Abstract
Wastewater surveillance is a promising tool for population-level monitoring of the spread of infectious diseases, such as the coronavirus disease 2019 (COVID-19). Different from clinical specimens, viruses in community-scale wastewater samples need to be concentrated before detection because viral RNA is highly diluted. The present study evaluated eleven different virus concentration methods for the detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in wastewater. First, eight concentration methods of different principles were compared using spiked wastewater at a starting volume of 30 mL. Ultracentrifugation was the most effective method with a viral recovery efficiency of 25 ± 6%. The second-best option, AlCl3 precipitation method, yielded a lower recovery efficiency, only approximately half that of the ultracentrifugation method. Second, the potential of increasing method sensitivity was explored using three concentration methods starting with a larger volume of 1000 mL. Although ultracentrifugation using a large volume outperformed the other two large-volume methods, it only yielded a comparable method sensitivity as the ultracentrifugation using a small volume (30 mL). Thus, ultracentrifugation using less volume of wastewater is more preferable considering the sample processing throughput. Third, a comparison of two viral RNA extraction methods showed that the lysis-buffer-based extraction method resulted in higher viral recovery efficiencies, with cycle threshold (Ct) values 0.9–4.2 lower than those obtained for the acid-guanidinium-phenol-based method using spiked samples. These results were further confirmed by using positive wastewater samples concentrated by ultracentrifugation and extracted separately by the two viral RNA extraction methods. In summary, concentration using ultracentrifugation followed by the lysis buffer-based extraction method enables sensitive and robust detection of SARS-CoV-2 for wastewater surveillance.
Keywords: Ultracentrifugation, Concentration, Recovery efficiency, SARS-CoV-2
Graphical abstract
1. Introduction
The coronavirus disease 2019 (COVID-19) outbreak caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection has severely affected the global public health and economy. During the onset of the pandemic, increasing screening and testing is important for the timely implementation of control policies (Mercer and Salit, 2021). However, screening and testing have challenges, particularly when the clinical testing capacity is underutilized or unavailable. The emergence of new strains, such as alpha, beta, delta, and omicron, shows rapid viral evolution and challenges community-level testing and control of the pandemic (Hrudey and Conant, 2021).
Wastewater-based epidemiology (WBE) tool has been effective for community monitoring of gastrointestinal pathogenic viruses, such as poliovirus, norovirus, and hepatitis A virus (Brouwer et al., 2018; Hellmer et al., 2014). Recent studies have indicated that SARS-CoV-2 RNA fragments can be detected in fecal samples of patients of COVID-19, with a viral load ranging 2–8 log10 copies/mL (Cheung et al., 2020; Guo et al., 2021; Wolfel et al., 2020; Zheng et al., 2020). The application of the WBE tool in SARS-CoV-2 detection is effective and efficient, providing early warning signals before community outbreaks (Medema et al., 2020; Nemudryi et al., 2020; Xu et al., 2021). Furthermore, it enables the monitoring of the progression of new strains ahead of clinical testing (Graber et al., 2021; Jahn et al., 2021). In addition, it can serve as an unbiased and non-invasive surveillance tool for identifying previously unknown symptomatic or asymptomatic patients (Wu et al., 2020). Effective and robust wastewater testing for SARS-CoV-2 can help provide important and timely information to stakeholders and decision-makers for making prompt control measures to fight against COVID-19, such as evaluation of the lockdown effect on pandemic dynamics (Hillary et al., 2021; Wurtzer et al., 2020), uncovering hidden cases on college campuses (Gibas et al., 2021), and assessment of the effectiveness of vaccination (Bivins and Bibby, 2021).
Compared with the clinical testing, the detection of SARS-CoV-2 in wastewater is difficult due to low viral amounts and matrix effects on detection imposed by the complex components in the wastewater. Therefore, effective virus concentration methods are essential for the sensitive detection of low concentration SARS-CoV-2 in wastewater. Up to now, researchers adopted various concentration methods, such as polyethylene glycol (PEG) precipitation (Wu et al., 2020), AlCl3 precipitation (Randazzo et al., 2020), ultrafiltration with centrifugal filters (Medema et al., 2020), ultracentrifugation (Wurtzer et al., 2020), and membrane adsorption (Haramoto et al., 2020).
To date, there are limited systematic comparison studies on concentration methods for SARS-CoV-2 virus in wastewater. Reported studies have focused on the performance of virus concentration methods using different concentration principles (Ahmed et al., 2020; LaTurner et al., 2020), inter-laboratory comparisons (Pecson et al., 2021), or different surrogate viruses (Jafferali et al., 2021; Philo et al., 2021). However, most of the published virus concentration methods are processed with 30–250 mL wastewater because of the practicality of sample handling in the laboratory and the availability of some special instruments. A few studies have proposed increasing detection sensitivity by increasing wastewater sample volume (Gerrity et al., 2021; McMinn et al., 2021; Xu et al., 2021). In addition, wastewater characteristics and matrices should be carefully considered when selecting a virus concentration method for SARS-CoV-2 (Pecson et al., 2021).
The present study conducted a comprehensive evaluation of the concentration methods commonly used for SARS-CoV-2 in wastewater. Specifically, the objectives of this study were to 1) compare the performance of eight concentration methods using small-volume (30 mL) wastewater, 2) compare the performance of small-volume (30 mL) and large-volume (1000 mL) wastewater concentration methods, and 3) evaluate the effects of two viral RNA extraction methods on SARS-CoV-2 detection.
2. Material and methods
2.1. Sampling
Twelve raw wastewater samples were collected at the inlets of the Shatin Sewage Treatment Works (Shatin STW) from September to November 2020 for the spiking experiments, including four samples for small-volume concentration method comparison, three samples for large-volume and small-volume comparison, two samples for spiked wastewater at marginal levels, and three samples for serial dilution experiments. In addition, the virus was spiked into four raw wastewater samples from four sewage pumping stations (SPS), including Ho Pong Street SPS (HP), Sham Shui Po SSP No. 1 SPS (SSP1), Siu Hong SPS (SH), and Yau Tong SPS (YT), and used for comparison of extraction methods.
Furthermore, 66 wastewater samples were collected from manholes, SPS, and wastewater treatment plants during a COVID-19 outbreak in Hong Kong (November to December 2020). Among them, 35 were used for comparison of starting sample volumes and 31 for comparison of extraction methods.
Wastewater samples were transported to the laboratory on ice and processed within 24 h. All wastewater samples were heat-inactivated at 60 °C for 30 min to ensure lab safety before sample processing (Chin et al., 2020). The effect of heat-inactivation on detection and method comparison was not evaluated in this study. Previous studies showed that heat-inactivation at 56 °C for 30 or 60 min could inactivate the infection of the SARS-CoV-2 virus but insignificantly affect the detection of Ct values using reverse transcription-quantitative polymerase chain reaction (RT-qPCR) (Auerswald et al., 2021; Batejat et al., 2021).
