Abstract
Objective:
To determine whether single-cell and plasma proteomic elements of the host’s immune response to surgery accurately identifies patients who develop a Surgical Site Complication (SSC) after major abdominal surgery.
Summary Background Data:
SSCs may occur in up to 25% of patients undergoing bowel resection, resulting in significant morbidity and economic burden. However, the accurate prediction of SSCs remains clinically challenging. Leveraging high-content proteomic technologies to comprehensively profile patients’ immune response to surgery is a promising approach to identify predictive biological factors of SSCs.
Methods:
Forty-one patients undergoing non-cancer bowel resection were prospectively enrolled. Blood samples collected before surgery and on post-operative day one (POD1) were analyzed using a combination of single-cell mass cytometry and plasma proteomics. The primary outcome was the occurrence of an SSC, including surgical site infection, anastomotic leak, or wound dehiscence within 30 days of surgery.
Results:
A multiomic model integrating the single-cell and plasma proteomic data collected on POD1 accurately differentiated patients with (n=11) and without (n=30) an SSC (AUC = 0.86). Model features included co-regulated pro-inflammatory (e.g. IL-6- and MyD88- signaling responses in myeloid cells) and immunosuppressive (e.g. JAK/STAT signaling responses in M-MDSCs and Tregs) events preceding an SSC. Importantly, analysis of the immunological data obtained before surgery also yielded a model accurately predicting SSCs (AUC = 0.82).
Conclusions:
The multiomic analysis of patients’ immune response after surgery and immune state before surgery revealed systemic immune signatures preceding the development of SSCs. Our results suggest that integrating immunological data in perioperative risk assessment paradigms is a plausible strategy to guide individualized clinical care.
MINI ABSTRACT:
The integrated analysis of single-cell immune responses and plasma proteins 24 hours after surgery reveals an immune signature of surgical site complications (SSCs) after abdominal surgery. Furthermore, a predictive model built on the immunological data obtained before surgery accurately identifies patients who would later develop an SSC.
INTRODUCTION
Over 300 million operations are performed annually worldwide, a number that is expected to increase.1 Surgical complications including infection, protracted pain, functional impairment and end-organ damage occur in 10–60% of surgeries, causing personal suffering, longer hospital stays, readmissions, and significant socioeconomic burden.2 After major abdominal operations, surgical site complications (SSCs) including superficial or deep wound infections, organ space infections, anastomotic leaks, fascial dehiscence, and incisional hernias are some of the most devastating, costly, and common surgical complications.3, 4
The accurate prediction of SSC risk for individual patients is critically important to guide high-quality surgical decision making, including optimizing preoperative interventions and timing of surgery. Existing risk prediction tools are based on clinical parameters and while they may accurately predict risk of mortality or discharge to nursing facility, they may not be sufficient to estimate an individual patient’s risk for SSCs.5–9 As such, integration of biological parameters echoing mechanisms that drive the pathogenesis of SSCs is a highly plausible approach to increase risk prediction accuracy and to facilitate the implementation of such risk prediction in individual patient’s care planning and delivery.8, 10
Surgery triggers a profound and highly-coordinated immune response engaging innate and adaptive immune cells11–14 and involving pro- and anti-inflammatory (pro-resolving) processes for pathogen defense,15 tissue remodeling, and the resolution of pain.16 Complications, such as SSCs, arise as pro-inflammatory and immunosuppressive responses tilt out of balance.
Prior attempts to detect biological markers predicting the risk for SSCs focused on secreted humoral factors (e.g. IL-6 and IL-10),17 surface marker expression on select immune cells (e.g. HLA-DR expression) or transcriptional analysis of pooled circulating leukocytes.18–20 However, detected associations are insufficient to accurately predict the risk of SSCs for individual patients. A major impediment has been the lack of high-content, functional assays that can characterize the complex, multicellular inflammatory response to surgery with single-cell resolution.21, 22 Strong signals were likely undetected as immune cell subsets and signaling pathways were phenotypically or functionally under-evaluated.
In this study, we employed an integrated approach combining the functional analysis of immune cell subsets using mass cytometry23 with the highly-multiplexed assessment of inflammatory plasma proteins to quantify the dynamic changes of over 2,388 single-cell and plasma proteomic events in patients before and after major abdominal surgery. The primary aim of the study was to identify elements of the immune response to surgery that differentiate patients with or without an SSC with sufficient predictive performance. We also examined whether patient-specific immune states before surgery could differentiate these patient groups.
METHODS
Study design
Patients undergoing non-urgent major abdominal colorectal surgery were enrolled in this prospective cohort study after approval by the Institutional Review Board of Stanford University and obtaining written informed consent (IRB 48298). Inclusion criteria were patients over 18 years of age who were willing and able to sign written consent. Exclusion criteria were a diagnosis of cancer within the previous year, surgery that did not include resection of bowel, or a history of inflammatory/autoimmune conditions not related to the indication for colorectal surgery (e.g. inflammatory bowel disease [IBD] was included). The primary outcome was whether a patient developed an SSC within 30 days of surgery, defined as superficial, deep, or organ space surgical site infection, anastomotic leak, or dehiscence of the surgical incision. SSC was diagnosed clinically by the patients’ treating providers, and three members of the research team reviewed medical records prospectively to identify documentation of these outcomes. Other clinical variables included age, sex, body mass index (BMI), comorbidities, perioperative medications, and surgical characteristics (study flow chart, Supp. Fig. 1).
Sample collection for single-cell and plasma proteomic analysis
Whole blood and plasma samples were collected on the day of surgery (DOS) before induction of anesthesia and on the first postoperative day (POD1). Whole blood samples were either left unstimulated (to quantify cell frequency and endogenous cellular activities) or stimulated with a series of receptor-specific ligands eliciting key intracellular signaling responses implicated in the host’s immune response to trauma/injury13, 24 [including, lipopolysaccharide (LPS), PMA/Ionomycin (PI), interleukin (IL)-1β, interferon (IFN)α, tumor necrosis factor (TNF)α, and a combination of IL-2,4,6]. From each sample, 2,116 single-cell proteomic features were extracted using a 47-parameter single-cell mass cytometry immunoassay (Supp Table 1), including the frequency of 23 major innate and adaptive immune cells (Supp Fig. 2) and their intracellular signaling activities (e.g. the phosphorylation state of 11 proteins).25, 26 In parallel, the plasma concentrations of 272 proteins were quantified using the Olink platform (Olink proteomics, Sweden).27
Statistical methods
A Stacked Generalization (SG) predictive modeling approach28 that accounts for the high-dimensionality of the dataset was employed to detect differences in single-cell and plasma proteomic features between patients with or without an SSC on POD1 (primary analysis) or on DOS before surgery (secondary analysis). Elastic net (EN) logistic regression predictive models28 were trained for individual data layers (including one plasma proteomic and eight single-cell proteomic layers) and cross-validated using a leave-one-out cross validation (LOOCV) procedure with SSC used as the binary predictor variable. Individual data layers were then integrated using an SG method adapted from Ghaemi et al.29 to yield a final predictive SG model. The final SG model was built as a weighted average of the individual data layer models using a second LOOCV procedure. SG model performance was evaluated using the Area Under the Receiver Operating Characteristics Curve (AUC) and significance was tested using an unpaired Mann-Whitney test on the model cross-validated values. A bootstrap procedure provided estimates of the fold-enrichment and ranking of individual model features based on the robustness of selection with the EN.
