Abstract
Over 80% of women with high-grade serous ovarian cancer develop tumor resistance to chemotherapy and die of their disease. There are currently no FDA-approved agents to improve sensitivity to first-line platinum- and taxane-based chemotherapy or to poly (ADP-ribose) polymerase (PARP) inhibitors. Here, we tested the hypothesis that expression of growth arrest-specific 6 (GAS6), the ligand of receptor tyrosine kinase AXL, is associated with chemotherapy response and that sequestration of GAS6 with AVB-S6-500 (AVB-500) could improve tumor response to chemotherapy and PARP inhibitors. We found that GAS6 levels in patient tumor and serum samples collected before chemotherapy correlated with ovarian cancer chemoresponse and patient survival. Compared to chemotherapy alone, AVB-500 plus carboplatin and/or paclitaxel led to decreased ovarian cancer cell survival in vitro and tumor burden in vivo. Cells treated with AVB-500 plus carboplatin had more DNA damage, slower DNA replication fork progression, and fewer RAD51 foci than cells treated with carboplatin alone, indicating AVB-500 impaired homologous recombination. Finally, treatment with the PARP inhibitor olaparib plus AVB-500 led to decreased ovarian cancer cell survival in vitro and less tumor burden in vivo. Importantly, this effect was seen in homologous recombination-proficient and homologous recombination-deficient ovarian cancer cells. Collectively, our findings suggest that GAS6 levels could be used to predict response to carboplatin and AVB-500 could be used to treat platinum-resistant, homologous recombination-proficient high-grade serous ovarian cancer.
Keywords: replication stress, GAS6, ovarian cancer, DNA damage, carboplatin, synergism
Introduction
High-grade serous ovarian cancer (HGSOC) affects over 22,000 women in the United States annually and is the most deadly gynecologic malignancy. Patients with HGSOC are traditionally treated with either neoadjuvant taxane- and platinum-based chemotherapy followed by interval cytoreductive surgery or primary cytoreductive surgery followed by chemotherapy (1). Although the majority of patients respond to initial chemotherapy, over 80% of women develop resistance and die within five years of diagnosis (2, 3). Carboplatin, which causes DNA damage and cell death by introducing platinum-DNA adducts, is most effective in HGSOCs with increased genetic instability caused by deficiencies in homologous recombination (HR) (4). Additionally, patients with HR-deficient tumors can be treated with a combination of chemotherapy plus poly (ADP-ribose) polymerase (PARP) inhibitors such as olaparib. This combination is effective because PARP inhibitors in the setting of HR defects result in synthetic lethality (5). However, 50% of ovarian cancers are HR proficient (6, 7) and therefore are not susceptible to this approach. Thus, to improve care for HGSOC patients, we need to identify therapeutic agents that can improve response to chemotherapy or PARP inhibitors in HR-proficient tumors.
One candidate target in ovarian and other cancers is growth arrest-specific 6 (GAS6), which binds to and activates the receptor tyrosine kinases Tyro3, AXL, and Mer (8, 9). AXL mRNA expression is upregulated in several ovarian cancer cell lines, and data from The Cancer Genome Atlas as well as other sources indicate that AXL and GAS6 overexpression was associated with poor prognosis in ovarian cancer (10–13). Furthermore, Buehler et al. found that epithelial ovarian cancers commonly had high GAS6 mRNA and protein expression, and ovarian cancers from patients with residual disease after primary cytoreductive surgery had higher GAS6 expression than those without residual disease (14, 15).
One promising GAS6/AXL inhibitor is AVB-S6-500 (AVB-500), a highly selective AXL decoy receptor that binds to GAS6 with high affinity. Given the safety profile in healthy volunteers, AVB-500 was recently granted FDA fast-track designation for potential treatment of platinum-resistant ovarian cancer. Here, our objectives were to determine the utility of AVB-500 in combination with standard of care treatments in ovarian cancer and to determine whether GAS6 could be associated with treatment response. We show that ovarian cancer patients with high tumor expression of GAS6 had poor response to neoadjuvant chemotherapy and shorter progression-free as well as overall survival than those with low tumor expression of GAS6. In cell culture and in vivo mouse models, treatment with carboplatin and paclitaxel plus AVB-500 was more effective than chemotherapy alone. Mechanistically, this response was accompanied by stalled replication fork progression, increased DNA damage, and decreased HR repair (HRR) in both HR-deficient and HR-proficient chemoresistant cell lines. Finally, AVB-500 synergized with olaparib and increased replication fork progression in HR-proficient ovarian cancer cells. Together, our data support combining AVB-500 with carboplatin or olaparib to improve sensitivity to these treatments for HGSOC patients, including those with HR-proficient tumors.
Materials and Methods
Patient population
This study was approved by the Washington University in St. Louis Institutional Review Board (#201407156). Patients were included if they had stage III-IV ovarian cancer of serous, endometrioid, or clear cell histology and were planning to undergo neoadjuvant chemotherapy and interval cytoreductive surgery. Patients were excluded if they had previously undergone chemotherapy. Blood and tissue specimens were collected before and after three cycles of neoadjuvant chemotherapy.
Cases were evaluated for response to neoadjuvant chemotherapy according to a modified form of a published pathologic scoring system (16). All cases were scored for histological response to neoadjuvant chemotherapy and given a chemotherapy response score (CRS) of 1–3 as previously published (16). Additionally, they were given a score of 0–3 (0= no response, 3= complete response) for radiologic and surgical response(Supplementary Table S1). Radiologic scores were calculated based on RECIST criteria. Specifically, a score of 0 was associated with progressive disease, 1 with no progressive disease but less than a partial response, 2 with a partial response (≥ 30% decrease in disease, no new lesions), and 3 with a complete response or disappearance of all lesions. Surgical response was extrapolated from the operative report by two separate researchers (one physician, one research assistant). A score of 0 was assigned if there was persistent large volume disease and ascites, 1 for mild treatment effects but large volume disease, 3 if there was no or minimal residual disease, and 2 for a response not otherwise specified. The sum of each response was subtracted from 10 for a final chemoresponse score between 1 and 10. Cases were considered to have a good response if the score was 5 or lower and a poor response if the score was 6 or higher. This cutoff was representative of a moderate response or higher in two of the three categories evaluated. This novel score was utilized due to its comprehensive nature and clinical applicability. Progression-free and overall survival were calculated from the time of interval cytoreductive surgery.
GAS6 serum testing
Serum was collected from patients before three cycles of neoadjuvant chemotherapy. The concentration of free, unbound GAS6 was measured by an ELISA in which immobilized AVB-500 was used as a capture reagent. Briefly, the plate was coated with 1 μg/mL AVB-500 in PBS and GAS6 was detected on a Bio Tek 2 plate reader with a biotinylated polyclonal anti-human GAS6 antibody (BAF885, R&D Systems) and streptavidin-conjugated to horseradish peroxidase (DY998, R&D systems). The lower and upper limits of detection were 2 ng/mL and 128 ng/mL, respectively.
Immunohistochemistry
Formalin-fixed paraffin-embedded human and xenograft tumor slides were deparaffinized and immunohistochemistry performed as previously published with antibodies listed in Supplementary Table S2 (17). For patients, both pretreatment and post treatment slides obtained at the time of cytoreductive surgery were stained. Each slide was blindly scored for quantity and intensity of positive staining by two trained members of the research team, and the scores were averaged.
Cell lines
OVCAR5 and OVCAR8 cells were obtained from the National Cancer Institute. OVCAR3-TPMES cells were isolated from mouse mesentery tumor nodules in an OVCAR3TP intraperitoneal model (18). The OVCAR3TP cells were a generous gift from B. Sikic (19). These four cell lines were cultured in RPMI plus 10% fetal bovine serum (FBS) and 1% penicillin and streptomycin. To obtain POV71-hTERT, ascites from a patient with chemoresistant ovarian cancer was cultured, and tumor cells were identified morphologically. The primary cells were then immortalized via transfection with the lentiviral plasmid hTERT-hygro-pBABE and cultured in DMEM (Sigma-Aldrich) supplemented with 20% FBS and 1% penicillin and streptomycin. COV362 (Sigma-Aldrich) and CAOV3 (ATCC) cells lines were cultured in DMEM with 10% FBS and 1% penicillin and streptomycin. All cells were maintained at 37 °C in a 5% CO2 incubator. Cell line identities were confirmed by short tandem repeat profiling using the CellCheck9-Human test through IDEXX BioAnalaytics. Cell lines were confirmed mycoplasma negative with MycoAlert Mycoplasma Detection Kit (Lonza) before experiments.
Western blot analysis
Cultured cells were lysed in 9M urea, 0.075 M Tris, pH 7.6, and proteins were quantified by the Bradford assay. Protein lysates (60–100 μg) were subjected to reducing SDS-PAGE by standard methods and transferred to a nitrocellulose membrane. Each membrane was probed with primary antibody at 4°C for 1–3 nights, washed, and probed with corresponding horseradish peroxidase-conjugated secondary antibodies (all antibodies are listed in Supplementary Table S2). Signal was detected with the Pierce ECL Western Blotting Substrate, and chemiluminescence was measured on a ChemiDoc (Bio-Rad Laboratories, Hercules, CA).
