Abstract
Prions, misfolded proteins that aggregate, cause an array of progressively deteriorating conditions to which currently there are no effective treatments. The presently accepted model indicates that the soluble non-prion forms of prion-forming proteins, such as the well-studied SUP35, do not exist in large aggregated molecular complexes. Here, we show using analytical ultracentrifugation with fluorescent detection that the non-prion form of SUP35 exists in a range of discretely sized soluble complexes (19S, 28S, 39S, 57S, and 70S-200S). Similar to the [PSI+] aggregated complexes, each of these [psi-] complexes associates at stoichiometric levels with a large variety of molecular chaperones: HSP70 proteins comprise the major component. Another yeast prion-forming protein, RNQ1 (known to promote the production of the prion SUP35 state), is also present in SUP35 complexes. These results establish that the non-prion SUP35, like its prion form, is predisposed to form large molecular complexes containing chaperones and other prion-forming proteins. These results agree with our previous studies on the huntingtin protein. That the normal forms for aggregation-prone proteins may preexist in large molecular complexes has important ramifications for the progression of diseases involving protein aggregation.
Keywords: prions, SUP35, aggregates, chaperones, non-prion state
INTRODUCTION
Prions are misfolded infectious agents that join together to form protein aggregates. Both prions and other related protein aggregates lead to a variety of neurodegenerative disorders, including Alzheimer’s, Parkinson’s, and Huntington’s diseases.1,2 These diseases cause progressive deterioration to which currently there are no effective treatments.
Prion diseases (like that of Kuru and Creutzfeldt-Jakob disease in humans and Mad Cow’s disease in cows) are self-propagating and even transmissible.3 For prions, the normal prion protein (PrPc) in an alpha-helix-rich cellular form converts into abnormal misfolded prions (PrPsc) characterized by insoluble β-sheet polymers that trigger an autocatalytic reaction which leads to the accumulation of amyloid fibrils.4,5 As more insoluble aggregates are created, they serve as a corrupted template or seed, transmitting and converting normal non-toxic prions into abnormal masses of amyloids. This novel mechanism is also manifested in the propagation of pathogenic protein aggregation in numerous neurodegenerative diseases, including amyotrophic lateral sclerosis and some cancers.6 These results suggest that all these diseases have a common mechanism— an abnormal tendency of proteins to aggregate due to protein misfolding.7 These aggregates can interfere with normal cellular processes, leading to cell death.
The prion-forming protein SUP35 (involved in mRNA translation termination) from the yeast Saccharomyces cerevisiae has long been a model for prion formation.8 In yeast, the prion form of SUP35 is termed the [PSI+] state. While abnormal prions in the [PSI+] state have been extensively studied,9,10 in the [psi-] state (non-prion state) SUP35 molecular complexes have not been well characterized, although soluble complexes are known to exist.11
Recently, using the novel technique of analytical ultracentrifugation with fluorescent detection (AU-FDS),12–15 we identified in yeast in a [psi-] state a high presence of SUP35 protein in very large macromolecular complexes sized at about 19S, 28S, 39S, and 57S (estimated to be minimally from 500 KDa to at least 2 MDa, if spherical, but much larger if elongated).16 Consistent with this finding, the 19S and 28S complexes consisted essentially of protein, whereas the 39S and 57S complexes had some RNA components. AU-FDS is particularly amenable to detecting such specific, unique, and hitherto unacknowledged or unknown large protein complexes. For example, AU-FDS has recently established that the 60S ribosomal subunit by itself can bind mRNA and the closed-loop translation factors,15 detected protein translation complexes (e.g., the 77S monosomal translation complex) known to exist but not previously studied,12 and provided precise stoichiometric determinations and changes of the components of various translation complexes throughout the translation process.13 Most germane to protein aggregation, AU-FDS also characterized the variety and regulation of large molecular complexes formed from both the non-lethal and lethal forms of the huntingtin protein.17
The AU-FDS results on SUP35 suggest that even in the non-prion state SUP35 forms large molecular complexes of diverse sizes. Although these complexes are non-toxic, it was possible that they were the precursors to the prion aggregates that are formed in the [PSI+] state. Previous studies on the components of the SUP35 aggregates present in the [PSI+] state have shown that chaperone proteins are abundantly present.18,19 In one case, mass spectrometric analysis estimated that for every SUP35 protein there were 0.3 molecules of SSA1 and 0.3 molecules of SSA2.18 SSA1 and SSA2 are major components of the HSP70 chaperone complex and are often associated with aggregated proteins.20 Their binding to such aggregates may be one way the cell tries to clear such aggregates from the cell.21 Other chaperones were also found associated with the prion SUP35 aggregates but in lesser abundance (HSP60 proteins were present at about 10% the level of SUP35 and HSP104 was 2%).18
Studies in yeast have been conducted to elucidate the mechanism by which these molecular chaperones affect prion formation. However, it has been particularly challenging since chaperones can have inverse effects on folding and assembly.22 In yeast, any changes in activities of some chaperones and cochaperones can offset these opposing effects.23 The types of chaperones present could be critical to consequent effects on cell function.
In addition to these molecular chaperones, several other prion-forming proteins play an integral role in modulating the propagation and transmission of aggregates in a variety of yeast prions.24 For example, the prion form of RNQ1 ([PIN+]) promotes de novo production of prion SUP35 through genetic and physical interaction in a cross-seeding model where aggregated RNQ1 facilitates the misfolding of heterologous protein SUP35.25,26 In an alternate inhibitor-titration model, RNQ1 may prevent an inhibitor from binding to SUP35 by limiting its access, thereby allowing SUP35 aggregates to form.27
Because we have used AU-FDS to detect possible various complexes of SUP35 in the non-prion state,16 we subsequently determined which, if any, chaperones were present within the discretely sized soluble molecular complexes formed by SUP35 in this non-prion state. AU-FDS --being a technique that allows quantitative determination of components within complexes without resorting to the less precise mass spectrometric and western analysis normally used --was additionally used to calculate the stoichiometric abundance of each chaperone molecule and other yeast prion-forming proteins11,28 in each SUP35 complex.
Our results indicate that the 19S and 28S SUP35 complexes (as well as the larger SUP35 complexes) observed in the non-prion state are associated primarily with the SSA1 and SSA2 proteins (although other chaperones are also present). Thus, even in the non-prion state SUP35 forms large molecular complexes recognizable by the chaperone system. RNQ1 was the only other prion-forming protein found in these SUP35 aggregates, confirming that its known genetic effects with SUP35 may be a consequence of its association with SUP35 prior to SUP35 prion formation. These experiments further suggest that the formation of prion aggregates may depend on the composition and abundance of SUP35 and these other proteins (chaperones and others) in the non-prion state. Because other prion-forming proteins may interact with SUP35 and affect aggregation behavior, the composition of variant prion states and cytotoxicity may differ based on which cellular proteins are incorporated or misincorporated into these non-prion complexes. Most critically, since we have already demonstrated that the non-lethal form of huntingtin protein forms abundant amounts of large molecular complexes, it is highly likely that all aggregation-prone proteins in their “normal form” are already existing in large chaperone-containing complexes.
RESULTS
Identification of protein complexes containing eRF1 in the [psi-] state
Previously, using Flag-tagged eRF1 to purify complexes in which eRF1 was present, we identified a number of complexes (Figure 1A, identifying complexes containing primarily protein using AU-A230 analysis, and Figure 1B, identifying complexes containing primarily RNA using AU-A260 analysis).16 Several of these complexes were known. They included a monomeric complex (about 8S in size), the 80S ribosomal complex, and polysomal complexes (greater than 80S). Complexes that ranged in size from 19S to 57S, however, appeared to be novel.16 The 39S complex contained substantial amounts of protein in addition to the 40S ribosome whereas the 57S complex contained the 60S ribosome and probably other proteins. In contrast, the 19S and 28S complexes appeared to lack RNA and were essentially protein complexes (compare Figure 1B that identified primarily RNA to Figure 1A that identified primarily protein). All of these complexes existed prior to cell lysis.16
Figure 1.
