Abstract
Lectin protein families are diverse and multi-functional in crustaceans. The carbohydrate-binding domains (CRDs) of lectins recognize the molecular patterns associated with pathogens and orchestrate important roles in crustacean defense. In this study, two lectin homologs, a single CRD containing C-type lectin (CTL) and an L-type lectin (LTL) domain containing endoplasmic reticulum Golgi intermediate compartment 53 kDa protein (ERGIC-53) were identified from the freshwater prawn, Macrobrachium rosenbergii. The open reading frames of MrCTL and MrERGIC-53 were 654 and 1,515 bp, encoding polypeptides of 217 and 504 amino acids, respectively. Further, MrCTL showed a 20-amino acid transmembrane helix region and 10 carbohydrate-binding residues within the CRD. MrERGIC-53 showed a signal peptide region, a type-I transmembrane region, and a coiled-coil region at the C-terminus. Phylogenetic analysis revealed a close relationship between MrCTL and MrLectin and M. nipponense CTL (MnCTL), whereas MrERGIC-53 shared high sequence identity with Eriocheir sinensis ERGIC-53 and Penaeus vannamei MBL-1. A homology-based model predicted small carbohydrate-combining sites with a metal-binding site for ligand binding (Ca2+ binding site) in MrCTL and beta-sheets connected by short loops and beta-bends forming a dome-shaped beta-barrel structure representing the LTL domain of MrERGIC-53. Quantitative real-time polymerase chain reaction detected MrCTL and MrERGIC-53 transcripts in all examined tissues, with particularly high levels observed in hemocytes, hepatopancreas, and mucosal-associated tissues, such as the stomach and intestine. Further, the expression levels of MrCTL and MrERGIC-53 transcripts were remarkably altered after V. harveyi challenge, suggesting putative function in host innate immunity.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10499-022-00845-3.
Keywords: Macrobrachium rosenbergii, CTL; ERGIC-53, Immune response, V. harveyi
Introduction
The giant freshwater prawn Macrobrachium rosenbergii is an economically important crustacean under semi-intensive farming in aquaculture practicing countries. Due to its high value, research is focused on improving the growth performance of M. rosenbergii by reducing its vulnerability to pathogenic infections caused by bacteria and viruses (Miller et al. 2005; See et al. 2009). Indeed, the pathogenic load caused by viral infections in M. rosenbergii hatcheries has multiplied, with mass mortality of larvae reducing the production by almost 80% over the past decade (Alam et al. 2019). While opportunistic bacteria such as Vibrio sp., Aeromonas sp., and Pseudomonas sp. can cause infections at all life stages (reviewed by Pillai and Bonami 2012), viruses such as the M. rosenbergii nodavirus (MrNV) and white spot syndrome virus (WSSV) can cause mass mortality in the post-larval (PL) stages of the host (Sahul Hameed et al. 2000; Gangnonngiw et al. 2020; Huang and Ren 2021).
Detection and discrimination of invading pathogens as non-self by host pathogen recognition receptors (PRRs) are central to the activation of the innate immune response. While PRRs recognize pathogen-associated molecular patterns (PAMPs) on the cell surface of pathogens, processes such as antimicrobial peptide (AMP) activation, phagocytosis, and autophagy limit or stall a load of pathogens, thereby allowing the cells to recover from the infections (Basset et al. 2003). Further, PRRs show great diversity in the recognition of PAMPs, leading to the modulation of diverse signaling cascades and culminating in the evasion of pathogens from the host (Akira et al. 2006; Pedraza et al. 2010). PAMPs function as ligands to initiate PRR-dependent signaling cascades, including the indispensable nuclear factor kappa-B (NF-κB) response, which induces effector AMPs (Pedraza et al. 2010). Moreover, multiple PRRs can be engaged by a given pathogen concerning to their PAMPs, securing a rapid and robust inflammatory response. PRRs expressed by cells include the transmembrane Toll-like receptors (TLRs), lectin receptors, the intracellular activating Nod-like receptors (NLRs) and retinoic acid-inducible gene I-like helicase receptors (RLRs) (Jang et al. 2015). PRRs are also enriched with leucine-rich repeats (LRR) at their N-terminal (as in the case of TLRs) or C-terminal (as in the case of NLRs) ends to recognize the ligands as well as one or more protein–protein interaction domain(s).
Lectins are PRRs that play crucial roles in the innate immunity of crustaceans by differentiating between self and non-self. These molecules specifically recognize and bind to the carbohydrates at the cell surface of pathogens and ultimately target them for uptake and phagocytosis (Ewart et al. 2001). Based on their structure and functions, lectins have been categorized into many families, including C-, F-, I-, L-, M-, P-, R-, and F-type lectins, chitinase-like lectins, ficolins, calnexin/calreticulin, galectins, and intelectins (X-type lectins) (Dodd and Drickamer 2001; Wang and Wang 2013; Singrang et al. 2021). The lectin family lineage has further diversified with high-throughput transcriptome-sequencing strategies in crustaceans, especially in shrimps (Aweya et al. 2021). Lectins can also be classified as mannose-, fructose-, rhamnose-, and galactose-binding lectins (Lis and Sharon 1998). The most diverse and well-studied among the lectin families are the C-type lectin (CTLs), which are distinguished from other lectins by Ca2+-dependent (C-type) carbohydrate-binding proteins (Janeway and Medzhitov 2002). The carbohydrate recognition domain (CRD) is composed of four highly conserved cysteines which are formed by two disulfide bonds to preserve the atypical double-loop characteristics, where the second loop is involved in Ca2+-dependent carbohydrate binding. These loops represent two unique specific sites for carbohydrate and calcium-binding, a Ca2+ binding site and other distinct amino acid motifs, such as a “QPD” (Gln-Pro-Asp) or an “EPN” (Glu-Pro-Asn) motif, which specifically interact with galactose or mannose, respectively (Weis et al. 1991). Although many CTLs have a promiscuous presence in the hepatopancreas tissue of shrimps, recent evidence suggests that CTLs in the digestive tract elicit roles in the regulation of intestinal homeostasis (Zhang et al. 2021). The L-type lectins are characterized by luminal CRDs, also known as LTL-like domain (LTLD) (Zhu et al. 2013), which can bind to high-mannose-type oligosaccharides. Although LTLD-containing proteins are found in plants and animals, the animal LTL family are intracellular luminal proteins involved in the trafficking, sorting, and targeting of maturing glycoprotein distributions and dynamics in the luminal endoplasmic reticulum (ER)-Golgi compartments of animal cells (Nufer et al. 2003). While four members of LTL family proteins have been reported in vertebrates, namely endoplasmic reticulum-Golgi intermediate compartment-53 kDa protein (ERGIC-53), ERGIC-53 like protein (ERGL), 36 kDa vesicular integral membrane protein (VIP36), and VIP36 like protein (VIPL) (Itin et al. 1995), only two members, viz. ERGIC-53 and VIP36, have been identified in invertebrates (Itin et al. 1996; Kamiya et al. 2005). The M. rosenbergii VIP36 (MrVIP36) is required for the clearance of bacteria and inhibition of white spot syndrome virus (WSSV) in vivo where it serves to elicit antibacterial and antiviral responses in the host (Huang et al. 2018).
This study identified novel CTL and LTL homologs from M. rosenbergii (designated as MrCTL and MrERGIC-53, respectively). The novel lectins were characterized based on their sequence features and homology model-based structural identification. Furthermore, the tissue distribution and temporal expression of MrCTL and MrERGIC-53 mRNA were examined after the challenge of the host with V. harveyi.
Materials and methods
Ethics statement
The study was conducted following approval from the Institutional Biosafety Committee (IBSC) and was performed strictly according to the manual of the Review Committee of Genetic Manipulations (RCGM) under the purview of the Department of Biotechnology, Govt. of India. All of the surgeries were performed on ice to minimize the pain to the experimental animals.
Experimental animals and microorganisms
Healthy freshwater prawn M. rosenbergii (body length and weight of 12–15 cm and 15–17 g, respectively) were obtained from Rajdhani Aqua Farm, Pubasasan, Kausalyaganga, Bhubaneswar, Odisha, India. The collected prawns were acclimatized to laboratory conditions (room temperature of 25 °C ± 2 °C) in well-aerated water containers for 1 week before use as experimental animals. The prawns were fed daily with an artificial diet. The Gram-negative bacteria V. harveyi (local strain) was procured from the Fish Genetics and Biotechnology Division, ICAR-Central Institute of Freshwater Aquaculture (CIFA), Kausalyaganga, Bhubaneswar, Odisha, India. For expression analysis of MrCTL and MrERGIC-53 mRNA, tissues including the gill, hepatopancreas, muscle, stomach, and intestine were harvested from healthy M. rosenbergii. The hepatopancreas is a large orange-yellow mass in which we first remove the dorsal wall of the cephalothorax, pericardial sinus, gonads, and the renal sac. The hepatopancreas is removed by proceeding from the dorsal to the ventral surface to expose the whole part of the cardiac stomach, the pyloric stomach, and the anterior part of the midgut (intestine) is embedded. Further, to collect hemocytes, hemolymph (1 mL) was collected into pre-cooled Alsever’s solution (450 mM NaCl, 10 mM KCl, 10 mM EDTA, 10 mM HEPES, pH 7.45) as an anticoagulant at a 1:1 ratio. Subsequently, the mixture was centrifuged at 800 g for 10 min to obtain hemocyte pellets. The dissected tissues were stabilized in RNAlater solution (Thermo Fisher Scientific, USA) and stored at − 40 °C until further use.
Preparation of V. harveyi-challenged M. rosenbergii
For immune stimulation experiments, the prawns were randomly separated into two groups (injected and mock control groups: n = 25 per group). The mock control group of prawns received an intramuscular injection (50 µL) of phosphate-buffered saline (PBS; 0.14 M NaCl, 3 mM KCl, 8 mM Na2HPO4, 1.5 mM KH2PO4, pH 7.4). The challenged group of prawns received an intramuscular injection (50 µL) of V. harveyi suspended in PBS (1 × 106 colony-forming units (CFU)/mL). Tissues, including the stomach, hemocyte, and hepatopancreas that are representative of gut-specific and systemic immune stimulation, were harvested (n = 3) from the mock control (IC) group and the V. harveyi challenged group at different time-points (6 h, 12 h, 24 h, and 48 h). The harvested tissues were stored in RNAlater solution at − 40 °C until further use in relative gene expression experiments.
Cloning and identification of full-length cDNA of MrCTL and MrERGIC-53
The M. rosenbergii hepatopancreas expressed sequence tag (EST) (M. rosenbergii clone SMbr00361; Accession ID: KJ631055.1) partial sequence (242 bp), representative of full-length MrCTL was extracted from the National Center for Biotechnology Information (NCBI) database. The partial sequence (397 bp), representative of full-length MrERGIC-53, was extracted from the hepatopancreas cDNA library using semi-quantitative PCR primers, as described in Table 2. To clone the partial transcript of MrCTL and MrERGIC-53, total RNA was isolated from hepatopancreas tissue according to the method of Yaffe et al. 2012, with minor modifications. Briefly, 30 mg of tissue was homogenized at 3000 g for 5 min in 500 µL of RNA lysis buffer (6 M Guanidine hydrochloride, 0.5 M EDTA, 1 M MES buffer, 40 mM Phenol red, and 250 µL acetic acid in 50 µL distilled water). After incubation at room temperature (RT) for 3 min, the samples were centrifuged at 13,000 g for 5 min at 4 °C. To the upper layer, 99% ethanol was added (1 volume) and mixed by inversion, and after incubation at RT for 1 min, the samples were centrifuged at 13,000 g for 30 s at 4 °C. To eliminate genomic DNA contamination, the samples were treated with DNase (1 U/µL) and incubated at 37 °C for 15 min at RT. Subsequently, the samples were washed with 3 M sodium acetate, followed by washing with 80% ethanol. After centrifugation at 13,000 g for 2 min at 4 °C, total RNA was eluted with 50 µL of distilled water. The concentration and purity of RNA were determined using an Epoch Multi-plate Reader (Biotek Instruments Pvt. Ltd., Winooski, VT, USA) by measuring the absorbance at 260 and 280 nm wavelengths. For cDNA synthesis, 1 µg total RNA was used as the template. In the reverse transcription-1 (RT-1) reaction, the total RNA was complexed with OligodT (1 µL) in a reaction, incubated for 3 min at 72 °C, followed by 2 min at 42 °C. Subsequently, the RT-1 reaction components were mixed with RT-2 premix (Accupower RT Pre-mix, Bioneer, Daejeon, Korea) and incubated for 90 min at 42 °C and 10 min at 70 °C. The cDNA was stored at − 20 °C until further use in PCR experiments.