Drug treatment and cell-viability assays
Cells (5,000 per well) were plated in each well of a 96-well plate and incubated overnight. Cells were then serum starved and treated with 0.7–1000 nM paclitaxel (Sigma-Aldrich), 0.01–108 μM carboplatin (Teva Pharmaceuticals, Sellersville, PA), 1–5 μM AVB-500 (Aravive), or a combination of these drugs. Carboplatin and AVB-500 were dissolved in saline. Paclitaxel was dissolved in DMSO for all experiments. After 72 hours, MTS (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium) solution (Promega, Madison, WI) was added to the cells (20μL/well), incubated for 2 hours, and then the absorbance was measured at 490 nm with a 96-well plate reader (Tecan infinite M200 Pro). The drug combination index (CI) (20) was calculated using CompuSyn software to assess for synergy between drugs..
Clonogenic assay
Cells (5,000 per well) were seeded in six-well plates, serum starved, and treated with vehicle, 0.5 μM carboplatin, 50 nM paclitaxel, 2 μM olaparib, 1 μM AVB-500, or combinations of these drugs for 72 hours. As described previously (21), this assay required a lower carboplatin concentration than that required for the DNA damage assays. Cells were then incubated in media with 10% FBS until vehicle-treated cells formed colonies optimal for visualization (14–21 days). Cells were then washed with PBS, fixed with methanol, and stained with 25% v/v methanol containing 0.05% w/v crystal violet for 10 min. Residual staining solution was removed, and plates were washed with water and air-dried. Images were captured with a Bio-Rad Imager System. For quantification, retained crystal violet was dissolved in 10% acetic acid, the solution was diluted eight-fold with water, and absorbance was measured at 590 nm. Combenefit software was used to calculate synergism from clonogenic assays according to the Loewe model (22).
Mouse ethical approval and tumor model
All animal experiments were done in accordance with the guidelines of the American Association for Accreditation for laboratory Animal Care and the U.S. Public Health Service Policy on Humane Care and Use of Laboratory Animals and were approved by the Washington University Institutional Animal Care and Use Committee in accordance with the Animal Welfare Act, the Guide for the Care and Use of Laboratory Animals, and NIH guidelines. For established cancer cell line xenograft models, six-week-old female nude (nu/nu) mice (Jackson Laboratory) were used. For patient-derived xenograft (PDX) models, seven-week-old female NSG (NOD.Bg-Prkdcscid Il2rgtm1Wjl/SzJ) mice (Jackson Laboratory) were used.
To establish subcutaneous cell line tumors, 1.5×106 OVCAR8 cells were implanted subcutaneously into the flank of the mouse, and the mouse was allowed to rest until tumor engraftment and growth were observed. For the subcutaneous PDX models, flash frozen PB1 or ICS051 tumors were thawed and implanted subcutaneously (18). Treatment was started when the PB1 tumor volume reached 300 mm3 and when the ICS051 tumor volume reached 2 cm3. We used different tumor sizes before beginning treatment to determine whether established, larger tumors could also respond to treatment compared to smaller tumors. Intraperitoneal tumor models were established by IP injection of 2.5×106 OVCAR5 or OVCAR8 cells. Treatment was initiated on day 14. After initiating treatment, tumors were measured every three days with calipers, and tumor volume was calculated with the equation V= l*w*π/6 (l=longest diameter, w=shortest perpendicular diameter).
All mice (subcutaneous or intraperitoneal tumor models) were treated as follows: 20 mg/kg paclitaxel intraperitoneal (IP) weekly, 80 mg/kg carboplatin IP weekly, 30 mg/kg AVB-500 IP every 3 days, or 200 μL saline weekly. After 14 days of treatment mice were sacrificed two or four hours after receiving a single IP injection of 30 mg/kg AVB-500. Tumors were weighed, formalin fixed, and embedded in paraffin.
For the PARP inhibitor plus AVB-500 experiments, treatment was as follows: 30 mg/kg AVB-500 IP every 3 days, 50 mg/kg olaparib by oral gavage every 3 days, or 200 μL saline IP weekly. Mice were sacrificed on day 21, and tumor burden was evaluated via dissection.
Immunofluorescence microscopy
OVCAR8, COV362, CAOV3, and OVCAR3-TPMES cells were plated (10,000 cells per well) in an 8-well chamber slide. Cells were then treated with 500 μM carboplatin +/− 1 μM AVB-500 for 4 hours, 2 μM Olaparib +/− 1 μM AVB-500 for 96 hours refreshing drug treatment daily, or treated with 10 Gy ionizing radiation. The carboplatin dose was based on the literature (23) and optimized in multiple experiments evaluating DNA damage in OVCAR8 cells. Notably, a significantly higher dose of carboplatin was necessary in order to exaggerate the DNA damage response that is caused as a result of acute treatment. After treatment, the cells were washed with cold PBS, fixed with 4% paraformaldehyde for 10 min, washed with immunofluorescence (IF) Wash Buffer (25 mM Tris-HCl, pH 7.5, 250 mM NaCl, 0.1% Tween 20), and blocked with IF Blocking Buffer (PBS, 5% goat serum, 0.5% Triton-X-100) for 60 minutes. Cells were incubated overnight at 4 °C with primary antibodies (listed in Supplementary Table S2) in 1% goat serum, 0.1% Tween 20. Cells were then stained with secondary antibodies (1:500, Alexa Fluor 647- or FITC-conjugated; Invitrogen) and DAPI (Sigma) and mounted with Vectashield (Vector Laboratories). Cells were imaged on a Leica TCS SPE inverted confocal microscope. Raw images were exported into ImageJ and adjusted for contrast and brightness (24). Foci were counted with FociCounter (25). At least 100 cells were analyzed for each treatment group in duplicate. Student’s two-tailed t-test was used to determine statistical significance.
To classify cell lines as HR-proficient or -deficient, the method described by Graeser et al. (26) and the HR RePair CAPacity (RECAP) test (27) were used. Briefly, all cells with ≥5 RAD51 foci were considered RAD51 positive. Cells lines were considered HR-proficient if >50% of geminin-positive cells were RAD51 positive, HR-deficient if <20% were RAD51 positive, and HR intermediate if >20% but <50% were RAD51 positive (28). This assay was used to confirm that OVCAR8 cells were HR-deficient and that COV362, CAOV3, OVCAR3-TPMES cells were HR-proficient (Supplementary Figure S1) as previously described (29–31). Although COV362 cells were derived from a BRCA1 mutated tumor, they were confirmed to be HR-proficient. This is likely due to residual BRCA1 function as published by Pettitt et al (31) Additionally, OVCAR3-TPMES and CAOV3 were confirmed to be HR-proficient (28).
Flow cytometry
A BD FACS LSRII flow cytometer was used for all flow cytometry experiments, and FlowJo software was used for all data analysis. To measure chromatin-bound replication protein A (RPA), cells were treated with 0.5 mM carboplatin +/− 1 μM AVB-500 for 4 hours at 37 °C, then washed with PBS + 2% FBS and fixed in cytofix/cytoperm buffer (BD Biosciences) at room temperature. They were then washed with Perm/Wash buffer (BD Biosciences), stained with rat anti-RPA (Supplementary Table S2) overnight, at 4 °C, washed, and stained with secondary antibody (1:1000; Alexa Fluor488; Invitrogen). Cells were washed again with Perm/Wash buffer and resuspended in PBS + 2% FBS containing 7-AAD for analysis (32).
Cell cycle was analyzed by propidium iodide staining (33). Cells were treated with carboplatin (0.5 mM), AVB-500 (1 uM) alone or in combination for 24 hours. Cells were harvested by trypsinization, washed with PBS, and fixed with 70% ethanol for 2 hours on ice. After washing with PBS, cells were resuspended in PBS containing 100 μg/mL RNase A and 50 μg/mL propidium iodide, and incubated overnight at 4 °C before analysis. Fifty thousand events were measured per sample. Each assay was performed in triplicate in two independent experiments.
