Abstract
Circular RNAs (circRNAs) have gained growing attention in participating in various biological processes and referring to multiply kinds of diseases. Although differentially expressed circRNA profiling in Alzheimer’s disease (AD) has been established, little is known about the precise characteristic and functions of key circRNAs with direct relevance to AD in gene expression and disease-related cognition. Herein, we screened and identified circCwc27 as a novel circRNA implicated in AD. CircCwc27 was a neuronal-enriched circRNA that abundantly expressed in the brain and significantly upregulated in AD mice and patients. Knockdown of circCwc27 markedly improved AD-related pathological traits and ameliorated cognitive dysfunctions. Mechanistically, we excluded the miRNA decoy mechanism and focused on the important function of circRNA-RNA-binding protein (RBP) interaction in AD. CircCwc27 directly bound to purine-rich element-binding protein A (Pur-α), increased retention of cytoplasmic Pur-α, and suppressed Pur-α recruitment to the promoters of a cluster of AD genes, including amyloid precursor protein (APP), dopamine receptor D1 (Drd1), protein phosphatase 1, regulatory inhibitor subunit1B (Ppp1r1b), neurotrophic tyrosine kinase, receptor, type 1 (Ntrk1), and LIM homeobox 8 (Lhx8). Downregulation of circCwc27 enhanced the affinity of Pur-α binding to these promoters, leading to altered transcription of Pur-α targets. Moreover, Pur-α overexpression largely phenocopied circCwc27 knockdown in preventing Aβ deposition and cognitive decline. Together, our findings suggest significant functional consequences of a circRNA–protein interaction, that circCwc27, by associating with the regulatory protein Pur-α, may act as a crucial player in AD pathogenesis and represent a promising AD therapeutic target with clinical translational potential.
Subject terms: Cognitive ageing, Neurological disorders
Introduction
Alzheimer’s disease (AD) is pathologically characterized by extracellular neuritic plaques and intracellular neurofibrillary tangles, as well as neuroinflammation and synaptic dysfunction in the cortices and hippocampi of patients [1, 2]. Although many efforts have been put to prevent or slow down the pathological process of AD, there are no effective treatments to prevent or cure this disease so far [3]. Therefore, further investigating the underlying mechanisms of AD and facilitating the development of new effective therapeutic strategies are extremely urgent. Actually, transcriptional regulation underlies the complexity of central nervous system (CNS), and its dysregulation may be a major cause of neurodegeneration. More than 90% of the transcriptional output of mammalian genome is non-coding RNAs (ncRNAs) [4], and these products, especially microRNAs (miRNAs) and long non-coding RNAs (lncRNAs), have been reported to participate in modulating AD pathology by regulating key genes of the disease such as APP, BACE1, or MAPT [5]. Nevertheless, the present knowledge of the regulatory mechanisms of circular RNAs (circRNAs) is limited, which restricts our understanding of brain transcriptome diversity and the discovery of effective therapeutic targets for AD.
CircRNAs are a class of novel non-coding RNAs that lack canonical 5′ caps and 3′ poly-A tails and form continuous loops by covalent bonds [6]. These endogenous RNAs have attracted growing attention recently due to their high homology, stability, and tissue/developmental-stage-expression specificity [7]. Multiple researchers have found diverse roles of circRNAs such as regulating transcription of parental genes in the nucleus [8], sponging miRNAs and RNA-binding proteins [9–12], and exhibiting translation potential to generate peptides in the cytoplasm [13, 14]. Dysregulation of circRNAs has been associated with different pathological processes, including myocardial infarction [15], diabetes mellitus [16], and various forms of cancer [17–20]. Compared with other organs, circRNAs are highly represented in mammalian brains and show functional roles in nervous development and synaptic activity [21, 22], which play a crucial part in some neuropsychiatric disorders [23, 24]. Possibly because of their absence of free hydroxyl ends conferring resistance to exonucleases, circRNAs exhibit incredible stability with a longer half-life than linear RNAs and tend to accumulate during brain aging [25, 26]. The stability and specific expression profile of circRNAs have strongly indicated these molecules as promising candidates for the treatment of age-related neurodegenerative diseases like AD. Recently, circRNAs have been revealed to be differentially expressed in AD patients and animal models compared with healthy controls [27–29], and a number of these circRNAs co-express with known AD-related genes and pathways, indicating that these dysregulated circRNAs are likely to be promising therapeutic targets of this most prevalent neurodegenerative disorder worldwide. However, the precise role of most circRNAs in AD progression remains largely elusive, and it is still unclear whether regulation of specific circRNAs in vivo can ameliorate AD-related histopathological traits and cognitive decline. Comprehensively identifying and validating the important function and regulatory mechanism of key circRNAs is of great significance for developing new therapeutic methods such as circRNA-associated nucleotide drug in AD.
In the present study, we found that a poorly characterized mammalian circRNA-circular RNA Cwc27 (circCwc27) owned highly abundant expression in neurons, and was upregulated in the brains of APP/PS1 mice, and also in the temporal cortex and plasma of AD patients compared with healthy controls. Knockdown of circCwc27 improved AD pathology and ameliorated cognitive deficits. Furthermore, we elucidated a novel RNA-binding protein-dependent regulatory axis, that is circCwc27 interacted with Pur-α and trapped it in the cytoplasm, resulting in its inactivation and transcriptional alteration of APP and other genes associated with AD. Taken together, we revealed a novel regulating axis that is consist of circCwc27 and Pur-α and suggested that circCwc27 may represent a novel therapeutic target for AD.
Methods and materials
Study approval and human plasma samples
This research protocol was approved by the ethics committee at Shanghai Mental Health Center, Shanghai Jiao Tong University School of Medicine. Participants or their legally authorized representatives provided written informed consent. Patients with AD (n = 6) were enrolled through Shanghai Mental Health Center. Healthy control subjects (n = 6) were recruited through network publicity and community outreach. Detailed information of patients and non-demented control is presented in Supplementary Table 1. All AD patients had no evidence of other neurological diseases based on neuropathological examination.
Human brain samples
Brain tissue was obtained from the Netherlands Brain Bank (NBB), Netherlands Institute for Neuroscience, Amsterdam (open access: www.brianbank.nl). All materials have been collected from donors for or from whom a written informed consent for a brain autopsy and the use of the material and clinical information for research purposes had been obtained by the NBB. Temporal cortical regions were used for our experiments. Non-demented control cases (n = 8) did not suffer from any psychiatric or metabolic diseases. AD cases (n = 13) had no evidence of other neurological diseases according to neuropathological examination. Detailed information of AD patients and non-demented control is presented in Supplementary Table 2.
Mice and ethics statement
APP/PS1 transgenic mice (expressing human mutant APP695 carrying the Swedish mutations and human PS1 mutations) and wild-type (WT) C57BL/6J mice were purchased from Nanjing Model Animal Resource Center (Nanjing, China) and randomly allocated to different groups. All mice used in our experiments were male. The mice were kept in cages under a constant temperature and humidity and were exposed to a 12 h day/12 h night cycle with free access to tap water and food. All animal experiments in this study were reviewed and approved by the Ethics Committee of Shanghai Jiao Tong University School of Medicine. The number of animals used for each experiment is shown in the figure legends. Investigators were blinded to the group allocation.
Dual-luciferase reporter assay
APP, Drd1, Ppp1r1b, Ntrk1, and Lhx8 promoters were amplified from gDNA using PCR primers and subcloned into pGL3-Basic luciferase vector (Promega, USA). HEK293T cells were transfected with promoters for 24 h and transduced with circCwc27/Pur-α lentivirus for another 24 h. Dual-luciferase assay was performed to assess the luciferase activity according to the manufacturer’s instructions (Promega, USA). Renilla luciferase activity was normalized to a firefly luciferase signal and used as an internal control. The primer sequences are presented in Supplementary Table 3.
BASE-scope
The brain tissues were frozen and sectioned on a cryostat at 20 μm section after dehydration, followed by being mounted on SuperFrost plus slides (Fisher Scientific, USA) and stored at −80 °C. Each section was incubated in 2–4 drops of H2O2 for 10 min at room temperature and then exposed to boiled target repair reagent for 5 min. After that, brain sections were incubated in proteinase plus solution in HybEZ™ II hybridization system (Advanced Cell Diagnostics, catalog no. 321720) for 30 min at 40 °C. Fluorescent Basescope-ISH was performed using a BaseScope™ Reagent Kit v2-RED according to the manufacture’s protocol (Advanced Cell Diagnostics, catalog no. 323900). The circCwc27 probe was purchased from Advanced Cell Diagnostics USA. The circCwc27 probe sequence is shown in Supplementary Table 4. Fluorescent images were captured using a Leica SP8 confocal microscope (Leica, Germany).
BASE-scope in combination with immunostaining
After the Basescope-ISH procedure, the sections were washed three times in wash buffer at room temperature. The sections were blocked with a solution of 5% BSA in phosphate-buffered saline (PBS) for 1 h at room temperature and then incubated with primary antibody overnight at 4 °C. Primary antibodies to the following proteins were used: NeuN (rabbit, 1:500, Abcam, 177487), GFAP (rabbit, 1:500, Abcam, ab7260), Iba-1 (rabbit, 1:500, Abcam, ab178846). The next day, after washing three times with PBS, the sections were incubated with Goat Anti-Rabbit IgG H&L (Alexa Fluor® 488) (1:500, Abcam, ab150077) for 1 h at room temperature. The sections were washed three times in PBS and mounted with Prolong gold anti-fade reagent containing DAPI (Lifetech, USA). Immunofluorescence images were captured using a Leica SP8 confocal microscope (Leica, Germany).
High-throughput screening analysis
Three samples from the hippocampus of APP/PS1 and another three samples from the hippocampus of WT mice were screened with circRNA high-throughput screening analysis performed by Aksomics Incorporation (Shanghai, China). Briefly, total RNA extracted from hippocampus was tested and quantified by agarosaccharide electrophoresis and Nanodrop, and then mRNA was enriched with oligo (dT) beads. The first strand of cDNAs was generated by the RNA fragments after the RNA was broken into pieces at a high temperature, and the second strand DNA was synthesized in presence of dUTP subsequently. The constructed libraries were quality detected by Agilent 2100 and quantified by qPCR. Illumina Hiseq 4000 was used to perform paired-end sequencing. The backsplice junctions were determined by comparing the trimmed data produced after quality control with the reference transcriptome. The transcripts levels were calculated by the fragments per kilobase per million reads (GEO accession number: GSE158995).
