Abstract
Since the beginning of the COVID-19 pandemic, SARS-CoV-2 has mutated several times into new strains, with an increased infectivity. Infectivity of SARS-CoV-2 strains depends on binding affinity of the virus to its host cell receptor. In this paper, we quantified the binding affinity using Gibbs energy of binding and analyzed the competition between SARS-CoV-2 strains as an interference phenomenon. Gibbs energies of binding were calculated for several SARS-SoV-2 strains, including Hu-1 (wild type), B.1.1.7 (alpha), B.1.351 (beta), P.1 (Gamma), B.1.36 and B.1.617 (Delta). The least negative Gibbs energy of binding is that of Hu-1 strain, -37.97 kJ/mol. On the other hand, the most negative Gibbs energy of binding is that of the Delta strain, -49.50 kJ/mol. We used the more negative Gibbs energy of binding to explain the increased infectivity of newer SARS-CoV-2 strains compared to the wild type. Gibbs energies of binding was found to decrease chronologically, with appearance of new strains. The ratio of Gibbs energies of binding of mutated strains and wild type was used to define a susceptibility coefficient, which is an indicator of viral interference, where a virus can prevent or partially inhibit infection with another virus.
Keywords: Virus-virus-host interactions, SARS-CoV-2 threat, Gibbs energy of binding, SARS CoV-2 delta strain, COVID-19
Introduction
In late 2019, a new infectious disease appeared in Wuhan, China, which later became known as COVID-19. Soon after, the disease agent was isolated, named SARS-CoV-2, and chemically (Popovic and Minceva, 2020; Popovic, 2022; Degueldre, 2021; Şimşek et al., 2021) and thermodynamically characterized (Popovic & Minceva, 2020, 2021). The virus spread, causing a pandemic. The original strain was labelled Hu-1 (Islam et al., 2020). The pandemic has been active for 2 years, during which the virus has mutated several times. The strains have been labelled Hu-1 (wild type – Wuhan), B.1.1.7 (UK), B.1.351 (South Africa), P.1 (Brazil), B.1.36 (India, Canada and UK) and B.1.617 (India). The old stains are continuously suppressed by new strains. Obviously, interference has been taking place between various strains of SARS-CoV-2. Newer stains possess mutations giving them an advantage for survival. These changes are, from the chemical perspective, reflected in increased binding affinity of the mutant strains (Barton et al., 2021).
Infection is the result of interaction between a pathogen and its host organism, representing not only biological, but also a chemical and thermodynamic interaction (Kumar et al., 2021; Kruse et al., 2012; Popovic and Minceva 2020a, 2020b, 2021). Thus, due to this complexity, understanding infection demands a novel platform at the interface of virology, immunology, genetics, epidemiology, physical chemistry and biothermodynamics. If two pathogens simultaneously circulate in a population, the interaction becomes even more complex, as the pathogens compete not only with their host, but also with each other. When two viruses meet in a single host, the interaction is complex and threefold: each virus interacts with the host and the two viruses interact mutually (Popovic and Minceva, 2021). All three interactions are competitive, since both viruses and the host compete for a limited metabolic machinery and resources (i.e. amino-acids, nucleotides, energy sources etc.) (Popovic and Minceva 2020a, 2020b). Thus, the competition for resources represents the main mechanism of these interactions. The outcome of virus-virus-host interactions depends on two properties: susceptibility and permissiveness, both of which influence infectivity (Popovic and Minceva 2021). Susceptibility is the ability of a virus to enter the host cell (Duponchel and Fischer, 2019). On the other hand, once inside, permissiveness is the ability of a virus to multiply in the host cell (Duponchel and Fischer, 2019, Hou et al., 2017).
Infectivity is the ability of a pathogen to infect the host cell or organism. Infectivity varies between virus species. However, infectivity of SARS-CoV-2 differs between various strains of the same virus. Usually similar viruses do not show large differences in infectivity and perform coinfection (Popovic and Minceva 2021; Nickbakhsh et al., 2019). The infectivity of mutated strains of SARS-CoV-2 increases from the Wild type (Wuhan) to the new Delta strains (Thomas, 2021; Ramesh et al., 2021). What is the molecular basis for this trend? The evolutionary explanation states that this is the consequence of the tendency towards better adaptation to the host. This is the macroscopic explanation for the increased infectivity of the mutant strains. On the other hand, the microscopic explanation is based on changes on the receptor binding domain (RBD), making the antigen receptor binding more efficient. In particular, increased infectivity appears as the result of a decrease in the dissociation constant KD and thereby increased affinity of antigen for the receptor KB. All these parameters influence the susceptibility. On the other hand, infectivity also depends on permissiveness – the ability of the virus or its strain to multiply and thereby increase the size of its reservoir. Thus, both susceptibility and permissiveness influence infectivity (Popovic and Minceva, 2021; Sevenich et al., 2021). However, what is the driving force for these changes in infectivity? A tentative explanation comes from biothermodynamics.