2.2. Spiking experiments
2.2.1. Comparison of different small-volume concentration methods
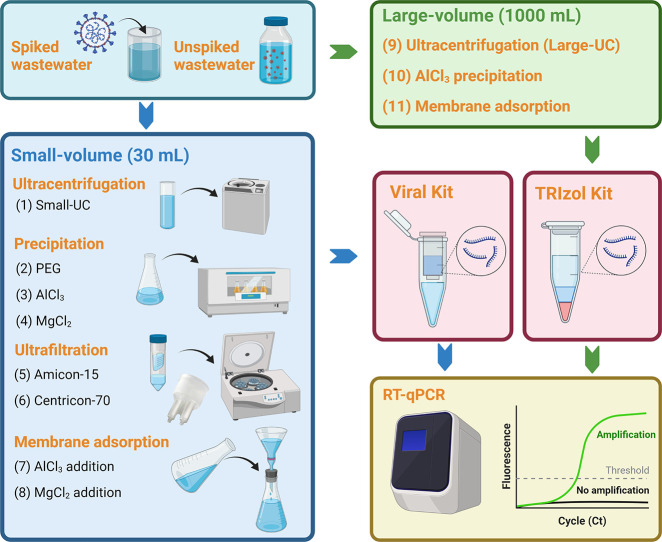
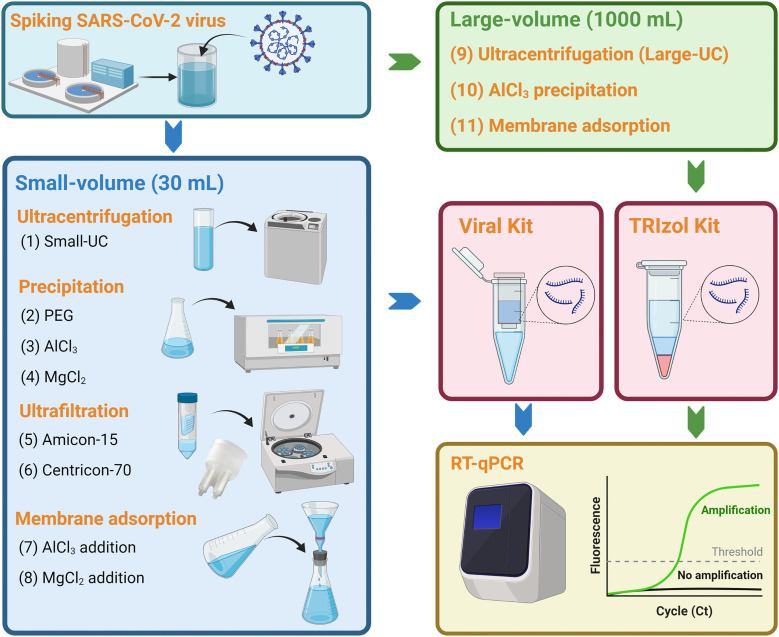
Eight concentration methods were compared using a starting volume of 30 mL (defined as “small-volume” in the present study). In detail, SARS-CoV-2 virus was collected from the cell culture and diluted to approximately 108 copies/mL (calculated based on the pretest experiment) after heat-inactivation. The accurate theoretical spiked virus concentration was calculated based on the triplicate extraction and detection of 10 μL of heat-inactivated SARS-CoV-2 virus in each batch of the spiking experiment. Next, 200 μL of diluted SARS-CoV-2 virus was spiked into 600 mL of supernatant collected after centrifugation of raw wastewater samples at 4750 ×g for 30 min. The spiked wastewater was further divided into aliquots of 30 mL for the evaluation of eight methods derived from four different concentration principles (Fig. 1 ): (1) ultracentrifugation, (2) precipitation (i.e. AlCl3, PEG, and MgCl2), (3) ultrafiltration (i.e. Amicon-15 and Centricon-70), and (4) membrane adsorption (using 0.45 μm electronegative membrane with the addition of MgCl2 or AlCl3).
Method 1 was ultracentrifugation to concentrate the virus in the samples. Samples were centrifuged at 150,000 ×g for 60 min at 4 °C on Optima XPN Ultracentrifuge (Beckman Coulter, Brea, CA, USA). The supernatant was removed without disturbing the pellet. The pellet was further resuspended in 200 μL phosphate buffered saline (PBS) and transferred into a new 1.5 mL micro-centrifugal tube for RNA extraction.
Methods 2–4 were precipitation methods using different flocculants. Method 2 began with adding PEG (10%, w/v) and NaCl (2%, w/v) into samples. Method 3 and Method 4 began with the flocculation of samples using 0.3 M AlCl3 (1%, v/v) or 2.5 M MgCl2 (1%, v/v). The resulting samples from Methods 2–4 were agitated at 150 rpm for 30 min on the Innova 44 Incubator Shaker Series (New Brunswick, Eppendorf, Hamburg, Germany). Next, they were centrifuged at 20,000 ×g for 30 min on the Sorvall LYNX 4000 Superspeed Centrifuge (Thermo Scientific, Waltham, MA, USA). The supernatant was discarded, and the pellet was used for RNA extraction.
Methods 5 and 6 were ultrafiltration methods using two kinds of centrifugal filters: Amicon-Ultra 15 Centrifugal Filter (Merck Millipore, Tullagreen, Cork, Ireland) with a molecular weight cut-off of 10 kDa, and Centricon Plus-70 centrifugal filter (Merck Millipore) with a cut-off of 30 kDa. Samples were centrifuged at 4500 ×g for 15 min at 4 °C. Concentrated samples (approximately 200 μL) were collected from the retentate and used for RNA extraction.
Methods 7 and 8 were membrane adsorption methods. The flocculants of 0.3 M AlCl3 (1%, v/v) or 2.5 M MgCl2 (1%, v/v) were added to wastewater samples. Samples were then passed through an electronegative membrane with 0.45 μm pore size and 47 mm diameter (Merck Millipore) through a filter funnel and a filter flask (Thermo Scientific). The membrane was removed and eluted with 4 mL of PBS to recover the virus. Viral particles were repeatedly blown and scraped from the membrane using a P200 pipette. Eluant was further concentrated at 20,000 ×g for 2 min at 4 °C to obtain a pellet (approximately 200 μL) for RNA extraction.
Fig. 1.
Workflow for the evaluation of eleven virus concentration methods in spiking experiments. Figure was created with BioRender.com.
Meanwhile, 30 mL of wastewater supernatant without spiking SARS-CoV-2 virus and 30 mL of double-distilled water (ddH2O) were concentrated using the eight methods as negative controls. All these wastewater and ddH2O samples were finally tested negative.
All the above comparison experiments were conducted for four times using four wastewater samples taken on different dates. All concentrated samples were extracted using QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany) following the manufacturer's instructions, with a final elution volume of 40 μL.