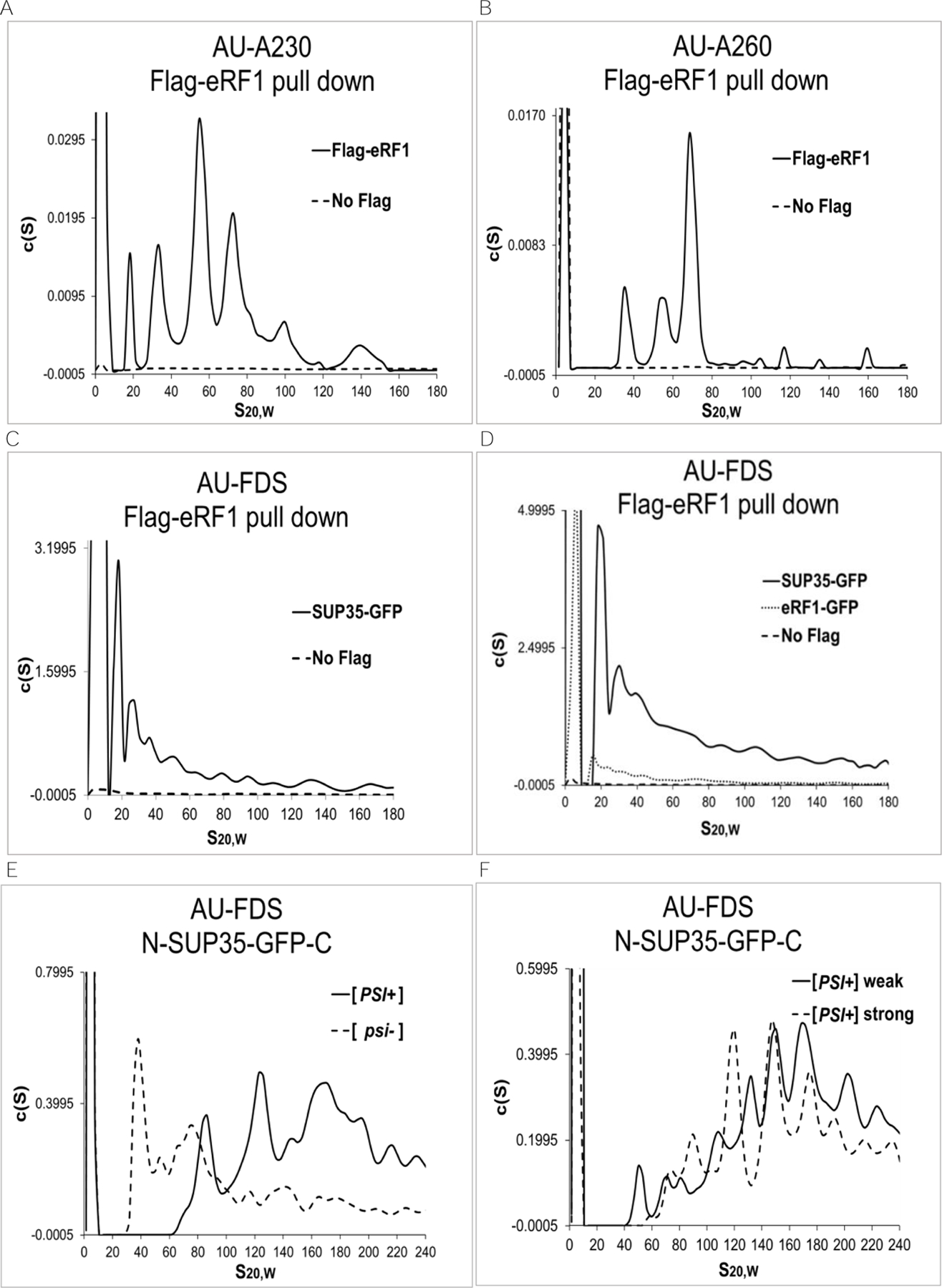
Identification of eRF1-containing complexes following AU analysis. A.-D. AU-A230, AU-A260, and AU-FDS analysis was conducted as previously described following immunoprecipitation of Flag-eRF1 in a strain containing SUP35-GFP.16 E.-F. AU-FDS analysis was conducted using strains containing GFP in the center of SUP35 and with the different [psi] phenotypes as indicated. A. AU-A230 analysis; B. AU-A260 analysis; C. AU-FDS analysis; D. AU-FDS analysis also using strain eRF1-GFP; E. and F. AU-FDS analysis.
In addition, the 19S complex --if spherical --would be at least about 500 KDa in size (and possibly much larger if elongated), which is much greater than the size of Flag-eRF1 alone (about 50 KDa). The 19S and 28S complexes, lacking ribosomal components, contained very low levels of a variety of translationally related proteins except for one translation factor, SUP35 (also known as eRF3), a dimerization partner of eRF1.16 AU-FDS analysis of Flag-eRF1 pull-downs in the presence of SUP35-GFP indicated that SUP35 was present in all Flag-eRF1 complexes (Figure 1C).16
The sizes of these discrete peaks that contained SUP35-GFP following AU-FDS analysis are summarized in Table 1. The 8S complex is presumed to be dimeric Flag-eRF1/SUP35-GFP.16 The discrete sizes of complexes greater than 100S could not be consistently distinguished by AU-FDS, and therefore, were not listed. Because the sizes of the SUP35-GFP complexes that ranged from 19S to 77S were very similar to the sizes of the eRF1-containing complexes identified by AU-A230 analysis (Figure 1A), we have used the following nomenclature to identify the sizes of these complexes: 19S, 28S, 39S, 57S, and 77S.
Table 1.
The average peak position of SUP35-GFP in Flag-eRF1 pull-downs.
| Peak | Average AU-FDS S Position | Estimated Minimal Mass (KDa) |
|---|---|---|
| 19S | 19±0.87 | 470 |
| 28S | 30±1.2 | 920 |
| 39S | 42±0.88 | 1400 |
| 57S | 55±1.5 | 2300 |
| 77S | 77±0.94 | 3800 |
Legend: Values for average peak positions are given with their standard error of the means (S.E.M.s). The corresponding predicted spherical molecular weight is calculated in KDa. A total of 25 samples were used for each of these analyses.
Because SUP35 is known to form aggregates in the [PSI+] state, these data suggested that the observed multiple eRF1-containing complexes represented large molecular complexes of SUP35 in the [psi-] state. Such soluble SUP35-containing complexes in the non-prion state have been shown to exist, but they have generally been under examined and are poorly understood.29–31 To address this issue, we further characterized the size of the SUP35-GFP-containing complexes identified following Flag-eRF1 pull-downs. Assuming all complexes were spherical, their corresponding molecular weights were calculated for the aggregates observed in the [psi-] state. For example, the estimated molecular weight of 19S would be approximately 465 KDa. Since the size of a single SUP35-GFP protein is about 103 KDa and the size of Flag-eRF1 is 50 KDa, the multiple peaks from 19S to 200S strongly suggest that the SUP35-GFP proteins are forming soluble complexes in the non-prion state containing multiple SUP35 polypeptides. It should also be noted that if any of these complexes were elongated, then the molecular weight sizes corresponding to a particular S value would be even greater (e.g., a 2:1 ratio of length to width would increase the calculated molecular weight by a factor of 1.4), and hence the number of proteins present in the complexes would be even greater.