Table 2.
Primers used for molecular cloning and expression analysis of MrERGIC-53
| Primers | Oligos name | Sequence (5′-3′) |
|---|---|---|
| qPCR | MrERGIC-53-Fw | TCCGAATCACACCATCCTTAC |
| MrERGIC-53-Rev | GAAAGGCTTATTGCGGAAATC | |
| Sequencing | M13-Fw | GTAAAACGACGGCCAG |
| M13-Rev | CAGGAAACAGCTATGAC | |
| RACE-PCR | MrERGIC-53 (3′-RACE) GSP1 | GAGTGGTGGGAGGTAGACTTCGTTT |
| MrERGIC-53 (3′-RACE) GSP2 | AGGAGTTGAAGGTCCAGTCTTTGG | |
| MrERGIC-53 (5′-RACE) GSP1 | CCAAAGACTGGACCTTCAACTCCT | |
| MrERGIC-53 (5-RACE) GSP2 | AAACGAAGTCTACCTCCCACCACTC | |
| UPM Long | CTAATACGACTCACTATAGGGCAAGCAGTGGTATCAACGCAGAGT | |
| UPM Short | CTAATACGACTCACTATAGGGC | |
| qRT-PCR | MrERGIC-53-Fw | TCCGAATCACACCATCCTTAC |
| MrERGIC-53-Rev | GAAAGGCTTATTGCGGAAATC | |
| β-actin-Fw | AATCGTGCGTGACATCAAGG | |
| β-actin-Rev | TCTCGTTACCGATGGTGATGAC |
Semi-quantitative PCR was performed using AccuPower® pfu PCR premix (Bioneer, Daejeon, Korea) to identify and amplify MrCTL and MrERGIC-53 partial sequences. Briefly, 10 µL of the premix was used in a 20 µL total reaction volume [containing 2 μL cDNA and 1 µL of forward and reverse primer mix (10 pmol each) and 7 µL of distilled water]. The primer information of MrCTL and MrERGIC-53 are shown in Tables 1 and 2, respectively. The PCR cycling condition was as follows: initial denaturation at 94 °C for 5 min, followed by 31 cycles of denaturation at 94 °C for 30 s, annealing at 52 °C for 30 s, and extension at 72 °C for 35 s and ending with a final extension at 72 °C for 5 min. The PCR product was purified using the AccuPrep PCR purification kit (Bioneer, Daejeon, Korea) according to the manufacturer’s protocol. The purified PCR product was cloned into the T-blunt vector using the T-Blunt PCR Cloning Kit (Solgent, Daejeon, Korea) according to the manufacturer’s protocol. The cloned product was transformed into competent E. coli (DH5α strain) cells, and positive recombinants were identified after overnight incubation (37 °C for 15 h) of the clones on Luria Bertani (LB)-Kanamycin (50 mg/ml) plates. After validation of positive clones by colony PCR using M13 universal primers (Tables 1 and 2), subculture was performed overnight to extract plasmid DNA. The plasmid DNA was sequenced using the ABI 3730xl capillary sequencer (Applied Biosystems). The partial gene sequences of MrCTL and MrERGIC-53 were identified using BLASTx, with all non-redundant CDS translations + protein data bank + SwissProt + protein identification resource + protein families’ database.
Table 1.
Primers used for molecular cloning and expression analysis of MrCTL
| Primers | Oligos name | Sequence (5′-3′) |
|---|---|---|
| Cloning | MrCTL-Fw | GCGACCATTCTTCTTCTTGC |
| MrCTL-Rev | TTCTCTGAAGCGACTGCTTG | |
| β-actin-Fw | AATCGTGCGTGACATCAAGG | |
| β-actin-Rev | TCTCGTTACCGATGGTGATGAC | |
| Sequencing | M13-Fw | GTAAAACGACGGCCAG |
| M13-Rev | CAGGAAACAGCTATGAC | |
| RACE-PCR | MrCTL (3′-RACE)-GSP1 | GTGAAGGGGAAGAAGACTGTATCC |
| MrCTL (3′-RACE)-GSP2 | CAACCTGGGTTGGAACTTTG | |
| MrCTL (5′-RACE)-GSP1 | GCAGGTTTCTCGCAGATGTA | |
| MrCTL (5′-RACE)-GSP2 | GTCTTCTTCCCCTTCACAGTTG | |
| UPM-Long | CTAATACGACTCACTATAGGGCAAGCAGTGGTATCAACGCAGAGT | |
| UPM-Short | CTAATACGACTCACTATAGGGC | |
| qRT-PCR | MrCTL-Fw-qRT-PCR | GCATGACGGGACCTACAGAT |
| MrCTL-Rev-qRT-PCR | ACTTTTCTGCAGGCGTTTGT | |
| β-actin-Fw | GAGACCTTCAACACCCCAGC | |
| β-actin-Rev | TAGGTGGTCTCGTGAATGCC |
To elucidate the full-length cDNA sequences of MrCTL and MrERGIC-53 from partial sequences, 5′- and 3′-rapid amplification of cDNA ends (RACE)-PCR was performed. Two pairs of gene-specific primers for 5′- and 3′-RACE-PCR (5′-GSP1, 5′-GSP2, and 3′-GSP1, 3′-GSP2) were designed and synthesized based on the partial sequences of MrCTL and MrERGIC-53. The primer sequence information for 5′- and 3′-RACE-PCR of MrCTL and MrERGIC-53 is provided in Tables 1 and 2, respectively. Briefly, the 5′- and 3′-RACE-ready cDNA was synthesized from 1 µg of total RNA. Next, taking the 5′- and 3′-RACE-ready cDNA as the template, 5′- and 3′-RACE (first PCR) was performed in separate reactions. A 50 μL total reaction volume was prepared for the first PCR reaction, comprising 2.5 μL of RACE-ready cDNA, 5 μL 10 × Universal Primer Mix (UPM), and 1 μL 5′-or 3′-gene-specific primers in 41.5 μL master mix (including 1.0 μL SeqAmp DNA polymerase). PCR was conducted for 25 cycles at 94 °C for 30 s, 68 °C for 30 s, and 72 °C for 3 min. The first PCR products (5 μL) were subsequently used as a template for 5′- and 3′-RACE (second PCR) reactions. The second PCR was also conducted in a 50 μL reaction volume, comprising 41.5 μL of master mix (including 1.0 μL SeqAmp DNA polymerase), having 1 μL UPM-short and 1 μL of 5′- or 3′-gene- specific primers (5′-GSP2 and 3′-GSP2). PCR was conducted for 25 cycles, as described for the first PCR. The second PCR product for MrCTL was cloned onto the T-blunt cloning vector (Solgent, Daejeon, Korea), and the obtained plasmid DNA was sequenced. In contrast to MrCTL, the 3′-second PCR product for MrERGIC-53 was directly sequenced using the primer-walking method using the BigDye™ Terminator ver3.1 cycle sequencing kit (Applied Biosystems). The vector-contaminated sequences were excised from the target sequence using the VecScreen tool at NCBI (https://www.ncbi.nlm.nih.gov/tools/vecscreen/). The CAP3 DNA sequence assembly program (http://doua.prabi.fr/software/cap3) was used to define non-overlapping contigs and obtain the full-length cDNA sequence. The full-length cDNA sequences of MrCTL and MrERGIC-53 are registered with GenBank with accession IDs MK864100 and MK864099, respectively.
Sequence features and phylogenetic analysis of MrCTL and MrERGIC-53
The full-length cDNA and deduced amino acid sequences of MrCTL and MrERGIC-53 were analyzed using BLASTx and BLASTp algorithms at NCBI (http://www.ncbi.nlm.nih.gov/blast). The Hidden Markov Model (HMM)-based gene structure prediction tool, FGENESH (Solovyev et al. 2006), was used to identify the ORF region and the translated amino acid sequences. The complete cDNA sequence and the deduced amino acid sequence of MrCTL and MrERGIC-53 were formatted using UltraEdit64-bit text-editor. The ExPASy Translate tool was used for the reverse complement of the nucleotide sequence (http://www.expasy.org). Prediction of the conserved protein domains was accomplished by the Simple Modular Architecture Research Tool (SMART) program (http://www.smart.emblheidelberg.de/). The multiple sequence alignment (MSA), percent identity, and distance matrix were analyzed by the ClustalX 2.1 program (Larkin et al. 2007). The phylogenetic tree was constructed based on the amino acid sequence alignments using the maximum-likelihood method with the bootstrap trial set to 1000. The phylogenetic tree was visualized using the Tree Viewer at Molecular Evolutionary Genetics Analysis (MEGA) software version 7.0 (https://www.megasoftware.net/) (Kumar et al. 2018). The homology-based SWISS-MODEL program (https://swissmodel.expasy.org/) was used to predict the structural characteristics of the MrCTL and MrERGIC-53 amino acid sequences. The secondary structure was predicted using PSIPRED 4.0 (http://bioinf.cs.ucl.ac.uk/psipred_new/). The Ramachandran plot was constructed using VADAR 1.8 software (http://vadar.wishartlab.com/). The prediction of the phosphorylation sites in the protein sequence was analyzed using the NetPhos server (http://www.cbs.dtu.dk/services/NetPhos/).
Expression analysis of MrCTL and MrERGIC-53 using qRT-PCR
The basal expression levels of MrCTL and MrERGIC-53 mRNA in the tissues, including the gill, hepatopancreas, muscle, stomach, intestine, and hemocyte, of M. rosenbergii were analyzed by using qRT-PCR using the Applied Biosystems QuantStudio 3 real-time system, 96-well 0.2 mL block system (Thermo Fisher Scientific, USA). The qRT-PCR mixture (20 μL) consisted of 10 μL of 2 X SYBR Green Master mix, 0.5 μL of forward and reverse primers (10 pmol each), and 2 μL of template cDNA (diluted 20 times with molecular biology grade water). PCR was performed at an initial denaturation of 95 °C for 5 min followed by 40 cycles of amplification at 95 °C for 15 s, and 60 °C for 30 s. The M. rosenbergii beta-actin (Mrβ-actin) gene served as an endogenous control to normalize the expression in all experimental groups. The relative gene expression was measured using the comparative Ct value method (Livak and Schmittgen 2001). The reactions were performed in triplicate, and the results are presented as the mean ± SE (n = 3) of three biological replicates. For the V. harveyi challenge experiments, the immune stimulated prawns were randomly selected for total RNA isolation, cDNA synthesis, and qRT-PCR analysis after harvesting of the hemocyte, stomach, and hepatopancreas at different time-points (please refer to “Preparation of V. harveyi-challenged M. rosenbergii” section). The reactions were performed in triplicate, and the results are presented as the mean ± SE (n = 3) of three biological replicates.
One-way analysis of variance (ANOVA) performed using SAS 9.1.3 for Windows (SAS Institute, Cary, NC) was used to estimate significant differences between the treated and control groups at each time-point.