Huge numbers of global SARS-CoV-2 infections and insufficient discipline in conducting anti-epidemic measures have resulted in the emergence of new virus variants, starting from original Wuhan, through the Alpha (B.1.1.7 UK), Beta (B.1.351 S. Africa), Gamma (P.1 Brazil), Epsilon (B.1.429 California), Iota (B.1.526 New York), and more recently, Delta and Kappa (B.1.617.2 and B.1.617.1 India) (Farinholt et al., 2021). Each of these strains gained advantageous mutations to become a dominant strain. Domination of one virus over other virus, or one strain over other strain of the same virus is the result of interference with other (less advantageous) strains of SARS CoV-2 virus and replacement of older with a new strain. Viral interference is a phenomenon where a virus can prevent or partially inhibit infection with another virus within the same host (Schultz-Cherry, 2015; Wu et al., 2020; Dianzani, 1975).
A strain becomes dominant through competitive interaction with other strains, in the host population (Popovic and Minceva 2021). Obviously, an acquired mutation gives an advantage to one strain, enabling it to dominate. In extreme cases, domination leads to exclusion (Popovic and Minceva 2021). For example, the Iota strain was first discovered November 2020, and through interference came to represent 45% of new cases in February 2021 (Farinholt et al., 2021). Moreover, dual SARS-CoV-2 infection has been studied with two phylogenetically distant strains. The initially dominant strain belonged to GH clade and was suppressed by another strain from the GR clade, in only eight days (Samoilov et al., 2021). The initial ratio of the GH to GR strain was 70:30, while 8 days later it became 3:97, obviously as a result of interference (Samoilov et al., 2021). Thus, we have observed interference between various strains of SARS-CoV-2 (Korber et al., 2020). In that case a question is raised about the mechanism and driving force for this interference.
Gibbs energy represents the driving force of all chemical and physical processes in nature (von Stockar, 2013b; Demirel, 2014; Atkins and de Paula, 2011). Gibbs energy is important because it can be used to estimate the spontaneity and rate of a chemical process (von Stockar, 2013b; Demirel, 2014; Atkins and de Paula, 2011). One such process is antigen-receptor binding (Gale, 2020, 2019). Thus, Gibbs energy of binding is a very significant property of SARS-CoV-2 (Ngo et al., 2021).
The aim of this paper is to explore the mechanism and the driving force of interference between various strains of SARS-CoV-2 viruses. A thermodynamic approach will be used because the thermodynamic property, namely Gibbs energy represents the driving force for chemical reactions performed by viruses (von Stockar, 2013a; Von Stockar, 2013b; Popovic and Minceva, 2021, 2020a, 2020b; Şimşek et al., 2021), enabling susceptibility and permissiveness. Virus-receptor binding and replication, transcription, translation and self-assembly are essentially chemical reactions driven by Gibbs energy. Faster virus multiplication leads to increase in the size of infective reservoir, causing the increase in infectivity. Influence of increased susceptibility has already been described in the literature (Ozono et al., 2021; Hasegawa et al., 2007).
Methods
A thermodynamic analysis is used to calculate Gibbs energies of binding of various strains of SARS-CoV-2. Gibbs energies of competing pairs of strains will be compared, to explain the phenomenon of interference of various pairs of virus strains.
Antigen-receptor binding represents a chemical process (Popovic and Minceva, 2021). The rate of this process is given by the phenomenological equation
| (1) |
Where rB is the rate of antigen-receptor binding, L a constant (known as phenomenological coefficient), T temperature and ΔBG Gibbs energy of antigen-receptor binding (Popovic and Minceva, 2021; von Stockar, 2013a; Demirel, 2014; Hellingwerf et al., 1982; Westerhoff et al., 1982). The rate of binding is proportional to the absolute value of the Gibbs energy of binding. It is well documented that mutations lead to change in Gibbs energy of binding (Barton et al., 2021). The ability of coronaviruses to infect humans is invariably associated with their binding strengths to human receptor proteins (Zou et al., 2020). Mutation induces significant conformational transitions in the spike glycoprotein (Istifli et al., 2021). Natural selection promotes mutations that increase the spike ACE2 binding affinity (Istifli et al., 2021). Gibbs energy of binding can be calculated from dissociation constants, through the equation
| (2) |
Where R is the universal gas constant and T temperature (Popovic and Minceva, 2021; Du et al., 2016). KB represents the binding constant, which is the reciprocal of the dissociation constant KD (Du et al., 2016).
| (3) |
The dissociation constant is the equilibrium constant of the dissociation reaction of the antigen-receptor complex into the receptor and antigen
| (4) |
where R represents the host cell receptor (ACE2), A the virus antigen (spike protein) and RA the receptor antigen complex (Du et al., 2016). Thus, KD is defined as the ratio of concentrations of the free receptor [R] and antigen [A] to the receptor antigen complex [RA] (Du et al., 2016)
| (5a) |
However, KD, can also be defined through kinetic parameters of the reaction: the association, kon, and dissociation, koff, rate constants.
| (5b) |
Dissociation constants of various SARS-CoV-2 strains have been reported by (Augusto et al., 2021; Barton et al., 2021; Ramanathan et al., 2021) and are given in Table 1 . The reported data are for binding between a single antigen (SGP trimer) and a single receptor (ACE2).