To test the hypothesis that the Flag-eRF1 pull-down material contained multiple components of eRF1 and eRF3, we additionally examined whether Flag-eRF1 could pull down eRF1-GFP. In this case, if the complexes contained multiple copies of eRF1, some of these molecules would be expected to be present as eRF1-GFP. As shown in Figure 1D, Flag-eRF1 pulled down eRF1-GFP in complexes that were similar to those of SUP35-GFP.
Presence and abundance of chaperone proteins in SUP35-GFP complexes
SUP35 aggregates in the [PSI+] state have been shown to contain substantial abundances of the SSA1 and SSA2 chaperones and lesser amounts of other chaperones.18 To determine if the non-prion state molecular chaperones were also present in the above described large SUP35 complexes, we conducted AU-FDS and AU-A260 analyses. We selected and tested the following molecular chaperone proteins: HSP26, HSP40 family members (SIS1, YDJ1), HSP60, HSP70 family members (SSA1, SSA2, SSB1, SSB2), HSP90 family members (HSC82, HSP82), HSP104, SSZ1, and STI1. In order to analyze these chaperone proteins, we used yeast strains each carrying Flag-eRF1 and a molecular chaperone protein tagged at its C-terminus with GFP. These strains were purified using the one-step purification method as previously described.12 As an internal control, pull-downs were also conducted with SUP35-GFP in a strain lacking Flag-eRF1 tag. This control was used to correct for variability in cell extract production and AU-FDS analysis between the sedimentation experiments and to determine how much fluorescent material was immunoprecipitating in the absence of Flag-eRF1.
Starting with SSA1- and SSA2-GFP, following purification of complexes with Flag-eRF1 (Figures 2A and, in an expanded version, 2B), the SSA1-GFP and SSA2-GFP proteins were identified in the same discrete sized complexes of 19S, 28S, 39S, and 57S as previously observed for SUP35. Table 2 summarizes the normalized fluorescent peak intensities of the 19S to 57S complexes (the 77S-200S fluorescent intensities were combined). While the highest fluorescent peak abundance corresponds to 19S for both SSA1 and SSA2 (Figure 2B), as it did for SUP35, the absolute normalized abundances of fluorescence in each of the 19S to 57S peaks (also 77S-200S) were fairly uniform for SSA1 and SSA2. These results suggest that SSA1 and SSA2 are present in all of these SUP35 molecular complexes.
Figure 2.
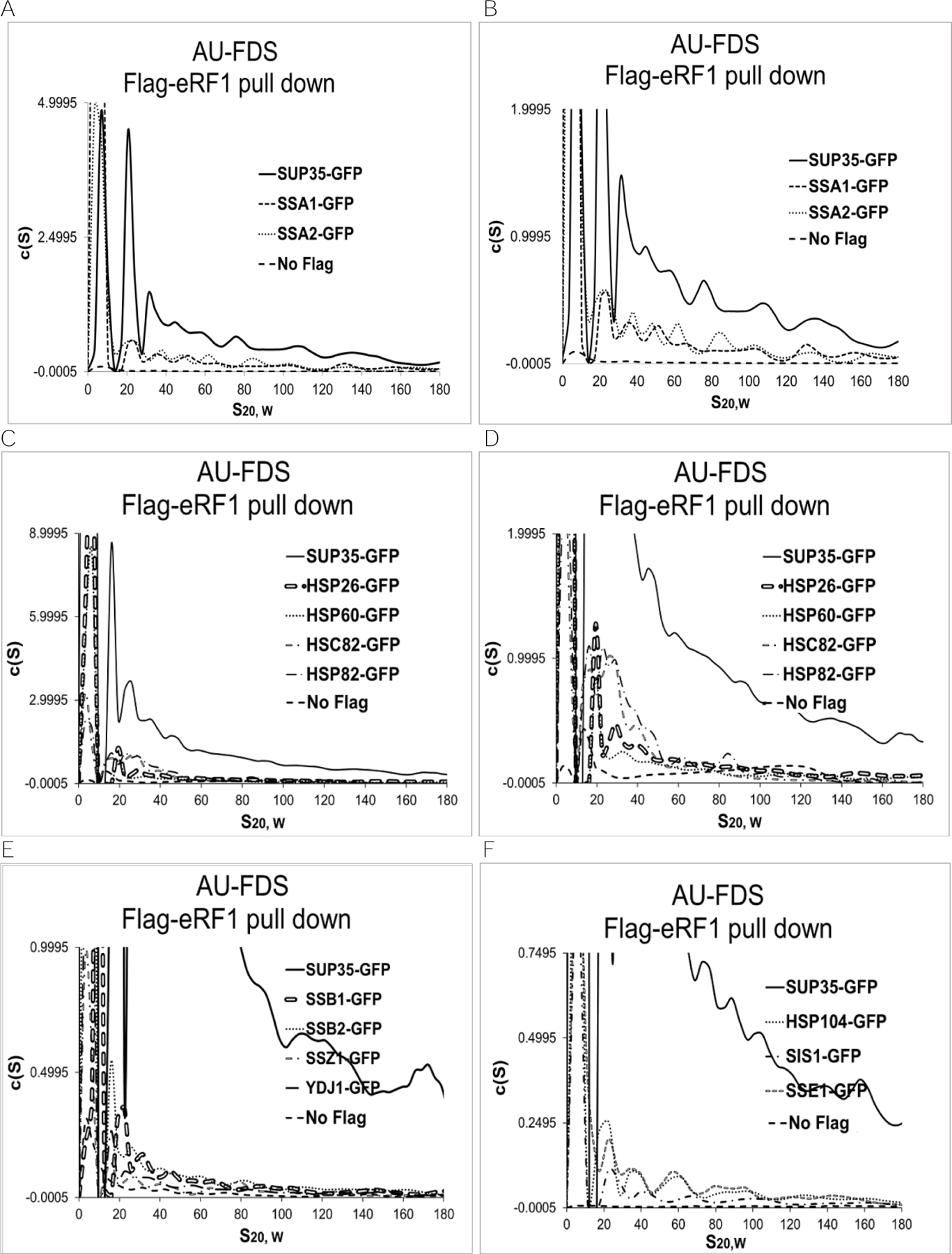
Flag-eRF1 pull-downs in the presence of GFP-tagged chaperones. AU-FDS analysis was conducted as described in Figure 1. A. and B.- same except with different scales; C. and D.- same except with different scales; E.- same as Suppl. Figure A, except with expanded scale; and F.- same as Suppl. Figure B, except with expanded scale.
Table 2.
Relative normalized AU-FDS intensities of molecular chaperones.
| 19S | 28S | 39S | 57S | 77S-200S | |
|---|---|---|---|---|---|
| SSA1 | 0.29±0.019 | 0.37±0.082 | 0.28±0.038 | 0.28±0.043 | 0.31±0.022 |
| SSA2 | 0.28±0.0041 | 0.35±0.012 | 0.36±0.045 | 0.27±0.044 | 0.38±0.15 |
| HSC82 | 0.080±0.023 | 0.088±0.010 | 0.11±0.042 | 0.11±0.046 | 0.083±0.034 |
| HSP26 | 0.087±0.023 | 0.12±0.020 | 0.056±0.0033 | 0.052±0.0062 | 0.065±0.038 |
| HSP60 | 0.069±0.0070 | 0.11±0.039 | 0.10±0.040 | 0.11±0.040 | 0.13±0.061 |
| HSP82 | 0.066±0.020 | 0.13±0.064 | 0.075±0.040 | 0.058±0.036 | 0.027±0.010 |
Legend: The values represent normalized AU-FDS intensities and (±S.E.M) from 19S out to 77S-200S relative to that of the comparable peak intensities of SUP35-GFP. SUP35-GFP was immunoprecipitated from a strain containing Flag-eRF1 on the same day as each particular chaperone was analyzed.
