Abstract
The circadian clock is an essential timekeeper that controls, for humans, the daily rhythm of biochemical, physiological, and behavioral functions. Irregular performance or disruption in circadian rhythms results in various diseases, including cancer. As a factor in cancer development, perturbations in circadian rhythms can affect circadian homeostasis in energy balance, lead to alterations in the cell cycle, and cause dysregulation of chromatin remodeling. However, knowledge gaps remain in our understanding of the relationship between the circadian clock and cancer. Therefore, a mechanistic understanding by which circadian disruption enhances cancer risk is needed. This review article outlines the importance of the circadian clock in tumorigenesis and summarizes underlying mechanisms in the clock and its carcinogenic mechanisms, highlighting advances in chronotherapy for cancer treatment.
Keywords: Biological Clock, Cancer, Molecular mechanisms, Chronotherapy, Metabolism
1. Introduction
As organisms have evolved, they have synchronized their biological processes to the daily light/dark cycle. Nearly all living organisms have an inherent time-keeping mechanism, an endogenous clock that synchronizes and controls their daily biological processes. The daily oscillations directed by endogenous clocks are circadian rhythms, one of the essential characteristics of all living organisms [1]. In mammals, a circadian timing system generates and regulates circadian rhythms with a central pacemaker located in a hypothalamic structure called the suprachiasmatic nucleus (SCN) in the central nervous system [2]. Circadian oscillators, known as peripheral oscillators, are located in all most all tissues of an organism. Together with these, a central clock generates circadian rhythms in the hypothalamic SCN of the brain, which constantly synchronizes with environmental cues and controls the various clocks through circadian output pathways [3, 4]. Since this endogenous time-keeping system is close to, but not precisely, 24 h, it must be synchronized to the 24-hour day on a regular basis. The circadian rhythms govern various biological functions, such as sleeping and awakening, rest and activity, cognitive performance, blood pressure, metabolic processes, body temperature, hormone secretions, and immune responses [5–7] (Fig 1). Circadian rhythms are necessary for the organism’s well-being and survival, and their disruption can lead to pathological conditions, including cancer (Fig 1). In the twenty-first century, the incidence and mortality of cancer have increased in essentially all countries [8]. People have irregular work shifts in the modern lifestyle, including working longer weekly hours, changing their transitions from day to night, and spending more time on activities such as electronic media. This prolonged light exposure, which results in disruption in circadian rhythmicity and has led to altered expression of clock genes, may increase abnormalities in DNA damage, cell metabolism, cell proliferation, and apoptosis, which can contribute to cancer development and progression [9, 10]. However, mechanistic understanding of cancer risk and its association with the circadian clock is incomplete. Therefore, this review discusses recent developments in mechanisms of circadian rhythms and their effects on signaling pathways related to cancer development and progression. At the molecular level, we further explore the future directions required to understand clock disruptions and their role in tumor initiation and progression.
Figure 1. Circadian disruption and its impact on major biological processes.

The breaking of circadian tolerance or disbalance in the circadian clock results in the outbreak of several diseases. The prime factors that play in a disbalance circadian clock are artificial light during day or night, unbalanced diet, work-life balance, and unbalanced lifestyle. (Created with BioRender.com)
2. Circadian Clock
The circadian time-keeping mechanism has three components: the central pacemaker, the input, and the out pathways [11] (Fig 2). Over the past four decades, the focus has been on identifying the structures that contain the circadian pacemaker. In 1972, the suprachiasmatic nucleus (SCN) in mammals was identified as the brain’s circadian clock [12]. The SCN, which acts as a core pacemaker to regulate the cycle of rhythms throughout the body, consists of ~15,000 neurons located in the basal hypothalamus, dorsal to the optic chiasm on both sides of the third ventricle [13–15]. Other clocks reside in peripheral organs, including the skin, lungs, liver, and kidneys. Core clock genes regulate both clocks (central and peripheral). Light and dark periods have substantial effects on the behavioral and physiological functions of mammals. On exposure to excessive light, the retina is activated and transmits signals to the SCN. It converts signals from the external environment to tissues through the autonomic nervous and endocrine systems, synchronizing circadian rhythms and oscillations. The SCN pacemaker generates self-sustaining, cell-autonomous circadian oscillations that directly or indirectly control the oscillations in the expression of various circadian clock genes [11]. Additional discoveries show separate clocks in the eyes and pineal glands of lower vertebrates such as reptiles, fish, and birds [16, 17]; in the eyes of marine mollusks [17]; and in the lateral neurons of Drosophila [18]. Thus, circadian clocks are usually found in neural or neuronal structures, and the brain SCN clock provides standard time for the peripheral clocks. The output pathways within the circadian system are responsible for the expression of circadian rhythms in hormonal (e.g., secretion of melatonin, cortisol, luteinizing hormone, and thyroxine), behavioral (e.g., locomotor activity and rhythms of feeding), neural (e.g., action potentials and rhythmic neuronal activity), and physiological (e.g., temperature and sleep-wake cycles) functions [19]. Thus, to keep the organism healthy, an interactive network of the master clock and peripheral clocks regulates various physiological and homeostatic functions.
Figure 2. Biological timekeeping system.
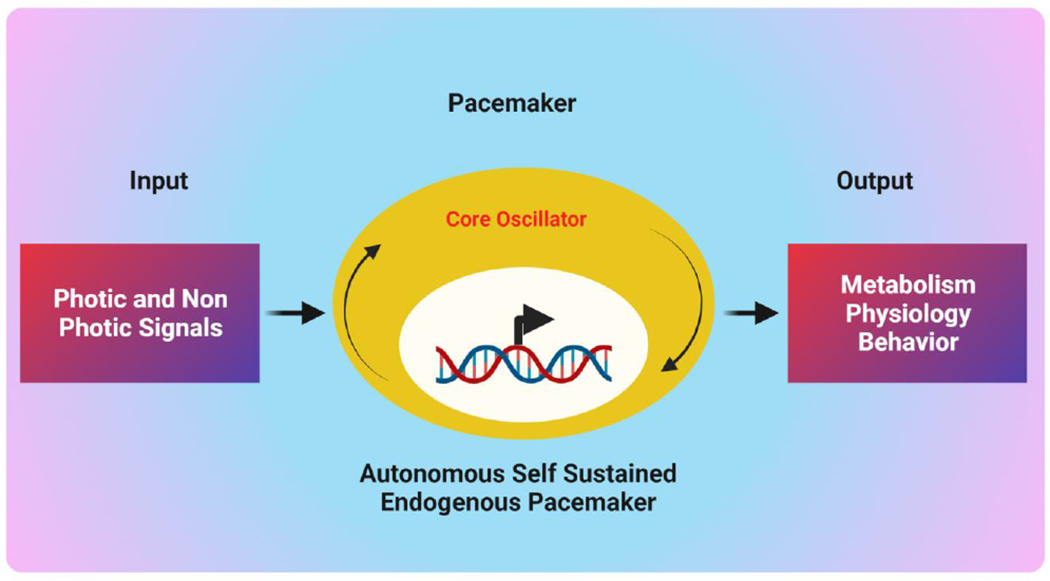
The circadian timing system is central to the organization of bodily functions. The input pathway receives environmental cues (hormone secretion, light, and temperature) and attunes the oscillator. The central oscillator generates rhythmicity by processing environmental inputs. The output pathways that generate rhythmic processes include various physiological processes, metabolic pathways, and neuronal functions. (Created with BioRender.com)
2.1. Molecular Regulation of the Circadian Clock
Circadian rhythms are produced by genetically-determined biological clocks [20]. In the mammalian core clock, the circadian oscillations function in a cell-autonomous manner, generated by circadian clock genes, such as Clock and Bmal1; period genes (Per3, Per2, and Per1); and cryptochrome genes (Cry2 and Cry1) [21]. These clock genes provide an auto-regulatory network with transcriptional-translational feedback loops that include genetic transcription of clock genes and the post-translational activities of “clock proteins” [20, 22, 23]. Their transcriptional activity peaks during the day and is inhibited at night by repressors Per and Cry [24]. In addition to the core clock genes, other genes are involved in the circadian gene network through clock elements, including E-boxes (5’-CACGTG-3’), ROR/REV-ERB (retinoic acid receptor-related orphan receptor/reverse erythroblastosis virus), and ROR response elements (ROREs) [25–27]. The transcriptional repressors (REV-ERBα/REV-ERBβ) and activators (RORα/RORβ/RORγ) bind to ROREs, and then REV-ERBα and RORα directly regulate the circadian expression of core clock genes (Bmal1, Clock, Cry1, and Npas2) via RORE [27]. These other pathways in the circadian gene network add robustness to the molecular clock mechanism [26], which involves at least two large, interconnected feedback loops [21, 22]. Positive regulators of the loop drive the clock. The proteins of CLOCK (circadian locomotor output cycles kaput) (or neuronal PAS-domain protein [NPAS2], mainly expressed in the forebrain), and Bmal1 (brain and muscle ARNT-like protein 1) heterodimers form the positive limb of the feedback loop [28, 29]. The three-period genes and two cryptochrome genes, which form the negative limb of the feedback loop, act as the primary initiators of the circadian rhythm [30]. The CLOCK/Bmal1 heterodimer initiates the transcription by binding to the E-box cis-elements in the promoter regions of Per3, Per2, Per1, Rorb, Rora, Rorc, Rev-Erba, and Rev-Erβ [3, 27]. Accumulation of the Cry and Per proteins results in the Per/Cry repressor complex and its nuclear translocation from the cytoplasm. Upon entry into the nucleus, the Per/Cry complex binds to CLOCK-Bmal1 at the E-box and suppresses its transcriptional activity by recruiting a SIN3-histone deacetylase complex (SIN3-HDAC) [31]. The time delay in the transcription-translation-based feedback loop within the clock generates daily oscillations with variations in clock genes and their protein products that indicate various forms of the circadian cycle. The amplitude of circadian oscillations is maintained by a cyclic proteolytic degradation of the repressors Per/Cer complex. The degradation/stability level of the Cry and Per proteins is necessary to set the period of the clock. Casein kinase 1 epsilon (CK1ε) and casein kinase 1 delta (CK1δ) phosphorylate PER proteins, which can lead to their ubiquitination by βTrCP and proteasomal degradation (26S proteasome) [21]. Similar to Per, Cry proteins are polyubiquitinated by FBXL3, targeting them for proteasomal degradation. Both Cry1 and Cry2 are targeted for ubiquitination. AMPK1 and Cry2 phosphorylate Cry1 by a subsequent dual-specificity Tyrosine Phosphorylation-Regulated Kinase 1A (DYRK1A)/Glycogen Synthase Kinase 3β (GSK-3β) cascade [32–35]. Collectively, these molecular events control the transcriptional feedback and allow the CLOCK-Bmal1 activator complex to start a new transcriptional cycle [36]. RORs and Rev-Erbs are transcribed during the subjective day, and their protein products regulate the transcription of Bmal1. REV-ERB and ROR proteins, which compete for the ROR regulatory element (RRE) binding sites of the Bmal1 promoter region, activate and repress the transcription of Bmal1 [37]. The circadian clock is regulated through a distinct set of transcriptional regulatory factors, and modifications of critical components of clock-regulatory factors contribute to cancer phenotypes (Fig 3).
Figure 3. Molecular design of the mammalian daily clock.
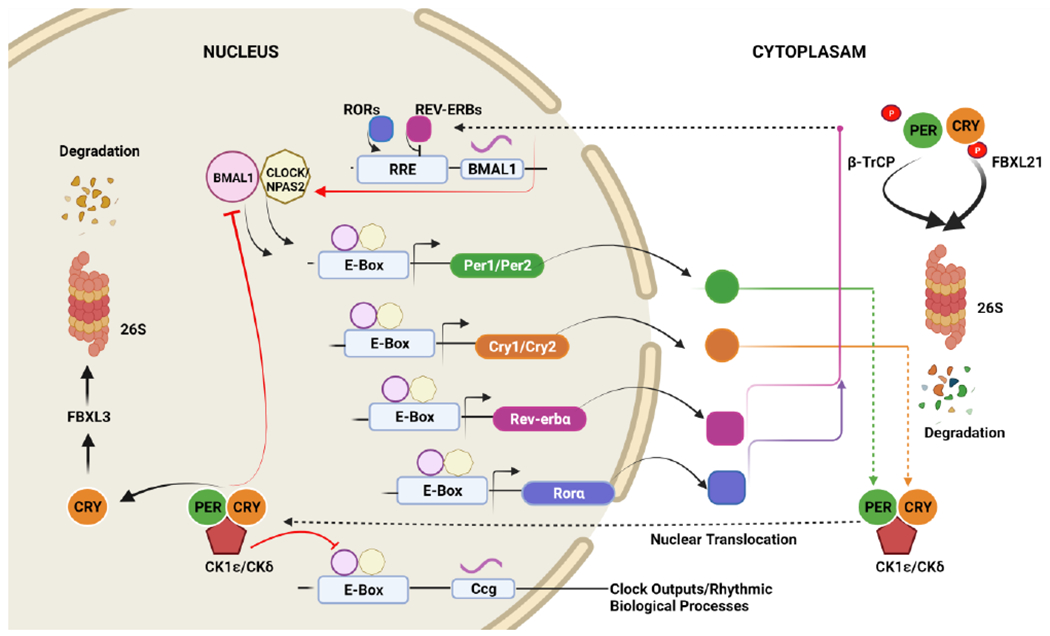
The core molecular clock is comprised of positive regulators (Bmal1 and CLOCK) and negative regulators (cryptochrome (Cry 1/2) and period-Per1/2). The protein clock Bmal1 heterodimerizes with CLOCK/NPAS2 to initiate the transcription of target genes through E-boxes present in the promoter region of target genes, resulting in the expression of Per (green circles) and Cry (orange circles) genes. Per and Cry dimerizes and translocates to the nucleus after being phosphorylated by casein kinase 1 epsilon (CK1ε) and casein kinase 1 delta (CK1δ), respectively, and repress their transcription. The Per and Cry proteins inhibit the expression of Bmal1 and CLOCK, creating the core negative feedback loop. In turn, CLOCK-Bmal1 generates another negative feedback loop that upregulates the negative transcription of REV-ERB proteins and negatively regulates Bmal1 transcription, and RORα binding to RORE elements increases the expression of Bmal1. Additionally, Per and Cry genes are regulated by proteins such as AMP-activated protein kinase (AMPK) and CK1ε/δ by phosphorylation and subsequent degradation. This phosphorylation targets Per genesfor polyubiquitination by βTrCP and, similarly, Cry1/Cry2 are phosphorylated by the Fox- proteins, FBXL3 and FBXL21, directing them to ubiquitin-mediated proteasomal degradation. Collectively, these feedback loops trigger circadian oscillations and subsequently regulate the circadian expression of downstream clock-controlled genes (CCGs). (Created with BioRender.com)
2.2. Circadian Clock Dysfunction Promotes Cancer
Circadian clock disruption is caused by various factors, including genetic, environmental, and internal factors [38]. Defects or disruption in normal circadian functioning and altered levels of clock gene expressions can increase the risk of prostate cancers, breast cancers, ovarian cancers, colorectal cancers, endometrial cancers, non-Hodgkin’s lymphoma, pancreatic cancers, osteosarcomas, head and neck squamous cell carcinomas, acute myeloid leukemia, and hepatocellular carcinomas (HCCs) [10, 39–43] (Table 1). Such alterations in the circadian system are risk factors for tumor incidence and may accelerate the progression of existing tumors. Circadian regulation can be a prerequisite for maintaining host defenses to prevent cancer [10]. Since circadian genes control the expression of genes, including cell cycle genes, tumor suppressor genes, and genes encoding caspases and transcription factors, they are involved in cancer-associated biological pathways [44]. Endocrine and metabolic hormones exhibit rhythmicity, and the levels of these hormones reciprocally influence circadian rhythms. Defects in this communication may lead to hormone-related pathological conditions, such as breast and endometrial cancers. This is evident by the higher prevalence of breast cancers and other malignancies for those who have been night-shift workers [45, 46]. Another line of evidence, which indicates an association of disrupted circadian functioning and development of breast cancer, comes from analysis of breast tumor tissues, which express lower levels of period genes, Per1 and Per2, than normal breast tissue [40, 47, 48]. Also, in HCCs, Bmal1 heterodimerizes with Neuronal PAS domain protein 2 (NPAS2) to facilitate NPAS2-mediated survival of cancer cells [49]. Polymorphisms in the circadian clock genes are associated with an elevated risk of cancer. For example, polymorphisms in the NPAS2 gene are associated with a higher risk of breast cancer [50]; NPAS2, Per1, and Per2 are associated with gastric cancer [51]; and CLOCK1 is related to the development of colorectal cancers [52]. As determined for a Spanish population, eating dinner before nine pm is associated with a lower risk of prostate and breast cancers [53]. Further, excess artificial light can cause a disruption in circadian rhythm, which can disturb sleep; delayed sleep may contribute to obesity, leading to cancer. Together with smoking, obesity accounts for 30% of cancer incidence [54].
Table 1.