Post-hoc confounder analysis was performed using age, sex, BMI, comorbidities, perioperative medications, and surgical characteristics. For each confounder, a simple logistic regression including the confounder and the SG model cross-validated values was fitted. F-statistics were computed to test whether each confounding variable explains out the model predictions.
The derivation of single-cell and plasma proteomic features, model performance, sample size estimate, feature ranking, correlation network analyses, and data visualization are described in detail in the supplemental online materials.
Chord diagram
We used a chord diagram representation to visualize inter-omic correlations between the nine data layers using the R library edgebundleR v.0.1.4 and igraph v.1.2.6. The correlation matrix of the features was computed based on the 41 POD1 samples. Correlations between model features (fold > 2) belonging to different data layers and with a Spearman correlation coefficient superior to 0.5 are depicted on the chord diagram.
RESULTS
Study participant characteristics
Forty-one patients undergoing major, non-cancer abdominal surgery involving bowel resection were prospectively enrolled. Eleven patients developed an SSC within 30 days after surgery, including surgical site infection (superficial, deep, or organ space, including anastomotic leak) or wound dehiscence in the absence of infection. Patient characteristics are listed in Table 1. Patients who developed an SSC had significantly higher BMIs, operative duration, and estimated intraoperative blood loss.
Table 1.
Patient characteristics.
| No Surgical Site Complication (73%, n=30) |
Surgical Site Complication (27%, n=11) |
p-value | |
|---|---|---|---|
| Female, % (n) | 60% (18) | 27.3% (3) | |
| Age, years mean ± SD | 44.2 ± 17.1 | 48.7± 14.1 | |
| BMI, mean ± SD | 23.2 ± 4.6 | 27.2 ± 3.9 | 7e-3 |
| Surgical Indication | |||
| Inflammatory Bowel Disease | 76.7% (23) | 72.7% (8) | |
| Crohn’s disease | 53.3% (16) | 36.4% (4) | |
| Ulcerative Colitis | 23.3% (7) | 36.4% (4) | |
| Other non-cancer diagnoses | 23.3% (7) | 27.3% (3) | |
| Preoperative Biologic Therapy | |||
| Anti-TNFα | 20% (6) | 9.1% (1) | |
| IL-12/23 Inhibitor | 6.7% (2) | 18.2% (2) | |
| Jak Inhibitor | 3.3% (1) | 0 | |
| a4b7 integrin blocker | 3.3% (1) | 0 | |
| Perioperative Systemic Steroids | 83.3% (25) | 63.6% (7) | |
| Preoperative Steroids | 10% (3) | 0 | |
| Intraoperative Dexamethasone | 83.3% (25) | 63.6% (7) | |
| Postoperative Steroids | 10% (3) | 0 | |
| Surgical Approach | |||
| Minimally Invasive | 46.7% (14) | 9.1% (1) | |
| Open | 53.3% (16) | 90.9% (10) | |
| Operative Duration, minutes, mean ± SD | 150.5 ± 98.9 | 285.1 ± 139.1 | 2e-3 |
| Wound Classification | |||
| Clean-Contaminated | 73.3% (22) | 36.4% (4) | |
| Contaminated | 23.3% (7) | 54.5% (6) | |
| Infected | 3.3% (1) | 9.1% (1) | |
| Intraoperative Blood Loss, mL, mean ± SD | 39.2 ± 34.5 | 118 ± 68.1 | 3e-4 |
| Intraoperative Blood Transfusion | 0 | 9.1% (1) | |
| Postoperative Blood Transfusion | 3.3% (1) | 0 | |
| Length of Hospitalization, days, mean ± SD | 4.5 ± 2.1 | 7.6 ± 6.7 |
Integrated modeling of immune responses 24h after surgery accurately classifies patients with a surgical site complication (SSC)
Blood samples were collected on the DOS (prior to induction of anesthesia) and on POD1. Samples were analyzed for soluble plasma proteins (plasma proteomics)27 and immune cell distribution and signaling responses (whole blood single-cell proteomics) with single-cell mass cytometry (Figure. 1a,b). The analysis yielded nine data layers (Fig. 1c), including a plasma proteomic data layer and eight mass cytometry data layers comprising immune cell frequencies, endogenous intracellular signaling, and signaling responses to the six ex vivo stimulations LPS, TNFα, IL-2,4,6 combination, P/I, IFNα, or IL-1β.
Figure 1. Integrated modeling of post-operative day 1 (POD1) immune responses categorizes patients with and without a surgical site complication (SSC).
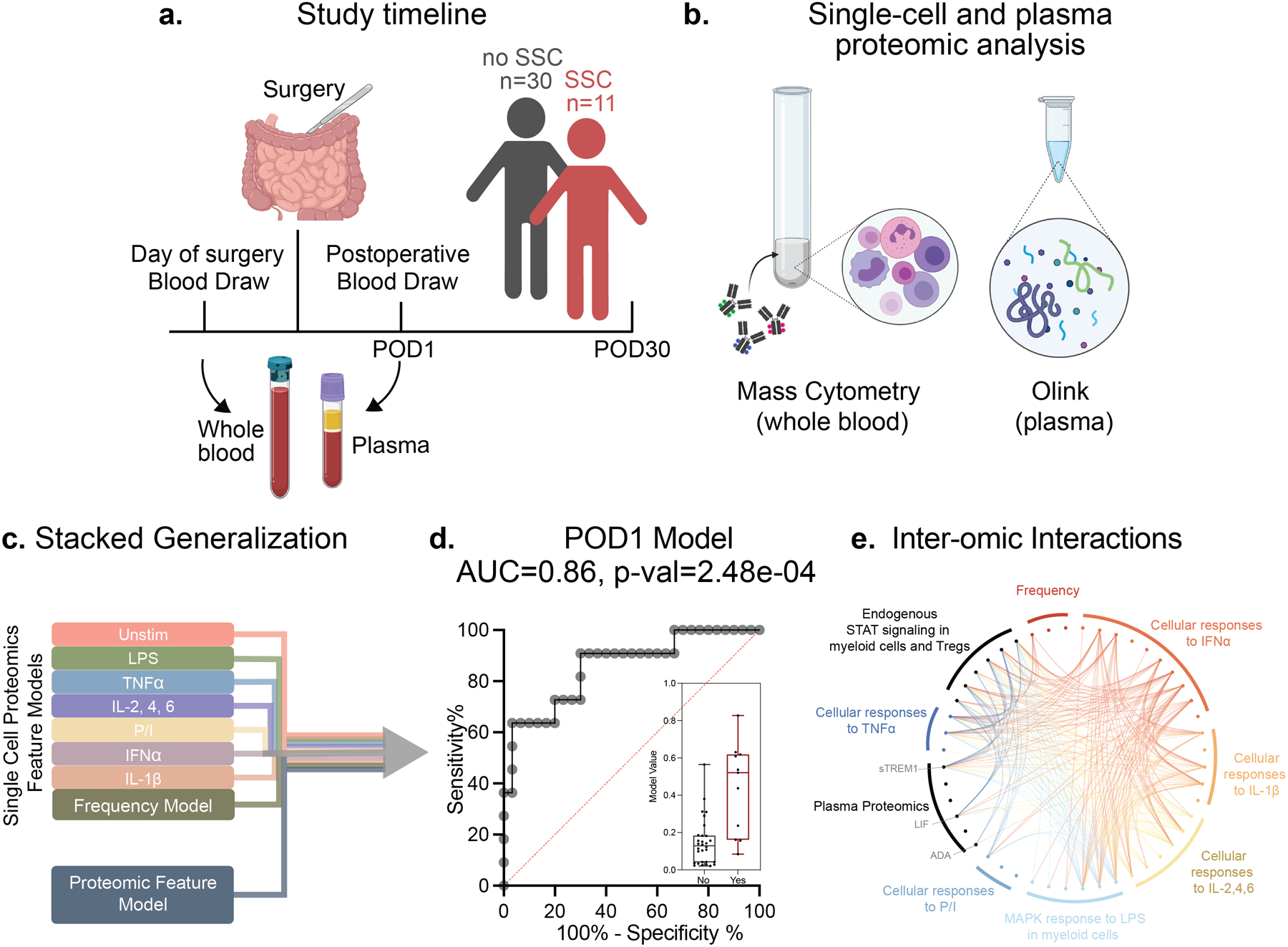