A similar pattern of discretely sized peaks ranging from 19S to 77S was found for HSC82, HSP26, HSP60, and HSP82 (Figure 2C and, in an expanded version, 2D). All of these chaperones had relatively high c(s) fluorescence intensities. In contrast, relatively low levels of SSB1, SSB2, SSZ1, SSE1, SIS1, STI1, and YDJ1 were present in SUP35 complexes, although they displayed a similar pattern of peaks (in expanded versions, Figures 2E and 2F, and Suppl. Figures A and B).
Determining the stoichiometry of chaperone proteins in complexes containing SUP35
To determine the abundance of a GFP-tagged protein in these complexes, the ratio of the abundance of the fluorescent signal was compared to the total protein analyzed in the centrifugation run. To accomplish this, Flag pull-down material was split and analyzed on the same day with AU-FDS analysis to determine GFP-tagged protein abundance and with AU-A260 analysis to determine how much total protein was present in the Flag pull-down material. For the AU-A260 analysis, the 77S peak abundance was used as the standard. The abundance of each individual protein complex from the AU-FDS analysis was calculated. Because the size distributions and normalized abundances between sizes were generally similar for a chaperone protein compared to SUP35 (Table 2), the absolute stoichiometry of chaperone proteins relative to SUP35 was determined using the total fluorescent intensity from 19S to 200S and not for individual peaks. The fluorescent intensity for the internal control (SUP35-GFP in the absence of Flag-eRF1) that was run on the same day was subtracted to quantify fluorescent intensity above this background level.
SUP35 was set to 100% and the relative abundance of each of the chaperone proteins is given in Table 3. Our analysis showed that the molecular chaperones such as SSA1, SSA2, HSC82, HSP26, HSP82, and HSP60 were found to be present at relatively high levels in the large SUP35 complexes. In particular, the members of the HSP70 family (SSA1 and SSA2) were about 30% relative to SUP35. In the case of SSA1 and SSA2, there would be about one molecule of SSA1 and SSA2 for every three molecules of SUP35 molecules in these complexes. The HSP90 family members (HSC82 and HSP82), HSP26, and HSP60 (Table 3) were around 10% relative to SUP35. Molecular chaperones, such as SIS1, SSE1, HSP26, HSP104 and YDJ1, were more marginally present in the SUP35 complexes (under 5%). These proteins along with SSB1, SSB2, SLA1, HSP104, SSZ1 and STI1 also displayed very low signal intensity (c(S) intensity below 0.1) as compared to the abundant proteins with a c(S) signal greater than 0.5. As a further comparison, the relative abundance of eRF1-GFP was about 13% of that of SUP35. While this value confirms that multiple eRF1 proteins are present in the 19S to 57S complexes, it was not possible to estimate this number because some of these copies of eRF1 could also be Flag-eRF1.
Table 3.
Percent abundance of molecular chaperones normalized to SUP35.
| Protein | % Abundance | % Control |
|---|---|---|
| SUP35 | 100 | 1.2 |
| SSA1 | 30±0.63 | 2.1 |
| SSA2 | 33±3.45 | 2.1 |
| HSP60 | 11±0.75 | 2.8 |
| HSC82 | 9.5±3.5 | 5.0 |
| HSP82 | 8.6±2.9 | 4.8 |
| HSP26 | 7.6±2.9 | 1.8 |
| SSB1 | 5.4±0.38 | 20 |
| SSB2 | 4.7±0.99 | 15 |
| SSE1 | 3.4±0.56 | 25 |
| SIS1 | 2.1±0.40 | 16 |
| HSP104 | 2.0±0.42 | 21 |
| SSZ1 | 2.1±0.59 | 48 |
| STI1 | 0.59±0.080 | 33 |
| SLA2 | 0.77±0.40 | 46 |
| YDJ1 | 3.6±0.59 | 23 |
| YEF3 | 11±0.82 | 28 |
| eRF1 | 13±4.5 | 16 |
Legend: Relative levels of each protein abundance (19S-200S) were normalized to that of the SUP35 aggregates and the levels of the background control (SUP35-GFP pull-down in a strain lacking Flag-eRF1) relative to fluorescent signal intensity for each protein.
In addition, the relative abundance of the control to that of each chaperone protein was calculated and evaluated (Table 3, right column). Overall, proteins such as SSA1 and SSA2 -- which were highly represented in the SUP35 complexes --had relatively low levels of control material migrating in the 19S to 200S region relative to the absolute abundance of the chaperones. This further supports the suggestion that the levels of abundance of such chaperones as SSA1 and SSA2 are significant. On the other hand, for low abundance proteins, the fluorescent abundances in the controls often represented a very high percentage of that observed for the chaperones. For example, the control had abundances in the 19S to 200S region that were 20% for SSB1, 32% for HSP104, and 48% for SSZ1. These data would suggest that for any absolute abundance level where the control was equal to or greater than about 20% relative to the chaperone’s abundance, the chaperone was only 5-fold or less above the typical control and its presence might not be significant.
Furthermore, we compared sedimentation profiles against the background control for each run. The sedimentation profiles suggested that only when the control abundance was below 20% of that of the chaperone abundance were clearly separated regions in the sedimentation coefficient distribution of the chaperone protein being analyzed. Conversely, nothing seemed to be sedimenting when the control abundance was at or above 20% of that of the chaperone. This observation indicates that when the control represented a substantial amount of fluorescence compared to that of the chaperone’s fluorescence there may not be a reasonable amount of chaperone material in the Flag pull-down.
Based on the above considerations, SSA1, SSA2, HSC82, HSP26, HSP60, and HSP82 --whose controls abundances were below 20% of the respective chaperone’s abundance --represent components that were present in eRF1 pull-down material and were thus part of these SUP35 complexes. Significantly, these chaperone proteins were present in SUP35 aggregates analyzed in the [PSI+] state.18 These correspondences strongly suggest that SUP35 is forming molecular complexes associated with chaperone proteins in the [psi-] state that may be related to the aggregates that are formed in the [PSI+] state. It should also be noted that eRF1-GFP had a control abundance below 20% (Table 3).
Prion-forming proteins in the SUP35 complexes
In addition to molecular chaperones, a number of prion-forming proteins in yeast may be incorporated into the SUP35 complexes. These prion-forming proteins are known to form multiple discrete aggregates, and the presence of these proteins may influence the structure and formation of SUP35 aggregates in the prion state. For example, RNQ1 displays genetic interactions with SUP35 in the [PSI+] state.27,32,33 For these studies, we screened and selected prion-forming proteins likely to be present in soluble aggregates: CYC8, MCA1, MOT3, NAB3, RNQ1, SPF1, SWI1 and URE2.34,35 Furthermore, we analyzed the presence of the non-Q/N rich prion-like protein, MOD5, in SUP35 complexes. We used the same methodology described above to determine the stoichiometry of these prion-forming proteins in SUP35 complexes in the [psi-] state.
The percent abundances of prion-forming proteins are reported in Table 4. Of these twelve proteins, only RNQ1 (Figure 3A) was above 5% relative to SUP35. The rest of the prion-forming proteins including CAF1, CYC8, HRP1, NAB3 (Figure 3B), MOD3, NAB3, SPF1, SWI1 and URE2 (Figures 3C to 3F) were about 1%, indicating that they were minimally present in the large SUP35 complexes. Again, it should be noted that among the prion-forming proteins only RNQ1-GFP had a control abundance below 20% of its abundance. It also had a clearly separated AU-FDS profile, suggesting protein complexes were separating. These observations indicate that the absolute abundance of RNQ1 in SUP35 aggregates is significant.