Deregulated Clock Genes in Cancers
| Type of Cancer | Deregulated Clock Genes | References |
|---|---|---|
| Breast Cancer | Bmal1, Clock, Tim, Cry1, Per1, Cry2, Per2, Per3, Npas2 | [39, 40, 46, 48, 55–57] |
| Colorectal Cancer | Per1, Per2, Per3, Clock, Npas2, Tim | [58–64] |
| Colon Cancer | Per2 | [9, 65] |
| Diffuse large B-cell Lymphoma | Clock, Bmal1 | [66] |
| Endometrial Cancer | Per1, Per2, Per3, Cry1 | [45, 67, 68] |
| Esophageal Cancer | Dec1, Dec2, Per, Cry | [69] |
| Glioma | Cry1, Per1, Per2, Per3, Cry2, | [70–72] |
| Gastric Cancer | Cry1 Per2 | [73] |
| HNSCC (Head and Neck Squamous Cell Carcinoma) | Cry1, Per1, CKIε, Per2, Per3, Bmal1, Cry2, Bmal1, Tim | [74–77] |
| Hepatocellular Carcinoma | Cry2, Per1, Per3, Per2, Tim | [78, 79] |
| Leukemia (Acute Lymphocytic Leukemia; Chronic Lymphocytic Leukemia, & Chronic Myeloid Leukemia) | Per3, Cry2, Clock, Per1, Cry1, CKIε, Per2, Bmal1 | [74, 80–83] |
| Lung Cancer | Per1, Per2, Per3, Clock | [60, 84–87] |
| Malignant Pleural Mesothelioma | Per1, Per3, Npas2, Bmal1, Cry2, Rev-erbα, Rev-erbβ, Tim | [88–90] |
| Non-Hodgkin’s Lymphoma | Cry2, Npas2, Bmal1 | [70, 91, 92] |
| Osteosarcoma | Per2, CK1ε | [88, 93] |
| Ovarian Cancer | Clock, Bmal1, Cry2, Cry1, Per2, Per3, Per1, CK1ε. | [94–96] |
| Pancreatic Cancer | Per2, Bmal1, Cry1, Cry2, Per1, Per3, CK1ε, Clock, Dec1, Tim | [56, 97–99] |
| Prostate Cancer | Cry1, Bmal1, Cry2, Clock, Per2, Per1, Per3, Npas2, CK1ε, | [100, 101] |
Studies with transgenic models of cancer reveal the impact of tumors on circadian rhythms. After γ-radiation of mice with thymocytes deficient in the mPer2 gene, the mice develop more tumors, and there is less apoptosis of tumor cells. γ-Radiation induces the core circadian genes. Thus, for mice, the Per2 gene regulates the circadian rhythm by controlling DNA damage-responsive pathways [44]. In p53−/− and KrasG12D mutant mice, the mutation in Per2 accelerates the initiation and progression of lung tumors [102]. Further, for KrasG12D mutant mice, deletion of Bmal1 promotes lung turnorigenesis, and mutation of Cry2 drives the development of aggressive lymphomas in EμMyc models [103]. Bmal1 mutant mice develop lymphomas, along with liver and ovarian tumors [104]. Similar results are observed for mice lacking Cry1 and Cry2 [104]. For mice, liver and breast cancers cause alterations in the expression of the core clock genes, Clock, Per2, Rev-erbα, and RORγ, suggesting that dysregulation from physiological oscillations results in inflammation, oxidative stress and polyploidy.
Similarly, for mice, alterations in RORγ lead to the development of T-cell lymphomas [105]. Therefore, disruption of circadian clock genes promotes cancer progression in rodents and humans (Table 1). However, further studies are needed to understand the connection between circadian rhythms and cancer incidence.
2.3. Post-translational Modifications and Cancer Risk
The precision of the circadian clock mechanism is controlled by multiple post-translational modifications (PTMs), including sumoylation, phosphorylation, ubiquitination, and acetylation [106, 107]. PTMs control the core circadian clock dynamics, facilitated by the regulation of localization, activity, and degradation pathways of components implicated in controlling the feedback loop of core clock proteins [3]. A cyclic regulation of transcriptional activity of clock proteins is necessary for the maintenance of daily oscillations of these proteins and 24-h functioning of the clock. To maintain circadian oscillatory movement, the clock proteins, Bmal1, CLOCK, Per, and Cry, undergo modifications, including phosphorylation, acetylation, sumoylation, ubiquitination, methylation, and poly-ADP-ribosylation catalyzed by specific enzymes [108]. However, there is limited information on the PTMs of these proteins and their role in cancer pathogenesis. This review focuses on understanding the various PTMs from the perspective of carcinogenesis. Several studies have illustrated the importance of PTMs in the cellular machinery for promoting cancer dormancy. For example, the transcription repressors, Stra13 and Sharp-1, undergo sumoylation to mediate cellular growth arrest and inhibit myogenic differentiation, respectively [109, 110]. Another transcription repressor, DEC1 (differentiated embryo-chondrocyte expressed gene 1 protein), potentially involves sensing light signals from the environment [111]. DEC1 regulates the mammalian circadian rhythms by suppressing Bmal/CLOCK activity in a histone deacetylase-dependent and -independent mechanism [69, 111, 112].
Additionally, DEC1 undergoes proteasomal degradation by USP17 ubiquitin protease to regulate the DNA damage response, which is implicated in the prevention of cancer development [113]. Sumoylation is a reversible PTM mediated by small ubiquitin-related modifier protein (SUMO), like the ubiquitin pathway [114–116]. Abnormal gene expression of BMAL1 in SUMO-deficient mice suggests the importance of sumoylation in regulating the Bmal1 circadian expression and controlling the core circadian clock [117]. These circadian clock genes coordinate various cell functions and help slow tumor growth [118]. As a clock regulator, DEC1 represses the CLOCK-Bmal1 heterodimer-mediated promotor activity via recruiting HDAC1 [26], thus feeding-forward regulation of circadian rhythms.
In MCF-7 breast cancer cells, sumoylation of DEC1 increases the suppression of Bmal1-CLOCK-mediated transcriptional activity via recruiting HDAC1 [119]. Thus, PTMs of DEC 1 appear to exert a substantial effect on regulating circadian transcription factors and mediating the effects in gastric cancers, non-small cell lung cancers, and breast cancers during their development [120–123]. Several hormone-related cancers are associated with disruptive circadian rhythms. For example, abnormal estrogen receptor α (ERα) signaling forms the basis of the development and progression of breast cancers [124]. Sumoylation of CLOCK results in elevated ERα transcriptional activity and thus mediates the estrogen signaling pathway [125]. This sumoylation is promoted by treatment with estradiol. The direct involvement of CLOCK via PTM in modulating hormone-related tumorigenesis underlies the advantages in manipulating the CLOCK-ERα-mediated signaling pathway [125]. Also, CLOCK exhibits intrinsic histone acetyltransferase (HAT) activity, leading to acetylation of specific non-histone proteins, which are most likely responsible for cell cycle regulation and thus linking the circadian clock and carcinogenesis [126]. In sum, PTMs provide an additional level of control of the circadian system based on regulating the transcriptional activity of core clock proteins. These processes appear to be susceptible to pharmacological interventions for cancer treatment. Nevertheless, it is necessary to understand the key mechanisms associated with PTMs and their pathways to develop the drugs to treat cancers.
3. Chromatin Remodeling and Cancer
Regulation of the circadian clock machinery is primarily based on a complex gene expression program, which involves dynamic changes in chromatin structure. Regulation of circadian machinery is achieved through chromatin remodeling and epigenetic control, resulting from modifications to gene expression rather than to changes in DNA sequence [127]. Chromatin structure, regulated through specific proteins, is necessary for synchronizing gene expression in cells, tissues, and organs. Environmental and extracellular signals cause alterations in chromatin structure [128]. These include disruption of DNA-histone interactions through the formation of DNA-loops and sliding nucleosomes; regulation by multiple-protein chromatin remodeling complexes, such as the limitation-switch (ISWI) family; switching to defective/sucrose non-fermentable (SWI/SNF); histone deacetylation (NuRD/CHD) of the inositol 80 (INO80) family; and Mi-2/nucleosome remodeling [129]. These proteins, which are chromatin structure-regulating proteins, are controlled by ATP-dependent, histone-modifying enzymes and chromatin-remodeling enzymes [130]. These families include one or two distinct SW12/SNF2-type catalytic ATPases and multiple associated subunits [127] (Table 2). The SWI/SNF complex comprises twelve to fifteen subunits, including catalytic ATPase subunits (BRM [SMARCA2] or BRG1 [SMARCA4], alternate subunits [BAF47, BAF250, BAF200, BAF170, and BAF155]) and other accessory subunits. The SWI/SNF family of the chromatin remodeling complex regulates cell death, cell cycle, genomic instability, and DNA repair [131]. Analysis of the genome in human cancers reveals mutated or inactivated genes encoding SWI/SNF proteins [132, 133]. Further, as determined by whole-exome sequencing of 24 HCCs, chromatin regulators are the most mutated genes [134]. In various cancers, the principal subunits of the SWI/SNF complex, ARID1A (BAF250A) and ARID1B (BAF250B), are most frequently mutated (Table 2). In eukaryotic cells, ISWI (Imitation SWItch) has a conserved role in chromatin remodeling [89, 135, 136]. and its complexes are essential for nuclear functions, including regulation of DNA replication transcription and repair and maintenance of chromosome structure [137–140]. The role of ISWI family genes in tumor suppression is evident because mutations in the subunits of the ISWI complex are associated with various types of human cancers (Table 2) and promote tumor aggressiveness [141]. As chromatin remodelers, the Mi-2NuRD/CHD family proteins contribute to the regulation of genomic stability and are associated with various cancers; however, the mechanistic role of these family remodelers in cancer remains poorly understood (Table 2). These proteins share similar features in their capacity to bind nucleosomes, recognize histone modifications, and regulate ATPase activity. The processes and proteins implicated in regulating chromatin structure maintain DNA transcription, synthesis, replication, repair response, and chromatin stability [142]. Dysfunctions of these regulatory elements are associated with perturbations in the transcription of genes involved in tumor growth.
Table 2.
Dysregulation of chromatin remodelers in various types of cancer.
| Chromatin Remodeler | Types of Tumors | References |
|---|---|---|
| SWI/SNF | Breast cancer; Renal cancer; Stomach adenocarcinoma; Ovarian serous cystadenocarcinoma; Uterine corpus endometrioid carcinoma; Stomach adenocarcinoma; Cholangiocarcinoma; Melanoma | [110, 143–147] |
| ISWI | Uterine corpus endometrioid carcinoma; Melanoma; Lung cancer; Prostate adenocarcinoma; Stomach adenocarcinoma; Adenocarcinoma; Cholangiocarcinoma | [110, 148–153] |
| Mi-2/NuRD | Melanoma; Colorectal adenocarcinoma; Stomach adenocarcinoma; Prostate cancer; Endometrial cancer; Uterine serous carcinomas; Uterine corpus endometrioid carcinomas; Head and neck squamous cell carcinoma; Lung cancer; Bladder urothelial carcinoma; Pancreatic cancer. | [109, 110, 148, 151, 153–160] |
Dysregulation of chromatin remodeling machinery causes an increase of epigenetic abnormalities, leading to the initiation and progression of cancer. Histone variants can profoundly modify chromatin organization and functioning, which control DNA-template-based processes and functions [161]. The capacity of histone variants to facilitate chromatin deposition and provide epigenetic information for biological functioning makes them essential in developmental and differentiation processes. In cancers, the dysregulation of histone variants, employing depletion, overexpression, or mutation, contributes to their pathophysiological roles [162]. Histone deacetylases (HDACs) mediate higher-order chromatin changes in ovarian cancer cells and DNA damage-induced responses.[163].
Further, chemical modifications of histone H3 acetyl K9 and histone H3 trimethyl K4 correlate with altered gene expression for cervical cancer specimens and with poor prognosis of patients [164]. A histone isoform, Hist2h2ac (histone H2A type 2-C), involved in cell proliferation and differentiation, is altered in 17% of breast cancers and likely contributes to the dysregulation of targeted genes [165]. CHD1L is a chromatin remodeler that promotes metastasis and progression of HCCs and has therapeutic applications [166]. Further, liver-specific deletion of histone deacetylase 3 (HDAC3) results in HCCs, and the nuclear receptor corepressor 1 (NcoR1)-HDAC3-Rev-Erb complex formation regulates the circadian control of gene expression [167, 168].
NAD+-dependent SIRT 1 is a modulator of circadian clock machinery (Fig. 4). SIRT1 deacetylates various histone and non-histone targets, including clock proteins Bmal1 and Per2, which modulate clock function and expression of clock-controlled genes [169, 170]. Similar to SIRT1, AMPK (metabolic sensor), which regulates NAD+, has been linked to clock function by targeting the Cry and Per proteins [171]. SIRT1 deacetylates another non-histone target, the enzyme, AceCS1, which synthesizes acetyl-CoA, resulting in the cyclic synthesis of acetyl-CoA and thus oscillating the availability of acetyl groups for global acetylation [172]. AceCS1 is acetylated at its Lys661 residue. SIRT1 regulates the HAT activity of CLOCK. Circadian changes of chromatin properties by H3K4me3 are necessary for the regulation of circadian genes. SIRT1 deacetylates MLL1, which is responsible for H3K4me3 and circadian genes [173]. Regulation of SIRT1 activity, a critical circadian change of chromatin properties, indicates that histone H3 Lys9 and Lys14 on circadian promoters and Bmal1 are targets of SIRT1 [169]. Accumulating evidence implicating the importance of chromatin remodelers in the development and progression of cancers has established the core chromatin proteins as therapeutic targets. It is encouraging to find, in various cancers, the involvement of chromatin regulators globally and locally. This situation warrants further investigation of epigenetic changes within tumors and evaluation of their potential in driving the development and progression of tumors.
Figure 4. The role of SIRT1 in deacetylation of various non-histone targets.
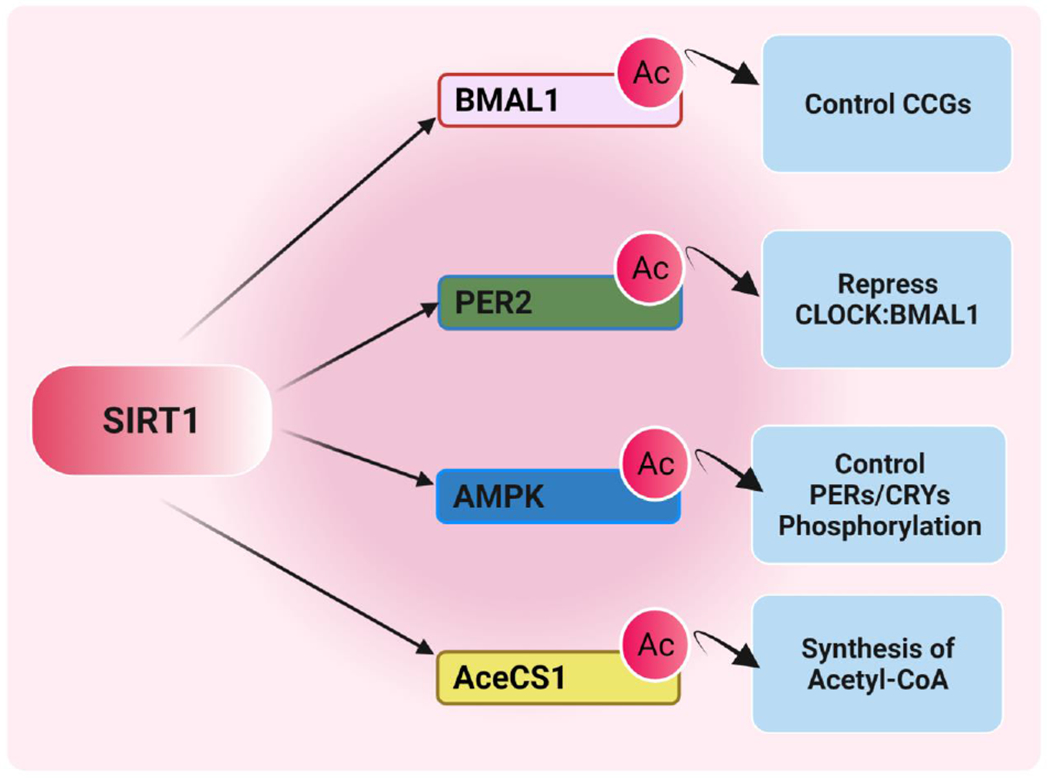
SIRT1 is an NAD+-dependent lysine deacetylase that deacetylates different histone and non-histone targets. SIRT1 deacetylates the core circadian transcription factor Bmal1 and core repressor Per2 to modulate CCG expression and a clock function. Similar to SIRT1, AMPK (metabolic sensor) is implicated in regulating NAD+, which has been associated with clock function by targeting CRY and PER proteins. Another non-histone target is the enzyme acetyl-CoA synthetase 1 (AceCS1). The synthesis of acetyl-CoA is activated by deacetylation of SIRT1, providing acetyl groups globally for acetylation reactions. (Created with BioRender.com)
4. The Circadian Clock in Cancer Cell Metabolism
The cross-talk between the circadian and metabolic systems is of significance. In healthy cells, an intimate association between metabolism and circadian rhythms is required to maintain physiologic homeostasis. Metabolic processes regulated by the circadian clock include gluconeogenesis, glycolysis, glycogen synthesis, glucose transport, and cholesterol metabolism [6, 38]. A misalignment between internal and external environments leads to disruption of circadian rhythms, associated with increased metabolic impairments and incidence of cancer [6, 174–177]. Since individual mammalian tissues cannot generate self-sustained metabolic rhythms, circadian metabolite oscillation is expected to affect the rhythmic expression of metabolic and clock genes. Metabolic signaling to the circadian pacemaker exerts an effect on these processes. The nutrients reset circadian and peripheral clocks. The peripheral clocks indirectly control the Warburg switch by regulating the expression of tumor suppressors and oncoproteins. Thus, the central nervous system and peripheral organs contribute to body homeostasis by regulating various metabolic and physiological responses [7, 8, 178, 179]. Previously, the focus has been on what foodstuffs we eat, but now there is a shift from what we eat to when we eat, i.e., meals’ frequency and circadian timing. Shifting food availability through restricted feeding of animals entrained to a standard light-dark schedule resets clock gene expression in the peripheral tissues [180]. Further, jet lag causes extensive changes in the expression of molecular clock genes and levels of metabolites.
