murine hindpaw μCT datasets.
Methods
In vivo μCT was performed on male (n = 4; 2–8-months) and female (n = 4; 2–5-months) C57BL/6 mice longitudinally each month. Additional 9.5-month-old male C57BL/6 hindpaws (n = 6 hindpaws) were imaged by ex vivo μCT to investigate the effects of resolution and integration time on analysis outcomes. The DICOMs were exported to Amira software for the watershed-based segmentation, and watershed markers were generated automatically at approximately 80% accuracy before user correction. The semi-automated segmentation method utilizes the original data, binary mask, and bone-specific markers that expand to the full volume of the bone using watershed algorithms.
Results
Compared to the conventional manual segmentation using Scanco software, the semi-automated approach produced similar raw bone volumes. The semi-automated segmentation also demonstrated a significant reduction in segmentation time for both experienced and novice users compared to standard manual segmentation. ICCs between experienced and novice users were >0.9 (excellent reliability) for all but 4 bones.
Discussion
The described semi-automated segmentation approach provides remarkable reliability and throughput advantages. Adoption of the semi-automated segmentation approach will provide standardization and reliability of bone volume measures across experienced and novice users and between institutions. The application of this model provides a considerable strategic advantage to accelerate various research opportunities in pre-clinical bone and joint analysis towards clinical translation.
Abbreviations: μCT, Micro-Computed Tomography; TNF-Tg, Tumor Necrosis Factor Transgenic; MTP, Metatarsophalangeal; MRI, Magnetic Resonance Imaging; HU, Hounsfield Unit; Gy, Gray
Keywords: Micro-CT, Image analysis, Automation, Segmentation, Classification, Foot & ankle
Graphical abstract
Highlights
-
•
High-throughput, semi-automated generation of bone seeds for watershed segmentation.
-
•
Segmentation method provided throughput benefits compared to conventional analysis.
-
•
Excellent inter-rater reliability and quick adoption by new users.
-
•
Segmentation results and accuracy are dependent on image resolution.
-
•
Future application to additional complex anatomical structures is warranted.
1. Introduction
Micro-computed tomography (μCT) is a valuable imaging modality for a variety of quantitative measures, such as evaluation of bone health in pre-clinical models (Stauber and Müller, 2008). Images can be acquired either longitudinally in vivo or ex vivo after tissue harvest, and various analytical processes have been developed to extract bone quality measures from the resulting datasets. Primarily, analytical approaches have focused on the evaluation of long bones to isolate the cortical and trabecular portions of the bones for detailed outcome measures. Various strategies have been developed to specifically evaluate trabecular and cortical components of bone, such as regions of interest placed within the trabecular bone, manual contouring of the trabecular and cortical boundary, or automated segmentation approaches (Bouxsein et al., 2010; Buie et al., 2007; Kohler et al., 2007; Newton et al., 2020). These well-described segmentation methods have allowed for establishment of various derived measures (i.e. trabecular number, cortical thickness, etc.), that together provide detailed insight on the health and quality of the bone (Bouxsein et al., 2010).
However, in complex structures such as the murine hindpaw, which contains 30 or 31 (as the fused navicular/lateral cuneiform (Richbourg et al., 2017) can also be variably fused with the intermediate cuneiform in C57BL/6 mice) distinct bones of various sizes and shapes, no strategies have been widely adopted to segment these individual bones for high-throughput and reproducible analysis. The conventional approaches to μCT analysis in structures such as the hindpaw are solely reliant on manual contouring or density-based thresholding approaches (Proulx et al., 2007) that are prone to inaccuracies at bone edges (Iassonov et al., 2009; Diaz et al., 2021; Rathnayaka et al., 2011). Given the intensive segmentation efforts involved in further analysis, data analysis is typically limited to a single (Proulx et al., 2007), small subset, and/or focal regions of bones (Cambre et al., 2018). Since each individual bone in the hindpaw experiences unique in vivo forces, tendon strains, and combinations of articulating surfaces, limiting the analysis to single bones reduces the capacity to identify reliable bone-specific biomarkers to guide future pre-clinical and clinical investigation. In addition, reliance on these conventional manual segmentation methods conducted by different users with various segmentation strategies increases the probability of inter-user variability in outcome measures. Thus, the analytical strategies for complex structures with multiple small bones, such as the hindpaw, have enormous possibility for improvement.
For example, in our previous work investigating inflammatory arthritis in the tumor necrosis factor transgenic (TNF-Tg) mouse model (Keffer et al., 1991), we performed μCT analysis on the hindpaw as a biomarker of arthritis onset and progression (Proulx et al., 2007). However, this analysis was limited to volume measurements of the talus, which articulates with the tibia in the ankle, primarily because of the ease in manual segmentation. To properly evaluate the relationship between experimental interventions and the effect on arthritis, μCT analysis is essential due to its comprehensive ability to evaluate complete bone volumes and capacity for longitudinal in vivo measurements of bone erosions. The reliance on manual segmentation strategies thus forces us and others to limit our analysis to single bones, such as the talus, while certain interventions may exhibit bone-specific effects that have yet to be investigated. Thus, establishment of a high-throughput data analysis approach would provide considerable benefit to validate or discover additional biomarkers of bone health for various pathologic conditions.
Herein, we developed a high-throughput and user-friendly semi-automated segmentation workflow for murine hindpaw μCT datasets using commercially available algorithms in Amira software. As one of the major roadblocks to widespread adoption of automated segmentation approaches is the intensive computer-based knowledge necessary for application, we validated that the segmentation strategy could be quickly adopted with minimal training by novice users with no prior experience using the Amira software. The described segmentation method demonstrated significant benefits in throughput compared to conventional μCT analysis performed by manual contouring, and the approach also showed excellent internal consistency and inter-user reliability. Thus, we have established an automated approach to isolate individual small bones in complex structures, such as the hindpaw, for μCT analysis with the capacity for widespread adoption.
2. Materials and methods
2.1. Mouse models and micro-CT data collection
All animal experiments were approved by the University Committee for Animal Resources at the University of Rochester. In vivo μCT datasets of the hindpaws (2 hindpaws/dataset) were acquired longitudinally at monthly intervals in male (n = 4; 2–8 month-old) and female (n = 4; 2–5 month-old) C57BL/6 mice. The C57BL/6 mice used in this study were littermates to TNF-Tg mice (Douni et al., 1995-1996; Kontoyiannis et al., 1999) originally obtained from Dr. George Kollias and maintained at the University of Rochester. The longitudinal measures of the females were terminated earlier than the males to compare with the TNF-Tg littermates in a separate study, where TNF-Tg females exhibit accelerated mortality at <6 months of age (Bell et al., 2019).
For the μCT data collection, the mice were anesthetized with isoflurane and placed on their left side in a Derlin plastic and clear acrylic tube. Adhesive tape was used to bind the hindpaws together and foam was placed above and below the hindlimbs to secure them. Micro-CT datasets were acquired using a VivaCT 40 (Scanco Medical, Bassersdorf, Switzerland), and all in vivo scans were collected with the following imaging parameters: 55 kV, 145 μA, 300 ms integration time, 2048 × 2048 pixels, 1000 projections over 180°, resolution 17.5 μm isotropic voxels. Image collection for the hindpaws was completed in 30-45 min. A total of 44 in vivo datasets were generated. Two datasets were excluded from the study due to considerable animal movement during the scan, and one additional calcaneus bone was excluded due to incorrect localization of image acquisition. In total, 42 datasets (84 hindpaws) were analyzed.