Table 4.
Percent abundance of prion-like protein normalized to SUP35.
| Protein | % Abundance | Control |
|---|---|---|
| SUP35 | 100 | |
| RNQ1 | 7.1±1.1 | 14 |
| CAF1 | 3.1±1.8 | 23 |
| CCR4 | 2.8±1.2 | 21 |
| SPF1 | 1.8±0.11 | 35 |
| MCA1 | 1.4±0.12 | 24 |
| MOT3 | 1.4±0.24 | 32 |
| CYC8 | 1.3±0.39 | 56 |
| HRP1 | 1.3±0.27 | 24 |
| NAB3 | 0.89±0.17 | 46 |
| SWI1 | 1.2±0.09 | 37 |
| URE2 | 1.1±0.25 | 33 |
| NAB3 | 0.89±0.17 | 46 |
Legend: Percent of prion-forming proteins relative to SUP35 and the percent of control relative to the prion-forming protein was determined as described in Table 3.
Figure 3.
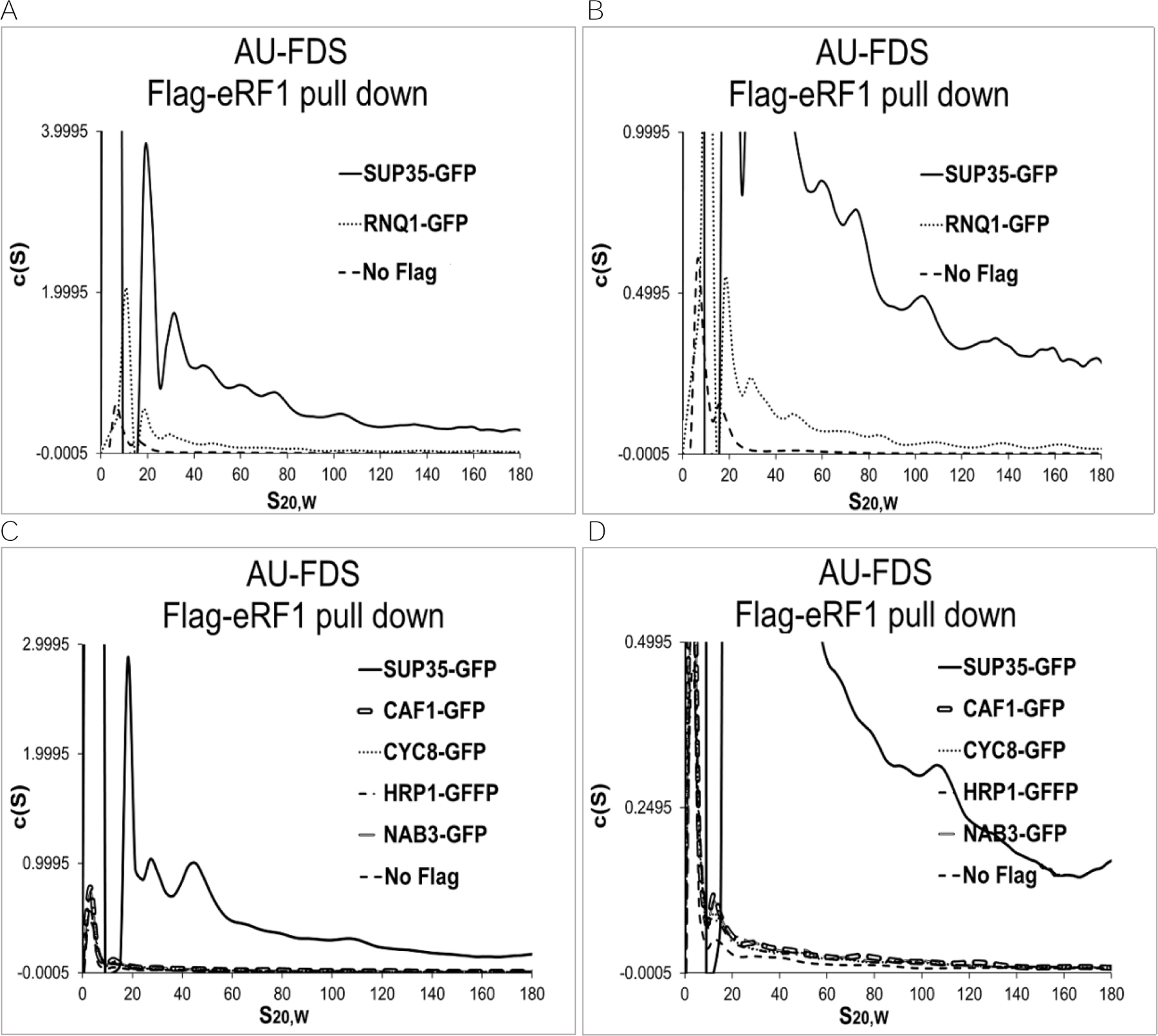
Additional Flag-eRF1 pull-downs. AU-FDS analysis was conducted as described in Figure 1. A. and B.- same except with different scales; C. and D.- same except with different scales.
[PSI+] aggregates are much larger than [psi-] complexes
To determine if the large molecular complexes we were observing in the [psi-] state were similar in size to those observed in the [PSI+] state, we analyzed an isogenic set of strains that were either [psi-], weak [PSI+], strong [PSI+], or [PSI+]. Each of these four strains carried a single copy of SUP35-GFP, although in this case the GFP entity was located between the N- and C-terminal domains of SUP35 (designated N-SUP35-GFP-C). AU-FDS analysis was conducted on each of these strains following the Flag-eRF1 pull-down. As shown in Figure 1D, in the [psi-] state SUP35 was found in complexes that ranged from about 40S to 240S, with the bulk of the complexes in the lower range from 40S to about 80S. While this overall pattern is similar to that observed for SUP35-GFP as described above (Figure 1C), in this strain background with N-SUP35-GFP-C, the smallest complex was noticeably larger, running at 40S. One possibility that might affect the sizes of the SUP35 complexes could be the different locations of the GFP moiety in the SUP35 proteins from the two strains. Most importantly, in the [PSI+] state the smallest complex observed migrated at 80S with the bulk of the material migrating at 125 to 200S. This result agrees with a previous analysis by sucrose gradient centrifugation which showed that SUP35 complexes in the [psi+] state were much larger than those present in the [psi-] state.31 Our analysis of the weak [PSI+] and strong [PSI+] strains further indicated that the SUP35 complexes ran in intermediary positions between the [psi-] and the [PSI+] states. These analyses with the four strains indicate that the sizes of SUP35 complexes increase dramatically as the strain becomes more [PSI+] and suggest that there may be a functional relationship between the SUP35 complexes in the [psi-] state and those in the [PSI+] state.
DISCUSSION
Identification of SUP35 molecular complexes in the [psi-] state
The primary focus of these studies has been to determine the character of the 19S to 57S complexes that Flag-eRF1 has previously been shown to pull down.16 Especially for the 19S and 28S complexes that lack RNA components, their large sizes suggested the presence of multiple proteins. The predicted molecular weight of the 19S and 28S complexes, if spherical, would be about 0.5 and 0.9 MDa, respectively, and much larger if elongated. Using Flag-eRF1 to immunoprecipitate complexes containing SUP35-GFP, we demonstrated that SUP35 even in its non-prion state is abundantly present in the 19S and 28S complexes and also in the larger complexes. These data suggest that SUP35 is forming complexes of various sizes, ranging from 19S to at least 200S. We also find that these complexes contained both Flag-eRF1 and eRF1-GFP. Since eRF1 efficiently dimerizes with SUP35, these data suggest multiple copies of eRF1 and SUP35 are present in these complexes. Whether eRF1 and SUP35 are always associating in a dimeric fashion in these complexes is not known.