(a) Of the 41 patients enrolled in the study 11 patients developed SSCs within 30 days of surgery while 30 patients did not. (b) Whole blood samples collected on the day of surgery (DOS) prior to induction of anesthesia and on POD1 were stimulated with lipopolysaccharide (LPS), tumor necrosis factor (TNF)α, interleukin (IL)-2,4,6 cocktail, PMA/Ionomycin (P/I), interferon (IFN)α, IL-1β, or left unstimulated (Unstim) for analysis by single-cell mass cytometry. Plasma samples were analyzed using the Olink multiplex proteomic platform. (c) Elastic Net (EN) penalized linear regression models differentiating patients with and without an SSC were trained for individual data layers (plasma proteome, immune cell frequency, single-cell endogenous signaling, and signaling responses to each of six ex vivo stimulations) before integration of all nine data layers using a stacked generalization (SG) method. (d) The SG model output was evaluated using a leave-one-out cross validation (LOOCV) procedure. The receiver operating characteristic (ROC) analysis (area under the curve, AUC = 0.86, p = 2.5e-04), median values, and interquartile range of model outputs (boxplot, p = 2.48e-04, two-sided Mann-Whitney rank-sum test) are shown. (e) Chord diagram depicting correlation between model features of different data layers (Spearman R>0.5). Data layers are highlighted using the color scheme in (a). All model features (fold >2) are listed
We employed an SG predictive modeling approach28, 29 to determine whether differences in immune responses between patients with or without an SSC can be detected on POD1, which is before SSCs become clinically apparent. This approach integrates individual data layers into a single multivariate model. The SG model classified patients with high accuracy (AUC = 0.86, p = 2.5e-04, unpaired Mann-Whitney rank-sum test on the SG model cross-validated values, Fig. 1d). To account for confounding clinical and demographic variables, a post-hoc confounder analysis was performed on the SG model cross-validated values. A generalized linear model built on confounders with the SG predictions led to a significantly better fit than a model not including the SG values (p = 2e-07, Chi-square test for the deviance between fits). Additionally, the SG model remained predictive of SSCs when accounting for patient group differences in age, sex, surgical approach, surgery length, preoperative diagnosis, perioperative steroid administration or biologic therapy (e.g. anti-TNFα treatment, Supp. Table 2a).
Post-operative Day 1 (POD1) immune signatures preceding the development of SSCs
To better understand the biological implications of the high-dimensional SG model, individual SG model features were ranked according to their relative contribution to the multivariate SG model (fold enrichment, see supplemental online methods) using an iterative bootstrap procedure30 (Supp. Fig. 3). The most informative plasma and single-cell proteomic features (fold>2) were organized on a chord diagram (Fig. 1e, Supp. Table 3) and examined further.
POD1 plasma proteomic signature of SSCs
A t-distributed Stochastic Neighbor Embedding (t-SNE)31 layout was computed to visually represent the plasma proteomic correlation network on POD1 (Fig. 2). The highest-ranking protein of the SG model was TGFα, a member of the epithelial growth factor family that promotes mucosal cell proliferation and has been previously associated with anastomotic leak,32 which was elevated in patients with an SSC. Other proteomic features of the POD1 model that were elevated in patients with SSC included ADA, a protein involved in purine metabolism pathways implicated in lymphocyte proliferation and epithelial cell differentiation, soluble (s) TREM1, the ectodomain of the TREM1 receptor that is cleaved in response to inflammatory stimulation33 and LIF, an IL-6-family cytokine that promotes the release of proinflammatory mediators by fibroblasts.34 In contrast, three of the most informative SG model proteomic features were decreased in patients with SSCs, including EIF5A, a conserved eukaryotic translation initiation factor implicated in T-cell activation and autophagy;35 NT-ProBNP, a myocardial-derived prohormone that has recently been investigated as a predictor of long-term outcomes in sepsis;36 and KRT19, a cytoskeletal protein involved in epithelial cell differentiation, particularly of the colonic mucosa.37
Figure 2. POD1 plasma proteomic features differentiating patients with and without SSC.
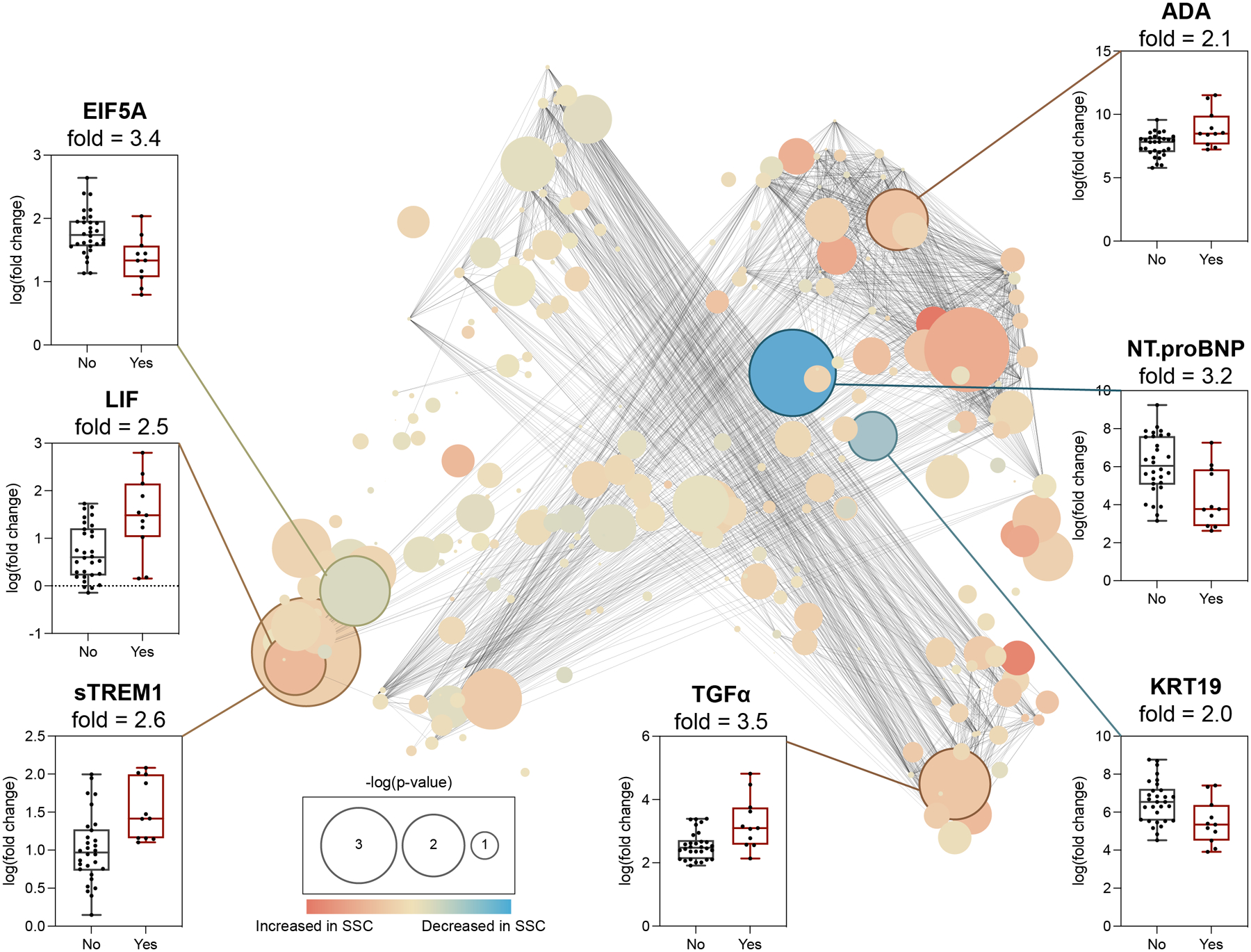
Center: Correlation network depicting plasma proteomic features of the SG model differentiating patients with and without SSC. Edges represent the correlation between features (Spearman coefficient > 0.9). Blue/orange nodes highlight positive/negative correlation with SSC. Node size reflects -log10 of p-value (Spearman). Periphery: Proteomic features with fold > 2.0 (bootstrap analysis, Supp. Fig. 3) are shown as box plots. Four features, ADA, TGFα, sTREM1, and LIF, were elevated while three features, KRT19, NT.proBNP, EIF5A, were decreased in patients with (red) compared to without (black) an SSC.