DNA fiber assay
Exponentially growing cells were pulse-labeled with 20 μM 5-Iodo-2’-deoxyuridine (IdU, Millipore Sigma) for 20 min, washed twice with PBS, pulse-labeled with 200 μM 5-Chloro-2’-deoxyuridine (CldU, Millipore Sigma) for 1 h, then washed twice with PBS (34, 35). In the case of cisplatin and carboplatin treatment, CldU was added concomitantly with 150 μM cisplatin or 500 μM carboplatin. Cisplatin was evaluated initially, as its effects have been well established in these assays(36, 37). In the case of AVB-500 treatment, cells were treated with 1 μM AVB-500 24 hours before the start of the fiber assay, and 1 μM AVB-500 was maintained in the cell media during the entire labeling period. In the case of olaparib treatment, cells were treated with 10 μM olaparib for 24 hours before the start of the fiber assay, and 10 μM olaparib was maintained in the cell media during the entire labeling period. This concentration of olaparib, used previously for DNA fiber assays (38, 39) was selected in order to maximize any potential fork perturbations caused by this drug. In all cases, cells were harvested, pelleted at 300 × g for 5 min at 4 °C, and resuspended in PBS at 1,500 cells/μL. Then, 2 μL of cells were mixed with 6 μL of lysis buffer (200 mM Tris-HCl, pH 7.5, 50 mM EDTA, 0.5% SDS in water) on top of a positively charged glass slide. After 5 min at RT, slides were tilted at a 20–60° angle to spread the fibers at a constant, low speed. After air drying for 15 min at RT, DNA was fixed onto the slides with a freshly prepared solution of methanol: glacial acetic acid (3:1) for 5 min, dried, then stored at 4 °C for at least overnight. For immuno-staining of DNA fibers, DNA was rehydrated in PBS twice for 5 min, then denatured with 2.5 M HCl for 1 h at RT. Slides were then washed with PBS three times and blocked with 5% BSA in PBS at 37 °C for 1 h. DNA fibers were immuno-stained with rat anti-BrdU (1:100, ab6326, abcam) and mouse-anti-BrdU (1:20, 347580, BD Biosciences) (Supplementary Table S2) for 1.5 h at RT, rinsed in PBS, washed three times with PBS-0.1%Tween-20 for 5 min, rinsed with PBS, then incubated with anti-rat Alexa Fluor 488 and anti-mouse Alexa Fluor 546 (1:100, A21470 and A21123, respectively, ThermoFisher Scientific) for 1 h at RT. After three washes with PBS-0.1% Tween-20 of 5 min each, slides were rinsed in PBS, then mounted with Prolong Gold Antifade Reagent (P36930, ThermoFisher Scientific). Images were acquired with red and green channels on a fluorescent microscope (LEICA DMU 4000B, 63X/1.4 oil immersion objective) with a LEICA DFC345FX camera or with LAS AF software using a TCS SP5 confocal microscope (LEICA, 63x/1.4 oil immersion objective). The fiber experiments were repeated for 2 or 3 independent biological replicates in CAOV3 and OVCAR3-TPMES cell lines, respectively. At least 15 images were taken across the whole slide using only one channel to select the regions for the images. At least 150–200 individual tracts were scored for each dataset. DNA fiber tracts were analyzed only if the IdU and CldU signals were contiguous and if the beginning and end of each color were unambiguously defined. The length of each tract was measured manually with the segmented line tool on ImageJ software (NIH). The pixel values were converted into μm by using the scale bar generated by the microscope software. Ratios (CldU/IdU) or total tract lengths (IdU + CldU) from individual DNA fibers were plotted as scatter dot plot with the horizontal line representing the median.
Statistics
GraphPad Prism 7 software was used for statistical analysis and IC50 calculations. Statistical significance was determined by the student t-test, chi-square test, or one-way ANOVA as appropriate. Significance was defined as P<0.05. For the DNA fiber assays, data were pooled from three independent experiments. Statistical differences in DNA fiber tract lengths were determined by a one-way ANOVA (Kruskal-Wallis test) followed by a Dunn’s multiple comparison test.
Results
Tumor GAS6 expression is associated with response to platinum-based chemotherapy in ovarian cancer patients
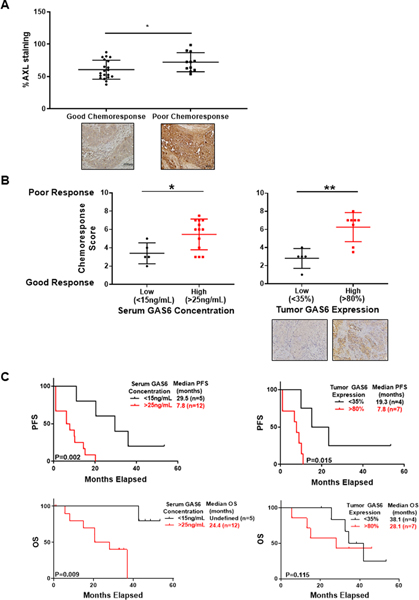
Given the associations between tumor AXL expression and outcomes in cancer patients (14, 15, 18), we asked whether AXL expression correlated with resistance to neoadjuvant chemotherapy in ovarian cancer patients. To answer this question, we performed immunohistochemistry on matched serum and tumor samples from 38 patients before neoadjuvant paclitaxel and carboplatin. Seventy-nine percent of patients had Stage III disease and 21% had Stage IV disease. All patients had at least mixed high grade serous histology. BRCA1/2 mutations were identified in 15.8% of patients. Median follow-up was 33 months. Supplementary Table S3 shows the characteristics of our study population stratified by response to neoadjuvant chemotherapy on a 1–10 scale in which a score of 5 or lower was considered a good response, and a score of 6 or higher was considered a poor response (see Methods for details). By this stratification, 24 (63.2%) of the patients had a good chemoresponse and 14 (36.8%) had a poor response. There were no significant differences in the demographics or tumor characteristics between these two groups. We assigned each tumor an AXL expression score between 0% and 100%. Consistent with previous findings, AXL expression was higher in tumors with poor chemoresponse than in those with a good chemoresponse (72% vs 60%, P=0.04) (Figure 1A).
Figure 1. AXL and GAS6 expression is associated with neoadjuvant chemoresponse and survival.
A, Percent of AXL staining in tumor samples with good and poor chemoresponse after neoadjuvant chemotherapy. Representative images of AXL staining in tumors before neoadjuvant chemotherapy and at time of interval cytoreductive surgery at 20X magnification. Horizontal lines indicate mean and inter-quartile range. *, P<0.05. B, Mean chemoresponse scores for patients with high and low serum and tumor GAS6 concentration. Chemoresponse score was a composite of radiologic, surgical, and pathologic response at time of interval cytoreductive surgery. Representative images of GAS6 staining in tumors before neoadjuvant chemotherapy at 40X magnification C, Kaplan Meier curve evaluating progression-free survival (PFS, top) and overall survival (OS, bottom) in patients stratified by serum (left) or tumor (right) GAS6 concentration.
We next measured AXL ligand, GAS6, in serum and tumors collected before neoadjuvant chemotherapy from the same patients. In an attempt to stratify patients into high- and low-risk groups based on GAS6 levels, we explored all observed GAS6 values, identified cutoff values that minimized the P-value, and made statistical adjustments to resolve the increased false positive rate (40). By this method, patients were grouped into high vs. low serum GAS6 concentrations (>25 ng/mL vs. <15ng/mL) and high vs. low tumor GAS6 expression (>80% vs. <35%). Supplementary Table S4 shows the characteristics of patients with high and low serum GAS6 concentrations. There were no differences in the demographics or tumor characteristics between these two groups. Patients with high serum GAS6 concentrations had worse chemoresponse scores than those with low serum GAS6 concentrations (mean score of 5.5 vs. 3.4, P=0.023) (Figure 1B). Similarly, patients with high tumor GAS6 expression had significantly worse chemoresponse scores than those with low tumor GAS6 expression (mean score of 6.3 vs 2.8, P=0.002) (Figure 1B). Moreover, patients with high serum GAS6 concentrations had significantly shorter progression-free survival (P=0.002) and shorter overall survival (P=0.002) than those with low serum GAS6 concentrations. Likewise, patients with high tumor GAS6 expression had shorter progression-free survival (P=0.015) than those with low tumor GAS6 expression (Figure 1C). Serum GAS6 concentrations obtained prior to treatment were not significantly different between patients with and without BRCA mutations (23.6 ± 8.7 vs 21.7 ± 7.14, P=0.615). However, tumor GAS6 levels were significantly lower in patients with BRCA mutations than without (59.6 ± 20.4 vs 41.3 ± 17.9, P=0.46). Thus, high GAS6 concentrations in serum and tumor before neoadjuvant chemotherapy correlated with poor chemoresponse, shorter progression-free survival, and shorter overall survival.
To better understand how GAS6 expression correlated with tumor burden we evaluated associations between both serum and tumor GAS6 levels and the administration of chemotherapy, the presence of ascites, and levels of CA 125, a prominent biomarker in the diagnosis and management of ovarian cancer. Serum GAS6 levels decreased significantly after 3 cycles of chemotherapy (23.5 ± 8.4 vs 18.5 ± 9.3, p<0.001), but tumor levels did not (55.3 ± 19.6 vs 52.5 ± 20.1, P=0.413). There was no correlation between serum or tumor GAS6 and CA 125 or ascites.