2.2.2. Comparison of large-volume and small-volume concentration methods
Heat-inactivated SARS-CoV-2 virus (approximately 108 copies/mL) of 1.5 mL were spiked into 4.5 L wastewater supernatant collected after centrifugation at 4750 ×g for 30 min, and mixed completely by shaking for 15 min. Triplicate aliquots of 334 μL heat-inactivated SARS-CoV-2 virus were extracted and detected directly to obtain the theoretical spiking virus concentration for calculating the method recovery efficiency in each batch experiment. Then, for large-volume concentration methods, 1000 mL aliquots were collected and processed separately using one of the following three methods (Fig. 1): (1) centrifugation at 20,000 ×g for 30 min and then ultracentrifugation at 150,000 ×g for 60 min, (2) AlCl3 precipitation, and (3) 0.45 μm electronegative membrane adsorption. In addition, 30 mL of the spiked wastewater sample was concentrated by ultracentrifugation at 150,000 ×g for 60 min to represent the small-volume method for comparison (Method 1).
Method 9 was based on a combination of centrifugation and ultracentrifugation. First, 1000 mL spiked wastewater was centrifuged at 20,000 ×g for 30 min via the Sorvall LYNX 4000 Superspeed Centrifuge (Thermo Scientific), and approximately 30 mL of the precipitate was retained for further ultracentrifugation at 150,000 ×g for 60 min using the Optima XPN Ultracentrifuge (Beckman Coulter). Next, the concentrated viral pellet was resuspended with approximately 400 μL PBS and used for further RNA extraction.
Method 10 was an AlCl3 precipitation-based concentration method. Ten milliliter AlCl3 solution (1%, 0.3 M, v/v) was added to 1000 mL spiked wastewater to precipitate the viral particles. After shaking at 150 rpm for 30 min and centrifuging at 20,000 ×g for 30 min using Sorvall LYNX 4000 Superspeed Centrifuge (Thermo Scientific), concentrated viral pellets were obtained and used for RNA extraction.
Method 11 was the membrane adsorption method. The spiked wastewater (1000 mL) was passed through a 0.45 μm pore-size, 47 mm diameter electronegative membrane (MF-Millipore, Darmstadt, Germany. Cat. No: HAWP09000). The membrane was collected and washed with 4 mL of PBS to obtain a filter pellet for RNA extraction. Visible film-forming viral particles can be easily washed off from the membrane in lumps.
Wastewater samples without spiking viruses and ddH2O of the same volume were used as negative controls. No SARS-CoV-2 signal was detected in these negative wastewater or ddH2O. The above comparisons of the three large-volume and one small-volume method were conducted three times using wastewater samples taken on different dates. RNA extraction for all the concentrated samples from large-volume and small-volume wastewater were conducted using TRIzol Plus RNA Purification Kit (Thermo Fisher) with an elution volume of 40 μL, as our pre-experiment showed that the QIAamp Viral RNA Mini Kit did not apply to large-volume wastewater due to spin column clogging issues.
2.2.3. Comparison of two extraction methods
To evaluate the performance of two viral RNA extraction methods (lysis-buffer-based vs acid-guanidinium-phenol-based), 80 μL of heat-inactivated SARS-CoV-2 (approximately 108 copies/mL) was spiked into 240 mL supernatant collected after raw wastewater was centrifugated at 4750 ×g for 30 min, to obtain a final concentration of approximately 105 copies/mL. Next, 30 mL samples were concentrated by ultracentrifugation methods at 150,000 ×g for 60 min, followed by RNA extraction using the QIAamp Viral RNA Mini Kit (lysis-buffer-based method, called “Viral Kit” in the following description) or TRIzol Plus RNA Purification Kit (acid-guanidinium-phenol-based method, called “TRIzol Kit”). For comparison, triplicate samples using wastewater from the same sampling site were used to test the processing variation. The comparison was repeated using wastewater samples from four sampling sites: HP, SSP1, SH, and YT in Hong Kong.
Reagent blanks using 200 μL AVE buffer or RNase-free water from the viral RNA extraction kits were used in each extraction batch as a quality control measure.
2.3. Method evaluations of spiked wastewater at marginal levels
After centrifugation at 4750 ×g for 30 min, the wastewater supernatant was spiked with inactivated SARS-CoV-2 virus to obtain a concentration of 1–100 copies/mL wastewater to test the performance of three methods (Method 1 with Viral Kit or TRIzol Kit, and Method 9 with TRIzol Kit) under the challenge of virus concentration at marginal levels. To be specific, 400 μL of virus at three concentrations (around 103–105 copies/mL) were spiked into 1.2 L wastewater samples to generate spiked wastewater of different concentrations respectively, naming “high-concentration spiked wastewater (approximately 100 copies/mL, called “High” in the following description), “middle-concentration spiked wastewater (approximately 10 copies/mL, called “Medium”), and “low-concentration spiked wastewater (approximately 1 copy/mL, called “Low”). For each virus concentration, two 30 mL spiked samples were concentrated by ultracentrifugation and extracted with two viral extraction methods, that is, TRIzol Kit and Viral Kit. In addition, a 1000 mL spiked wastewater sample was extracted using the TRIzol Kit after the large-volume concentration methods. Wastewater samples without spiking SARS-CoV-2 virus were used as negative controls. All experiments were performed in duplicate.
2.4. Serial dilution experiments
The heat-inactivated SARS-CoV-2 with a stock concentration of 6.09 × 108 copies/reaction (approximately 1010 copies/mL virus) was 10-fold diluted using either PBS or negative wastewater samples to obtain a virus concentration range of 6.09 × 102–6.09 × 107 copies/reaction (the range of log10 virus concentration was 3–8). For each virus concentration, 100 μL of PBS diluted virus or wastewater diluted virus was extracted using either the TRIzol Kit or Viral Kit, with a final elution of 50 μL. The entire process was repeated in triplicates.
2.5. Evaluation using unspiked wastewater samples
Wastewater samples without spiking were used to evaluate the detection sensitivity of the different concentration methods. The evaluation focused on the comparison of ultracentrifugation with small-volume or large-volume wastewater, and the comparison of extraction by Viral kit or TRIzol kit, based on detection rates and detected Ct values. These samples were taken from manholes, sewage pumping stations, and wastewater treatment plants in Hong Kong.
2.6. Detection methods
RT-qPCR were performed to quantify the extracted viral RNA. For spiked wastewater, the HKU-N probe and primers (Chu et al., 2020) was used for detection and quantification. For wastewater samples without spiking, N1 primers and probe from the United States Center for Disease Control (US CDC) (CDC, 2020) was used. Two fragments of 110 bp and 72 bp containing HKU-N and N1 targets, respectively, were synthesized and cloned into pUC-19 vectors by the Beijing Genomics Institute (BGI, Hong Kong, China). The plasmids were used to generate standard curves. The concentration of plasmids was quantified using the Qubit dsDNA HS assay kit (Thermo Fisher) and the copy number was calculated based on DNA length and Avogadro's number. Then, the RT-qPCR was carried out for 45 cycles in 20 μL reaction mixture using TaqMan Fast Virus One-step Master Mix (Thermo Fisher). The one-step RT-qPCR reaction mixtures contained 5 μL 4 × TaqMan Fast Virus 1-Step Master Mix (Thermo Fisher), 500 nM forward primer, 500 nM reverse primer, 250 nM probe, 4 μL template RNA, and DEPC-treated water to 20 μL. The RT-qPCR thermal cycling conditions were as follows: 50 °C for 5 min, 95 °C for 20 s, 45 cycles of 95 °C for 5 s and 58 °C (HKU-N) or 55 °C (N1) for 30 s on the Applied Biosystems ViiA7 qPCR system (Thermo Fisher). If the Ct value of the extracted RNA was less than 40, the sample was considered to have SARS-CoV-2 RNA signals.
