Abstract
Bmf contributes to the onset of anoikis by translocating from cytoskeleton to mitochondria when cells lose attachment to the extracellular matrix. However, the structural details of Bmf cytoskeleton tethering and the control of Bmf release upon loss of anchorage remained unknown. Here we showed that cell detachment induced rapid and sustained activation of p38 MAPK in mammary epithelial cell lines. Inhibition of p38 signaling or Bmf knockdown rescued anoikis. Activated p38 MAPK could directly phosphorylate Bmf at multiple sites including a non-proline-directed site threonine 72 (T72). Crystallographic studies revealed that Bmf T72 directly participated in DLC2 binding and its phosphorylation would block Bmf/DLC2 interaction through steric hindrance. Finally, we showed that phosphomimetic mutation of T72 enhanced Bmf apoptotic activity in vitro and in a knock-in mouse model. This work unraveled a novel regulatory mechanism of Bmf activity during anoikis and provided structural basis for Bmf cytoskeleton tethering and dissociation.
Subject terms: Cell biology, Molecular biology, X-ray crystallography
Introduction
Anoikis is a special type of programed cell death triggered when cells lose attachment to extracellular matrix (ECM) [1]. It regulates duct morphogenesis and lumen formation in mammary and saliva glands, and also participates in the maintenance of tissue homeostasis [1–3]. Deregulation of anoikis may cause anchorage-independent growth of cells and predispose malignant cells to metastasis during cancer development [4].
Two closely related Bcl-2-homology domain 3 (BH3)-only family proteins, Bim and Bmf have been implicated in the anoikis of mammary and intestinal epithelial cells [5–10]. Bim and Bmf expressions are induced upon loss of attachment and are required for anoikis in mammary epithelial cells and for lumen formation during mammary development [7, 10, 11]. Besides expression regulation, subcellular sequestration of Bim and Bmf represents another important regulatory mechanism for their apoptotic activities. Bim is sequestered to the dynein motor complex through direct binding to DLC1 [12]. The tethering of Bim to dynein motor complex can be blocked by Bim phosphorylation at threonine 56 (T56, in BimL) within the DLC1 binding sequence (DKSTQTP) by c-Jun N-terminal kinases (JNKs) [13]. By contrast, Bmf is sequestered to the myosin V motor complex through interacting with dynein light chain 2 (DLC2) protein [6]. Upon cell detachment from ECM, Bmf is unleashed from cytoskeleton and contributes to the onset of anoikis through its relocalization to mitochondria [6]. In melanoma cells, MAPK inhibition-induced Bmf cytosolic translocation correlates with MEK inhibitor sensitivity [14]. Based on the similarity between the DLC1/2 binding sequences of Bim and Bmf, Lei et al. proposed that Bmf might also be phosphorylated by JNK at serine 74 (S74) proximal to the DLC2 binding sequence (DKATQTLSP) and that S74 phosphorylation by JNK would release Bmf from the myosin V motor complex and promote apoptosis [13]. However, phosphomimetic mutation (S74D) of Bmf did not apparently alter its DLC2 binding capacity in vitro [15], nor its apoptotic activity in vivo [16]. Therefore, the signaling pathway that regulates Bmf release from myosin V complex in response to stress signals, in particular, loss of attachment remain enigmatic.
The mitogen-activated protein kinase (MAPK) superfamily consists of three major kinase families including extracellular signal-regulated kinases (ERK), JNK and p38 mitogen-activated protein kinases (p38 MAPK), which have been shown to be activated by various stress stimulus and function as mediators of cellular stress responses [17]. However, the potential roles and functional mechanisms of MAPKs in the regulation of anoikis have not been fully elucidated. In this study, we examined the activation kinetics of three MAPK pathways during the process of anoikis and investigated their roles in regulating the apoptotic activity of Bmf during anoikis.
Results
p38 signaling and Bmf contribute to anoikis in MCF-10A and MCF-7 cells
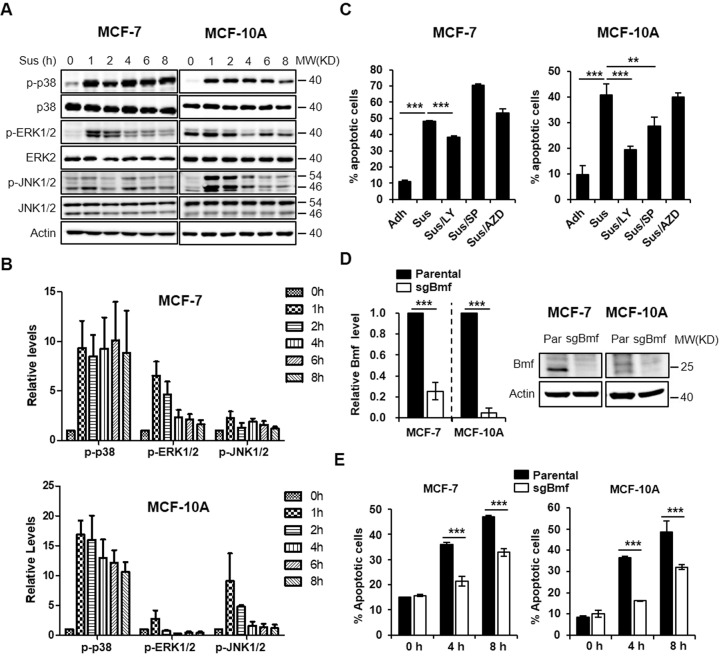
Activation of MAPK signaling pathways including ERK, p38 and JNK have been implicated in various stress-induced apoptosis [17, 18]. We examined the activation kinetics of ERK1/2, p38 and JNK1/2 in response to loss of attachment in two human mammary epithelial cell lines MCF-7 and MCF-10A (Fig. 1A). Matrix detachment induced rapid and sustained activation of p38 in MCF-7 and to a lesser extent, in MCF-10A cells (Fig. 1B). By contrast, ERK1/2 was transiently activated only in MCF-7 but not in MCF-10A cells, whereas JNK1/2 exhibited a reversed pattern (Fig. 1B). Therefore, p38 appeared to be the consistently and durably activated MAPK in response to loss of anchorage in both cell lines.
Fig. 1. p38 signaling and Bmf contribute to anoikis in mammary epithelial cells.
A MCF-7 or MCF-10A cells were trypsinized, resuspended and plated in poly-HEMA coated dishes and cultured for various time before lysed for western blot analysis. Attached cells were used as the “0 h” treatment groups. B Quantitation of the p-p38, p-ERK1/2 and p-JNK1/2 blots from (A), N = 3, error bars represent standard deviations. C MCF-7 or MCF-10A cells were replated in poly-HEMA coated dishes for 8 h in the presence or absence of indicated inhibitors (LY 2228820, 1 μM; AZD6244, 10 μM; SP 600125, 10 μM). Cells were then harvested for apoptosis analysis using annexin V-PI staining and flow cytometry. D qRT-PCR and western blot analysis of Bmf in MCF-7 and MCF-10A cells transduced with lentiviruses expressing Bmf sgRNA (sgBmf). E Parental or sgBmf lentivirus-transduced MCF-7/MCF-10A cells (polyclonal) were replated in poly-HEMA coated dish for 0, 4 and 8 h and assay for apoptosis using annexin V-PI staining and flow cytometry. All quantitative results were shown as mean ± STD from three independent experiments. Significance was determined by ANOVA one-way test, *p < 0.05; **p < 0.01, ***p < 0.001.
As previously reported [7, 19], loss of adhesion led to apoptosis in MCF-7 and MCF-10A cells (Fig. 1C, Supplementary Fig. S1A). Of note, inhibition of the p38 pathway using a p38 kinase inhibitor, LY 2228820 partially rescued cell death in both MCF-7 and MCF-10A cells. The JNK inhibitor, SP 600125 showed a weaker rescue effect in MCF-10A, but not in MCF-7 cells. The MEK inhibitor AZD6244 which blocks the ERK1/2 signaling, however, showed no protection in either cell line. These results suggested that activated p38 signaling contributes to anoikis in MCF-7 and MCF-10A cells.
The BH3-only protein Bmf has been deemed as one of the central regulators of anoikis [6, 7]. We performed CRISPR/Cas9-mediated Bmf knockout in MCF-7 and MCF-10A cells using a validated Bmf sgRNA [20] (Fig. 1D). Bmf depletion rescued detachment-induced apoptosis in MCF-7 and MCF-10A cells (Fig. 1E, Supplementary Fig. S1B). Similar results were observed using siRNA knockdown approach (Supplementary Fig. S2). These results indicated that Bmf participated in the regulation of anoikis in mammary epithelial cell lines.