Plasma proteomic findings indicate that patients who develop SSCs have an exaggerated proinflammatory response and altered epithelial cell function within 24h of surgery, which may precede the clinical detection of an SSC. However, the analysis provided only indirect evidence from the measurement of soluble protein concentrations, that cannot be linked directly to the activity of specific cells or cellular processes.
POD1 single-cell proteomic signature of SSCs
Examination of the single-cell proteomic features (mass cytometry) of the predictive model complemented the analysis of the plasma proteome by affording a direct assessment of immune cell distribution and signaling behavior. Vopo, an unsupervised clustering algorithm was used for mapping and visualization of statistical information for the single-cell proteomic dataset (Fig. 3, center, Supp. Fig. 4). Innate and adaptive immune cell clusters were first identified using canonical surface markers. Differences in cell frequency or functional features between patients with and without SSCs were then quantified and projected onto individual cell clusters. Results obtained using a traditional gating strategy for identification of immune cell subsets corroborated major differences visualized on the Vopo cluster plots (Fig.3 box plots, Supp. Fig. 4).
Figure 3. POD1 single-cell proteomic features differentiating patients with and without an SSC.
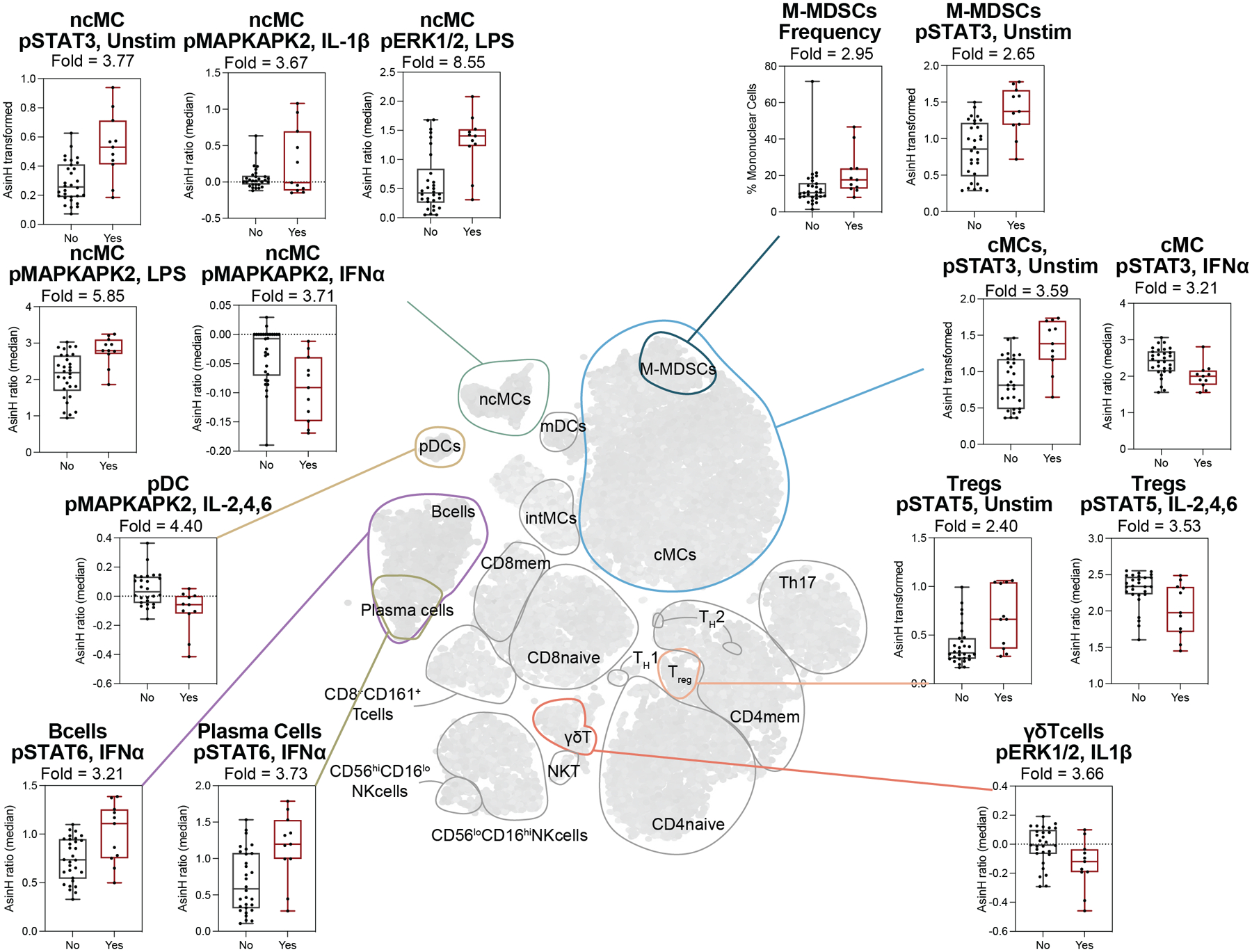
Center: Unsupervised clustering of mononuclear cell populations using the Vopo algorithm (see Supp. Fig. 4). Major innate and adaptive immune cell compartments are contoured and labeled. Periphery: Boxplots showing the most informative single-cell proteomic features (fold > 2.0, obtained from cell subsets identified using a manual gating strategy, Supp. Fig. 1) for patients with (red) and without (black) SSC. Median values and interquartile range are shown.