Wastewater samples were detected and separated into different batch experiments for comparison purposes, that is, detection of spiked wastewater samples in different batches of spiking experiments, and detection of unspiked wastewater samples for paired comparison. In this practice, compared to the virus concentration, the Ct value is more comparable to represent the difference between methods and more directly reflects the sensitivity of RT-qPCR. The Ct value can be transformed into virus concentrations for comparison using standard curves, but it may introduce other biases due to batch effects. Similarly, previous studies have used Ct values rather than virus concentrations for comparison (Jafferali et al., 2021; Perez-Cataluna et al., 2021; Philo et al., 2021).
2.7. Calculation of recovery efficiency
SARS-CoV-2 virus recovery efficiency was calculated based on the ratio of the detected viral concentration throughout the process and the theoretical spiked virus concentration, which was determined by RT-qPCR after direct RNA extraction.
2.8. Statistical analysis
One-way analysis of variance (ANOVA) was used to determine the significance of differences between the means of Ct values and recovery efficiency among different concentration methods. If the difference was significant (p < 0.05), a post-hoc t-test was used to determine which methods were significantly different. All analyses were performed using R Studio version 1.3.
3. Results and discussions
3.1. SARS-CoV-2 assays performances
The HKU-N and N1 detection assays were used in this study, and they showed similar dynamic linear ranges, with values of 1.056 × 10–1.056 × 107 copies/reaction and 1.052 × 10–1.052 × 107 copies/reaction, respectively. Standard curves of HKU-N and N1 assays showed strong linear fits, with R2 values ranging from 0.9956 to 0.9996 and 0.9942 to 0.9992, respectively. The Y-intercept of the standard curves for HKU-N and N1 assays ranged from 38.59 to 43.63 and 35.94 to 37.47, respectively. The slopes of the standard curves ranged from −3.295 to −3.487 and −3.111 to −3.427, and the amplification efficiencies ranged from 93.86 to 101.2% and 95.81 to 109.6% for the HKU-N and N1 assays, respectively. Detailed information was summarized in Table 1 .
Table 1.
Performance of SARS-CoV-2 RT-qPCR detection assays.
| Run | Assays | Efficiency (%) | R2 | Slope | Y-intercept |
|---|---|---|---|---|---|
| 1 | HKU-N |
95.37 | 0.9956 | −3.438 | 43.63 |
| 2 | 93.56 | 0.9995 | −3.487 | 41.52 | |
| 3 | 94.62 | 0.9996 | −3.458 | 41.25 | |
| 4 | 94.24 | 0.9967 | −3.468 | 40.55 | |
| 5 | 93.86 | 0.9995 | −3.478 | 39.32 | |
| 6 | 101.15 | 0.9991 | −3.295 | 38.59 | |
| 7 | 95.90 | 0.9970 | −3.424 | 39.09 | |
| 8 | N1 | 100.53 | 0.9989 | −3.309 | 37.26 |
| 9 | 101.78 | 0.9992 | −3.280 | 37.39 | |
| 10 | 109.64 | 0.9969 | −3.111 | 35.94 | |
| 11 | 99.70 | 0.9985 | −3.329 | 37.17 | |
| 12 | 95.81 | 0.9985 | −3.427 | 37.47 | |
| 13 | 104.23 | 0.9983 | −3.225 | 36.22 | |
| 14 | 97.67 | 0.9942 | −3.379 | 37.31 | |
3.2. Comparisons of different concentration methods using a small starting volume (30 mL)
We evaluated the performance of the eight concentrations methods using a small wastewater volume of 30 mL (Fig. 1). As shown in Fig. 2 , ultracentrifugation had the highest recovery efficiency of 25.4 ± 5.9%, significantly higher than other methods (p < 0.01), followed by precipitation-based methods, ultrafiltration methods, and finally membrane-adsorption methods. The recovery efficiency was similar to those from other studies on the bovine respiratory syncytial virus using ultracentrifugation (Prado et al., 2021), and higher than the values reported in studies on RNA bacteriophage (Prado et al., 2021) and inactivated SARS-CoV-2 virus (Green et al., 2020) (Table 2 ). The results imply that the method sensitivity of ultracentrifugation outperformed other methods for the detection of SARS-CoV-2. However, ultracentrifugation relies on the ultracentrifugation equipment, not readily available in many laboratories.
Fig. 2.
Comparison of eight small-volume concentration methods. (a) Ct values; (b) recovery efficiency (%). Experiments were repeated for four time (n = 4). UC: Ultracentrifuge at 150,000 ×g for 60 min; AlCl3: AlCl3 precipitation (1%, 0.3 M, v/v); PEG: PEG precipitation (10%, w/v) with the addition of NaCl (2%, w/v); MgCl2: MgCl2 precipitation (1%, 2.5 M, v/v); A15: using a Amicon-Ultra 15 Centrifugal Filter with a cut-off of 10 kDa; C70: using a Centricon Plus-70 centrifugal filter with a cut-off of 30 kDa; AlM: using a 0.45 μm electronegative membrane with the addition of AlCl3 solution (1%, 0.3 M, v/v); MgM: using a 0.45 μm electronegative membrane with the addition of MgCl2 solution (1%, 2.5 M, v/v).
Table 2.
Comparison of virus recovery efficiency between this study and other studies.