Table 1.
Gibbs energy of binding of SARS-CoV-2 strains. The values of the binding constant, KB, and Gibbs energy of binding, ΔBG, values were calculated using KD values from the literature. Mutations led to increase in binding affinity and decrease in Gibbs energy of binding, implying greater spontaneity of binding of mutated strains. More negative Gibbs energy makes the process of antigen-receptor binding more favorable. The values have been calculated at 37⁰C (310.15 K).
| Date of isolation | PANGO lineage | WHO label | First outbreak | Mutations | KD (M) | Reference | KB (M − 1) | ΔbG (kJ/mol) |
|---|---|---|---|---|---|---|---|---|
| Dec-19 | Hu-1 | Wild type | Wuhan | Wild type | 2.13E-08 | Augusto et al., 2021 | 4.69E+07 | −45.55 |
| Dec-19 | Hu-1 | Wild type | Wuhan | Wild type | 4.03E-07 | Ramanathan et al., 2021 | 2.48E+06 | −37.97 |
| 26-Jan-21 | B.1.1.7 | Alpha | United Kingdom | 8.76E-08 | Ramanathan et al., 2021 | 1.14E+07 | −41.90 | |
| May-20 | B.1.351 | Beta | South Africa | 2.04E-07 | Ramanathan et al., 2021 | 4.90E+06 | −39.72 | |
| May-20 | B.1.351 and P.1 | Beta and Gamma | South Africa and Brazil | E484K | 1.97E-08 | Augusto et al., 2021 | 5.08E+07 | −45.75 |
| B.1.36 | / | India, Canada, and UK | N440K | 9.90E-09 | Augusto et al., 2021 | 1.01E+08 | −47.53 | |
| 10/1/2020 | B.1.617 | Delta | India | L452R/E484Q | 4.60E-09 | Augusto et al., 2021 | 2.17E+08 | −49.50 |
| Dec-19 | Hu-1 | Wild type | Wuhan | WT | 6.26E-08 | Barton et al., 2021 | 1.60E+07 | −42.77 |
| 18-Dec-20 | B.1.1.7 | Alpha | United Kingdom | N501Y (Alpha) | 5.5E-09 | Barton et al., 2021 | 1.82E+08 | −49.04 |
| 26-Jan-21 | B.1.1.7 | Alpha | United Kingdom | E484K/N501Y (UK2) | 3.7E-09 | Barton et al., 2021 | 2.70E+08 | −50.06 |
| 14-Jan-21 | B.1.351 | Beta | South Africa | K417N | 3.49E-07 | Barton et al., 2021 | 2.87E+06 | −38.34 |
| 14-Jan-21 | B.1.351 | Beta | South Africa | K417N/E484K | 2.51E-07 | Barton et al., 2021 | 3.98E+06 | −39.19 |
| 14-Jan-21 | B.1.351 | Beta | South Africa | K417N/E484K/N501Y (Beta) | 1.74E-08 | Barton et al., 2021 | 5.75E+07 | −46.07 |
| Nov-20 | P.1 | Gamma | Brazil | K417T | 2.26E-07 | Barton et al., 2021 | 4.42E+06 | −39.46 |
| Nov-20 | P.1 | Gamma | Brazil | K417T/E484K | 1.47E-07 | Barton et al., 2021 | 6.80E+06 | −40.57 |
| Nov-20 | P.1 | Gamma | Brazil | K417T/E484K/N501Y (Gamma) | 1.22E-08 | Barton et al., 2021 | 8.20E+07 | −46.99 |
The binding constant, KB, was calculated using Eq. (3), from KD. The dissociation constant refers to reaction (4), between a single antigen (SGP trimer) and a single receptor (ACE2). Thus, the calculated KB is for a single antigen-receptor interaction. This might not be the case if there are multiple SGP-trimer/ACE2 interactions, during virus binding (Gale, 2021). In that case, KB for the entire virus might be much greater than that for a single SGP trimer/ACE2 binding (Gale, 2021). Moreover, KB will be reduced if entropy decreases during whole-virus binding (Gale, 2021). However, human coronaviruses share a very similar size and structure (Neuman and Buchmeier, 2016). Thus, an assumption can be made that the number of SGP/ACE2 interactions is the same for all SARS-CoV-2 strains and that the entropy change on virus binding is the same for each strain (Gale, 2021). In that case, KB for the SGP/ACE2 interaction is proportional to, or is at least an indication of, the relative magnitudes of KB for the whole virus (Gale, 2021). This is because the ΔBG values are additive for each SGP/ACE2 interaction (Gale, 2021), meaning that multiple SGP/ACE2 interactions will not change the conclusions of the paper.