2.2. Bone names and abbreviations
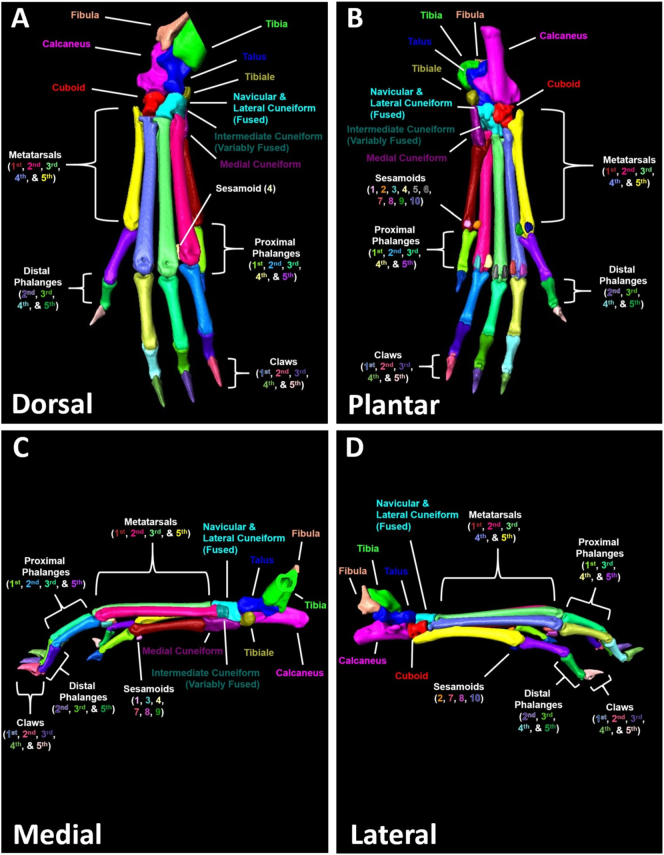
The bone nomenclature was primarily derived from Bab et al. (2007), and a fully labeled bone atlas annotating each of the individual bones in the hindpaw is provided in Figure 1. In the naming process, the bones were named according to shorthand bone codes followed by L (left) or R (right) for the corresponding hindpaws (Supplementary Table 1). For the mouse hindpaw, we confirmed previous studies where the navicular and lateral cuneiform were found to be ubiquitously fused in C57BL/6 mice (Richbourg et al., 2017). However, we also noted that the intermediate cuneiform is variably fused with these bones as well, which was consistent between animals and even within a single animal where the left ankle may be unfused, while the right ankle is fused, for example. For the digits, 1–5 represent the digit number with 1 being the most medial digit and 5 being the most lateral digit. The metatarsals, proximal phalanges, and distal phalanges are numbered according to the digits. Note, the first digit does not have a distal phalange. A small region of bone near the articulation of the distal phalanges and claw on the plantar surface variably segments as a separate material, and in these situations is added to the distal phalange. However, since this bony projection of the distal phalange near the claw articulation is spatially distinct from the majority of the distal phalange bone (hypodense space between objects), this was not considered a segmentation error. There are 2 sesamoid bones per digit (2 sesamoids × 5 digits = 10 sesamoids per hindpaw) located on the plantar surface near the articulation of the metatarsal and proximal phalange (MTP joint). On each digit, the odd numbered sesamoid is medial, while the even numbered sesamoid is lateral as the sesamoids are numbered from medial to lateral. In total, there are 30–31 bones per hindpaw (60–62 bones per dataset) depending on the variable fusion of navicular/lateral cuneiform with the intermediate cuneiform. There are also a few bones present in each dataset that were segmented, but not evaluated for analysis: 1) Tibia and Fibula (Abbreviation: FIBTIB; Segmented together as a single material, present in variable length depending on scan), 2) Extra Sesamoid (Abbreviation: FLOAT; Plantar surface near proximal end of metatarsal 5, appears “floating” below ankle), and 3) Claws (Abbreviation: CLAW; Segmented together as a single material, 1 Claw per digit). The shorthand bone codes are defined in Supplementary Table 1, and in Supplementary Video 4 – Mouse Ankle Anatomy.
Fig. 1.
Visual representation of the individual bones in murine hindpaws. A fully segmented hindpaw is shown with each bone identified by color and annotated in the dorsal (A), plantar (B), medial (C), and lateral (D) viewpoints. Note that this particular hindpaw has separated navicular/lateral cuneiform and intermediate cuneiform bones that are variably fused in the hindpaws of C57BL/6 mice. A detailed description of the hindpaw anatomy is provided in Supplementary Video 4, while shorthand bone codes are defined in Supplementary Table 1.
2.3. Segmentation method
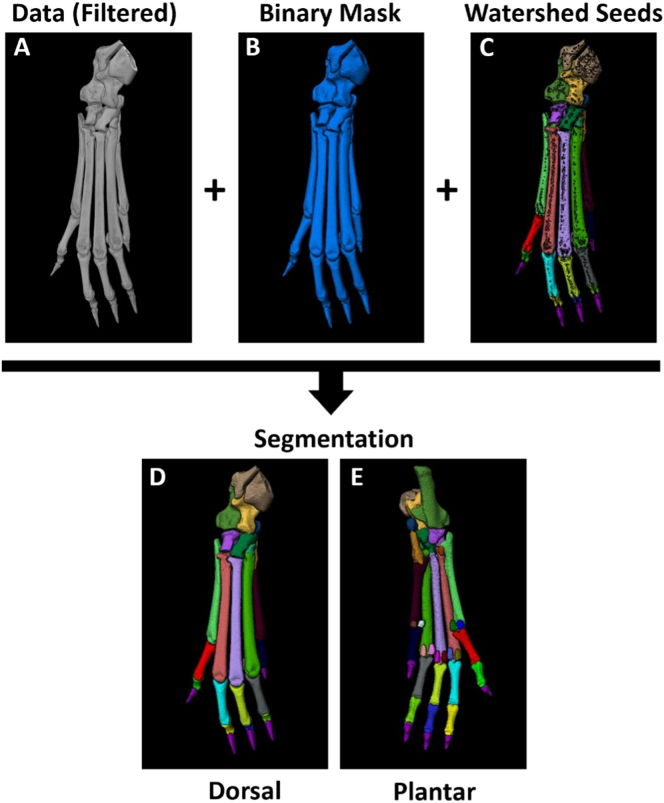
To generate a minimally supervised, reliable, and efficient segmentation method for analysis of each individual bone in murine hindpaw μCT datasets, we utilized established watershed-based algorithms available through the Amira computer software. Well-defined bone borders are essential for accurate bone separation, and thus a three-dimensional median filter was applied to the original dataset to denoise the image (Fig. 2A). A set threshold (>2500 Hounsfield Units (HU)) was then used to define bone relative to the surrounding air and soft tissue, and a top-hat representation of the image (>750 HU, defining steep valleys or large changes in signal intensity between adjacent voxels) was subtracted from the thresholded label field. The subtraction of the top-hat representation removes large pores in the bone and generates a binary mask of the bones (Fig. 2B). In all, placement of the watershed seeds, which are eroded versions of each individual bone identified as unique objects, is the most essential aspect of the segmentation process, and a variety of steps were used to create these watershed seeds to define and separate each distinct bone (Fig. 2C). Together, the original data, binary mask, and watershed seed data objects expand the bone-specific markers to the edges of each individual bone for a complete segmentation of the hindpaw μCT dataset (Figs. 2D,E).
Fig. 2.
Watershed-based semi-automated segmentation of murine hindpaw micro-CT datasets. 3D rendering of the μCT data was performed, and representative images of the hindpaw bones are shown to demonstrate the semi-automated segmentation method. This method utilizes the original μCT data after application of a three-dimensional median filter for edge detection (A), a binary mask with a set threshold of 2500 HU (B), and bone-specific watershed seeds (eroded versions of the bone-specific labels) (C). Together, these inputs generate bone-specific segmentations that expand to the full volume of the bone using the “Marker Based Watershed Inside Mask” algorithms available in Amira software. The complete segmentations of the 31 possible bones present in the murine hindpaw are shown from the dorsal (D) and plantar (E) viewpoints, and the volumes of each individual bone were subsequently extracted for downstream analysis.
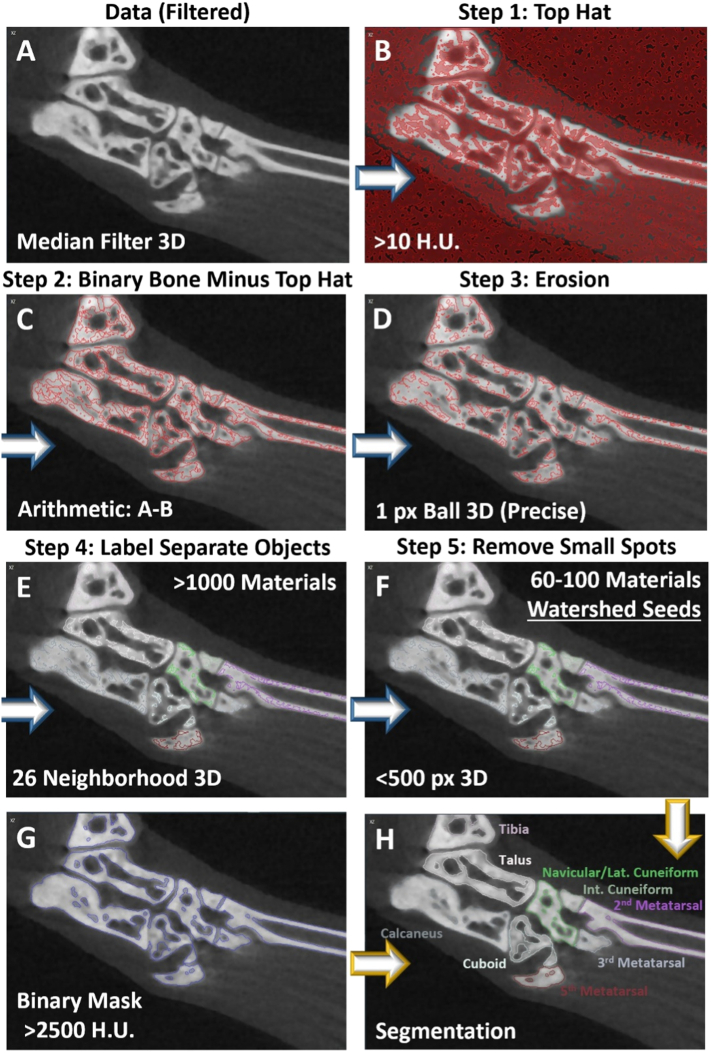
Similar to the creation of the binary mask, the automated workflow for generation of the watershed seeds begins with the filtered dataset, application of a set threshold (>2500 HU), and subtraction of a top-hat representation of the image. However, in the case of the watershed seeds, the top-hat was set at a much lower threshold (>10 HU) to also capture shallow valleys with limited change in signal intensity between adjacent voxels. As a result, the only voxels that remain segmented are those regions of bone with relatively constant signal intensity, which primarily isolates the central portions of the cortices for the individual bones (Figs. 3A-C). To further specify the central portions of each bone, the segmentation is then eroded by 1 additional pixel (Ball, 3D) to create small, connected seeds scattered throughout the bones (Fig. 3D, red contours). The various connected seeds are then labeled as separate objects, while objects remain connected if they share at least one common vertex (Fig. 3E; Connectivity: 26 Neighborhood in 3D). At this stage, many small objects (<500 pixels) remain that only define minute regions of particular bones, and thus these small objects are removed as the final step in the automated generation of the watershed seeds (Fig. 3F). Thus, these inputs to create the watershed seeds, together with the filtered data (Fig. 3A) and the binary mask (Fig. 3G), generate the fully segmented dataset (Fig. 3H).