Due to the induction of metabolic changes in rats, excessive exposure to constant light can lead to larger tumors than exposure to a standard light: dark cycle [181]. For mice, chronic jet lag leads to non-alcoholic fatty liver disease through metabolic disruptions in the liver [104]. In rodents and humans, the phases of liver lipogenesis and leptin concentrations in plasma are altered, reset with the timing of meals [182, 183]. This alteration in the nutritional and hormonal signals due to change in feeding times synchronizes peripheral clocks to the underlying metabolic rhythms. Thus, peripheral and central clocks are profoundly affected by meal timing and nutrients. Mice fed with a high-fat diet at the incorrect circadian time exhibit altered clock gene expression as well as behavioral perturbations [184] and accelerated weight gain compared with animals fed at their appropriate circadian time [185], suggesting that signals from metabolic pathways to the circadian pacemaker are transduced via dietary fats, which engage nutrient-responsive machinery. The dietary nutrients are linked to the clock pathway by nuclear hormone receptors and their cofactors. Exposure to a high-fat diet influences the metabolome and circadian transcriptome due to perturbation in Bmal1 and recruitment of PPAR (peroxisome proliferator-activated receptor) γ. For mice, deletion of PPARγ co-activator 1α (PPARGC1α) causes defects in body temperature regulation, locomotor activity, and metabolic rate because PPARGC1α has a circadian expression pattern in metabolic tissues [186]. Moreover, under nutrient-deprived conditions, the circadian rhythm is altered through phosphorylation of Per and Cry1 by a nutrient sensor, AMPK [171, 187]. Hepatocyte nuclear factor 4 α (HNF4α) is highly expressed in human HCCs. Increased expression of BMAL1 in HNF4α-positive HCCs inhibits tumor growth [188]. These results indicate that metabolic oscillators provide a mechanism of circadian timekeeping in addition to the already described transcription feedback loop, which serves as a conserved model of the circadian oscillator.
In general, cancer cells are distinct from normal cells and have specific metabolic characteristics. Therefore, perturbations in molecular clock machinery would change the metabolism homeostasis and energy balance in peripheral organs, favoring tumor progression and initiation. Cancer-associated metabolic abnormalities are characterized by higher NADPH levels, lower TCA cycle activity, and more remarkable fatty acid synthesis [189, 190]. Tumors typically have elevated levels of glycolytic metabolism, driven by various oncogenes, tumor suppressors, and transcriptional regulatory factors. Other regulators of metabolism, including the mTOR and P13K/AKT pathways, c-Myc, Ras, and nuclear hormone receptors, are also associated with circadian rhythms. Activation of the P13K/Pdk1/AKT pathway promotes the glycolytic program in cancer cells and affects the pace of the circadian clock [191]. Under low oxygen conditions, hypoxia-inducible factor-1(HIF1α and HIF1β) is involved in the cellular response to regulate gene expression, and HIFs bind to the hypoxia response element (HRE). Through transcriptional and post-translational mechanisms, the circadian clock enforces a circadian rhythm on HIF-1α for the rhythmic gene expression of hypoxia-dependent genes [192, 193]. In cancer cells, HIF-1α is also activated by low oxygen levels, which contributes to elevated glycolysis by regulating the circadian clock genes Rora, Cry, and Per2 [194, 195]. Oxygen levels control cellular respiration in a circadian manner, but how disruption of this metabolic rhythm drives tumor progression is unknown. There is a potential role of mTOR complexes, TORC1 and TORC2, in controlling the circadian machinery. mTORC1 is affected by cellular acidification, which deactivates mTORC1-dependent control of translation, resulting in suppression of clock regulators such as Per2, Bmal, and CLOCK [8]. Linked to mTOR activity, the circadian metabolic clock is rhythmic and follows food intake [196]. The RAS/MAPK pathway is active in various tumor types, and its activity modulates circadian rhythms through impairment of Bmal1/CLOCK activity. PI3 kinase (PI3K), an essential component of the RAS signaling pathway, is involved in the glucose metabolism of cancer cells [197, 198]. These results warrant further investigations to examine the cellular energy levels, amino acids, and growth factors sensed by the P13 kinase, mTOR, and RAS/MAPK pathways. This information will be helpful for therapeutic targeting for cancer.
MYC-MAX, a heterodimer of essential helix-loop-helix transcription factors, binds to the canonical E-box to drive rhythmic transcription of genes [199, 200]. In cancer cells, c-MYC supports cell proliferation and growth by regulating glycolytic, lipogenic, and mitochondrial genes to balance the regular loss of glucose as a mitochondrial substrate. c-MYC disrupts circadian clock function through E-box-dependent and -independent mechanisms [201]. Elevated expression of MYC is coupled with the expression of negative regulators of the clock, such as PERI, PER2, CRY1, and REV-ERBα, which suppress Bmal1 expression by binding to E-box sequences [174, 202]. The complex of c-MYC and MIZ1 inhibits Bmal1 expression, providing evidence for directly inhibiting the Bmal1 promoter (E-box independent) by forming transcriptionally repressive MYC-MIZ1 complexes [201]. By repressing the circadian clock, MYC/MIZ regulates cell proliferation and glucose and glutamine utilization [174, 201, 202]. In addition, in various cancers, the MondoA/MLX pathway is implicated in nutrient sensing, lipogenesis regulation, and glutaminolysis [203] (Fig. 5). There is a limited understanding of the relationship between the circadian clock and the nutrient-sensing pathway of MondoA. MYCN, a member of the MYC family, could alter the clock by activating NR1D1. In neuroblastomas, elevated expression of NR1D1 is associated with a poorer prognosis than that for neuroblastomas with lower expression [174, 202] (Fig. 5). However, the MYC superfamily may interfere with the circadian clock through various mechanisms to regulate gene expression programs that control cell proliferation and cellular metabolism. Since we lack a molecular understanding of clock proteins in the regulation of metabolism in multiple cancers, future studies should assess circadian disruption and its influence on cancer metabolism.
Figure 5. The circadian clock and its interplay with oncogenes and tumor suppressors.
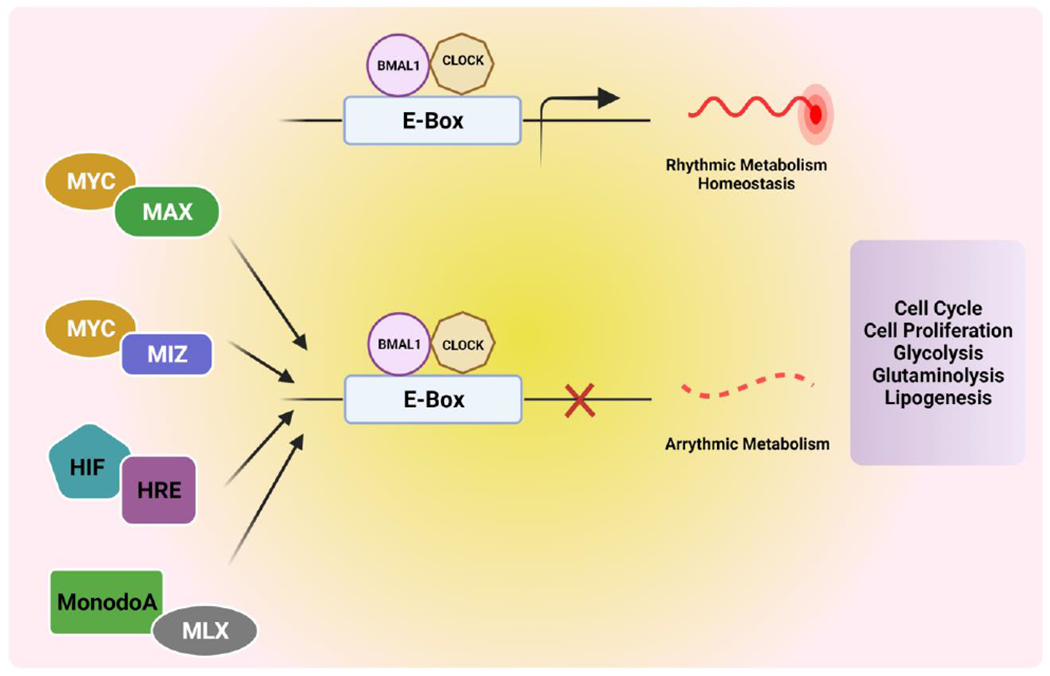
An outline of the critical pathways of tumor suppressors and oncogenes and their role in dysregulation of the circadian clock. The MYC-MAX heterodimer is a dimer of primary helix-loop-helix transcription factors and binds to the E-box engaged by CLOCK-Bmal1. High expression of oncogenic MYC disrupts the circadian clock and its rhythmic metabolism. Dimerization of MYC with MAX inhibits oscillation of the clock and rhythmic metabolism by engagement of the promoter region of Bmal1. MonodoA/MLX is also an E-box binding transcription factor that regulates tumor metabolism by transcriptionally uniting with CLOCK: Bmal1 activity. HRE (hypoxia response element) is an E-box-like sequence bound by HIF under low oxygen conditions and regulated by the CLOCK-Bmal1 complex. It can lead to the expression of glycolysis-related genes. These oncogenes and tumor suppressors regulate pathways involved in the cell cycle, cell proliferation, glycolysis, glutaminolysis, and lipogenesis. (Created with BioRender.com)
4.1. Role of Sirtuins in Metabolic Rhythms and Cancer Risk
Sirtuins, nicotinamide adenine dinucleotide (NAD+)-dependent histone deacetylases (HDACs), are implicated in numerous physiological functions, including metabolic control and cancer. There are seven mammalian sirtuins, SIRT1-SIRT7. Among the seven, three (SIRT1, SIRT3, and SIRT6) are functionally associated with control of the circadian clock and regulate cyclic outputs in response to metabolic cues [204–206]. SIRT1 localizes between the cytoplasm and the nucleus, whereas SIRT6 is exclusively localized in the nucleus and SIRT3 in the mitochondria [207]. These three sirtuins, through different mechanisms, coordinate the clock machinery. CLOCK-mediated chromatin remodeling and metabolic output are regulated by SIRT1 [208]. SIRT1, an energy and nutrition sensor activated by various stimuli such as low ATP, low nutrition, and exercise, converts cellular metabolites and nutrient signals to the circadian clock [209]. The nutrient-dependent activation of SIRT1 regulates Bmal1 expression and controls the expression of peptides and cofactors [170]. SIRT1 is implicated in energy metabolism by targeting PGC-1α in the SCN [210] (Fig. 6). Further, SIRT1 may be essential for the process of carcinogenesis, as it targets several acetylated proteins, including those for Wnt signaling, MYC, p53, and FQF. At present, there are contradictory ideas relating to the role of SIRT1 either as a tumor promoter or suppressor [211–214]. When SIRT1 acts as a tumor promoter, it reduces p53-mediated apoptosis and negatively regulates p53-induced cellular senescence [213], leading to uncontrolled cell division and the development of tumors. In addition, SIRT1-mediated deacetylation of p53 and forkhead box 03 alpha (Foxo3a) induces cell survival rather than apoptosis, which is favorable for tumor cells, leading to enhancing tumor growth [215]. SIRT1 regulates cell fate and stress response in mammalian cells and promotes cell survival by inhibiting apoptosis [101]. SIRT1 may also contribute to the nutrition of cancer cells, leading to enhanced growth and cellular survival [215]. Furthermore, SIRT1 has oncogenic activity, and the SIRT1 protein is highly expressed in breast cancer tissues. [216]. Further, crosstalk between SIRT1 and MYC oncogenic signaling regulates drug resistance of the FLT3 receptor in acute myeloid leukemia (AML) [217]. On the other hand, transgenic SIRT1 mice show a lower level of DNA damage and lower expression of p16 (Ink4a), an inhibitor of CDK4, suggesting that SIRT1 acts as a tumor suppressor through chromatin regulation and DNA repair. Further, SIRT1-transgenic mice have a lower chance of developing sarcomas and spontaneous carcinomas than wild-type mice. Thus, SIRT1 is involved in tumor suppression in metabolic syndrome-associated cancers [218]. Furthermore, SIRT1 deacetylates β-catenin, which leads, for APC min/+ mice, to reduce colon cancer development [219]. Similar to SIRT1, SIRT6 forms a complex with CLOCK: Bmal1 and regulates gene expression in a circadian manner, but its target genes are different. Chromatin recruitment of CLOCK/Bmal1 and SREBP1 controlled by SIRT6 regulates lipid and carbohydrate metabolic pathway genes [220]. Although SIRT3 regulates fatty acid oxidation and glucose oxidation rates in an NAD+-dependent manner (Fig. 6), it acts as a regulator of aerobic glycolysis for energy production (Fig. 6) [221]. Low expression of SIRT3 correlates with the upregulation of HIF-1α target genes, and overexpression of SIRT3 suppresses glycolysis and proliferation in breast cancer cells [222, 223]. This indicates that genomic partitioning of these sirtuins (SIRT1, SIRT6, and SIRT3) contributes to differential control of circadian metabolism (Fig. 6). Alterations in the circadian clock by sirtuins generate robust metabolic yields in various organs and likely drive tumor progression. However, understanding the mechanisms by which sirtuins regulate tumor progression is necessary for developing suitable sirtuin-based therapeutic approaches for cancer.
Figure 6. The role of sirtuins in controlling the circadian clock and energy metabolism.