RNA pull down and mass spectrometry
Biotin-labeled oligonucleotide probes targeting junction sites of circRNAs were synthesized by SunBio Biomedical Technology Co., Ltd (Shanghai, China). Hippocampal tissue samples of APP/PS1 mice were ground in lysis buffer and incubated with the biotinylated probes. After that, the cell lysates were incubated with streptavidin-coated magnetic beads and the RNA–protein complex was pulled down. The retrieved proteins in the complex were analyzed by subsequent western blotting or mass spectrometry (SunBio Biomedical Technology Co., Ltd, Shanghai, China). The bound RNA in the pull-down was detected by PCR. The circRNA probe sequences are listed in Supplementary Table 4.
siRNA and plasmid transfection
CircRNAs (circCwc27 and CDR1AS), Flag-Pur-α WT, and truncated plasmids and siRNAs targeting Pur-α were obtained from Genomeditech Co., Ltd (Shanghai, China). Cells were transfected with plasmid or siRNA using Lipofectamine 3000 (Invitrogen, USA) according to the manufacturers’ instructions. The primer sequences are listed in Supplementary Table 3. The sequences of siRNAs are listed in Supplementary Table 4.
RNA-binding protein immunoprecipitation (RIP)
RIP experiments were performed using a Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, USA) as the protocol described. Briefly, hippocampal tissues were washed and homogenized by a ultrasonic homogenizer in ice-cold PBS. SH-SY5Y and HEK293T cells were harvested in 48 h after transfection. After centrifugation, the cell pellets were resuspended in an equal volume of RIP lysis buffer, followed by being incubated with Protein A/G agarose beads (Roche, USA) and corresponding specific antibodies (Pur-α, Santa Cruz Biotechnology, sc-130397; Ago2, Abcam, ab32381; HNRNPK, Abcam, ab52600; TDP-43, Abcam, ab190963; HNRNPL, Abcam, ab6106; G3BP2, Invitrogen, PA5-53797). The immunoprecipitated RNAs were detected by quantitative reverse transcriptase PCR (qRT-PCR) using specific primers. The primer sequences are shown in Supplementary Table 5.
Chromatin immunoprecipitation (ChIP)
ChIP was performed using the MAGnify™ Chromatin Immunoprecipitation System (Thermo Fisher Scientific, USA) following the manufacturer’s instructions. Briefly, the used antibodies were first coupled to Dynabeads. Cells were harvested and incubated with formaldehyde for 10 min at room temperature to crosslink and DNAs were shredded by sonicated. Sonicated chromatin extracts were then immunoprecipitated using the antibody specific for Pur-α (Santa Cruz Biotechnology, sc-130397) and Dynabeads® Protein A/G at 4 °C for 2 h. Mouse IgG antibody was used as the negative control antibody. After that, the bound chromatin was washed and the crosslinking was reversed using the reverse crosslinking buffer. DNAs were then extracted and purified, followed by being analyzed by qRT-PCR. The primer sequences are shown in Supplementary Table 5.
Real-time PCR
Total RNA from cell and mouse tissue samples was extracted using Trizol reagent (Invitrogen, 15596026), and total RNA of human plasma was extracted using the RNeasy Mini Kit (Qiagen, 74104) following the manufacturer’s instructions. Two micrograms of RNA was then reverse-transcribed into cDNA using PrimeScript™ RT Master Mix (TAKARA, RR036A) and quantified using TB Green™ Premix Ex Taq™ (TAKARA, RR042A). Quantitative real-time PCR was run and analyzed by mean of LightCycler480 System (Roche, Switzerland). GAPDH was used as a housekeeping gene and the relative levels were shown as fold changes relative to the corresponding control group. When detecting the nuclear and cytoplasmic circCwc27 levels, we separated the nuclear and cytoplasmic fractions by using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Scientific, USA), and then the RNA was extracted with Trizol. To determine the absolute quantity of circCwc27 per cell, circCwc27 was amplified from cDNA and cloned into pcDNA3.1. The plasmid was serially diluted to generate a standard curve, and copy counts were determined from standard curve analysis. The primer sequences are listed in Supplementary Table 5.
Statistical analysis
All data were analyzed using GraphPad Prism 7 and were presented as mean standard error of the mean (SEM). Two-sided unpaired Student’s t-test and one-way ANOVA followed by the Holm–Sidak test were used to compare the difference between two independent samples. The comparisons among groups were analyzed by two-way analysis of variance (ANOVA) followed by Bonferroni’s post hoc multiple comparison tests. The correlations were calculated by Pearson correlation analysis. The statistical analyses used for the different experiments are described in the respective figure legends. All statistical analyses were deemed significant when P < 0.05. Sample size was determined by our previous works in reasonable sample sizes for different experiments.
Results
CircCwc27 is upregulated in AD
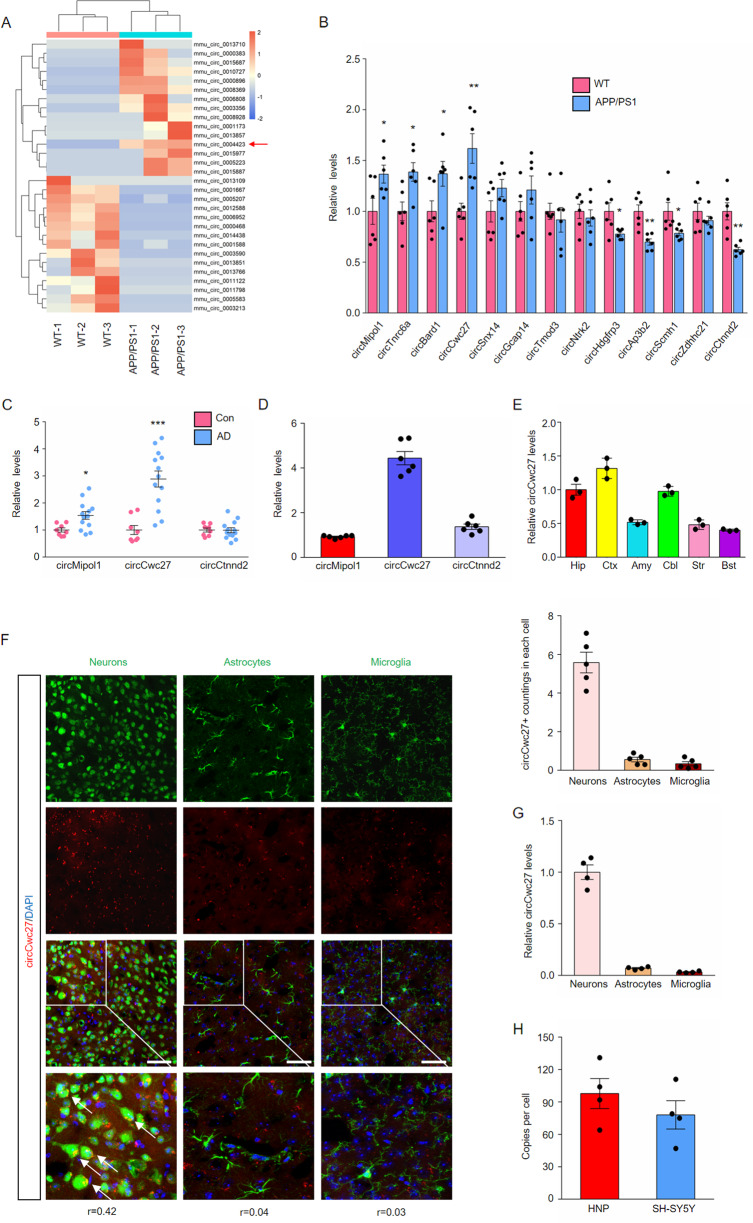
Changes in circRNA expression are found in the early stage of AD, and these dysregulated circRNAs may directly contribute to the disease [27]. Therefore, we aimed to screen the promising circRNA in the early stage of AD. We collected hippocampus from 6-month APP/PS1 transgenic and WT control mice, employed deep circRNA sequencing, and found that a total of 131 circRNAs were significantly dysregulated (fold change >1.5, P < 0.05), and the top 15 most upregulated and downregulated circRNAs are shown in the heatmap (Fig. 1A). Among these 30 circRNAs, 13 circRNAs were identified conserved between human and mouse by Rybak-Wolf et al. [21] (Supplementary Table 6), among which 4 circRNAs were significantly upregulated and 4 circRNAs significantly downregulated in the hippocampus of APP/PS1 mice compared to WT mice (Fig. 1B). We detected these eight circRNA levels in the plasma and temporal cortex of AD patients, and compared their relative abundance in the hippocampus of WT mice. Notably, circCwc27 (mmu_circ_0004423) was detectable and significantly elevated in the temporal cortex and plasma of AD patients as we had observed in the hippocampus of APP/PS1 mice (Fig. 1C and Supplementary Fig. 1A–H), and had the highest expression in the hippocampus among these circRNAs (Fig. 1D). The expression of linear Cwc27 stayed unchanged in both cortex and hippocampus (Supplementary Fig. 2A). Consequently, functional characterization of circCwc27 in AD became the focal point in our current study.
Fig. 1. CircCwc27 is a highly abundant and stable circRNA, that is upregulated in AD.
A A heatmap shows the top 15 most upregulated and top 15 most downregulated circRNAs in three samples of hippocampus from 6-month APP/PS1 vs WT mice by circRNA sequencing (orange color scale = higher expression. Blue color scale = lower expression. n = 3 samples/group. P value is determined by using Student’s t-test and corrected for multiple testing by using the Benjamini and Hochberg method). B qRT-PCR analysis was used to determine the expression levels of indicated circRNAs in the hippocampus from 6-month WT and APP/PS1 mice. n = 6. *P < 0.05, **P < 0.01 versus the WT group using Student’s t-test. C The expression of indicated circRNAs in the temporal cortex of healthy controls and AD patients were analyzed by qRT-PCR. n(Control) = 8, n(AD) = 13. *P < 0.05, ***P < 0.001 versus control group using Student’s t-test. D Comparing the relative abundance of indicated circRNAs in the hippocampus of WT mice by qRT-PCR. n = 6. E qRT-PCR was performed to determine the relative expression of circCwc27 in Hip (Hippocampus), Ctx (Cortex), Amy (Amygdala), Cbl (Cerebellum), Str (Striatum), Bst (Brainstem). n = 3. F BASE-scope in combination with immunostaining was performed to assess the colocalization between circCwc27 (red) and neurons (NeuN), astrocytes (GFAP), and microglia (Iba-1) in sections of cortex of APP/PS1 mice. The boxed areas were zoomed in the under panels. Nuclei were stained with DAPI (blue). The corresponding colocalization coefficients (Pearson’s R value) between circCwc27 and each cellular marker are shown in the merged images. The average circCwc27 + signal counts per neural cell are shown in the right. n = 5. Scar bar: 50 μm. G Relative expression of circCwc27 in day 14 mouse cortical neurons, astrocytes, and microglia was detected by qRT-PCR. n = 4. H Absolute quantification of circCwc27 copy number per cell, determined by qRT-PCR, in human neurons-hippocampal (HNP) and SH-SY5Y cells. Copy number was calculated from standard curves prepared from serial dilutions of circCwc27 oligonucleotide standards with known concentrations. n = 4. All data in the figure are shown as mean ± SEM.