AVB-500 improves response to chemotherapy and decreases ovarian cancer cell survival in vitro and tumor burden in vivo
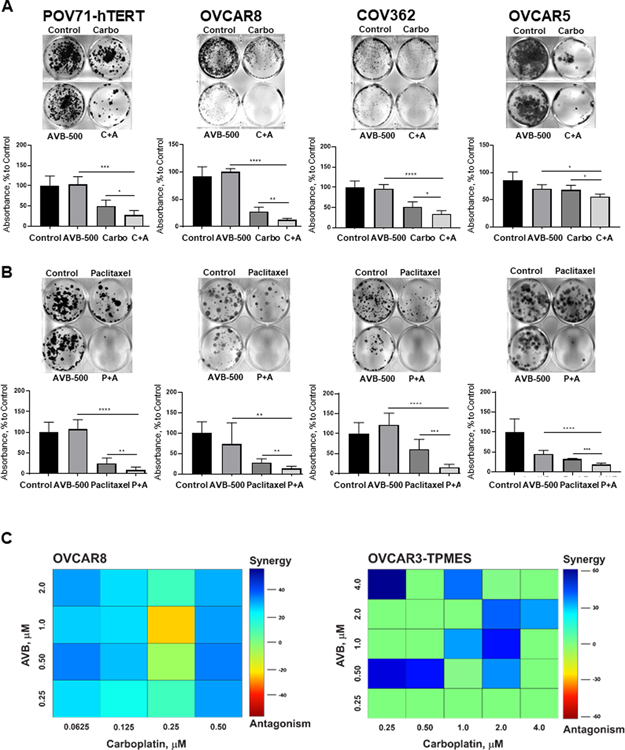
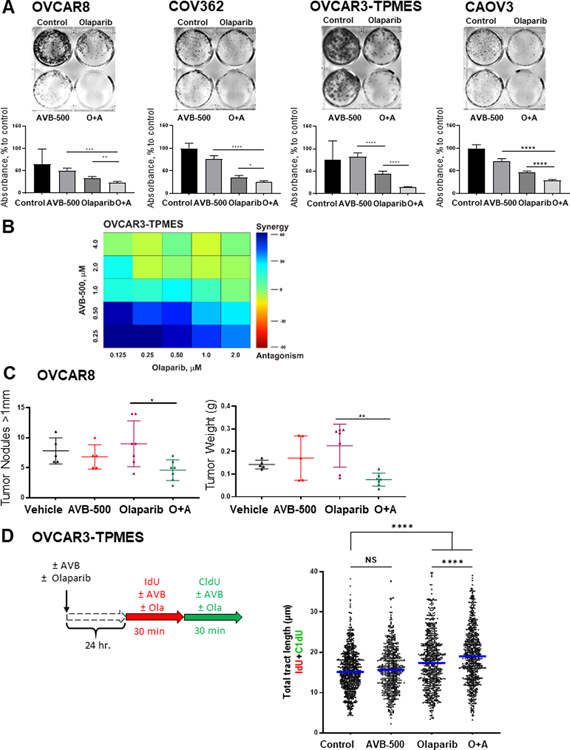
Given our finding that tumors with poor response to neoadjuvant chemotherapy had high GAS6 expression, we wondered whether a combination of the GAS6 inhibitor AVB-500 plus chemotherapy would be more effective than chemotherapy alone in reducing ovarian tumor cell growth. We evaluated AXL/GAS6 expression in multiple chemoresistant ovarian cancer cell lines (18) (Supplementary Figure S2). We then performed clonogenic growth assays in the chemoresistant cell lines OVCAR5, POV71-hTERT, OVCAR8, and COV362. In all four cell lines, cells treated with carboplatin or paclitaxel plus AVB-500 formed significantly fewer colonies than cells treated with chemotherapy alone (P<0.05) (Figure 2A–B). To evaluate the effects of drug combinations, we calculated combination indexes (CI) in which synergism was defined as CI<1, additive effect as CI=1, and antagonism as CI>1. In POV71-hTERT, OVCAR5, and OVCAR3-TPMES cells, AVB-500 synergized with paclitaxel (Supplementary Figure S3). In OVCAR5 cells, AVB-500 synergized with carboplatin (CI 0.31). We also evaluated synergism using Loewe analysis which revealed that AVB-500 synergized with carboplatin to inhibit colony formation in OVCAR8 and OVCAR3-TPMES cells (Figure 2C).
Figure 2. GAS6/AXL inhibitor AVB-500 improves response to carboplatin and paclitaxel in ovarian cancer cell lines.
A, Colony formation assay in cells treated with 0.5 μM Carboplatin (Carbo), 1 μM AVB-500, or a combination of carboplatin and AVB-500 (C+A). Top, representative images; bottom, quantitation of percent to control absorbance at 590nm. Cells were treated for 72 hours and then incubated in media with 10% FBS until vehicle-treated cells formed colonies optimal for visualization. B, Colony formation assay in cells treated with 50 nM Paclitaxel, 1 μM AVB-500, or a combination of paclitaxel and AVB-500 (P+A). Cells were treated for 72 hours and then incubated in media with 10% FBS until vehicle-treated cells formed colonies optimal for visualization. C, Loewe synergism analysis of OVCAR8 and OVCAR3-TPMES cells treated with varying doses of carboplatin and AVB-500.
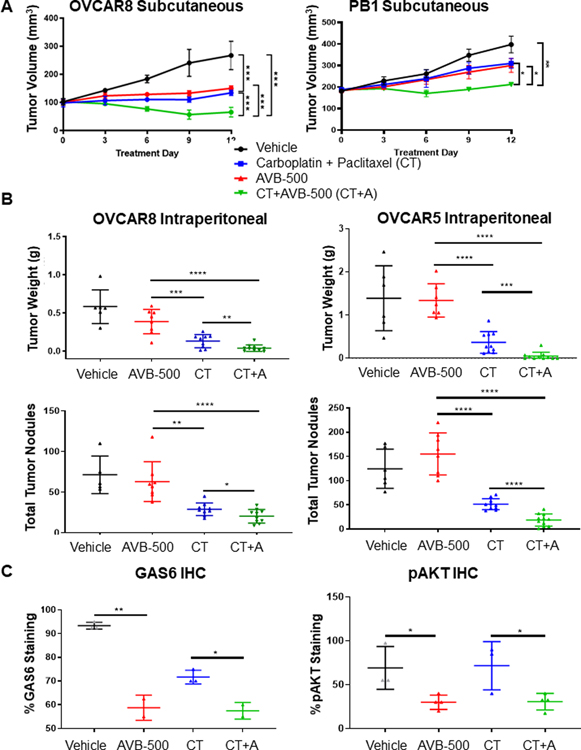
Given our in vitro findings, we next wanted to know whether AVB-500 plus chemotherapy was more effective than chemotherapy alone in in vivo ovarian cancer models. We first created three models in which we subcutaneously injected the chemoresistant, HR-proficient patient-derived xenograft (PDX) model PB1 or ICS051, or the chemoresistant, HR-deficient cell line OVCAR8 into mice. After the PB1 and OVCAR8 tumors reached ~300 mm3 and the ICS051 tumors reached 2 cm3, mice were treated with vehicle, carboplatin plus paclitaxel (chemotherapy), AVB-500 alone, or chemotherapy plus AVB-500 for 12 days. In mice that received chemotherapy plus AVB-500, tumors grew more slowly and reached lower volume and weight than those that received chemotherapy alone (Figure 3A, Supplementary Figure S4).
Figure 3. GAS6/AXL inhibitor AVB-500 improves response to carboplatin and decreases tumor burden in mouse models of ovarian cancer.
A, Volume of tumors over 12 days of treatment in mice engrafted with OVCAR8 or PB1 cells and treated with vehicle, 80mg/kg IP weekly carboplatin + 20mg/kg paclitaxel IP weekly paclitaxel (CT), 30mg/kg AVB-500, or chemotherapy and AVB-500 (CT+A). B, Tumor burden of mice bearing OVCAR8 or OVCAR5 tumors as quantified by tumor weight and number of tumor nodules after treatment with vehicle, CT, AVB-500, or CT+A. C, Quantitation of GAS6 and pAKT immunohistochemistry staining in OVCAR8 tumors in mice treated with vehicle, CT, AVB-500, or CT+A.
To further assess the effectiveness of AVB-500 plus chemotherapy, we created in vivo models in which we intraperitoneally implanted the chemoresistant cell lines OVCAR8 or OVCAR5 into mice. After 14 days, we treated the mice with either vehicle, carboplatin and paclitaxel (chemotherapy), AVB-500, or chemotherapy plus AVB-500 for 14 days and then assessed tumor burden. Mice that received chemotherapy plus AVB-500 had significantly fewer tumor nodules and smaller tumor mass than mice that received chemotherapy alone (Figure 3B). Immunohistochemical analysis revealed that tumors from the mice treated with AVB-500 or chemotherapy plus AVB-500 had significantly less staining for GAS6 and the downstream protein phospho-AKT (pAKT) than those treated with vehicle or chemotherapy alone (Figure 3C, Supplementary Figure S5).
Collectively, both in vivo and in vitro results suggest that chemotherapy plus AVB-500 was more effective at reducing ovarian tumor cell growth than chemotherapy alone in both HR-proficient and HR-deficient cells.