Fig. 3.
Automated workflow for generation of bone-specific watershed seeds. To create the bone-specific watershed seeds in an efficient and reproducible manner, an automated workflow was developed and packaged as a convenient “Recipe” where all steps are embedded within a single module in Amira. To visualize the changes at each step in the recipe, a 2D section near the tarsal region of a representative hindpaw is shown, while the modules function on the full 3D dataset. The recipe requires the input of the dataset after application of a three-dimensional median filter (A) and a binary representation of the bone (>2500 HU). A top-hat was then applied to identify local valleys in signal intensity, and a threshold >10 HU was set to define the valley depth (B). The top hat segmentation was then subtracted from the binary representation of the bone to define only high-intensity regions with limited signal change in adjacent voxels (C). The result was then eroded to further separate the intense regions of the individual bones (D), and the separate objects were then defined as individual materials with connected voxels sharing at least one common vertex (E). Small objects were then removed to isolate the larger connected materials defining the majority of the individual bones as the final step in the generation of the watershed seeds (F). An automated workflow was used to create the binary mask, which utilizes the same approach as shown in A-C, but with an increased threshold for valley depth at >750 HU in the top-hat step (G). The filtered dataset (A), watershed seeds (bone-specific, color-annotated contours) (F), and binary mask (blue contours) (G) were combined together in the “Marker Based Watershed Inside Mask” Amira module for the complete segmentation of the individual bones in the hindpaw (H), as described in Figure 2.
However, the automated portion of the watershed seed placement contains errors in most cases. On average for datasets collected in vivo (n = 84 hindpaws), 79.2% (bones segmented correctly/total bones) of the individual bones were segmented accurately using the automated workflow, while analysis of ex vivo datasets (n = 6 hindpaws) improved the accuracy to 91.1% on average. These findings suggest that slight movements in vivo may affect the segmentation accuracy. In fact, specific bones tended to be more prone to errors than others in analysis of in vivo datasets. While most bones were segmented accurately without user intervention, four bones (CUB, DP3, NAVLATINT (fused), and NAVLAT (separate)) demonstrated the highest error rates where these bones were accurately segmented in <50% of the hindpaws. The error rates for each of the individual bones are provided in Supplementary Table 2.
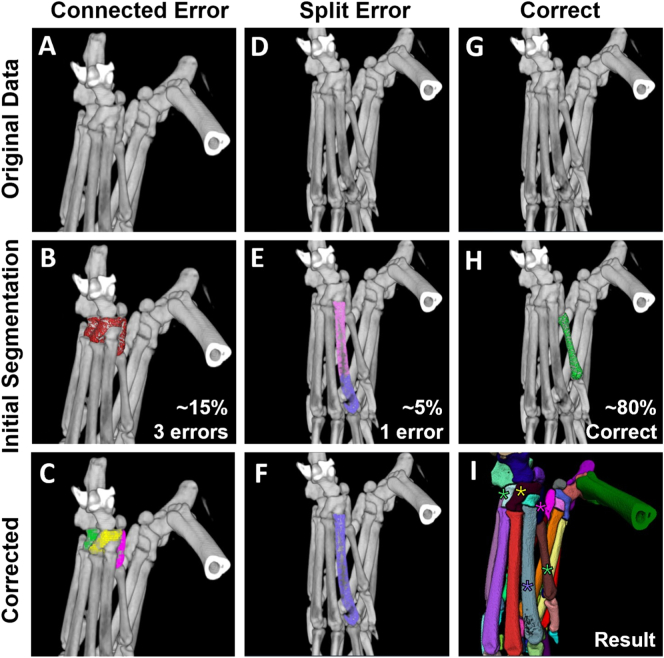
Despite the potential for error with the automated workflow, we developed methods to quickly fix the mistakes and generate accurate seed placements for the average of 20.8% (6 bones/hindpaw) of bones that exhibited errors in each dataset. The two primary types of errors are connected (~15% of bones; 2+ bones segmented as 1 material) or split (~5% of bones; 1 bone segmented as 2+ materials). Connected errors can be identified by selecting a given material, and the seeds expand across bone borders to cover multiple bones (Figs. 4A-C), while split errors are noted by a material that does not define the entire expanse of the bone (Figs. 4D-F). Together, the bones segmented accurately using the automated workflow (Figs. 4G,H) and the corrected bone segmentations generate the final watershed seeds that produce the complete segmentation (Fig. 4I).
Fig. 4.
Semi-automated segmentation produces connected or split errors prior to user correction. During the automated watershed seed development, approximately 15% of the seeds will be erroneously connected (A-C), 5% will be incorrectly split (D–F), and 80% will be correctly segmented without user intervention (G,H). After correction of the erroneous watershed seeds shown in B to C and E to F, the watershed seeds can then be used in the “Marker-Based Watershed Inside Mask” module for segmentation of the hindpaw, where the example bones are identified in the final segmentation by stars with corresponding colors (I). Note that in quantification of the error rate, all of the bones connected as in B are considered as individual errors (i.e. 3 bones connected means 3 errors), while the split bone as in E is considered a single error.
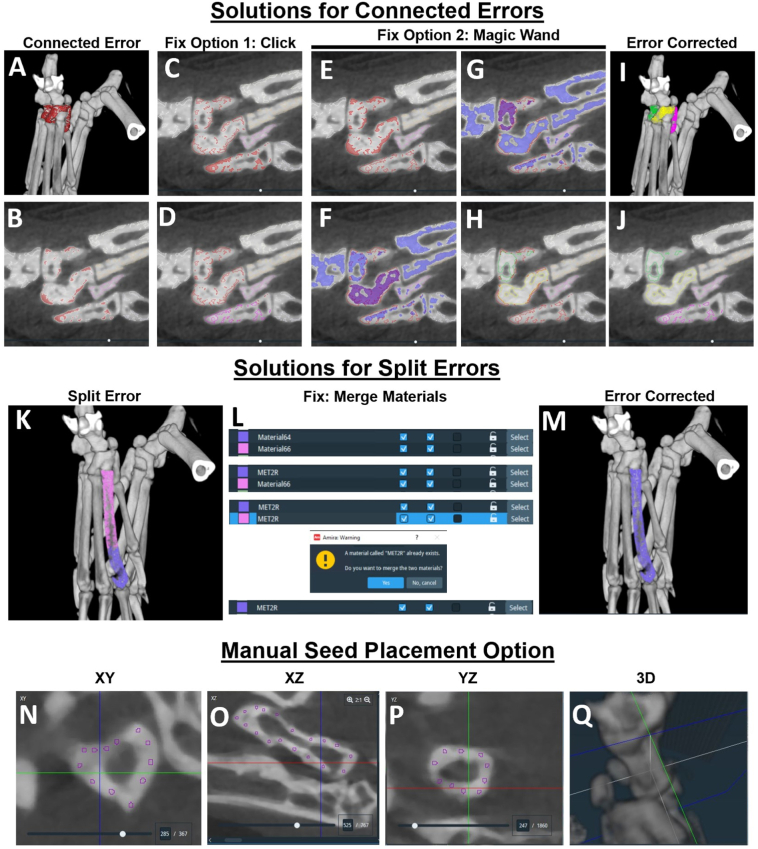
The connected errors (Figs. 5A,B) can be fixed in two different ways: 1) If the connected bones are only connected by a single vertex, the bones can be separated by selecting the individual seeds in 2D and generate new materials (Figs. 5C,D); or 2) New seeds are formed by the magic wand tool that selects connected voxels at a set threshold, and is able to similarly target the central regions of the bone with high signal intensity for separation from adjacent bones (Figs. 5E-H). Importantly, the final correction (Figs. 5I,J) requires the original erroneous and connected material to be deleted. On the other hand, split errors are simple to fix by naming the different split objects defining the particular bone as the same material name, which will then be automatically merged by the software to fix the error (Figs. 5K-M). As a last resort, the seeds can be placed manually using the brush tool by targeting the bone using 2D crosshairs and placing small seed points around the edges of the bone in each of the 3 planes of section with the bone target validated in 3D (Figs. 5N-Q). Once the segmentation process is complete, the user can extract each individual bone volume as a data table within Amira. Thus, we have developed various troubleshooting approaches to complete the final version of the watershed seeds for segmentation of the murine hindpaw μCT datasets using watershed algorithms in Amira. A detailed workflow for the watershed seed development, correction processes, and complete segmentation of the hindpaw datasets are provided in the Supplementary Methods and Supplementary Videos.