In the innate immune compartment, major differences between patients with and without SSCs were observed in the frequency and signaling activity of myeloid immune cell subsets, including neutrophils, classical monocytes (cMCs), monocytic-myeloid derived suppressor cells (M-MDSCs), non-classical (nc)MCs, and plasmacytoid dendritic cells (pDCs, Fig. 3, Supp. Fig. 4). These differences were particularly prominent in the endogenous STAT3 signaling responses, which were higher in patients with an SSC compared to controls (Supp. Fig. 4a–c). STAT3 is a major transcription factor activated by a wide range of inflammatory ligands, including the IL-6 cytokine family. Increased phosphorylated (p)STAT3 signal in myeloid cell subsets is paralleled by increased plasma levels of IL-6 and LIF in patients with SSCs suggesting that activation of canonical pro-inflammatory pathways is more pronounced in patients who will develop SCCs. Of note, increased pSTAT3 signal in M-MDSCs was accompanied by a pronounced expansion of this immunosuppressive cell population (Supp. Fig. 4d). In addition to basal signaling activities, several innate signaling responses to ex vivo stimulation differed between the two patient groups. In myeloid innate immune cells, the MyD88 signaling responses to LPS were exaggerated in patients with an SSC compared to controls (Supp. Fig. 4e–g), including increased pMAPKAPK2 and pERK signals in ncMCs, and decreased IκB signal in neutrophils, which mirrors the activation of the NFκB branch of the MyD88 pathway. In contrast, the MyD88 and JAK/STAT signaling responses to IL-2/4/6 or IFNα were dampened (Supp. Fig. 4h–j).
In adaptive immune compartments, key differences in cell frequencies and signaling responses between patients with and without an SSC were observed in CD4+T cell subsets, regulatory T cells (Tregs) and B cell subsets (Supp. Fig. 4k–p). Notably, the endogenous pSTAT5 signal in Tregs was increased in patients with an SSC on POD1, while the pSTAT5 response to external stimulation (IL-2/4/6) was decreased. Less pronounced differences between the two patient groups included an increase in pSTAT6 response to IFNα in B cells and plasma cells, a decreased pERK1/2 response to IL-1β in TCRγδ cells, and a decrease in naïve CD4+ T cell frequencies.
To complement the biological interpretation of the POD1 model, a chord diagram was built to highlight inter-omic correlations between model features from individual data layers (Fig. 1e, Spearman R >0.5). Correlations between the nine data layers included 520 moderate (R = 0.5 to 0.75), and 128 strong (R = 0.75 to 1) correlations. Most of the correlations were found within the single-cell proteomic dataset (94.8%), while 5.2% were inter-omic correlations between the plasma and the single-cell proteome. Notably, sTREM1 and LIF were the most interconnected plasma proteomic features and correlated with several single-cell signaling responses including MyD88 signaling responses to LPS in ncMC, MDSCs and granulocytes as well as basal JAK/STAT3 signaling activities in cMCs, ncMCs, MDSCs, granulocytes and Tregs, consistent with the coordinated increase observed for these plasma and single-cell proteomic events in patients with SSCs.
In summary, the complex interplay between pro-inflammatory and immunosuppressive cellular mechanisms after surgery differed between patients with and without an SSC. The combined analysis of the plasma proteome and single cell proteomic features highlight that sentinel pro-inflammatory responses are more pronounced in patients with an SSC on POD1 and precede the clinical manifestation of SSCs. Importantly, the single-cell data further revealed differences regarding the simultaneous engagement of innate (M-MDSC) and adaptive (Treg) immunosuppressive cell responses. These results suggest the balance between pro- and anti-inflammatory responses may be tilted towards an immunosuppressive state in patients at risk for developing an SSC.
Pre-operative immune states differentiate patients with and without an SSC
The analysis of immune responses to surgery on POD1 identified immune features separating patients who developed an SSC from those who did not, thereby highlighting biological differences in the response to traumatic injury that may drive the pathogenesis of SSCs. Identification of these features on POD1 that precede the onset of an SSC is clinically relevant as it allows for preemptive interventions potentially preventing SSCs. However, the ability to identify patients at risk for SSCs ahead of surgery may allow for more effective preemptive strategies. To determine whether the pre-surgical immune state of patients who later suffer from an SSC differ from those with normal recovery, the SG analysis algorithm was applied to the immunological data derived from blood samples collected before surgery (Fig. 4a). The resulting SG model accurately differentiated the two patient groups (AUC = 0.82, p = 1.56e-03, unpaired Mann-Whitney rank-sum test on the SG model cross-validated values, Fig. 4b).
Figure 4. Integrated modeling of preoperative immune responses categorizes patients with and without an SSC.
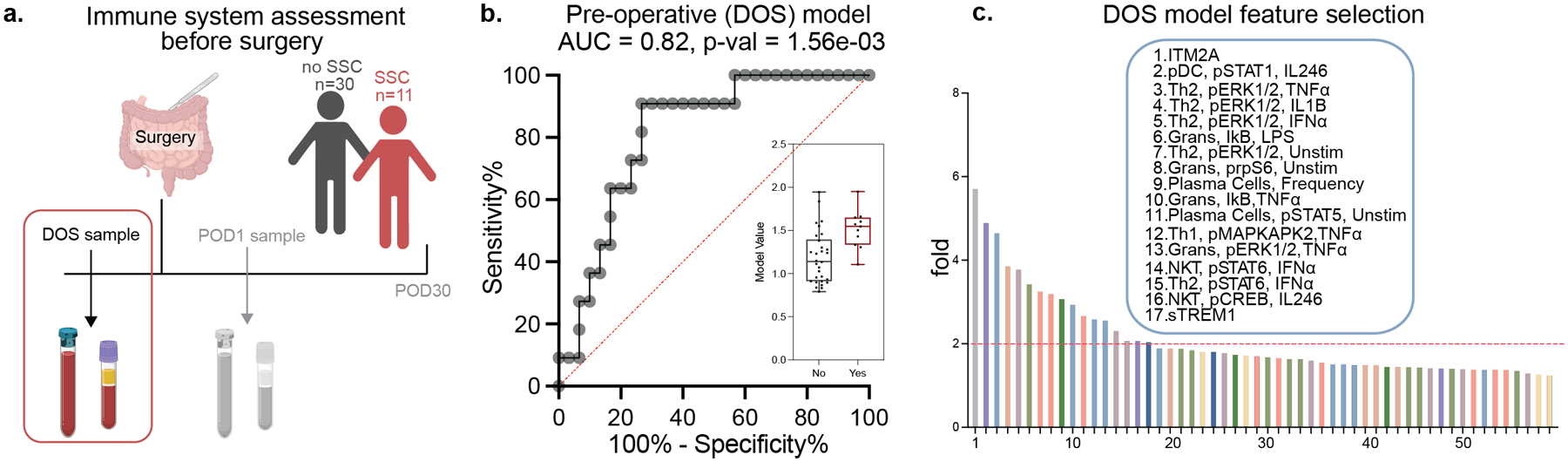
(a) Blood samples collected before surgery, on the DOS were processed for single-cell (mass cytometry) and plasma (Olink) analysis as in Fig. 1. (b) An SG analysis of the combined single-cell and plasma proteomic dataset identified a predictive model accurately differentiating patients with and without an SSC (AUC = 0.82, p = 1.56e-03, Mann Whitney rank-sum test). (c) DOS model features were ranked using a bootstrap analysis method, and the most informative features differentiating patients with and without an SSC (bootstrap fold > 2.0) were examined in detail (Supp. Fig. 5, Supp. Fig. 6).