| Methodology | Methods | Surrogate | Recovery efficiency (%) | References |
|---|---|---|---|---|
| Ultracentrifugation |
Ultracentrifugation | inactivated SARS-CoV-2 virus |
range: 20.5% - 33.4% mean: 25.4 ± 5.91% |
this study |
| Bovine respiratory syncytial virus (BRSV) |
range: 11.4% - 40.2% mean: 27.4 ± 8.64% |
(Prado et al., 2021) |
||
| RNA bacteriophage (PP7) |
range: 6.7% - 24.9% mean: 18.5 ± 7.46% |
(Prado et al., 2021) |
||
| inactivated SARS-CoV-2 virus |
12% |
(Green et al., 2020) |
||
| Precipitation |
AlCl3 | inactivated SARS-CoV-2 virus |
range: 4.9% - 14.8% mean: 11.0 ± 4.36% |
this study |
| Porcine Epidemic Diarrhea Virus (PEDV) |
10 ± 3.5% |
(Randazzo et al., 2020) |
||
| mengovirus (MgV) |
10 ± 2.1% |
(Randazzo et al., 2020) |
||
| PEG | inactivated SARS-CoV-2 virus |
range: 5.8% - 23.6% mean: 11.8 ± 8.38% |
this study |
|
| gamma-irradiated SARS-CoV-2 virus |
52.8 ± 18.2% |
(Randazzo et al., 2020) |
||
| Porcine Epidemic Diarrhea Virus (PEDV) |
27.5 ± 14.3% |
(Randazzo et al., 2020) |
||
| Bovine coronavirus (BCoV) |
0.08% |
(LaTurner et al., 2020) |
||
| MgCl2 |
inactivated SARS-CoV-2 virus |
range: 6.1% - 23.8% mean: 12.4 ± 8.34% |
this study |
|
| Ultrafiltration |
Amicon Ultra-15 | inactivated SARS-CoV-2 virus |
range: 6.5% - 17.5% mean: 9.6 ± 5.23% |
this study |
| murine hepatitis virus (MHV) |
56.0 ± 32.3% |
(Ahmed et al., 2020) |
||
| Bovine coronavirus (BCoV) |
0.36% |
(LaTurner et al., 2020) |
||
| Centricon Plus-70 | inactivated SARS-CoV-2 virus |
range: 3.5% - 13.0% mean: 7.9 ± 4.09% |
this study |
|
| F-specific RNA phages |
73 ± 50% |
(Medema et al., 2020) |
||
| Bovine coronavirus (BCoV) |
55% ± 38% |
(Gerrity et al., 2021) |
||
| murine hepatitis virus (MHV) |
28.0 ± 9.1% |
(Ahmed et al., 2020) |
||
| Membrane-adsorption | AlCl3 |
inactivated SARS-CoV-2 virus |
range: 1.6% - 17.1% mean: 11.0 ± 8.25% |
this study |
| MgCl2 | inactivated SARS-CoV-2 virus |
range: 0.7% - 8.5% mean: 4.1 ± 3.33% |
this study |
|
| murine hepatitis virus (MHV) |
56.0 ± 32.3% |
(Ahmed et al., 2020) |
||
| MS2 |
mean 1.0% - 9.5% |
(Torii et al., 2021) |
||
| φ6 | mean 1.6% - 9.7% | (Torii et al., 2021) |
The mean recovery efficiency of different precipitants (AlCl3, PEG, or MgCl2) showed no significant differences, but AlCl3 precipitation had the lowest variability. The recovery efficiency of AlCl3 precipitation (11.0 ± 4.36%) concurred with other studies using the Porcine Epidemic Diarrhea Virus and Mengovirus as the surrogate viruses (Randazzo et al., 2020) (Table 2). Therefore, AlCl3 precipitation was considered a preferable substitute method considering the less availability of the special equipment needed in the ultracentrifugation method, although its recovery efficiency was only half of that of the ultracentrifugation method.
The recovery efficiencies of ultrafiltration-based and membrane-adsorption methods, ranging from 4 to 11%, were lower than those in other studies (Table 2). A previous report obtained the highest recovery efficiency using MgCl2 membrane adsorption compared to the other methods for concentrating murine hepatitis virus (MHV) from wastewater influents (Ahmed et al., 2020), but MgCl2 membrane adsorption showed the most inferior performance for the SARS-CoV-2 virus in this study. Such inconsistency could probably result from the difference in the detachment of viral particles from the electronegative membrane. Bead beating of the entire membrane was used in the study of Ahmed et al (Ahmed et al., 2020), while viral particles were only detached from the membrane by repeated washing and scraping using a P200 pipette in the present study. Except for these operational differences, the differences in matrix components in wastewater samples (Cashdollar and Wymer, 2013) and utilization of different surrogate viruses (MHV vs inactivated SARS-CoV-2) may also contribute to difference. Compared to surrogate viruses, directly spiking with the SARS-CoV-2 virus is more representative to evaluate the performance of virus concentration methods for SARS-CoV-2 wastewater surveillance. However, there is still a limitation that spiking inactivated virus for evaluation cannot totally reflect actural behaviors of SARS-CoV-2 virus in sewage samples, which may affect the performances of virus concentration methods in practical applications.
3.3. Comparisons of large-volume and small-volume concentration methods
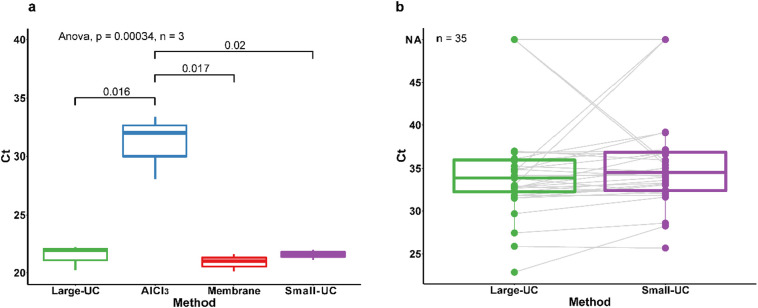
To explore the potential of increasing the method sensitivity by using a larger sample volume, three large-volume methods were compared and evaluated. As shown in Fig. 3a, large-volume samples concentrated using ultracentrifugation and membrane adsorption showed lower Ct values than using AlCl3 precipitation. Because the membrane adsorption method includes a time-consuming step to filter 1000 mL of wastewater samples through the membrane (approximately 3 h per sample), which is more time-consuming than the ultracentrifugation method (less than 2 h per six samples), the large-volume ultracentrifugation method was chosen for further comparison with the ultracentrifugation-based small-volume method which had the highest recovery efficiency among the eight small-volume methods.
Fig. 3.
Comparison of large-volume concentration methods and the small-volume ultracentrifugation method. (a) Ct values of three large-volume concentration methods and one small-volume ultracentrifugation concentration method; Experiments were repeated in triplicate (n = 3). (b) Ct values of ultracentrifugation methods based on 35 unspiked wastewater samples (n = 35). Large-UC: centrifuge at 20,000 ×g for 30 min and then ultracentrifuge at 150,000 ×g for 60 min; AlCl3: AlCl3 precipitation (1%, 0.3 M, v/v); Membrane: using a 0.45 μm electronegative membrane; Small-UC: Ultracentrifuge at 150,000 ×g for 60 min.
Similar Ct values were obtained for the small-volume and large-volume ultracentrifugation methods (Fig. 3b). For viral concentrations at a marginal level of 1–100 copies/mL wastewater, both the large- and small-volume methods reached the same virus concentration range of 10 copies/mL wastewater (medium concentration) (Table 3 ). In addition, the detection rates of the small- and large-volume methods were the same for 35 wastewater samples without spiking (Table 3). Only slight lower Ct values were observed in the large-volume method than in the small-volume method (Fig. 3b). The results show that increasing the sample volume did not significantly increase the detection sensitivity, probably because the wastewater matrix was co-enriched with viral particles using the large-volume method, unfavorable for the viral RNA extraction and detection. This implies the need to quantify the effects of the wastewater matrix on viral RNA extraction and detection for a more comprehensive understanding of the quantification of SARS-CoV-2 in wastewater samples.