Results
Characterization of MrCTL and MrERGIC-53 cDNA
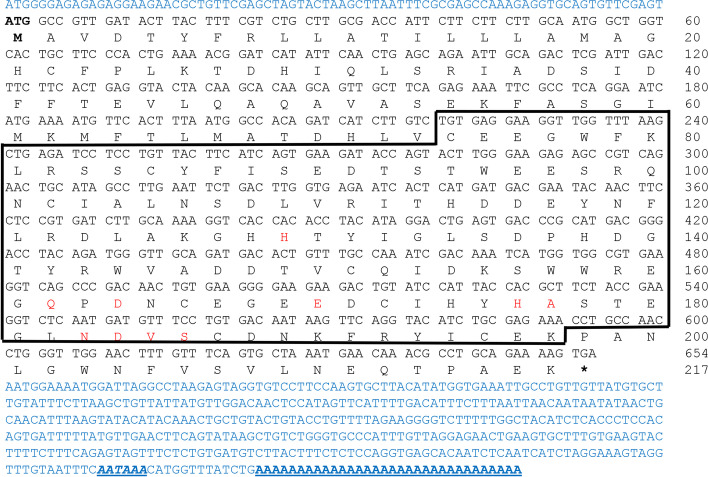
The cDNA sequences of MrCTL and MrERGIC-53 were cloned from the giant freshwater prawn, M. rosenbergii, and confirmed using PCR and sequencing. The full-length cDNA of MrCTL comprises a 5′-UTR of 79 bp, a 3′-UTR of 458 bp, and an open reading frame (ORF) of 654 bp, encoding a 217 amino acid protein. Further, the 3′-UTR contains a poly(A) sequence of 32 bp and a poly(A) recognition sequence (AATAAA) 13 bp upstream of the poly(A) sequence (Fig. 1). A single carbohydrate recognition domain (CRD), representing the CTL family domain of 124 amino acid residues (residues 74–197), with 10 carbohydrate-binding sites (H129, Q162, D164, E170, H176, A177, N183, D184, V185, and S186) was predicted in the deduced protein. A 20-amino acid transmembrane helix region (residues 6–25) was predicted in the deduced protein. The MrCTL protein had a theoretical pI of 4.75 and a molecular weight (MW) of 24.86 kDa. However, MrERGIC-53 contained an ORF of 1,515 bp, encoding a putative protein of 504 amino acid residues, with a predicted MW and pI of 56.91 kDa and 6.01, respectively. The polyadenylation signal (AATAAA) was found in the 3′-UTR region of MrERGIC-53 cDNA. A typical signal peptide sequence of 19 amino acid residues (cleavage between A19 and Q20, with a likelihood probability of 0.9675) was found at the N-terminus of MrERGIC-53. Domain analysis predicted a 225 amino acid L-type lectin (LTL) domain (residues 24–247) in the deduced protein. A 23 amino acid type-I transmembrane region (residues 470–492) was also localized at the C-terminus of the deduced protein. Further, a coiled-coil region of 34 amino acid residues was present between the LTL domain and the type-I transmembrane region (Fig. 2). A schematic representation of the MrERGIC-53 precursor and mature protein is illustrated in Fig. 3.
Fig. 1.
Nucleotide and deduced amino acid sequence of M. rosenbergii CTL (MrCTL). The deduced amino acids are denoted as one-letter codes. The C-type lectin domain (CTLD) is boxed. The carbohydrate-binding amino acid residues within CTLD are shown in red text. The 5′ and 3′ UTR regions are indicated in blue text. The translation start (ATG) is indicated in bold, and the stop codon is shown by an asterisk (*). The 3′-polyadenylation recognition sequence (AATAAA) is shown in bold and italics. The 3′-poly A sequence is underlined, and the polyadenylation recognition sequence (AATAAA) is shown in italics and bold
Fig. 2.
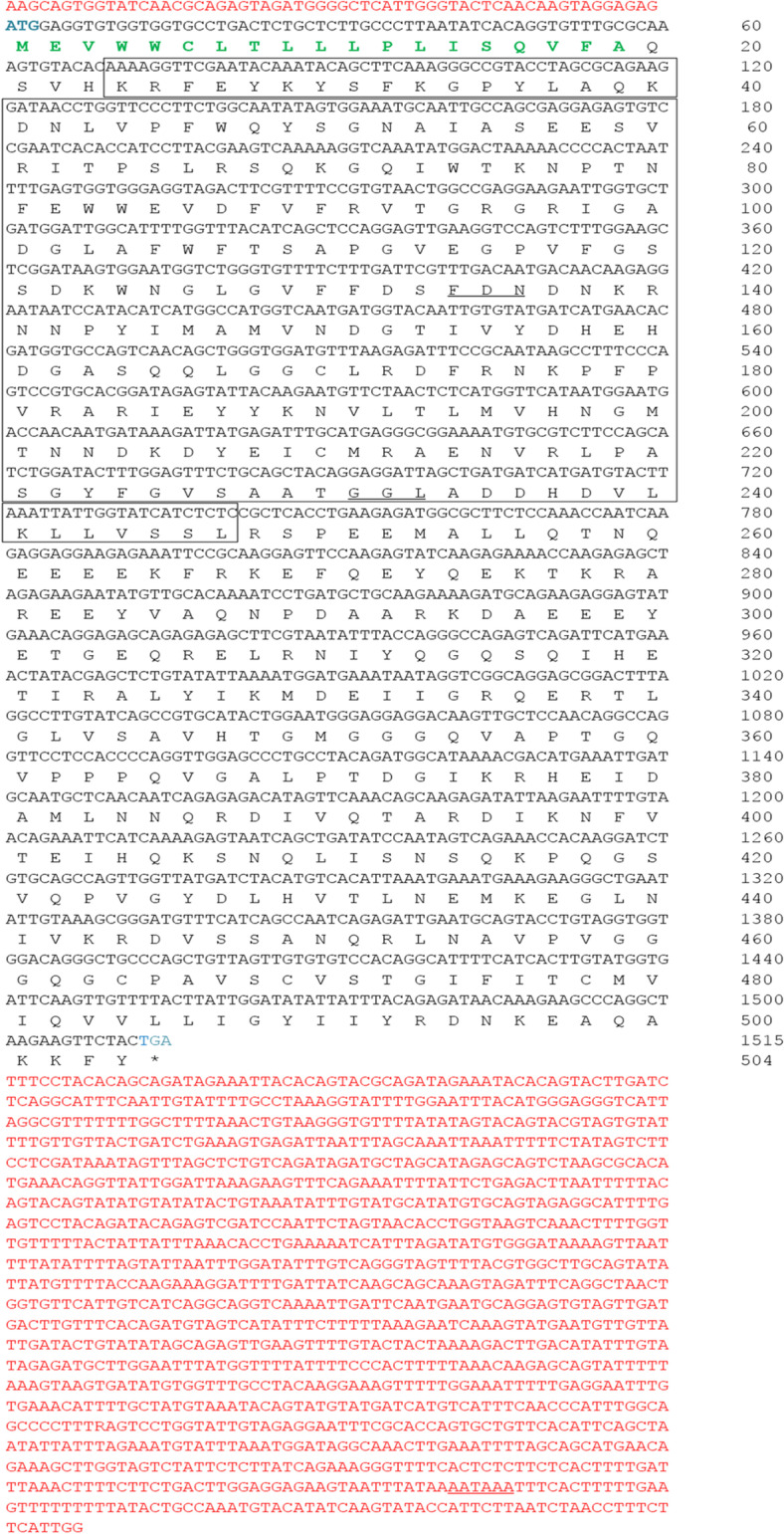
Nucleotide and deduced amino acid sequence of M. rosenbergii ERGIC-53 (MrERGIC-53). The deduced amino acids are denoted as one-letter codes. The lectin L-type superfamily domain (LTLD) is boxed. The carbohydrate binding regions within LTLD are shown in italics. The signal peptide sequence predicted between amino acid positions 19 and 20 (probability score of 0.9675) is shown in bold and green text. The 5′-UTR and the 3′-UTR regions are represented in red text. The 3′-polyadenylation recognition sequence (AATAAA) is underlined. The translation start (ATG) and the stop (TGA) codons representing the beginning and end of the ORF, respectively, are marked in blue text
Fig. 3.
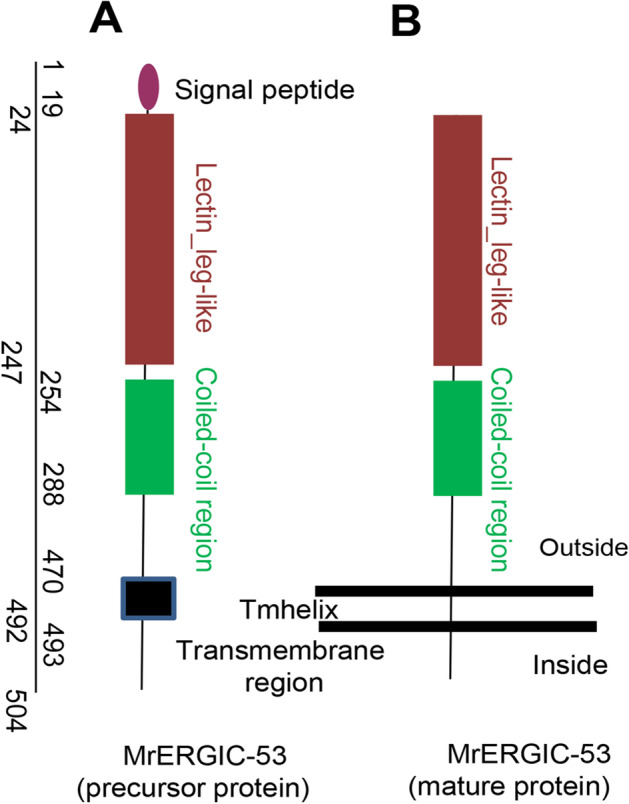
Schematic representation of MrERGIC-53 sequence features. (A) The precursor protein contains an N-terminal signal peptide sequence of 19 amino acids, which is absent in the mature protein. (B) Further, the L-type lectin domain (LTLD) (amino acid nos. 24 to 247) and coiled-coil region (amino acid nos. 254 to 288) are characteristic features of the protein. A transmembrane domain (amino acid nos. 470–493) and a short cytoplasmic domain (amino acid nos. 492–504) are also found toward the C-terminus
Sequence analysis of MrCTL and MrERGIC-53
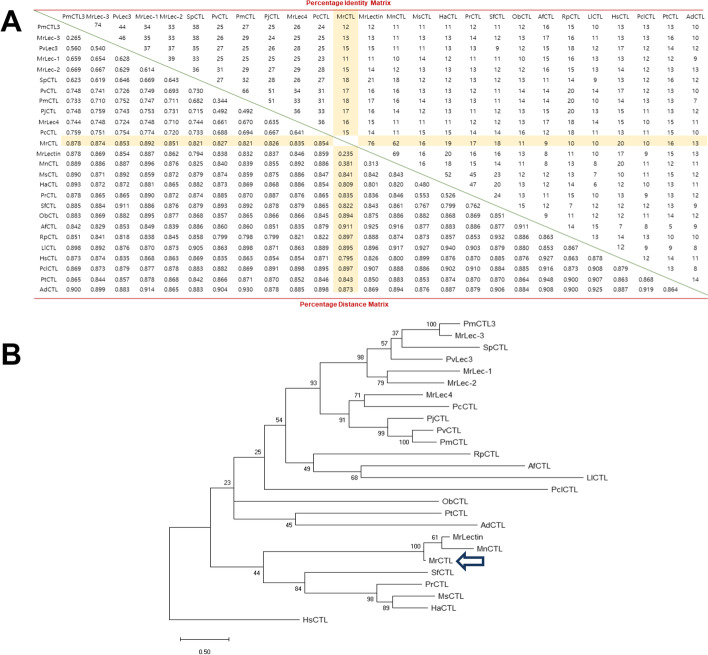
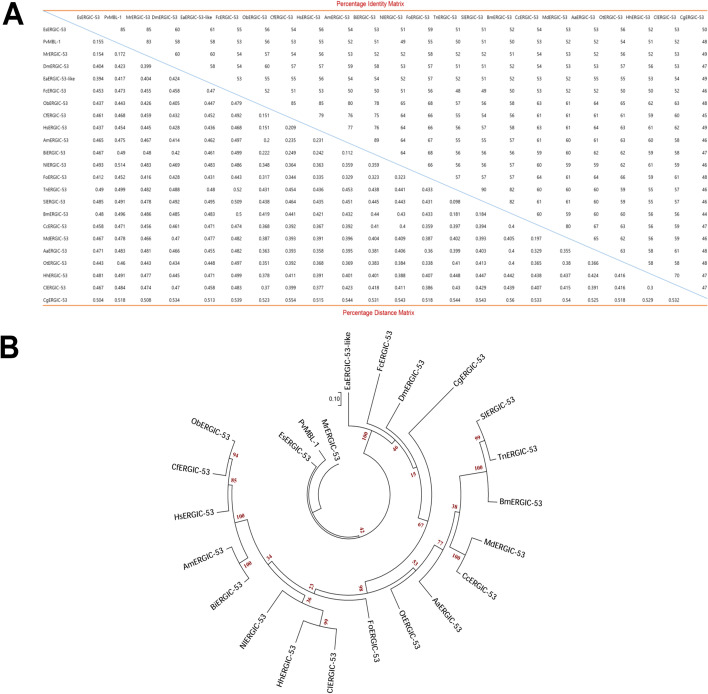
Multiple alignment analysis followed by percent distance and identity matrix revealed that MrCTL shows a maximum identity of 76% with M. rosenbergii lectin (MrLectin), followed by 62% with M. nipponense CTL (MnCTL) (Fig. 4A). The identity or relatedness of MrCTL with lectin sequences from representative species from invertebrates was found to be in the range of 9–20%. The distance matrix also showed a similar trend with the shortest distance observed between amino acid sequences shared between MrCTL and MrLectin, followed by MnCTL. Phylogenetic relationships between CTL homolog proteins revealed the close clustering of MrCTL with MrLectin and MnCTL (Fig. 4B). Homo sapiens CTL (HsCTL) formed as an outgroup in the phylogenetic analysis. Sequences of insect CTLs, such as Spodoptera frugiperda CTL (SfCTL), Mythimna separata CTL (MsCTL), Pieris rapae CTL (PrCTL), and Helicoverpa armigera CTL (HaCTL), belonged to a separate close cluster that was remarkably different from CTLs originating from crustaceans (Fig. 4B). The MrERGIC-53 amino acid sequence shared maximum identity of 85% with Eriocheir sinensis ERGIC-53 (EsERGIC-53), followed by 83%, 60%, and 58% with Penaeus vannamei mannose-binding lectin 1 (PvMBL1), Daphna magna ERGIC-53 (DmERGIC-53), and Eurytemora affinis ERGIC-53 (EaERGIC-53), respectively (Fig. 5A). The relatedness of MrERGIC-53 with other representative ERGIC-53 homologs was in the range of 50–60%. The distance matrix also showed a similar trend, with less distance between sequences shared with PvMBL1, followed by DmERGIC-53 and EaERGIC-53. Further, from an evolutionary perspective, MrERGIC-53 showed close clustering with EsERGIC-53 and PvMBL-1. The ERGIC-53 from the molluscan species, Crassostrea gigas (CgERGIC-53), formed a completely separate branch in the phylogenetic analysis. The ERGIC-53 proteins belonging to the insect order Lepidoptera, including Spodoptera litura ERGIC-53 (SlERGIC-53), Trichoplusia ni ERGIC-53 (TnERGIC-53), and Bombyx mori ERGIC-53 (BmERGIC-53), formed a separate clade (Fig. 5B).
Fig. 4.