Fig. 5.
Methods for correcting errors in the automated watershed seed placement. To demonstrate the approach for correcting the connected and split errors, the datasets shown in Figure 4 are used as examples. When connected errors occur as with the CUB, NAVLAT, and MED shown in 3D (A; red selection from left to right) and 2D (B; red filled in contours from top to bottom), there are 2 primary methods to fix these mistakes. First, the edges of the predominant watershed seeds of a specific bone may already be correctly split, but labeled as the same material because of a connected vertex in 3D. By clicking inside the seeds within a particular bone (C, i.e. MED), the bone may segment independently from the connected bones and can be added as a new and separate material (note red seeds converted to pink seeds) (D). However, if clicking within the bone-specific seeds does not separate the bones (E; CUB and NAVLAT remain connected with the red contours filled in), new seeds can be quickly generated using the magic wand tool. Starting with a mask of 4500 HU, click within the bone of interest and sequentially increase the mask by 250 HU until the bone separates (F; NAVLAT is selected as purple, while CUB is not selected as blue). The NAVLAT can then be added as a new and separate material then the process repeated for CUB (G) to create the new watershed seeds (H). Importantly, the material that represented the original, erroneously connected watershed seeds must be deleted to finalize the corrected watershed seeds as shown in 3D (I), and 2D (J). On the other hand, split errors, as in the example of the 2nd metatarsal (K; MET2), can be fixed by naming the two components of the bone as MET2, which will then be merged (L) to correct the error (M). As a last resort, the watershed seeds can be placed manually by visualizing the 2D crosshairs in the center of the bone to be segmented, and small dots placed around the bone using the brush tool in the XY (N), XZ (O), and YZ (P) planes. The crosshairs can also be visualized in 3D to confirm the identification of the bone being segmented (Q). The manual seed placement is also helpful in situations where the automated watershed seeds do not adequately segment the articulating surfaces of two bones and generate an unclearly segmented border. A detailed description of these correction processes is also provided in the Supplementary Methods and Supplementary Video 3.
2.4. Reproducibility and efficiency measures
To correlate the semi-automated segmentation approach with conventional μCT analysis where individual bones are manually contoured, we evaluated the bone volume measurements from a subset of bones (n = 60 bones) across 6 different μCT datasets. An experienced Scanco user manually contoured these 60 bones, which represented the expected distribution of segmentation accuracy in the semi-automated workflow. The volume measurements derived from manual segmentation were then compared to these outcomes from the Amira software by linear regression and Bland-Altman agreement analyses. The experienced Scanco user also provided the segmentation time for each dataset where the manual contouring time for each bone was quantified. The total time necessary to segment an entire dataset was extrapolated by multiplying the segmentation time for a particular bone and the total number of bones in the associated bone compartment (i.e. tarsals, metatarsals, proximal phalanges, distal phalanges, or sesamoids).
To evaluate the feasibility for a novice user (with no prior Amira experience) to adopt the semi-automated segmentation method, 3 previously untrained graduate students were tasked with analyzing the same 6 datasets. In order to train the users on the segmentation method, 5 training videos were developed (<2-h total, Supplementary Videos 1–5). The novice users then analyzed the 6 datasets once for practice, and then analyzed the 6 datasets again to evaluate the segmentation time for each dataset. Analysis time for each dataset was defined from the application of the median filter to the final segmentation after double checking and fixing any errors.
2.5. Training materials
Novice users were provided 5 training videos (Supplementary Videos 1–5, <2-hours total) and 6 practice datasets from the in vivo scans. The training videos cover the following topics: 1) Introduction to Amira, 2) Segmentation Process, 3) Correction Processes, 4) Mouse Ankle Anatomy, and 5) Example Segmentation. The practice datasets represent hindpaws from 5-month-old mice (n = 4 male, n = 2 female), and were selected to demonstrate the range of potential segmentation accuracy before user correction. Thus, the training datasets include the following distribution of accuracy determined by percent of bones correct without user intervention: >85% (n = 2), 85–75% (n = 2), and < 75% (n = 2), with the average of 80.3% closely mirroring the accuracy across all datasets at 79.2%. The training videos (Supplementary Videos 1–5), Amira recipes, and an excel sheet indicating the expected volume measurements for each bone are provided in the Supplementary Materials, while the training datasets can be found at the following reference (Kenney & Awad, 2021), doi: 10.17632/7sm9wznp6d.1). Together, all of these materials represent the same training materials provided to the novice users in this study.
2.6. Image acquisition parameters
To evaluate the effects of changing image resolution and integration time on the segmentation method, hindpaws were also harvested from 9.5-month-old C57BL/6 males (n = 3 mice or 6 hindpaws). The hindpaws were fixed in 10% neutral buffered formalin, rocking for 24-h at room temperature. Sequential ex vivo μCT scans were performed on the hindpaws to directly compare changes in the volume measurements and segmentation accuracy of 9 different image resolution and integration time combinations. These combinations consisted of the following imaging parameters: High (10.5 μm isotropic voxel size), medium (17.5 μm), and low (35.0 μm) image resolutions; high (300 ms), medium (200 ms), and low (120 ms) integration times. The standard parameters used for the development of this segmentation method were medium resolution and high integration time (17.5 μm; 300 ms).
2.7. Computer and software specifications
The segmentation workflow described in this study was performed on a computer running Windows 10 (Microsoft Corporation, Redmond, WA; operating system build: 19042.1288) with the following specifications: Processor: Intel® Core™ i7-7700K CPU at 4.20 GHz (Intel, Santa Clara, CA); RAM: 52.0 GB; System type: 64-bit operating system; Graphics card: NVIDIA Quadro M4000 (NVIDIA, Santa Clara, CA). Amira software (v2020.2; ThermoFisher Scientific, FEI, Hillsboro, OR, USA) was used for the segmentation, with the following extensions: XImagePAQ (Modules: Interactive Thresholding, Interactive Top-Hat, Erosion, Labeling, Remove Small Spots, Marker Based Watershed Inside Mask, Recipe Player) and XRecipe (Module: Recipe Player).
2.8. Statistics
Linear regression, Bland-Altman agreement, and ANOVA statistical analyses were performed in GraphPad Prism (v9.1.0, San Diego, CA, USA). The intraclass correlation coefficients (ICCs) were calculated in SPSS (v.28.0.0.0; IBM, Armok, NY, USA) using a two-way mixed model and consistency type analysis.