If these SUP35 complexes were related to SUP35 aggregates formed in the [PSI+] state, it might be expected that they should correspondingly have chaperone proteins associated with them. Molecular chaperones may play many roles in aggregate formation, including breaking down aggregates of denatured proteins into smaller parts.36 As hypothesized, we identified several chaperone factors present in the SUP35 complexes in the non-prion state. The stoichiometric analysis revealed that the molecular chaperones SSA1 and SSA2 comprised the major components. The abundances of SSA1 and SSA2 were about one-third that of SUP35 in the different complexes detected between 19S and 200S. Related to these results, mass spectrometric analysis showed that SSA1/2 interacts preferentially with SUP35 in the aggregated prion state (again at a ratio of about one molecule SSA1 and one molecule of SSA2 for every three SUP35 molecules).18 These data suggest that molecular chaperones such as SSA1 and SSA2 are recognizing a feature of these non-prion SUP35 aggregates that is common to that of the [PSI+] aggregates.
Of the thirteen other chaperones analyzed for presence in the SUP35 non-prion complexes, only four displayed a reasonably high abundance: HSP26, HSP60, HSP82, and HSC82. For every 100 SUP35 molecules, these four chaperones were found to range from 7.6 to 11 molecules. Thus, in looking at the chaperone results in total, there appear to be 0.6 SSA1/SSA2 proteins for every SUP35 protein and similarly about 0.4 HSP26/HSP60/HSP82/HSC82 proteins for each SUP35 protein (see Figure 4). Because the 19S and 28S complexes may each be diverse in nature, it appears that on average there is one chaperone present for each SUP35 protein present (with, of course, SSA1/SSA2 being the most likely chaperone present).
Figure 4.
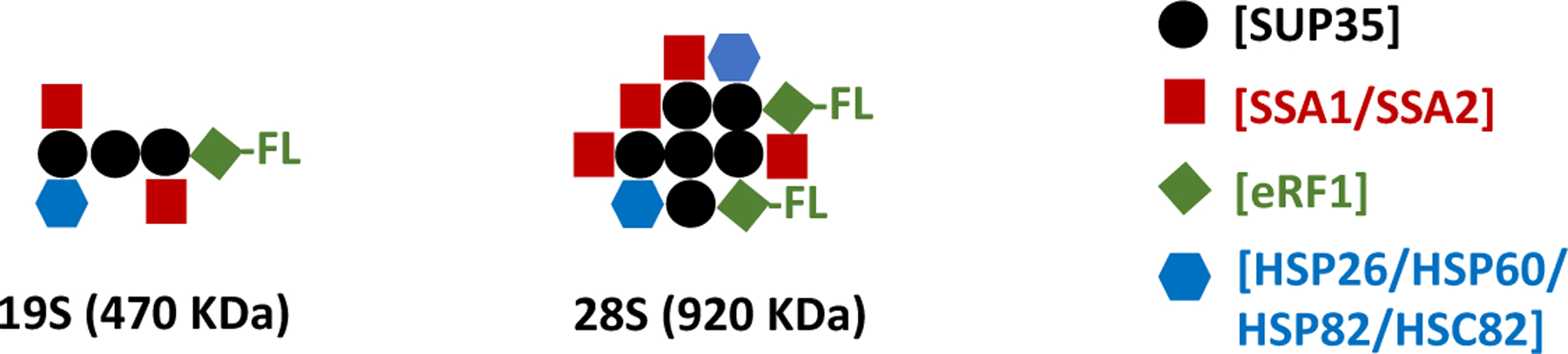
Model for protein components of 19S and 28S SUP35 complexes in the non-prion state. The contacts between particular pairs of proteins are only suggested, as the AUC experiments were not able to clarify such interactions.
The minimal character of the 19S and 28S complexes can therefore be estimated from the above analyses. Each complex would contain at least one Flag-eRF1 (about 50 KDa), one or more copies of SUP35 (approximately 70 KDa), and one or more copies of chaperones (about 80 KDa, based on the relative frequencies of the six different chaperones). For a spherical 19S complex, then, one Flag-eRF1, three SUP35, and three chaperones would provide a 500 KDa molecular weight. In contrast, two Flag-eRF1, two SUP35, and two chaperones would provide a 400 KDa molecular weight, slightly less than the predicted molecular weight of a 19S complex. As mentioned above, if the complex were elongated, the molecular weight would be substantially larger, and there would be even more copies of SUP35 present. For the 28S complex, migrating minimally at around 900 KDa, a minimum of 5–6 SUP35 proteins might be expected to be present. In either of these cases, these considerations indicate that in the non-prion state SUP35 is forming large molecular complexes in the presence of chaperones.
With respect to the larger 39S and 57S complexes, the presence of the respective 40S and 60S ribosomes complicates the ability to determine the character of these complexes. For example, our analyses may be detecting complexes containing very diverse forms. That is, there may be complexes with SUP35 aggregates and complexes with primarily Flag-eRF1 bound respectively to the 40S and 60S subunits. Because of this possible heterogeneity, it is difficult to determine the character of the SUP35 aggregates in these larger complexes. On the other hand, if no ribosomal subunit were to be present in the 39S and 57S complexes, then the number of SUP35 and chaperone molecules would be high.
These results establishing the presence of large SUP35 molecular complexes in the [psi-] non-prion state are similar to our previous observations that the huntingtin protein in its non-toxic state also forms large molecular complexes.17 The fact that two such non-related aggregation-prone proteins in their normal non-toxic form exist as a variety of large molecular forms targeted by chaperones (at least in the case of SUP35) suggests that other proteins that form toxic aggregates may be behaving similarly. At least in the cases of SUP35 and the huntingtin protein, these behaviors suggest that it is not the ability to form complexes of large size per se that causes their lethal effects. Instead, it appears that cell toxicity and the consequent neurodegenerative effects that are observed with aggregating proteins are dependent on the particular type of aggregates and structures that are created in the toxic state.
Comparison of [psi-] SUP35 aggregates to those found in the [PSI+] state
Three features of the [psi-] SUP35 complexes that we have observed can be compared to the aggregates found in the [PSI+] state. First, of particular interest is our finding that the pattern and the size of SUP35 complexes in the [psi-] state appears to be different and smaller than that observed in the [PSI+] state. Previous studies on prion polymers in the [PSI+] state showed that the smallest size observed is about ~600 KDa.36 In gel filtration experiments, SUP35 from a [PSI+] corresponded to intermediate amyloid structure with molecular weight higher than 40S (~1000 KDa).37 However, neither of these studies compared [psi-] complexes directly to those found in the [PSI+] state. One analysis that did compare the complexes in the two different prion states employed a sucrose gradient separation to identify large complexes.18 In this case, they isolated [PSI+] aggregates by sucrose gradient centrifugation, taking the material below the 30% sucrose layer that contained [PSI+] aggregates (approximately aggregates greater than 80S). In this case, however, most soluble protein complexes would have been excluded and certainly the bulk of the [psi-] complexes we studied by AUC analysis would not have been present. As a result, this methodology was probably unable to detect the bulk of the non-prion complexes of SUP35, as they migrate smaller than 80S. However, another previous study using sucrose gradient sedimentation to compare the relative sizes of [PSI+] and [psi-] SUP35 complexes did show that about 30% of SUP35 in the [psi-] state migrated at 40S to 80S in size.31 In contrast, in the [PSI+] state almost all of the SUP35 migrated from 40S to at least 200S.31 These results are generally similar to those found in our AUC analyses.