Bmf is phosphorylated by p38 at multiple sites during loss of anchorage
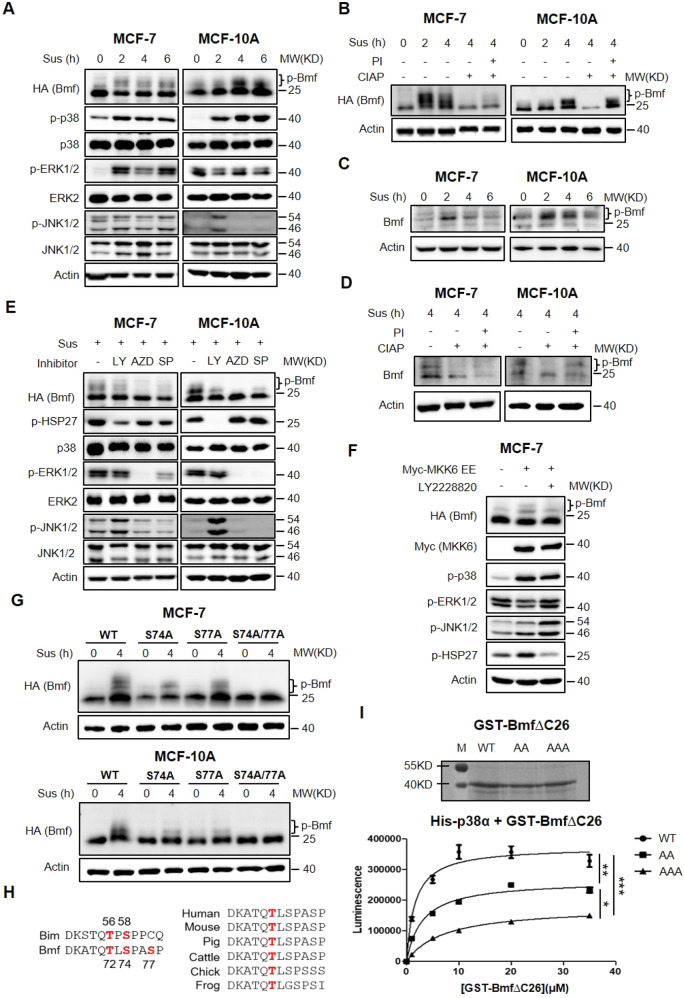
Since both MAPKs and Bmf were implicated in the regulation of anoikis, we speculated that activated MAPKs, in particular, p38 might regulate anoikis by modulating the apoptotic activity of Bmf through phosphorylation. Phosphorylation of Bmf can be detected through protein mobility shift in SDS PAGE gels [15]. When overexpressed in attached MCF-7 cells, HA-tagged Bmf proteins were detected by western blot as a major fast-migrating band followed by a small population of slow-migrating bands (Fig. 2A). Loss of adhesion induced rapid accumulation of slow-migrating HA-Bmf bands that were well correlated with the activation of p38 and/or ERK1/2 in MCF-7 cells (Fig. 2A). MCF-10A cells are highly sensitive to Bmf induced apoptosis, so we expressed a truncated Bmf (HA-Bmf∆C26) without the C-terminus transmembrane domain (a.a. 159–184) that contains no serine or threonine residues [21]. Consistently, the slow-migrating HA-Bmf∆C26 bands accumulated upon detachment and their levels were associated with p38 and/or JNK1/2 activation in MCF-10A cells (Fig. 2A). These observations were further confirmed in MCF-7 cells expressing HA-Bmf∆C26 (Supplementary Fig. S3A). Importantly, calf intestine alkaline phosphatase (CIAP) treatment reduced the suspension-induced slow-migrating Bmf population and co-treatment of phosphatase inhibitor (PI) reversed this effect (Fig. 2B). Similar effects were observed on endogenous Bmf (Fig. 2C, D). These results demonstrated that Bmf underwent phosphorylation in response to loss of anchorage.
Fig. 2. p38 MAPK phosphorylates Bmf at multiple sites.
A MCF-7 HA-Bmf cells (pre-induced with 100 ng/mL doxycycline for 24 h) and MCF-10A HA-Bmf∆C26 cells (constitutive Bmf expression) were cultured in suspension for 0, 2, 4, 6 h before lysed for western blot analysis. B Lysates of MCF-7 HA-Bmf∆C26 and MCF-10A HA-Bmf∆C26 cells were treated with calf intestine alkaline phosphatase (CIAP) and/or phosphatase inhibitor (PI), followed by western blot analysis. C MCF-7 and MCF-10A cells were cultured in suspension for 0, 2, 4, 6 h and expression of endogenous Bmf was analyzed by western blotting. D MCF-7 and MCF-10A cells were cultured in suspension for 4 h and lysed. Lysates were treated with −/+ CIAP/ PI and expression of endogenous Bmf was analyzed by western blotting. E MCF-7 HA-Bmf cells (pre-treated with 100 ng/mL doxycycline for 24 h) and MCF-10A HA-Bmf∆C26 cells were cultured in suspension in the presence of indicated inhibitors (LY 2228820, 1 μM; AZD6244, 10 μM; SP 600125, 10 μM) for 4 h. Cells were harvested and lysed for western blot analysis. F MCF-7 HA-Bmf∆C26 cells were transfected with empty vector or Myc-MKK6 EE expressing plasmids for 48 h in the presence or absence of p38α inhibitor, LY 2228820 (1 μM). Cells were then lysed for western blot analysis. G MCF-7 or MCF-10A cells transduced with lentiviruses expressing HA-Bmf∆C26 variants (WT, S74A, S77A, S74A/S77A) were individually cultured in suspension for 4 h and lysed for western blot analysis. H Alignment of the DLC1/2 binding domain of Bmf with that of Bim (Upper panel) and among different species (Bottom panel). I In vitro kinase assay was carried out using purified GST-Bmf∆C26 proteins and activated recombinant His-p38α. Top: SDS PAGE of purified GST-Bmf∆C26 and its mutant analogues. Bottom: quantitation of the luminescence as an indicator of phosphorylation level. Quantitative results were shown as mean ± STD from three independent experiments. Significance was determined by ANOVA one-way test, *p < 0.05, **p < 0.01, ***p < 0.001.
Inhibition of the ERK1/2, p38 or JNK1/2 pathways using corresponding inhibitors all resulted in diminishing of the slow-migrating Bmf (Fig. 2E, Supplementary Fig. S3B), suggesting that all three MAPKs either at basal or activated levels can phosphorylate Bmf at multiple sites. To further confirm that p38 can phosphorylate Bmf, we co-expressed in MCF-7 cells HA-Bmf and Myc-tagged MKK6-EE, a constitutively active MKK6 mutant that specifically activates the p38 MAPK [22]. As expected, expression of MKK6-EE potently improved p38 activity as manifested by the increased phosphorylation of p38 and its substrate, HSP27, but had mild impacts on ERK1/2 or JNK1/2 activities (Fig. 2F). Furthermore, MKK6-EE expression enhanced Bmf phosphorylation in a p38-dependent manner because treatment of p38 inhibitor reduced Bmf phosphorylation.
MAPKs generally phosphorylate proline-directed serine or threonine residues in protein substrates [23]. Analysis of Bmf protein sequence revealed only two proline-directed “SP” sites: S74P and S77P. Interestingly, these two sites are located proximal to the DLC2 binding sequence (DKATQTLSPASP) and have been shown to be phosphorylated by ERK2 in melanoma cells [15]. To test whether S74 and/or S77 are phosphorylated in response to loss of anchorage, we generated MCF-7 and MCF-10A cell lines that constitutively express HA-Bmf∆C26 variants including wild type (WT), S74A, S77A and S74A/S77A (AA). WT HA-Bmf∆C26 was phosphorylated at multiple sites after cells were cultured in suspension for 4 h (Fig. 2G). Replacement of S74 or S77 alone with alanine reduced the phosphorylation of Bmf as evidenced by the disappearance of some slow-migrating bands. However, the multiple shifted bands (at least 3) in WT Bmf and the remaining two slow-migrating bands observed in either S74A or S77A Bmf clearly indicated the presence of extra phosphorylated site(s) other than S74 and S77. Surprisingly, when both S74 and S77 were mutated at the same time, no apparent slow-migrating bands were observed. We speculated that either phosphorylation of the additional site(s) is dependent on S74/S77 phosphorylation, or phosphorylation of those additional site(s) alone do not cause mobility shift, as seen in many other phosphorylated proteins.
Since S74 and S77 are the only two proline-directed sites in Bmf, the discovery of non-proline-directed phosphorylation sites delivered a surprise. Unlike ERK1/2 and JNK1/2 kinases which strictly phosphorylate SP/TP sites, p38 is a rather promiscuous kinase that can phosphorylate many non-proline-directed sites in various substrates [24–26]. Lei et al. reported that JNK mediated UV-induced apoptosis by phosphorylating Bim at T56 residue (in BimL) in the DLC1 binding domain [13]. Sequence alignment revealed a high degree of similarity between the DLC binding domains of Bim and Bmf with T56 of Bim matching T72 of Bmf except that Bim T56 is followed by a proline while Bmf T72 is not (Fig. 2H). In addition, T72 of Bmf is evolutionary conserved (Fig. 2H). These observations inspired us to investigate whether T72 of Bmf is likely a non-canonical phosphorylation site of p38. In vitro kinase assay revealed that activated p38α (His-tagged) could directly phosphorylate WT GST-Bmf∆C26 (Fig. 2I). The S74A/S77A double mutation (AA Bmf) significantly reduced Bmf phosphorylation level, consolidating that these two sites are phosphorylated by p38. Intriguingly, the AA Bmf still retained decent level of phosphorylation by p38, implying the presence of other p38 phosphorylation sites. When T72 was mutated in AA Bmf, the phosphorylation signal was further reduced, supporting that T72 was also phosphorylated by p38 and its phosphorylation was independent of S74/S77. This result was confirmed by in vitro kinase assay using a different recombinant p38α kinase (GST-tagged) and WT or T72A GST-Bmf∆C26 (Supplementary Fig. S4A, B). Unlike S74 and S77, T72 phosphorylation didn’t seem to influence Bmf’s gel mobility shift as demonstrated by T72A and T72A/S77A mutants (Supplementary Fig. S4C). Together, the above results supported that p38 is at least partially responsible for the phosphorylation of Bmf at multiple sites upon loss of adhesion.