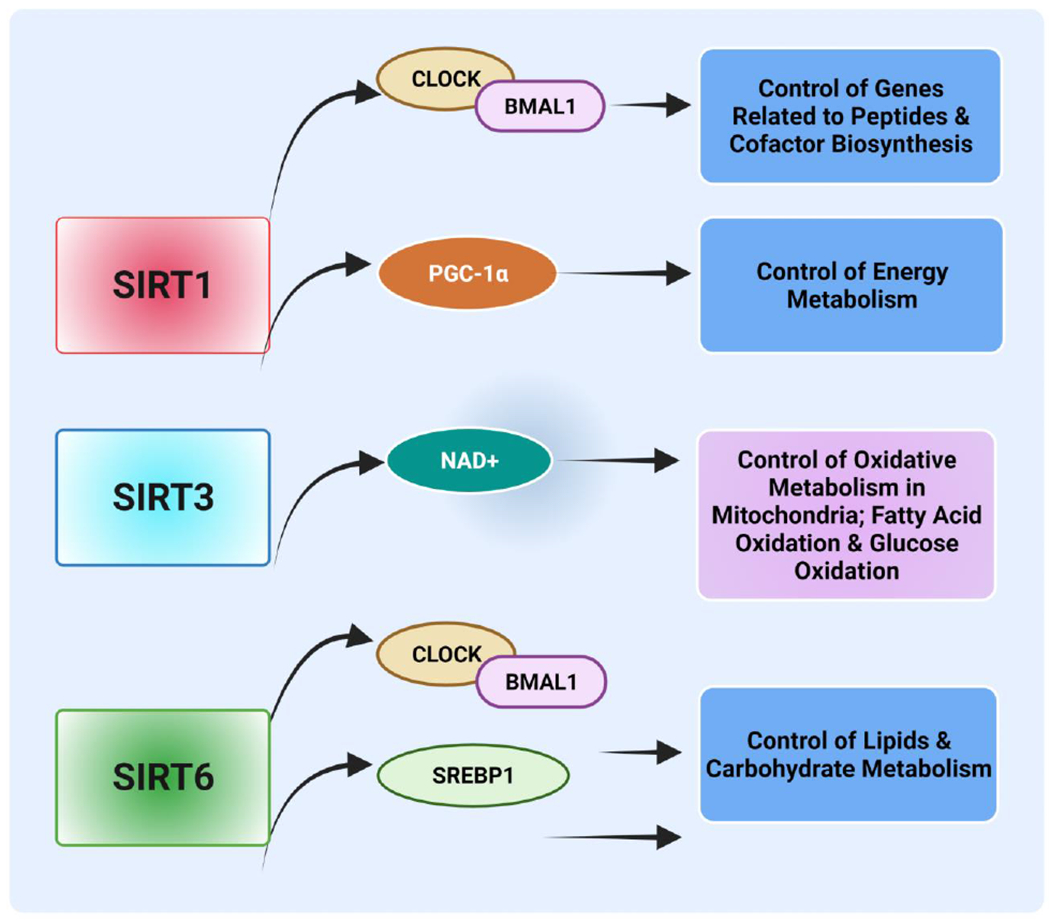
Sirtuins are regulators of energy homeostasis. SIRT1 regulates genes related to the synthesis of peptides and cofactors by controlling CLOCK/Bmal1 circadian transcription factors. The nutrient-dependent activation of SIRT1 regulates energy metabolism by influencing Bmal1 expression through PGC-1α in the suprachiasmatic nucleus. Deacetylation of mitochondrial oxidative enzymes by SIRT3 controls mitochondrial oxidative metabolism. Regulation of chromatin recruitment of CLOCK/Bmal1 via SIRT6 through SREBP1 regulates lipid and carbohydrate metabolism genes. (Created with BioRender.com)
5. Cell Cycle: Clock Genes and Cancer
A prominent feature of cancer cells is uncontrolled cell proliferation, which can be connected to the machinery of the circadian clock [224]. The cell cycle comprises a sequence of events leading to DNA duplication and the division of cells. Of the four phases, two are considered critical: the S phase, in which the cell duplicates its DNA, and the M (mitosis) phase, in which the cell divides. These two stages are preceded by growth phases G1 and G2. The cellular interphase is comprised of G1 and S, followed by G2 [225]. G0 represents the quiescent state wherein cells are in the non-dividing (somatic) phase. The cells may enter in G1 phase after exposure to environmental stimuli, such as growth factors [226] (Fig. 7). The cell cycle phases are regulated by the circadian clock [227–229]. However, the bond between the cell cycle and circadian rhythms is not evident. The expressions of β-catenin, c-Myc, and Mdm2 (oncogenes); CCND1, CCNB, and CCN1(cyclins); and Cdk4, Wnt3, and Tcf4 (cell-cycle regulators) are controlled by the circadian clock [104, 230] (Fig.7). Cell cycle progression depends on subsequent and transient activation of CDKs (cyclin-dependent kinases) forming complexes with CCNs (cyclins) such as CCND/CDK4–6 (G1), CCNE/CDK2 (G1/S transition), CCNA/ CDK2 (S), CCNA/CDK1 (S/G2 transition), and CCNB/CDK1 (M) [231, 232]. The CCN/CDK activity is inhibited across the cycle either by CDK inhibitors (CKI-P16, P27, and P21) or by phosphorylation by a kinase. Phosphatases such as CDC25A–B–C activate CCN/CDK [233, 234]. In response to DNA damage, these activators or inhibitors are involved in DNA repair and delay cell cycle progression by regulating cell cycle proteins and directly interacting with clock proteins or checkpoint proteins. The clock-controlled genes NONO, p20, and p21 are involved in the G1-S transition. NONO regulates the expression of the CDK inhibitor p16 at the G1–S transition by regulating the PER protein. Another clock gene, Per1, is involved in checkpoint 2 (CHK2) and the ataxia-telangiectasia–mutated (ATM) signaling pathways. The G1–S transition is inhibited by the circadian genes Per1 and Timeless (TIM) through interaction with ATM and CHK2, causing cell-cycle arrest [56]. Circadian clock mechanisms affect proliferating cells in the cell cycle by controlling the expression of Weel, which regulates Cdc2, transitioning from the G2 to the M-phase of the cell cycle [235]. In mammals, CLOCK/Bmal1 or Bmal1/NPAS2 regulates the anti-mitotic kinase Wee1 [105, 235, 236]. Per1, requiring p53, stabilizes c-Myc and alters transcription of Cdc2, Ccnb1, and Wee1. In tumor cells, deletion of Per1 affects the regulatory component of the cell cycle [56, 97]. Deregulation of c-Myc, a regulator of cell-cycle progression, is common in lymphoproliferative cancers [237]. In Per2 mutant mice, c-Myc transcription is elevated, and it downregulates the expression of CLOCK and BMAL1. c-Myc, phosphorylated by Cry2, recruits FBXL3-E3 ligase and promotes its degradation and ubiquitination; in the absence of Cry2, un-ubiquitinated cMyc enhances the incidence of lymphoma [238]. The pivotal tumor suppressor p53 controls multiple cell cycle checkpoints and regulates Per2 expression by blocking CLOCK: Bmal1 [239]. In Per2-mutated mice, c-Myc is upregulated, and p53 is downregulated, which leads to a general dysregulation of the cell cycle [236]. Overall loss of Per2 can lead to alterations in p53 function, thereby accelerating tumorigenesis and suggesting that similar clock mechanisms inhibit tumorigenesis or increase tumor progression depending on the clock genes involved. Hence, in cancer cells, it is essential to determine alterations of CLOCK: BmalL1-dependent genes in response to the activation of oncogenic Myc or failure of p53 expression. Additional studies are required to comprehend the functions of the cell cycle in various cancers mediated by circadian disruption.
Figure 7. The cell cycle and its molecular interaction with the circadian clock:
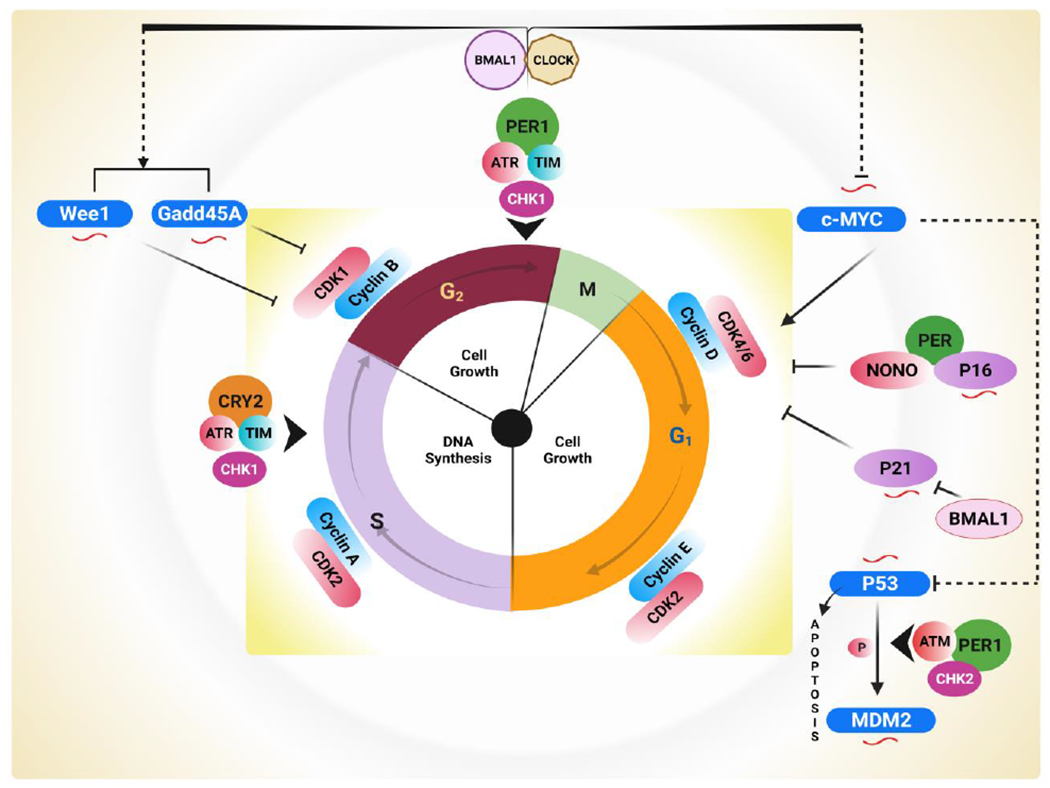
The cell cycle, comprised of several stages regulated by CDK/Cyclin complexes, is involved in the control of the progression of cells. CLOCK transcriptionally governs the expression of Wee1 and Gadd45A proteins. Bmal1, through the CDK1/Cyclin B complex, is involved in the inhibition of the G2/M transition of the cell cycle. Further, activation of the c-Myc oncogene promotes the expression of Cyclin D and cell proliferation. Transcription of the tumor suppressor genes p21 and p16 is under circadian control through checkpoints G1 and G1/S. The circadian output effector NONO connects to p16 and promotes transcription with the association of Per. Another CDK inhibitor, p21, is negatively regulated by BMal1. In addition, the interaction between p53 and Per results in increased activation of p53 target genes, including p21 and mdm2, which are regulated by the ATM–Per1–CHK2 complex. In contrast, ATR–Chk1–CRY–TIM pathways regulate the early response to DNA damage by interacting with Cry and Per clock proteins during the S phase and G2/M checkpoints. NONO: Nuclear RNA II binding protein or GADD45A; p54nrb; Growth Arrest; and DNA Damage inducible 45; TIM: Timeless mammalian protein; CHK2: Checkpoint kinase 2; CHK1: Checkpoint kinase 1; ATR: Ataxia telangiectasia and rad3-related; ATM: Ataxia-telangiectasia mutated. Circadian clock-regulated cell cycle genes are symbolized by ( ). (Created with BioRender.com)
). (Created with BioRender.com)
6. Chronotherapy for Cancer
Chronotherapy for cancer involves optimal timing of drug delivery based on individual circadian times, which can improve treatment tolerability and efficacy of anti-cancer medications [240]. Since a chronomodulated chemotherapy approach has treated various cancers, some researchers believe that, in the coming years, chronotherapy will be employed as an advantageous therapeutic option for cancer and other diseases [241–247]. Although normal cells usually display well-synchronized circadian variations within a tissue, tumor cells often have disrupted circadian rhythms, becoming a hallmark of cancer. This escape from circadian control allows for increased cellular proliferation [248] and may be linked with lesser sensitivity to anti-cancer drugs [249]. Although cancer chronotherapy relies on strong circadian rhythms in healthy tissues, efforts must be made to minimize clock disruptions. The application of chronotherapy to treat gastrointestinal and gynecological cancers showed an excellent response [250]. Recently, Li et al. demonstrated that the REV-ERBα and Bmal1 regulatory transcription loop could enhance the acceptability of chemotherapy. Circadian timing is especially relevant for anti-cancer drugs, often utilized at maximally tolerated doses and responsible for severe toxicities [251]. Mormont and Levi [252] showed that cancer patients exhibit altered circadian rhythms based on the tumor stage and that the same dose of the chemotherapeutic drug becomes fatally toxic only when administered at certain times.
In contrast, at other times, a 10-fold increase in dose was tolerated. Adler et al. found, however, that circadian timing may not always allow the safe application of high doses [253]. Hence, a current clinical challenge lies in reducing their life-threatening and dose-limiting side effects. In addition to reducing the toxicity, the side effects of increasing the dose intensity could also be decreased by chronotherapy. After patients with colorectal carcinomas received chronomodulated 5-fluorouracil, leucovorin daily for 14 days at 04:00 hours, and oxaliplatin at 16:00 hours for four days every 14 days, there was an improvement in cancer treatment, as evident by less toxicity and increased median survival of 50% [243]. The lower toxicities arising from treatment chronomodulation may be explained by different molecular statuses of healthy cells over the 24-h span, protecting them from the drug effect at particular times of the day. A Phase III clinical trial in Europe with 278 patients showed, relative to the conventional treatment group, no beneficial effect of chronotherapy after dosing with leucovorin, fluorouracil, and oxaliplatin. Close to traditional administration, there was a beneficial trend for chronotherapy for men, decreasing their risk of death by 25% but increasing mortality by 38% for women [254, 255]. These studies suggest the need for individualization of drug combinations and the timing of chemotherapy to improve treatment outcomes, considering factors such as patient gender, chronotype, and genetic background. Indeed, clinical trials show that proper chemotherapy timing lowers the toxicities of anti-cancer drugs. However, chronotherapy is not consistently beneficial for treating cancer. Therefore, mechanism-based studies are necessary to understand the potential of a chronotherapeutic approach for cancer treatment.
7. Conclusions and Future Directions
In this review, we have discussed recent reports and developments that support the idea that cell metabolism, the cell cycle, and chromatin remodeling regulate the circadian clock. Disruption of these regulatory pathways can affect tumor growth. Despite efforts for understanding the molecular mechanisms of the role of the circadian clock in cancer progression, challenges remain. These reports suggest that inherited genetic mutations cause only 8-10% of all cancers; however, an unhealthy lifestyle is a significant risk factor. Disruption of the circadian cycle by environmental factors, including nighttime work, jetlag, and nighttime light exposure, increases cancer risk. Therefore, more studies on light exposure and lifestyle interventions combining food, physical activity, nutrition, and sleep are needed to reduce cancer incidence. In addition, investigations are required to examine the relationship between circadian rhythm disruption and other confounding factors, including diet choices, smoking, alcohol consumption, and exposure to radiation and other carcinogens. Properly regulated, rhythmic metabolism can contribute to cancer prevention.
In several cancers, circadian metabolic pathways are deregulated. SIRT1, which exists as a complex with CLOCK, is involved in circadian clock-mediated metabolism. Metabolic regulation of the circadian clock controls the acetyltransferase activity of CLOCK and the counterbalancing deacetylase activity of SIRT1. NAD+ metabolism has a close relationship with SIRT1, glycolysis (metabolism), and the clock. However, no evidence has demonstrated that SIRT1 is the link between clock-mediated cancer and NAD+ availability. Furthermore, research is needed to realize how, in cancers, circadian rhythms influence NAD+ levels and their interaction with SIRT1. Nevertheless, SIRT1 should be considered a target for therapeutics. A more extensive understanding of the cellular and molecular connections between circadian rhythm and cancer cell metabolism offers new therapeutic strategies for cancer.
Circadian genes regulate diverse components of physiology. Circadian endocrine perturbations are linked with higher concentrations of sex hormones, facilitating the development of cancers. Disruption of the central clock affects the peripheral clocks in various tissues, which can be involved in tumorigenesis and cancer progression. Thus, there is a need to assess the impact of circadian factors on tumorigenesis in a tissue- and tumor type-specific and age-dependent manner. Disruption of the cell cycle linked to clock proteins may interfere with DNA repair, increasing cancer risk. However, the role of the circadian clock in cell cycle regulation remains relatively unexplored. Finally, the circadian function is based on a complex gene expression program, which involves dynamic changes in chromatin structure. The molecular mechanism of clock-controlled chromatin remodeling that responds to environmental stimuli has yet to be determined. Growing evidence indicates that chromatin-modifying enzymes and chromatin remodelers are deregulated in human cancers. However, further understanding is needed regarding how histone modifications and chromatin remodeling complexes are involved in malignancy. These changes provide promising targets for treating human cancers. However, previous studies or clinical trials of chronochemotherapy have not shown significantly increased efficacy for cancer patients relative to conventional therapies. Therefore, a better understanding of chronotherapy could benefit cancer patients.
In conclusion, the circadian clock is a unique system. In cancers, the clock genes regulate various signaling pathways, including cell cycle, chromatin remodeling, metabolism, cell proliferation, cell death, and DNA damage repair. These interlinking processes require mechanistic insights at the molecular and cellular levels that will help develop treatment strategies. Deregulation of circadian homeostasis stimulates cancer initiation and development and is associated with inadequate anticancer treatments; however, restoring normal circadian rhythmicity reduces tumor growth in tumor models.
Highlights.
The endogenous circadian clock regulates various daily physiological and behavioral rhythms.
Circadian rhythm disruption (e.g., irregular work shifts and frequent travel across time zones) increases cancer initiation and progression risk.
Understanding the relationship between cancer and the circadian clock offers possibilities for new and effective cancer treatments and would help develop chronotherapeutic approaches to overcome cancer progression.
9. Acknowledgements
We thank Dr. Donald Hill, the University of Alabama at Birmingham, Birmingham, Alabama, USA, for his help in editing this article. Ms. Hanan Musa and Dr. Chand Davuljigari provided technical assistance.
10. Funding
This work was supported by the National Institutes of Health (NIH) (Grant No. UO1GM132769), National Science Foundation (NSF) (Grant No. 1912322), and Cancer Biology Research and Training at Alabama State University, USA. Dr. Malik’s effort was supported by Raman postdoctoral fellowship funded by the University Grants Commission, Government of India.