Table 3.
Detection rates and Ct values of spiked wastewater at marginal levels and unspiked wastewater samples.
| Spiked wastewater |
Unspiked wastewater |
|||||||
|---|---|---|---|---|---|---|---|---|
| Blank | Low | Medium | High | Total detection rates | Single detection rates | Lower Ct | Average Ct values | |
| Small-UC | 0 (0/2) | 0 (0/2) | 50% (1/2) | 100% (2/2) | 85.7% (30/35) | 5.7% (2/35) | 28.6% (8/28) | 33.88 ± 3.01 |
| Large-UC | 0 (0/2) | 0 (0/2) | 100% (2/2) | 100% (2/2) | 85.7% (30/35) | 5.7% (2/35) | 71.4% (20/28) | 32.85 ± 3.15 |
| TRIzol Kit | 0 (0/2) | 0 (0/2) | 50% (1/2) | 100% (2/2) | 67.7% (21/31) | 3.2% (1/31) | 20% (4/20) | 34.39 ± 2.68 |
| Viral Kit | 0 (0/2) | 50% (1/2) | 100% (2/2) | 100% (2/2) | 77.4% (24/31) | 12.9% (4/31) | 80% (16/20) | 33.46 ± 2.67 |
Blank: raw wastewater without spiking SARS-CoV-2 virus;
Low: SARS-CoV-2 virus-spiked wastewater with a concentration of approximately 1 copy/mL wastewater;
Medium: SARS-CoV-2 virus-spiked wastewater with a concentration of approximately 10 copies/mL wastewater;
High: SARS-CoV-2 virus-spiked wastewater with a concentration of approximately 100 copies/mL wastewater;
Total detection rates: the percentage of detectable samples for the chosen method;
Single detection rates: the percentage of samples which were only detectable for the chosen method;
Lower Ct: the percentage of samples with lower Ct values when detectable using both methods.
3.4. Comparisons of two extraction methods
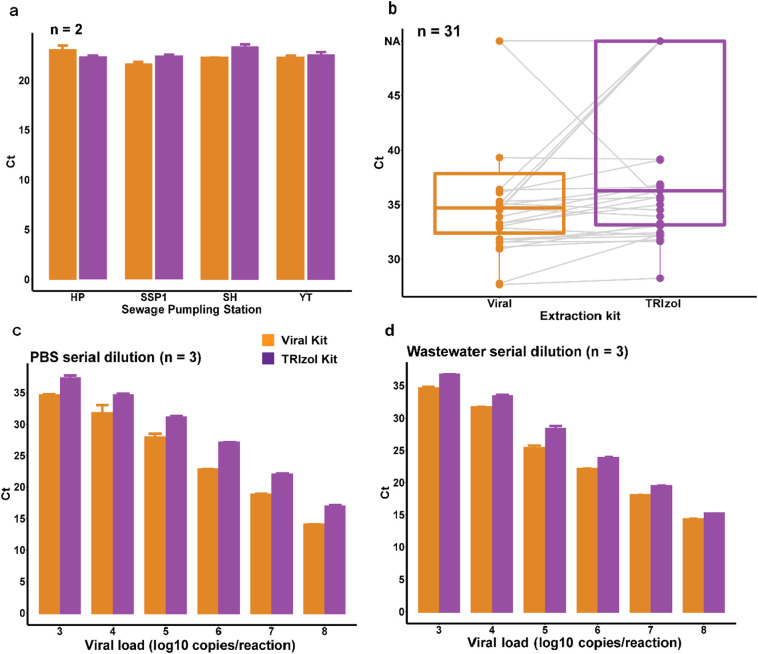
Two viral RNA extraction methods were compared using SARS-CoV-2 spiked wastewater samples and 31 unspiked wastewater samples using the small-volume ultracentrifugation method. Lower Ct values were observed using the lysis-buffer-based method (Viral Kit) than the acid-guanidinium-phenol-based method (TRIzol Kit) for spiked wastewater from different sampling sites, which represented varied matrix compositions (Fig. 4a). In addition, for spiked wastewater at marginal levels, the Viral Kit can detect SARS-CoV-2 RNA at “Low” concentration (1 copy/mL wastewater), representing good sensitivity using Viral Kit (Table 3). In contrast, TRIzol Kit could only detect RNA low to “Medium” concentration (10 copies/mL wastewater) level. Finally, the Viral Kit yielded higher detection rates and lower Ct values than the TRIzol Kit in 31 unspiked wastewater samples (Table 3, Fig. 4b). These results indicate that the Viral Kit may perform better while both of the two kits had good performances.
Fig. 4.
Comparison of two extraction methods. (a) Ct values of spiked wastewater from four sewage pumping stations (SPS); Experiments were repeated in duplicate (n = 2). (b) Ct values of ultracentrifugation methods based on 31 unspiked wastewater samples (n = 31). (c) Ct values of PBS serial dilution SARS-CoV-2 virus; Experiments were repeated in triplicate (n = 3). (d) Ct values of wastewater serial dilution SARS-CoV-2 virus; Experiments were repeated in triplicate (n = 3). HP: Ho Pong Street SPS; SSP1: Sham Shui Po SSP No. 1 SPS; SH: Siu Hong SPS; YT: Yau Tong SPS. Viral Kit: a lysis-buffer-based method using QIAamp Viral RNA Mini Kit (Qiagen); TRIzol Kit: an acid-guanidinium-phenol-based method using TRIzol Plus RNA Purification Kit (Thermo Fisher). Error bars represent standard deviation.
The differences in detection rates and Ct values may be due to higher virus extraction efficiency in the Viral Kit rather than in the TRIzol Kit, especially when processing relatively particle-free samples. To confirm this hypothesis, serial dilutions of SARS-CoV-2 virus in PBS or wastewater were extracted using two viral extraction methods. As shown in Fig. 4c and d, the Viral Kit consistently obtained lower Ct values than the TRIzol Kit for a dynamic range of 3–8 log10 copies/reaction, irrespective of the sample type. The Ct values differences ranged 2.86–4.18 and 0.86–2.99 for PBS solution and wastewater samples, respectively. These results suggest the Viral Kit obtain more viral RNA for detection than the TRIzol Kit in the same samples.
Compared with the TRIzol Kit, the Viral Kit is more rapid in experimental procedures and more commonly used in SARS-CoV-2 WBE studies (Haramoto et al., 2020; Hata et al., 2021). The Viral Kit is applicable to automated extraction machines such as QIAcube Connect (Goncalves et al., 2021), permitting high-throughput viral RNA extraction in the rapid implementation of wastewater surveillance during a pandemic. However, the Viral Kit has a disadvantage of spin column clogging issues, whereas the TRIzol Kit present superior performance in turbid samples, such as large-volume wastewater samples in this study, and wastewater samples concentrated by PEG precipitation (Torii et al., 2021).