AVB-500 increases DNA damage and slows DNA replication in the presence of carboplatin
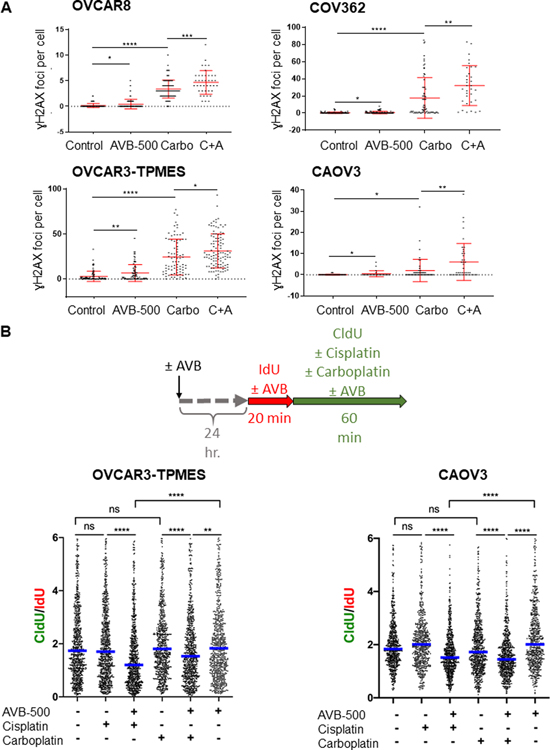
We next sought to determine the mechanism by which AVB-500 plus chemotherapy was more effective than chemotherapy alone. Paclitaxel causes cell death by inhibiting microtubule assembly, whereas carboplatin causes cell death by causing platinum-DNA adducts that lead to DNA double-strand breaks. Because increased DNA damage has been associated with increased platinum sensitivity and improved efficacy of DNA damaging agents, we hypothesized that AVB-500 would increase the DNA damage caused by carboplatin in both HR-proficient and HR-deficient cell lines. To test this hypothesis, we treated OVCAR8, COV362, CAOV3, and OVCAR3-TPMES cell lines with vehicle, AVB-500, carboplatin, or carboplatin plus AVB-500 for four hours and then performed immunofluorescence (IF) assays. Cleaved caspase 3, a marker of apoptosis, was not detectable at this time point (Supplementary Figure S6) excluding apoptosis as the cause of DNA damage. To assess the extent of DNA damage, we performed IF for the DNA damage marker ɣH2AX, as formation of ɣH2AX foci is a validated approach to evaluate DNA damage that occurs as a result of platinum therapy (23). As expected, cells treated with carboplatin had more γH2AX foci than cells treated with AVB-500 or vehicle. Moreover, consistent with previous results (41), cells from all four lines treated with AVB-500 alone had more γH2AX foci than cells treated with vehicle. Additionally, cells treated with carboplatin plus AVB-500 had significantly more γH2AX foci than cells treated with carboplatin alone (Figure 4A). These results suggest that AVB-500 increased DNA damage independently and enhanced carboplatin-induced DNA damage. To confirm this finding, we performed comet assays in which cells with damaged DNA have comet “tails”. The intensity of the tails reflect the extent of DNA damage. We confirmed that OVCAR8 cells treated with AVB-500 plus carboplatin had longer tails than cells treated with carboplatin alone (Supplementary Figure S7).
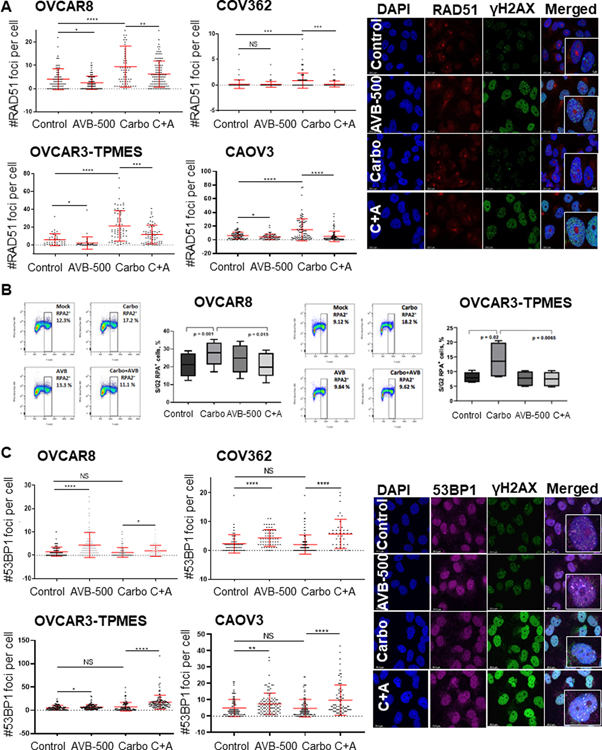
Figure 4. AVB-500 alone or in combination with chemotherapy increases DNA damage and slows replication fork progression.
A, Number of γH2AX foci per nucleus induced by treatment with vehicle, 1 μM AVB-500, 500 μM carboplatin (Carbo), or carboplatin plus AVB-500 (C+A) in OVCAR8, COV362, OVCAR3-TPMES, and CAOV3 cells. Cells were treated for 4 hours. Foci were quantified from two technical replicates with n>100 cells per experiment. Error bars indicate ± SD. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001 by student’s two-tailed t-test. B, Top, Schematic for DNA fiber assays. Bottom, CldU/IdU ratios in OVCAR3-TPMES cells ± 1 μM AVB-500 ± 150μM cisplatin ± 500 μM carboplatin (n=3) and CAOV3 cells ± AVB-500 ± cisplatin ± carboplatin (n=2) cells treated as indicated. Horizontal lines indicate medians. CldU was added concomitantly with 150 μM cisplatin or 500 μM carboplatin. In the case of AVB-500 treatment, cells were treated with 1 μM AVB-500 24 hours before the start of the fiber assay, and 1 μM AVB-500 was maintained in the cell media during the entire labeling period. **P < 0.01, **** P < 0.0001.
Because increased DNA damage can impair replication, we next asked whether replication fork progression was impaired in ovarian cancer cells treated with carboplatin plus AVB-500. We used the single-molecule DNA fiber approach to monitor fork progression in OVCAR3-TPMES cells treated with AVB-500 alone, AVB-500 plus carboplatin, or AVB-500 plus cisplatin. Cells were pretreated with 1 μM AVB-500 for 24 hours because downregulation of pAKT, a downstream target of GAS6, was seen after 24 hours (Supplementary Figure S8). Cells were then incubated with the thymidine analog IdU for 20 minutes followed by the thymidine analog CldU and concomitantly treated with carboplatin or cisplatin for 60 minutes (Figure 4B). In AVB-500 treated samples, AVB-500 was maintained in the cell culture media for the entire duration of the thymidine analog incorporation. The CldU/IdU ratios were similar in cells treated with vehicle, carboplatin, cisplatin, or AVB-500 alone, indicating that these single agents did not affect fork progression in OVCAR3-TPMES cells, at least at the concentrations used in these experiments. However, combined treatment of the OVCAR3-TPMES cells with carboplatin plus AVB-500 or cisplatin plus AVB-500 led to a significant reduction of CldU incorporation and overall lower CldU/IdU ratios (Figure 4B). We saw a similar effect in the HR-proficient cell line CAOV3 (Figure 4B). These findings suggest that treatment with carboplatin plus AVB-500 causes DNA damage that significantly hinders replication fork progression, which may contribute to the increased chemoresponse observed in cells co-treated with AVB-500 and carboplatin.
AVB-500 treatment alters the DNA damage repair response of chemotherapy-treated ovarian cancer cells
DNA damage caused by platinum chemotherapy results in activation of a signaling cascade referred to as the DNA damage response (DDR). The goal of the DDR is to maintain genomic stability by activating DNA repair, cell cycle arrest, or apoptosis (42). The type of cell response depends on the amount of DNA damage. Enhanced DDR is a well-established mechanism for chemotherapy resistance. Therefore, DDR inhibition is a valuable target to sensitize cancer cells to platinum chemotherapy. Given that cells treated with AVB-500 plus chemotherapy had more DNA damage than cells treated with chemotherapy alone, we sought to define the effects of AVB-500 on key modulators of DNA damage. We first evaluated expression of key proteins involved in DNA damage repair and found that treatment of OVCAR8 cells with carboplatin plus AVB-500 resulted in decreased expression of phospho-ATM and increased expression of phospho-Chk1 (Supplementary Figure S9). We then treated OVCAR8, COV362, OVCAR3-TPMES, and CAOV3 cells with vehicle, AVB-500, carboplatin, or carboplatin plus AVB-500 and performed IF and quantified the number of RAD51 foci per cell. In all four cell types, cells treated with carboplatin plus AVB-500 had significantly fewer RAD51 foci than cells treated with carboplatin alone, suggesting that AVB-500 treatment reduced HRR (Figure 5A). To confirm this finding, we used flow cytometry to measure the number of replication protein A (RPA)-positive cells in S/G2 phase in each treatment condition. In all four cell lines, a smaller percentage of S/G2 phase cells treated with carboplatin plus AVB-500 were RPA-positive than cells treated with carboplatin alone (Figure 5B, Supplementary Figure S10A). Importantly, cell cycle analysis confirmed that the treatment regimens did not alter the proportion of cells in different phases of the cell cycle (Supplementary Figure S10B), suggesting that the effects on DNA damage repair were not due to cell cycle disruption.
Figure 5. AVB-500 in combination with carboplatin decreases RAD51 foci, increases 53BP1 foci, and decreases RPA binding.