Abbreviations
- ARID1A
AT-rich Interactive Domain 1A
- AMPK
AMP-Activated Protein Kinase
- BMAL1
Brain and Muscle ARNT-like Protein-1
- CDK4/6
Cyclin-Dependent Kinase 4/6
- Cdkn1a
Cyclin-Dependent Kinase Inhibitor 1A
- CK1δ
Casein Kinase 1 Delta
- CLOCK
Circadian Locomotor Output Cycles Kaput
- CRY1/2
Cryptochrome
- CK1ε
Casein Kinase 1 Epsilon
- Dyrk1A
Dual-Specificity Tyrosine Phosphorylation-Regulated Kinase 1A
- FBXL3
F-Box and Leucine-Rich Repeat Protein 3
- GSK-3β
Glycogen Synthase Kinase -3 Beta
- HDACs
Histone Deacetylases
- HIF-1α
Hypoxia-inducible Factor 1-Alpha
- HRE
Hypoxia Response Element
- HIF
Hypoxia Inducible Factor
- KrasG12D
Kras Mutation, and the Single-Site Mutation G12D
- NPAS2
Neuronal PAS Domain Protein 2
- NR1D1
Nuclear Receptor Subfamily 1, Group D, Member 1
- PGC-1alpha
Peroxisome Proliferator-Activated Receptor-Gamma Coactivator-1alpha
- PERK
Protein Kinase RNA-like Endoplasmic Reticulum Kinase
- PER1/2/3
Period1, Period2, Period3
- ROR
Retinoic Acid Receptor-Related Orphan Receptor
- SCN
Suprachiasmatic Nucleus
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
The authors declare that they do not have any conflict of interest.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
12. References
- [1].Patke A, Young MW, Axelrod S, Molecular mechanisms and physiological importance of circadian rhythms, Nat Rev Mol Cell Biol, 21 (2020) 67–84. [DOI] [PubMed] [Google Scholar]
- [2].Rivkees SA, The Development of Circadian Rhythms: From Animals To Humans, Sleep Med Clin, 2 (2007) 331–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Reppert SM, Weaver DR, Coordination of circadian timing in mammals, Nature, 418 (2002) 935–941. [DOI] [PubMed] [Google Scholar]
- [4].Buijs RM, Scheer FA, Kreier F, Yi C, Bos N, Goncharuk VD, Kalsbeek A, Organization of circadian functions: interaction with the body, Prog Brain Res, 153 (2006) 341–360. [DOI] [PubMed] [Google Scholar]
- [5].Bunney WE, Bunney BG, Molecular clock genes in man and lower animals: possible implications for circadian abnormalities in depression, Neuropsychopharmacology, 22 (2000) 335–345. [DOI] [PubMed] [Google Scholar]
- [6].Sahar S, Sassone-Corsi P, Metabolism and cancer: the circadian clock connection, Nat Rev Cancer, 9 (2009) 886–896. [DOI] [PubMed] [Google Scholar]
- [7].Fu L, Kettner NM, The circadian clock in cancer development and therapy, Prog Mol Biol Transl Sci, 119 (2013) 221–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Walton ZE, Patel CH, Brooks RC, Yu Y, Ibrahim-Hashim A, Riddle M, Porcu A, Jiang T, Ecker BL, Tameire F, Koumenis C, Weeraratna AT, Welsh DK, Gillies R, Alwine JC, Zhang L, Powell JD, Dang CV, Acid Suspends the Circadian Clock in Hypoxia through Inhibition of mTOR, Cell, 174 (2018) 72–87.e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wood PA, Yang X, Hrushesky WJ, Clock genes and cancer, Integr Cancer Ther, 8 (2009) 303–308. [DOI] [PubMed] [Google Scholar]
- [10].Evans JA, Davidson AJ, Health consequences of circadian disruption in humans and animal models, Prog Mol Biol Transl Sci, 119 (2013) 283–323. [DOI] [PubMed] [Google Scholar]
- [11].Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB, Coordinated transcription of key pathways in the mouse by the circadian clock, Cell, 109 (2002) 307–320. [DOI] [PubMed] [Google Scholar]
- [12].Rusak B, Zucker I, Neural regulation of circadian rhythms, Physiol Rev, 59 (1979) 449–526. [DOI] [PubMed] [Google Scholar]
- [13].Ralph MR, Foster RG, Davis FC, Menaker M, Transplanted suprachiasmatic nucleus determines circadian period, Science, 247 (1990) 975–978. [DOI] [PubMed] [Google Scholar]
- [14].Welsh DK, Logothetis DE, Meister M, Reppert SM, Individual neurons dissociated from rat suprachiasmatic nucleus express independently phased circadian firing rhythms, Neuron, 14 (1995) 697–706. [DOI] [PubMed] [Google Scholar]
- [15].Weaver DR, The suprachiasmatic nucleus: a 25-year retrospective, J Biol Rhythms, 13 (1998) 100–112. [DOI] [PubMed] [Google Scholar]
- [16].Menaker M, Moreira LF, Tosini G, Evolution of circadian organization in vertebrates, Braz J Med Biol Res, 30 (1997) 305–313. [DOI] [PubMed] [Google Scholar]
- [17].Block GD, Geusz M, Khalsa SB, Michel S, Whitmore D, Circadian rhythm generation, expression and entrainment in a molluscan model system, Prog Brain Res, 111 (1996) 93–102. [DOI] [PubMed] [Google Scholar]
- [18].Ewer J, Frisch B, Hamblen-Coyle MJ, Rosbash M, Hall JC, Expression of the period clock gene within different cell types in the brain of Drosophila adults and mosaic analysis of these cells’ influence on circadian behavioral rhythms, J Neurosci, 12 (1992) 3321–3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Vinod Kumar BPSSR, The Bird Clock: A Complex, Multi-Oscillatory and Highly Diversified System Biological Rhythm Research 35, (2004) 121–144. [Google Scholar]
- [20].Dunlap JC, Molecular bases for circadian clocks, Cell, 96 (1999) 271–290. [DOI] [PubMed] [Google Scholar]
- [21].Ko CH, Takahashi JS, Molecular components of the mammalian circadian clock, Hum Mol Genet, 15 Spec No 2 (2006) R271–277. [DOI] [PubMed] [Google Scholar]
- [22].Okamura H, Yamaguchi S, Yagita K, Molecular machinery of the circadian clock in mammals, Cell Tissue Res, 309 (2002) 47–56. [DOI] [PubMed] [Google Scholar]
- [23].Glossop NR, Hardin PE, Central and peripheral circadian oscillator mechanisms in flies and mammals, J Cell Sci, 115 (2002) 3369–3377. [DOI] [PubMed] [Google Scholar]
- [24].Masri S, Sassone-Corsi P, The emerging link between cancer, metabolism, and circadian rhythms, Nat Med, 24 (2018) 1795–1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Roenneberg T, Wirz-Justice A, Merrow M, Life between clocks: daily temporal patterns of human chronotypes, J Biol Rhythms, 18 (2003) 80–90. [DOI] [PubMed] [Google Scholar]
- [26].Sato F, Kawamoto T, Fujimoto K, Noshiro M, Honda KK, Honma S, Honma K, Kato Y, Functional analysis of the basic helix-loop-helix transcription factor DEC1 in circadian regulation. Interaction with BMAL1, Eur J Biochem, 271 (2004) 4409–4419. [DOI] [PubMed] [Google Scholar]
- [27].Ueda HR, Chen W, Adachi A, Wakamatsu H, Hayashi S, Takasugi T, Nagano M, Nakahama K, Suzuki Y, Sugano S, Iino M, Shigeyoshi Y, Hashimoto S, A transcription factor response element for gene expression during circadian night, Nature, 418 (2002) 534–539. [DOI] [PubMed] [Google Scholar]
- [28].Vitaterna MH, King DP, Chang AM, Kornhauser JM, Lowrey PL, McDonald JD, Dove WF, Pinto LH, Turek FW, Takahashi JS, Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior, Science, 264 (1994) 719–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Gekakis N, Staknis D, Nguyen HB, Davis FC, Wilsbacher LD, King DP, Takahashi JS, Weitz CJ, Role of the CLOCK protein in the mammalian circadian mechanism, Science, 280 (1998) 1564–1569. [DOI] [PubMed] [Google Scholar]
- [30].Zhang EE, Kay SA, Clocks not winding down: unravelling circadian networks, Nat Rev Mol Cell Biol, 11 (2010) 764–776. [DOI] [PubMed] [Google Scholar]
- [31].Duong HA, Robles MS, Knutti D, Weitz CJ, A molecular mechanism for circadian clock negative feedback, Science, 332 (2011) 1436–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Shirogane T, Jin J, Ang XL, Harper JW, SCFbeta-TRCP controls clock-dependent transcription via casein kinase 1-dependent degradation of the mammalian period-1 (Per1) protein, J Biol Chem, 280 (2005) 26863–26872. [DOI] [PubMed] [Google Scholar]
- [33].Busino L, Bassermann F, Maiolica A, Lee C, Nolan PM, Godinho SI, Draetta GF, Pagano M, SCFFbxl3 controls the oscillation of the circadian clock by directing the degradation of cryptochrome proteins, Science, 316 (2007) 900–904. [DOI] [PubMed] [Google Scholar]
- [34].Siepka SM, Yoo SH, Park J, Song W, Kumar V, Hu Y, Lee C, Takahashi JS, Circadian mutant Overtime reveals F-box protein FBXL3 regulation of cryptochrome and period gene expression, Cell, 129 (2007) 1011–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Nangle SN, Rosensweig C, Koike N, Tei H, Takahashi JS, Green CB, Zheng N, Molecular assembly of the period-cryptochrome circadian transcriptional repressor complex, Elife, 3 (2014) e03674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Eide EJ, Woolf MF, Kang H, Woolf P, Hurst W, Camacho F, Vielhaber EL, Giovanni A, Virshup DM, Control of mammalian circadian rhythm by CKIepsilon-regulated proteasome-mediated PER2 degradation, Mol Cell Biol, 25 (2005) 2795–2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Guillaumond F, Dardente H, Giguere V, Cermakian N, Differential control of Bmal1 circadian transcription by REV-ERB and ROR nuclear receptors, J Biol Rhythms, 20 (2005) 391–403. [DOI] [PubMed] [Google Scholar]
- [38].Sulli G, Lam MTY, Panda S, Interplay between Circadian Clock and Cancer: New Frontiers for Cancer Treatment, Trends Cancer, 5 (2019) 475–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Chen ST, Choo KB, Hou MF, Yeh KT, Kuo SJ, Chang JG, Deregulated expression of the PER1, PER2 and PER3 genes in breast cancers, Carcinogenesis, 26 (2005) 1241–1246. [DOI] [PubMed] [Google Scholar]
- [40].Kuo SJ, Chen ST, Yeh KT, Hou MF, Chang YS, Hsu NC, Chang JG, Disturbance of circadian gene expression in breast cancer, Virchows Arch, 454 (2009) 467–474. [DOI] [PubMed] [Google Scholar]
- [41].Krugluger W, Brandstaetter A, Kallay E, Schueller J, Krexner E, Kriwanek S, Bonner E, Cross HS, Regulation of genes of the circadian clock in human colon cancer: reduced period-1 and dihydropyrimidine dehydrogenase transcription correlates in high-grade tumors, Cancer Res, 67 (2007) 7917–7922. [DOI] [PubMed] [Google Scholar]
- [42].Mostafaie N, Kallay E, Sauerzapf E, Bonner E, Kriwanek S, Cross HS, Huber KR, Krugluger W, Correlated downregulation of estrogen receptor beta and the circadian clock gene Per1 in human colorectal cancer, Mol Carcinog, 48 (2009) 642–647. [DOI] [PubMed] [Google Scholar]
- [43].Xia HC, Niu ZF, Ma H, Cao SZ, Hao SC, Liu ZT, Wang F, Deregulated expression of the Per1 and Per2 in human gliomas, Can J Neurol Sci, 37 (2010) 365–370. [DOI] [PubMed] [Google Scholar]
- [44].Fu L, Lee CC, The circadian clock: pacemaker and tumour suppressor, Nat Rev Cancer, 3 (2003) 350–361. [DOI] [PubMed] [Google Scholar]
- [45].Viswanathan AN, Schernhammer ES, Circulating melatonin and the risk of breast and endometrial cancer in women, Cancer Lett, 281 (2009) 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Hoffman AE, Yi CH, Zheng T, Stevens RG, Leaderer D, Zhang Y, Holford TR, Hansen J, Paulson J, Zhu Y, CLOCK in breast tumorigenesis: genetic, epigenetic, and transcriptional profiling analyses, Cancer Res, 70 (2010) 1459–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Lee CC, Tumor suppression by the mammalian Period genes, Cancer Causes Control, 17 (2006) 525–530. [DOI] [PubMed] [Google Scholar]
- [48].Winter SL, Bosnoyan-Collins L, Pinnaduwage D, Andrulis IL, Expression of the circadian clock genes Per1 and Per2 in sporadic and familial breast tumors, Neoplasia, 9 (2007) 797–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Yuan P, Li J, Zhou F, Huang Q, Zhang J, Guo X, Lyu Z, Zhang H, Xing J, NPAS2 promotes cell survival of hepatocellular carcinoma by transactivating CDC25A, Cell Death Dis, 8 (2017) e2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Benna C, Rajendran S, Spiro G, Tropea S, Del Fiore P, Rossi CR, Mocellin S, Associations of clock genes polymorphisms with soft tissue sarcoma susceptibility and prognosis, J Transl Med, 16 (2018) 338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Rajendran S, Benna C, Marchet A, Nitti D, Mocellin S, Germline polymorphisms of circadian genes and gastric cancer predisposition, Cancer Commun (Lond), 40 (2020) 234–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Karantanos T, Theodoropoulos G, Gazouli M, Vaiopoulou A, Karantanou C, Stravopodis DJ, Bramis K, Lymperi M, Pektasidis D, Association of the clock genes polymorphisms with colorectal cancer susceptibility, J Surg Oncol, 108 (2013) 563–567. [DOI] [PubMed] [Google Scholar]
- [53].Kogevinas M, Espinosa A, Castelló A, Gómez-Acebo I, Guevara M, Martin V, Amiano P, Alguacil J, Peiro R, Moreno V, Costas L, Fernández-Tardón G, Jimenez JJ, Marcos-Gragera R, Perez-Gomez B, Llorca J, Moreno-Iribas C, Fernández-Villa T, Oribe M, Aragones N, Papantoniou K, Pollan M, Castano-Vinyals G, Romaguera D, Effect of mistimed eating patterns on breast and prostate cancer risk (MCC-Spain Study), Int J Cancer, 143 (2018) 2380–2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Whiteman DC, Wilson LF, The fractions of cancer attributable to modifiable factors: A global review, Cancer Epidemiol, 44 (2016) 203–221. [DOI] [PubMed] [Google Scholar]
- [55].Zhu Y, Stevens RG, Leaderer D, Hoffman A, Holford T, Zhang Y, Brown HN, Zheng T, Non-synonymous polymorphisms in the circadian gene NPAS2 and breast cancer risk, Breast Cancer Res Treat, 107 (2008) 421–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Gery S, Komatsu N, Baldjyan L, Yu A, Koo D, Koeffler HP, The circadian gene per1 plays an important role in cell growth and DNA damage control in human cancer cells, Mol Cell, 22 (2006) 375–382. [DOI] [PubMed] [Google Scholar]
- [57].Hua H, Wang Y, Wan C, Liu Y, Zhu B, Yang C, Wang X, Wang Z, Cornelissen-Guillaume G, Halberg F, Circadian gene mPer2 overexpression induces cancer cell apoptosis, Cancer Sci, 97 (2006) 589–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Xue X, Liu F, Han Y, Li P, Yuan B, Wang X, Chen Y, Kuang Y, Zhi Q, Zhao H, Silencing NPAS2 promotes cell growth and invasion in DLD-1 cells and correlated with poor prognosis of colorectal cancer, Biochem Biophys Res Commun, 450 (2014) 1058–1062. [DOI] [PubMed] [Google Scholar]
- [59].Schernhammer ES, Laden F, Speizer FE, Willett WC, Hunter DJ, Kawachi I, Fuchs CS, Colditz GA, Night-shift work and risk of colorectal cancer in the nurses’ health study, J Natl Cancer Inst, 95 (2003) 825–828. [DOI] [PubMed] [Google Scholar]
- [60].Kloog I, Haim A, Stevens RG, Portnov BA, Global co-distribution of light at night (LAN) and cancers of prostate, colon, and lung in men, Chronobiol Int, 26 (2009) 108–125. [DOI] [PubMed] [Google Scholar]
- [61].Innominato PF, Giacchetti S, Bjarnason GA, Focan C, Garufi C, Coudert B, Iacobelli S, Tampellini M, Durando X, Mormont MC, Waterhouse J, Levi FA, Prediction of overall survival through circadian rest-activity monitoring during chemotherapy for metastatic colorectal cancer, Int J Cancer, 131 (2012) 2684–2692. [DOI] [PubMed] [Google Scholar]
- [62].Dai H, Zhang L, Cao M, Song F, Zheng H, Zhu X, Wei Q, Zhang W, Chen K, The role of polymorphisms in circadian pathway genes in breast tumorigenesis, Breast Cancer Res Treat, 127 (2011) 531–540. [DOI] [PubMed] [Google Scholar]
- [63].Zhu Y, Stevens RG, Hoffman AE, Fitzgerald LM, Kwon EM, Ostrander EA, Davis S, Zheng T, Stanford JL, Testing the circadian gene hypothesis in prostate cancer: a population-based case-control study, Cancer Res, 69 (2009) 9315–9322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Elshazley M, Sato M, Hase T, Yamashita R, Yoshida K, Toyokuni S, Ishiguro F, Osada H, Sekido Y, Yokoi K, Usami N, Shames DS, Kondo M, Gazdar AF, Minna JD, Hasegawa Y, The circadian clock gene BMAL1 is a novel therapeutic target for malignant pleural mesothelioma, Int J Cancer, 131 (2012) 2820–2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Faustino RS, Cheung P, Richard MN, Dibrov E, Kneesch AL, Deniset JF, Chahine MN, Lee K, Blackwood D, Pierce GN, Ceramide regulation of nuclear protein import, J Lipid Res, 49 (2008) 654–662. [DOI] [PubMed] [Google Scholar]