Bmf T72 phosphorylation disrupts Bmf/DLC2 binding
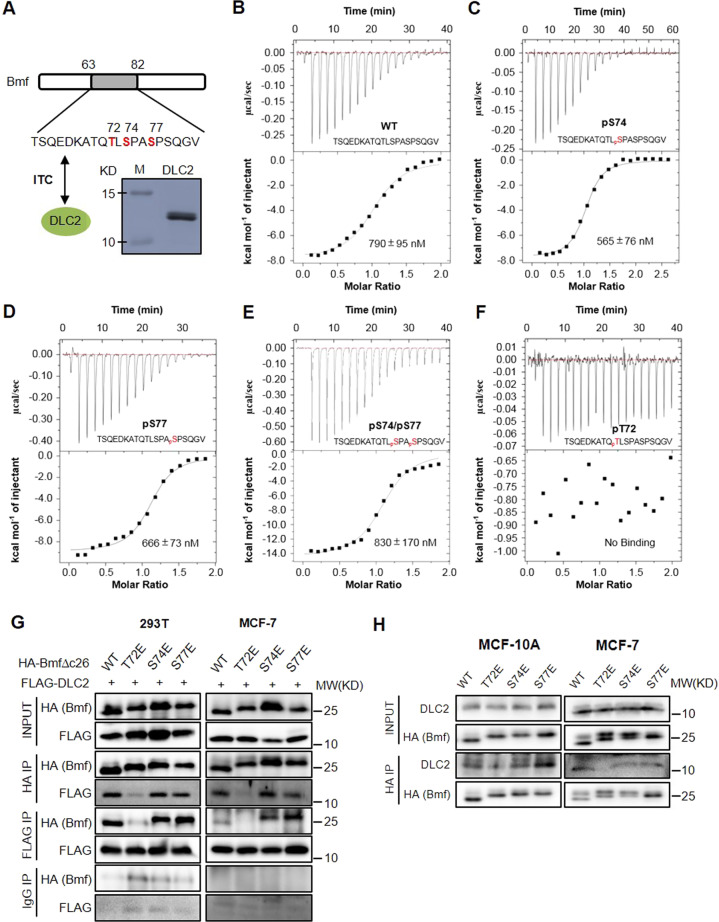
Since all three identified p38 phosphorylation sites (T72, S74 and S77) are located within or proximal to the DLC2 binding domain of Bmf, we interrogated the impact of Bmf phosphorylation on its binding to DLC2 (Fig. 3A). Isothermal titration calorimetric (ITC) assays revealed that the non-phosphorylated Bmf peptide encompassing the DLC2 binding domain interacted with recombinant DLC2 at an affinity of 0.79 µM (Fig. 3B). S74/S77-single or dual phosphorylated Bmf peptides bound DLC2 with similar affinities (Fig. 3C–E). Remarkably, T72-phosphorylated Bmf peptides completely lost DLC2 binding ability (Fig. 3F). These in vitro results were further validated by co-immunoprecipitation experiments showing the binding of WT, S74E and S77E HA-Bmf, but not the T72E analog to both exogeneous and endogenous DLC2 (Fig. 3G, H). Therefore, phosphorylation of T72, but not of S74 and S77 could compromise the Bmf/DLC2 interaction.
Fig. 3. Bmf T72 phosphorylation disrupts Bmf/DLC2 binding.
A Top: schematic show of DLC2 binding domain of Bmf protein; Bottom: SDS PAGE of purified 6xHis-DLC2 protein. B–F ITC analysis of interaction between 6xHis-DLC2 and non-phosphorylated (B), pS74 (C), pS77 (D), pS74/pS77 (E) or pT72 (F) Bmf peptides. G 293 T or MCF-7 cells were co-transfected with HA-Bmf∆C26 or FLAG-DLC2 expressing plasmids for 48 h. Cells were lysed, and reciprocal co-immunoprecipitations were performed using HA- or FLAG-tag antibodies with IgG being the negative control. H Lentivirus-transduced MCF-7 or MCF-10A cells inducibly expressing full length HA-Bmf variants (WT, T72E, S74E and S77E) were induced with doxycycline (100 ng/mL) for 4 h and lysed for immunoprecipitation using HA antibody. Immunoprecipitants were analyzed by HA and DLC2 antibodies.
Phosphomimetic mutation of T72 alters Bmf subcellular distribution
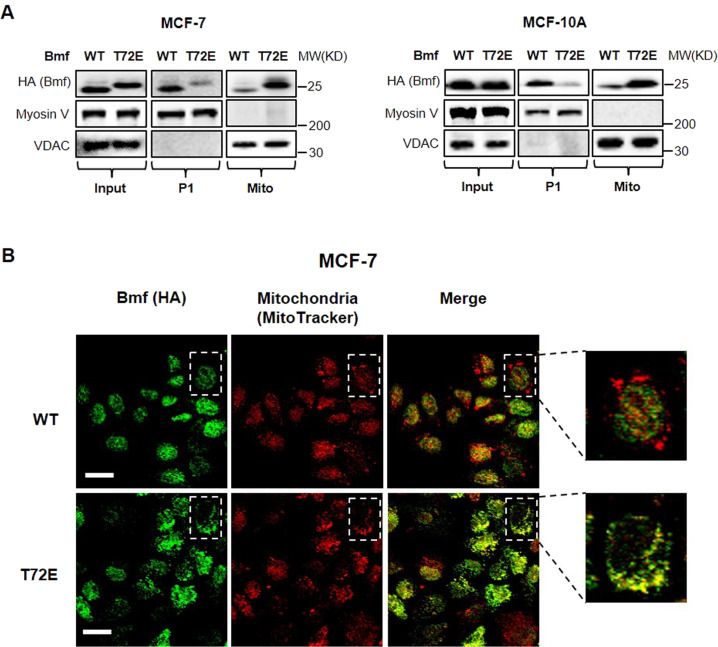
To determine whether T72 phosphorylation alters Bmf subcellular localization during anoikis, we comparatively analyzed the expressions of WT and T72E Bmf in the myosin V-rich fraction (P1) [6] and the mitochondria fraction. In accordance with its cytoskeletal sequestration, WT Bmf was predominantly detected in the P1 fraction, with a small amount in the mitochondria fraction (Fig. 4A). T72E Bmf, however, exhibited a reversed distribution pattern, high in the mitochondrial fraction but low in the P1 fraction. In addition, immunofluorescence staining revealed that T72E Bmf was more strongly associated with mitochondria than WT Bmf (Fig. 4B). These results supported our hypothesis that phosphorylation of Bmf T72 led to the translocation of Bmf from myosin V complex to mitochondria.
Fig. 4. Bmf T72E mutation promotes mitochondria translocation.
A MCF-7 HA-Bmf (WT/T72E) or MCF-10A HA-Bmf (WT/T72E) cells were induced with doxycycline (100 ng/mL) for 4 h. Myosin V-rich P1 fraction and mitochondria fraction were separately isolated and analyzed by western blotting. B MCF-7 HA-Bmf (WT/T72E) cells was induced by doxycycline for 1 h and immunofluorescence staining of HA-Bmf (green) and mitochondria (red, labeled by mitotracker) was performed. Scale bar = 20 μm.
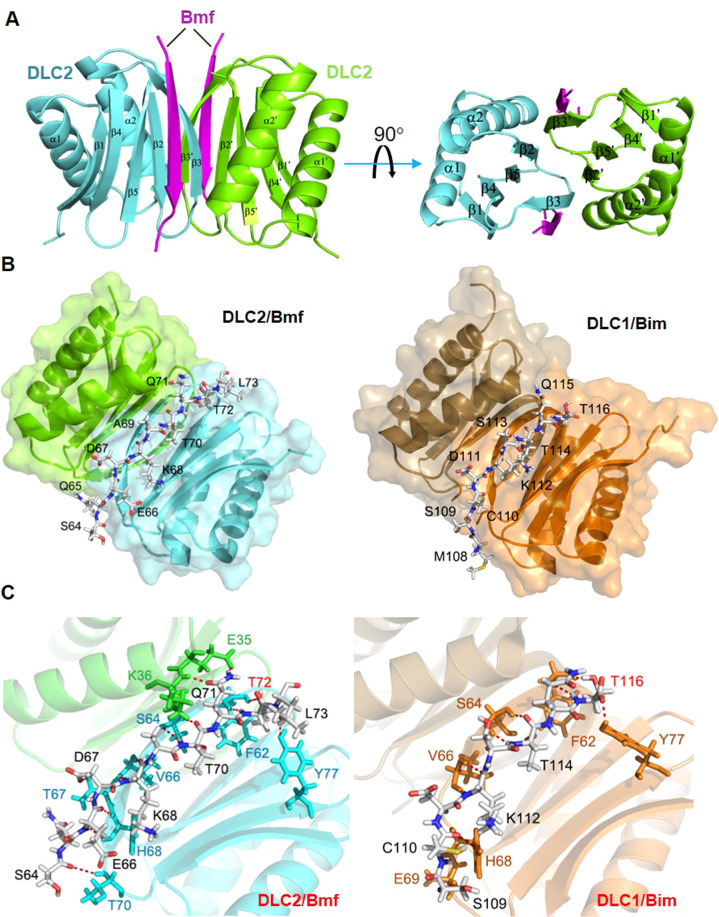
Structural basis of Bmf/DLC2 interaction
To understand the molecular mechanism of how T72 phosphorylation would interfere with Bmf/DLC2 interaction, we solved the crystal structure of the Bmf/DLC2 complex at a resolution of 2.0 Å (Fig. 5). Consistent with the solution (NMR) and crystal structures of DLC1/2 dimers [27–29], two DLC2 monomers assemble into a dimer through stacking of two 5-stranded β-sheets, each of which consists of the β1, β2, β4, β5 strands from one monomer and the β3 strand from the other (Fig. 5A). The DLC2/Bmf complex adopts a dimer/dimer stoichiometry, in which two Bmf peptides separately fit into two binding grooves formed by the β1, β3 and β4 strands of one DLC2 monomer and the α2 helix of the partner, at either side of the DLC2 dimer (Fig. 5A). The Bmf peptide also assumes a β-strand conformation in the complex and packs with the β3 strand of DLC2 in an anti-parallel manner, extending the 5-stranded β-sheet of DLC2 to a 6-stranded one (Fig. 5A). This binding mode is highly similar to that of DLC1/Bim complex solved by NMR spectroscopy [27] (Fig. 5B). The DLC2/Bmf complex is stabilized by hydrophobic interactions between Bmf peptide and the binding groove of DLC2 as well as numerous backbone and side chain polar contacts between Bmf peptide and the DLC2 β3 strand (Fig. 5B, C and Supplementary Fig. S5). Comparison of contact details between DLC2/Bmf and DLC1/Bim complexes revealed several conserved contacts involving DLC1/2 residues F62, S64, V66, H68 and Y77 (Fig. 5C and Supplementary Fig. S5A). Nevertheless, Q71 of Bmf makes extra contacts with E35 and K36 from the α2 helix of DLC2, which is not observed in DLC1/Bim complex, albeit with the presence of the conserved Q115 residue in Bim peptide (Fig. 5C and Supplementary Fig. S5A). According to the DLC2/Bmf complex structure, Bmf residue T72 contributes to the complex stability by forming two polar contacts with F62 of DLC2 using both its backbone and side chain (Fig. 5C and Supplementary Fig. S5A). Interestingly, the contact details of T72 (Bmf)/F62 (DLC2) interaction is mirrored in DLC1/Bim complex (T116 in BimEL, equivalent to T56 in BimL and F62 in DLC1) (Fig. 5C and Supplementary Fig. S5A), highlighting the importance of these threonine residues in Bim/Bmf-DLC binding. To further confirm the essential role of T72 in DLC2/Bmf interaction, we found that T72A mutation completely negated DLC2 binding of Bmf, while S74A/S77A double mutant Bmf retained DLC2 binding capacity close to the WT level (Supplementary Fig. S5B, C). These results clearly demonstrated that T72 residue participates in the binding of Bmf to DLC2 and therefore, phosphorylation of T72 would likely disrupt Bmf/DLC2 interaction by steric hindrance.