3. Results
3.1. The semi-automated segmentation method provides reproducibility and efficiency benefits compared to conventional micro-CT analysis
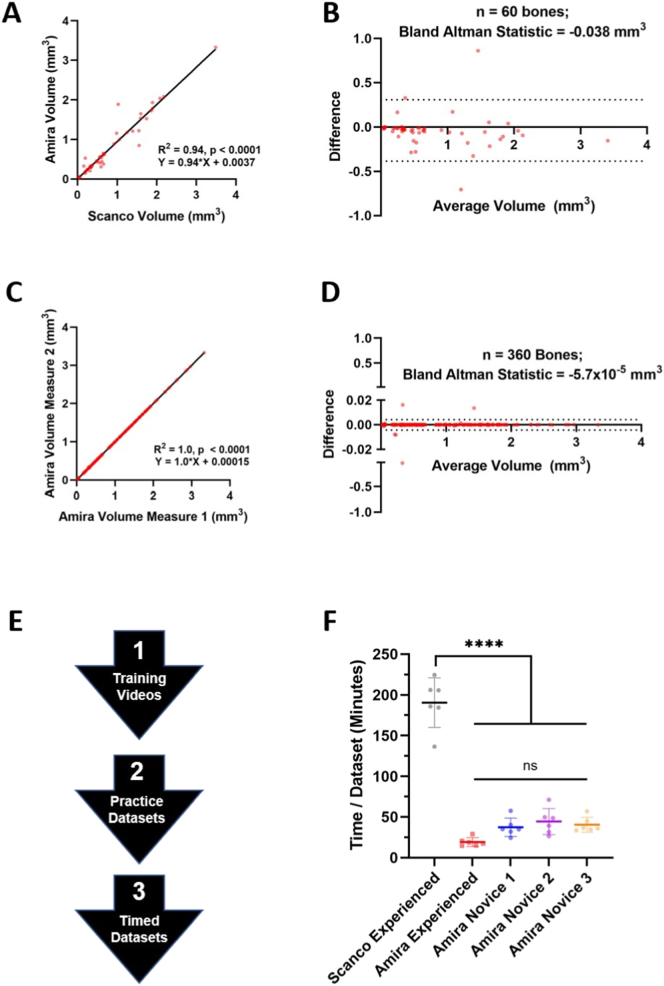
In order for the described semi-automated segmentation method to be utilized by others, we first had to confirm that the analysis: 1) produced similar results to conventional μCT analysis, 2) demonstrated strong intra-user consistency, and 3) exhibited excellent inter-user reliability, especially when the analysis was performed by novice users. Linear regression (R2 = 0.94, p < 0.0001) and Bland-Altman agreement (−0.038mm3 average difference) analyses demonstrated a strong correlation between the semi-automated Amira segmentation and the conventional Scanco manual contouring (Figs. 6A,B). Moreover, when an experienced Amira user analyzed the same 6 datasets twice for segmentation (360 bones/replicate), linear regression (R2 = 1.0, p < 0.0001) and Bland-Altman agreement (−5.7 × 10−5 mm3 average difference) analyses also demonstrated strong internal consistency in the performance of the semi-automated segmentation approach (Fig. 6C,D).
Fig. 6.
The semi-automated segmentation method provides reproducibility and efficiency benefits compared to conventional micro-CT analysis. To validate the outcomes from the semi-automated segmentation approach, we compared these Amira-derived volumes to those of conventional manual contouring of bones using the Scanco system based on segmentation of 60 total bones from 6 representative datasets collected in vivo. From these 6 datasets, 10 representative bones were selected that were evenly distributed across the bone compartments (i.e. tarsals, metatarsals, proximal phalanges, distal phalanges, and sesamoids) and approximated the error rate for the dataset. Linear regression (A; R2 = 0.94, p < 0.0001) and Bland-Altman agreement (B; −0.038 mm3 avg. difference, dashed lines represent 95% confidence intervals) analyses were performed between the volume measurements derived from an experienced Amira (semi-automated) and Scanco (manual, conventional) users. The experienced Amira user then analyzed the same 6 datasets a second time where linear regression (C; R2 = 1.0, p < 0.0001) and Bland-Altman agreement (D; −5.7 × 10−5 mm3 avg. difference) analyses between the replicates were performed on all bones (n = 360), which demonstrated excellent internal consistency in the volume measurements. Note the increased range of the 95% confidence intervals when the Amira segmentation is compared to Scanco (B) relative to the Amira replicate (D), suggesting the variability shown in B derives from the manual contouring process. To evaluate adoption of this method by new users of the Amira software, novice users were recruited and trained to analyze the same 6 datasets (E). Both experienced and novice Amira users showed a significant reduction in segmentation time compared to the conventional manual contouring in Scanco where data is presented as mean ± SEM (F). Statistics: Linear regression (A,C), Bland-Altman analysis (B,D), and One-way ANOVA with Tukey's multiple comparisons, **** p < 0.0001 (F).
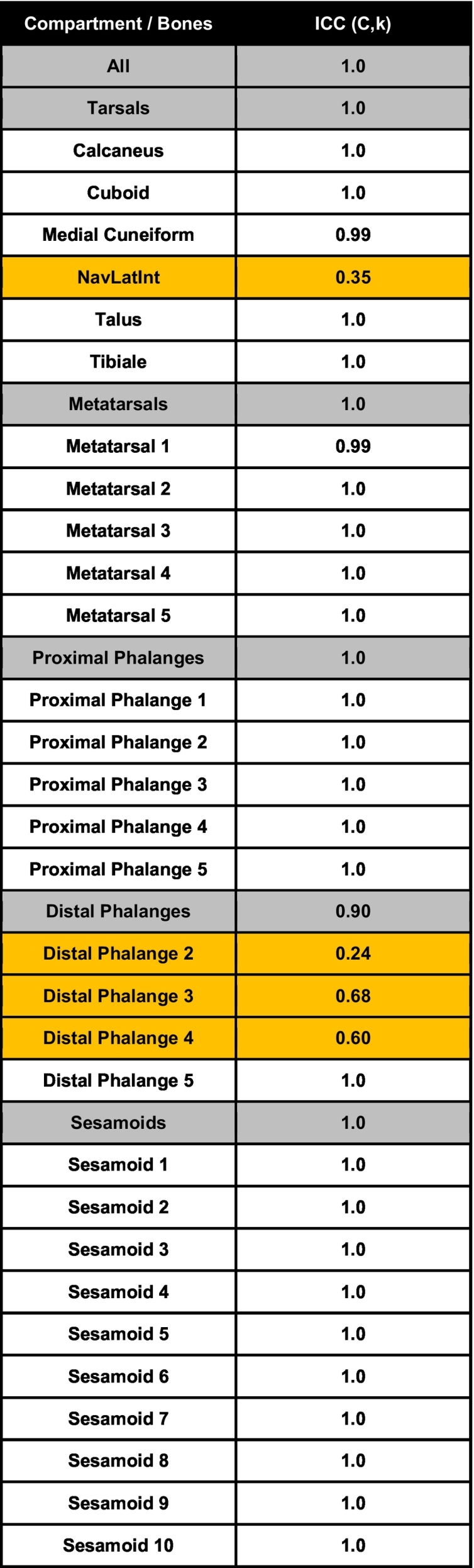
Following training in the semi-automated Amira method (Fig. 6E), segmentation time was quantified by both experienced and novice Amira users for comparison with the conventional manual segmentation in Scanco. Remarkably, both experienced (19.3 ± 5.34 min/dataset) and novice (User 1 = 37.4 ± 11.2, User 2 = 44.5 ± 15.9, User 3 = 40.5 ± 9.06 min/dataset) Amira users showed significantly reduced segmentation time compared to the conventional Scanco analysis (190.6 ± 30.4 min/dataset). In addition, after relatively limited training and practice, all 3 novice Amira users showed no significant change in segmentation time compared to the experienced Amira user (Fig. 6F). The volume measurements between all 4 users (1 experienced and 3 novice) also demonstrated excellent inter-user reliability through ICC quantification (ICC > 0.9). The DP2–4 bones showed variation between users due to unidentified connected errors most common between the distal phalange and claw. In addition, the NAVLATINT exhibited poor inter-user reliability, which is intuitive given the variable fusion of these bones in C57BL/6 hindpaws (Table 1). Along with the excellent inter-user reliability overall, each of the novice users also demonstrated strong internal consistency in the volume measurements from the datasets used for practice and timing analysis (Supplementary Fig. 1). Thus, the process from no Amira experience to data segmentation could be completed in 1–2 days of practice, where high-throughput and reliable data analysis can be performed thereafter.
Table 1.
Intraclass correlation coefficients of bone-specific volume measurements identify common segmentation errors. Four different users (1 experienced, 3 novice) performed the semi-automated segmentation method on the same datasets (n = 6 datasets = 12 hindpaws). The ICCs (C,k: Two-way mixed, average score) were calculated in SPSS for the volumes of all bones (n = 360 bones), each bone compartment (i.e. tarsals, metatarsals, proximal phalanges, distal phalanges, and sesamoids; n = number of bones per compartment * 12 replicates of each bone), and each individual bone (n = 12 replicates of each bone). Together, 86.7% (26 total) of the individual bones showed excellent reliability between users (ICC ≥ 0.9), while the NAVLATINT and DP2–4 were bones with the greatest potential disagreement between users (orange rows). Note that the NAVLAT and INT were not included in the analysis, and when separated were added together to match the datasets with the bones fused as NAVLATINT.