- [66].Yang MY, Chang JG, Lin PM, Tang KP, Chen YH, Lin HY, Liu TC, Hsiao HH, Liu YC, Lin SF, Downregulation of circadian clock genes in chronic myeloid leukemia: alternative methylation pattern of hPER3, Cancer Sci, 97 (2006) 1298–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Viswanathan AN, Hankinson SE, Schernhammer ES, Night shift work and the risk of endometrial cancer, Cancer Res, 67 (2007) 10618–10622. [DOI] [PubMed] [Google Scholar]
- [68].Tokunaga H, Takebayashi Y, Utsunomiya H, Akahira J, Higashimoto M, Mashiko M, Ito K, Niikura H, Takenoshita S, Yaegashi N, Clinicopathological significance of circadian rhythm-related gene expression levels in patients with epithelial ovarian cancer, Acta Obstet Gynecol Scand, 87 (2008) 1060–1070. [DOI] [PubMed] [Google Scholar]
- [69].Honma S, Kawamoto T, Takagi Y, Fujimoto K, Sato F, Noshiro M, Kato Y, Honma K, Dec1 and Dec2 are regulators of the mammalian molecular clock, Nature, 419 (2002) 841–844. [DOI] [PubMed] [Google Scholar]
- [70].Hsu CM, Lin SF, Lu CT, Lin PM, Yang MY, Altered expression of circadian clock genes in head and neck squamous cell carcinoma, Tumour Biol, 33 (2012) 149–155. [DOI] [PubMed] [Google Scholar]
- [71].Lewintre EJ, Martin CR, Ballesteros CG, Montaner D, Rivera RF, Mayans JR, Garcia-Conde J, Cryptochrome-1 expression: a new prognostic marker in B-cell chronic lymphocytic leukemia, Haematologica, 94 (2009) 280–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Roe OD, Anderssen E, Helge E, Pettersen CH, Olsen KS, Sandeck H, Haaverstad R, Lundgren S, Larsson E, Genome-wide profile of pleural mesothelioma versus parietal and visceral pleura: the emerging gene portrait of the mesothelioma phenotype, PLoS One, 4 (2009) e6554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Gapstur SM, Diver WR, Stevens VL, Carter BD, Teras LR, Jacobs EJ, Work schedule, sleep duration, insomnia, and risk of fatal prostate cancer, Am J Prev Med, 46 (2014) S26–33. [DOI] [PubMed] [Google Scholar]
- [74].Yang WS, Stockwell BR, Inhibition of casein kinase 1-epsilon induces cancer-cell-selective, PERIOD2-dependent growth arrest, Genome Biol, 9 (2008) R92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Relles D, Sendecki J, Chipitsyna G, Hyslop T, Yeo CJ, Arafat HA, Circadian gene expression and clinicopathologic correlates in pancreatic cancer, J Gastrointest Surg, 17 (2013) 443–450. [DOI] [PubMed] [Google Scholar]
- [76].Lin YM, Chang JH, Yeh KT, Yang MY, Liu TC, Lin SF, Su WW, Chang JG, Disturbance of circadian gene expression in hepatocellular carcinoma, Mol Carcinog, 47 (2008) 925–933. [DOI] [PubMed] [Google Scholar]
- [77].Pukkala E, Aspholm R, Auvinen A, Eliasch H, Gundestrup M, Haldorsen T, Hammar N, Hrafnkelsson J, Kyyrönen P, Linnersjö A, Rafnsson V, Storm H, Tveten U, Cancer incidence among 10,211 airline pilots: a Nordic study, Aviat Space Environ Med, 74 (2003) 699–706. [PubMed] [Google Scholar]
- [78].Buzzelli G, Dattolo P, Pinzani M, Brocchi A, Romano S, Gentilini P, Circulating growth hormone and insulin-like growth factor-I in nonalcoholic liver cirrhosis with or without superimposed hepatocarcinoma: evidence of an altered circadian rhythm, Am J Gastroenterol, 88 (1993) 1744–1748. [PubMed] [Google Scholar]
- [79].Luo Y, Wang F, Chen LA, Chen XW, Chen ZJ, Liu PF, li FF, Li CY, Liang W, Deregulated expression of cry1 and cry2 in human gliomas, Asian Pac J Cancer Prev, 13 (2012) 5725–5728. [DOI] [PubMed] [Google Scholar]
- [80].Shih MC, Yeh KT, Tang KP, Chen JC, Chang JG, Promoter methylation in circadian genes of endometrial cancers detected by methylation-specific PCR, Mol Carcinog, 45 (2006) 732–740. [DOI] [PubMed] [Google Scholar]
- [81].Suzuki T, Sato F, Kondo J, Liu Y, Kusumi T, Fujimoto K, Kato Y, Sato T, Kijima H, Period is involved in the proliferation of human pancreatic MIA-PaCa2 cancer cells by TNF-alpha, Biomed Res, 29 (2008) 99–103. [DOI] [PubMed] [Google Scholar]
- [82].Taniguchi H, Fernandez AF, Setien F, Ropero S, Ballestar E, Villanueva A, Yamamoto H, Imai K, Shinomura Y, Esteller M, Epigenetic inactivation of the circadian clock gene BMAL1 in hematologic malignancies, Cancer Res, 69 (2009) 8447–8454. [DOI] [PubMed] [Google Scholar]
- [83].Zhu Y, Leaderer D, Guss C, Brown HN, Zhang Y, Boyle P, Stevens RG, Hoffman A, Qin Q, Han X, Zheng T, Ala394Thr polymorphism in the clock gene NPAS2: a circadian modifier for the risk of non-Hodgkin’s lymphoma, Int J Cancer, 120 (2007) 432–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Eisele L, Prinz R, Klein-Hitpass L, Nückel H, Lowinski K, Thomale J, Moeller LC, Dührsen U, Dürig J, Combined PER2 and CRY1 expression predicts outcome in chronic lymphocytic leukemia, Eur J Haematol, 83 (2009) 320–327. [DOI] [PubMed] [Google Scholar]
- [85].Kim KS, Kim YC, Oh IJ, Kim SS, Choi JY, Ahn RS, Association of worse prognosis with an aberrant diurnal cortisol rhythm in patients with advanced lung cancer, Chronobiol Int, 29 (2012) 1109–1120. [DOI] [PubMed] [Google Scholar]
- [86].Sephton SE, Lush E, Dedert EA, Floyd AR, Rebholz WN, Dhabhar FS, Spiegel D, Salmon P, Diurnal cortisol rhythm as a predictor of lung cancer survival, Brain Behav Immun, 30 Suppl (2013) S163–170. [DOI] [PubMed] [Google Scholar]
- [87].Zhou F, He X, Liu H, Zhu Y, Jin T, Chen C, Qu F, Li Y, Bao G, Chen Z, Xing J, Functional polymorphisms of circadian positive feedback regulation genes and clinical outcome of Chinese patients with resected colorectal cancer, Cancer, 118 (2012) 937–946. [DOI] [PubMed] [Google Scholar]
- [88].Blask DE, Hill SM, Dauchy RT, Xiang S, Yuan L, Duplessis T, Mao L, Dauchy E, Sauer LA, Circadian regulation of molecular, dietary, and metabolic signaling mechanisms of human breast cancer growth by the nocturnal melatonin signal and the consequences of its disruption by light at night, J Pineal Res, 51 (2011) 259–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Aihara T, Miyoshi Y, Koyama K, Suzuki M, Takahashi E, Monden M, Nakamura Y, Cloning and mapping of SMARCA5 encoding hSNF2H, a novel human homologue of Drosophila ISWI, Cytogenet Cell Genet, 81 (1998) 191–193. [DOI] [PubMed] [Google Scholar]
- [90].Narasimamurthy R, Hatori M, Nayak SK, Liu F, Panda S, Verma IM, Circadian clock protein cryptochrome regulates the expression of proinflammatory cytokines, Proc Natl Acad Sci U S A, 109 (2012) 12662–12667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Lahti TA, Partonen T, Kyyrönen P, Kauppinen T, Pukkala E, Night-time work predisposes to non-Hodgkin lymphoma, Int J Cancer, 123 (2008) 2148–2151. [DOI] [PubMed] [Google Scholar]
- [92].Sun CM, Huang SF, Zeng JM, Liu DB, Xiao Q, Tian WJ, Zhu XD, Huang ZG, Feng WL, Per2 inhibits k562 leukemia cell growth in vitro and in vivo through cell cycle arrest and apoptosis induction, Pathol Oncol Res, 16 (2010) 403–411. [DOI] [PubMed] [Google Scholar]
- [93].Panzer A, Melatonin in osteosarcoma: an effective drug?, Med Hypotheses, 48 (1997) 523–525. [DOI] [PubMed] [Google Scholar]
- [94].Touitou Y, Bogdan A, Lévi F, Benavides M, Auzéby A, Disruption of the circadian patterns of serum cortisol in breast and ovarian cancer patients: relationships with tumour marker antigens, Br J Cancer, 74 (1996) 1248–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Xiang S, Coffelt SB, Mao L, Yuan L, Cheng Q, Hill SM, Period-2: a tumor suppressor gene in breast cancer, J Circadian Rhythms, 6 (2008) 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Jung-Hynes B, Schmit TL, Reagan-Shaw SR, Siddiqui IA, Mukhtar H, Ahmad N, Melatonin, a novel Sirt1 inhibitor, imparts antiproliferative effects against prostate cancer in vitro in culture and in vivo in TRAMP model, J Pineal Res, 50 (2011) 140–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Gery S, Koeffler HP, The role of circadian regulation in cancer, Cold Spring Harb Symp Quant Biol, 72 (2007) 459–464. [DOI] [PubMed] [Google Scholar]
- [98].Mullenders J, Fabius AW, Madiredjo M, Bernards R, Beijersbergen RL, A large scale shRNA barcode screen identifies the circadian clock component ARNTL as putative regulator of the p53 tumor suppressor pathway, PLoS One, 4 (2009) e4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Brandi G, Calabrese C, Pantaleo MA, Morselli Labate A, Di Febo G, Hakim R, De Vivo A, Di Marco MC, Biasco G, Circadian variations of rectal cell proliferation in patients affected by advanced colorectal cancer, Cancer Lett, 208 (2004) 193–196. [DOI] [PubMed] [Google Scholar]
- [100].Cao Q, Gery S, Dashti A, Yin D, Zhou Y, Gu J, Koeffler HP, A role for the clock gene per1 in prostate cancer, Cancer Res, 69 (2009) 7619–7625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Jung-Hynes B, Huang W, Reiter RJ, Ahmad N, Melatonin resynchronizes dysregulated circadian rhythm circuitry in human prostate cancer cells, J Pineal Res, 49 (2010) 60–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Filipski E, Li XM, Lévi F, Disruption of circadian coordination and malignant growth, Cancer Causes Control, 17 (2006) 509–514. [DOI] [PubMed] [Google Scholar]
- [103].Lee S, Donehower LA, Herron AJ, Moore DD, Fu L, Disrupting circadian homeostasis of sympathetic signaling promotes tumor development in mice, PLoS One, 5 (2010) e10995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Kettner NM, Voicu H, Finegold MJ, Coarfa C, Sreekumar A, Putluri N, Katchy CA, Lee C, Moore DD, Fu L, Circadian Homeostasis of Liver Metabolism Suppresses Hepatocarcinogenesis, Cancer Cell, 30 (2016) 909–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Gauger MA, Sancar A, Cryptochrome, circadian cycle, cell cycle checkpoints, and cancer, Cancer Res, 65 (2005) 6828–6834. [DOI] [PubMed] [Google Scholar]
- [106].Lee C, Etchegaray JP, Cagampang FR, Loudon AS, Reppert SM, Posttranslational mechanisms regulate the mammalian circadian clock, Cell, 107 (2001) 855–867. [DOI] [PubMed] [Google Scholar]
- [107].Gallego M, Virshup DM, Post-translational modifications regulate the ticking of the circadian clock, Nat Rev Mol Cell Biol, 8 (2007) 139–148. [DOI] [PubMed] [Google Scholar]
- [108].Harms E, Kivimae S, Young MW, Saez L, Posttranscriptional and posttranslational regulation of clock genes, J Biol Rhythms, 19 (2004) 361–373. [DOI] [PubMed] [Google Scholar]
- [109].Wang Y, Rao VK, Kok WK, Roy DN, Sethi S, Ling BM, Lee MB, Taneja R, SUMO modification of Stra13 is required for repression of cyclin D1 expression and cellular growth arrest, PLoS One, 7 (2012) e43137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Wang Y, Shankar SR, Kher D, Ling BM, Taneja R, Sumoylation of the basic helix-loop-helix transcription factor sharp-1 regulates recruitment of the histone methyltransferase G9a and function in myogenesis, J Biol Chem, 288 (2013) 17654–17662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Kon N, Hirota T, Kawamoto T, Kato Y, Tsubota T, Fukada Y, Activation of TGF-beta/activin signalling resets the circadian clock through rapid induction of Dec1 transcripts, Nat Cell Biol, 10 (2008) 1463–1469. [DOI] [PubMed] [Google Scholar]
- [112].Nakashima A, Kawamoto T, Honda KK, Ueshima T, Noshiro M, Iwata T, Fujimoto K, Kubo H, Honma S, Yorioka N, Kohno N, Kato Y, DEC1 modulates the circadian phase of clock gene expression, Mol Cell Biol, 28 (2008) 4080–4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Kim J, D’Annibale S, Magliozzi R, Low TY, Jansen P, Shaltiel IA, Mohammed S, Heck AJ, Medema RH, Guardavaccaro D, USP17- and SCFbetaTrCP--regulated degradation of DEC1 controls the DNA damage response, Mol Cell Biol, 34 (2014) 4177–4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Muller S, Hoege C, Pyrowolakis G, Jentsch S, SUMO, ubiquitin’s mysterious cousin, Nat Rev Mol Cell Biol, 2 (2001) 202–210. [DOI] [PubMed] [Google Scholar]
- [115].Gill G, Post-translational modification by the small ubiquitin-related modifier SUMO has big effects on transcription factor activity, Curr Opin Genet Dev, 13 (2003) 108–113. [DOI] [PubMed] [Google Scholar]
- [116].Gill G, SUMO and ubiquitin in the nucleus: different functions, similar mechanisms?, Genes Dev, 18 (2004) 2046–2059. [DOI] [PubMed] [Google Scholar]
- [117].Cardone L, Hirayama J, Giordano F, Tamaru T, Palvimo JJ, Sassone-Corsi P, Circadian clock control by SUMOylation of BMAL1, Science, 309 (2005) 1390–1394. [DOI] [PubMed] [Google Scholar]
- [118].You S, Wood PA, Xiong Y, Kobayashi M, Du-Quiton J, Hrushesky WJ, Daily coordination of cancer growth and circadian clock gene expression, Breast Cancer Res Treat, 91 (2005) 47–60. [DOI] [PubMed] [Google Scholar]
- [119].Hong Y, Xing X, Li S, Bi H, Yang C, Zhao F, Liu Y, Ao X, Chang AK, Wu H, SUMOylation of DEC1 protein regulates its transcriptional activity and enhances its stability, PLoS One, 6 (2011) e23046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Giatromanolaki A, Koukourakis MI, Sivridis E, Turley H, Wykoff CC, Gatter KC, Harris AL, DEC1 (STRA13) protein expression relates to hypoxia- inducible factor 1-alpha and carbonic anhydrase-9 overexpression in non-small cell lung cancer, J Pathol, 200 (2003) 222–228. [DOI] [PubMed] [Google Scholar]
- [121].Chakrabarti J, Turley H, Campo L, Han C, Harris AL, Gatter KC, Fox SB, The transcription factor DEC1 (stra13, SHARP2) is associated with the hypoxic response and high tumour grade in human breast cancers, Br J Cancer, 91 (2004) 954–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Turley H, Wykoff CC, Troup S, Watson PH, Gatter KC, Harris AL, The hypoxia-regulated transcription factor DEC1 (Stra13, SHARP-2) and its expression in human tissues and tumours, J Pathol, 203 (2004) 808–813. [DOI] [PubMed] [Google Scholar]
- [123].Yunokawa M, Tanimoto K, Nakamura H, Nagai N, Kudo Y, Kawamoto T, Kato Y, Hiyama E, Hiyama K, Nishiyama M, Differential regulation of DEC2 among hypoxia-inducible genes in endometrial carcinomas, Oncol Rep, 17 (2007) 871–878. [PubMed] [Google Scholar]
- [124].Yager JD, Davidson NE, Estrogen carcinogenesis in breast cancer, N Engl J Med, 354 (2006) 270–282. [DOI] [PubMed] [Google Scholar]
- [125].Li S, Wang M, Ao X, Chang AK, Yang C, Zhao F, Bi H, Liu Y, Xiao L, Wu H, CLOCK is a substrate of SUMO and sumoylation of CLOCK upregulates the transcriptional activity of estrogen receptor-alpha, Oncogene, 32 (2013) 4883–4891. [DOI] [PubMed] [Google Scholar]
- [126].Doi M, Hirayama J, Sassone-Corsi P, Circadian regulator CLOCK is a histone acetyltransferase, Cell, 125 (2006) 497–508. [DOI] [PubMed] [Google Scholar]
- [127].Kumar R, Li DQ, Muller S, Knapp S, Epigenomic regulation of oncogenesis by chromatin remodeling, Oncogene, 35 (2016) 4423–4436. [DOI] [PubMed] [Google Scholar]
- [128].Ho L, Crabtree GR, Chromatin remodelling during development, Nature, 463 (2010) 474–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Manelyte L, Chromatin remodelers, their implication in cancer and therapeutic potential, Journal of Rare Diseases Research & Treatment, 2 (2017) 34–40. [Google Scholar]
- [130].Clapier CR, Cairns BR, The biology of chromatin remodeling complexes, Annu Rev Biochem, 78 (2009) 273–304. [DOI] [PubMed] [Google Scholar]