Alignment of MrCTL with CTL family members of representative species. (A) Percentage identity and distance matrix of MrCTL and the representative CTL proteins sequences from various species. The analysis was performed using ClustalX2 (Version 2.0). (B) Phylogenetic analysis of MrCTL with other CTL proteins. The consensus maximum likelihood tree was built using MEGA (Version 7.0) with the bootstrap trials set to 1000. The CTL sequences were selected from the following representative species: Macrobrachium nipponense (MnCTL, ARH56436.1), Macrobrachium rosenbergii (MrCTL, MK854100; MrLectin, AOF80290.1; MrLec-3, AFN20599.1; MrLec-1, AFN20597.1; MrLec-2, AFN20598.1; MrLec-4, AFN20600.1), Palaemon modestus (PmCTL3, AGZ95687.1), Scylla paramamosain (SpCTL, AEO92001.1), Penaeus vannamei (PvLec-3, ROT70826.1), Palaemon carinicauda (PcCTL, AIS36112.1), Penaeus japonicus (PjCTL, AHF21000.1), Penaeus vannamei (PvCTL, ABI97374.1), Penaeus merguiensis (PmCTL, ACR56805.1), Homo sapiens (HsCTL, NP_057593.3), Portunus trituberculatus (PtCTL, ATE51203.1), Ruditapes philippinarum (RpCTL, BAO49754.1), Littorina littorea (LlCTL, AJA37850.1), Azumapecten farreri (AfCTL, AAT77680.1), Oryctes borbonicus (ObCTL, KRT82901.1), Procambarus clarkii (PclCTL, AVP26799.1), Anopheles dirus (AdCTL, AFK83719.1), Spodoptera frugiperda (SfCTL, CRN04268.1), Mythimna separata (MsCTL, BBC20960.1), Helicoverpa armigera (HaCTL, ABF83203.1), and Pieris rapae (PrCTL, AEO52696.1). MrCTL shows 76% identity with MrLectin and 62% identity with MnCTL
Fig. 5.
Alignment of MrERGIC-53 with orthologs from representative species. (A) Percentage identity and distance matrix of MrERGIC-53 and representative ERGIC-53 protein sequences from representative species. The analysis was performed using ClustalX2 (Version 2.0). The percentage identity matrix is shown from left to right, and the percentage distance matrix is shown from top to bottom. (B) Phylogenetic analysis of MrERGIC-53 with ERGIC-53 protein sequences from representative species. The consensus maximum likelihood tree (Tree Explorer, circular original tree) was built on the basis of the JTT matrix-based model using MEGA (Version 7.0) software. The bootstrap values are indicated by the numbers at the nodes. The ERGIC-53 sequences were selected from the following representative species: Eriocheir sinensis (EsERGIC-53, AIM45537.1), Penaeus vannamei (PvMBL-1, ROT63057.1), Daphnia magna (DmERGIC-53, JAN85016.1), Eurytemora affinis (EaERGIC-53-like, XP_023340026.1), Folsomia candida (FaERGIC-53, OXA55457.1), Ooceraea biroi (ObERGIC-53, XP_011335262.2), Camponotus floridanus (CfERGIC-53), Harpegnathos saltator (HsERGIC-53, XP_011154813.1), Apis mellifera (AmERGIC-53, XP_006565093.1), Bombus impatiens (BiERGIC-53, XP_012246683.1), Nilaparvata lugens (NlERGIC-53, XP_022197547.1), Frankliniella occidentalis (FoERGIC-53, XP_026275094.1), Trichoplusia ni (TnERGIC-53, XP_02673666.1), Spodoptera litura (SlERGIC-53, XP_022817189.1), Bombyx mori (BmERGIC-53, XP_012546331.1), Ceratitis capitata (CcERGIC-53, XP_004525276.2), Musca domestica (MdERGIC-53, XP_005179276.1), Aedes aegypti (AaERGIC-53, XP_021697592.1), Onthophagus taurus (OtERGIC-53, XP_022911466.1), Halyomorpha halys (HhERGIC-53, XP_014294482.1), Cimex lectularius (ClERGIC-53, XP_014245666.1), and Crassostrea gigas (CgERGIC-53, XP_011441168.1)
Structural prediction analysis of MrCTL and MrERGIC-53
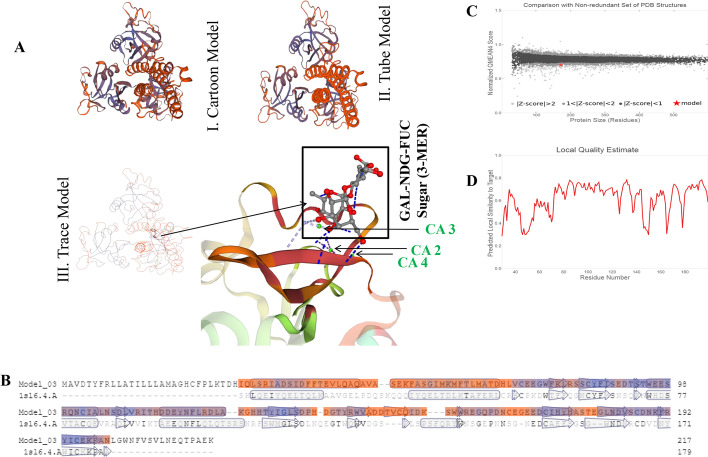
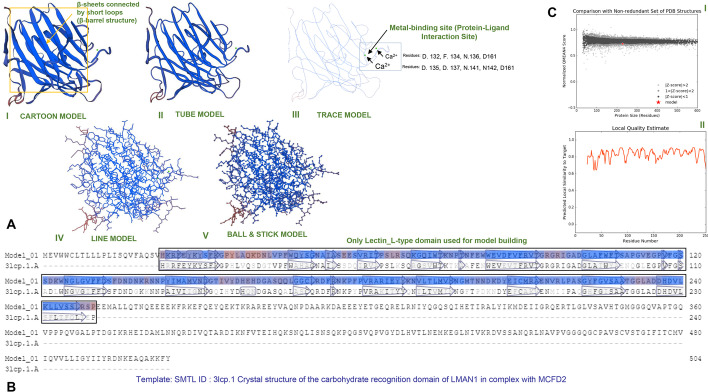
A homology-based model of MrCTL and MrERGIC-53 was predicted using SWISS-MODEL molecular modeling software. The predicted structure for MrCTL was constructed based on the conserved CTL domain sequence of MrCTL (Fig. 6A). The carbohydrate-binding residues were found to be located in the CTL domain. The subunits of the protein have small carbohydrate-combining sites, with a metal-binding site for ligand-binding (Ca2+ binding site). A 3-mer sugar moiety (the β-D-Galactose (GAL), 2-(Acetylamino)-2-Deoxy-A-D-Glucopyranose (NDG), and Alpha-L-Fucose (FUC)) is predicted to bind with MrCTL at the Ca+ binding sites (CA2, CA3, and CA4) (Fig. 6). The Ramachandran plot matrix of the predicted MrCTL structure designed using VADAR 1.8 software shows the allowed combination of conformational angles, such as psi (ψ) and phi (φ) (S1 Figure). The plot demonstrates an “allowed” region for alpha-helix and beta-sheet conformation where the residues are favored. Approximately 93.49% of the residues of the predicted MrCTL structure was favored with regard to the Ramachandran plot, and only two residues (A109 and A110) were predicted to be Ramachandran outlier(s). Further, a clash score of 4.9 for residues A174-A185 was predicted. Although no bad angles were predicted, 18 of 1923 angles were not favored in the predicted structural model of MrCTL. Further, prediction based on position-specific scoring matrices found that alpha-helices with coiled-coil regions dominated the C-terminus of MrCTL. In the homology-based-predicted MrERGIC-53 structure, only the conserved LTL domain sequence was considered. The homology-based model was derived by considering the crystal structure of the CRD of a glycoprotein receptor LMAN1 in complex with the co-receptor MCFD2 as the reference model (Wigren et al. 2010). The LTL domain was found to be composed of beta-sheets connected by short loops and beta-bends, forming a dome-shaped beta-barrel structure (Fig. 7). The carbohydrate-binding site is generally localized toward the apex of the dome-shaped beta-barrel structure. A metal-binding site for ligand-binding (Ca2+) was also predicted for MrERGIC-53. The amino acid residues predicted to be involved at the interface of the protein–ligand interaction site include D132, F134, D135, N136, D137, N141, N142, and D161. The predicted model of MrERGIC-53 was also assessed using the Ramachandran plot diagram ( Figure S2). Approximately 96.9% residues was favored by the Ramachandran plot, while two residues (S65 and N42) were classified as Ramachandran outlier(s). A clash score of 5.05 for residues L127-L192 and P105-P131 was also predicted. Further, 15 of 2,548 angles were not favored by the Ramachandran plot in the homology-based structure of MrERGIC-53 (with no bad angles predicted).