Finally, we will introduce a new property – susceptibility coefficient Ƨ, which is defined as the ratio of rates of binding of two viruses or in this case a mutant strain and wild type virus.
| (6) |
Combining this equation with the phenomenological Eq. (1) gives
| (7) |
The L coefficient and T are equal for both strains, since both attack the same host at the same physiological temperature. The phenomenological coefficient is proportional to the equilibrium forward and backward half-reaction rate (Demirel, 2014). These in turn depend on the forward and backward the association, kon, and dissociation, koff, rate constants (Du et al., 2016). Since kon, and koff vary between different virus strains (Barton et al., 2021), it might be possible that L differs between virus strains. This will be a subject of our future research.
Results
Based on dissociation constants, Gibbs energies of binding were calculated for various SARS-CoV-2 strains. The results are presented in Table 1. The analyzed SARS-CoV-2 strains include Hu-1 (wild type), B.1.1.7 (alpha), B.1.351 (beta), P.1 (Gamma), B.1.36 and B.1.617 (Delta). The original Hu-1 strain has been designated as wild type and was isolated in Wuhan, China in December 2019. The strain B.1.1.7 has been designated Alpha and was first isolated in the United Kingdom in late January 2021. The strain B.1.351 was named Beta, isolated in South Africa in May 2020. The strain P.1. was labeled Gamma and was first isolated Brazil in November 2020. The strain B.1.36 has been initially reported from India, Canada, and UK (Basheer and Zahoor, 2021). Finally, the strain B.1.617 has been labeled Delta and was first isolated in India, in October 2020.
The analyzed strains exhibit a variation in binding constants and hence Gibbs energies of binding according to Eq. (2). The B.1.1.7 (Alpha) has the most negative Gibbs energy of binding, of −50.06 kJ/mol. On the other hand, the least negative Gibbs energy of binding is that of Hu-1 strain, −37.97 kJ/mol. Moreover, Gibbs energies of binding decrease chronologically, with appearance of new strains. Similarly, the binding constants span the range between 2.48 ∙ 106 M and 2.70 ∙ 108 M. All the data in Table 1 has been calculated at the physiological temperature of 37⁰C (310.15 K).
Discussion
We hypothesize that mutations that appeared during time have led to increase in binding constant and more negative Gibbs energy of binding. According to the phenomenological Eq. (1), this leads to a greater binding rate, which in turn leads to more rapid cell entry and multiplication of one of the strains. Finally, this results in a greater infectious reservoir and greater infectivity.
Virus-virus interactions influence the epidemiology of respiratory infections (Dee et al., 2021; Popovic and Minceva 2021). After the initial development of the pandemic caused by the Hu-1 wild type strain, mutations have developed with time, in various countries. Chronologically, pairs of strains compete for resources, through indirect interaction of strain pairs (Popovic and Minceva, 2021). Thus, the discussion will be represented as an arena, where the strains compete. In the beginning, the Hu-1 strain spread across the planet and had no competition, since no other strains existed. With the appearance of the Alpha and other strains, except for the direct interaction with the host, an interaction between two strains of SARS-CoV-2 occurred.
Wild type vs Alpha (Hu-1 vs B.1.1.7)
The Hu-1 strain is characterized by a Gibbs energy of binding of −37.97 kJ/mol. Simultaneously, the mutated B.1.1.7 strain is characterized by a Gibbs energy of binding of −41.90 kJ/mol. Several authors reported various values of dissociation constants for the same strain. Thus, several values can be found in the table. Having in mind that Gibbs energy of the B.1.1.7 strain is more negative, one can conclude that the binding rate for this strain will be greater. Thus, during competition, the strain B.1.1.7 will enter the host cell faster and thereby gain an advantage while hijacking the host cell metabolism. This advantage will lead to suppression of the Hu-1 strain.
Wild type vs. Beta (Hu-1 vs B.1.351)
In South Africa, simultaneously in circulation, appeared the Wild type and Beta strains. From epidemiological studies, it is known that there had been interference and the Wild type strain was suppressed (Korber et al., 2020). Thus, we can expect that Gibbs energy of the Beta strain will be more negative than that of the Wild type. Indeed, Gibbs energy of binding of the Wild type is −37.97 kJ/mol, while that of the Beta strain is −39.72 kJ/mol. Like in the previous case, due to more negative Gibbs energy of binding, the Beta strain has an advantage to enter the host cell faster and hence multiplies faster and increases the infectious reservoir, making Beta strain more infective.
Wild type vs Gamma (Hu-1 vs P.1)
In Brazil, simultaneously in circulation, appeared the Wild type and Gamma strains. The Wild type has a Gibbs energy of binding is −37.97 kJ/mol. The Gamma strain has a Gibbs energy of binding of −39.46 kJ/mol. Since the Gamma strain has a more negative Gibbs energy of binding and greater affinity for the receptor, it enters the host cell and multiplies faster, leading to interference and suppression of the Hu-1 strain.