3.5. Perspectives
In the present study, ultracentrifugation outperformed other concentration methods when processing 30 mL of wastewater, but we cannot overlook the sensitivity increase potential of other concentration methods when processing larger sample volumes. In addition, low-speed centrifugation (4750 ×g for 30 min) was conducted before the virus concentration, which could enhance method sensitivity by removing debris but it may also influence the performance of precipitation-based methods evaluated in this study. Meanwhile, different recovery efficiencies of small-volume ultracentrifugation methods were observed among different sampling sites due to matrix effects imposed by the wastewater samples. The performance of different virus concentration methods will be influenced by their tolerance to the matrix effect of the wastewater samples, which needs to be re-considered and re-evaluated when selecting a suitable concentration method for wastewater samples from other sampling sites in different countries.
Furthermore, the sensitivity of virus concentration methods could be increased by modifying their method operational parameters or combining them with other virus RNA extraction kits. Therefore, future studies should explore the optimal operational parameters for a specific method and the selection of virus RNA extraction kits.
Finally, in addition to the method sensitivity evaluated in the present study using Hong Kong wastewater, the selection of virus concentration methods relied on equipment accessibility, method familiarity, labor requirement, supply availability, and throughput of the sample processing in the laboratory.
4. Conclusions
Using heat-inactivated SARS-CoV-2 virus spiked wastewater and unspiked wastewater samples, the present study assessed the performance of eleven virus concentration methods for detecting SARS-CoV-2 virus. The comparison of eight small-volume (30 mL) methods showed that ultracentrifugation was the most sensitive method, with the lowest Ct values and highest recovery efficiency (25.4 ± 5.9%). To explore the potential to increase method sensitivity, larger-volume concentration methods were compared with the small-volume method using ultracentrifugation. The sensitivity of the evaluated large-volume methods was similar to that of the small-volume method regarding detection rates and Ct values. Considering operational feasibility and throughput, processing small-volume wastewater samples is a cost-effective, sensitive, and robust strategy. Furthermore, using the small-volume ultracentrifugation method, two viral RNA extraction methods, i.e. lysis-buffer-based method vs acid-guanidinium-phenol-based method, were compared, and the results showed that the lysis-buffer-based was more sensitive, with a higher processing capacity and less time demand than the acid-guanidinium-phenol-based method. Overall, the present study found that the combination of ultracentrifugation and lysis buffer-based RNA extraction method could be used for the rapid and sensitive detection of SARS-CoV-2 in wastewater surveillance. The findings of our study provide recommendations and references for the future applications of existing and newly developed virus concentration methods for SARS-CoV-2 wastewater surveillance.
CRediT authorship contribution statement
Xiawan Zheng: Methodology, Experiment, Analysis, Writing – original draft, Writing – review & editing. Yu Deng: Methodology, Writing – review & editing. Xiaoqing Xu: Methodology, Experiment. Shuxian Li: Experiment. Yulin Zhang: Experiment. Jiahui Ding: Experiment. Hei Yin On: Experiment. Jimmy C.C. Lai: Methodology, Resources. Chung In Yau: Methodology. Alex W.H. Chin: Resources. Leo L.M. Poon: Methodology, Resources, Writing – review & editing. Hein Min Tun: Resources, Writing – review & editing. Tong Zhang: Conceptualization, Methodology, Analysis, Resources, Supervision, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This study was financially supported by Health and Medical Research Fund (HMRF) (COVID1903015 and COVID190116), the Food and Health Bureau, The Government of the Hong Kong Special Administrative Region (SAR), China. We thank the Environmental Protection Department (EPD) and Drainage Services Department (DSD) of Hong Kong SAR Government for the wastewater sample collection. Xiawan Zheng, Xiaoqing Xu, Shuxian Li, Yulin Zhang and Jiahui Ding would like to thank for The University of Hong Kong for the Postgraduate Studentship (PGS).
Editor: Warish Ahmed
References
- Ahmed W., Bertsch P.M., Bivins A., Bibby K., Farkas K., Gathercole A., et al. Comparison of virus concentration methods for the RT-qPCR-based recovery of murine hepatitis virus, a surrogate for SARS-CoV-2 from untreated wastewater. Sci. Total Environ. 2020;739 doi: 10.1016/j.scitotenv.2020.139960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerswald H., Yann S., Dul S., In S., Dussart P., Martin N.J., et al. Assessment of inactivation procedures for SARS-CoV-2. J. Gen. Virol. 2021;102 doi: 10.1099/jgv.0.001539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batejat C., Grassin Q., Manuguerra J.C., Leclercq I. Heat inactivation of the severe acute respiratory syndrome coronavirus 2. J. Biosaf. Biosecur. 2021;3:1–3. doi: 10.1016/j.jobb.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bivins A., Bibby K. Wastewater surveillance during mass COVID-19 vaccination on a college campus. Environ. Sci. Technol. Lett. 2021;8:792–798. doi: 10.1021/acs.estlett.1c00519. [DOI] [PubMed] [Google Scholar]
- Brouwer A.F., Eisenberg J.N.S., Pomeroy C.D., Shulman L.M., Hindiyeh M., Manor Y., et al. Epidemiology of the silent polio outbreak in Rahat, Israel, based on modeling of environmental surveillance data. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E10625–E10633. doi: 10.1073/pnas.1808798115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashdollar J.L., Wymer L. Methods for primary concentration of viruses from water samples: a review and meta-analysis of recent studies. J. Appl. Microbiol. 2013;115:1–11. doi: 10.1111/jam.12143. [DOI] [PubMed] [Google Scholar]
- CDC . 2020. 2019-Novel Coronavirus (2019-nCoV) Real-time RT-PCR Primers and Probes. [Google Scholar]
- Cheung K.S., Hung I.F.N., Chan P.P.Y., Lung K.C., Tso E., Liu R., et al. Gastrointestinal manifestations of SARS-CoV-2 infection and virus load in fecal samples from a Hong Kong cohort: systematic review and meta-analysis. Gastroenterology. 2020;159:81–95. doi: 10.1053/j.gastro.2020.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin A.W.H., Chu J.T.S., Perera M.R.A., Hui K.P.Y., Yen H.-L., Chan M.C.W., et al. Stability of SARS-CoV-2 in different environmental conditions. LancetMicrobe. 2020;1 doi: 10.1016/S2666-5247(20)30003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu D.K.W., Pan Y., Cheng S.M.S., Hui K.P.Y., Krishnan P., Liu Y., et al. Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin. Chem. 2020;66:549–555. doi: 10.1093/clinchem/hvaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerrity D., Papp K., Stoker M., Sims A., Frehner W. Early-pandemic wastewater surveillance of SARS-CoV-2 in Southern Nevada: methodology, occurrence, and incidence/prevalence considerations. Water Res. X. 2021;10 doi: 10.1016/j.wroa.2020.100086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibas C., Lambirth K., Mittal N., Juel M.A.I., Barua V.B., Roppolo Brazell L., et al. Implementing building-level SARS-CoV-2 wastewater surveillance on a university campus. Sci. Total Environ. 