Fig. 6.
Predicted model of MrCTL using the SWISS-MODEL program. (A) Homology model of MrCTL based on the reference model (SMTL ID: 1sl6.4.A, Crystal structure of a fragment of DC-SIGNR (containing the carbohydrate recognition domain and two repeats of the neck) complexed with Lewis-x)). I. Cartoon model. II. Tube model. III. Trace model showing the metal binding sites (protein–ligand interaction site). The GAL-NDG-FUC sugars (3-MER) are boxed, and the Ca+ binding sites (CA2, CA3, CA4) are shown by green arrows. (B) Sequence alignment of the MrCTL template generated by the SWISS-MODEL. (C) QMEAN4 of MrCTL predicted model showing the Z-score value. (D) Local quality estimation of MrCTL
Fig. 7.
Homology model of MrERGIC-53 predicted using the SWISS-MODEL interface. (A) MrERGIC-53 protein predicted structure in the cartoon (beta-barrel structure boxed) (I), tube (II), trace (protein–ligand interaction site shown) (III), line (IV), and ball and stick (IV) models. (B) Alignment of the predicted MrERGIC-53 model with the most-close template (SMTL ID: 3lcp.1, crystal structure of the carbohydrate recognition domain of LMAN1 in complex with MCFD2). (C) The statistical reference of the predicted model shown using the QMEAN4 score and the local quality estimate
Tissue distribution of MrCTL and MrERGIC-53 in healthy M. rosenbergii
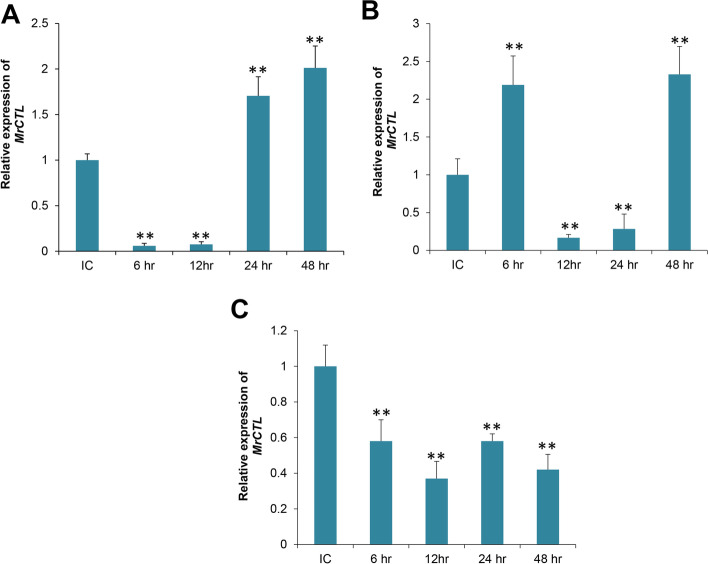
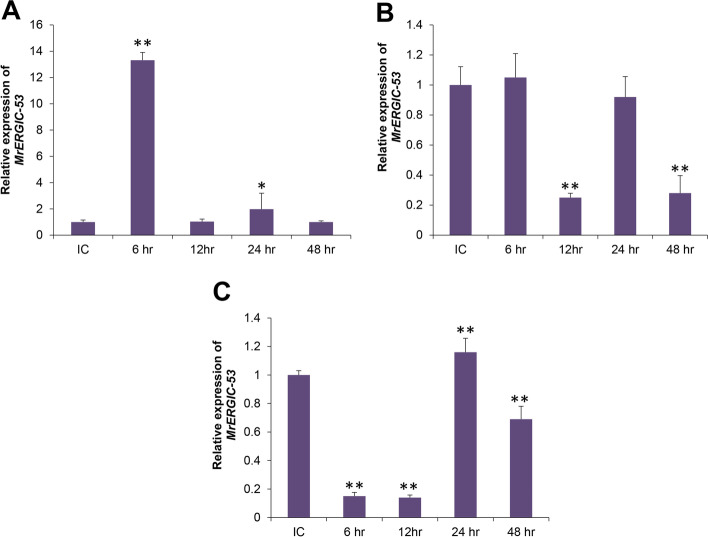
The distribution of MrCTL and MrERGIC-53 transcripts in different tissues of the freshwater prawn, M. rosenbergii, was examined by qRT-PCR. The expression of MrCTL mRNA was found to be considerably higher in healthy M. rosenbergii than in MrERGIC-53 mRNA. MrERGIC-53 mRNA expression in hemocytes was considerably higher than in the other tissues analyzed. Moreover, the expression of MrERGIC-53 mRNA in gut-associated tissues was significantly lower than that in hemocytes and the hepatopancreas. Among the tissues analyzed, the expression of MrERGIC-53 mRNA was greater in the hemocytes and hepatopancreas than in the stomach, intestine, and muscle of healthy M. rosenbergii (Fig. 8A). The expression of MrCTL mRNA in the tissues of healthy M. rosenbergii was found to be considerably greater in the hepatopancreas, followed by the gut-associated tissues, such as the stomach and intestine (Fig. 8B). Surprisingly, the expression of MrCTL mRNA was found to be significantly lower in the hemocytes, gills, and muscle tissues of healthy M. rosenbergii. This might reveal distribution of CTLs in the hepatopancreas and gut-associated tissues of M. rosenbergii.
Fig. 8.
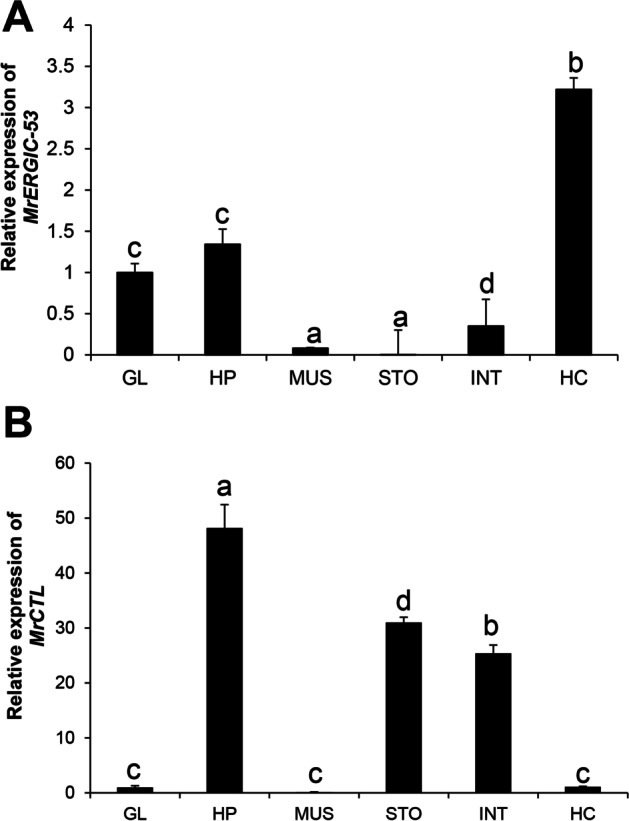
Tissue-specific expression of MrERGIC-53 and MrCTL mRNA using qPCR. (A) Relative expression of MrERGIC-53 in M. rosenbergii tissues. (B) Relative expression of MrCTL in M. rosenbergii tissues. GL gill, HP hepatopancreas, MUS muscle, STO stomach, INT intestine, HC hemocytes. β-actin (M. rosenbergii) served as the internal control to normalize RNA levels between samples. Vertical bars represent standard error (n = 3). Different letters above the bars represent significant differences between groups
Time course expression profiles of MrCTL and MrERGIC-53 after immune challenge with V. harveyi
To understand the putative role of MrCTL and MrERGIC-53 in the immune response to pathogens, we challenged a group of healthy M. rosenbergii with the Gram-negative bacterium, V. harveyi. The results of this study will address the role of these novel lectins in the innate immune response of the host against pathogens. MrCTL mRNA was significantly upregulated in hemocytes and stomach tissue but was downregulated in the hepatopancreas tissue as suggested by the time-course results post V. harveyi challenge. After V. harveyi challenge, the levels of MrCTL mRNA increased to the highest level at 48 h in hemocytes compared to the injected control (Fig. 9A). In the stomach, the expression of MrCTL mRNA was considered highest compared to other time-points at 48 h post-challenge (Fig. 9B). MrCTL mRNA was continuously downregulated in the hepatopancreas tissue of the host after V. harveyi challenge (Fig. 9C). In summary, the immune induction of MrCTL mRNA was observed only in the hemocytes and stomach tissue post-challenge with the Gram-negative bacterium V. harveyi. MrERGIC-53 mRNA was also expressed in the hemocytes of healthy M. rosenbergii challenged with V. harveyi (Fig. 10). The MrERGIC-53 mRNA expression was found to be greatest at 6 h post-challenge and subsequently reduced at higher time-points, with expression levels closer to the injected control (Fig. 10A). Moreover, in the stomach tissue, immune induction of MrERGIC-53 mRNA was not noticed after V. harveyi challenge, rather, there was a significant downregulation at 12 h and 48 h (Fig. 10B). A similar observation was noticed in the hepatopancreas tissue after V. harveyi challenge, with no noticeable induction of MrERGIC53 mRNA (Fig. 10C). In summary, the immune induction of MrERGIC-53 mRNA was confirmed only in the hemocytes post-challenge with the Gram-negative bacterium V. harveyi.
Fig. 9.
Temporal expression patterns of MrCTL mRNA in the hemocytes (A), stomach (B), and hepatopancreas (C) of M. rosenbergii challenged with V. harveyi. Total RNA was extracted from M. rosenbergii 6, 12, 24, and 48 h post-injection and profiled using qPCR. The mock control group of prawns received intramuscular injection (50 µL) of phosphate buffered saline (PBS, pH 7.4) and acted as mock (injected) control. β-actin (M. rosenbergii) was used as an internal control. Results of triplicate experiments are shown with standard errors. * P < 0.05, ** P < 0.01 (SAS, ANOVA)
Fig. 10.