CircCwc27 was derived from exon 8, 9, and 10 of the Cwc27 gene (Supplementary Fig. 2B) and had 79.2% nucleotide identity between human and mouse (Supplementary Fig. 2C). Head-to-tail junction-specific primers could only amplify circCwc27 in cDNA but not in genomic DNA, confirming that these RNA molecules had circular structures (Supplementary Fig. 2D). PCR products were further analyzed by Sanger sequencing and the results were completely in accord with the circCwc27 sequence presented in circBase (Supplementary Fig. 2E). Besides, the expression of circCwc27 was the highest in the brain instead of other organs such as heart, liver, spleen, lung, and kidney (Supplementary Fig. 2F). Cerebral circRNA expression usually depends on developmental age [30], but there was only a slight increase of circCwc27 levels in the hippocampus of WT mice from 3 to 12 months (Supplementary Fig. 2G). However, circCwc27 started to increase at 3 months before the onset of Aβ burden, and significantly upregulated with age in APP/PS1 mice (Supplementary Fig. 2G). Taken together, these results indicated that circCwc27 might participate in the pathogenesis of AD and have clinical significance.
CircCwc27 is an abundant, stable circRNA that mainly localized in the cytoplasm of neuron
To better characterize the role of circCwc27, BASEscope-ISH and qRT-PCR assays were used to assess the expression of circCwc27 in the brain. We observed that circCwc27 was highly represented in the cortex and hippocampus, two vulnerable cerebral regions in AD (Fig. 1E and Supplementary Fig. 4A). Moreover, circCwc27 was mainly expressed in neurons rather than glial cells in the brain of both APP/PS1 mice and WT mice (Fig. 1F, G and Supplementary Fig. 3A, B). Absolute copy number of circCwc27 was also detected, and the results showed that an average of 98 copies of circCwc27 were presented in every human hippocampal neuron and 78 copies of circCwc27 were presented in every SH-SY5Y cell (Fig. 1H). BASEscope-ISH in combination with immunostaining assay further confirmed the upregulation of neuronal circCwc27 in APP/PS1 mice (Supplementary Fig. 4B).
Next, we investigated the stability and subcellular location of circCwc27 in vitro. RNase R [31] resistance of circCwc27 was quantified by qRT-PCR, and circCwc27 was much more resistant to RNase R digestion than linear Cwc27 (Supplementary Fig. 4C). Similar to this, circCwc27 was more stable than linear Cwc27 in SH-SY5Y cells treated with actinomycin D (Supplementary Fig. 4D). Moreover, circCwc27 was predominantly expressed in the cytoplasm (Supplementary Fig. 4E, F). Taken together, we demonstrated that circCwc27 was a circRNA with high abundance and stability that mainly localized in the cytoplasm of neurons.
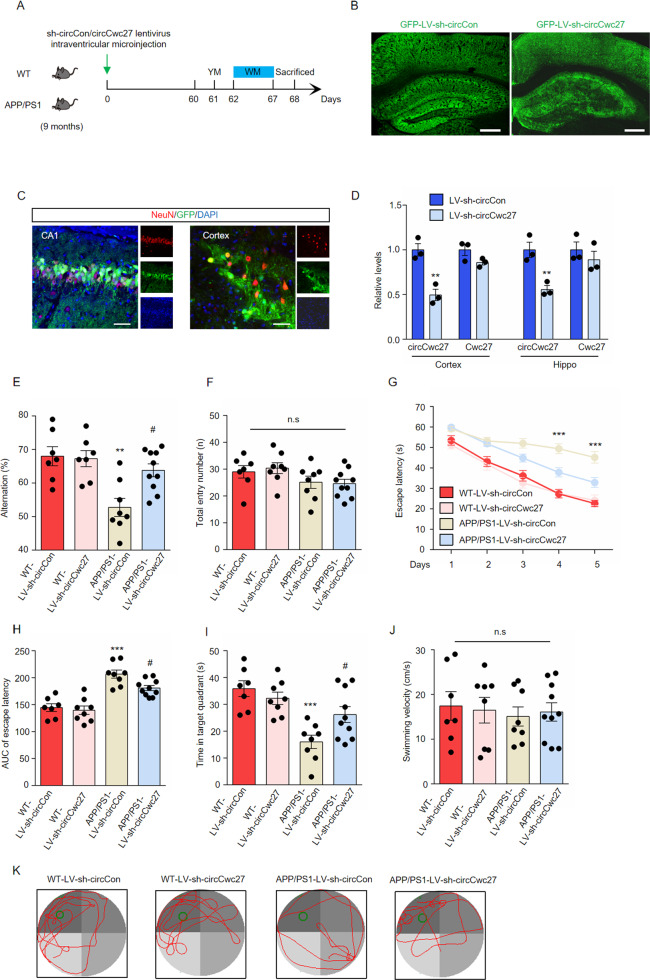
Knockdown of circCwc27 ameliorates cognitive decline in the APP/PS1 mouse
To determine whether the increase of circCwc27 influenced behavioral deficits and pathological traits in AD, we microinjected lentiviral preparations expressing either shRNA circCon-GFP (LV-sh-circCon) or shRNA circCwc27-GFP (LV-sh-circCwc27) into the lateral ventricle of 9-month-old mice and analyzed their memory and spatial learning ability after 2 months (Fig. 2A). Green fluorescent protein (GFP) signal was largely distributed and co-localized with neurons in the whole cortex and hippocampus (Fig. 2B, C). CircCwc27 expression was significantly downregulated while Cwc27 levels kept unchanged after lentiviral transduction (Fig. 2D). Y maze spontaneous alternation behavioral tests showed that APP/PS1 mice microinjected with LV-sh-circCon showed an obvious decline in alternation whereas circCwc27 inhibition significantly prevented this decline (Fig. 2E). No deficits in motor function were supported by the same total entry numbers among groups (Fig. 2F).
Fig. 2. Knockdown of circCwc27 improves spatial learning and memory in the APP/PS1 mouse model.
A Timeline of experimental procedure in our research. Spatial learning and memory ability were examined 60 days after lentivirus injection. B Confocal images of brain sections injected with lentivirus were shown, and GFP (green) was used to visualize viral diffusion. Scale bar, 200 μm. C Colocalization of NeuN (red) and GFP (green) in the CA1 region and cortex of injected mice. Nuclei were stained with DAPI (blue). Scale bar, 50 μm. D qRT-PCR was used to determine the expression of circCwc27 and Cwc27 in the cortex and hippocampus after LV-sh-circCon or LV-sh-circCwc27 injection. n = 3. **P < 0.01 versus corresponding LV-sh-circCon group. E, F Y maze spontaneous alternation behavioral test was performed in wild type (WT) or APP/PS1 mice 2 months after injection of either control LV-sh-circCon or LV-sh-circCwc27 (n = 7 mice for WT-sh-circCon, n = 8 mice for WT-sh-circCwc27, n = 8 mice for APP/PS1-sh-circCon, and n = 10 mice for APP/PS1-sh-circCwc27). The percentage of spontaneous alternations (E) and total entries (F) were measured. **P < 0.01 versus WT-LV-sh-circCon group; #P < 0.05 versus APP/PS1-LV-sh-circCon group. Data are analyzed with two-way ANOVA followed by Bonferroni test. G–K Morris water maze (MWM) task was carried out to assess spatial learning and memory of WT-sh-circCon (n = 7), WT-sh-circCwc27 (n = 8), APP/PS1-sh-circCon (n = 8), and APP/PS1-sh-circCwc27 (n = 10) mice. Data are analyzed with two-way ANOVA followed by Bonferroni test. G Latency to escape to a hidden platform in the MWM task during a 5-day training phase. Day4: APP/PS1-sh-circCwc27 vs. APP/PS1-sh-circCon; ***P < 0.001, Day5: APP/PS1-sh-circCwc27 vs. APP/PS1-sh-circCon; ***P < 0.001. H The area under the curve (AUC) of escape latency during the training phase. ***P < 0.001 versus WT-LV-sh-circCon group; #P < 0.05 versus APP/PS1-LV-sh-circCon group. I Probe trial was performed 24 h after the last training day and the time spent in the quadrant containing the hidden platform was recorded. ***P < 0.001 versus WT-LV-sh-circCon group; #P < 0.05 versus APP/PS1-LV-sh-circCon group. J The swimming velocity of each group was recorded in the probe trial. K Representative swimming trajectories from different group of mice were shown. The green circle represented the hidden platform. All data in the figure are shown as mean ± SEM.
We also performed the Morris water maze task to assess spatial learning and memory of mice. During the training phase, LV-sh-circCon-microinjected APP/PS1 mice exhibited an obviously longer latency to find the hidden platform, while LV-sh-circCwc27 microinjection improved learning ability (Fig. 2G, H). In the probe trials, memory retention was measured by the time spent in the quadrant containing the hidden platform. Our results suggested that downregulation of circCwc27 improved spatial memory in APP/PS1 mice (Fig. 2I, K). All groups of mice had similar swimming velocity in our experiments (Fig. 2J). Together, these data indicated that knockdown of circCwc27 largely ameliorated cognitive decline in AD.