3.2. Volume measurements and segmentation accuracy depend on image resolution with minimal effects of integration time
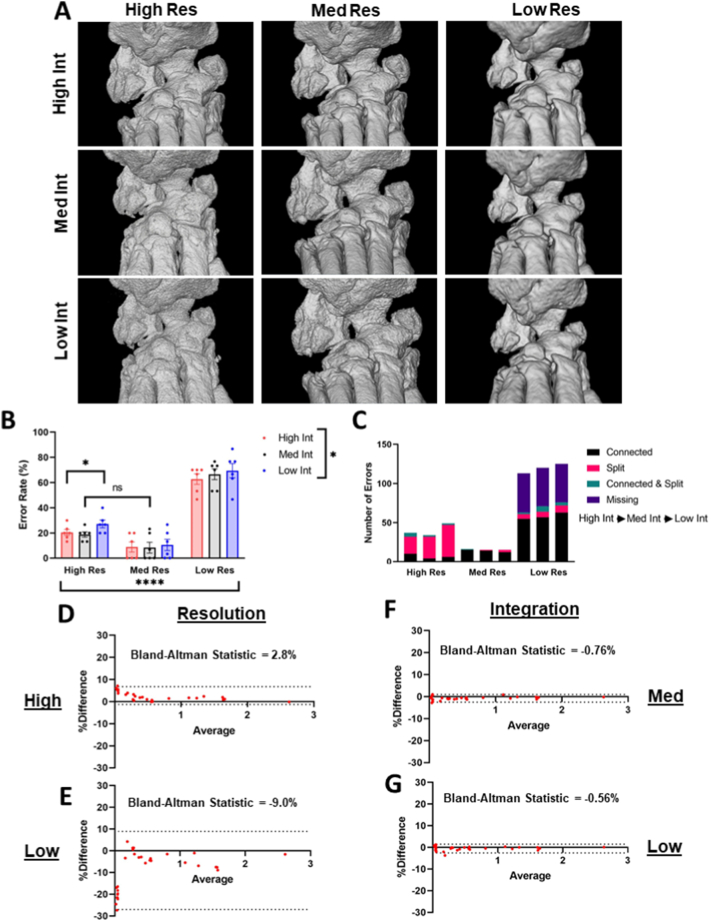
As the data analysis and validation of the semi-automated segmentation method was developed using set imaging parameters, we evaluated the performance of the segmentation approach when image resolution and integration time were modified. Representative images of the hindpaw tarsal region at 9 different combinations of image resolution (high/medium/low) and integration time (high/medium/low) are shown, where medium resolution and high integration time represent the parameters used in the development of the semi-automated segmentation workflow (Fig. 7A). Modulation of image resolution demonstrated a significant effect on segmentation accuracy accounting for 87.3% of the variance between conditions (p < 0.0001) with a significant difference in all comparisons of image resolution (p < 0.05) except one (high vs medium resolution at medium integration time). Thus, both increasing or decreasing image resolution significantly reduced the accuracy of the segmentation model, although the increase in error rate was more dramatic with reduced image resolution. Changes in integration time also demonstrated a significant interaction between conditions (p < 0.05), but only accounted for 0.74% of the variance with only one significant comparison (high vs low integration time at high resolution) (Fig. 7B). The increased errors with high resolution were associated with a higher proportion of split errors, while the increased errors with low resolution showed increased connected errors and bones completely missing from the final segmentation. On the other hand, changes in integration time demonstrated no appreciable change in the types of errors (Fig. 7C). For the volume measurements at a constant high integration time, increasing resolution showed a trend towards higher bone volumes (2.8% average difference) (Fig. 7D), and reducing resolution tended towards lower bone volumes (−9.0% average difference) (Fig. 7E) compared to the measurements of the standard medium resolution images. At constant medium resolution, medium integration time (−0.76% average difference) (Fig. 7F) and low integration time (−0.56% average difference) (Fig. 7G) exhibited limited change in bone volume measurements when compared to the standard high integration time. Overall, image resolution is essential for accuracy of the segmentation method and consistency in bone volume measurements, while modulation of integration time shows minimal effects on these outcome measures.
Fig. 7.
Volume measurements and segmentation accuracy depend on image resolution with minimal effects of integration time. As the semi-automated segmentation method was developed on datasets imaged using a single combination of medium resolution and high integration time (17.5 μm, 300 ms), we investigated the effects of modulating the resolution and integration time on the segmentation accuracy and volume measurements. Sequential μCT on ex vivo hindpaws (n = 6) was performed at 9 combinations (high/medium/low) of image resolution and integration time, as described in the Methods. Representative images are shown for all combinations of resolution and integration time in a 3 × 3 pattern (A). The error rates between all combinations were then evaluated with a significant interaction of resolution accounting for 87.3% of the variance between conditions (**** p < 0.0001). All comparisons between changing resolutions were significantly different (p < 0.05) except for one noted on the graph between high and medium resolution at medium integration time. Integration time also showed a significant interaction, but only accounted for 0.74% of the variance between conditions (* p < 0.05) driven by the single significant comparison noted between high and low integration time at high resolution. All other comparisons between integration times were not significantly different (B). The proportions in types of errors between the conditions were also compared, where high resolution datasets showed a propensity towards split errors (pink) and low resolution datasets showed increased connected (black) and missing (purple) errors where bones were completely unsegmented. Integration time showed minimal effect in types of errors (C). At a constant integration time (high as standard), high resolution datasets tended towards increased bone volumes (D), while low resolution images tended towards decreased bone volumes (E) compared to the medium resolution datasets (dashed lines represent 95% confidence intervals). At a constant image resolution (medium as standard), medium (F) and low (G) integration time showed limited difference in bone volume measurements when compared to datasets imaged at high integration times. Note the increased range of the 95% confidence intervals when image resolution is modified (D,E) compared to changes in the integration time (F,G). Data is presented as mean ± SEM (B). Statistics: 2-Way ANOVA with Tukey's multiple comparisons (B) and Bland-Altman analysis (D-G).
4. Discussion
In this work, we have developed and validated a high-throughput and user-friendly semi-automated segmentation method for each individual bone in the complex structure of the murine hindpaw with potential for widespread adoption in future μCT analysis. The novelty in the study is the establishment of an automated workflow that generates watershed seeds at an average of ~80% accuracy for in vivo and >90% accuracy for ex vivo hindpaw μCT datasets prior to minimal user supervision to quickly fix any errors using prescribed protocols. The minimally supervised generation of the watershed seeds is then applied together with the filtered data and a binary mask defining bone to expand the watershed seeds to the bone borders for accurate and reproducible segmentations using established watershed algorithms available commercially via Amira software.
Importantly, this novel segmentation approach dramatically reduces the potential for inter-user bias and inaccuracies inherent with the conventional analysis methods of manual or density-based segmentation. As manual segmentation places the responsibility on the user to accurately detect the edge across the bone volume, this process is both time-consuming and prone to user error (Iassonov et al., 2009; Diaz et al., 2021). On the other hand, the density-based approach relies on strict parameters for threshold selection to separate structures that may similarly introduce inaccuracies at the edges (Rathnayaka et al., 2011), and the stringent parameters make the adoption for comparable datasets difficult. Through the segmentation approach described in this work, a majority of the bones are segmented accurately without user intervention, which ensures agreement between users for these structures. Even when user corrections are required, the correction processes do not require a specific approach for watershed seed placement, but instead a variety of user-dependent methods can be employed to place the markers as the computerized watershed algorithms will automatically detect the bone edges regardless of minor differences between users. While a set threshold ought to be used for accurate comparisons within an experiment (Bouxsein et al., 2010), the semi-automated watershed segmentation approach also provides the flexibility to utilize any threshold selection on a wide range of structures with variable material densities as the set threshold is primarily used to constrain the segmentation processes to a binary representation of the data.
In addition, one of the major benefits of the semi-automated watershed segmentation approach is the remarkable increase in throughput compared to conventional analysis methods, even for novice users with no prior experience using the Amira software. We demonstrated that new users could adopt the method with excellent inter-user reliability and internal consistency after only 1–2 days of practice with a significant reduction in segmentation time per dataset compared to manual contouring. In fact, one of the primary benefits to this workflow in Amira is the potential for adoption without requiring the knowledge of any programming languages, which can be a barrier to many. However, we acknowledge that those with experience in alternative open-source software options, or with extensive knowledge in computer programming, may prefer to utilize this workflow on these platforms. For those users, Amira provides the capabilities of two-way exchange of data objects with open-source programs to allow image processing external to the Amira software. In addition, we have provided an extensive description of the segmentation approach in the Methods and Supplementary Materials, and we encourage others to replicate and adopt this workflow in their preferred software options.
The capacity for widespread adoption with this segmentation approach increases the potential to establish fully automated machine learning models that require no user input or time investment. In fact, recent advances and impressive utilization of machine learning models have demonstrated the utility of automated segmentations in analyzing specific organs (Zhou, 2020; Skourt et al., 2018) and regions of bone (Buie et al., 2007; Kohler et al., 2007; Newton et al., 2020; Hamwood et al., 2021). As the ability to train and validate machine learning models requires an abundance of previously segmented datasets, the high-throughput generation of these segmentations using the semi-automated watershed approach allows the development of these inputs to be efficient and practical. However, even if fully automated machine learning processes are eventually established for the segmentation of complex structures such as the hindpaw, the adoption of these methods may be limited to those individuals with specialized skills in computer programming, and ultimately reduce the widespread use of these tools. As demonstrated in this work, the described semi-automated watershed-based segmentation approach has the benefit of quick and easy adoption by new users without the need for extensive training, and thus these methods can be utilized widely and immediately.