Temporal expression patterns of MrERGIC-53 mRNA in the hemocytes (A), stomach (B), and hepatopancreas (C) of M. rosenbergii challenged with V. harveyi. Total RNA was extracted from M. rosenbergii after 6, 12, 24, and 48 h post-injection and profiled using qPCR. The mock control group of prawns received intramuscular injection (50 µL) of phosphate buffered saline (PBS, pH 7.4) and acted as mock (injected) control. β-actin (M. rosenbergii) was used as an internal control. Results of triplicate experiments have been shown with standard errors. * P < 0.05, ** P < 0.01 (SAS, ANOVA)
Discussion
Lectins are known to participate in the non-self-recognition process as immune surveillance molecules (Marques and Barracco 2000). The lectin family, as PRRs, is well-diversified in crustaceans, and many reports have suggested their promiscuous role in innate immunity and processes leading to pathogen evasion (reviewed by Cerenius et al. 2010; Denis et al. 2016; Sanchez-Salgado et al. 2017). Lectins have been studied from the perspective of prophenoloxidase activation, enhancement of encapsulation and nodulation of hemocytes, and opsonization (Sun et al. 2008). Lectins have also been implicated in the humoral innate immune mechanism where they assist the modulation of signaling cascades leading to the transcriptional activation of AMPs (Sanchez-Salgado et al. 2021). Considering the dynamism in innate immune recognition against a plethora of microorganisms, the lectin family of proteins has been characterized in economically important crustaceans, such as shrimps and prawns (Preetham et al. 2019). The overall health of shrimps and prawns in semi-intensive cultures and farming practices is compelling as it caters to the overall needs of marine shellfish-processing industries for export and revenue generation. In prawns and shrimps, lectins are regarded as bi-functional molecules involved in the sensing of pathogenic infections, where they possess potent antibacterial, antifungal, and antiviral properties, orchestrating an indispensable role in innate immunity (Sun et al. 2008; Wang et al. 2014; Mohanty et al. 2020). CTLs and LTL domain lectins have been at the forefront among the lectin family members contributing to host immune surveillance, thus serving as a biotherapeutant for shrimps or prawns (Wei et al. 2012; Wang et al. 2014; Huang et al. 2019). Therefore, the characterization of CTLs and LTL domain lectins from the freshwater prawn, M. rosenbergii, will enable us to unravel the function and specificity of this important component of the prawn immune system. Moreover, as information on the function of LTL in the innate immune system of M. rosenbergii is limited, we characterized a novel CTL (MrCTL) and LTL domain-containing protein (MrERGIC-53) from M. rosenbergii.
The LTL domain gene characterized for the first time from M. rosenbergii was designated as ER-Golgi intermediate compartment 53 kDa protein (MrERGIC-53). ERGIC-53 is mainly located in the ER and Golgi of cells and serves as a regulator of glycoprotein export (Hauri et al. 2000). As known for ERGIC-53 homologs in other invertebrate and vertebrate species (Zhang et al. 2009, 2012), MrERGIC-53 is a type I transmembrane protein, with an N-terminal signal sequence, a LTL domain (for carbohydrate recognition), a stalk region, and a short cytoplasmic region. The homolog of ERGIC-53 in humans, LMAN1 in complex with MCFD2 (lectin, mannose binding 1/multiple coagulation factor deficiency protein 2), forms a specific cargo receptor for the ER-to-Golgi transport of selected proteins (Zheng et al. 2010). Further, as reported earlier, LTL domain proteins are implicated in the modulation of innate immunity in hemocytes after Staphylococcus aureus, Vibrio parahaemolyticus, and WSSV infections (Huang et al. 2018). Therefore, we hypothesized that MrERGIC-53 could putatively regulate the pathogenic load in M. rosenbergii. Moreover, it has been reported that ERGIC-53 is likely candidate in the propagation of highly pathogenic RNA genome family viruses, such as arenavirus, hantavirus, coronavirus, and filovirus (Klaus et al. 2013). The hepatitis B virus (HBV) also exploits the functions of ERGIC-53 for its trafficking and egress, illustrating the ability to manipulate the viral life cycle for therapeutic purposes (Zeyen et al. 2020).
The MrERGIC-53 gene was found to comprise a 1,515 bp ORF, encoding a polypeptide of 504 amino acid residues. MrVIP36 is another LTL family protein identified from M. rosenbergii, comprising a 972 bp ORF and encoding a polypeptide of 323 amino acid residues (Huang et al. 2018). Further, the LvLTLC1 cDNA from L. vannamei was 1,184 bp (ORF of 990 bp, encoding a polypeptide of 329 amino acid residues), comprising a 17 bp 5′-UTR and a 177 bp 3′-UTR (Tian et al. 2018). The complete cDNA of PcL-lectin from the red swamp crayfish (P. clarkii) was 1,680 bp, with a 1,506 bp ORF, encoding a polypeptide of 501 amino acid residues. As observed in the case of MrERGIC-53, PcL-lectin contained a putative signal peptide of 19 amino acids, and one lectin leg-like domain located at the N-terminus, followed by a coiled-coil region and a transmembrane domain at the C-terminus (Dai et al. 2016). In the Chinese mitten crab, Eriocheir sinensis, two LTLs have been identified, namely, EsERGIC-53 and EsVIP36. While EsERGIC-53 cDNA was 1,955 bp, containing an ORF of 1,506 bp and encoding a putative protein of 501 amino acid residues, EsVIP36 comprised a 984 bp ORF, encoding a protein of 327 amino acid residues (Huang et al. 2014).
Multiple sequence alignment of MrERGIC-53 with LTL homologs (not shown) revealed the promiscuous presence of LTL and coiled-coil domains. ClustalX analysis demonstrated that the MrERGIC-53 amino acid sequence has high homology with EsERGIC-53 and PvMBL1 sharing 85% and 83% identity, respectively. In an earlier study, EsERGIC-53 was found to share 53–56% homology with other invertebrate LTL family homologs. Moreover, EsVIP-36 shared 80%, 78%, and 66% identity with P. clarkii LTL (PcLTL), Marsupenaeus japonicus LTL (MjLTL), and Nasonia vitripennis VIP36L (NvVIP36L), respectively (Huang et al. 2014). The phylogenetic analysis provided insights into the evolution of MrERGIC-53 with respect to LTL family homologs from other species. The ERGIC-53 sequences (including MrERGIC-53 and ERGIC-53 of other species) were clustered in a single clade. Previous phylogenetic analysis revealed the classification of VIP36 (36 kDa) and ERGIC-53 (53 kDa) into separate clades, originally identified in animals (Qin et al. 2012).
Most of the LTLs consist of two or four subunits (each 200–300 amino acid subunit is 25–30 kDa), each with a single, small carbohydrate combining site with the same specificity and a metal binding site for calcium and manganese (protein–ligand interaction site) (Gupta 2012). From a structural perspective, as revealed from the homology-based model, MrERGIC-53 was found to possess a conserved LTL domain (carbohydrate binding site) and a transmembrane region. Further, MrERGIC-53 possessed a protein–ligand interaction site (preferentially for binding calcium ions). The LTL superfamily domain of MrERGIC-53 is composed of beta-sheets connected by short loops and beta bands forming a dome-shaped beta-barrel carbohydrate recognition domain. The MrERGIC-53 Ca2+ binding domain is the globular tail domain which is putatively involved in the direct interaction with the carbohydrate. The structural features, phylogenetic tree analysis, and sequence alignment suggest that MrERGIC-53 is a member of the LTL superfamily.
To understand the putative role of MrERGIC-53 transcripts in host immunity, we challenged prawns with V. harveyi. The time-course gene expression analysis showed the regulation of MrERGIC-53 mRNA post-bacterial challenge, especially in the hemocytes. Previous studies have shown that LTLs play an important role in the sorting and transportation of mature glycoproteins in crustaceans, but their functions during infection are not well documented (Denis et al. 2017). In another context, our findings are also consistent with studies on MjLTL1, which is highly expressed in the hepatopancreas and upregulated following bacterial challenge (Xu et al. 2014). The results of qRT-PCR analysis showed that MrERGIC-53 mRNA is significantly expressed in hepatopancreas and hemocyte tissues relative to other tissues. MrVIP-36 mRNA is expressed in the intestine and hepatopancreas (Huang et al. 2018), while PcLTL mRNA is expressed in hemocytes, heart, and stomach (Dai et al. 2016). These findings suggest that diverse LTL domain proteins are ubiquitously distributed in crustaceans and that their tissue-specific and temporal distribution patterns are distinct.
CTLs (Ca2+-dependent sugar-recognition proteins) are probably the largest family of lectins, with multiple functions, including participation in cell adhesion, endocytosis, pathogen neutralization, glycoprotein clearance, and phagocytosis (Wang and Wang 2013; Dambuza and Brown 2015). The function of CTL family proteins is well conserved from invertebrates to vertebrates. Specifically, CTLs in shrimp were found to display agglutinating, antimicrobial, and anti-WSSV effects (Xiu et al. 2016). In this study, we identified a novel CTL homolog from the freshwater prawn, M. rosenbergii, designated as MrCTL. The identification of another MrCTL homolog in M. rosenbergii provides compelling evidence on the diversity of CTLs in prawns and shrimps. Furthermore, the high expression of MrCTL in gut-associated and immune tissues of M. rosenbergii challenged with V. harveyi may be related to the important roles of these tissues in immune responses. Notably, CTL domain proteins were downregulated in M. rosenbergii and in other shrimps after challenge with V. harveyi (Baliarsingh et al. 2020), V. parahaemolyticus (Rao et al. 2015), and WSSV infection (Peruzza et al. 2019) suggesting that they sabotage the internal defense mechanisms of the host under pathogenic stress. A hepatopancreas-specific lectin with a conserved CRD was identified in E. sinensis, which showed an antibacterial response by binding to lipopolysaccharide (LPS) (Jin et al. 2013). Moreover, the CTLs, SpLec1 and SpLec2 from the mud crab Scylla paramamosain, were highly expressed in hemocytes, muscle, and hepatopancreas (Jiang et al. 2012). The MrCTL transcript identified in this study was present in high-copy numbers in the hepatopancreas, stomach, and intestinal tissue of M. rosenbergii. Another study also reported the presence of CTL domain proteins in the hepatopancreas of Portunus trituberculatus and E. sinensis after challenge with V. alginolyticus and S. aureus, respectively, making the hepatopancreas a target for immune surveillance in the host (Huang et al. 2014; Zhang et al. 2018).
The full-length cDNA of MrCTL was 1,191 bp (ORF of 654 bp, encoding a polypeptide of 217 amino acid residues) with a polyadenylated tail. MrLec1, another CTL family protein from M. rosenbergii, comprised an ORF of 969 bp, encoding a polypeptide of 322 amino acid residues (Ren et al. 2012). Further, EsCTL1 cDNA from E. sinensis was 915 bp (ORF of 510 bp encoding polypeptide of 169 amino acid residues) containing a 111 bp 5′-UTR and a 294 bp 3′-UTR (Huang et al. 2014). The P. trituberculatus CTL4 (PtCTL4) comprised a 654 bp complete cDNA (480 bp ORF, encoding 159 amino acids) containing an 80 bp 5′-UTR and a 94 bp 3′-UTR with a poly(A) tail (Zhang et al. 2018). Consistent with the MrCTL ORF and deduced amino acid sequence obtained in this study, the puffer fish, Takifugu obscurus CTL1 (ToCTL1), comprised an ORF of 687 bp, encoding a polypeptide of 228 amino acids (Huang et al. 2020). Similar to CTL1, the MrCTL obtained in this study showed a single transmembrane domain and one typical CRD domain. These structural features were conserved among CTL homologs in vertebrates and invertebrates (Alenton et al. 2017; Du et al. 2018; Angulo et al. 2019; Huang et al. 2020). Within the CRD, the “EPN" and “QPD” motifs are conspicuously present in CTLs and bind to mannose and galactose, respectively. However, divergence of such motifs, or the existence of a new motif (for instance “QPD” to “QAP” motif), does not alter the specificity of the carbohydrate binding of CTLs (Alenton et al. 2017). The “QPD” motif was available in the deduced amino acid sequence of MrCTL obtained in this study, suggesting galactose recognition and non-self discrimination.