Application of the iterative bootstrap procedures selected 17 features that contributed most to the multivariate DOS model (Fig 4c), including two plasma proteins (ITM2A and TREM1, Supp. Fig. 5) that were increased in patients who later developed an SSC, and 15 single-cell proteomic features. For the innate immune compartment, pSTAT1 in response to stimulation with IL-2/4/6 was decreased in pDCs in patients who developed an SSC (Supp. Fig. 6). In contrast, higher endogenous prpS6 activity in neutrophils along with a decrease in inhibitory factor IκB in response to LPS suggested increased proinflammatory activities. For the adaptive immune compartment, the frequency of plasma cells was mildly increased in patients who developed SCCs. Interestingly, while endogenous MAPK/ERK signaling activity in T helper (Th)2 cells was elevated in patients who developed SCCs, their response to ex vivo stimulation with IL-1β, TNFα, or IFNα was decreased (Supp. Fig 6). A confounder analysis, including clinical and demographic variables that differed between the two patient groups, showed that the predictive power of the DOS SG model increased significantly when accounting for differences in age, BMI, and preoperative diagnostics, perioperative steroid administration, wound classification, anti-TNF⍺ or other biologic therapy, or surgical approach (Supp. Table 2b). Comparing a generalized linear model with or without the SG predictions led to a significantly better fit of the model with the SG values (p = 8e-05, Chi-squared test). Furthermore, we evaluated the relationship between model prediction and the timing of SSC development in patients with SSC but found no correlation for either predictive model (POD1 model: R = - 0.37, p-value = 0.23; DOS model: R = - 0.17, p-value = - 0.62). Taken together, the results suggest that the multi-modal interrogation of patients’ proteome and immunome before surgery has a strong predictive potential for identifying patients at risk for developing an SSC.
DISCUSSION
We performed an integrated plasma and single-cell proteomic analysis of patients’ immune response to major abdominal surgery to identify immunological events associated with post-operative SSCs. Multiple biological dimensions (including plasma proteins, immune cell composition, and intracellular signaling responses) were assessed to capture relevant aspects of the complex biology that may separate patients without complications from patients who develop an SSC. Derived models identified patients with SSCs with high performance as indicated by an AUC of 0.82 for the preoperative DOS model and an AUC of 0.86 for the POD1 model. Model performance was favorable compared to that of other prognostic models including the ACS-NSQIP Risk Calculator score.7
The analysis of plasma-based and single-cell immune events before and shortly after surgery provided a systems level view of trauma-related immune mechanisms associated with the development of an SCC. Two major themes emerged characterizing the early immune response to surgery in patients who later developed an SCC: 1) an exacerbation of pro-inflammatory IL-6R and TLR-related signaling responses and 2) an increase in immunosuppressive cell responses, including M-MDSC and Treg responses.
Key elements of the POD1 SG model integrate well with prior knowledge regarding immune mechanisms predisposing to SSCs. Previous reports indicate that elevated IL-6 plasma concentrations early after surgery correlate with an increased risk of post-operative complications, including infections.20 Consistent with prior findings, the increased STAT3 signaling activity in cMCs (canonically activated by IL-6) was one of the most informative single-cell features associated with SSCs. Similarly, exacerbation of MyD88 signaling responses to LPS in innate myeloid cells in patients who later developed an SSC echoes prior results indicating that unchecked, systemic activation of pro-inflammatory innate immune cells in response to surgical site injury may contribute to the development of an SSC. As such, an excessive local immune response to inflammation can amplify the release of DAMPs and PAMPs from the surgery site in a cycle of intensifying MyD88-related TLR signaling, induction of barrier breakdown, and additional tissue damage.15 In this context it is also noteworthy that overstimulation of TLR signaling can produce a state of endotoxin tolerance, which may increase a patient’s susceptibility to infection.38
The single-cell resolution afforded by mass cytometry provided new insight into cell-type specific responses that may contribute to the pathogenesis of SSCs. Increased STAT3 signaling in M-MDSCs and increased M-MDSC frequencies at 24h after surgery were among the most informative features of the POD1 model. The results dovetail with prior studies of patients undergoing orthopedic surgery that show a strong correlation between STAT3 signaling in MDSCs and delayed surgical recovery.13, 39 MDSCs are a heterogenous subset of immature myeloid cells with immunosuppressive function that are mobilized in the context of acute and chronic inflammatory diseases.40 In previous investigations of the immune response to trauma and sepsis, MDSCs have been identified as important players in a counter-inflammatory program that represses the adaptive immune system, particularly antigen-specific CD8+ and CD4+ T cell responses.41 In patients who later developed an SSC, elevated STAT3 signaling, which is required for MDSC’s proliferation and immunosuppressive function,40 could synergistically promote MDSC expansion and, therefore, aggravate a state of immunosuppression.
We also observed the upregulation of endogenous STAT5 signaling in immunosuppressive Tregs in patients who developed an SSC. In contrast, the pSTAT5 response to ex vivo stimulation with IL-2/4/6 was lower in patients who developed an SSC, which may indicate that higher endogenous pSTAT5 signaling tone may prevent further ex vivo activation. IL-2R-dependent activation of STAT5 in Tregs is essential for mature Tregs to maintain FoxP3 expression levels and to exert their immunosuppressive function.42 Reportedly, FoxP3 expression and Treg-lineage-specific transcription is further promoted by the IL-6 family cytokine LIF.43 The regulatory functions of LIF in the induction of Treg development and maturation are indicative of the ambiguous role of IL-6-family cytokines in the context of inflammation and trauma. Overall, excessive endogenous Treg signaling could synergize with the observed exaggerated MDSC response and initiate a sustained immunosuppressive state that dampens the response to invading pathogens in patients who develop an SSC.
While the POD1 model provided important information as to surgery-induced mechanisms implicated in the pathogenesis of SSC, the DOS SG model pointed at single-cell features and plasma proteomic factors differentiating the two patient groups before surgery. The most informative features of the DOS SG model were the proteomic features sTREM1 and ITM2A. Our result showing that sTREM1 is elevated on DOS and on POD1 in patients who later develop SSC is reminiscent of previous studies showing increased sTREM1 plasma concentration in patients with bacterial infection44 and sepsis.45 From a mechanistic standpoint, sTREM1 is the metalloprotease-cleaved product of membrane-bound TREM1, an amplifier of pattern recognition receptors on myeloid cells46. sTREM1 can function as a decoy receptor that antagonizes TREM146. However, microbial products such as LPS can both increase the membrane expression of TREM1 and stimulate the release of sTREM133, thereby increasing sTREM1 plasma concentration. Whether elevated sTREM1 in patients with SSC parallels TREM1 expression on myeloid cells, or results in the functional inhibition of TREM1 is an important question that warrants further investigation.