Wild type vs Alpha vs Delta (Hu-1 vs B.1.1.7 vs B.1.617)
In 2021, in Europe, three strains met simultaneously. Through the same competition mechanism, the strain with the most negative Gibbs energy of binding will have the greatest rate of binding, enter the cell the fastest and multiply the most rapidly. This leads to the interference phenomenon, resulting in suppression of the two other strains. Indeed, the Wild type has a Gibbs energy of −37.97 kJ/mol, the Alpha strain has a Gibbs energy of binding of −41.90 kJ/mol, while the for the Delta strain it is −49.50 kJ/mol. The Delta strain has the most negative Gibbs energy of binding. Thus, it was able to suppress the other two strains. Notice that every wave of the pandemic has been followed by appearance of a new strain. Every new strain had a greater infectivity and hence the number of infected grew with each wave of the pandemic. Newly emerging variants of SARS-CoV-2 have contributed to successive waves of COVID-19. The new variants increased disease severity and viral transmissibility, thus increasing the morbidity and mortality of COVID-19 (Thomas, 2021; Ramesh et al., 2021). The increase in wave size is the consequence of faster entry of the virus into its host cells and multiplication inside the cell.
The anti-epidemic measures of wearing protective masks, distancing, isolation of the diseased and their contacts, as well as vaccination, are making a selective pressure on the virus. All these measures attempt to decrease the infectious reservoir and the possibility of its transmission. The virus reacts to this selective pressure by evolving towards greater infectivity and transmission rate. This is reflected in increase in binding affinity and decrease in Gibbs energy of binding. The strains are not fighting each other, but each strain tends to develop a more efficient mechanism for its survival. Natural selection can act upon rare but favorable mutations (Korber et al., 2020). This unfortunately means that the fight against SARS-CoV-2 will have to continue with development of new vaccines and medicines, adapted to new strains.
Competition of SARS-CoV-2 strains greatly depends on susceptibility, the ability of the strains to infect host cells. A question is raised of how to compare susceptibility of two strains? It seems that the susceptibility coefficient, defined by Eq. (7) is a good parameter, including the ratio of driving forces for the SGP-trimer/ACE2 binding reaction, which influence the binding rates. Thus, the susceptibility coefficient represents the ratio of SGP-trimer/ACE2 binding rates.
The susceptibility coefficient shows how much binding of one strain is faster than that of another strain. The susceptibility coefficient is greater than unity if the Gibbs energy of binding of the mutant strain is more negative. If the wild type has a more negative Gibbs energy, the susceptibility coefficient will be lower than unity. In this way, one can explain why some mutations give an advantage to the mutant and lead to suppression of the wild type. However, most randomly occurring mutations do not give the advantage to the mutant and is suppressed by the wild type. As was shown here, Gibbs energies of binding have more negative values for the Alpha, Beta, Gamma and Delta mutants. They hence have an advantage and are able to suppress the wild type. On the other hand, most other mutations have not led to a more negative Gibbs energy and such strains have disappeared from the population.
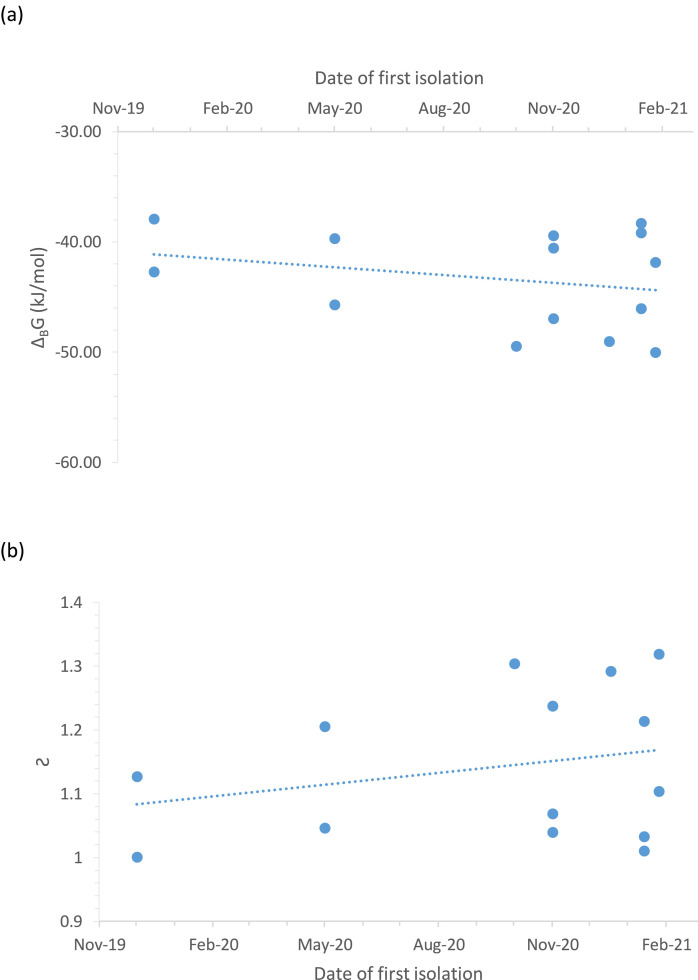
Fig. 1a shows Gibbs energies of binding of various SARS-CoV-2 strains through time, from the appearance on the wild type in late 2019 until 2021. Gibbs energy of binding decreases with the appearance of every new strain, which is in agreement with the observation that a selective pressure exists towards greater infectivity. Fig. 1b shows that the susceptibility coefficient increases, indicating greater entry rate of the mutant strains into the host cells. This leads to increased infectivity.