ClustalX analysis demonstrated that MrCTL shared high homology with MrLectin and MnCTL (76% and 62%, respectively). Another CTL protein from M. rosenbergii “MrLec” shared identities of 35%, 34%, 31%, and 31%, with CTLs from Maylandia zebra (MzCD, XP004540734), Oreochromis niloticus (OnCD, XP003453111), Procambarus clarkii (PcCTL), and Fenneropenaeus chinensis (FcCTL), respectively (Huang et al. 2016). Further, MrLec1, another CTLD-domain protein from M. rosenbergii, shared 28% identity with CTLs from F. merguiensis (FmLec) and 27% identity with FcLec5-1 and LvLec (Ren et al. 2012). The evolutionary relationship of the MrCTL amino acid sequence obtained in this study highlighted sequence conservation at the level of CRD. The phylogenetic tree accounted for the clustering of CTLs into distinct clades, one of which included the CTLsequences (M. rosenbergii CTLand CTLof other species) that were grouped together belonging to the CTLsuperfamily. The sequence features, phylogenetic tree analysis, and sequence alignment suggest MrCTL to be a CTLsuperfamily member. Structurally, most of the CTLs consist of one or more CRDs, which form a characteristic double-loop structure, disulfide bond positions, and metal-binding sites for calcium (protein–ligand interaction sites). Our secondary structure prediction analysis was useful to define an initial hypothesis regarding the local folds present in the MrCTL structural assessment. Further, each CRD of the CTLs contains 1–4 Ca2+ binding sites, in which the second Ca2+ binding site is associated with the sugar-binding specificity of CTLs. This is more so related to the presence of conserved motifs, such as the “WND,” “QPD,” or “EPN” motifs (Drickamer 1988; Zelensky and Gready 2005). In the MrCTL obtained in this study, a 3-mer sugar moiety of β-D-Galactose (GAL), 2-(Acetylamino)-2-Deoxy-α-D-Glucopyranose (NDG), and Alpha-L-Fucose (FUC) is predicted to bind to the protein molecule at Ca+ binding sites, such as CA2, CA3, and CA4. This study was crucial for addressing the structural and sequence-level features of a single CRD CTL family protein and an LTL domain containing protein MrERGIC-53 involved in the transport of cargo glycoproteins from the ER to ER-Golgi intermediate complexes. Both the proteins were shown to be immune-related.
Conclusion
This study was conducted to explore the innate immune defense molecules in the freshwater prawn, M. rosenbergii challenged by V. harveyi. Two novel lectin family proteins—a single CRD containing CTL (MrCTL) and an LTL domain containing ERGIC-53 (MrERGIC-53)—were successfully identified and characterized at the sequence and structural level. The mRNA producing MrCTL was predominantly distributed in the hepatopancreas and gut-associated tissues, such as the stomach and intestine, while MrERGIC-53 mRNA was found distributed in the hemocytes. Following bacterial challenge, the expression of MrCTL and MrERGIC-53 mRNA was upregulated in vivo. Collectively, the results indicated that MrCTL and MrERGIC-53 might be required for an effective innate immune response of the host against the bacteria.
Supplementary Information
Below is the link to the electronic supplementary material.
Author contribution
Snigdha Baliarsingh designed methodology, performed the experiments and data analysis, and wrote the original draft of the manuscript; Sonalina Sahoo performed the experiments and data analysis; Yong Hun Jo designed the experiments, conceptualized the data, and reviewed the draft manuscript; Yeon Soo Han conceptualized the data, discussed the results, and reviewed the draft manuscript; Arup Sarkar co-supervised the experiments and validated the data; Yong Seok Lee discussed the results and reviewed the draft manuscript; Jyotirmaya Mohanty co-supervised the experiments, designed methodology, and reviewed the draft manuscript; Bharat Bhusan Patnaik supervised the experiments and reviewed the draft manuscript to publication standards.
Funding
This work was supported by the Department of Biotechnology (DBT), Government of India, Grant No. BT/PR12710/10/AAQ/3/713/2015 under the “Aquaculture and Marine Biotechnology” Category.
Data availability
The full-length cDNA sequences of MrCTL and MrERGIC-53 are registered with GenBank, with accession IDs MK864100 and MK864099, respectively.
Declarations
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Alam MS, Rahman-Al-Mamun MA, Hossain MT, Yeasin M Hossain MM (2019) Mass larval mortality in a giant freshwater prawn Macrobrachium rosenbergii hatchery: an attempt to detect microbes in the berried and larvae. Bangladesh J Fisheries 31:41-48
- Alenton RRR, Koiwai K, Miyaguchi K, Kondo H Hirono I (2017) Pathogen recognition of a novel C-type lectin from Marsupenaeus japonicus reveal the divergent sugar-binding specificity of QAP motif. Sci Rep 7:45818. 10.1038/srep45818 [DOI] [PMC free article] [PubMed]
- Angulo C, Sanchez V, Delgado K Reyes-Becerril M (2019) C-type lectin 17A and macrophage-expressed receptor genes are magnified by fungal β-glucan after V. parahaemolyticus infection in Totoaba macdonaldi cells. Immunobiol 224:102-109. 10.1016/j.imbio.2018.10.003 [DOI] [PubMed]
- Aweya JJ, Zheng J, Zheng X, Yao D Zhang Y (2021) The expanding repertoire of immune-related molecules with antimicrobial activity in penaeid shrimps: a review. Rev Aquacult. 10.1111/raq.12551
- Baliarsingh S, Chung JM, Sahoo S, Sarkar A, Mohanty J, Han YS, Lee YS Patnaik BB (2020) Transcriptome analysis of Macrobrachium rosenbergii hepatopancreas in response to Vibrio harveyi infection. Aquacult Res 52:1855-1875. 10.1111/are.15034
- Basset C, Holton J, O' Mahony R, Roitt I (2003) Innate immunity and pathogen-host interaction. Vaccine 21:S12-S23. 10.1016/s0264-410x(03)00195-6 [DOI] [PubMed]
- Cerenius L, Jiravanichpaisal P, Liu HP, Soderhall I. Crustacean immunity. Adv Exp Med Biol. 2010;708:239–259. doi: 10.1007/978-1-4419-8059-5_13. [DOI] [PubMed] [Google Scholar]
- Dai Y, Wang Y, Zhao L, Qin Z, Yuan J, Qin Q, Lin L, Lan J. A novel L-type lectin was required for the multiplication of WSSV in red swamp crayfish (Procambarus clarkii) Fish Shellfish Immunol. 2016;55:48–55. doi: 10.1016/j.fsi.2016.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dambuza IM, Brown GD. C-type lectins in immunity; recent developments. Curr Opin Immunol. 2015;32:21–27. doi: 10.1016/j.coi.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis M, Thayappan K, Ramasamy SM, Munusamy A. Lectin in innate immunity of Crustacea. Austin Biol. 2016;1:1001. [Google Scholar]
- Denis M, Mullaivanam SR, Thayappan K, Munusamy A. Immune response of anti-lectin Pjlec antibody in freshwater crab Paratelphusa jacquemontii. Int J Biol Macromol. 2017;104:1212–1222. doi: 10.1016/ijbiomac.2017.07.034. [DOI] [PubMed] [Google Scholar]
- Dodd RB, Drickamer K. Lectin- like proteins in model organisms: implications for evolution of carbohydrate-binding activity. Glycobiol. 2001;11:71R–R79. doi: 10.1093/glycob/11.5.71r. [DOI] [PubMed] [Google Scholar]
- Drickamer K. Two distinct classes of carbohydrate-recognition domains in animal lectins. J Biol Chem. 1988;263:9557–9560. doi: 10.1016/S0021-9258(19)81549-1. [DOI] [PubMed] [Google Scholar]
- Du X, Wang GH, Su YL, Zhang M, Hu YH. Black rockfish C-type lectin, SsCTL4: a pattern recognition receptor that promotes bactericidal activity and virus escape from host immune defense. Fish Shellfish Immunol. 2018;79:340–350. doi: 10.1016/j.fsi.2018.05.033. [DOI] [PubMed] [Google Scholar]
- Ewart KV, Johnson SC, Ross NW. Lectins of the innate immune system and the irrelevance to fish health. ICES J Mar Sci. 2001;58:380–385. doi: 10.1006/jmsc.2000.1020. [DOI] [Google Scholar]
- Gangnonngiw W, Bunnontae M, Phiwsaiya M, Dhar SS, AK, In experimental challenge with infectious clones of Macrobrachium rosenbergii nodavirus (MrNV) and extra-small virus (XSV), MrNV alone can cause mortality in freshwater prawn (Macrobrachium rosenbergii) Virol. 2020;540:30–37. doi: 10.1016/j.virol.2019.11.004. [DOI] [PubMed] [Google Scholar]
- Gupta GS. Animal lectins: form, function, and clinical applications. Springer, Vienna. 2012 doi: 10.1007/978-3-7091-1065-2. [DOI] [Google Scholar]
- Hauri HP, Appenzeller C, Kuhn F Nufer O (2000) Lectins and traffic in the secretory pathway. FEBS Lett 476:32-37. 10.1016/S0014-5793(00)01665-3 [DOI] [PubMed]
- Huang X, Feng JL, Jin M, Ren Q, Wang W. C-type lectin (MrCTL) from the giant freshwater prawn, Macrobrachium rosenbergii participates in innate immunity. Fish Shellfish Immunol. 2016;58:136–144. doi: 10.1016/j.fsi.2016.08.006. [DOI] [PubMed] [Google Scholar]
- Huang X, Han K, Li T, Wang W, Ren Q (2018) Novel L-type lectin from freshwater prawn, Macrobrachium rosenbergii participates in antibacterial and antiviral immune responses. Fish Shellfish Immunol 77:304–311. j.fsi.2018.03.061 [DOI] [PubMed]
- Huang Y, Ren Q. Innate immune responses against viral pathogens in Macrobrachium. Dev Comp Immunol. 2021;117:103966. doi: 10.1016/j.dci.2020.103966. [DOI] [PubMed] [Google Scholar]
- Huang Y, Tan JM, Wang Z, Yin SW, Huang X, Wang W Ren Q (2014) Cloning and characterization of two different L-type lectin genes from the Chinese mitten crab Eriocheir sinensis. Dev Comp Immunol 46:255-266. 10.1016/j.dci.2014.04.015 [DOI] [PubMed]
- Huang Y, Shi Y, Hu S, Wu T Zhao Z (2020) Characterization and functional analysis of two transmembrane C-type lectins in obscure puffer (Takifugu obscurus). Front Immunol 11:436. 10.3389/fimmu.2020.00436 [DOI] [PMC free article] [PubMed]
- Huang Y, Zhang R, Gao T, Xu H, Wu T Ren Q (2019) 2-Transmembrane C-type lectin from Oriental River Prawn, Macrobrachium nipponense participates in antibacterial immune response. Fish Shellfish Immunol 91:58-67. 10.1016/j.fsi.2019.05.029 [DOI] [PubMed]
- Itin C, Roche AC, Monsigny M, Hauri HP. ERGIC-53 is a functional mannose-selective and calcium dependent human homologue of leguminous lectins. Mol Biol Cell. 1996;7:483–493. doi: 10.1091/mbc.7.3.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itin C, Schindler R, Hauri HP. Targeting of protein ERGIC-53 to the ER/ ERGIC/cis Golgi recycling pathway. J Cell Biol. 1995;131:57–67. doi: 10.1083/jcb.131.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janeway CA, Medzhitov R. Innate immune recognition. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- Jang JH, Shin HW, Lee JM, Lee HW, Kim EC, Park SH (2015) An overview of pathogen recognition receptors for innate immunity in dental pulp. Mediators of Inflamm 1-12. 10.1155/2015/794143 [DOI] [PMC free article] [PubMed]