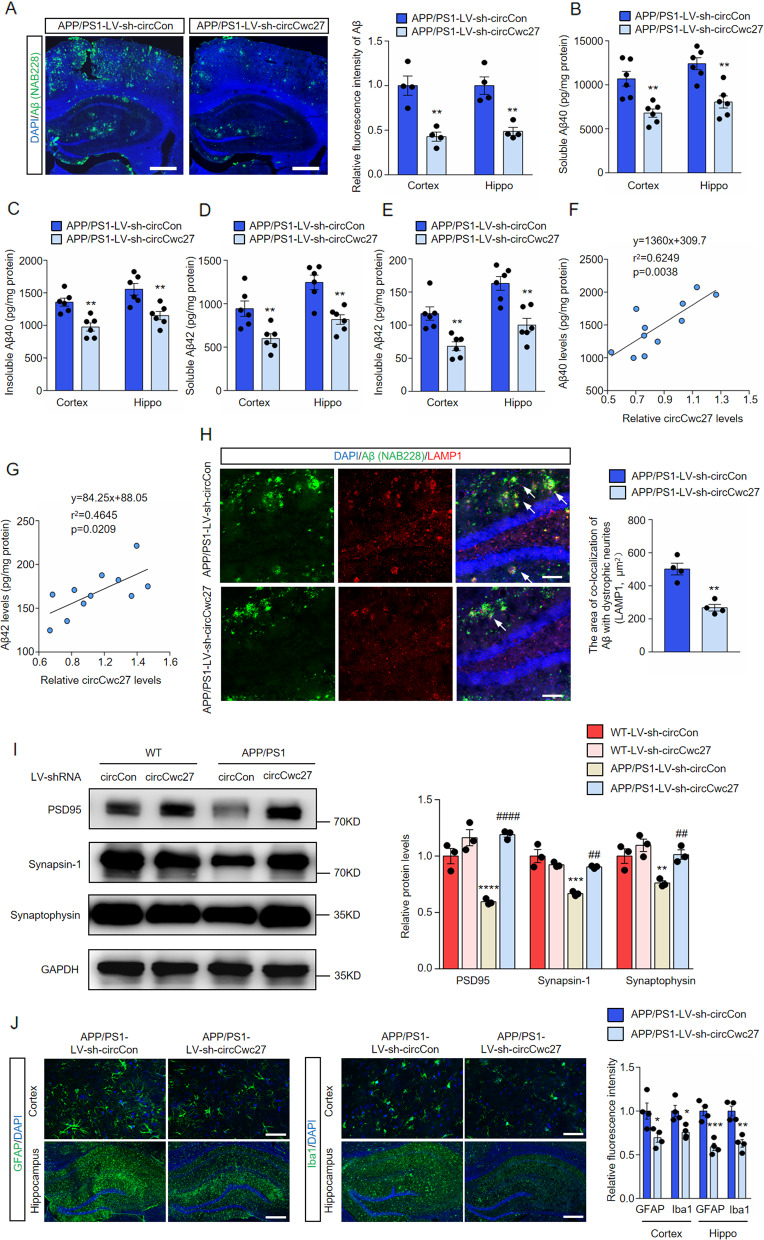
Downregulation of circCwc27 prevents Aβ deposition and related neurodegenerative effects in APP/PS1 mice
After behavioral tests, we further unraveled the influence of circCwc27 in AD pathology. We stained brain sections of APP/PS1 mice with anti-Aβ antibody (NAB228) and showed that LV-sh-circCwc27 microinjection obviously reduced plaque burden (Fig. 3A). We also showed a consistent reduction of cortical and hippocampal soluble and insoluble Aβ40 and Aβ42 after LV-sh-circCwc27 injection (Fig. 3B–E). Furthermore, we observed a positive correlation between the expression of circCwc27 and Aβ40 (Fig. 3F) or Aβ42 levels (Fig. 3G) in a cohort of APP/PS1 mice. Given the protective effects of LV-sh-circCwc27 injection on plaque deposition, we examined whether circCwc27 knockdown affects axonal dystrophy by immunostaining with an antibody against LAMP1, since LAMP1-positive dystrophic neurites are closely related to Aβ plaques in AD mouse models [32]. We showed that the area of colocalization of Aβ with dystrophic neurites was significantly reduced in the hippocampus of LV-sh-circCwc27-microinjected mice (Fig. 3H).
Fig. 3. Knockdown of circCwc27 improves AD pathology in APP/PS1 mice.
A Confocal images showing DAPI (blue) for nuclei, NAB228 (green) for an amyloid plaque in the cortex, and hippocampus from the APP/PS1 mice injected with LV-sh-circCon or LV-sh-circCwc27. Scar bar: 200 μm. Amyloid plaque deposition was quantified and normalized to APP/PS1-LV-sh-circCon group. n = 4. **P < 0.01 versus APP/PS1-LV-sh-circCon group using Student’s t-test. B–E Soluble and insoluble Aβ40 and Aβ42 levels in cortical and hippocampal tissues of APP/PS1-LV-sh-circCon or APP/PS1-LV-sh-circCwc27 injected mice were quantified by ELISA. n = 6. **P < 0.01 versus APP/PS1-LV-sh-circCon group using Student’s t-test. F The expression of Aβ40 and circCwc27 in the hippocampus of APP/PS1 mice were determined by ELISA and qRT-PCR, respectively. Scatter plot of Aβ40 versus circCwc27 levels was shown. The data were analyzed with a linear regression method. n = 11. G The expression of Aβ42 and circCwc27 in the hippocampus of APP/PS1 mice were determined by ELISA and qRT-PCR, respectively. Scatter plot of Aβ42 versus circCwc27 levels was shown. The data were analyzed with a linear regression method. n = 11. H Confocal images showing DAPI (blue) for nuclei, NAB228 (green) for an amyloid plaque, and LAMP1 (red) for dystrophic neurites in the hippocampus from the APP/PS1 mice injected with LV-sh-circCon or LV-sh-circCwc27. Scar bar: 50 μm. The area of Lamp1-positive dystrophic neurites in each plaque was quantified. n = 4. **P < 0.01 versus APP/PS1-LV-sh-circCon group using Student’s t-test. I Immunoblotting analysis and quantification of MAP2, PSD95, Synapsin-1, and Synaptophysin in cortex of mice injected with LV-sh-circCon or LV-sh-circCwc27. GAPDH was used as an inner control. n = 3. **P < 0.01, ***P < 0.001, ****P < 0.0001 versus WT-LV-sh-circCon group; ##P < 0.01, ####P < 0.0001 versus APP/PS1-LV-sh-circCon group. Data are analyzed with two-way ANOVA followed by Bonferroni test. J Representative immunofluorescence images and quantification of GFAP and Iba-1 in the cortex (upper row) and hippocampus (lower row) of APP/PS1 mice injected with LV-sh-circCon or LV-sh-circCwc27. Nuclei were stained with DAPI (blue). Scar bar: 50 μm for (upper row) and 200 μm for (lower row). n = 4. *P < 0.05, **P < 0.01, ***P < 0.001 versus APP/PS1-LV-sh-circCon group using Student’s t-test. All data in the figure are shown as mean ± SEM.
We also determined the role of circCwc27 on synaptic integrity since downregulation of circCwc27 confers protection against cognitive defects in AD (Fig. 2). LV-sh-circCwc27 microinjection did not affect the expression of synapse-associated proteins [33, 34] in WT mice, while markedly prevented their decrease in APP/PS1 mice (Fig. 3I). These findings were further confirmed by immunofluorescent staining of mice brain sections (Supplementary Fig. 5A).
Neuroinflammation is another important pathological hallmark in AD [2, 35]. We showed that LV-sh-circCwc27 microinjection significantly reduced the activation of microglia and astrocytes in APP/PS1 mice (Fig. 3J). In addition, we profiled some cytokines implicated in neuroinflammation by Quantibody® Mouse Interleukin Array and showed a dramatic decrease in the expression of several well-known pro-inflammatory markers in LV-sh-circCwc27-microinjected APP/PS1 mice (Supplementary Fig. 5B, C). In concordance with what we observed in protein assay, downregulation of circCwc27 led to significant reduction of some glial cell activation markers and pro-inflammatory cytokines at transcriptional level (Supplementary Fig. 5D). The above results demonstrated that knockdown of circCwc27 exerted a protective role in the neuroinflammatory and neurodegenerative effects of AD.
CircCwc27 directly binds to Pur-α protein and influences its distribution
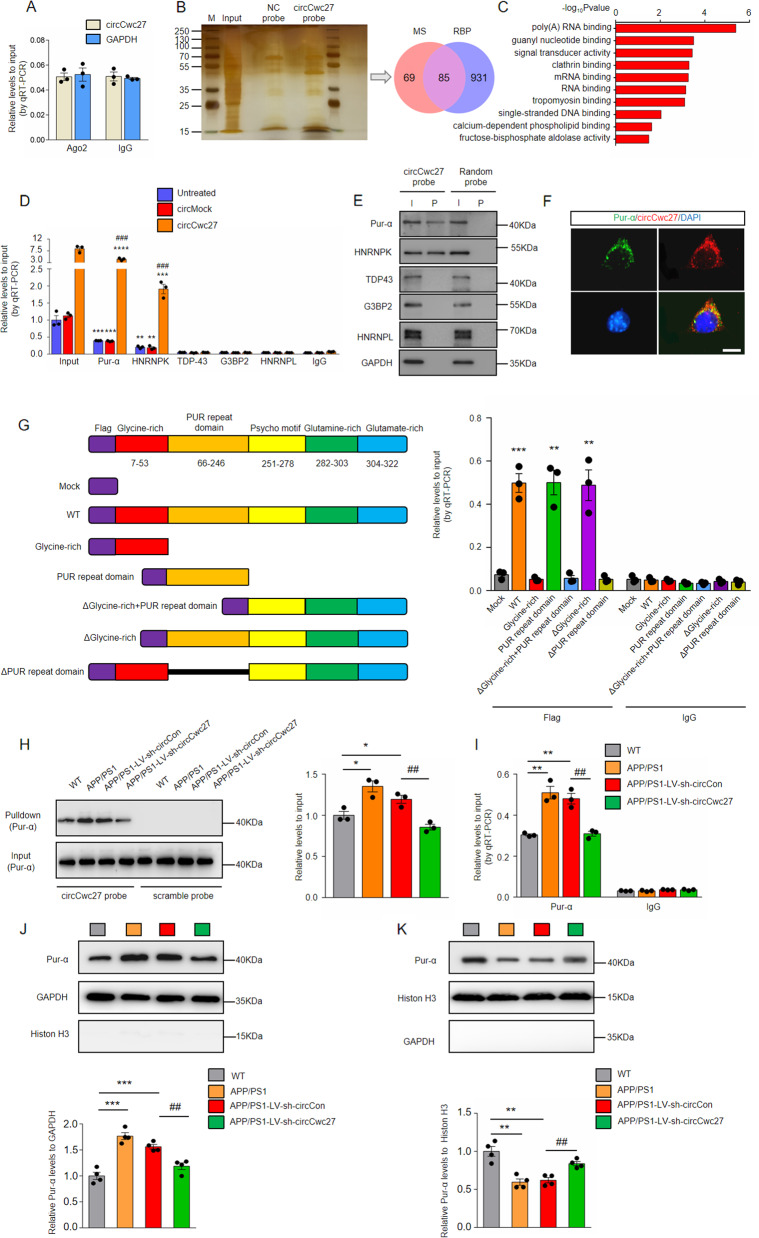
We next aimed to elucidate the molecular mechanism circCwc27 causing AD pathology. In order to explore the possibility of circCwc27 translation [13], we utilized Regrna 2.0 (http://regrna2.mbc.nctu.edu.tw/) to analysis if there was an open reading frame (ORF) in the sequence of circCwc27. The results showed that circCwc27 did not contain an ORF, suggesting that circCwc27 was not able to encode for short peptides. Considering that accumulating evidence has revealed the ability of cerebral circRNAs sponging miRNAs in the cytoplasm [9, 24, 36], we tested whether cytoplasmic circCwc27 could bind to miRNAs in neurons. AGO2, a core component of the RNA-induced silencing complex, is requisite for circRNAs to sponge miRNAs [10]. We performed RNA immunoprecipitation (RIP) and observed that endogenous circCwc27 was not enriched upon AGO2 (Fig. 4A), suggesting that circCwc27 does not act as a miRNA sponge. Besides interacting with miRNAs, circRNAs also function by binding to RNA-binding proteins in the brain [23, 37]. We therefore performed RNA pull-down assays followed by mass spectrometry (MS) analysis to search for potential circCwc27-interacting proteins in the hippocampus of APP/PS1 mice (Fig. 4B). A total of 154 proteins were pulled down by circCwc27 junction probe, and these proteins were mostly enriched in the brain (Supplementary Fig. 6A). Gene Ontology (GO) enrichment analysis showed that most of these proteins were associated with RNA/DNA binding (Fig. 4C).