While the segmentation method was developed and validated using hindpaws as an anatomical model, the segmentation approach is not specific to the murine hindpaw and has the capacity to be applied to other complex structures of the mouse or other species. In fact, the workflow is flexible by modifying various settings within the steps described in Figure 3 to adjust the process for different structures with the primary consideration being the size of the bones. The segmentation approach was established on bones of a particular size that reside within the hindpaw of a mouse, within the range of approximately 0.2 (sesamoids) – 3.5mm3 (calcaneus). While the current settings may be applied successfully to other structures without need for modification, the primary steps to adjust if needed are the top-hat threshold (Fig. 3B) and pixel size for erosion (Fig. 3D) in the generation of the watershed seeds. Another consideration is the change in material density of bones or other structures that would require adjusting the threshold selection. In fact, the application of this workflow has been successfully tested on murine forepaws (unpublished data), and further application of this segmentation method on forepaw datasets is an active area of investigation.
To improve upon this work, users may develop alternative creative solutions to improve the accuracy and throughput of the described segmentation method for complex anatomical structures, such as the mouse hindpaw. For instance, we considered another approach for automated segmentation of the mouse hindpaws by exploiting the benefits of longitudinal scans for 3D registration and bone mapping from related datasets. Similarly, certain datasets could serve as anatomical models for overlay on experimental images to perform automated segmentation, similar to previous studies (Newton et al., 2020). However, the datasets used in this study proved to be too variable in paw placement where bones associated with highly mobile joints, such as the digits, demonstrated dramatic changes in location across timepoints and datasets. As a result, 3D registration models were unable to colocalize each bone in 3D space. For those only interested in the tarsal bones of the ankle, exclusion of the digits, tibia, and fibula from the datasets may allow for a reliable 3D registration process given the more rigid articulations between the tarsal bones. Future studies may also consider alternative approaches to securing the hindpaws for in vivo datasets to retain the foot and ankle in a reproducible alignment.
Moreover, in future studies the described semi-automated segmentation approach ought to be tested in hindpaws or other complex structures with notable pathology, such as bone erosions during arthritis. An important application of this work is the potential to discover novel bone-specific biomarkers through extensive analysis of μCT datasets that was not previously possible. Through the identification of these biomarkers, in silico experimentation with finite element modeling could be utilized to predict the implications of bone-specific injury and indicate potential bone stabilization strategies for therapeutic intervention. These types of experiments could evolve into investigation of gait analysis for animals with arthritis or weight-bearing conditions with bone-specific pathology for association with changes in ambulation. Investigation into bone-specific biomarkers in various conditions may also catalyze a focused analysis of treatment, time, and sex dependent effects to elucidate the related biomechanical mechanisms. While such findings would provide considerable pre-clinical benefit for future experimentation, there is also an important capacity for clinical translation. Identification of bone-specific pathology that can be visualized and monitored longitudinally may serve as valuable screening procedures or evaluation of treatment effectiveness in the clinic. Thus, adoption of this high-throughput segmentation approach provides many further possibilities for pre-clinical and clinical studies on bone homeostasis and pathology.
Although the segmentation approach demonstrated excellent inter-rater reliability and accuracy for most bones analyzed, a select few bones were more prone to error. Namely, the NAVLATINT and DP2–4 were the only bones with ICCs <0.9 (excellent reliability), and these bones corresponded to bones with the highest error rates (>50% of datasets with error across 84 hindpaws). As such, while the protocols for fixing errors are effective for some bones (i.e. CUB with >50% error rate, but ICC > 0.9), other bones may be more problematic and demonstrate a limitation of the segmentation method.
We further investigated the imaging parameters for the acquisition of the μCT datasets to evaluate the role of image resolution and integration time on segmentation accuracy. While image resolution and integration time should typically increase or decrease together, we assessed theoretical combinations of these parameters at high, medium, or low values. Our findings indicate that regardless of image resolution, integration time has minimal effect on segmentation quality. However, the accuracy of the segmentation method is dependent on image resolution at nearly all integration times. From the image resolutions we tested, we found that a 17.5 μm isotropic voxel size was optimal for segmentation accuracy. Reducing the image resolution (increased voxel size) showed a significant reduction in segmentation quality with more connected errors and unsegmented bones, while increasing the image resolution (decreased voxel size) was similarly detrimental to the segmentation workflow with more split errors. Thus, future investigations using this workflow should consider image resolution as an integral aspect to the success of the described method.
Given the dependence on the segmentation workflow on image resolution and the benefit of longitudinal scans for evaluation of changes in bone volume, radiation dose to the animal ought to be considered for animal health and effects on outcome measures. As noted, since increasing image resolution typically coincides with increased integration times, modulation of image resolution will exhibit non-linear changes in radiation dose to the animal. A previous study measured the proximal tibia of rats by μCT for 8-consecutive-weeks using higher resolution, integration time, and total scan time than used in our work. This study measured local CT dose, and concluded that the radiation dose was between 441 and 939 mGray (Gy) where 441 mGy was measured in the center of a cylinder phantom, 939 mGy was measured in the air, and the penetrance to the bone was considered between these values (Brouwers et al., 2007). Thus, the radiation dose in our study is perceived to be within or below this range for each scan at monthly intervals. In another well-controlled study where one limb of a mouse was imaged by μCT weekly and the contralateral limb served as a non-imaged control, the radiation-exposed limb was found to have significantly reduced bone volumes (Klinck et al., 2008). Thus, the radiation exposure in longitudinal studies must be accounted for when interpreting the results of bone volume measures, especially when comparing between time points. Beyond the local environment of the bone being imaged, the potential harmful effects to the whole animal must also be carefully considered. In the VivaCT40 system (Scanco Medical) used for our study, the beam has been precisely collimated to minimize exposure to the animal beyond the imaging area. Shielding has also been strategically placed over potential areas of radiation leaks to avoid stray radiation exposure to the animal. Thus, similar precautions are encouraged to avoid or limit the potential detrimental effects of radiation exposure to animals involved in future studies.
Beyond bone, application of similar segmentation strategies has the potential to generate detailed structural segmentations of specific organs, or detailed structures within these tissues. For example, multiple automated or semi-automated approaches have been utilized to accurately identify the edges defining the aerated portions of lung by CT (Mansoor et al., 2015; Bell et al., 2018; Walsh et al., 2021), or sinus regions within the skull by magnetic resonance imaging (MRI) (Andersen et al., 2018). The common features of these structures include the necessity for accurate edge detection between air and adjacent tissue, while complicated by the similarity in density with the surrounding air. Although a binary mask was utilized for segmentation of bone in this study to avoid interaction between the exterior air and inner marrow of each small bone, the watershed algorithms do not require the use of a set threshold, and actually benefit from changes in material density for edge detection. Thus, development of strategies to place watershed seeds within interior aerated structures, surrounding tissue, and the exterior may offer the potential to generate accurate segmentations in a user-friendly manner. This approach is also not limited to CT analysis and could also be utilized for other imaging techniques, such as MRI, where changes in signal intensity are used to define particular structures. Altogether, the hindpaw segmentation approach described in this work has considerable potential to benefit multiple aspects of image analysis for μCT and other imaging modalities in various pre-clinical and clinical areas of investigation.
5. Conclusion
In this work, we generated a novel high-throughput semi-automated segmentation method for volume measurements of individual bones in murine hindpaws imaged by μCT using a watershed-based approach in Amira software. The segmentation strategy was shown to be user-friendly with quick adoption by new users and provided excellent intra- and inter-user reliability. Compared to conventional μCT analysis approaches, the described segmentation method also demonstrated remarkable throughput benefits for both experienced and novice users. In conclusion, this segmentation strategy has the potential for widespread adoption and the capacity to be applied towards the segmentation of various additional complex structures in future studies.
The following are the supplementary data related to this article.
Training Video 1 - Introduction to Amira.
Training Video 2 - Segmentation Process.
Training Video 3 - Correction Processes.
Training Video 4 - Mouse Ankle Anatomy.
Training Video 5 - Example Segmentation.
Supplementary Figures
Supplementary Methods
Mask Generation Recipe
Markers Generation Recipe
Paw Analysis Test
Author contributions
HMK conceptualized and designed the segmentation method with input from RWW, EMS, and HAA. YP, KLC, RA, and LS assisted in the validation measures for the segmentation method. All authors contributed to the acquisition, analysis, and interpretation of the data along with drafting and revising the work.
Funding
This work was supported by the National Institutes of Health: T32GM007356, T32AR076950, R01AR056702, and P30AR069655.
Data statement
As mentioned in the Materials and Methods section, training content is available in the Supplementary Materials, and the training datasets are accessible at the following reference (Kenney & Awad, 2021), doi: 10.17632/7sm9wznp6d.1). Any additional data can be made available upon reasonable request.