- Jiang K, Zhang D, Zhang F, Sun M, Qi L, Zhang S, Qiao Z, Ma L. Isolation of the C-type lectin like-domain cDNAs from the mud crab, Scylla paramamosain ESTAMPADOR, 1949, and its expression profiles in various tissues during larval development, and under Vibrio challenge. Crustaceana. 2012;85:817–834. doi: 10.1163/156854012X650269. [DOI] [Google Scholar]
- Jin XK, Guo XN, Li S, Wu MH, Zhu YT, Yu AQ, Tan SJ, Li WW, Zhang P Wang Q (2013) Association of a hepatopancreas-specific C-type lectin with the antibacterial response of Eriocheir sinensis. PLoS One 8:e76132. 10.1371/journal.pone.0076132 [DOI] [PMC free article] [PubMed]
- Kamiya Y, Yamaguchi Y, Takahashi N, Arata Y, Kasai K, Ihara Y, Matsuo I, Ito Y, Yamamoto K, Kato K. Sugar-binding properties of VIP36, an intracellular animal lectin operating as a cargo receptor. J Biol Chem. 2005;280:37178–37182. doi: 10.1074/jbc.M505757200. [DOI] [PubMed] [Google Scholar]
- Klaus JP, Eisenhauer P, Russo J, Mason AB, Do D, King B, Taatjes D, Cornillez-Ty C, Boyson JE, Thali M, Zheng C, Liao L, Yates JR, III, Zhang B, Ballif BF, Botten JW. The intracellular cargo receptor ERGIC-53 is required for the production of infectious arenavirus, coronavirus, and filovirus particles. Cell Host Microbe. 2013;14:522–534. doi: 10.1016/j.chom.2013.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lis H, Sharon N. Lectins: carbohydrate-specific proteins that mediate cellular recognition. Chem Rev. 1998;98:637–674. doi: 10.1021/cr940413g. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Marques MRF, Barraco MA. Lectins as non-self recognition factors in crustaceans. Aquaculture. 2000;191:23–44. doi: 10.1016/s0044-8486(00)00417-8. [DOI] [Google Scholar]
- Miller RF, Bates JD, Svejcar TJ, Pierson FB, Eddleman LE. Biology, ecology, and management of western juniper (Juniperus occidentalis) Oregon State University Agricultural Experiment Station Technical Bulletin. 2005;152:82. [Google Scholar]
- Mohanty J, Sahoo S, Badhe MR, Pillai BR, Sahoo PK, Suryawanshi AK Patnaik, BB (2020) Lectin-like activity of hemocyanin in freshwater prawn, Macrobrachium rosenbergii. The Prot J 39:358-36510.1007/s10930-020-09912-1 [DOI] [PubMed]
- Nufer O, Mitrovic S, Hauri HP. Profile-based data base scanning for animal L-type lectins and characterization on of VIPL, a novel VIP36-like endoplasmic reticulum protein. J Biol Chem. 2003;278:15886–15896. doi: 10.1074/jbc.M211199200. [DOI] [PubMed] [Google Scholar]
- Pedraza ST, Betancur JG, Urcuqui-Inchima S. Viral recognition by the innate immune system: the role of pattern recognition receptors. Colomb Med. 2010;41:377–387. doi: 10.25100/cm.v41i4.731. [DOI] [Google Scholar]
- Peruzza S, Shekhar MS, Vinaya Kumar K, Swathi A, Karthic K, Hauton C Vijayan KK (2019) Temporal changes in transcriptome profile provides insights of White Spot Syndrome Virus infection in Litopenaeus vannamei. Sci Rep 9:13509. 10.1038/s41598-019-49836-0 [DOI] [PMC free article] [PubMed]
- Pillai D, Bonami JR. A review of the diseases of freshwater prawns with special focus on white tail disease of Macrobrachium rosenbergii. Aquacult Res. 2012;43:1029–1037. doi: 10.1111/j.1365-2109.2011.03061.x. [DOI] [Google Scholar]
- Preetham E, Rubeena AS, Vaseeharan B, Chaurasia MK, Arockiaraj J Olsen RE (2019) Anti-biofilm properties and immunological response of an immune molecule lectin isolated from shrimp Metapenaeus monoceros. Fish Shellfish Immunol 94:896-906. 10.1016/j.fsi.2019.09.032 [DOI] [PubMed]
- Qin SY, Kawasaki N, Hu D, Tozawa H, Matsumoto N Yamamoto K (2012) Subcellular localization of ERGIC-53 under endoplasmic reticulum stress conditions. Glycobiol 22:1709-1720. 10.1093/glycob/cws114 [DOI] [PubMed]
- Rao XJ, Shahzad T, Liu S, Wu P, He YT, Sun WJ, Fan XY, Yang YF, Shi Q Yu XQ (2015) Identification of C-type lectin-domain proteins (CTLDPs) in Silkworm, Bombyx mori. Dev Comp Immunol 53:328-338. 10.1016/j.dci.2015.07.005 [DOI] [PubMed]
- Ren Q, Li M, Du J, Zhang CY Wang W (2012) Immune response of four dual-CRD C-type lectins to microbial challenges in giant freshwater prawn, Macrobrachium rosenbergii. Fish Shellfish Immunol 33:155-167. 10.1016/j.fsi.2012.03.009 [DOI] [PubMed]
- Sahul Hameed AS, Xavier Charles M, Anilkumar M. Tolerance of Macrobrachium rosenbergii to white spot syndrome virus. Aquaculture. 2000;183:207–213. doi: 10.1016/S0044/8486(99)00305-1. [DOI] [Google Scholar]
- Sanchez-Salgado JL, Pereyra MA, Alpuche-Osorno JJ Zenteno E (2021) Pattern recognition receptors in the crustacean immune response against bacterial infections. Aquaculture 532:735998. 10.1016/j.aquaculture.2020.735998
- Sanchez-Salgado JL, Pereyra MA, Agundis C, Vivanco-Rojas O, Sierra-Castillo C, Alpuche-Osorno JJ, Zenteno E. Participation of lectins in crustacean immune system. Aquacult Res. 2017;48:4001–4011. doi: 10.1111/are.13394. [DOI] [Google Scholar]
- See LM, Tan SG, Hassan R, Siraj SS Bhassu S (2009) Development of microsatellite markers from an enriched genomic library for the genetic analysis of the Malaysian giant freshwater prawn, Macrobrachium rosenbergii. Biochem Genet 47:722-726. 10.1007/s.10528-009-9270-2 [DOI] [PubMed]
- Singrang N, Laophetsakunchai S, Tran BN, Matsudaira PT, Tassanakajon A Wangkanont K (2021) Biochemical and structural characterization of a recombinant fibrinogen-related lectin from Penaeus monodon. Sci Rep 11:2934. 10.1038/s41598-021-82301-5 [DOI] [PMC free article] [PubMed]
- Solovyev V, Kosarev P, Seledsov I Vorobyev D (2006) Automatic annotation of eukaryotic genes, pseudogenes, and promoters. Genome Biol 7:S10. 10.1186/gb-2006-7-s1-s10 [DOI] [PMC free article] [PubMed]
- Sun YD, Fu LD, Jia YP, Du XJ, Wang Q, Wang YH, Zhao XF, Yu XQ Wang JX (2008) A hepatopancreas-specific C-type lectin from the Chinese shrimp Fenneropenaeus chinensis exhibits antimicrobial activity. Mol Immunol 45:348-36110.1016/j.molimm.2007.06.355 [DOI] [PubMed]
- Tian, Y, Chen T, Huang W, Luo P, Huo D, Yun L, Hu C Cai Y (2018) A new L-type lectin (LvLTLC1) from the shrimp Litopenaeus vannamei facilitates the clearance of Vibrio harveyi. Fish Shellfish Immunol 73: 185–191. j.fsi.2017.12.011 [DOI] [PubMed]
- Wang XW, Wang JX. Diversity and multiple functions of lectins in shrimp immunity. Dev Comp Immunol. 2013;39:27–38. doi: 10.1016/j.dci.2012.04.009. [DOI] [PubMed] [Google Scholar]
- Wang XW, Xu YH, Xu JD, Zhao XF Wang JX (2014) Collaboration between a soluble C-type lectin and calreticulin facilitates white spot syndrome virus infection in shrimp. J Immunol 193:2106-2117. 10.4049/jimmunol.1400552 [DOI] [PubMed]
- Wei X, Liu X, Yang J, Fang J, Qiao H, Zhang Y Yang J (2012) Two C-type lectins from shrimp Litopenaeus vannamei that might be involved in immune response against bacteria and virus. Fish Shellfish Immunol 32:132-140. 10.1016/j.fsi.2011.11.001 [DOI] [PubMed]
- Weis WI, Kahn R, Fourme R, Drickamer K Hendrickson WA (1991) Structure of the calcium binding protein determined by MAD phasing. Science 254:1608–1615. 10.1126/science.172141 [DOI] [PubMed]
- Wigren E, Bourhis JM, Kursula I, Guy JE, Lindqvist Y. Crystal structure of the LMAN1-CRD/MCFD2 transport receptor complex provides insight into combined deficiency of factor V and factor VIII. FEBS Lett. 2010;584:878–882. doi: 10.1016/j.febslett.2010.02.009. [DOI] [PubMed] [Google Scholar]
- Xiu Y, Wang Y, Bi J, Liu Y, Ning M, Liu H, Li S, Gu W, Wang W Meng Q (2016) A novel C-type lectin is involved in the innate immunity of Marsupenaeus japonicus. Fish Shellfish Immunol 50:117-126. 10.1016/j.fsi.2016/01.026 [DOI] [PubMed]
- Xu S, Wang L, Wang XW, Zhao YR, Bi WJ, Zhao XF, Wang JX. L-type lectin from the kuruma shrimp Marsupenaeus japonicus promotes hemocyte phagocytosis. Dev Comp Immunol. 2014;44:397–405. doi: 10.1016/j.dci.2014.01.016. [DOI] [PubMed] [Google Scholar]
- Yaffe H, Buxdorf K, Shapira I, Ein-Gedi S, Moyal ben Zvi M, Fridman E, Moshelion M, Levy M (2012) LogSpin: a simple, economical and fast method for RNA isolation from infected or healthy plants and other eukaryotic tissues. BMC Res Notes 5: 45. 10.1186/1756-0500-5-45 [DOI] [PMC free article] [PubMed]
- Zelensky AN, Gready JE. The C-type lectin-like domain superfamily. FEBS J. 2005;272:6179–6217. doi: 10.1111/j1742-4658.2005.05031.x. [DOI] [PubMed] [Google Scholar]
- Zeyen L, Doring T Prange R (2020) Hepatitis B virus exploits ERGIC-53 in conjunction with COPII to exit cells. Cells 9:1889. 10.3390/cells9081889 [DOI] [PMC free article] [PubMed]
- Zhang H, Peatman E, Liu H, Feng T, Chen L Liu Z (2012) Molecular characterization of three L-type lectin genes from channel catfish, Ictalurus punctatus and their responses to Edwardsiella ictaluri challenge. Fish Shellfish Immunol 32:598-608. 10.1016/j.fsi.2011.12.009 [DOI] [PubMed]
- Zhang YX, Zhang ML Wang XW (2021) C-type lectin maintains the homeostasis of intestinal microbiota and mediates biofilm formation by intestinal bacteria in shrimp. J Immunol 206. 10.4049/jimmunol.2000116 [DOI] [PubMed]
- Zhang YC, Zhou Y, Yang CZ, Xiong DS (2009) A review of ERGIC-53: its structure, functions, regulation and relations with diseases. Histol Histopathol 24, 1193–1204. 10.14670/HH-24.1193 [DOI] [PubMed]
- Zheng C, Liu HH, Yuan S, Zhou J Zhang B (2010) Molecular basis of LMAN1 in coordinating LMAN1-MCFD2 cargo receptor formation and ER- to Golgi transport of FV/FVIII. Blood 116:5698-5706. 10.1182/blood-2010-04-278325 [DOI] [PMC free article] [PubMed]
- Zhang X, Lu J, Mu C, Li R, Song W, Ye Y, Shi C, Liu L Wang C (2018) Molecular cloning of a C-type lectin from Portunus trituberculatus, which might be involved in the innate immune response. Fish Shellfish Immunol. 76, 216-223. 10.1016/j.fsi.2018.01.051 [DOI] [PubMed]
- Zhu H, Du J, Hui KM, Liu P, Chen J, Xiu Y, Yao W, Wu T, Meng Q, Gu W, Ren Q, Wang W. Diversity of lectins in Macrobrachium rosenbergii and their expression patterns under Spiroplasma MR-1008 stimulation. Fish Shellfish Immunol. 2013;35:300–309. doi: 10.1016/j.fsi.2013.04.036. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The full-length cDNA sequences of MrCTL and MrERGIC-53 are registered with GenBank, with accession IDs MK864100 and MK864099, respectively.