Fig. 4. CircCwc27 interacts with Pur-α protein and influences its distribution.
A RNA-binding protein immunoprecipitation (RIP) assays were carried out in SH-SY5Y cells using anti-AGO2 or IgG control antibodies, and circCwc27 levels were quantified using qRT-PCR. n = 3. Data were analyzed with Student’s t-test. B Silver staining followed by mass spectrometry assay was used to identify the circCwc27-protein complex pulled down by circCwc27 probe in protein extracts from hippocampus tissues of APP/PS1 mice. RNA-binding proteins (RBPs) were further identified according to RBPDB (http://rbpdb.ccbr.utoronto.ca). MS mass spectrometry, RBP RNA-binding protein. C GO functional categories of the proteins pulled down by circCwc27. D RIP and qRT-PCR assays were performed to assess the interaction of Pur-α, HNRNPK, TDP43, G3BP2, HNRNPL with circCwc27 in HEK293T cells that transfected with circMock or circCwc27. IgG was used as a negative control. n = 3. **P < 0.01, ***P < 0.001, ****P < 0.0001 versus IgG group; ###P < 0.001 versus circMock group using Student’s t-test. E Pur-α, HNRNPK, TDP-43, G3BP2, and HNRNPL levels were detected by immunoblotting after pulled down by circCwc27 probe or scramble probe in SH-SY5Y cells. The experiments were repeated three times independently with similar results. I input, P pulldown. F Confocal image showed that the endogenous circCwc27 (red) and Pur-α (green) were mainly bound in the cytoplasm in SH-SY5Y cells. Nuclei were stained with DAPI (blue). The experiments were repeated three times independently with similar results. Scar bar: 25 μm. G Left, a summary of Pur-α truncations. Right, relative enrichment of circCwc27 levels associated with WT or truncated Pur-α. IgG was used as a negative control. n = 3. **P < 0.01, ***P < 0.001 versus IgG group using Student’s t-test. H Immunoblotting assays were performed to assess Pur-α expression in the hippocampus of WT, APP/PS1, APP/PS1-LV-sh-circCon, and APP/PS1-LV-sh-circCwc27 mice following biotinylated circCwc27 probe pull-down assay. n = 3. *P < 0.05 versus WT group. ##P < 0.01 versus APP/PS1-LV-sh-circCon group. Data are analyzed with one-way ANOVA. I RIP and qRT-PCR assays were performed to assess the interaction of Pur-α with circCwc27 in the hippocampus of WT, APP/PS1, APP/PS1-LV-sh-circCon, and APP/PS1-LV-sh-circCwc27 mice. IgG was used as a negative control. n = 3. **P < 0.01 versus WT group. ##P < 0.01 versus APP/PS1-LV-sh-circCon group. Data are analyzed with one-way ANOVA. All data in the figure are shown as mean ± SEM. J, K Cytoplasmic and nuclear proteins were separated and the levels of Pur-α in the cytoplasm (J) and nucleus (K) were determined by immunoblotting. n = 4. **P < 0.01, ***P < 0.001 versus WT group. ##P < 0.01 versus APP/PS1-LV-sh-circCon group. Data are analyzed with one-way ANOVA.
We found 85 of 154 proteins pulled down by circCwc27 were RNA-binding proteins (RBPs) according to RBPDB (http://rbpdb.ccbr.utoronto.ca) (Fig. 4B), and selected five RBPs that displaying maximum peptide counts for further study. RIP assay showed that circCwc27 was pulled down by antibodies against Pur-α and HNRNPK, rather than the other three candidates in SH-SY5Y cells (Fig. 4D). Moreover, Pur-α and HNRNPK were able to pull down significantly higher levels of circCwc27 from the circCwc27-transfected cells than those from the circMock-transfected cells (Fig. 4D). Importantly, RIP assay suggested that Pur-α showed the greatest binding capacity to circCwc27, which was consistent with the MS results (Pur-α displays the most peptide counts of all proteins pulled down by circCwc27), indicating a significant role of Pur-α in mediating circCwc27 functions (Fig. 4D). RNA pull-down results also validated the direct interaction of circCwc27 with Pur-α [38, 39] (Fig. 4E). In contrast, Pur-α antibody could not pull down CDR1AS, another brain enriched circRNA (Supplementary Fig. 6B). We further validated the endogenous colocalization between circCwc27 and Pur-α (Fig. 4F). Besides, Pur-α was highly expressed in neurons, also supporting the possible interaction between circCwc27 and Pur-α (Supplementary Fig. 6C). Previous reports have shown that glycine-rich domain and PUR repeat domain of Pur-α could be responsible for recruiting RNA molecules [40], we therefore constructed Flag-labeled Pur-α full-length WT and truncated plasmids. The results of RIP assays indicated that PUR repeat domain (66–246aa) but not other domains directly bound to circCwc27 (Fig. 4G). Of note, PUR repeat domain is 100% identical in amino acid sequence between human and mouse [41, 42]. Our data collectively indicated that circCwc27 directly bound to Pur-α protein in neurons.
We next investigated the regulating role of circCwc27 on Pur-α. Downregulation of circCwc27 did not influence total Pur-α mRNA and protein levels (Supplementary Fig. 6D, E), but the levels of Pur-α precipitated by circCwc27 was significantly upregulated in the hippocampus of APP/PS1 and APP/PS1-LV-sh-circCon mice compared with WT mice (Fig. 4H). CircCwc27 knockdown in APP/PS1 mice caused a significant decrease in the binding of circCwc27 and Pur-α (Fig. 4H). Similar results were obtained in RIP assays (Fig. 4I). Because circCwc27 was mainly localized in the cytoplasm and was upregulated in AD, we explored whether increased interaction between circCwc27 and Pur-α influenced the cytoplasmic and nuclear distribution of Pur-α in AD. Immunoblotting results showed that the expression of Pur-α was notably decreased in the nucleus but increased in the cytoplasm in APP/PS1 mice (Fig. 4J, K). Injection of LV-sh-circCwc27 significantly promoted nuclear distribution of Pur-α (Fig. 4J, K). By confocal microscopy in vitro, we also detected mainly cytoplasmic Pur-α in APPswe-SH-SY5Y cells and nuclear translocation of Pur-α after circCwc27 siRNA transfection (Supplementary Fig. 6F).
Previous studies have shown that Pur-α regulated cytoplasmic stress granule dynamics upon cellular stress [43]. We assessed the colocalization of Pur-α or circCwc27 with a known stress granule marker-G3BP in SH-SY5Y cells. The colocalization coefficient between Pur-α and G3BP is 0.65 (analyzed by Coloc 2), much higher than that between circCwc27 and G3BP (the colocalization coefficient between circCwc27 and G3BP is 0.41, that between circCwc27 and Pur-α is 0.81) (Supplementary Fig. 7A, B). In addition, upregulation of circCwc27 does not influence the expression of G3BP in SH-SY5Y cells (Supplementary Fig. 7C). These results indicate that circCwc27 may not directly cause the formation of stress granule in SH-SY5Y cells. Furthermore, there is no evidence that Pur-α participates in the formation of Aβ-mediated stress granule. Therefore, we suppose that the relocalization of Pur-α is directly influenced by circCwc27 in our system. Together, our results suggested that the increased circCwc27 interacted with Pur-α and trapped it in the cytoplasm, while knockdown of circCwc27 promoted the nuclear translocation of Pur-α.
Pur-α reduces Aβ load and prevents cognitive decline in AD
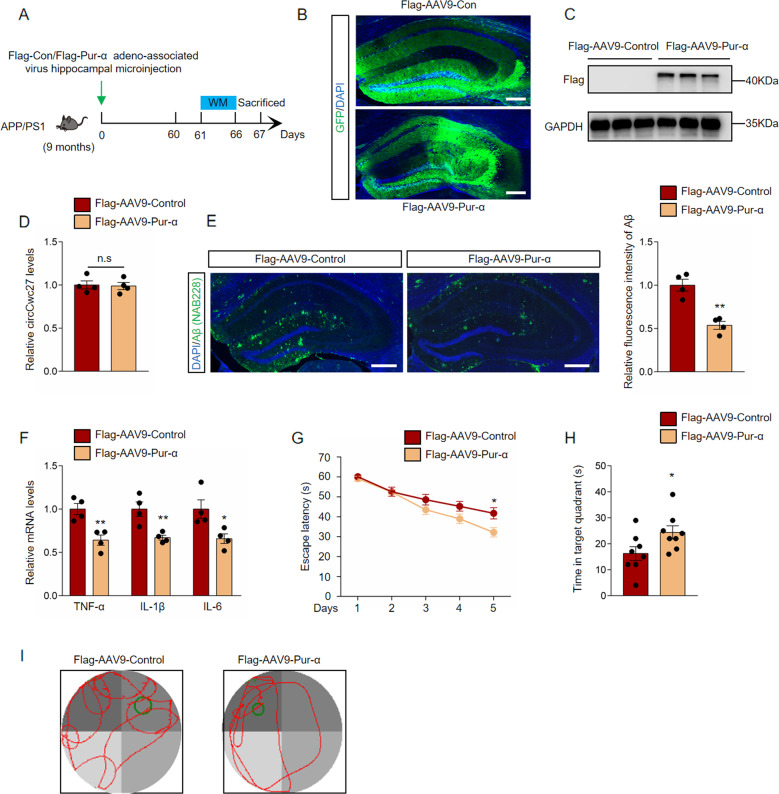
Emerging evidence has suggested that Pur-α plays a crucial part in amyotrophic lateral sclerosis and frontotemporal dementia [43, 44]. But little is known about the role of Pur-α in AD. Since Pur-α was a direct target of circCwc27, we microinjected adeno-associated virus preparations carrying either Flag-labeled Control-GFP (Flag-AAV9-Control) or Pur-α (Flag-AAV9-Pur-α) into the hippocampus of APP/PS1 mice to investigate the effect of Pur-α on AD pathology. Two months after stereotactic injection, the presence of marked green fluorescence and detectable Flag expression at about 42 kDa in the hippocampus indicated an effective transduction (Fig. 5A–C). Notably, circCwc27 expression was unchanged in Pur-α-overexpressed brains (Fig. 5D). Compared with the control group, overexpression of Pur-α significantly reduced Aβ deposition and pro-inflammatory cytokines in the hippocampus of APP/PS1 mice (Fig. 5E, F). Flag-AAV9-Pur-α microinjection largely protected APP/PS1 mice from severe deficits in memory formation (Fig. 5G–I). Collectively, these results revealed that Pur-α could reduce Aβ deposition and rescue spatial memory impairments.