- [131].Orvis T, Hepperla A, Walter V, Song S, Simon J, Parker J, Wilkerson MD, Desai N, Major MB, Hayes DN, Davis IJ, Weissman B, BRG1/SMARCA4 inactivation promotes non-small cell lung cancer aggressiveness by altering chromatin organization, Cancer Res, 74 (2014) 6486–6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Helming KC, Wang X, Roberts CWM, Vulnerabilities of mutant SWI/SNF complexes in cancer, Cancer Cell, 26 (2014) 309–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Biegel JA, Busse TM, Weissman BE, SWI/SNF chromatin remodeling complexes and cancer, Am J Med Genet C Semin Med Genet, 166C (2014) 350–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Guichard C, Amaddeo G, Imbeaud S, Ladeiro Y, Pelletier L, Maad IB, Calderaro J, Bioulac-Sage P, Letexier M, Degos F, Clement B, Balabaud C, Chevet E, Laurent A, Couchy G, Letouze E, Calvo F, Zucman-Rossi J, Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma, Nat Genet, 44 (2012) 694–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Okabe I, Bailey LC, Attree O, Srinivasan S, Perkel JM, Laurent BC, Carlson M, Nelson DL, Nussbaum RL, Cloning of human and bovine homologs of SNF2/SWI2: a global activator of transcription in yeast S. cerevisiae, Nucleic Acids Res, 20 (1992) 4649–4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Tsukiyama T, Wu C, Purification and properties of an ATP-dependent nucleosome remodeling factor, Cell, 83 (1995) 1011–1020. [DOI] [PubMed] [Google Scholar]
- [137].Loyola A, LeRoy G, Wang YH, Reinberg D, Reconstitution of recombinant chromatin establishes a requirement for histone-tail modifications during chromatin assembly and transcription, Genes Dev, 15 (2001) 2837–2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Corona DF, Tamkun JW, Multiple roles for ISWI in transcription, chromosome organization and DNA replication, Biochim Biophys Acta, 1677 (2004) 113–119. [DOI] [PubMed] [Google Scholar]
- [139].Perpelescu M, Nozaki N, Obuse C, Yang H, Yoda K, Active establishment of centromeric CENP-A chromatin by RSF complex, J Cell Biol, 185 (2009) 397–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Yadon AN, Tsukiyama T, SnapShot: Chromatin remodeling: ISWI, Cell, 144 (2011) 453–453 e451. [DOI] [PubMed] [Google Scholar]
- [141].Sheu JJ, Choi JH, Guan B, Tsai FJ, Hua CH, Lai MT, Wang TL, Shih Ie M, Rsf-1, a chromatin remodelling protein, interacts with cyclin E1 and promotes tumour development, J Pathol, 229 (2013) 559–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Wilson BG, Roberts CW, SWI/SNF nucleosome remodellers and cancer, Nat Rev Cancer, 11 (2011) 481–492. [DOI] [PubMed] [Google Scholar]
- [143].Shah V, Stokes J, Sutton M, Inequalities in health-related quality of life: repeated cross-sectional study of trends in general practice survey data, Br J Gen Pract, 71 (2021) e178–e184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].N. Cancer Genome Atlas Research, Kandoth C, Schultz N, Cherniack AD, Akbani R, Liu Y, Shen H, Robertson AG, Pashtan I, Shen R, Benz CC, Yau C, Laird PW, Ding L, Zhang W, Mills GB, Kucherlapati R, Mardis ER, Levine DA, Integrated genomic characterization of endometrial carcinoma, Nature, 497 (2013) 67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Chan-On W, Nairismagi ML, Ong CK, Lim WK, Dima S, Pairojkul C, Lim KH, McPherson JR, Cutcutache I, Heng HL, Ooi L, Chung A, Chow P, Cheow PC, Lee SY, Choo SP, Tan IB, Duda D, Nastase A, Myint SS, Wong BH, Gan A, Rajasegaran V, Ng CC, Nagarajan S, Jusakul A, Zhang S, Vohra P, Yu W, Huang D, Sithithaworn P, Yongvanit P, Wongkham S, Khuntikeo N, Bhudhisawasdi V, Popescu I, Rozen SG, Tan P, Teh BT, Exome sequencing identifies distinct mutational patterns in liver fluke-related and non-infection-related bile duct cancers, Nat Genet, 45 (2013) 1474–1478. [DOI] [PubMed] [Google Scholar]
- [146].Kakiuchi M, Nishizawa T, Ueda H, Gotoh K, Tanaka A, Hayashi A, Yamamoto S, Tatsuno K, Katoh H, Watanabe Y, Ichimura T, Ushiku T, Funahashi S, Tateishi K, Wada I, Shimizu N, Nomura S, Koike K, Seto Y, Fukayama M, Aburatani H, Ishikawa S, Recurrent gain-of-function mutations of RHOA in diffuse-type gastric carcinoma, Nat Genet, 46 (2014) 583–587. [DOI] [PubMed] [Google Scholar]
- [147].Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, Nickerson E, Auclair D, Li L, Place C, Dicara D, Ramos AH, Lawrence MS, Cibulskis K, Sivachenko A, Voet D, Saksena G, Stransky N, Onofrio RC, Winckler W, Ardlie K, Wagle N, Wargo J, Chong K, Morton DL, Stemke-Hale K, Chen G, Noble M, Meyerson M, Ladbury JE, Davies MA, Gershenwald JE, Wagner SN, Hoon DS, Schadendorf D, Lander ES, Gabriel SB, Getz G, Garraway LA, Chin L, A landscape of driver mutations in melanoma, Cell, 150 (2012) 251–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].N. Cancer Genome Atlas Research, Comprehensive molecular characterization of clear cell renal cell carcinoma, Nature, 499 (2013) 43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Berger MF, Hodis E, Heffernan TP, Deribe YL, Lawrence MS, Protopopov A, Ivanova E, Watson IR, Nickerson E, Ghosh P, Zhang H, Zeid R, Ren X, Cibulskis K, Sivachenko AY, Wagle N, Sucker A, Sougnez C, Onofrio R, Ambrogio L, Auclair D, Fennell T, Carter SL, Drier Y, Stojanov P, Singer MA, Voet D, Jing R, Saksena G, Barretina J, Ramos AH, Pugh TJ, Stransky N, Parkin M, Winckler W, Mahan S, Ardlie K, Baldwin J, Wargo J, Schadendorf D, Meyerson M, Gabriel SB, Golub TR, Wagner SN, Lander ES, Getz G, Chin L, Garraway LA, Melanoma genome sequencing reveals frequent PREX2 mutations, Nature, 485 (2012) 502–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Imielinski M, Berger AH, Hammerman PS, Hernandez B, Pugh TJ, Hodis E, Cho J, Suh J, Capelletti M, Sivachenko A, Sougnez C, Auclair D, Lawrence MS, Stojanov P, Cibulskis K, Choi K, de Waal L, Sharifnia T, Brooks A, Greulich H, Banerji S, Zander T, Seidel D, Leenders F, Ansen S, Ludwig C, Engel-Riedel W, Stoelben E, Wolf J, Goparju C, Thompson K, Winckler W, Kwiatkowski D, Johnson BE, Janne PA, Miller VA, Pao W, Travis WD, Pass HI, Gabriel SB, Lander ES, Thomas RK, Garraway LA, Getz G, Meyerson M, Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing, Cell, 150 (2012) 1107–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Rudin CM, Durinck S, Stawiski EW, Poirier JT, Modrusan Z, Shames DS, Bergbower EA, Guan Y, Shin J, Guillory J, Rivers CS, Foo CK, Bhatt D, Stinson J, Gnad F, Haverty PM, Gentleman R, Chaudhuri S, Janakiraman V, Jaiswal BS, Parikh C, Yuan W, Zhang Z, Koeppen H, Wu TD, Stern HM, Yauch RL, Huffman KE, Paskulin DD, Illei PB, Varella-Garcia M, Gazdar AF, de Sauvage FJ, Bourgon R, Minna JD, Brock MV, Seshagiri S, Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer, Nat Genet, 44 (2012) 1111–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Ong CK, Subimerb C, Pairojkul C, Wongkham S, Cutcutache I, Yu W, McPherson JR, Allen GE, Ng CC, Wong BH, Myint SS, Rajasegaran V, Heng HL, Gan A, Zang ZJ, Wu Y, Wu J, Lee MH, Huang D, Ong P, Chan-on W, Cao Y, Qian CN, Lim KH, Ooi A, Dykema K, Furge K, Kukongviriyapan V, Sripa B, Wongkham C, Yongvanit P, Futreal PA, Bhudhisawasdi V, Rozen S, Tan P, Teh BT, Exome sequencing of liver fluke-associated cholangiocarcinoma, Nat Genet, 44 (2012) 690–693. [DOI] [PubMed] [Google Scholar]
- [153].Seshagiri S, Stawiski EW, Durinck S, Modrusan Z, Storm EE, Conboy CB, Chaudhuri S, Guan Y, Janakiraman V, Jaiswal BS, Guillory J, Ha C, Dijkgraaf GJ, Stinson J, Gnad F, Huntley MA, Degenhardt JD, Haverty PM, Bourgon R, Wang W, Koeppen H, Gentleman R, Starr TK, Zhang Z, Largaespada DA, Wu TD, de Sauvage FJ, Recurrent R-spondin fusions in colon cancer, Nature, 488 (2012) 660–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Damaschke NA, Yang B, Blute ML Jr., Lin CP, Huang W, Jarrard DF, Frequent disruption of chromodomain helicase DNA-binding protein 8 (CHD8) and functionally associated chromatin regulators in prostate cancer, Neoplasia, 16 (2014) 1018–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].N. Cancer Genome Atlas Research, Comprehensive molecular profiling of lung adenocarcinoma, Nature, 511 (2014) 543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Le Gallo M, O’Hara AJ, Rudd ML, Urick ME, Hansen NF, O’Neil NJ, Price JC, Zhang S, England BM, Godwin AK, Sgroi DC, N.I.H.I.S.C.C.S. Program, Hieter P, Mullikin JC, Merino MJ, Bell DW, Exome sequencing of serous endometrial tumors identifies recurrent somatic mutations in chromatin-remodeling and ubiquitin ligase complex genes, Nat Genet, 44 (2012) 1310–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Krauthammer M, Kong Y, Ha BH, Evans P, Bacchiocchi A, McCusker JP, Cheng E, Davis MJ, Goh G, Choi M, Ariyan S, Narayan D, Dutton-Regester K, Capatana A, Holman EC, Bosenberg M, Sznol M, Kluger HM, Brash DE, Stern DF, Materin MA, Lo RS, Mane S, Ma S, Kidd KK, Hayward NK, Lifton RP, Schlessinger J, Boggon TJ, Halaban R, Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma, Nat Genet, 44 (2012) 1006–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Colbert LE, Petrova AV, Fisher SB, Pantazides BG, Madden MZ, Hardy CW, Warren MD, Pan Y, Nagaraju GP, Liu EA, Saka B, Hall WA, Shelton JW, Gandhi K, Pauly R, Kowalski J, Kooby DA, El-Rayes BF, Staley CA 3rd, Adsay NV, Curran WJ Jr., Landry JC, Maithel SK, Yu DS, CHD7 expression predicts survival outcomes in patients with resected pancreatic cancer, Cancer Res, 74 (2014) 2677–2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Wang X, Reece SP, Van Scott MR, Brown JM, A circadian clock in murine bone marrow-derived mast cells modulates IgE-dependent activation in vitro, Brain Behav Immun, 25 (2011) 127–134. [DOI] [PubMed] [Google Scholar]
- [160].Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, Kryukov GV, Lawrence MS, Sougnez C, McKenna A, Shefler E, Ramos AH, Stojanov P, Carter SL, Voet D, Cortes ML, Auclair D, Berger MF, Saksena G, Guiducci C, Onofrio RC, Parkin M, Romkes M, Weissfeld JL, Seethala RR, Wang L, Rangel-Escareno C, Fernandez-Lopez JC, Hidalgo-Miranda A, Melendez-Zajgla J, Winckler W, Ardlie K, Gabriel SB, Meyerson M, Lander ES, Getz G, Golub TR, Garraway LA, Grandis JR, The mutational landscape of head and neck squamous cell carcinoma, Science, 333 (2011) 1157–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Mattiroli F, D’Arcy S, Luger K, The right place at the right time: chaperoning core histone variants, EMBO Rep, 16 (2015) 1454–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Buschbeck M, Hake SB, Variants of core histones and their roles in cell fate decisions, development and cancer, Nat Rev Mol Cell Biol, 18 (2017) 299–314. [DOI] [PubMed] [Google Scholar]
- [163].Huang R, Langdon SP, Tse M, Mullen P, Um IH, Faratian D, Harrison DJ, The role of HDAC2 in chromatin remodelling and response to chemotherapy in ovarian cancer, Oncotarget, 7 (2016) 4695–4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Beyer S, Zhu J, Mayr D, Kuhn C, Schulze S, Hofmann S, Dannecker C, Jeschke U, Kost BP, Histone H3 Acetyl K9 and Histone H3 Tri Methyl K4 as Prognostic Markers for Patients with Cervical Cancer, Int J Mol Sci, 18 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Monteiro FL, Vitorino R, Wang J, Cardoso H, Laranjeira H, Simoes J, Caldas M, Henrique R, Amado F, Williams C, Jeronimo C, Helguero LA, The histone H2A isoform Hist2h2ac is a novel regulator of proliferation and epithelial-mesenchymal transition in mammary epithelial and in breast cancer cells, Cancer Lett, 396 (2017) 42–52. [DOI] [PubMed] [Google Scholar]
- [166].Liu M, Chen L, Ma NF, Chow RK, Li Y, Song Y, Chan TH, Fang S, Yang X, Xi S, Jiang L, Li Y, Zeng TT, Li Y, Yuan YF, Guan XY, CHD1L promotes lineage reversion of hepatocellular carcinoma through opening chromatin for key developmental transcription factors, Hepatology, 63 (2016) 1544–1559. [DOI] [PubMed] [Google Scholar]
- [167].Alenghat T, Meyers K, Mullican SE, Leitner K, Adeniji-Adele A, Avila J, Bućan M, Ahima RS, Kaestner KH, Lazar MA, Nuclear receptor corepressor and histone deacetylase 3 govern circadian metabolic physiology, Nature, 456 (2008) 997–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Feng D, Liu T, Sun Z, Bugge A, Mullican SE, Alenghat T, Liu XS, Lazar MA, A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism, Science, 331 (2011) 1315–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente LP, Sassone-Corsi P, The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control, Cell, 134 (2008) 329–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, Schibler U, SIRT1 regulates circadian clock gene expression through PER2 deacetylation, Cell, 134 (2008) 317–328. [DOI] [PubMed] [Google Scholar]
- [171].Lamia KA, Sachdeva UM, DiTacchio L, Williams EC, Alvarez JG, Egan DF, Vasquez DS, Juguilon H, Panda S, Shaw RJ, Thompson CB, Evans RM, AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation, Science, 326 (2009) 437–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Sahar S, Masubuchi S, Eckel-Mahan K, Vollmer S, Galla L, Ceglia N, Masri S, Barth TK, Grimaldi B, Oluyemi O, Astarita G, Hallows WC, Piomelli D, Imhof A, Baldi P, Denu JM, Sassone-Corsi P, Circadian control of fatty acid elongation by SIRT1 protein-mediated deacetylation of acetyl-coenzyme A synthetase 1, J Biol Chem, 289 (2014) 6091–6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Aguilar-Arnal L, Katada S, Orozco-Solis R, Sassone-Corsi P, NAD(+)-SIRT1 control of H3K4 trimethylation through circadian deacetylation of MLL1, Nat Struct Mol Biol, 22 (2015) 312–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Altman BJ, Cancer Clocks Out for Lunch: Disruption of Circadian Rhythm and Metabolic Oscillation in Cancer, Front Cell Dev Biol, 4 (2016) 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Green CB, Takahashi JS, Bass J, The meter of metabolism, Cell, 134 (2008) 728–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Salgado-Delgado R, Tapia Osorio A, Saderi N, Escobar C, Disruption of circadian rhythms: a crucial factor in the etiology of depression, Depress Res Treat, 2011 (2011) 839743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].M. RK Arble DM, Bass J, Turek FW., Circadian disruption and metabolic disease: findings from animal models, Best Pract. Res. Clin. Endocrinol. Metab, 24 (2010) 785–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Verlande A, Masri S, Circadian Clocks and Cancer: Timekeeping Governs Cellular Metabolism, Trends Endocrinol Metab, 30 (2019) 445–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Shafi AA, AlHarthi B, Riaz MM, AlBagir A, Gaint phyllodes tumour with axillary & interpectoral lymph node metastasis; A rare presentation, Int J Surg Case Rep, 66 (2020) 350–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Damiola F, Le Minh N, Preitner N, Kornmann B, Fleury-Olela F, Schibler U, Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus, Genes Dev, 14 (2000) 2950–2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Guerrero-Vargas NN, Navarro-Espindola R, Guzman-Ruiz MA, Basualdo MDC, Espitia-Bautista E, Lopez-Bago A, Lascurain R, Cordoba-Manilla C, Buijs RM, Escobar C, Circadian disruption promotes tumor growth by anabolic host metabolism; experimental evidence in a rat model, BMC Cancer, 17 (2017) 625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Celia LK, Van Cauter E, Schoeller DA, Effect of meal timing on diurnal rhythm of human cholesterol synthesis, Am J Physiol, 269 (1995) E878–883. [DOI] [PubMed] [Google Scholar]
- [183].Schoeller DA, Celia LK, Sinha MK, Caro JF, Entrainment of the diurnal rhythm of plasma leptin to meal timing, J Clin Invest, 100 (1997) 1882–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Kohsaka A, Laposky AD, Ramsey KM, Estrada C, Joshu C, Kobayashi Y, Turek FW, Bass J, High-fat diet disrupts behavioral and molecular circadian rhythms in mice, Cell Metab, 6 (2007) 414–421. [DOI] [PubMed] [Google Scholar]