Fig. 5. Structural comparison of the Bmf/DLC2 and Bim/DLC1 complexes.
A Overall crystal structure of the Bmf/DLC2 complex. Two Bmf peptides separately fit into two binding grooves formed by the β1, β3 and β4 strands of one DLC2 monomer and the α2 helix of the partner, at either side of the DLC2 dimer, adopting a dimer/dimer stoichiometry. B Comparison of overall structures between Bmf/DCL2 complex and Bim/DLC1 complex (PDB: 1F95). DLC1 and DLC2 dimers were shown in ribbon diagram (DLC1: orange and brown; DLC2: green and teal) and Bim/Bmf peptides were shown in backbone diagram with each amino acid labeled. Bmf and Bim peptides shared highly similar modes of DLC binding. C Detailed interaction interfaces of Bmf/DLC2 complex (Left) and Bim/DLC1 complex (Right). Bim/Bmf peptides and DLC residues involved in polar contacts were shown in backbone diagram and color labeled. Green and teal: residues from DLC2; Orange: residues from DLC1. T72 (Bmf) and T116 (BimEL) were highlighted in red. Polar contacts were denoted by dashed red lines.
Phosphomimetic mutation of T72 enhances apoptotic activity of Bmf in vitro and in vivo
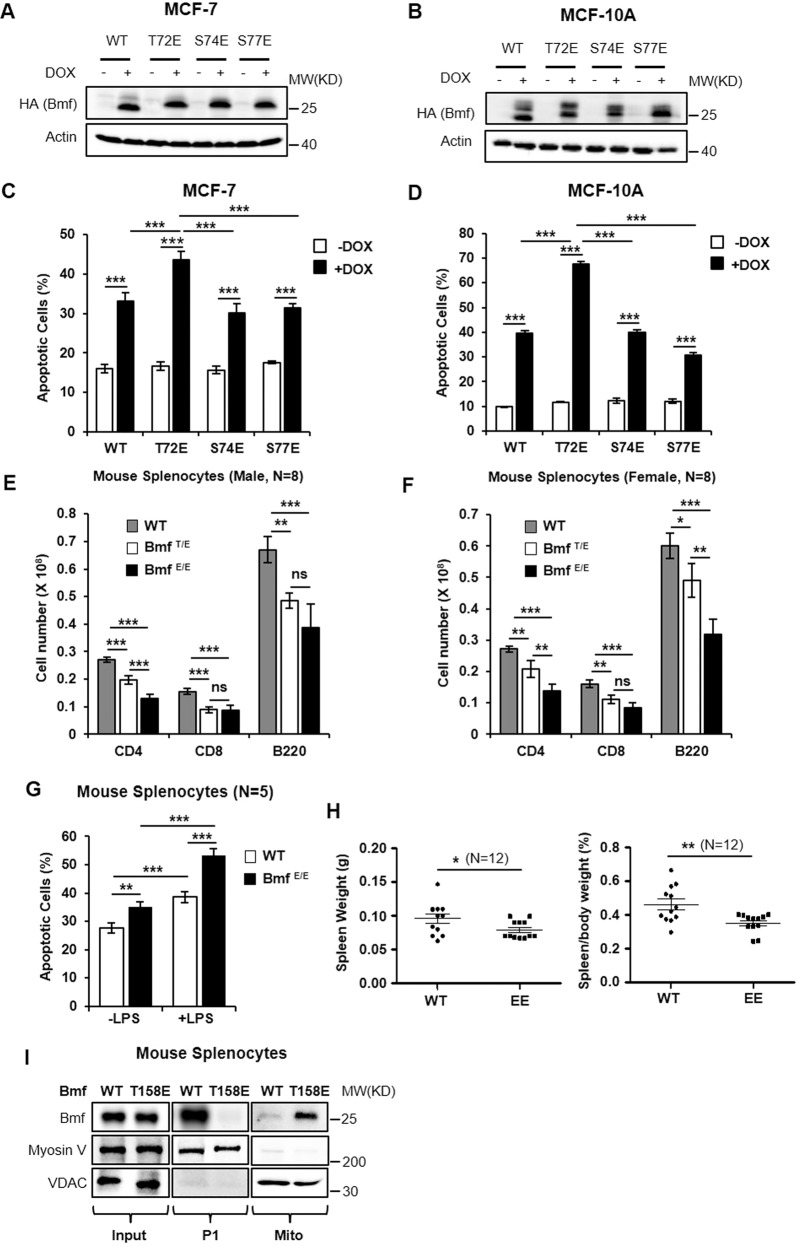
We constructed MCF-10A and MCF-7 cell lines that inducibly express WT or phosphomimetic variants of Bmf (full length) to investigate the influence of Bmf phosphorylation on its apoptotic activity. Western blot analysis confirmed that doxycycline induced expression of Bmf variants at close levels (Fig. 6A, B). Of note, while expression of S74E and S77E Bmf caused similar levels of cell death as WT Bmf, expression of T72E Bmf led to a higher death rate than the other three analogs (Fig. 6C, D and Supplementary Fig. S6). Therefore, phosphomimetic mutation of T72 could enhance Bmf apoptotic activity in human mammary epithelial cells in vitro.
Fig. 6. Phosphomimetic mutation of T72 enhances Bmf apoptotic activity in vitro and in vivo.
A, B Lentivirus-transduced MCF-7 (A) or MCF-10A (B) cells that inducibly express Bmf variants (including WT, T72E, S74E and S77E) were treated with −/+ doxycycline (100 ng/mL) for 24 h and cells were lysed for western blot analysis. C, D Lentiviral-transduced MCF-7 (C) or MCF-10A (D) cells inducibly expressing Bmf variants were treated with or without doxycycline for 24 h and cells were harvest for apoptosis assay using annexin V-PI staining and flow cytometry. E, F Quantitation of the numbers of CD4+ , CD8+ and B220+ splenocytes in male (E) and female (F) WT, Bmf+/T158E and BmfT158E/T158E mice, respectively. Quantitative results were shown as mean ± STD from 8 mice. Significance was determined by ANOVA one-way test, *p < 0.05; **p < 0.01; ***p < 0.001. G Apoptosis analysis on splenocytes from WT and BmfT158E/T158E mice with or without LPS stimulation using annexin V-PI staining and flow cytometry. Quantitative results were shown as mean ± STD from 5 mice. Significance was determined by ANOVA one-way test, **p < 0.01; ***p < 0.001. H Absolute (left) or relative (right) spleen weights of WT and BmfT158E/T158E mice (N = 12). Significance was determined by student T test, two tailed; *p < 0.05; **p < 0.01. I Myosin V-rich P1 fraction and mitochondria fraction were separately isolated from the splenocytes of WT or BmfT158E/T158E mice and analyzed by western blotting.
We next generated a BmfT158E (equivalent to BmfT72E in human) knock-in mouse model to interrogate the in vivo activity of this phosphomimetic mutation (Supplementary Fig. S7). No apparent developmental defects were observed in BmfT158E/T158E mice (Supplementary Fig. S8). Since Bmf regulates the homeostasis of lymphocytes [16, 30], we first analyzed the spleen lymphocytes from BmfT158E knock-in mice. Compared to the WT mice, Heterozygous Bmf+/T158E mice exhibited reduced CD4+ , CD8+ and B220+ splenocytes, and homozygous mice (BmfT158E/T158E) had further decreased populations of CD4+ and B220+ splenocytes (Fig. 6E, F). Accordingly, splenocytes from BmfT158E/T158E mice showed elevated basal or LPS-induced apoptosis levels when compared to those from WT mice (Fig. 6G and Supplementary Fig. S9). The average spleen weight was also lower in BmfT158E/T158E mice (Fig. 6H). The enhanced apoptotic activity of T158E Bmf in mouse splenocytes was associated with its mitochondria translocation (Fig. 6I). Importantly, T158E mutation did not cause significant changes on Bmf expression (Supplementary Fig. S10).
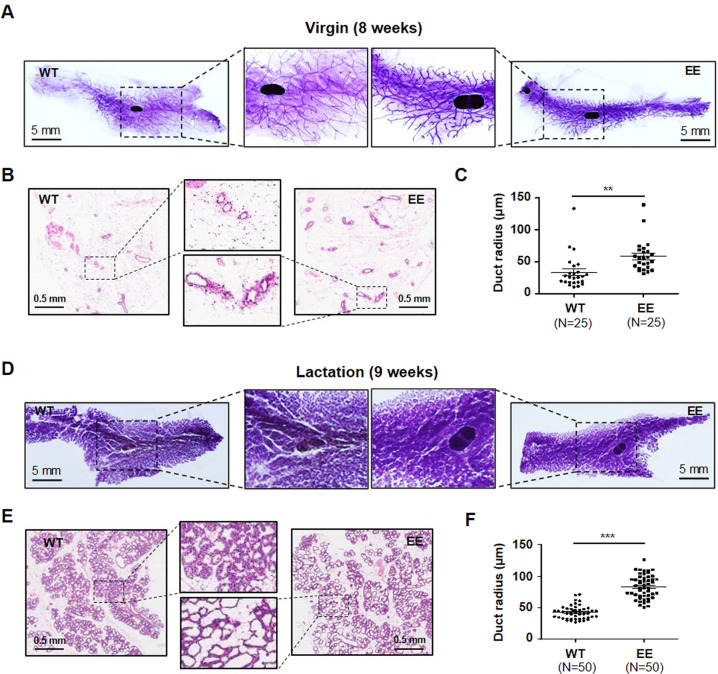
Since Bmf is essential for mammary cell anoikis and morphogenesis [7], we also investigated the influence of Bmf T158E mutation on mouse mammary cell apoptosis and mammary gland development. Mammary epithelial cells from BmfT158E/T158E mice exhibited higher apoptosis rate than those from WT mice (Supplementary Fig. S11). The mammary glands of BmfT158E/T158E mice appeared to develop normally except that dilated mammary ducts were observed (Fig. 7 and Supplementary Fig. S12), a finding in accordance with the enhanced apoptotic activity of T158E Bmf. In lactating mice, apoptotic mammary cells were associated with increased phospho-p38 levels (Supplementary Fig. S13A). In addition, higher level of apoptosis (as demonstrated by TUNEL staining) was observed in mammary tissues of BmfT158E/T158E mice than WT mice (Supplementary Fig. S13B).