Fig. 5. Pur-α reduces Aβ deposition and prevents cognitive decline in AD.
A Timeline of experimental procedure in our research. B Confocal images of hippocampus were shown, and GFP (green) were used to visualize viral diffusion. Scale bar, 200 μm. C Flag levels were determined by immunoblotting in the hippocampus of APP/PS1 mice injected with Flag-AAV9-Control or Flag-AAV9-Pur-α. GAPDH was used as an inner control. n = 3. D qRT-PCR was used to detect the relative circCwc27 levels in the hippocampus of Flag-AAV9-Control or Flag-AAV9-Pur-α injected mice. n = 4. E Confocal images indicating DAPI (blue) for nuclei, NAB228 (green) for an amyloid plaque in the hippocampus from the APP/PS1 mice injected with Flag-AAV9-Control or Flag-AAV9-Pur-α. Scar bar: 200 μm. Amyloid plaque deposition was quantified and normalized to Flag-AAV9-Control group. n = 4. **P < 0.001 versus APP/PS1-Flag-AAV9-Control group using Student’s t-test. F qRT-PCR was used to detect cytokines levels in the hippocampus of APP/PS1-Flag-AAV9-Control or APP/PS1-Flag-AAV9-Pur-α injected mice. n = 4. *P < 0.05, **P < 0.01 versus APP/PS1-Flag-AAV9-Control group using Student’s t-test. G Latency to escape to a hidden platform in the MWM task during a 5-day training phase. Day 5: APP/PS1-Flag-AAV9-Pur-α (n = 8) versus APP/PS1-Flag-AAV9-control group (n = 8); *P < 0.05. Data are analyzed with Student’s t-test. H Probe trial was performed 24 h after the last training day and the time spent in the quadrant containing the hidden platform was recorded. n = 8. *P < 0.05 versus APP/PS1-Flag-AAV9-Control group using Student’s t-test. I Representative swimming trajectories from different groups of mice were shown. The green circle represented the hidden platform. All data in the figure are shown as mean ± SEM.
CircCwc27 regulates AD genes transcription by binding to Pur-α
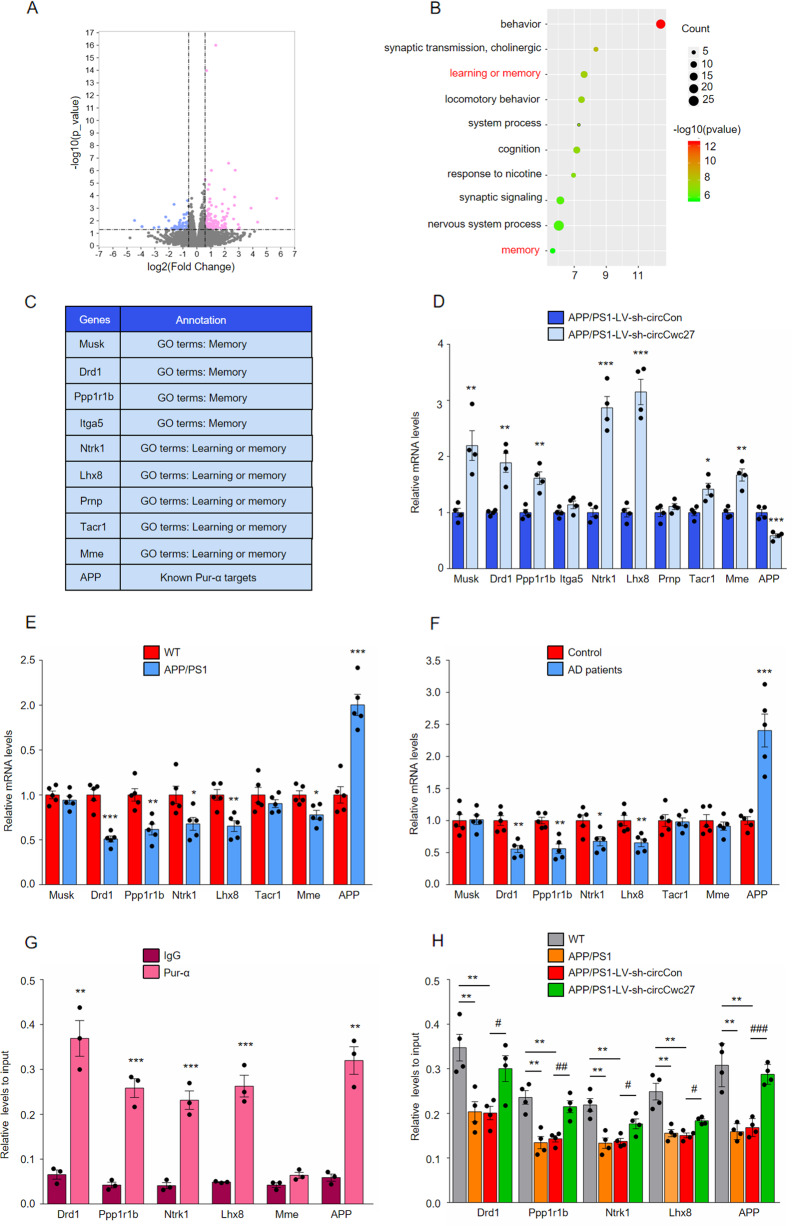
Previous studies have reported that Pur-α contributed to memory retention and learning ability [45, 46], and negatively controlled APP expression [47]. Combined with our results that circCwc27 bound to Pur-α, and overexpressing Pur-α largely phenocopied circCwc27 knockdown in improving Aβ burden and cognitive decline (Fig. 5), we supposed that Pur-α was a critical downstream mediator of circCwc27 in regulating Aβ and memory-related genes in AD. To address this hypothesis, we performed RNA-seq in the hippocampus of LV-sh-circCon-microinjected and LV-sh-circCwc27-microinjected APP/PS1 mice and identified 339 genes that were differentially expressed (fold change>1.5, P < 0.05) (Fig. 6A). Intriguingly, APP and membrane metallo endopeptidase (Mme), two genes responsible for Aβ metabolism, were regulated after circCwc27 knockdown. Furthermore, genes enriched in learning and memory were significantly upregulated (Fig. 6B–D) (Supplementary Table 7). Our results further showed that the expression of most circCwc27-regulated genes was changed in both APP/PS1 mice and AD patients (Fig. 6E, F). Notably, besides APP and Mme, many of these targets, such as dopamine receptor D1 (Drd1) [48, 49], protein phosphatase 1, regulatory inhibitor subunit1B (Ppp1r1b) [50], neurotrophic tyrosine kinase, receptor, type 1 (Ntrk1) [51, 52], and LIM homeobox 8 (Lhx8) [53], have been reported to be involved in AD pathology.
Fig. 6. CircCwc27 regulates AD genes transcription by binding to Pur-α.
A Volcano plots indicate the differentially expressed mRNAs in the hippocampus of APP/PS1-LV-sh-circCwc27 vs APP/PS1-LV-sh-circCon mice (x-axis = relative to APP/PS1-LV-sh-circCon mice log2 fold changes; y-axis: negative log10 of the P values. Vertical lines represent >1.5-fold changes, and the horizontal line represents P < 0.05). B Gene ontology (GO) annotation of differentially expressed genes in the hippocampus of APP/PS1-LV-sh-circCwc27 vs APP/PS1-LV-sh-circCon mice. Top 10 GO biological process terms are shown. The size of circle represents the number of enriched genes. The larger the circle, the more genes are enriched. The color depth indicates the enrichment degree. Red color = higher expression. Green color = lower expression. C List of genes enriched in “learning and memory” as well as “memory” categories in Gene Ontology (GO) annotation in B and known Pur-α targets that were differentially expressed in the hippocampus of APP/PS1-LV-sh-circCwc27 versus APP/PS1-LV-sh-circCon mice. D qRT-PCR analysis was used to determine the expression levels of mRNAs in the hippocampus from APP/PS1-LV-sh-circCon and APP/PS1-LV-sh-circCwc27 mice. n = 4. *P < 0.05, **P < 0.01, ***P < 0.001 versus the APP/PS1-LV-sh-circCon group using Student’s t-test. E qRT-PCR analysis was used to determine the expression levels of mRNAs in the hippocampus from 9-month WT and APP/PS1 mice. n = 5. *P < 0.05, **P < 0.01, ***P < 0.001 versus the WT group using Student’s t-test. F The expression of indicated mRNAs in the temporal cortex of healthy controls and AD patients were analyzed by qRT-PCR. n = 5. *P < 0.05, **P < 0.01, ***P < 0.001 versus control group using Student’s t-test. G ChIP-qPCR assays were performed to assess the enrichment of Pur-α on promoters of genes in SH-SY5Y cells. Expression of each gene was changed by circRNA knockdown as well as in APP/PS1 mice and AD patients. n = 3. **P < 0.01, ***P < 0.001 versus IgG group using Student’s t-test. H ChIP-qPCR assays were performed to assess the enrichment of Pur-α on genes promoter in the hippocampus of WT, APP/PS1, APP/PS1-LV-sh-circCon, and APP/PS1-LV-sh-circCwc27 mice. n = 4. **P < 0.01 versus WT group. #P < 0.05, ##P < 0.01, ###P < 0.001 versus APP/PS1-LV-sh-circCon group. Data are analyzed with one-way ANOVA. All data in the figure are shown as mean ± SEM.
Besides interacting with RNA molecules, Pur-α can also bind to DNA sequences and participate in the transcriptional control of many cellular genes [54, 55]. To examine whether circCwc27 regulated gene expression by mediating Pur-α recruitment to their promoters, we performed ChIP followed by qPCR (ChIP-qPCR) and verified that Pur-α directly bind to the promoters of the genes that were dysregulated in AD and controlled by circCwc27 (Fig. 6G). Ectopic expression of Pur-α changed the promoter activity and mRNA levels of these genes (Supplementary Fig. 8A–C). We next determined the effect of circCwc27 knockdown on Pur-α enrichment at promoters of its targets. ChIP-qPCR experiments showed that the decreased binding of Pur-α to the promoters of these genes was reversed by circCwc27 knockdown (Fig. 6H). In addition, ectopic expression of Pur-α counteracted the inhibitory effects of overexpressing circCwc27 on the promoter activity of these genes (Supplementary Fig. 8D). Taken together, our data demonstrated that circCwc27 reduced the recruitment of Pur-α to the promoters of AD-related genes by binding to Pur-α.