Fig. 1.
Evolution of Gibbs energy of binding and susceptibility coefficient of SARS-CoV-2. (a) The SARS-CoV-2 mutant strains are characterized by a more negative Gibbs energy of binding compared to the wild type. (b) The susceptibility coefficient has been increasing with new mutations on the SARS-CoV-2 virus.
Conclusion
The basic mechanism of the interaction between various strains of SARS-CoV-2 is competition. The competition is reflected at the susceptibility and permissiveness levels. This paper analyzes only susceptibility, since only data on binding constants have been available in the literature. It would be possible to determine the permissiveness for various strains, if their elemental composition were known. This is unfortunately currently not the case. Since the counter-epidemic measures are making a selective pressure on the virus, the virus evolves towards an increased binding affinity and more negative Gibbs energy. This has been shown by the data given in Table 1. Gibbs energies of binding decrease chronologically, with appearance of new strains. The Delta strain has the most negative Gibbs energy of binding. SARS-CoV-2 is also expected to evolve towards a changed permissiveness, also through mutations on other parts of the viral nucleic acid. The test of this hypothesis can be made in the future, once the elemental composition of the SARS-CoV-2 strains is known.
Conflict of interest statement
The authors declare no conflict of interest.
Literature
- Şimşek B., Özilgen M., Utku F.Ş. How much energy is stored in SARS-CoV-2 and its structural elements? Energy Storage. 2021:e298. doi: 10.1002/est2.298. [DOI] [Google Scholar]
- Atkins, P.W., & de Paula, J. (2011). Physical Chemistry for the Life Sciences (2nd Edition), W. H. Freeman and Company.
- Augusto G., Mohsen M.O., Zinkhan S., Liu X., Vogel M., Bachmann M.F. In vitro data suggest that Indian delta variant B.1.617 of SARS-CoV-2 escapes neutralization by both receptor affinity and immune evasion. Allergy. 2021 doi: 10.1111/all.15065. 10.1111/all.15065Advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton, M.I., MacGowan, S.A., Kutuzov, M.A., Dushek, O., Barton, G.J., & van der Merwe, P.A. (2021). Effects of common mutations in the SARS-CoV-2 Spike RBD and its ligand, the human ACE2 receptor on binding affinity and kinetics. eLife, 10, e70658. https://doi.org/10.7554/eLife.70658. [DOI] [PMC free article] [PubMed]
- Basheer A., Zahoor I. Genomic epidemiology of SARS-CoV-2 divulge B. 1, B. 1.36, and B. 1.1. 7 as the most dominant lineages in first, second, and third wave of SARS-CoV-2 infections in Pakistan. medRxiv. 2021 doi: 10.1101/2021.07.28.21261233. 2021.07.28.21261233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dee Kieran, Goldfarb Daniel M, Haney Joanne, Amat Julien A R, Herder Vanessa, Stewart Meredith, Szemiel Agnieszka M, Baguelin Marc, Murcia Pablo R. Human rhinovirus infection blocks severe acute respiratory syndrome coronavirus 2 replication within the respiratory epithelium: implications for COVID-19 epidemiology. J. Infect. Dis. 2021;224(1):31–38. doi: 10.1093/infdis/jiab147. 1 July. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degueldre C. Single virus inductively coupled plasma mass spectroscopy analysis: A comprehensive study. Talanta. 2021;228 doi: 10.1016/j.talanta.2021.122211. [DOI] [PubMed] [Google Scholar]
- Demirel, Y. (2014). Nonequilibrium Thermodynamics: Transport and Rate Processes in Physical, Chemical and Biological Systems, 3rd ed. Amsterdam: Elsevier.