A, Left, number of RAD51 foci per nucleus induced by treatment with vehicle, 1 μM AVB-500, 500 μM carboplatin (Carbo), or carboplatin plus AVB-500 (C+A) in OVCAR8, COV362, OVCAR3-TPMES, and CAOV3 cells. Right, representative images of γH2AX, RAD51, and colocalization of γH2AX/RAD51 foci in OVCAR8 cells at 10X with 63X. B, RPA binding in S/G2 in OVCAR8 and OVCAR-TPMES cells treated with vehicle, carboplatin, AVB-500, or carboplatin plus AVB-500. C, Left, number of 53BP1 foci per nucleus induced by treatment with vehicle, AVB-500, Carbo, or C+A in OVCAR8, COV362, OVCAR3-TPMES, and CAOV3 cells. Right, representative images of γH2AX, 53BP1, and colocalization of γH2AX/53BP1 in OVCAR8 cells at 10X with 63X insets. Foci were quantified from two technical replicates with n>100 cells per experiment. Error bars indicate ± SD. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001 by two-tailed student’s t-test.
It was possible that this functional decrease of HRR efficiency was a result of increased 53BP1 binding, which inhibits DNA end resection and thus HRR. To test this idea, we treated cells with vehicle, AVB-500 alone, carboplatin alone, or AVB-500 plus carboplatin, performed IF, and counted γH2AX and 53BP1 foci. We found that HR-deficient and HR-proficient cells treated with carboplatin plus AVB-500 had significantly more 53BP1 foci than cells treated with carboplatin alone (Figure 5C), and the 53BP1 foci appeared to co-localize with γH2AX foci (Figure 5C). Western blot analysis demonstrated that total amounts of RAD51 and 53BP1 were similar in all treatment groups, indicating that the observed differences in RAD51 and 53BP1 foci were not due to variable protein expression (Supplementary Figure S8). Taken together, these findings suggested that AVB-500 impaired HRR in ovarian cancer cells treated with chemotherapy.
AVB-500 imparts PARP inhibitor sensitivity to HR-proficient and -deficient cells both in vitro and in vivo
Given that AVB-500 appeared to impair HRR in response to DNA damage, we asked whether treatment with AVB-500 would increase sensitivity to the PARP inhibitor olaparib. First, we performed in vitro colony formation assays and found that both HR-deficient (OVCAR8) and HR-proficient (COV362, OVCAR3-TPMES, and CAOV3) ovarian cancer cells were more sensitive to AVB-500 plus olaparib than to olaparib alone (Figure 6A). Moreover, Loewe analysis revealed that AVB-500 synergized with olaparib to inhibit colony formation in OVCAR3-TPMES cells (Figure 6B). These in vitro findings led us to perform an in vivo experiment in which we intraperitoneally injected OVCAR8 cells into mice, allowed the tumors to grow for 7 days, and then treated the mice with vehicle, AVB-500, olaparib, or AVB-500 plus olaparib until day 21. Mice treated with olaparib plus AVB-500 had fewer tumor nodules and lower total tumor weight than mice treated with olaparib alone (Figure 6C).
Figure 6. AXL inhibitor AVB-500 improves response to olaparib in vitro and in vivo.
A, Colony formation assay in cells treated with vehicle, 2μM Olaparib, 1μM AVB-500, or olaparib plus AVB-500 (O+A). Top, representative images; bottom, quantitation of percent to control absorbance at 590nm. Cells were treated for 72 hours and then incubated in media with 10% FBS until vehicle-treated cells formed colonies optimal for visualization. B, Loewe synergism analysis of OVCAR3-TPMES cells treated with varying doses of olaparib and AVB-500. C, Tumor burden of mice intraperitoneally injected with OVCAR8 as quantified by tumor nodule number and weight after treatment with vehicle, olaparib, AVB-500, or O+A. D, Schematic for DNA fiber assays. Total tract length of IdU+CldU in OVCAR3-TPMES cells treated with vehicle, 1 μM AVB-500, 10 μM olaparib, or O+A. Horizontal lines indicate medians. Cells were treated with 1 μM AVB-500 and 10 μM olaparib 24 hours before the start of the fiber assay. AVB-500 and olaparib were maintained in the cell media during the entire labeling period. **** P < 0.0001.
IF analysis demonstrated that OVCAR8 and OVCAR3-TPMES cells treated with AVB-500 with olaparib had more ɣH2AX foci and fewer RAD51 foci than cells treated with olaparib alone (Supplementary Figure S11). We then asked whether replication fork progression was impaired in ovarian cancer cells treated with olaparib plus AVB-500. We used the single-molecule DNA fiber approach to monitor fork progression in OVCAR3-TPMES cells treated with AVB-500 alone, olaparib alone, or AVB-500 plus olaparib. Cells were pretreated with 1 μM AVB-500 and 10 μM olaparib for 24 hours. Cells were then incubated with the thymidine analog IdU for 30 minutes followed by the thymidine analog CldU for 30 minutes (Figure 6D). AVB-500 and olaparib was maintained in the cell culture media for the entire duration of the thymidine analog incorporation. The IdU+CldU total tract lengths (μm) were similar in cells treated with vehicle or AVB-500 alone indicating that these single agents did not affect fork progression in OVCAR3-TPMES cells. However, single agent olaparib as well as the combined treatment of the OVCAR3-TPMES cells with olaparib plus AVB-500 led to a significant increase in total tract length (Figure 6D). Our findings support those previously published by Maya-Mendoza et al. which suggest that PARP inhibition increases the speed of replication forks (39) and suggest that the combined treatment of olaparib plus AVB-500 augments this increase in replication fork speed resulting in increased DNA damage and genomic instability. Together, these findings indicated that AVB-500 increased sensitivity to a PARP inhibitor in both HR-deficient and HR-proficient ovarian cancer cells.
Discussion
In this paper, we present several lines of evidence that the GAS6 inhibitor AVB-500 improved ovarian cancer cell sensitivity to both traditional chemotherapy and PARP inhibitors. First, we demonstrated that AVB-500 improved ovarian tumor cell response to chemotherapy both in vitro and in vivo. This is due to increased DNA damage, replication fork perturbation, and decreased HRR. Accordingly, AVB-500 improved in vitro and in vivo response to the PARP inhibitor olaparib in both HR-deficient and HR-proficient ovarian cancer cells. Lastly, our data suggests that GAS6 in both patient serum and tumor is associated with outcomes in women with ovarian cancer.
AVB-500 is a recombinant protein containing a truncated, modified portion of the extracellular region of human AXL fused to the human immunoglobulin G1 heavy chain to induce dimerization. Because AVB-500 binds GAS6 with 200-fold higher affinity than the soluble receptor AXL, GAS6’s highest affinity receptor (43), AVB-500 is less likely to have off-target effects than traditional small molecule tyrosine kinase inhibitors. Consistent with this idea, AVB-500 caused minimal side effects at therapeutic doses in a healthy volunteer trial (44). Thus, AVB-500 is currently being evaluated in a phase 1b/2 trial in platinum-resistant, recurrent ovarian cancer (NCT03639246).
In our work, AVB-500 improved response to traditional chemotherapy, leading to decreased cell viability and tumor burden. These findings align with previous reports that AXL inhibition improves sensitivity to chemotherapy (11, 45). Our data are consistent with data indicating that AXL inhibition prevented the normal DNA damage response (41) and gives insight into the mechanism. Specifically, we show that cells treated with chemotherapy plus AVB-500 had more γH2AX foci, more 53BP1 foci, fewer RAD51 foci, less RPA binding, and slower replication fork progression than cells treated with chemotherapy alone, indicating that AVB-500 impaired HRR. Our findings suggest that the etiology of the HRR deficiency lies in increased 53BP1 binding and thus prevention of DNA end resection and subsequent HRR. Our proposed model is presented in Supplementary Figure S12 (46, 47). Increases in DNA damage upon AVB-500 and platinum chemotherapy treatment likely also underlie the replication fork perturbations we observed by the DNA fiber assay. Given our findings, AVB-500 may improve ovarian cancer response to traditional taxane- and carboplatin-based chemotherapy.
Our finding that cells treated with AVB-500 plus chemotherapy had fewer RAD51 foci than cells treated with chemotherapy alone has important clinical implications, as HRR deficiencies arecorrelated with response to chemotherapy and PARP inhibitors in ovarian cancer (48). Decreased formation of RAD51 foci is strongly predictive of a complete pathologic response to chemotherapy (26). Additionally, RAD51 foci formation defects have been correlated with platinum sensitivity, platinum-free interval, time to progression, and overall survival in ovarian cancer (26, 28, 49). Importantly, we found that AVB-500 caused decreased RAD51 foci formation in both HR-deficient (OVCAR8) and HR-proficient (CAOV, COV362, and OVCAR3-TPMES) ovarian cancer cell lines. Thus, AVB-500 may be broadly applicable in treating ovarian cancer and other solid tumors.