Discussion
In the present study, we for the first time not only report the characterization of a novel exon-derived circRNA, circCwc27, that is highly conserved and abundantly expressed in neurons, but also demonstrated the effects of circCwc27 on cognition, neuropathology, and transcriptomic changes in APP/PS1 mice. Moreover, we illuminated that Pur-α, a DNA/RNA binding protein, was an important downstream mediator of circCwc27-regulated genes expression. Prominent among Pur-α targets are a cluster of genes that function in Aβ production as well as learning and memory. Knockdown of circCwc27 alters Pur-α nuclear–cytoplasmic shuttling, transcriptionally regulating Pur-α targets including APP, Drd1, Ppp1r1b, Ntrk1, and Lhx8. These results collectively provide novel evidence indicating that circCwc27 regulates AD-related genes by interacting with Pur-α, shedding light on the unexplored role of circCwc27 in AD.
With the advances in the next-generation sequencing (NGS) and bioinformatics, circRNAs are no longer considered merely as by-products of mis-splicing or transcriptional noise [56, 57]. Herein, we focused on the AD-associated circular transcriptome and found that circCwc27 began to elevate in presymptomatic 3-month-old APP/PS1 mice and further increased with age, suggesting that circCwc27 might be causally linked to AD progression. Furthermore, circRNAs with higher expression levels usually are more likely to be conserved among species [21], and these circRNAs are more possible to play an essential function in the regulation of pathophysiological processes [9, 58]. The high expression and evolutionary conservation of circCwc27 raised the possibility that it might exhibit a remarkable regulatory role in AD and become a promising biomarker. This possibility was further validated by our study indicating the function of circCwc27 on pathological traits and cognitive decline in AD mouse model. Together, our present study carried out a relatively comprehensive analysis on the function of a novel AD-related circRNA.
CircRNAs have been indicated to regulate gene expression through several molecular mechanisms [8–14]. The most well-characterized is “miRNA sponge”, that circRNAs can sequester homologous miRNAs through miRNA binding sites. In our research, however, circCwc27 did not exhibit miRNA sponge activity, possibly due to its relatively short length or lack of sufficient numbers of binding sites for certain miRNAs. In addition, circCwc27 was not able to encode for peptides since it did not contain an ORF. Besides binding to miRNAs or translating, circRNAs can interact with RNA-binding proteins and affect their distribution and function, which act as prominent regulators of gene expression [11, 23, 37]. Our results illustrated a novel RBP-associated regulatory axis, in which circCwc27 acted as a multivalent binding platform for Pur-α. Pur-α is a multifunctional RBP involved in brain development, synaptic plasticity, and memory retention, and serves as a pivotal role in the regulation of gene transcription [44, 47]. We focused on a set of genes in the LV-sh-circCon-microinjected and LV-sh-circCwc27-microinjected APP/PS1 mice, which may indicate mechanisms relevant to Aβ as well as learning and memory. CircCwc27 knockdown decreased APP levels, leading to reduced Aβ production while increased the expression of Mme, a gene encodes Neprilysin which is a critical driver of Aβ degradation, eventually ameliorated the deposition of misfolded Aβ peptide and related neurodegenerative effects. In addition, we showed that genes implicated in learning and memory were upregulated in LV-sh-circCwc27-microinjected APP/PS1 mice. Drd1 mediates dopaminergic neurotransmission and is critical for hippocampal synaptic function and memory [49]. Ppp1r1b decreases the expression of PP1 by stabilizing the PP1-DARPP-32 complex and inducing the phosphorylation of cAMP-response element-binding protein (CREB), thereby contributing to cognitive function [50]. Also, it is involved in dopaminergic signaling [59]. Ntrk1 binds to nerve growth factor (NGF) and plays an important role in neurotrophic support to cholinergic neurons and cholinergic neurotransmission, with downstream effects on cognition [60]. Lhx8 regulates Ntrk1 expression and acetylcholine release, thus playing a pivotal role in learning and memory [53]. Intriguingly, most of these circCwc27 targets were transcriptionally controlled by Pur-α, indicating a role of Pur-α as an important regulator in AD.
In APP/PS1 mice, the interaction between Pur-α and circCwc27 was remarkedly increased, leading to a less nuclear distribution of Pur-α in neurons and loss of its function. This was significantly attenuated by circCwc27 knockdown. Therefore, association of Pur-α with circCwc27, overproduced in AD brains, may account for the functional inactivation of this transcription factor. Of note, circCwc27 is capable to sequester a significant fraction of Pur-α proteins, thereby suppressing their ability to regulate genes expression transcriptionally due to its high abundance in neurons, which makes the regulatory role of circCwc27 appealing. We also found Pur-β and Pur-γ, the other two Pur protein family members, in the mass spectrometry results, but both of them exhibited low abundance in mouse cortex and hippocampus [43, 44]. We therefore focused our attention on Pur-α, and demonstrated a direct molecular connection between Pur-α and AD.
Taken together, the elucidation of this circCwc27/Pur-α regulatory interaction has expanded our understanding of cerebral circRNA functions and has uncovered a heretofore-unknown role for Pur-α in the regulation of AD. However, although overexpression of Pur-α largely phenocopied circCwc27 knockdown, it still did not completely reach the level of protection provided by downregulating circCwc27 in Aβ deposition and cognitive decline, suggesting that there might be other potential targets under the control of circCwc27. Other circCwc27-interacting proteins like HNRNPK need to be further investigated.
Supplementary information
Acknowledgements
We thank Prof. Xiaoling Gao from Shanghai Jiao Tong University School of Medicine for research information. We are grateful to Dr. Jianrong Xu from Shanghai University of Traditional Chinese Medicine and Dr. Lanxue Zhao from Shanghai Jiao Tong University School of Medicine for critical reading of the paper.
Author contributions
HW designed the research and revised the manuscript. HC planned and designed the research. CS performed the behavior test, immunostaining, qRT-PCR, and wrote the manuscript. YZ performed the western blot and qRT-PCR. WH performed the RNA pulldown and RNA immunoprecipitation. JS designed the primers and performed chromatin immunoprecipitation (CHIP). QH performed the behavior test. MJ performed the BASE-scope. YQ performed the luciferase reporter gene assay. TW performed the ChIP assay.
Funding information
This work was funded by the National Natural Science Foundation of China (Grant Nos. 81973297 [to HW], 81571298 [to TW], 81872841 [to HC], and 82073836 [to HC]), Key project of Shanghai Science and Technology Commission (Grant No. 19JC1413100 [to HW]), Research Foundation of Translational Medicine, Shanghai Jiao Tong University (Grant No. ZH2018ZDA23 [to HW]).
Data availability
All data are available from the corresponding author upon reasonable request.
Competing interests
The authors declare no competing interests.
ETHICS STATEMENT
The human plasma samples research was approved by the ethics committee at Shanghai Mental Health Center, Shanghai Jiao Tong University School of Medicine. Participants or their legally authorized representatives provided written informed consent. All animal experiments conducted were compliant with Ethics Committee Shanghai Jiao Tong University School of Medicine.
Footnotes
Edited by G. Melino
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Chenghuan Song, Yongfang Zhang.
Contributor Information
Hongzhuan Chen, Email: hongzhuan_chen@hotmail.com.
Hao Wang, Email: angela_wanghao@sjtu.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41418-021-00865-1.
References
- 1.Braak H, Alafuzof I, Arzberger T, Kretzschmar H, Tredici K. Staging of Alzheimer disease-associated neurofibrillary pathology using paraffin sections and immunocytochemistry. Acta Neuropathol. 2006;112:389–404. doi: 10.1007/s00401-006-0127-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ballard C, Gauthier S, Corbett A, Brayne C, Aarsland D, Jones E. Alzheimer’s disease. Lancet. 2011;9770:1019–31. doi: 10.1016/S0140-6736(10)61349-9. [DOI] [PubMed] [Google Scholar]
- 3.Sevigny J, Chiao P, Bussière T, Weinreb P, Williams L, Maier M, et al. The antibody Aducanumab reduces Aβ plaques in Alzheimer’s disease. Nature. 2016;7618:50–56. doi: 10.1038/nature19323. [DOI] [PubMed] [Google Scholar]
- 4.Mehtaa SL, Dempseya RJ, Vemuganti R. Role of circular RNAs in brain development and CNS diseases. Prog Neurobiol. 2020;186:101746. doi: 10.1016/j.pneurobio.2020.101746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Idda ML, Munk R, Abdelmohsen K, Gorospe M. Noncoding RNAs in Alzheimer’s disease. RNA. 2018;9:e1463. doi: 10.1002/wrna.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li X, Yang L, Chen LL. The biogenesis, functions, and challenges of circular RNAs. Mol Cell. 2018;71:428–42. doi: 10.1016/j.molcel.2018.06.034. [DOI] [PubMed] [Google Scholar]
- 7.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–8. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 8.Li Z, Huang C, Bao C, Liang C, Lin M, Wang X, et al. Exonintron circular RNAs regulate transcription in the nucleus. Nat Struct. 2015;22:256–64. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 9.Monika P, Petar G, Luis R, Hernandez M, Sebastian M, Wolf S, et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science. 2017;357:1254. doi: 10.1126/science.aam8526. [DOI] [PubMed] [Google Scholar]