Second, although the size of the [psi-] complexes is smaller than those found in the [PSI+] state, the SSA1/SSA2 chaperone composition of the [psi-] complexes is similar to that observed for those found in the [PSI+] state. Previously, immunoblot analysis of [PSI+] aggregates found that such aggregates contained about 0.5 molecules of SSA1/SSA2 for every SUP35 protein.18 This value is very similar to the 0.6 value we found for SSA1/SSA2 abundance relative to that of SUP35 in the [psi-] complexes. While lesser concentrations of other chaperones (SIS2, HSP104, SSE1, SSB1/2, YDJ1, and SIS1) were detected in the [PSI+] aggregates, no estimate of their abundance was given.18 We analyzed by AUC analysis these chaperone proteins for presence in the [psi-] complexes, but none of them were present above the 5% level relative to that of SUP35, the approximate cut off for detecting significant presence of a protein in the SUP35 aggregates (Table 3). Overall, these comparisons suggest that both the [psi-] and [PSI+] SUP35 complexes equivalently interact with SSA1/SSA2, but that the [PSI+] aggregates may be distinctly different due to the presence of a number of other chaperone proteins. If different molecular chaperones and other endogenous aggregated proteins are present in the [PSI+] aggregates, as also suggested previously,18 these chaperones could influence formation of SUP35 aggregates and such alterations could induce changes in the structures of the aggregates.
Third, we found a significant level of the yeast prion RNQ1 in the SUP35 [psi-] complexes (Table 3). In contrast, RNQ1 was not present in the [PSI+] aggregates previously resolved by sucrose gradient centrifugation analysis.18 However, other studies indicate RNQ1 constitutes a relatively high abundance in SUP35 aggregates.38 The exact role of RNQ1 in the [psi-] state is unknown, but RNQ1 is required for de novo induction of [PSI+]. According to a cross-seeding mechanism, RNQ1 transiently enhances initiation of SUP35 polymerization.39 Once SUP35 prion aggregates are formed, RNQ1 assumes an independent structure and is no longer needed for SUP35 polymerization. Furthermore, the formation of SUP35 amyloid polymers is dependent upon RNQ1 aggregates. Hence, the interaction between SUP35 and RNQ1 suggests that RNQ1 facilitates the conversion of the normal conformation of SUP35 into its prion form.
While these large non-prion complexes may be precursors to the prions, our AUC analysis was not able to confirm this. Defining the factors that cause toxicity has been particularly challenging, and it is still not clear whether single prion polymers or their higher-order complexes affect the conversion to prions and the propagation of the toxic form. It has been proposed that size and composition of aggregates are linked to cell toxicity, transmissibility, propagation and insolubility.40,41 In fact, small soluble oligomers about the size of 300–660 KDa with masses equivalent to 14–28 monomeric units (if homogeneous) are considered most infectious and highly toxic in prion disease.42,43 Alternatively, SUP35 can become lethal by disrupting and limiting essential pathways. For instance, overexpression of the SUP35 prion-determining domain suppresses chaperone protein activity, which in turn increases the rate of aggregation and the size of prion aggregation.44 Increase in polymer size, e.g., full-length SUP35, would influence the overall level of termination activity by altering its composition in [PSI+] cells and decreasing its availability to the ribosome, causing cell toxicity.44,45
MATERIALS and METHODS
Yeast strains and growth conditions
Yeast strains carrying GFP fusions to particular factors have been previously described.16 In almost all cases the GFP fusion was at the C-terminus of the protein and marked with the HIS3 gene. These strains were [psi-] and isogenic with the genotype Matα ura3 his3 leu2 met15. As indicated in the text, four strains had the GFP protein in the center of SUP35 and were isogenic to each other except for the [psi] phenotype. Their genotype was Matα ade1–14 trp1–289 his3–200 ura3–52 leu2–3,112. No differences in growth rate were observed between any of the strains carrying GFP attachments to different proteins. Strains were transformed with the TK41 plasmid (Flag-eRF1-URA3; pEAU-Flag-SUP45).16 Cells were grown at 30oC to mid-log phase in synthetic complete medium with appropriate amino acids as described before.16 Generally, 400 mL of cells were used for AU-FDS analyses. Cell lysis and Flag pull-downs were done as described.16 The protein concentration of samples analyzed by AU ranged from 0.1–0.3 mg/ml.
AUC analyses
Flag eluted samples (350 µL) were subjected to AU analysis using A230 and A260 absorption or a fluorescence detection system (AU-FDS)46,47 to detect GFP-fusion proteins.12,13 All analytical ultracentrifugation experiments were conducted at 20oC and a rotor speed of 25,000 rpm. For AU-FDS experiments 400 scans were obtained and for AU- A230 and AU-A260 analysis at least 100 scans were obtained. Data were analyzed by SEDFIT software as described previously with regularization of 0.68 (one standard deviation).12 The absolute abundance of a particular peak was determined by summing all the c(S) values within the peak identified by the AU-FDS analysis.16 The total protein abundance in the sample was calculated from the AU-A260 result by taking the sum of the c(S) value of the highest peak, the 77S complex. The stoichiometric abundance of a GFP-tagged protein in a particular peak was calculated by dividing the ratio of the AU-FDS/ total AU-A260 for the protein (e.g., SSA1-GFP) by the corresponding ratio of the standard protein (SUP35-GFP). The control SUP35-GFP analysis was always done on the same day as the analysis for the sample protein.
Supplementary Material
ACKNOWLEDGEMENTS
We wish to thank R. Parker and S. Liebman for providing strains used in these analyses. This research was supported by NIH grant GM106286 to C.L.D. Partial funding was provided by the New Hampshire Agricultural Experiment Station. This is Scientific Contribution Number 2830. This work was supported by the USDA National Institute of Food and Agriculture Hatch Project 00589 to C.L.D. None of the co-authors have a conflict of interest.