ITM2A, another proteomic feature of the DOS model, is upregulated by PKA-CREB signaling and leads to an accumulation of autophagosomes and inhibition of autolysosomal formation.47 Effective autophagy is essential for many physiological functions including tissue differentiation, cell cycle regulation, and immune cell maturation, particularly Th cell development. Other informative features of the DOS model included differences across multiple innate and adaptive cell subsets, such as neutrophils, pDCs, and Th2 cells. Notably, in patients who developed SSC, the signaling responses to multiple stimulations (including IL-1β, TNFα, and IFNα) were dampened in CRTH2+ Th2-like CD4+ T cells, which play important roles in defensive immunity against extracellular pathogens and tissue repair. Our results suggests that patient-specific immune states before surgery may increase the risk for developing an SSC. As such, the preoperative assessment of specific immune markers may assist in risk stratifying patients along with applying interventions to attenuate the risk for developing an SSC.
This study has certain limitations. The cohort included patients from one center who underwent major abdominal surgery. The two patient groups differed with respect to known risk factors for SSCs, such as BMI, surgery length, and blood loss (all of which were increased in patients with SSCs)3, 4. In addition, our patient population included patients with IBD, some undergoing treatment with immune-modulatory therapies that may have influenced their immune response to surgery. However, post-hoc analyses showed that our predictive models of SSC remained significant when accounting for these potentially confounding variables suggesting that differences in immune responses between the two patient groups are not predominantly due to these clinical or demographic risk factors. Nonetheless, future studies in a larger population undergoing a broader variety of preoperative diagnoses and surgical interventions will be required to establish the generalizability of our results. We did not assess local immune responses nor did we measure differences in patients gut microbiota, which will be important to determine the relationship between local and systemic immune events that are associated with SSCs. This will be particularly relevant for patients with IBD on immunotherapy, who are prone to gut microbiome alteration. While mass cytometry allows for the simultaneous detection of up to 50 parameters on a single-cell level, the technology requires the a priori selection of cell surface and intracellular markers. Similarly, the proteomic platform included a selected panel enriched for inflammatory mediators. Future studies, including non-targeted transcriptomic, proteomic, or metabolic approaches will help determine whether a deeper profile of patients’ inflammatory, metabolic, and hormonal responses to traumatic injury will improve the power of the prediction models.
In summary, estimating a patient’s risk for surgical complications remains a clinical challenge. Our combined single-cell and plasma proteomic analysis of the systemic immune response to surgery provides clinically relevant biological insights into trauma-related immune alterations that precede the development of an SSC. As importantly, the model derived from samples collected before surgery suggests that sufficiently powerful predictive algorithms can be developed to risk stratify individual patients and to assign them to patient-specific care pathways.
Supplementary Material
Supplemental Materials and Methods
Supp. Figure 1: Study flow chart according to CONSORT guidelines
Supp. Figure 2: Immune cell gating strategy
Supp. Figure 3: Bootstrap procedure for model feature ranking and selection.
Supp. Figure 4: Single-cell proteomic features differentiating patients with and without an SSC on POD1
Supp. Figure 5: Plasma proteomic features differentiating patients with and without an SSC on DOS
Supp. Figure 6: Single-cell proteomic features differentiating patients with and without an SSC on DOS
Supp. Table 1: Mass Cytometry antibody panel
Supp. Table 2: Confounder analysis
Supp. Table 3: POD1 model features
Supp. Table 4: DOS model features
Conflicts of Interest and Source of Funding:
This work was supported by the Stanford Department of Anesthesiology, Pain and Perioperative Medicine, the Stanford Department of Surgery, the national institute of health (NIH) R35GM137936 (BG), R35GM138353 (NA), NS114926 (MSA), AG065744 (MSA), the Fluegel Research Fund (KR), and the Center for Human Systems Immunology (BG). A provisional patent application that covers aspects of the subject matter of the paper has been filed (S31-07151.PRO, title: systems and methods to generate a surgical risk score and uses thereof, co-inventors: BG, JH, KR, NA, MSA).
REFERENCES
- 1.Meara JG, Leather AJ, Hagander L, et al. Global Surgery 2030: evidence and solutions for achieving health, welfare, and economic development. Lancet 2015; 386(9993):569–624. [DOI] [PubMed] [Google Scholar]
- 2.Healy MA, Mullard AJ, Campbell DA Jr., et al. Hospital and Payer Costs Associated With Surgical Complications. JAMA Surg 2016; 151(9):823–30. [DOI] [PubMed] [Google Scholar]
- 3.Nikolian VC, Kamdar NS, Regenbogen SE, et al. Anastomotic leak after colorectal resection: A population-based study of risk factors and hospital variation. Surgery 2017; 161(6):1619–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gantz O, Zagadailov P, Merchant AM. The Cost of Surgical Site Infections after Colorectal Surgery in the United States from 2001 to 2012: A Longitudinal Analysis. Am Surg 2019; 85(2):142–149. [PubMed] [Google Scholar]
- 5.Eamer G, Al-Amoodi MJH, Holroyd-Leduc J, et al. Review of risk assessment tools to predict morbidity and mortality in elderly surgical patients. Am J Surg 2018; 216(3):585–594. [DOI] [PubMed] [Google Scholar]
- 6.Cohen ME, Liu Y, Ko CY, et al. An Examination of American College of Surgeons NSQIP Surgical Risk Calculator Accuracy. J Am Coll Surg 2017; 224(5):787–795 e1. [DOI] [PubMed] [Google Scholar]
- 7.Bilimoria KY, Liu Y, Paruch JL, et al. Development and evaluation of the universal ACS NSQIP surgical risk calculator: a decision aid and informed consent tool for patients and surgeons. J Am Coll Surg 2013; 217(5):833–42 e1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bihorac A, Ozrazgat-Baslanti T, Ebadi A, et al. MySurgeryRisk: Development and Validation of a Machine-learning Risk Algorithm for Major Complications and Death After Surgery. Ann Surg 2019; 269(4):652–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vos EL, Russo AE, Hohmann A, et al. Performance of the American College of Surgeons NSQIP Surgical Risk Calculator for Total Gastrectomy. J Am Coll Surg 2020; 231(6):650–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baek B, Lee H. Prediction of survival and recurrence in patients with pancreatic cancer by integrating multi-omics data. Sci Rep 2020; 10(1):18951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Angele MK, Faist E. Clinical review: immunodepression in the surgical patient and increased susceptibility to infection. Crit Care 2002; 6(4):298–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stoecklein VM, Osuka A, Lederer JA. Trauma equals danger--damage control by the immune system. J Leukoc Biol 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaudillière B, Fragiadakis GK, Bruggner RV, et al. Clinical recovery from surgery correlates with single-cell immune signatures. Sci Transl Med 2014; 6(255):255ra131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ganio EA, Stanley N, Lindberg-Larsen V, et al. Preferential inhibition of adaptive immune system dynamics by glucocorticoids in patients after acute surgical trauma. Nat Commun 2020; 11(1):3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huber-Lang M, Lambris JD, Ward PA. Innate immune responses to trauma. Nat Immunol 2018; 19(4):327–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sugimoto MA, Vago JP, Perretti M, et al. Mediators of the Resolution of the Inflammatory Response. Trends Immunol 2019; 40(3):212–227. [DOI] [PubMed] [Google Scholar]