CRediT authorship contribution statement
H. Mark Kenney: Conceptualization, Methodology, Writing – original draft. Yue Peng: Validation, Writing – review & editing. Kiana L. Chen: Validation, Writing – review & editing. Raquel Ajalik: Validation, Writing – review & editing. Lindsay Schnur: Validation, Writing – review & editing. Ronald W. Wood: Supervision, Writing – review & editing. Edward M. Schwarz: Supervision, Writing – review & editing. Hani A. Awad: Supervision, Writing – review & editing.
Declaration of competing interest
The authors have no conflict of interest to disclose.
Acknowledgements
We would like to thank the staff and faculty of the Biomechanics and Multimodal Tissue Imaging Core at the University of Rochester.
Contributor Information
H. Mark Kenney, Email: Howard_Kenney@urmc.rochester.edu.
Yue Peng, Email: Yue_Peng@urmc.rochester.edu.
Kiana L. Chen, Email: Kiana_Chen@urmc.rochester.edu.
Raquel Ajalik, Email: Raquel_Ajalik@urmc.rochester.edu.
Lindsay Schnur, Email: Lindsay_Schnur@urmc.rochester.edu.
Ronald W. Wood, Email: Ronald_Wood@urmc.rochester.edu.
Edward M. Schwarz, Email: Edward_Schwarz@urmc.rochester.edu.
Hani A. Awad, Email: Hani_Awad@urmc.rochester.edu.
References
- Andersen T., Darvann T., Murakami S., Larsen P., Senda Y., Bilde A., et al. Accuracy and precision of manual segmentation of the maxillary sinus in MR images—a method study. Br. J. Radiol. 2018;91(1085):1. doi: 10.1259/bjr.20170663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bab I., Hajbi-Yonissi C., Gabet Y., Müller R. Hindfoot. Micro-tomographic Atlas of the Mouse Skeleton. Springer Science + Business Media; 2007. pp. 183–188. [Google Scholar]
- Bell R., Rudmann C., Wood R., Schwarz E., Rahimi H. Longitudinal micro-CT as an outcome measure of interstitial lung disease in TNF-transgenic mice. PLoS One. 2018;13(1):1. doi: 10.1371/journal.pone.0190678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell R., Wu E., Rudmann C., Forney M., Kaiser C., Wood R., et al. Selective sexual dimorphisms in musculoskeletal and cardiopulmonary pathologic manifestations and mortality incidence in the tumor necrosis factor-transgenic mouse model of rheumatoid arthritis. Arthritis Rheumatol. 2019;71(9):12. doi: 10.1002/art.40903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouxsein M., Boyd S., Christiansen B., Guldberg R., Jepsen K., Müller R. Guidelines for assessment of bone microstructure in rodents using micro-computed tomography. J. Bone Miner. Res. 2010;25(7):19. doi: 10.1002/jbmr.141. [DOI] [PubMed] [Google Scholar]
- Brouwers J., van Rietbergen B., Huiskes R. No effects of in vivo micro-CT radiation on structural parameters and bone marrow cells in proximal tibia of wistar rats detected after eight weekly scans. J. Orthop. Res. 2007;25(10):8. doi: 10.1002/jor.20439. [DOI] [PubMed] [Google Scholar]
- Buie H., Campbell G., Klinck R., MacNeil J., Boyd S. Automatic segmentation of cortical and trabecular compartments based on a dual threshold technique for in vivo micro-CT bone analysis. Bone. 2007;41(4):11. doi: 10.1016/j.bone.2007.07.007. [DOI] [PubMed] [Google Scholar]
- Cambre I., Gaublomme D., Burssens A., Jacques P., Schryvers N., Myunck A., et al. Mechanical strain determines the site-specific localization of inflammation and tissue damage in arthritis. Nat. Commun. 2018;9(1):1. doi: 10.1038/s41467-018-06933-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz S., H'ng C., Qu X., Doube M., Nguyen J., de Veer M., et al. A new pipeline to automatically segment and semi-automatically measure bone length on 3D models obtained by computed tomography. Front. Cell Dev. Biol. 2021;9(736574) doi: 10.3389/fcell.2021.736574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douni E., Akassoglou K., Alexopoulou L., Georgopoulos S., Haralambous S., Hill S., et al. Transgenic and knockout analyses of the role of TNF in immune regulation and disease pathogenesis. J. Inflamm. 1995-1996;47(1–2):10. [PubMed] [Google Scholar]
- Hamwood J., Schmutz B., Collins M., Allenby M., Alonso-Caneiro D. A deep learning method for automatic segmentation of the bony orbit in MRI and CT images. Sci. Rep. 2021;11(13693) doi: 10.1038/s41598-021-93227-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iassonov P., Gebrenegus T., Tuller M. Segmentation of X-ray computed tomography images of porous materials: a crucial step for characterization and quantitative analysis of pore structures. Water Resour. Res. 2009;45(9) [Google Scholar]
- Keffer J., Probert L., Cazlaris H., Georgopoulos S., Kaslaris E., Kioussis D., et al. Transgenic mice expressing human tumour necrosis factor: a predictive genetic model of arthritis. EMBO J. 1991;10(13):4025–4031. doi: 10.1002/j.1460-2075.1991.tb04978.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney H., Awad H. Training datasets: a high-throughput semi-automated bone segmentation workflow for murine hindpaw micro-CT datasets. Mendeley Data. 2021 doi: 10.17632/7sm9wznp6d.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinck R., Campbell G., Boyd S. Radiation effects on bone architecture in mice and rats resulting from in vivo micro-computed tomography scanning. Med. Eng. Phys. 2008;30(7):8. doi: 10.1016/j.medengphy.2007.11.004. [DOI] [PubMed] [Google Scholar]
- Kohler T., Stauber M., Donahue L., Müller R. Automated compartmental analysis for high-throughput skeletal phenotyping in femora of genetic mouse models. Bone. 2007;41(4):9. doi: 10.1016/j.bone.2007.05.018. [DOI] [PubMed] [Google Scholar]
- Kontoyiannis D., Pasparakis M., Pizarro T., Cominelli F., Kollias G. Impaired on/off regulation of TNF biosynthesis in mice lacking TNF AU-rich elements: implications for joint and gut-associated immunopathologies. Immunity. 1999;10(3):12. doi: 10.1016/s1074-7613(00)80038-2. [DOI] [PubMed] [Google Scholar]
- Mansoor A., Bagci U., Foster B., Xu Z., Papadakin G., Folio L., et al. Segmentation and image analysis of abnormal lungs at CT: current approaches, challenges, and future trends. Radiographics. 2015;35(4):21. doi: 10.1148/rg.2015140232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton M., Junginger L., Maerz T. Automated MicroCT-based bone and articular cartilage analysis using iterative shape averaging and atlas-based registration. Bone. 2020;137(115417) doi: 10.1016/j.bone.2020.115417. [DOI] [PubMed] [Google Scholar]
- Proulx S., Kwok E., You Z., Papuga M., Beck C., Shealy D., et al. Longitudinal assessment of synovial, lymph node, and bone volumes in inflammatory arthritis in mice by in vivo magnetic resonance imaging and microfocal computed tomography. Arthritis Rheumatol. 2007;56(12):14. doi: 10.1002/art.23128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathnayaka K., Sahama T., Schuetz M., Schmutz B. Effects of CT image segmentation methods on the accuracy of long bone 3D reconstructions. Med. Eng. Phys. 2011;33(2):8. doi: 10.1016/j.medengphy.2010.10.002. [DOI] [PubMed] [Google Scholar]
- Richbourg H., Martin M., Schachner E., McNulty M. Anatomical variation of the Tarsus in common inbred mouse strains. Anat. Rec. (Hoboken) 2017;300(3):10. doi: 10.1002/ar.23493. [DOI] [PubMed] [Google Scholar]
- Skourt B., Hassani A., Majd A. Lung CT image segmentation using deep neural networks. Procedia Comput. Sci. 2018;127:5. [Google Scholar]
- Stauber M., Müller R. Micro-computed tomography: a method for the non-destructive evaluation of the three-dimensional structure of biological specimens. Methods Mol. Biol. 2008;45:20. doi: 10.1007/978-1-59745-104-8_19. [DOI] [PubMed] [Google Scholar]
- Walsh C., Tafforeau P., Wagner W., Jafree D., Bellier A., Werlein C., et al. Imaging intact human organs with local resolution of cellular structures using hierarchical phase-contrast tomography. Nat. Methods. 2021;18(12):10. doi: 10.1038/s41592-021-01317-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X. In: Deep Learning in Medical Image Analysis Advances in Experimental Medicine and Biology. Lee G., Fujita H., editors. Vol. 1213. Springer; Cham: 2020. Automatic segmentation of multiple organs on 3D CT images by using deep learning approaches; pp. 135–147. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Training Video 1 - Introduction to Amira.
Training Video 2 - Segmentation Process.
Training Video 3 - Correction Processes.
Training Video 4 - Mouse Ankle Anatomy.
Training Video 5 - Example Segmentation.
Supplementary Figures
Supplementary Methods
Mask Generation Recipe
Markers Generation Recipe
Paw Analysis Test