Fig. 7. The effects of BmfT158E/T158E on mice mammary gland development.
A Mammary gland whole mounts of WT (left) and BmfT158E/T158E virgin mice (right). B H&E staining of mammary gland sections of WT and BmfT158E/T158E virgin mice. C Quantitation of the internal radii of mammary ducts in WT and BmfT158E/T158E virgin mice. Significance was determined by student T test, two tailed; **p < 0.01. D Mammary gland whole mounts of WT (left) and BmfT158E/T158E lactating mice (right). E H&E staining of mammary gland sections of WT and BmfT158E/T158E lactating mice. F Quantitation of the internal radii of mammary ducts in WT and BmfT158E/T158E lactating mice. Significance was determined by student T test, two tailed; ***p < 0.001.
Discussion
Stress-induced apoptosis is an important protective machinery of organisms in response to deleterious intracellular or extracellular stimuli such as UV or toxins exposure, viral infection, hypoxic, metabolic or oxidative stresses and loss of anchorage etc. While various stresses eventually lead to a converged caspase-dependent apoptotic endpoint, the upstream signaling that trigger apoptosis appear to be stress type-specific. Anoikis is a special type of apoptosis that occurs when cells are deprived of ECM adhesion. The BH3-only protein, Bmf functions as an important mediator of anoikis. Puthalakath et al. elegantly demonstrated that Bmf is sequestered to myosin V motor complex through binding to DLC2 in attached cells, but translocates to mitochondria to initiate anoikis when cells lose attachment [6]. However, the signaling pathway that prompts the release of Bmf from cytoskeleton upon loss of anchorage has not been characterized. We found that p38 MAPK was activated rapidly and durably in mammary epithelial cells deprived of attachment. Activated p38 directly phosphorylated Bmf at multiple sites including T72, S74 and S77. While phosphorylation of S74/S77 had little impact on Bmf activity, phosphorylation of T72 disrupted Bmf/DLC2 binding, promoted Bmf mitochondria translocation and enhanced its apoptotic activity in vitro and in vivo. Thus, we identified a new regulatory mechanism of anoikis by p38, which involved p38-mediated phosphorylation of Bmf. Interestingly, p38 has also been shown to regulate the anoikis of mammary epithelial cells through activating Bax [31] or BimEL [32]. These results established a central role of p38 in the regulation of anoikis.
The activity and stability of BH3-only proteins are intensely regulated by phosphorylation [33, 34]. We have previously shown that Bmf could be phosphorylated by ERK2 at S74 and S77 in melanoma cells [15]. Lei et al. proposed that Bmf S74 could be phosphorylated by JNK and modulate Bmf apoptotic activity in response to stress signals [13]. However, a follow-up study by Hübner et al. found no development defects in BmfS74D/S74D knock-in mice [16]. Only when combined with Bim−/−, BmfS74D displayed mildly increased apoptotic activity [16]. These results argued against a strong function of S74 phosphorylation and left a question of whether there exist other phosphorylation sites that may regulate Bmf activity during anoikis. According to sequence alignment (Fig. 2H), Bmf T72 residue (when compared to S74) seems to be a more plausible counterpart of BimL T56, except that it is not followed by a proline residue and thus not deemed as a potential MAPK phosphorylation site. Our in vitro kinase assay showed that T72, in addition to S74 and S77, could be phosphorylated by p38. This non-classic phosphorylation of Bmf T72 by p38 may therefore transduce an apoptotic signal that is unique to loss of adhesion. Of note, we found that T72A/S74A/S77A Bmf still retained moderate phosphorylation signal in p38 kinase assay (Fig. 2I), implying the presence of additional non-proline directed p38 phosphorylation site(s). Further studies are required to identify these sites and investigate their functions.
In addition to the cytoskeleton sequestration model, Häcker group recently proposed that DLC1/2 not only induces homo-dimerization of Bim and Bmf but also triggers the formation of ternary complexes of Bim-DLC1/2-Bmf [35, 36]. Importantly, these DLC1/2-mediated Bim/Bmf homo- or hetero-complexes, instead of being sequestered to cytoskeleton, rather localize on the mitochondria membrane. Our subcellular fractionation experiments revealed that WT Bmf was mainly detected in the myosin V fraction with a minor portion residing in the mitochondrial fraction (Figs. 4 and 6I). These data comply more with the cytoskeleton sequestration model, while not ruling out the possibility that the small population of mitochondrial Bmf regulates apoptosis through formation of DLC1/2: Bim/Bmf complexes. Hacker’s model also suggested that formation of the mitochondrial DLC1/2: Bim/Bmf complexes could stabilize Bim, Bmf and the associated Bcl-2 family proteins. In this context, p38-mediated T72 phosphorylation or phosphomimetic mutation of Bmf, which blocked DLC1/2 binding, would dissolve Bmf-containing complexes and foster DLC1: Bim complex assembly, promoting Bim protein stability and destabilizing Bmf. However, in splenocytes from Bmf T158E knock-in mice, Bim and Bmf protein levels were not apparently affected (Supplementary Fig. S10). Finally, both models suggested that Bmf release from the DLC1/2 complexes would promote apoptosis, which was supported by our data that Bmf phosphorylation increased its apoptotic activity. Future studies are required to fully understand the functions of these differentially located DLC1/2: Bim/Bmf complexes.
Acquisition of anoikis resistance is an essential step for the development of cancer metastasis. While this work and other studies have provided substantial evidences for p38’s central role in the regulation of anoikis [31, 32], it will be of further interest to unravel the upstream signals of p38 during anoikis, the kinetics of p38 activity and the status of Bmf expression/phosphorylation during different stages of cancer metastasis.
Materials and methods
Reagent
LY 2228820, SP 600125 and AZD6244 were purchased from Selleck Chemicals LLC (Houston, TX, USA). Doxycycline was purchased from Thermo Fisher Scientific (Rockford, IL, USA). Poly-HEMA was purchased from Sigma–Aldrich (St. Louis, MO, USA).
Cell culture
MCF-10A, MCF-7 and 293 T cells were purchased from ATCC (Manassas, VA, USA). MCF-10A cells were cultured in DMEM-Ham’s F12 with 5% horse serum, 20 ng/mL EGF, 0.5 μg/mL hydrocortisone, 100 ng/mL cholera toxin, 10 μg/mL insulin and penicillin‐streptomycin (200 µg/mL). MCF-7 and 293 T cells were cultured in high‐glucose DMEM with 10% fetal bovine serum (FBS) and penicillin‐streptomycin (200 µg/mL). All cells were mycoplasma free. For cell suspension culture, poly-2-hydroxyethyl methacrylate (Poly-HEMA) powders were dissolved in 95% EtOH preheated to 65 °C to make 20 mg/mL solution. Culture dishes were then coated with 20 mg/mL poly-HEMA solution at 37 °C overnight or till the plates dried out. After washing the plates twice with PBS, cells were typsinized, resuspended, and added to the poly-HEMA coated dishes at a desired density in culture medium.
CRISPR/Cas9-mediated knockdown of Bmf
Oligonucleotides harboring guide RNA (gRNA) sequences to BMF (5′ GAAGAGCTGAAGTCGGCTGA 3′) [20] were cloned into the lentiCRISPRV2 vector (Addgene, #52961). Lentiviruses were produced in 293FT cells and MCF-7 or MCF-10A cells were transduced with lentivirus for 24 h before selected with puromycin (1 μg/mL) for one week. The selected cells were used as a pool for subsequent anoikis assays.
Western blotting
Western blotting was performed as previously described [37]. Antibodies against Phospho-p38 MAPK (Thr180/Tyr182, #9211), p38 MAPK (#9212), Phospho-p44/42 MAPK (Thr202/Tyr204, clone 197G2, #4377), Phospho-SAPK/JNK (Thr183/Tyr185, clone 81E11, #4668), SAPK/JNK (#9252), HA-tag (clone 6E2, #2367; clone C29F4, #3724), Myc-tag (clone 71D10, #2278), Myosin V (#3402) and VDAC (#4866) were purchased from Cell Signaling Technology (Beverley, MA, USA). Anti-β-actin (#A2066) and anti-FLAG-tag (clone M2, #F3165) were from Sigma Aldrich. Anti-ERK2 (C-14, clone sc-154, #F2013) were from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Anti-Phospho-HSP27 (Ser82, #YP0136) was purchased from ImmunoWay Biotechnology (Plano, TX, USA). Anti-Bmf antibody (clone 9G10, #ALX-804-343-C100) was from ENZO Life Sciences (Ann Arbor, MI, USA). Anti-DLC2 antibody (#ABT140) was from Millipore (Billerica, MA, USA). Anti-GAPDH (#TA802519) and β-Tubulin antibody (#TA503129) were from Origene (Rockville, MD, USA). All antibodies were diluted to 1 µg/mL for usage unless otherwise stated.