- 17.Rettig TC, Verwijmeren L, Dijkstra IM, et al. Postoperative Interleukin-6 Level and Early Detection of Complications After Elective Major Abdominal Surgery. Ann Surg 2016; 263(6):1207–12. [DOI] [PubMed] [Google Scholar]
- 18.Veenhof AA, Vlug MS, van der Pas MH, et al. Surgical stress response and postoperative immune function after laparoscopy or open surgery with fast track or standard perioperative care: a randomized trial. Ann Surg 2012; 255(2):216–21. [DOI] [PubMed] [Google Scholar]
- 19.Brown JR, Jacobs JP, Alam SS, et al. Utility of Biomarkers to Improve Prediction of Readmission or Mortality After Cardiac Surgery. Ann Thorac Surg 2018; 106(5):1294–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fong TG, Chan NY, Dillon ST, et al. Identification of Plasma Proteome Signatures Associated With Surgery Using SOMAscan. Ann Surg 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tarnok A Revisiting the crystal ball--high content single cells analysis as predictor of recovery. Cytometry A 2015; 87(2):97–8. [DOI] [PubMed] [Google Scholar]
- 22.Seshadri A, Brat GA, Yorkgitis BK, et al. Phenotyping the Immune Response to Trauma: A Multiparametric Systems Immunology Approach. Crit Care Med 2017; 45(9):1523–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bendall SC, Simonds EF, Qiu P, et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science 2011; 332(6030):687–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fragiadakis GK, Gaudillière B, Ganio EA, et al. Patient-specific Immune States before Surgery Are Strong Correlates of Surgical Recovery. Anesthesiology 2015; 123(6):1241–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Culos A, Tsai AS, Stanley N, et al. Integration of mechanistic immunological knowledge into a machine learning pipeline improves predictions. Nat Mach Intell 2020; 2(10):619–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stanley N, Stelzer IA, Tsai AS, et al. VoPo leverages cellular heterogeneity for predictive modeling of single-cell data. Nat Commun 2020; 11(1):3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Assarsson E, Lundberg M, Holmquist G, et al. Homogenous 96-plex PEA immunoassay exhibiting high sensitivity, specificity, and excellent scalability. PLoS One 2014; 9(4):e95192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hastie T, Tibshirani R, Friedman JH. The elements of statistical learning : data mining, inference, and prediction. 2nd ed. New York, NY: Springer, 2009. [Google Scholar]
- 29.Ghaemi MS, DiGiulio DB, Contrepois K, et al. Multiomics modeling of the immunome, transcriptome, microbiome, proteome and metabolome adaptations during human pregnancy. Bioinformatics 2019; 35(1):95–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chatterjee A, Lahiri SN. Bootstrapping Lasso Estimators. Journal of the American Statistical Association 2011; 106(494):608–625. [Google Scholar]
- 31.van der Maaten L, Hinton G. Visualizing Data using t-SNE. Journal of Machine Learning Research 2008; 9:2579–2605. [Google Scholar]
- 32.Matthiessen P, Strand I, Jansson K, et al. Is early detection of anastomotic leakage possible by intraperitoneal microdialysis and intraperitoneal cytokines after anterior resection of the rectum for cancer? Dis Colon Rectum 2007; 50(11):1918–27. [DOI] [PubMed] [Google Scholar]
- 33.Gomez-Pina V, Soares-Schanoski A, Rodriguez-Rojas A, et al. Metalloproteinases shed TREM-1 ectodomain from lipopolysaccharide-stimulated human monocytes. J Immunol 2007; 179(6):4065–73. [DOI] [PubMed] [Google Scholar]
- 34.Nguyen HN, Noss EH, Mizoguchi F, et al. Autocrine Loop Involving IL-6 Family Member LIF, LIF Receptor, and STAT4 Drives Sustained Fibroblast Production of Inflammatory Mediators. Immunity 2017; 46(2):220–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bevec D, Klier H, Holter W, et al. Induced gene expression of the hypusine-containing protein eukaryotic initiation factor 5A in activated human T lymphocytes. Proc Natl Acad Sci U S A 1994; 91(23):10829–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Custodero C, Wu Q, Ghita GL, et al. Prognostic value of NT-proBNP levels in the acute phase of sepsis on lower long-term physical function and muscle strength in sepsis survivors. Crit Care 2019; 23(1):230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Majumdar D, Tiernan JP, Lobo AJ, et al. Keratins in colorectal epithelial function and disease. Int J Exp Pathol 2012; 93(5):305–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bagchi A, Herrup EA, Warren HS, et al. MyD88-dependent and MyD88-independent pathways in synergy, priming, and tolerance between TLR agonists. J Immunol 2007; 178(2):1164–71. [DOI] [PubMed] [Google Scholar]
- 39.Fallahzadeh R, Verdonk F, Ganio E, et al. Objective Activity Parameters Track Patient-Specific Physical Recovery Trajectories After Surgery and Link With Individual Preoperative Immune States. Annals of Surgery 9000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gabrilovich DI, Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol 2009; 9(3):162–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mathias B, Delmas AL, Ozrazgat-Baslanti T, et al. Human Myeloid-derived Suppressor Cells are Associated With Chronic Immune Suppression After Severe Sepsis/Septic Shock. Ann Surg 2017; 265(4):827–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chinen T, Kannan AK, Levine AG, et al. An essential role for the IL-2 receptor in Treg cell function. Nat Immunol 2016; 17(11):1322–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Metcalfe SM. LIF in the regulation of T-cell fate and as a potential therapeutic. Genes Immun 2011; 12(3):157–68. [DOI] [PubMed] [Google Scholar]
- 44.Gibot S, Cravoisy A, Levy B, et al. Soluble triggering receptor expressed on myeloid cells and the diagnosis of pneumonia. N Engl J Med 2004; 350(5):451–8. [DOI] [PubMed] [Google Scholar]
- 45.Gibot S, Kolopp-Sarda MN, Bene MC, et al. Plasma level of a triggering receptor expressed on myeloid cells-1: its diagnostic accuracy in patients with suspected sepsis. Ann Intern Med 2004; 141(1):9–15. [DOI] [PubMed] [Google Scholar]
- 46.Klesney-Tait J, Turnbull IR, Colonna M. The TREM receptor family and signal integration. Nat Immunol 2006; 7(12):1266–73. [DOI] [PubMed] [Google Scholar]
- 47.Namkoong S, Lee KI, Lee JI, et al. The integral membrane protein ITM2A, a transcriptional target of PKA-CREB, regulates autophagic flux via interaction with the vacuolar ATPase. Autophagy 2015; 11(5):756–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stelzer IA, Ghaemi MS, Han X, et al. Integrated trajectories of the maternal metabolome, proteome, and immunome predict labor onset. Sci Transl Med 2021; 13(592). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zunder ER, Finck R, Behbehani GK, et al. Palladium-based mass tag cell barcoding with a doublet-filtering scheme and single-cell deconvolution algorithm. Nat Protoc 2015; 10(2):316–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hanley JA, McNeil BJ. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology 1982; 143(1):29–36. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Materials and Methods
Supp. Figure 1: Study flow chart according to CONSORT guidelines
Supp. Figure 2: Immune cell gating strategy
Supp. Figure 3: Bootstrap procedure for model feature ranking and selection.
Supp. Figure 4: Single-cell proteomic features differentiating patients with and without an SSC on POD1
Supp. Figure 5: Plasma proteomic features differentiating patients with and without an SSC on DOS
Supp. Figure 6: Single-cell proteomic features differentiating patients with and without an SSC on DOS
Supp. Table 1: Mass Cytometry antibody panel
Supp. Table 2: Confounder analysis
Supp. Table 3: POD1 model features
Supp. Table 4: DOS model features