- [185].Arble DM, Bass J, Laposky AD, Vitaterna MH, Turek FW, Circadian timing of food intake contributes to weight gain, Obesity (Silver Spring), 17 (2009) 2100–2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Liu C, Li S, Liu T, Borjigin J, Lin JD, Transcriptional coactivator PGC-1alpha integrates the mammalian clock and energy metabolism, Nature, 447 (2007) 477–481. [DOI] [PubMed] [Google Scholar]
- [187].Um JH, Yang S, Yamazaki S, Kang H, Viollet B, Foretz M, Chung JH, Activation of 5’-AMP-activated kinase with diabetes drug metformin induces casein kinase Iepsilon (CKIepsilon)-dependent degradation of clock protein mPer2, J Biol Chem, 282 (2007) 20794–20798. [DOI] [PubMed] [Google Scholar]
- [188].Fekry B, Ribas-Latre A, Baumgartner C, Deans JR, Kwok C, Patel P, Fu L, Berdeaux R, Sun K, Kolonin MG, Wang SH, Yoo SH, Sladek FM, Eckel-Mahan K, Incompatibility of the circadian protein BMAL1 and HNF4α in hepatocellular carcinoma, Nat Commun, 9 (2018) 4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].Srivastava A, Mannam P, Warburg revisited: lessons for innate immunity and sepsis, Front Physiol, 6 (2015) 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].Sun F, Dai C, Xie J, Hu X, Biochemical issues in estimation of cytosolic free NAD/NADH ratio, PLoS One, 7 (2012) e34525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].Pavlova NN, Thompson CB, The Emerging Hallmarks of Cancer Metabolism, Cell Metab, 23 (2016) 27–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Eckle T, Hartmann K, Bonney S, Reithel S, Mittelbronn M, Walker LA, Lowes BD, Han J, Borchers CH, Buttrick PM, Kominsky DJ, Colgan SP, Eltzschig HK, Adora2b-elicited Per2 stabilization promotes a HIF-dependent metabolic switch crucial for myocardial adaptation to ischemia, Nat Med, 18 (2012) 774–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [193].Peek CB, Levine DC, Cedernaes J, Taguchi A, Kobayashi Y, Tsai SJ, Bonar NA, McNulty MR, Ramsey KM, Bass J, Circadian Clock Interaction with HIF1α Mediates Oxygenic Metabolism and Anaerobic Glycolysis in Skeletal Muscle, Cell Metab, 25 (2017) 86–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [194].Wu Y, Tang D, Liu N, Xiong W, Huang H, Li Y, Ma Z, Zhao H, Chen P, Qi X, Zhang EE, Reciprocal Regulation between the Circadian Clock and Hypoxia Signaling at the Genome Level in Mammals, Cell Metab, 25 (2017) 73–85. [DOI] [PubMed] [Google Scholar]
- [195].Adamovich Y, Ladeuix B, Golik M, Koeners MP, Asher G, Rhythmic Oxygen Levels Reset Circadian Clocks through HIF1α, Cell Metab, 25 (2017) 93–101. [DOI] [PubMed] [Google Scholar]
- [196].Khapre RV, Patel SA, Kondratova AA, Chaudhary A, Velingkaar N, Antoch MP, Kondratov RV, Metabolic clock generates nutrient anticipation rhythms in mTOR signaling, Aging (Albany NY), 6 (2014) 675–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [197].Serchov T, Heumann R, Ras Activity Tunes the Period and Modulates the Entrainment of the Suprachiasmatic Clock, Front Neurol, 8 (2017) 264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [198].Morishita Y, Miura D, Kida S, PI3K regulates BMAL1/CLOCK-mediated circadian transcription from the Dbp promoter, Biosci Biotechnol Biochem, 80 (2016) 1131–1140. [DOI] [PubMed] [Google Scholar]
- [199].Walhout AJ, Gubbels JM, Bernards R, van der Vliet PC, Timmers HT, c-Myc/Max heterodimers bind cooperatively to the E-box sequences located in the first intron of the rat ornithine decarboxylase (ODC) gene, Nucleic Acids Res, 25 (1997) 1493–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Ripperger JA, Schibler U, Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions, Nat Genet, 38 (2006) 369–374. [DOI] [PubMed] [Google Scholar]
- [201].Shostak A, Ruppert B, Ha N, Bruns P, Toprak UH, Eils R, Schlesner M, Diernfellner A, Brunner M, MYC/MIZ1-dependent gene repression inversely coordinates the circadian clock with cell cycle and proliferation, Nat Commun, 7 (2016) 11807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].Altman BJ, Hsieh AL, Sengupta A, Krishnanaiah SY, Stine ZE, Walton ZE, Gouw AM, Venkataraman A, Li B, Goraksha-Hicks P, Diskin SJ, Bellovin DI, Simon MC, Rathmell JC, Lazar MA, Maris JM, Felsher DW, Hogenesch JB, Weljie AM, Dang CV, MYC Disrupts the Circadian Clock and Metabolism in Cancer Cells, Cell Metab, 22 (2015) 1009–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [203].Carroll PA, Diolaiti D, McFerrin L, Gu H, Djukovic D, Du J, Cheng PF, Anderson S, Ulrich M, Hurley JB, Raftery D, Ayer DE, Eisenman RN, Deregulated Myc requires MondoA/MIx for metabolic reprogramming and tumorigenesis, Cancer Cell, 27 (2015) 271–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].Dali-Youcef N, Lagouge M, Froelich S, Koehl C, Schoonjans K, Auwerx J, Sirtuins: the ‘magnificent seven’, function, metabolism and longevity, Ann Med, 39 (2007) 335–345. [DOI] [PubMed] [Google Scholar]
- [205].Michan S, Sinclair D, Sirtuins in mammals: insights into their biological function, Biochem J, 404 (2007) 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].Schwer B, Verdin E, Conserved metabolic regulatory functions of sirtuins, Cell Metab, 7 (2008) 104–112. [DOI] [PubMed] [Google Scholar]
- [207].Masri S, Rigor P, Cervantes M, Ceglia N, Sebastian C, Xiao C, Roqueta-Rivera M, Deng C, Osborne TF, Mostoslavsky R, Baldi P, Sassone-Corsi P, Partitioning circadian transcription by SIRT6 leads to segregated control of cellular metabolism, Cell, 158 (2014) 659–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [208].Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P, Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1, Science, 324 (2009) 654–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [209].Guarente L, Sirtuins, aging, and metabolism, Cold Spring Harb Symp Quant Biol, 76 (2011) 81–90. [DOI] [PubMed] [Google Scholar]
- [210].Chang HC, Guarente L, SIRT1 mediates central circadian control in the SCN by a mechanism that decays with aging, Cell, 153 (2013) 1448–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [211].Duong HA, Weitz CJ, Temporal orchestration of repressive chromatin modifiers by circadian clock Period complexes, Nat Struct Mol Biol, 21 (2014) 126–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [212].Roth M, Chen WY, Sorting out functions of sirtuins in cancer, Oncogene, 33 (2014) 1609–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [213].Lin Z, Fang D, The Roles of SIRT1 in Cancer, Genes Cancer, 4 (2013) 97–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [214].Menssen A, Hydbring P, Kapelle K, Vervoorts J, Diebold J, Luscher B, Larsson LG, Hermeking H, The c-MYC oncoprotein, the NAMPT enzyme, the SIRT1-inhibitor DBC1, and the SIRT1 deacetylase form a positive feedback loop, Proc Natl Acad Sci U S A, 109 (2012) E187–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [215].Islam S, Abiko Y, Uehara O, Chiba I, Sirtuin 1 and oral cancer, Oncol Lett, 17 (2019) 729–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [216].Jin X, Wei Y, Xu F, Zhao M, Dai K, Shen R, Yang S, Zhang N, SIRT1 promotes formation of breast cancer through modulating Akt activity, J Cancer, 9 (2018) 2012–2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [217].Li L, Osdal T, Ho Y, Chun S, McDonald T, Agarwal P, Lin A, Chu S, Qi J, Li L, Hsieh YT, Dos Santos C, Yuan H, Ha TQ, Popa M, Hovland R, Bruserud O, Gjertsen BT, Kuo YH, Chen W, Lain S, McCormack E, Bhatia R, SIRT1 activation by a c-MYC oncogenic network promotes the maintenance and drug resistance of human FLT3-ITD acute myeloid leukemia stem cells, Cell Stem Cell, 15 (2014) 431–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [218].Bosch-Presegue L, Vaquero A, The dual role of sirtuins in cancer, Genes Cancer, 2 (2011) 648–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [219].Deng CX, SIRT1, is it a tumor promoter or tumor suppressor?, Int J Biol Sci, 5 (2009) 147–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [220].Masri S, Kinouchi K, Sassone-Corsi P, Circadian clocks, epigenetics, and cancer, Curr Opin Oncol, 27 (2015) 50–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [221].Sebastián C, Zwaans BM, Silberman DM, Gymrek M, Goren A, Zhong L, Ram O, Truelove J, Guimaraes AR, Toiber D, Cosentino C, Greenson JK, MacDonald AI, McGlynn L, Maxwell F, Edwards J, Giacosa S, Guccione E, Weissleder R, Bernstein BE, Regev A, Shiels PG, Lombard DB, Mostoslavsky R, The histone deacetylase SIRT6 is a tumor suppressor that controls cancer metabolism, Cell, 151 (2012) 1185–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [222].Haigis MC, Sinclair DA, Mammalian sirtuins: biological insights and disease relevance, Annu Rev Pathol, 5 (2010)253–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [223].Finley LW, Haas W, Desquiret-Dumas V, Wallace DC, Procaccio V, Gygi SP, Haigis MC, Succinate dehydrogenase is a direct target of sirtuin 3 deacetylase activity, PLoS One, 6 (2011) e23295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [224].Siffroi-Fernandez S, Dulong S, Li XM, Filipski E, Grechez-Cassiau A, Peteri-Brunback B, Meijer L, Levi F, Teboul M, Delaunay F, Functional genomics identify Birc5/survivin as a candidate gene involved in the chronotoxicity of cyclin-dependent kinase inhibitors, Cell Cycle, 13 (2014) 984–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [225].Schafer KA, The cell cycle: a review, Vet Pathol, 35 (1998) 461–478. [DOI] [PubMed] [Google Scholar]
- [226].Pardee AB, A restriction point for control of normal animal cell proliferation, Proc Natl Acad Sci U S A, 71 (1974) 1286–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [227].Levens D, Disentangling the MYC web, Proc Natl Acad Sci U S A, 99 (2002) 5757–5759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [228].Bhattacharya A, Schmitz U, Wolkenhauer O, Schonherr M, Raatz Y, Kunz M, Regulation of cell cycle checkpoint kinase WEE1 by miR-195 in malignant melanoma, Oncogene, 32 (2013) 3175–3183. [DOI] [PubMed] [Google Scholar]
- [229].Abbas T, Dutta A, p21 in cancer: intricate networks and multiple activities, Nat Rev Cancer, 9 (2009) 400–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [230].Massague J, G1 cell-cycle control and cancer, Nature, 432 (2004) 298–306. [DOI] [PubMed] [Google Scholar]
- [231].Nurse P, Cyclin dependent kinases and cell cycle control (nobel lecture), Chembiochem, 3 (2002) 596–603. [DOI] [PubMed] [Google Scholar]
- [232].Malumbres M, Barbacid M, Mammalian cyclin-dependent kinases, Trends Biochem Sci, 30 (2005) 630–641. [DOI] [PubMed] [Google Scholar]
- [233].Morgan DO, Principles of CDK regulation, Nature, 374 (1995) 131–134. [DOI] [PubMed] [Google Scholar]
- [234].Morgan MN, Dvuchbabny S, Martinez CA, Kerr B, Cistulli PA, Cook KM, The Cancer Clock Is (Not) Ticking: Links between Circadian Rhythms and Cancer, Clocks Sleep, 1 (2019) 435–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [235].Matsuo T, Yamaguchi S, Mitsui S, Emi A, Shimoda F, Okamura H, Control mechanism of the circadian clock for timing of cell division in vivo, Science, 302 (2003) 255–259. [DOI] [PubMed] [Google Scholar]
- [236].Fu L, Pelicano H, Liu J, Huang P, Lee C, The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo, Cell, 111 (2002) 41–50. [DOI] [PubMed] [Google Scholar]
- [237].Bouchard C, Thieke K, Maier A, Saffrich R, Hanley-Hyde J, Ansorge W, Reed S, Sicinski P, Bartek J, Eilers M, Direct induction of cyclin D2 by Myc contributes to cell cycle progression and sequestration of p27, EMBO J, 18 (1999) 5321–5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [238].Huber AL, Papp SJ, Chan AB, Henriksson E, Jordan SD, Kriebs A, Nguyen M, Wallace M, Li Z, Metallo CM, Lamia KA, CRY2 and FBXL3 Cooperatively Degrade c-MYC, Mol Cell, 64 (2016) 774–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [239].Miki T, Matsumoto T, Zhao Z, Lee CC, p53 regulates Period2 expression and the circadian clock, Nat Commun, 4 (2013) 2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [240].Savvidis C, Koutsilieris M, Circadian rhythm disruption in cancer biology, Mol Med, 18 (2012) 1249–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [241].Eriguchi M, Levi F, Hisa T, Yanagie H, Nonaka Y, Takeda Y, Chronotherapy for cancer, Biomed Pharmacother, 57 Suppl 1 (2003) 92s–95s. [DOI] [PubMed] [Google Scholar]
- [242].Levi F, [Chronopharmacology and chronotherapy of cancers], Pathol Biol (Paris), 44 (1996) 631–644. [PubMed] [Google Scholar]
- [243].Levi F, Cancer prevention: epidemiology and perspectives, Eur J Cancer, 35 (1999) 1912–1924. [DOI] [PubMed] [Google Scholar]
- [244].Levi F, Circadian chronotherapy for human cancers, Lancet Oncol, 2 (2001) 307–315. [DOI] [PubMed] [Google Scholar]
- [245].Levi F, From circadian rhythms to cancer chronotherapeutics, Chronobiol Int, 19 (2002) 1–19. [DOI] [PubMed] [Google Scholar]
- [246].Hill RJW, Innominato PF, Levi F, Ballesta A, Optimizing circadian drug infusion schedules towards personalized cancer chronotherapy, PLoS Comput Biol, 16 (2020) e1007218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [247].Kuo TT, Ladurner AG, Exploiting the Circadian Clock for Improved Cancer Therapy: Perspective From a Cell Biologist, Front Genet, 10 (2019) 1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [248].Levi F, Okyar A, Dulong S, Innominato PF, Clairambault J, Circadian timing in cancer treatments, Annu Rev Pharmacol Toxicol, 50 (2010) 377–421. [DOI] [PubMed] [Google Scholar]
- [249].Ballesta A, Innominato PF, Dallmann R, Rand DA, Levi FA, Systems Chronotherapeutics, Pharmacol Rev, 69 (2017) 161–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [250].Hrushesky WJ, Grutsch J, Wood P, Yang X, Oh EY, Ansell C, Kidder S, Ferrans C, Quiton DF, Reynolds J, Du-Quiton J, Levin R, Lis C, Braun D, Circadian clock manipulation for cancer prevention and control and the relief of cancer symptoms, Integr Cancer Ther, 8 (2009) 387–397. [DOI] [PubMed] [Google Scholar]
- [251].Li XM, Mohammad-Djafari A, Dumitru M, Dulong S, Filipski E, Siffroi-Fernandez S, Mteyrek A, Scaglione F, Guettier C, Delaunay F, Levi F, A circadian clock transcription model for the personalization of cancer chronotherapy, Cancer Res, 73 (2013) 7176–7188. [DOI] [PubMed] [Google Scholar]
- [252].Mormont MC, Levi F, Cancer chronotherapy: principles, applications, and perspectives, Cancer, 97 (2003) 155–169. [DOI] [PubMed] [Google Scholar]
- [253].Adler S, Lang S, Langenmayer I, Eibl-Eibesfeldt B, Rump W, Emmerich B, Hallek M, Chronotherapy with 5-fluorouracil and folinic acid in advanced colorectal carcinoma. Results of a chronopharmacologic phase I trial, Cancer, 73 (1994) 2905–2912. [DOI] [PubMed] [Google Scholar]
- [254].Giacchetti S, Bjarnason G, Garufi C, Genet D, lacobelli S, Tampellini M, Smaaland R, Focan C, Coudert B, Humblet Y, Canon JL, Adenis A, Lo Re G, Carvalho C, Schueller J, Anciaux N, Lentz MA, Baron B, Gorlia T, Levi F, R. European Organisation for, G. Treatment of Cancer Chronotherapy, Phase III trial comparing 4-day chronomodulated therapy versus 2-day conventional delivery of fluorouracil, leucovorin, and oxaliplatin as first-line chemotherapy of metastatic colorectal cancer: the European Organisation for Research and Treatment of Cancer Chronotherapy Group, J Clin Oncol, 24 (2006) 3562–3569. [DOI] [PubMed] [Google Scholar]
- [255].Giacchetti S, Dugue PA, Innominato PF, Bjarnason GA, Focan C, Garufi C, Tumolo S, Coudert B, Iacobelli S, Smaaland R, Tampellini M, Adam R, Moreau T, Levi F, A.I.C. Group, Sex moderates circadian chemotherapy effects on survival of patients with metastatic colorectal cancer: a meta-analysis, Ann Oncol, 23 (2012) 3110–3116. [DOI] [PubMed] [Google Scholar]