Construction of lentiviral vectors and cell lines
HA-Bmf∆C26 cDNA was cloned into pENTR™/D-TOPO vector (Thermo Fisher Scientific, Waltham, MA, USA) to generate the entry plasmid. Entry plasmids of HA-Bmf∆C26 mutants (T72E, S74E, S77E) were constructed using Quickchange site-directed mutagenesis kit (Agilent Technologies Inc., Santa Clara, CA, USA). Entry plasmids were then recombined with pLentipuro/TO/V5-DEST plasmid using Gateway™ LR Clonase™ (Thermo Fisher Scientific) to generate lentiviral plasmids. Lentiviruses were produced in 293FT cells and MCF-7 or MCF-10A cells were infected with lentivirus for 72 h before selected with puromycin. For doxycycline-inducible expression, full length HA-Bmf cDNA was cloned into pENTRTM1A Dual Selection Vector (Thermo Fisher Scientific). The resulting plasmid was recombined with pInducer20 (Addgene, Watertown, MA, USA) to generate the lentiviral plasmid. Lentiviruses were produced in 293FT cells and MCF-10A/MCF-7 cells were transduced as described above before selected with G418.
Annexin V/PI apoptosis assay
Cells were collected, washed by PBS and stained using the APC Annexin V Apoptosis Detection Kit with PI (Biolegend, San Diego, CA, USA) according to manufacturer’s protocol. Stained cells were analyzed by flow cytometry on a CytoFLEX system (Beckman Coulter, Indianapolis IN, USA). Data were analyzed using CytExpert software (Beckman).
Quantitative RT-PCR
Total RNA was isolated using TRIzolTM reagent (Thermo Fisher Scientific) and reverse transcribed into cDNA using iScript cDNA Synthesis Kit (BioRad, Hercules, CA, USA). PCR reactions were performed using iQ SYBR Green Supermix (BioRad) and analyzed by a CFX Connect real-time PCR detection system (BioRad). Relative mRNA levels were calculated using the comparative Ct (ΔCt) method. The following primers were used: β-actin (forward, 5′TACCTCATGAAGATCCTCACC 3′; reverse, 5′TTTCGTGGATGCCACAGGAC 3′), human Bmf (forward: 5′GAGGTACAGATTGCCCGAAAG 3′; reverse: 5′TTCAAAGCAAGGTTGTGCA 3′), mouse Bmf (forward: 5′CCCATAAGCCAGGAAGACAA 3′; reverse: 5′CTGAAGCTTTCTGGCGATCT 3′).
Protein purification
The E.coli BL21 (DE3) cells transformed with pET28a-6 x His DLC2 plasmid were grown to an OD600 of 0.7 at 37 °C in LB medium with 100 μg/mL Kanamycin. The cultures were induced with 1 mM IPTG at 28 °C for 8 h and harvested by centrifugation at 7000 g, 4 °C. The cell pellets were resuspended and lysed by sonication in lysis buffer (20 mM Tris, pH 8.0, 150 mM NaCl) at 4 °C. The lysate was centrifuged at 25,000 g to remove cell debris. Filtered lysate supernatant was loaded onto a HisTrapTMHP Ni Sepharose column (GE Healthcare, Hilden, Germany) pre-equilibrated with loading buffer (20 mM Tris, pH 8.0, 150 mM NaCl) using an ÄKTA purifier system (GE Healthcare). Column-bound samples were then washed with washing buffer (20 mM Tris, pH 8.0, 150 mM NaCl, 20 mM imidazole) and eluted using elution buffer (20 mM Tris, pH 8.0, 150 mM NaCl, 500 mM imidazole). The eluents were concentrated by ultrafiltration and loaded on Superdex75 16/60 (GE Healthcare) size exclusion chromatography column in 20 mM Tris, pH 8.0, 150 mM NaCl, 5% glycerol for further purification. The main protein fractions were collected and concentrated for further usage. The protein concentration was measured with a NanoDrop® spectrometer (Thermo Fisher) using the extinction coefficient generated from ExPASy ProtParam program.
For GST-Bmf∆C26 purification, E.coli BL21 cells harboring pGEX-Bmf∆C26 WT plasmid were grown to an OD600 of 0.7 at 37 °C in LB medium supplemented with 100 μg/mL Ampicillin and induced with 1 mM IPTG for 8 h at 28 °C. Cell lysates prepared as above in 20 mM Tris, pH 8.0, 150 mM NaCl were loaded on a GSTrap™ HP Column using an ÄKTA purifier system. After washing (20 mM Tris, pH 8.0, 150 mM NaCl), proteins were eluted using a buffer containing 20 mM Tris, pH 8.0, 150 mM NaCl, 10 mM reduced glutathione. Eluted proteins were concentrated for following applications.
In vitro kinase assay
Different amount of the purified Bmf substrates (0, 1, 5, 10, 20 and 34 μM) were incubated with 0.35 mM activated recombinant His-p38α (ENZO Life Sciences, Farmingdale, NY, USA) or GST-p38α (Promega, Madison, WI, USA) and 0.5 mM ATP in 1 x NEBufferTM (50 mM Tris-HCl, 10 mM MgCl2, 0.1 mM EDTA, 2 mM DTT, 0.01% Brij 35, pH 7.5, NEB, Ipswich, MA, USA) at 30 °C for 40 min. The reaction products were analyzed by the ADP-Glo™ Kinase Assay Kit (Promega) following the manufacturer’s instruction.
Subcellular fractionation
For P1 fraction isolation, 107 MCF-7 or MCF-10A cells were lysed in 500 µL extraction buffer [0.05 M PIPES/NaOH, 0.05 M HEPES, pH 7.0, 2 mM MgCl2, 1 mM EDTA, protease inhibitors (CompleteTM tablets; Roche) and 1% Triton X-100] on ice for 30 min. Cellular debris was removed by centrifugation (10,000 g, 4 °C, 15 min) and the resultant supernatant was further centrifuged at 125,000 g for 1 h at 4 °C in a Beckman bench-top ultracentrifuge. The pellet (P1) is lysed with 1 x laemmli sample buffer (BioRad, USA, #1610737) for subsequent western blot assay.
For mitochondria fraction isolation, one confluent 10 cm dish cells were washed twice with cold 1x PBS. Cells were scraped and suspended in 1 mL cold 1 x PBS. The cells were pelleted by centrifugation (1000 g, 4 °C), resuspended in 0.5 mL hypotonic buffer (10 mM Tris, 10 mM NaCl, 2.5 mM MgCl2 and 1% PMSF at a pH of 7.4), incubated on ice for 10 min, and then harvested in a glass homogenizer for homogenizing. The homogenates were collected in a new tube with 165 μL 4x isotonic buffer (20 mM Tris, 0.84 M Mannitol, 0.25 M sucrose and 4 mM EDTA, pH 7.9), centrifuged at 1300 g for 5 min at 4 °C. The supernatants were collected and centrifuged at 17,000 g for 15 min at 4 °C to precipitate the mitochondria. The mitochondria pellets were washed with 1x isotonic buffer twice and lysed with 1x laemmli sample buffer for subsequent western blot assay.
Calf intestine alkaline phosphatase (CIAP) treatment
MCF-7 and MCF-10A HA-Bmf∆C26 WT cells were cultured in suspension for 2–4 h and lysed in NP40 buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% NP40). The cell lysates were incubated with CIAP (Invitrogen) alone or in combination with phosphatase inhibitor cocktail (Roche) for 1 h at 37 °C. The samples were then diluted with 5x SDS sample buffer for western blot analysis.
Immunofluorescence
MCF-7 HA-Bmf WT or T72E cells were grown on coverslips in 6-well plate and simultaneously treated with 100 ng/mL doxycycline and 200 nM mitotracker Red CMXRos (Invitrogen, #M7512) for 1 h at 37 °C. Cells were then fixed with 3.7% formaldehyde at room temperature for 15 min, followed by incubating in PBS containing 0.2% TritonX-100 for 5 min. After blocking with 2% BSA for 1 h, the cells were incubated with HA-tag antibody (clone 6E2, #2367) at 4 °C overnight. Next day, the cells were incubated with Alexa Fluor 488 goat anti-mouse IgG (Thermo Fisher, #A11029) for 1 h at room temperature. Fluorescence was visualized using an Olympus confocal microscopy system (FV1200).
Isothermal titration calorimetry (ITC)
WT (TSQEDKATQTLSPASPSQGV), pS74 (TSQEDKATQTLpSPASPSQGV), and pS77 (TSQEDKATQTLSPApSPSQGV), pT72 (TSQEDKATQpTLSPApSPSQGV) and pS74/pS77 (TSQEDKATQTLpSPApSPSQGV) Bmf peptides were purchased from CSBio Ltd. (Shanghai, China). All the ITC experiments were performed with a Microcal ITC200 calorimeter (GE Healthcare) at 25 °C. The His-tagged DLC2 protein samples were buffer exchanged to 20 mM Tris pH 8.0, 150 mM NaCl, 10% glycerol, using size-exclusion chromatography and the Bmf peptides were dissolved in the same buffer prior to the experiments. The experiments were conducted by injecting peptides (300 μM) into DLC2 solution (10 mM). The injections contained 0.4 μL for the first injection and 2 μL for the second to 19th injection, with 120 s intervals. All the ITC data were analyzed with the supplemented Microcal ITC data analysis package under the one binding site mode.
Crystallization and data collection
The DLC2 recombinant protein purified from affinity and size exclusion chromatography was concentrated and incubated with BMF peptides (1:6 molar ratio) for complex crystallization. Initial crystallization trails were carried out in 384-well plates using sitting drop vapor diffusion method and commercial sparse matrix crystallization kits (Hampton Research and Molecular Dimensions) at room temperature. The DLC2/BMF complex was crystallized in 0.2 M Ammonium chloride, 0.1 M HEPES, pH 7.0, 20% w/v PEG 6000. Crystals were flashed frozen in liquid nitrogen and cryoprotected with the reservoir solution supplemented with 15% glycerol. The X-ray diffraction data was collected on Beamline 18U1 at Shanghai Synchrotron Radiation Facility, processed and indexed with XDS package [38]. Data collection details and statistics are shown in Table S1.