- Dianzani F. Viral interference and interferon. Ric. Clin. Lab. 1975;5(3):196–213. doi: 10.1007/BF02908284. [DOI] [PubMed] [Google Scholar]
- Du X., Li Y., Xia Y.-.L., Ai S.-.M., Liang J., Sang P., Ji X.-.L., et al. Insights into Protein–Ligand Interactions: Mechanisms, Models, and Methods. Int. J. Mol. Sci. 2016;17(2):144. doi: 10.3390/ijms17020144. MDPI AG. Retrieved from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duponchel S., Fischer M.G. Viva lavidaviruses! Five features of virophages that parasitize giant DNA viruses. PLoS Pathog. 2019;15(3) doi: 10.1371/journal.ppat.1007592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farinholt T., Doddapaneni H., Qin X., Menon V., Meng Q., Metcalf G., Chao H., Gingras M.C., Avadhanula V., Farinholt P., Agrawal C., Muzny D.M., Piedra P.A., Gibbs R.A., Petrosino J. Transmission event of SARS-CoV-2 delta variant reveals multiple vaccine breakthrough infections. BMC Med. 2021;19(1):255. doi: 10.1186/s12916-021-02103-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale P. Towards a thermodynamic mechanistic model for the effect of temperature on arthropod vector competence for transmission of arboviruses. Microb. Risk Anal. 2019;12:27–43. doi: 10.1016/j.mran.2019.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale P. How virus size and attachment parameters affect the temperature sensitivity of virus binding to host cells: Predictions of a thermodynamic model for arboviruses and HIV. Microb. Risk Anal. 2020;15 doi: 10.1016/j.mran.2020.100104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale P. Using thermodynamic equilibrium models to predict the effect of antiviral agents on infectivity: Theoretical application to SARS-CoV-2 and other viruses. Microb. Risk Anal. 2021 doi: 10.1016/j.mran.2021.100198. Advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa K., Hu C., Nakamura T., Marks J.D., Russell S.J., Peng K.W. Affinity thresholds for membrane fusion triggering by viral glycoproteins. J. Virol. 2007;81(23):13149–13157. doi: 10.1128/JVI.01415-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellingwerf K.J., Lolkema J.S., Otto R., Neijssel O.M., Stouthamer A.H., Harder W., ... &, Westerhoff H.V. Energetics of microbial growth: an analysis of the relationship between growth and its mechanistic basis by mosaic non-equilibrium thermodynamics. FEMS Microbiol. Lett. 1982;15(1):7–17. doi: 10.1111/j.1574-6968.1982.tb00028.x. [DOI] [Google Scholar]
- Hou W., Armstrong N., Obwolo L.A., et al. Determination of the Cell Permissiveness Spectrum, Mode of RNA Replication, and RNA-Protein Interaction of Zika Virus. BMC Infect. Dis. 2017;17:239. doi: 10.1186/s12879-017-2338-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam M.R., Hoque M.N., Rahman M.S., Alam A., Akther M., Puspo J.A., Akter S., Sultana M., Crandall K.A., Hossain M.A. Genome-wide analysis of SARS-CoV-2 virus strains circulating worldwide implicates heterogeneity. Sci. Rep. 2020;10(1):14004. doi: 10.1038/s41598-020-70812-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Istifli E.S., Netz P.A., Sihoglu Tepe A., Sarikurkcu C., Tepe B. Understanding the molecular interaction of SARS-CoV-2 spike mutants with ACE2 (angiotensin converting enzyme 2) J. Biomol. Struct. Dyn. 2021:1–12. doi: 10.1080/07391102.2021.1975569. Advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182(4) doi: 10.1016/j.cell.2020.06.043. 812-827.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse H., Goerigk L., Grimme S. Why the standard B3LYP/6-31G* model chemistry should not be used in DFT calculations of molecular thermochemistry: understanding and correcting the problem. J. Org. Chem. 2012;77:10824–10834. doi: 10.1021/jo302156p. [DOI] [PubMed] [Google Scholar]
- Kumar P., Sobhanan J., Takano Y., et al. Molecular recognition in the infection, replication, and transmission of COVID-19-causing SARS-CoV-2: an emerging interface of infectious disease, biological chemistry, and nanoscience. NPG Asia Mater. 2021;13:14. doi: 10.1038/s41427-020-00275-8. [DOI] [Google Scholar]
- Neuman B.W., Buchmeier M.J. Supramolecular architecture of the coronavirus particle. Adv. Virus Res. 2016;96:1–27. doi: 10.1016/bs.aivir.2016.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo S.T., Tam N.M., Pham M.Q., Nguyen T.H. Benchmark of popular free energy approaches revealing the inhibitors binding to SARS-CoV-2 Mpro. J. Chem. Inf. Model. 2021;61(5):2302–2312. doi: 10.1021/acs.jcim.1c00159. [DOI] [PubMed] [Google Scholar]