2021;782 doi: 10.1016/j.scitotenv.2021.146749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncalves J., Koritnik T., Mioc V., Trkov M., Boljesic M., Berginc N., et al. Detection of SARS-CoV-2 RNA in hospital wastewater from a low COVID-19 disease prevalence area. Sci. Total Environ. 2021;755 doi: 10.1016/j.scitotenv.2020.143226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graber T.E., Mercier E., Bhatnagar K., Fuzzen M., D'Aoust P.M., Hoang H.D., et al. Near real-time determination of B.1.1.7 in proportion to total SARS-CoV-2 viral load in wastewater using an allele-specific primer extension PCR strategy. Water Res. 2021;205 doi: 10.1016/j.watres.2021.117681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green H., Wilder M., Middleton F.A., Collins M., Fenty A., Gentile K., et al. medRxiv; 2020. Quantification of SARS-CoV-2 and Cross-assembly Phage (crAssphage) From Wastewater to Monitor Coronavirus Transmission Within Communities. [Google Scholar]
- Guo M., Tao W., Flavell R.A., Zhu S. Potential intestinal infection and faecal-oral transmission of SARS-CoV-2. Nat. Rev. Gastroenterol. Hepatol. 2021;18(4):269–283. doi: 10.1038/s41575-021-00416-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haramoto E., Malla B., Thakali O., Kitajima M. First environmental surveillance for the presence of SARS-CoV-2 RNA in wastewater and river water in Japan. Sci. Total Environ. 2020;737 doi: 10.1016/j.scitotenv.2020.140405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata A., Hara-Yamamura H., Meuchi Y., Imai S., Honda R. Detection of SARS-CoV-2 in wastewater in Japan during a COVID-19 outbreak. Sci. Total Environ. 2021;758 doi: 10.1016/j.scitotenv.2020.143578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmer M., Paxeus N., Magnius L., Enache L., Arnholm B., Johansson A., et al. Detection of pathogenic viruses in sewage provided early warnings of hepatitis A virus and norovirus outbreaks. Appl. Environ. Microbiol. 2014;80:6771–6781. doi: 10.1128/AEM.01981-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillary L.S., Farkas K., Maher K.H., Lucaci A., Thorpe J., Distaso M.A., et al. Monitoring SARS-CoV-2 in municipal wastewater to evaluate the success of lockdown measures for controlling COVID-19 in the UK. Water Res. 2021;200 doi: 10.1016/j.watres.2021.117214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrudey S.E., Conant B. The devil is in the details: emerging insights on the relevance of wastewater surveillance for SARS-CoV-2 to public health. J. Water Health. 2021;20(1):246–270. doi: 10.2166/wh.2021.186. [DOI] [PubMed] [Google Scholar]
- Jafferali M.H., Khatami K., Atasoy M., Birgersson M., Williams C., Cetecioglu Z. Benchmarking virus concentration methods for quantification of SARS-CoV-2 in raw wastewater. Sci. Total Environ. 2021;755 doi: 10.1016/j.scitotenv.2020.142939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn K., Dreifuss D., Topolsky I., Kull A., Ganesanandamoorthy P., Fernandez-Cassi X., et al. Detection of SARS-CoV-2 variants in Switzerland by genomic analysis of wastewater samples. medRxiv. 2021 [Google Scholar]
- LaTurner Z.W., Zong D.M., Kalvapalle P., Gamas K.R., Terwilliger A., Crosby T., et al. Evaluating recovery, cost, and throughput of different concentration methods for SARS-CoV-2 wastewater-based epidemiology. Water Res. 2020;197:117043. doi: 10.1016/j.watres.2021.117043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMinn B.R., Korajkic A., Kelleher J., Herrmann M.P., Pemberton A.C., Ahmed W., et al. Development of a large volume concentration method for recovery of coronavirus from wastewater. Sci. Total Environ. 2021;774 doi: 10.1016/j.scitotenv.2021.145727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ. Sci. Technol. Lett. 2020;7:511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Mercer T.R., Salit M. Testing at scale during the COVID-19 pandemic. Nat. Rev. Genet. 2021;22:415–426. doi: 10.1038/s41576-021-00360-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemudryi A., Nemudraia A., Wiegand T., Surya K., Buyukyoruk M., Cicha C., et al. Temporal detection and phylogenetic assessment of SARS-CoV-2 in municipal wastewater. Cell Rep. Med. 2020;1 doi: 10.1016/j.xcrm.2020.100098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecson B.M., Darby E., Haas C.N., Amha Y.M., Bartolo M., Danielson R., et al. Reproducibility and sensitivity of 36 methods to quantify the SARS-CoV-2 genetic signal in raw wastewater: findings from an interlaboratory methods evaluation in the U.S. Environ. Sci.: Water Res. Technol. 2021;7:504–520. doi: 10.1039/d0ew00946f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Cataluna A., Cuevas-Ferrando E., Randazzo W., Falco I., Allende A., Sanchez G. Comparing analytical methods to detect SARS-CoV-2 in wastewater. Sci. Total Environ. 2021;758 doi: 10.1016/j.scitotenv.2020.143870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philo S.E., Keim E.K., Swanstrom R., Ong A.Q.W., Burnor E.A., Kossik A.L., et al. A comparison of SARS-CoV-2 wastewater concentration methods for environmental surveillance. Sci. Total Environ. 2021;760 doi: 10.1016/j.scitotenv.2020.144215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado T., Fumian T.M., Mannarino C.F., Resende P.C., Motta F.C., Eppinghaus A.L.F., et al. Wastewater-based epidemiology as a useful tool to track SARS-CoV-2 and support public health policies at municipal level in Brazil. Water Res. 2021;191 doi: 10.1016/j.watres.2021.116810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo W., Truchado P., Cuevas-Ferrando E., Simon P., Allende A., Sanchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;181 doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torii S., Furumai H., Katayama H. Applicability of polyethylene glycol precipitation followed by acid guanidinium thiocyanate-phenol-chloroform extraction for the detection of SARS-CoV-2 RNA from municipal wastewater. Sci. Total Environ. 2021;756 doi: 10.1016/j.scitotenv.2020.143067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Muller M.A., et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Wu F., Zhang J., Xiao A., Gu X., Lee W.L., Armas F., et al. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. mSystems. 2020;5 doi: 10.1128/mSystems.00614-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzer S., Marechal V., Mouchel J.M., Maday Y., Teyssou R., Richard E., et al. Evaluation of lockdown effect on SARS-CoV-2 dynamics through viral genome quantification in waste water, Greater Paris, France, 5 March to 23 April 2020. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.50.2000776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Zheng X., Li S., Lam N.S., Wang Y., Chu D.K.W., et al. The first case study of wastewater-based epidemiology of COVID-19 in Hong Kong. Sci. Total Environ. 2021;790 doi: 10.1016/j.scitotenv.2021.148000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S., Fan J., Yu F., Feng B., Lou B., Zou Q., et al. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January-March 2020: retrospective cohort study. BMJ. 2020;369 doi: 10.1136/bmj.m1443. [DOI] [PMC free article] [PubMed] [Google Scholar]