Structure determination and refinement
The Structure of DLC2/BMF complex was solved by molecular replacement using the DLC2 structure (PDB: 2XQQ) as search model [29, 39]. The BMF peptide was manually built with COOT and the further model refinement was carried out with PHENIX suite [40, 41]. The DLC2/BMF complex structure was solved to 2 Å in the space group of C2 with the unit cell parameter of a = 166.2 Å, b = 38.7 Å, c = 44.7 Å and α = γ = 90°, β = 99.874°. The crystal model was refined to R work/R free of 0.1933/0.2256 and contains 3 DLC2 molecules in the asymmetric unit cell with one of the DLC2 molecule in complex with BMF peptide, 67 water molecules. The structure showed no residues in the Ramachandran plot disallowed region and exhibited favorable stereochemistry. The dimer DLC2/BMF complex can be generated with symmetry mates and the PyMOL program was used to visualize and generate all the structural figures [42]. Detailed refinement statistics has been summarized in table S1.
Co-immunoprecipitation
Cells were transfected with indicated plasmids for 48 h. Cells were then washed in 1x cold PBS and lysed in lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% NP40) supplemented with protease and phosphatase inhibitor cocktails (Roche, Basel, Switzerland). Lysates were centrifuged to collect the supernatants. Immunoprecipitation was performed overnight at 4 °C using IgG (SC-2025, Santa Cruz, CA, USA), HA-tag or FLAG-tag antibodies. Antibody-protein complexes were captured by the protein A/G Plus-Agarose beads (Santa Cruz, CA, SUA), eluted with SDS sample buffer and subjected to western blot analysis.
Mice
C57BL/6 mice harboring Bmf T158E mutation (equivalent to Bmf T72E in human) were generated by Cas9/CRISPR-mediated genome editing (Cyagen Biosciences, Guangzhou, China). Briefly, DNA fragment carrying Bmf targeting gRNA (AGCTGGACTGAGGGTCTGAGTGG) was inserted into the gRNA cloning vector. The oligo donor with targeting sequence, flanked by 120 bp homologous sequences combined on both sides (5′CTC ACC CAT TGC TGT GGT CCC GGA CTC CGG CCC ATA AGC CAG GAA GAC AAG GCC ACT CAG GAG CTC AGT CCA GCT TCC CCA AGC CAG GGT GTC ATG CTG CCT TGT GGG GTG ACA GAG GAA CCC 3′) was synthesized (Integrated DNA Technologies). The Bmf gRNA and Cas9 mRNA were generated by in vitro transcription and co-injected with oligo donor into fertilized eggs. The pups were genotyped by PCR (primers: T158E-F, 5′TCTGACAGGAGAGATGGAGCCACC 3′; T158E-R, 5′GCAGAGCCACTGAAACGACATAGG 3′) and DNA sequencing analysis (primer: 5′GCAGAGCCACTGAAACGACATAGG 3′). Mice harboring BmfT158E allele were mated with WT C57BL/6 mice to acquire heterozygous progenies, which were then intercrossed to generate homozygous BmfT158E/T158E mice. All mice were maintained in pathogen-free conditions and housed separately during the experiment. Both male and female mice of 8–16 weeks’ old were used in this study. Mice died of unnatural causes during experiments were excluded from the analysis. Animal studies were not blinded. All animal experiments were performed under the approval of the Institutional Animal Care and Use Committee of Xi’an Jiaotong University.
Analysis of mouse splenocytes
Eight to twelve-week old female or male mice were randomly divided into three groups according to their genotypes (homozygote, heterozygote, wild type). Mice were euthanized to harvest the spleens. The spleens were fully grinded with a syringe piston in 1x PBS and the cells were filtrated through a 74 μm nylon membrane. Spleen cells were collected by centrifugation (400 g, 4 °C, 15 min) and resuspended in 2 mL 1x PBS. Red blood cells were lysed by adding 750 μL red blood cell lysis buffer (#EA0001, Sparkjade, China) to 250 μL spleen cell suspension and incubating on ice for 15 min with occasional agitation. Splenocytes were harvested by centrifugation (450 g, 4 °C, 10 min) and the red blood cell lysis step was repeated with addition of 500 μL red blood cell lysis buffer. After centrifugation, the splenocytes were resuspended in PBS, counted using a hemocytometer and the composition of spleen lymphocytes were further analyzed by flow cytometry. Briefly, splenocytes (1 × 106) were incubated (30 min at 4 °C) with R-phycoerythrin-conjugated anti-CD4 (L3T4), allophycocyanin-conjugated anti-CD8α (Ly-2), and fluorescein isothiocyanate-conjugated anti-CD45R/B220 antibodies (Biolegend) respectively, washed with PBS plus 2% bovine serum albumin and examined by flow cytometry. Apoptosis of splenocytes were analyzed by annexin V/PI staining and flow cytometry.
Lipopolysaccharide (LPS) administration
LPS from Escherichia coli 055: B5 (Sigma-Aldrich, #L2880) was diluted at 2 mg/mL with PBS. Eight-week old female mice were divided into two groups according to their genotypes. Mice were injected with LPS (20 mg/kg) or PBS intraperitoneally. After 24 h, mice were euthanized to harvest the spleens for subsequent assays.
Isolation and analysis of mouse mammary epithelial cells
The fourth pair of mammary glands were collected from female mice and cut into small pieces. Tissues were minced and digested in 5 mL collagenase solution (DMEM/F12, 5% FBS, 1% Penicillin/Streptomycin, 10 ng/mL EGF, 500 ng/mL hydrocortisone, 5 μg/mL insulin, 20 ng/mL cholera toxin, 1 mg/mL collagenase, 0.2 mg/mL hyaluronidase) at 37 °C for 1 h with thorough agitation every 10 min. Organoids were collected by centrifugation at 500 g for 5 min, resuspended in 1 mL 0.25% trypsin-EDTA solution and digested for 1.5 min at 37 °C. After addition of 3 mL medium (DMEM/F12, 5% FBS, 1% Penicillin/Streptomycin) to neutralize the trypsin, organoids were collected by centrifugation for 5 min at 500 g. Organoids were further digested in 2 mL medium (DMEM/F12, 5% FBS, 1% Penicillin/Streptomycin) containing 5 mg/mL dispase and 0.1 mg/mL DNase I for 5 min at 37 °C. After collection by centrifugation, organoids were incubated in 2 mL 0.64% NH4Cl solution for 5 min at 37 °C, followed by spin-collection again. The cells were resuspended in 1 mL FACS buffer (95% PBS, 5% FBS) and filtered through a 40 mm nylon membrane. The cells were incubated with CD31-biotin (BD Biosciences, #558737), CD45-biotin (BD Biosciences, #553078), Streptavidin-PE/Cy7 (Biolegend, #405206), CD24-PE (BD Bioscience, #553662) and subjected to flow cytometry analysis. The Lin- (CD31−/CD45−)/CD24+ population was designated as the mammary epithelial cells.
Mouse mammary gland whole mount preparation and analysis
The 4/5th mammary gland tissues were harvested from euthanized mice and transferred to a slide. The tissues were fully spread out using blunt end forceps. After fixing in Carnoy’s fixative for 2 h at room temperature, the tissues were washed in 70% ethanol for 15 min and in distilled water for 5 min. Tissues were then stained in carmine alum solution overnight at 4 °C followed by washing in 70, 95 and 100% ethanol sequentially, 15 min each. The slides were then soaked in xylene overnight at room temperature. The preparation was covered with a coverslip and sealed with neutral balsam. Images were visualized using a dissecting microscope.
Statistical analysis
The quantitative data were presented as mean ± SDM unless otherwise stated. Quantitative results of qRT-PCR, flow cytometry and western blot analysis were from three independent experiments. For animal studies, the number of mice used were indicated in relevant figures. The statistical differences were determined by unpaired Student’s t test (two-tailed) for two group comparison or one-way ANOVA test assuming unequal variance for multi-group comparison. Significant P values were denoted by *P < 0.05, **P < 0.01, ***P < 0.001. For animal studies, no statistical methods were used to pre-determine sample size.
Supplementary information
Acknowledgements
We thank the staffs from BL18U1/BL19U1 beamline for technical support during data collection in National Center for Protein Sciences Shanghai (NCPSS) at Shanghai Synchrotron Radiation Facility. We also thank Dr. Hanqiu Zheng and Jingyu Pan at Tsinghua University for technical assistance in animal works.
Author contributions
YS conceived and designed the research. YS and YW provided research funding. ZZ, ZO, YR, YC and PL performed the experiments and analyzed the data. YW analyzed the crystallography data. YS, ZZ, and YW wrote the paper. All authors reviewed and approved the paper for publication.
Funding
This work was supported by the National Natural Science Foundation of China (31970724, 31771557 to YS, 82072237, 31870132 to YW).
Data and materials availability
All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The crystallography data have been deposited to the Protein Data Bank (http://www.pdb.org) with PDB ID “7CNU”.
Competing interests
The authors declare no competing interests.
Ethical approval
This study did not involve human participants, human data or tissues. The animal studies were approved by the Institutional Animal Care and Use Committee of Xi’an Jiaotong University.
Footnotes
Edited by S. Kumar
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41418-021-00855-3.