Our data suggest that AVB-500 can improve ovarian cancer response to PARP inhibitors. In the upfront clinical treatment of advanced HGSOC patients with HR deficiency caused by BRCA mutations, PARP inhibitor maintenance after traditional chemotherapy offered a strong survival benefit (hazard ratio of 0.3) (6). However, this upfront clinical benefit has not been demonstrated in patients with HR-proficient tumors. Hill et al. and Castroviejo-Bermejo et al. showed that RAD51 foci formation is likely more predictive of response to PARP inhibitors and DNA damaging agents than mutations in HR genes such as BRCA given the risk of gene reversions and the fact that only half of HR-deficient tumors harbor mutations in HR genes (50–54). Specifically, in breast and ovarian cancers, decreased RAD51 foci formation is strongly correlated with improved PARP inhibitor response (50–54). Thus, our findings that treatment with AVB-500 resulted in decreased RAD51 foci formation and that AVB-500 plus olaparib decreased in vitro cell viability in both HR-proficient and HR-deficient tumor cells and in vivo tumor burden suggest that AVB-500 could be effectively combined with PARP inhibitors in ovarian cancer patients with and without BRCA mutations. Balaji et al. demonstrated similar findings in small cell lung, breast, and head and neck cancer further validating our results (41). Additionally, our findings that the addition of AVB-500 to olaparib increases replication fork speed are novel. It has been suggested that PARP inhibitor works primarily by preventing single-stranded breaks (SSB) and therefore the cell must rely on HR. Further research demonstrates that PARP inhibitor is also involved in replication fork progression (55). Specifically, PARP inhibition has been associated with increased speed of replication forks and as a result increased SSB and heavier reliance of HR. Our findings not only confirm these results, but also suggest that AVB-500 in addition to PARP inhibition further increases replication fork speed and thus genome instability. If our findings are validated in clinical trials, combined treatment with AVB-500 and a PARP inhibitor could significantly improve outcomes for ovarian cancer patients, particularly those with HR-proficient tumors.
Lastly, previous reports have shown that GAS6 expression correlates with survival in ovarian cancer (14, 15, 18), but whether GAS6 concentration could be used to predict response to carboplatin and paclitaxel has not been fully addressed. Our data demonstrates that high GAS6 levels in both serum and tumor tissue is associated with poor chemoresponse. Future studies evaluating GAS6 as a biomarker to predict which patients will have tumors that are resistant to platinum-chemotherapy and thus might benefit from inhibition of GAS6/AXL will be investigated.
Supplementary Material
Implications:
GAS6/AXL is a novel target to sensitize ovarian cancers to carboplatin and olaparib. Additionally, GAS6 levels can be associated with response to carboplatin treatment.
Acknowledgements
Support for this work was provided by a Reproductive Scientist Development Program award (NICHD 2K12HD000849-28) to KF and to MM, and a NCI F30 fellowship to EC (F30CA254215). Mary Mullen holds a Dean’s Scholars Award from the Washington University Division of Physician-Scientists, which is funded by a Burroughs Wellcome Fund Physician-Scientist Institutional Award (1020047). We thank Deborah Frank, PhD and Anthony Bartley for scientific editing and Andrea Byrum, PhD, for technical support.
Financial Support: NICHD Reproductive Scientist Development Program grant 2K12HD000849-28-KF.
Footnotes
- ER holds patents at Stanford University for Inhibition of AXL signaling in anti-metastatic therapy (US Patent PCT/US2011/022125) and Modified AXL peptides and their use in inhibition of AXL signaling in anti-metastatic therapy (US Patent PCT/US2013/074786).
- PT reports personal fees from Astra Zeneca and Aravive.
- MP has received consultancy fees from AstraZeneca.
- KF has received consultancy fees Aravive and holds patents at Stanford University for Inhibition of AXL signaling in anti-metastatic therapy (US Patent PCT/US2011/022125) and Modified AXL peptides and their use in inhibition of AXL signaling in anti-metastatic therapy (US Patent PCT/US2013/074786).
References
- 1.Fagotti A, Ferrandina G, Vizzielli G, Fanfani F, Gallotta V, Chiantera V, et al. Phase III randomised clinical trial comparing primary surgery versus neoadjuvant chemotherapy in advanced epithelial ovarian cancer with high tumour load (SCORPION trial): Final analysis of peri-operative outcome. Eur J Cancer. 2016;59:22–33. [DOI] [PubMed] [Google Scholar]
- 2.Parmar MK, Ledermann JA, Colombo N, du Bois A, Delaloye JF, Kristensen GB, et al. Paclitaxel plus platinum-based chemotherapy versus conventional platinum-based chemotherapy in women with relapsed ovarian cancer: the ICON4/AGO-OVAR-2.2 trial. Lancet. 2003;361(9375):2099–106. [DOI] [PubMed] [Google Scholar]
- 3.Paclitaxel plus carboplatin versus standard chemotherapy with either single-agent carboplatin or cyclophosphamide, doxorubicin, and cisplatin in women with ovarian cancer: the ICON3 randomised trial. Lancet. 2002;360(9332):505–15. [DOI] [PubMed] [Google Scholar]
- 4.Matulonis UA, Sood AK, Fallowfield L, Howitt BE, Sehouli J, Karlan BY. Ovarian cancer. Nature Reviews Disease Primers. 2016;2(1):16061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Helleday T The underlying mechanism for the PARP and BRCA synthetic lethality: clearing up the misunderstandings. Mol Oncol. 2011;5(4):387–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moore K, Colombo N, Scambia G, Kim BG, Oaknin A, Friedlander M, et al. Maintenance Olaparib in Patients with Newly Diagnosed Advanced Ovarian Cancer. N Engl J Med. 2018;379(26):2495–505. [DOI] [PubMed] [Google Scholar]
- 7.Frey MK, Pothuri B. Homologous recombination deficiency (HRD) testing in ovarian cancer clinical practice: a review of the literature. Gynecol Oncol Res Pract. 2017;4:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mark MR, Chen J, Hammonds RG, Sadick M, Godowsk PJ. Characterization of Gas6, a member of the superfamily of G domain-containing proteins, as a ligand for Rse and Axl. J Biol Chem. 1996;271(16):9785–9. [DOI] [PubMed] [Google Scholar]
- 9.Lew ED, Oh J, Burrola PG, Lax I, Zagórska A, Través PG, et al. Differential TAM receptor-ligand-phospholipid interactions delimit differential TAM bioactivities. Elife. 2014;3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kanlikilicer P, Ozpolat B, Aslan B, Bayraktar R, Gurbuz N, Rodriguez-Aguayo C, et al. Therapeutic Targeting of AXL Receptor Tyrosine Kinase Inhibits Tumor Growth and Intraperitoneal Metastasis in Ovarian Cancer Models. Mol Ther Nucleic Acids. 2017;9:251–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Macleod K, Mullen P, Sewell J, Rabiasz G, Lawrie S, Miller E, et al. Altered ErbB receptor signaling and gene expression in cisplatin-resistant ovarian cancer. Cancer Res. 2005;65(15):6789–800. [DOI] [PubMed] [Google Scholar]
- 12.Lozneanu L, Pinciroli P, Ciobanu DA, Carcangiu ML, Canevari S, Tomassetti A, et al. Computational and Immunohistochemical Analyses Highlight AXL as a Potential Prognostic Marker for Ovarian Cancer Patients. Anticancer Res. 2016;36(8):4155–63. [PubMed] [Google Scholar]
- 13.Rea K, Pinciroli P, Sensi M, Alciato F, Bisaro B, Lozneanu L, et al. Novel Axl-driven signaling pathway and molecular signature characterize high-grade ovarian cancer patients with poor clinical outcome. Oncotarget. 2015;6(31):30859–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buehler M, Tse B, Leboucq A, Jacob F, Caduff R, Fink D, et al. Meta-analysis of microarray data identifies GAS6 expression as an independent predictor of poor survival in ovarian cancer. Biomed Res Int. 2013;2013:238284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu G, Ma Z, Hu W, Wang D, Gong B, Fan C, et al. Molecular insights of Gas6/TAM in cancer development and therapy. Cell Death Dis. 2017;8(3):e2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bohm S, Faruqi A, Said I, Lockley M, Brockbank E, Jeyarajah A, et al. Chemotherapy Response Score: Development and Validation of a System to Quantify Histopathologic Response to Neoadjuvant Chemotherapy in Tubo-Ovarian High-Grade Serous Carcinoma. J Clin Oncol. 2015;33(22):2457–63. [DOI] [PubMed] [Google Scholar]