REFERENCES
- 1.Asante EA, Smidak M, Grimshaw A, Houghton R, Tomlinson A, Jeelani A, Brandner S. A naturally occurring variant of the human prion protein completely prevents prion disease. Nature 2015;522:478–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernis ME, Babila JT, Breid S, Wuston KA, Wullner U, Tamguney G. Prion-like propagation of human brain-derived alpha-synuclein in transgenic mice expressing human wild-type alpha-synuclein. Acta Neuropathol Commun 2015;3:75. doi: 10.1186/s40478-015-0254-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abbott A Autopsies reveal signs of Alzheimer’s in growth-hormone patients. Nature 2015;525:165–166. [DOI] [PubMed] [Google Scholar]
- 4.Gorkovskiy A, Thurber KR, Tycko R, Wickner RB. Locating folds of the in-register parallel β-sheet of the Sup35p prion domain infectious amyloid. Proc Natl Acad Sci USA 2014;11:4615–4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kupfer L, Hinrichs W, Groschup M. Prion Protein Misfolding. Curr Mol Med 2009;9:826–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brodsky JL. The threads that tie protein-folding diseases. Disease Models Mech 2014;7:3–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Valastyan JS, Lindquist S. Mechanisms of protein-folding diseases at a glance. Disease Models Mech 2014;7:9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wickner RB, Shewmaker FP, Bateman DA, Edskes HK, Gorkovkiy A, Dayani Y, Bezxonov EE. Yeast prions: structure, biology, and prion-handling systems. Microbiol Mol Biol Rev 2015;79:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Danilov LG, Matveenko AG, Ryzhkova VE, Belousov MV, Poleshchuk OI, Likholetova DV, Sokolov PA, Kasyanenko NA, Kajava AV, Zhouravleva GA, Bondarev SA. Design of a new [PSI+]-no-more mutation in SUP35 with strong inhibitory effect on the [PSI+] prion propagation. Front Mol Neurosci 2019;12:274. doi: 10.3389/fnmol.2019.00274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dergalev AA, Alexandrov AI, Ivannikov RI, Ter-Avanesyan MD, Kushnirov VV. Yeast Sup35 prion structure: two types, four parts, many variants. Int J Mol Sci 2019;.20:2633. doi: 10.3390/ijms20112633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liebman SW, Chernoff YO. Prions in yeast. Genetics 2012;191:1041–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Denis CL, Laue TM, Wang W. Identification of a 57S translation complex containing closed-loop factors and the 60S ribosome subunit. Sci Rep 2018;8:11468. doi: 10.1038/s41598-018-29832-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang X, Xi W, Toomey S, Chiang Y-C, Hasek J, Laue TM, Denis CL. Stoichiometry and change of the mRNA closed-loop factors as translating ribosomes transit from initiation to elongation. PLoS ONE 2016;11:e0150616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang X, Zhang C, Chiang Y-C, Toomey S, Power MP, Granoff ME, Richardson R, Xi W, Lee DL, Chase S, Laue TM, Denis CL. Use of the novel technique of analytical ultracentrifugation with fluorescence detection system identifies a 77S monosomal translation complex. Prot Sci 2012;21:1253–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang C, Wang X, Park S, Chiang Y-C, Xi W, Laue TM, Denis CL. Only a subset of the PAB1-mRNP proteome is present in mRNA translation complexes. Prot Sci 2014;23:1036–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Denis CL, Richardson R, Park S, Zhang C, Xi W, Laue TM, Wang X. Defining the protein complexome of translation termination factor eRF1: identification of four novel eRF1-containing complexes that range from 20S to 57S in size. Proteins: Structure, Function, and Bioinformatics 2017; 10.1002/prot.25422. [DOI] [PMC free article] [PubMed]
- 17.Xi W, Wang X, Laue TM, Denis CL. Multiple discrete soluble aggregates influence polyglutamine toxicity in a Huntington’s disease model system. Sci Rep 2016;6:e-34916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bagriantsev SN, Gracheva EO, Richmond JE, Liebman SW. Variant-specific [PSI+] infection is transmitted by Sup35 polymers within [PSI+] aggregates with heterogeneous protein composition. Mol Biol Cell 2008;19:2433–2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Winkler J, Tyedmers J, Bukau B, Mogk A. Hsp70 targets Hsp100 chaperones to substrates for protein disaggregation and prion fragmentation. J Cell Biol 2012;198:387–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hasin N, Cusack SA, Ali SS, Fitzpatrick DA, Jones GW. Global transcript and phenotypic analysis of yeast cells expressing Ssa1, Ssa2, Ssa3 or Ssa4 as sole source of cytosolic Hsp70-Ssa chaperone activity BMC Genomics 2014;15:194. doi:101186/1471-2164-15-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arai C, Kurahashi H, Pack CG, Sako Y, Nakamura Y. Clearance of yeast eRF3 prion [PSI+] by amyloid enlargement due to the imbalance between chaperone Ssa1 and cochaperone Sgt2. Translation 2013;1:e26574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ellis RJ. Assembly chaperones: a perspective. Phil Trans Royal Society B: Biol Sci 2013;368:e20110398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Masison DC, Reidy M. Yeast prions are useful for studying protein chaperones and protein quality control. Prion 2015;9:174–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fraser PE. Prions and prion-like proteins. J Biol Chem 2014;289:19839–19840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choe Y, Ryu Y, Kim H-J, Seok YJ. Increased appearance by fusion of Rnq1 with the prion domain of Sup35 in Saccharomyces cerevisiae. Eukaryotic Cell 2009;8:968–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vitrenko YA, Pavon ME, Stone SI, Liebman SW. Propagation of the [PIN+] prion by fragments of Rnq1 fused to GFP. Curr Genet 2007;51:309–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keefer KM, Stein KC, True HL. Heterologous prion-forming proteins interact to cross-seed aggregation in Saccharomyces cerevisiae. Sci Rep 2017;7:5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.MacLea KS, Ross ED. Strategies for identifying new prions in yeast. Prion 2011;5:263–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dulle JE, Bouttenot RE, Underwood LA, True HL. Soluble oligomers are sufficient for transmission of a yeast prion but do not confer phenotype. J Cell Biol 2013;203:197–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O’Driscoll J, Clare D, Saibil H. Prion aggregate structure in yeast cells is determined by the Hsp104-Hsp110 disaggregase machinery. J Cell Biol 2015;211:145–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paushkin SV, Kushnirov VV, Smirnov VN, Ter-Avanesyan MD. Propagation of the yeast prion-like [psi+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J 1996;15:3127–3134. [PMC free article] [PubMed] [Google Scholar]
- 32.Sharma J, Liebman SW. Exploring the basis of [PIN(+)] variant differences in [PSI(+)] induction. J Mol Biol 2013:425:3046–3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Derkatch IL, Bradley ME, Masse SV, Zadorsky P., Polozhov GV, Inge-Vechtomov SG, Liebman SW. Dependence and independence of [PSI(+)] and [PIN(+)]: a two-prion system in yeast? EMBO J 2000;19:1942–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alberti S, Halfmann R, King O, Kapila A, Lindquist S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell 2009;137:146–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Du Z, Li L. Investigating the interactions of yeast prions: [SWI+], [PSI+], and [PIN+]. Genetics 2014;197:685–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kryndushkin DS, Alexandrov IM, Ter-Avanesyan MD, Kushnirov VV. J Biol Chem 2004;278:49636–49643. [DOI] [PubMed] [Google Scholar]
- 37.Wetzel R, Kheterpal I. Amyloid, prions, and other protein aggregates San Diego, CA: Academic; 1999. [Google Scholar]
- 38.Kurahashi H, Pack C-G, Shibata S, Oishi K, Sako Y, Nakamura Y. [PSI+] aggregate enlargement in rnq1 nonprion domain mutants, leading to a loss of prion in yeast. Genes to Cells 2011;16:576–589. [DOI] [PubMed] [Google Scholar]
- 39.Arslan F, Hong JY, Kanneganti V, Park S-K, Liebman SW. Heterologous aggregates promote de novo prion appearance via more than one mechanism. PLoS Genetics 2015;11:e1004814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kushnirov VV, Vishnevskaya AB, Alexandrov IM, Ter-Avanesyan MD. Prion and nonprion amyloids: a comparison inspired by the yeast Sup35 protein. Prion 2007;1:179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morales R, Hu PP, Duran-Aniotz C, Moda F, Diaz-Espinoza R, Chen B, Bravo-Alegria J, Makarava N, Baskakov IV, Soto C. Strain-dependent profile of misfolded prion protein aggregates. Sci Rep 2016;6:e20526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Silveira JR, Raymond GJ, Hughson AG, Race RE, Sim VL, Hayes SF, Caughey B. The most infectious prion protein particles. Nature 2005;437:257–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Verma M, Vats A, Taneja V. Toxic species in amyloid disorders: Oligomers or mature fibrils. Ann Indian Acad Neurol 2015;18:138–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pezza JA, Villali J, Sindi SS, Serio TR. Amyloid-associated activity contributes to the severity and toxicity of a prion phenotype. Nature Comm 2014;5:4384. doi: 10.1038/ncomms5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Toombs JA, Liss NM, Cobble KR, Ben-Musa Z, Ross ED. [PSI+] maintenance is dependent on the composition, not primary sequence, of the oligopeptide repeat domain. PLoS ONE 2011;6:e21953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kroe RR, Laue TM. NUTS and BOLTS: Applications of fluorescence detected sedimentation. Anal Biochem 2009;390:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.MacGregor I, Anderson AL, Laue TM. Fluorescence detection for the XLI analytical ultracentrifuge. Biophys Chem 2004;108:165–185. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