References
- 1.Gilmore AP. Anoikis. Cell Death Differ. 2005;12:1473–7. doi: 10.1038/sj.cdd.4401723. [DOI] [PubMed] [Google Scholar]
- 2.Mailleux AA, Overholtzer M, Brugge JS. Lumen formation during mammary epithelial morphogenesis: insights from in vitro and in vivo models. Cell Cycle. 2008;7:57–62. doi: 10.4161/cc.7.1.5150. [DOI] [PubMed] [Google Scholar]
- 3.Sisto M, D’Amore M, Lofrumento DD, Scagliusi P, D’Amore S, Mitolo V, et al. Fibulin-6 expression and anoikis in human salivary gland epithelial cells: implications in Sjogren’s syndrome. Int Immunol. 2009;21:303–11. doi: 10.1093/intimm/dxp001. [DOI] [PubMed] [Google Scholar]
- 4.Liotta LA, Kohn E. Anoikis: cancer and the homeless cell. Nature. 2004;430:973–4. doi: 10.1038/430973a. [DOI] [PubMed] [Google Scholar]
- 5.Puthalakath H, Strasser A. Keeping killers on a tight leash: transcriptional and post-translational control of the pro-apoptotic activity of BH3-only proteins. Cell Death Differ. 2002;9:505–12. doi: 10.1038/sj.cdd.4400998. [DOI] [PubMed] [Google Scholar]
- 6.Puthalakath H, Villunger A, O’Reilly LA, Beaumont JG, Coultas L, Cheney RE, et al. Bmf: a proapoptotic BH3-only protein regulated by interaction with the myosin V actin motor complex, activated by anoikis. Science. 2001;293:1829–32. doi: 10.1126/science.1062257. [DOI] [PubMed] [Google Scholar]
- 7.Schmelzle T, Mailleux AA, Overholtzer M, Carroll JS, Solimini NL, Lightcap ES, et al. Functional role and oncogene-regulated expression of the BH3-only factor Bmf in mammary epithelial anoikis and morphogenesis. Proc Natl Acad Sci USA. 2007;104:3787–92. doi: 10.1073/pnas.0700115104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Whelan KA, Caldwell SA, Shahriari KS, Jackson SR, Franchetti LD, Johannes GJ, et al. Hypoxia suppression of Bim and Bmf blocks anoikis and luminal clearing during mammary morphogenesis. Mol Biol Cell. 2010;21:3829–37. doi: 10.1091/mbc.e10-04-0353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hausmann M, Leucht K, Ploner C, Kiessling S, Villunger A, Becker H, et al. BCL-2 modifying factor (BMF) is a central regulator of anoikis in human intestinal epithelial cells. J Biol Chem. 2011;286:26533–40. doi: 10.1074/jbc.M111.265322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Girnius N, Davis RJ. JNK promotes epithelial cell anoikis by transcriptional and post-translational regulation of BH3-only proteins. Cell Rep. 2017;21:1910–21. doi: 10.1016/j.celrep.2017.10.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reginato MJ, Mills KR, Paulus JK, Lynch DK, Sgroi DC, Debnath J, et al. Integrins and EGFR coordinately regulate the pro-apoptotic protein Bim to prevent anoikis. Nat Cell Biol. 2003;5:733–40. doi: 10.1038/ncb1026. [DOI] [PubMed] [Google Scholar]
- 12.Puthalakath H, Huang DC, O’Reilly LA, King SM, Strasser A. The proapoptotic activity of the Bcl-2 family member Bim is regulated by interaction with the dynein motor complex. Mol Cell. 1999;3:287–96. doi: 10.1016/S1097-2765(00)80456-6. [DOI] [PubMed] [Google Scholar]
- 13.Lei K, Davis RJ. JNK phosphorylation of Bim-related members of the Bcl2 family induces Bax-dependent apoptosis. Proc Natl Acad Sci USA. 2003;100:2432–7. doi: 10.1073/pnas.0438011100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.VanBrocklin MW, Verhaegen M, Soengas MS, Holmen SL. Mitogen-activated protein kinase inhibition induces translocation of Bmf to promote apoptosis in melanoma. Cancer Res. 2009;69:1985–94. doi: 10.1158/0008-5472.CAN-08-3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shao Y, Aplin AE. ERK2 phosphorylation of serine 77 regulates Bmf pro-apoptotic activity. Cell Death Dis. 2012;3:e253. doi: 10.1038/cddis.2011.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hubner A, Cavanagh-Kyros J, Rincon M, Flavell RA, Davis RJ. Functional cooperation of the proapoptotic Bcl2 family proteins Bmf and Bim in vivo. Mol Cell Biol. 2010;30:98–105. doi: 10.1128/MCB.01155-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cowan KJ, Storey KB. Mitogen-activated protein kinases: new signaling pathways functioning in cellular responses to environmental stress. J Exp Biol. 2003;206:1107–15. doi: 10.1242/jeb.00220. [DOI] [PubMed] [Google Scholar]
- 18.Wada T, Penninger JM. Mitogen-activated protein kinases in apoptosis regulation. Oncogene. 2004;23:2838–49. doi: 10.1038/sj.onc.1207556. [DOI] [PubMed] [Google Scholar]
- 19.Fiucci G, Ravid D, Reich R, Liscovitch M. Caveolin-1 inhibits anchorage-independent growth, anoikis and invasiveness in MCF-7 human breast cancer cells. Oncogene. 2002;21:2365–75. doi: 10.1038/sj.onc.1205300. [DOI] [PubMed] [Google Scholar]
- 20.Sale MJ, Minihane E, Monks NR, Gilley R, Richards FM, Schifferli KP, et al. Targeting melanoma’s MCL1 bias unleashes the apoptotic potential of BRAF and ERK1/2 pathway inhibitors. Nat Commun. 2019;10:5167. doi: 10.1038/s41467-019-12409-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hinds MG, Smits C, Fredericks-Short R, Risk JM, Bailey M, Huang DC, et al. Bim, Bad and Bmf: intrinsically unstructured BH3-only proteins that undergo a localized conformational change upon binding to prosurvival Bcl-2 targets. Cell Death Differ. 2007;14:128–36. doi: 10.1038/sj.cdd.4401934. [DOI] [PubMed] [Google Scholar]
- 22.Raingeaud JWA, Barrett T, Dérijard B, Davis RJ. MKK3- and MKK6-regulated gene expression is mediated by the p38 mitogen-activated protein kinase signal transduction pathway. Mol Cell Biol. 1996;16:1247–55. doi: 10.1128/MCB.16.3.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bardwell L. Mechanisms of MAPK signalling specificity. Biochem Soc Trans. 2006;34:837–41. doi: 10.1042/BST0340837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alam MS, Gaida MM, Debnath S, Tagad HD, Miller Jenkins LM, Appella E, et al. Unique properties of TCR-activated p38 are necessary for NFAT-dependent T-cell activation. PLoS Biol. 2018;16:e2004111. doi: 10.1371/journal.pbio.2004111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alvarado-Kristensson M, Melander F, Leandersson K, Ronnstrand L, Wernstedt C, Andersson T. p38-MAPK signals survival by phosphorylation of caspase-8 and caspase-3 in human neutrophils. J Exp Med. 2004;199:449–58. doi: 10.1084/jem.20031771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reynolds CH, Betts JC, Blackstock WP, Nebreda AR, Anderton BH. Phosphorylation sites on tau identified by nanoelectrospray mass spectrometry: differences in vitro between the mitogen-activated protein kinases ERK2, c-Jun N-terminal kinase and P38, and glycogen synthase kinase-3beta. J Neurochem. 2000;74:1587–95. doi: 10.1046/j.1471-4159.2000.0741587.x. [DOI] [PubMed] [Google Scholar]
- 27.Fan J, Zhang Q, Tochio H, Li M, Zhang M. Structural basis of diverse sequence-dependent target recognition by the 8 kDa dynein light chain. J Mol Biol. 2001;306:97–108. doi: 10.1006/jmbi.2000.4374. [DOI] [PubMed] [Google Scholar]
- 28.Day CL, Puthalakath H, Skea G, Strasser A, Barsukov I, Lian LY, et al. Localization of dynein light chains 1 and 2 and their pro-apoptotic ligands. Biochem J. 2004;377:597–605. doi: 10.1042/bj20031251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rapali P, Radnai L, Suveges D, Harmat V, Tolgyesi F, Wahlgren WY, et al. Directed evolution reveals the binding motif preference of the LC8/DYNLL hub protein and predicts large numbers of novel binders in the human proteome. PLoS ONE. 2011;6:e18818. doi: 10.1371/journal.pone.0018818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Labi V, Erlacher M, Kiessling S, Manzl C, Frenzel A, O’Reilly L, et al. Loss of the BH3-only protein Bmf impairs B cell homeostasis and accelerates gamma irradiation-induced thymic lymphoma development. J Exp Med. 2008;205:641–55. doi: 10.1084/jem.20071658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Owens TW, Valentijn AJ, Upton JP, Keeble J, Zhang L, Lindsay J, et al. Apoptosis commitment and activation of mitochondrial Bax during anoikis is regulated by p38MAPK. Cell Death Differ. 2009;16:1551–62. doi: 10.1038/cdd.2009.102. [DOI] [PubMed] [Google Scholar]
- 32.Wen HC, Avivar-Valderas A, Sosa MS, Girnius N, Farias EF, Davis RJ, et al. p38alpha Signaling induces anoikis and lumen formation during mammary morphogenesis. Sci Signal. 2011;4:ra34. doi: 10.1126/scisignal.2001684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shamas-Din A, Brahmbhatt H, Leber B, Andrews DW. BH3-only proteins: orchestrators of apoptosis. Biochim Biophys Acta. 2011;1813:508–20. doi: 10.1016/j.bbamcr.2010.11.024. [DOI] [PubMed] [Google Scholar]
- 34.Lomonosova E, Chinnadurai G. BH3-only proteins in apoptosis and beyond: an overview. Oncogene. 2008;27:S2–19. doi: 10.1038/onc.2009.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Singh PK, Roukounakis A, Frank DO, Kirschnek S, Das KK, Neumann S, et al. Dynein light chain 1 induces assembly of large Bim complexes on mitochondria that stabilize Mcl-1 and regulate apoptosis. Genes Dev. 2017;31:1754–69. doi: 10.1101/gad.302497.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singh PK, Roukounakis A, Weber A, Das KK, Sohm B, Villunger A, et al. Dynein light chain binding determines complex formation and posttranslational stability of the Bcl-2 family members Bmf and Bim. Cell Death Differ. 2020;27:434–50. doi: 10.1038/s41418-019-0365-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Han S, Ren Y, He W, Liu H, Zhi Z, Zhu X, et al. ERK-mediated phosphorylation regulates SOX10 sumoylation and targets expression in mutant BRAF melanoma. Nat Commun. 2018;9:28. doi: 10.1038/s41467-017-02354-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–32. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D Biol Crystallogr. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–21. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 42.The PyMOL Molecular Graphics System, Version 1.8 Schrödinger, LLC.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The crystallography data have been deposited to the Protein Data Bank (http://www.pdb.org) with PDB ID “7CNU”.