- 17.Rankin EB, Fuh KC, Taylor TE, Krieg AJ, Musser M, Yuan J, et al. AXL is an essential factor and therapeutic target for metastatic ovarian cancer. Cancer Res. 2010;70(19):7570–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quinn JM, Greenwade MM, Palisoul ML, Opara G, Massad K, Guo L, et al. Therapeutic Inhibition of the Receptor Tyrosine Kinase AXL Improves Sensitivity to Platinum and Taxane in Ovarian Cancer. Mol Cancer Ther. 2019;18(2):389–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moisan F, Francisco EB, Brozovic A, Duran GE, Wang YC, Chaturvedi S, et al. Enhancement of paclitaxel and carboplatin therapies by CCL2 blockade in ovarian cancers. Mol Oncol. 2014;8(7):1231–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chou TC. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol Rev. 2006;58(3):621–81. [DOI] [PubMed] [Google Scholar]
- 21.Deng M, Sun J, Xie S, Zhen H, Wang Y, Zhong A, et al. Inhibition of MCM2 enhances the sensitivity of ovarian cancer cell to carboplatin. Mol Med Rep. 2019;20(3):2258–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Di Veroli GY, Fornari C, Wang D, Mollard S, Bramhall JL, Richards FM, et al. Combenefit: an interactive platform for the analysis and visualization of drug combinations. Bioinformatics. 2016;32(18):2866–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stefanou DT, Bamias A, Episkopou H, Kyrtopoulos SA, Likka M, Kalampokas T, et al. Aberrant DNA damage response pathways may predict the outcome of platinum chemotherapy in ovarian cancer. PLoS One. 2015;10(2):e0117654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jucha A, Wegierek-Ciuk A, Koza Z, Lisowska H, Wojcik A, Wojewodzka M, et al. FociCounter: a freely available PC programme for quantitative and qualitative analysis of gamma-H2AX foci. Mutat Res. 2010;696(1):16–20. [DOI] [PubMed] [Google Scholar]
- 26.Graeser M, McCarthy A, Lord CJ, Savage K, Hills M, Salter J, et al. A marker of homologous recombination predicts pathologic complete response to neoadjuvant chemotherapy in primary breast cancer. Clin Cancer Res. 2010;16(24):6159–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meijer TG, Verkaik NS, Sieuwerts AM, van Riet J, Naipal KAT, van Deurzen CHM, et al. Functional Ex Vivo Assay Reveals Homologous Recombination Deficiency in Breast Cancer Beyond BRCA Gene Defects. Clin Cancer Res. 2018;24(24):6277–87. [DOI] [PubMed] [Google Scholar]
- 28.Tumiati M, Hietanen S, Hynninen J, Pietilä E, Färkkilä A, Kaipio K, et al. A Functional Homologous Recombination Assay Predicts Primary Chemotherapy Response and Long-Term Survival in Ovarian Cancer Patients. Clin Cancer Res. 2018;24(18):4482–93. [DOI] [PubMed] [Google Scholar]
- 29.Stordal B, Timms K, Farrelly A, Gallagher D, Busschots S, Renaud M, et al. BRCA1/2 mutation analysis in 41 ovarian cell lines reveals only one functionally deleterious BRCA1 mutation. Mol Oncol. 2013;7(3):567–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beaufort CM, Helmijr JC, Piskorz AM, Hoogstraat M, Ruigrok-Ritstier K, Besselink N, et al. Ovarian cancer cell line panel (OCCP): clinical importance of in vitro morphological subtypes. PLoS One. 2014;9(9):e103988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pettitt SJ, Krastev DB, Brandsma I, Dréan A, Song F, Aleksandrov R, et al. Genome-wide and high-density CRISPR-Cas9 screens identify point mutations in PARP1 causing PARP inhibitor resistance. Nat Commun. 2018;9(1):1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Forment JV, Walker RV, Jackson SP. A high-throughput, flow cytometry-based method to quantify DNA-end resection in mammalian cells. Cytometry A. 2012;81(10):922–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krishan A Rapid flow cytofluorometric analysis of mammalian cell cycle by propidium iodide staining. J Cell Biol. 1975;66(1):188–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quinet A, Carvajal-Maldonado D, Lemacon D, Vindigni A. DNA Fiber Analysis: Mind the Gap! Methods Enzymol. 2017;591:55–82. [DOI] [PubMed] [Google Scholar]
- 35.Nieminuszczy J, Schwab RA, Niedzwiedz W. The DNA fibre technique - tracking helicases at work. Methods. 2016;108:92–8. [DOI] [PubMed] [Google Scholar]
- 36.Gomes LR, Rocha CRR, Martins DJ, Fiore APZP, Kinker GS, Bruni-Cardoso A, et al. ATR mediates cisplatin resistance in 3D-cultured breast cancer cells via translesion DNA synthesis modulation. Cell Death Dis. 2019;10(6):459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quinet A, Tirman S, Jackson J, Šviković S, Lemaçon D, Carvajal-Maldonado D, et al. PRIMPOL-Mediated Adaptive Response Suppresses Replication Fork Reversal in BRCA-Deficient Cells. Mol Cell. 2020;77(3):461–74.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berti M, Ray Chaudhuri A, Thangavel S, Gomathinayagam S, Kenig S, Vujanovic M, et al. Human RECQ1 promotes restart of replication forks reversed by DNA topoisomerase I inhibition. Nat Struct Mol Biol. 2013;20(3):347–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maya-Mendoza A, Moudry P, Merchut-Maya JM, Lee M, Strauss R, Bartek J. High speed of fork progression induces DNA replication stress and genomic instability. Nature. 2018;559(7713):279–84. [DOI] [PubMed] [Google Scholar]
- 40.Woo SY, Kim S. Determination of cutoff values for biomarkers in clinical studies. Precis Future Med. 2020;4(1):2–8. [Google Scholar]
- 41.Balaji K, Vijayaraghavan S, Diao L, Tong P, Fan Y, Carey JP, et al. AXL Inhibition Suppresses the DNA Damage Response and Sensitizes Cells to PARP Inhibition in Multiple Cancers. Mol Cancer Res. 2017;15(1):45–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mirza-Aghazadeh-Attari M, Ostadian C, Saei AA, Mihanfar A, Darband SG, Sadighparvar S, et al. DNA damage response and repair in ovarian cancer: Potential targets for therapeutic strategies. DNA Repair (Amst). 2019;80:59–84. [DOI] [PubMed] [Google Scholar]
- 43.Kariolis MS, Miao YR, Diep A, Nash SE, Olcina MM, Jiang D, et al. Inhibition of the GAS6/AXL pathway augments the efficacy of chemotherapies. J Clin Invest. 2017;127(1):183–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bonifacio L, Dodds M, Prohaska D, Moss A, Giaccia A, Tabibiazar R, et al. Target-Mediated Drug Disposition Pharmacokinetic/Pharmacodynamic Model-Informed Dose Selection for the First-in-Human Study of AVB-S6-500. Clin Transl Sci. 2020;13(1):204–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brand TM, Iida M, Stein AP, Corrigan KL, Braverman CM, Coan JP, et al. AXL Is a Logical Molecular Target in Head and Neck Squamous Cell Carcinoma. Clin Cancer Res. 2015;21(11):2601–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hurley RM, Wahner Hendrickson AE, Visscher DW, Ansell P, Harrell MI, Wagner JM, et al. 53BP1 as a potential predictor of response in PARP inhibitor-treated homologous recombination-deficient ovarian cancer. Gynecol Oncol. 2019;153(1):127–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Daley JM, Sung P. 53BP1, BRCA1, and the choice between recombination and end joining at DNA double-strand breaks. Mol Cell Biol. 2014;34(8):1380–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wong-Brown MW et al. TDRiOCTR, Clinical Oncology, 10.1016/j.clon.2020.03.005. [DOI] [Google Scholar]
- 49.Spring L, Greenup R, Niemierko A, Schapira L, Haddad S, Jimenez R, et al. Pathologic Complete Response After Neoadjuvant Chemotherapy and Long-Term Outcomes Among Young Women With Breast Cancer. J Natl Compr Canc Netw. 2017;15(10):1216–23. [DOI] [PubMed] [Google Scholar]
- 50.Hill SJ, Decker B, Roberts EA, Horowitz NS, Muto MG, Worley MJ Jr., et al. Prediction of DNA Repair Inhibitor Response in Short-Term Patient-Derived Ovarian Cancer Organoids. Cancer Discov. 2018;8(11):1404–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mukhopadhyay A, Elattar A, Cerbinskaite A, Wilkinson SJ, Drew Y, Kyle S, et al. Development of a functional assay for homologous recombination status in primary cultures of epithelial ovarian tumor and correlation with sensitivity to poly(ADP-ribose) polymerase inhibitors. Clin Cancer Res. 2010;16(8):2344–51. [DOI] [PubMed] [Google Scholar]
- 52.Shah MM, Dobbin ZC, Nowsheen S, Wielgos M, Katre AA, Alvarez RD, et al. An ex vivo assay of XRT-induced Rad51 foci formation predicts response to PARP-inhibition in ovarian cancer. Gynecol Oncol. 2014;134(2):331–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Castroviejo-Bermejo M, Cruz C, Llop-Guevara A, Gutiérrez-Enríquez S, Ducy M, Ibrahim YH, et al. A RAD51 assay feasible in routine tumor samples calls PARP inhibitor response beyond BRCA mutation. EMBO Mol Med. 2018;10(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu Y, Burness ML, Martin-Trevino R, Guy J, Bai S, Harouaka R, et al. RAD51 Mediates Resistance of Cancer Stem Cells to PARP Inhibition in Triple-Negative Breast Cancer. Clin Cancer Res. 2017;23(2):514–22. [DOI] [PubMed] [Google Scholar]
- 55.van Wietmarschen N, Nussenzweig A. Mechanism for Synthetic Lethality in BRCA-Deficient Cancers: No Longer Lagging Behind. Mol Cell. 2018;71(6):877–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.