- Nickbakhsh, S.; Mair, C.; Matthews, L.; Reeve, R.; Johnson, P.; Thorburn, F.; von Wissmann, B.; Reynolds, A.; McMenamin, J.; Gunson, R.N.; et al. Virus-virus interactions impact the population dynamics of influenza and the common cold. Proc. Natl. Acad. Sci. USA 2019, 116, 27142–27150. Advance online publication. https://doi.org/10.1073/pnas.1911083116. (1) (PDF) Coinfection and Interference Phenomena Are the Results of Multiple Thermodynamic Competitive Interactions. Available from: https://www.researchgate.net/publication/354956185_Coinfection_and_Interference_Phenomena_Are_the_Results_of_Multiple_Thermodynamic_Competitive_Interactions [accessed Nov 22 2021]. [DOI] [PMC free article] [PubMed]
- Ozono S., Zhang Y., Ode H., Sano K., Tan T.S., Imai K., Miyoshi K., Kishigami S., Ueno T., Iwatani Y., Suzuki T., Tokunaga K. SARS-CoV-2 D614G spike mutation increases entry efficiency with enhanced ACE2-binding affinity. Nat. Commun. 2021;12(1):848. doi: 10.1038/s41467-021-21118-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovic M., Minceva M. A thermodynamic insight into viral infections: do viruses in a lytic cycle hijack cell metabolism due to their low Gibbs energy? Heliyon. 2020;6(5):e03933. doi: 10.1016/j.heliyon.2020.e03933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovic M., Minceva M. Thermodynamic insight into viral infections 2: empirical formulas, molecular compositions and thermodynamic properties of SARS, MERS and SARS-CoV-2 (COVID-19) viruses. Heliyon. 2020;6(9):e04943. doi: 10.1016/j.heliyon.2020.e04943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovic M., Minceva M. Coinfection and interference phenomena are the results of multiple thermodynamic competitive interactions. Microorganisms. 2021;9(10):2060. doi: 10.3390/microorganisms9102060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovic M. Atom counting method for determining elemental composition of viruses and its applications in biothermodynamics and environmental science. Comput. Biol. Chem. 2022;96 doi: 10.1016/j.compbiolchem.2022.107621. [DOI] [PubMed] [Google Scholar]
- Ramanathan, M., Ferguson, I.D., Miao, W., & Khavari, P.A. (2021). SARS-CoV-2 B.1.1.7 and B.1.351 Spike variants bind human ACE2 with increased affinity. bioRxiv: the preprint server for biology, 2021.02.22.432359. https://doi.org/10.1101/2021.02.22.432359. [DOI] [PMC free article] [PubMed]
- Ramesh, S.; Govindarajulu, M.; Parise, R.S.; Neel, L.; Shankar, T.; Patel, S.; Lowery, P.; Smith, F.; Dhanasekaran, M.; Moore, T. Emerging SARS-CoV-2 variants: a review of its mutations, its implications and vaccine efficacy. Vaccines 2021, 9, 1195. https://doi.org/10.3390/vaccines9101195. [DOI] [PMC free article] [PubMed]
- Samoilov A.E., Kaptelova V.V., Bukharina A.Y., et al. Case report: change of dominant strain during dual SARS-CoV-2 infection. BMC Infect. Dis. 2021;21:959. doi: 10.1186/s12879-021-06664-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz-Cherry S. Viral interference: the case of influenza viruses. J. Infect. Dis. 2015;212(11):1690–1691. doi: 10.1093/infdis/jiv261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevenich M.M., van den Heuvel J., Gering I., Mohrlüder J., Willbold D. Conversion rate to the secondary conformation state in the binding mode of SARS-CoV-2 spike protein to human ACE2 may predict infectivity efficacy of the underlying virus mutant. bioRxiv. 2021 doi: 10.1101/2021.07.14.452313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas L. The impact of SARS-CoV-2 variants on the COVID-19 epidemic in South Africa. News-Medical. Retrieved on. 2021 https://www.news-medical.net/news/20211031/The-impact-of-SARS-CoV-2-variants-on-the-COVID-19-epidemic-in-South-Africa.aspx November 22, 2021 from. [Google Scholar]
- Von Stockar, U. (2013a). Live cells as open non-equilibrium systems. In: Biothermodynamics: The Role of Thermodynamics in Biochemical Engineering, Urs von Stockar, ed., Lausanne: EPFL Press, pp. 399–421. https://doi.org/10.1201/b15428-23.
- Von Stockar, U. (2013b). Biothermodynamics of live cells: Energy disipation and heat generation in cellular cultures. In Biothermodynamics: The Role of Thermodynamics in Biochemical Engineering, Urs von Stockar, ed., Lausanne: EPFL Press, pp. 475–534. https://doi.org/10.1201/b15428-26.
- Westerhoff H.V., Lolkema J.S., Otto R., Hellingwerf K.J. Thermodynamics of growth. Non-equilibrium thermodynamics of bacterial growth. The phenomenological and the mosaic approach. Biochim. Biophys. Acta. 1982;683(3–4):181–220. doi: 10.1016/0304-4173(82)90001-5. [DOI] [PubMed] [Google Scholar]
- Wu A., Mihaylova V.T., Landry M.L., Foxman E.F. Interference between rhinovirus and influenza A virus: a clinical data analysis and experimental infection study. The Lancet. Microbe. 2020;1(6):e254–e262. doi: 10.1016/s2666-5247(20)30114-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J., Yin J., Fang L., Yang M., Wang T., Wu W., Bellucci M.A., Zhang P. Computational prediction of mutational effects on SARS-CoV-2 binding by relative free energy calculations. J. Chem. Inf. Model. 2020;60(12):5794–5802. doi: 10.1021/acs.jcim.0c00679. [DOI] [PubMed] [Google Scholar]