- 10.Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ri XC, Xin C, Liang PX, Zhang JX, Pan ZZ, Ma XD, et al. N6-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat Commun. 2019;10:4695. doi: 10.1038/s41467-019-12651-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qi Y, Du WW, Nan W, Yang WN, Awan MA, Fang L, et al. A circular RNA promotes tumorigenesis by inducing c-myc nuclear translocation. Cell Death Differ. 2017;24:1609–20. doi: 10.1038/cdd.2017.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pamudurti NR, Bartok O, Jens M, Fluss RA, Stottmeister C, Ruhe L, et al. Translation of CircRNAs. Mol Cell. 2017;66:9–21. doi: 10.1016/j.molcel.2017.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell. 2017;66:22–37. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang SL, Li XZ, Zheng H, Si XY, Li B, Wei GQ, et al. Loss of super-enhancer-regulated circRNA Nfix induces cardiac regeneration after myocardial infarction in adult mice. Circulation. 2019;139:2857–76. doi: 10.1161/CIRCULATIONAHA.118.038361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shan K, Liu C, Liu BH, Chen X, Dong R, Liu X, et al. Circular noncoding RNA HIPK3 mediates retinal vascular dysfunction in diabetes mellitus. Circulation. 2017;136:1629–42. doi: 10.1161/CIRCULATIONAHA.117.029004. [DOI] [PubMed] [Google Scholar]
- 17.Wang RJ, Zhang S, Chen XY, Li JW, Jia RC, Pan YQ, et al. EIF4A3-induced circular RNA MMP9 (circMMP9) acts as a sponge of miR-124 and promotes glioblastoma multiforme cell tumorigenesis. Mol Cancer. 2018;17:166. doi: 10.1186/s12943-018-0911-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng Z, Yu CT, Cui SH, Wang H, Jin HJ, Wang C, et al. CircTP63 functions as a ceRNA to promote lung squamous cell carcinoma progression by upregulating FOXM1. Nat Commun. 2019;10:3200. doi: 10.1038/s41467-019-11162-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu HW, Bi JM, Dong W, Yang MH, Shi JY, Jiang N, et al. Invasion-related circular RNA circFNDC3B inhibits bladder cancer progression through the miR-1178-3p/G3BP2/SRC/FAK axis. Mol Cancer. 2018;17:161. doi: 10.1186/s12943-018-0908-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qiu M, Xia W, Chen R, Wang SW, Xu YT, Ma ZF, et al. The circular RNA circPRKCI promotes tumor growth in lung adenocarcinoma. Cancer Res. 2018;78:2839–51. doi: 10.1158/0008-5472.CAN-17-2808. [DOI] [PubMed] [Google Scholar]
- 21.Rybak-Wolf A, Stottmeister C, Glazar P, Jens M, Pino N, Giusti S, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. 2015;58:870–85. doi: 10.1016/j.molcel.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 22.You X, Vlatkovic I, Babic A, Will T, Epstein I, Tushev G, et al. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci. 2015;18:603–10. doi: 10.1038/nn.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang RR, Zhang Y, Bai Y, Han B, Ju MZ, Chen BL, et al. N6 methyladenosine modification of fatty acid amide hydrolase messenger RNA in circular RNA STAG1-regulated astrocyte dysfunction and depressive-like behaviors. Biol Psychiatry. 2020;20:30113. doi: 10.1016/j.biopsych.2020.02.018. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y, Du LF, Bai Y, Han B, He CC, Gong L, et al. CircDYM ameliorates depressive-like behavior by targeting miR-9 to regulate microglial activation via HSP90 ubiquitination. Mol Psychiatry. 2020;6:1175–90. doi: 10.1038/s41380-018-0285-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Westholm JO, Miura P, Olson S, Shenker S, Joseph B, Sanfilippo P, et al. Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep. 2014;9:1966–80. doi: 10.1016/j.celrep.2014.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, et al. Circular intronic long noncoding RNAs. Mol Cell. 2013;51:792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 27.Dube U, Del-Aguila JL, Li ZR, Budde JP, Jiang S, Hsu S, et al. An atlas of cortical circular RNA expression in Alzheimer disease brains demonstrates clinical and pathological associations. Nat Neurosci. 2019;11:1903–12. doi: 10.1038/s41593-019-0501-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang S, Zhu D, Li H, Li HJ, Feng CQ, Zhang WS. Characterization of circRNA-associated-ceRNA networks in a senescence-accelerated mouse prone 8 brain. Mol Ther. 2017;9:2053–61. doi: 10.1016/j.ymthe.2017.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma NN, Pan J, Ye XY, Yu B, Zhang W, Wan J. Whole-transcriptome analysis of APP/PS1 mouse brain and identification of circRNA-miRNA-mRNA networks to investigate AD pathogenesis. Mol Ther Nucleic Acids. 2019;18:1049–62. doi: 10.1016/j.omtn.2019.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen J, Zou Q, Lv D, Raza MA, Wang X, Li PL, et al. Comprehensive transcriptional profiling of porcine brain aging. Gene. 2019;693:1–9. doi: 10.1016/j.gene.2019.01.019. [DOI] [PubMed] [Google Scholar]
- 31.Suzuki H, Zuo Y, Wang J, Zhang MQ, Malhotra A, Mayeda A. Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res. 2006;8:e63. doi: 10.1093/nar/gkl151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gowrishankar S, Yuan P, Wu Y, Schrag M, Paradise S, Grutzendler J, et al. Massive accumulation of luminal protease-deficient axonal lysosomes at Alzheimer’s disease amyloid plaques. Proc Natl Acad Sci USA. 2015;112:E3699–3708. doi: 10.1073/pnas.1510329112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Head E, Corrada MM, Kahle-Wrobleski K, Kim RC, Sarsoza F, Goodus M, et al. Synaptic proteins, neuropathology and cognitive status in the oldest-old. Neurobiol Aging. 2009;30:1125–34. doi: 10.1016/j.neurobiolaging.2007.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.VanGuilder HD, Yan H, Farley JA, Sonntag WE, Freeman WM. Aging alters the expression of neurotransmission-regulating proteins in the hippocampal synaptoproteome. J Neurochem. 2010;113:1577–88. doi: 10.1111/j.1471-4159.2010.06719.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cuello AClaudio. Early and late CNS inflammation in Alzheimer’s disease: two extremes of a continuum? Trends Pharmacol Sci. 2017;11:956–66. doi: 10.1016/j.tips.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 36.Han B, Zhang Y, Zhang YH, Bai Y, Chen XF, Huang RR, et al. Novel insight into circular RNA HECTD1 in astrocyte activation via autophagy by targeting MIR142-TIPARP: implications for cerebral ischemic stroke. Autophagy. 2018;7:1164–84. doi: 10.1080/15548627.2018.1458173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang L, Han B, Zhang ZT, Wang SG, Bai Y, Zhang Y, et al. Extracellular vesicle-mediated delivery of CircSCMH1 promotes functional recovery in rodent and nonhuman primate ischemic stroke models. Circulation. 2020;10.1161. [DOI] [PubMed]
- 38.Petkovic S, Muller S. RNA circularization strategies in vivo and in vitro. Nucleic Acids Res. 2015;43:2454–65. doi: 10.1093/nar/gkv045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li HH, Yang F, Hu AP, Wang XJ, Fang E, Chen YJ, et al. Therapeutic targeting of circ-CUX1/EWSR1/MAZ axis inhibits glycolysis and neuroblastoma progression. EMBO Mol Med. 2019;12:e10835. doi: 10.15252/emmm.201910835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tretiakova A, Gallia GL, Shcherbik N, Jameson B, Johnson E, Amini S, et al. Association of pura with RNAs homologous to 7 SL determines its binding ability to the myelin basic protein promoter DNA sequence. J Biol Chem. 1998;35:22241–7. doi: 10.1074/jbc.273.35.22241. [DOI] [PubMed] [Google Scholar]
- 41.Gallia GL, Johnson EM, Khalili K. Puralpha: a multifunctional single-stranded DNA- and RNA-binding protein. Nucleic Acids Res. 2000;28:3197–205. doi: 10.1093/nar/28.17.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gallia GL, Darbinian N, Tretiakova A, Ansari Sameer A, Rappaport J, Brady John, et al. Association of HIV-1 Tat with the cellular protein, Pura, is mediated by RNA. Proc Natl Acad Sci USA. 1999;96:11572–7. doi: 10.1073/pnas.96.20.11572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu Z, Poidevin M, Li X, Li YJ, Shu LQ, Nelson DL, et al. Expanded GGGGCC repeat RNA associated with amyotrophic lateral sclerosis and frontotemporal dementia causes neurodegeneration. Proc Natl Acad Sci USA. 2013;110:7778–83. doi: 10.1073/pnas.1219643110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Daniel DC, Johnson EM. PURA, the gene encoding Pur-alpha, member of an ancient nucleic acid-binding protein family with mammalian neurological functions. Gene. 2018;643:133–43. doi: 10.1016/j.gene.2017.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barbe MF, Krueger JJ, Loomis R, Otte J, Gordon J. Memory deficits, gait ataxia and neuronal loss in the hippocampus and cerebellum in mice that are heterozygous for Pur-alpha. Neuroscience. 2016;337:177–90. doi: 10.1016/j.neuroscience.2016.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hunt D, Leventer RJ, Simons C, Taft R, Swoboda KJ, Gawne-Cain M, et al. Whole exome sequencing in family trios reveals de novo mutations in PURA as a cause of severe neurodevelopmental delay and learning disability. J Med Genet. 2014;51:806–13. doi: 10.1136/jmedgenet-2014-102798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Darbiniana N, Cuia JQ, Basile A, Valle LD, Otte J, Miklossy J, et al. Negative regulation of AβPP gene expression by Pur-alpha. J Alzheimers Dis. 2008;15:71–82. doi: 10.3233/jad-2008-15106. [DOI] [PubMed] [Google Scholar]
- 48.Cheng ZY, Xia QP, Hu YH, Wang C, He L. Dopamine D1 receptor agonist A-68930 ameliorates Aβ1-42-induced cognitive impairment and neuroinflammation in mice. Int Immunopharmacol. 2020;88:106963. doi: 10.1016/j.intimp.2020.106963. [DOI] [PubMed] [Google Scholar]
- 49.Tsang J, Fullard JF, Giakoumaki SG, Katsel P, Katsel P, Karagiorga VE, et al. The relationship between dopamine receptor D1 and cognitive performance. NPJ Schizophr. 2015;1:14002. doi: 10.1038/npjschz.2014.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cho K, Cho MH, Seo JH, Peak J, Kong KH, Yoon SY, et al. Calpain-mediated cleavage of DARPP-32 in Alzheimer’s disease. Aging Cell. 2015;14:878–86. doi: 10.1111/acel.12374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen YC, Chiu YJ, Lin CH, Hsu WC, Wu JL, Huang CH, et al. Indole compound NC009-1 augments APOE and TRKA in Alzheimer’s disease cell and mouse models for neuroprotection and cognitive improvement. J Alzheimers Dis. 2019;67:737–56. doi: 10.3233/JAD-180643. [DOI] [PubMed] [Google Scholar]
- 52.Ginsberg DS, Malek-Ahmadi HM, Alldred JM, Che SL, Elarova RI, Chen YH, et al. Selective decline of neurotrophin and neurotrophin receptor genes within CA1 pyramidal neurons and hippocampus proper: correlation with cognitive performance and neuropathology in mild cognitive impairment and Alzheimer’s disease. Hippocampus. 2019;29:422–39. doi: 10.1002/hipo.22802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tomioka T, Shimazaki T, Yamauchi T, Oki T, Ohgoh M, Okano H. LIM homeobox 8 (Lhx8) is a key regulator of the cholinergic neuronal function via a tropomyosin receptor kinase A (TrkA)-mediated positive feedback loop. J Biol Chem. 2014;289:1000–10. doi: 10.1074/jbc.M113.494385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tretiakova A, Steplewski A, Johnson EM, Khalili K, Amini S. Regulation of myelin basic protein gene transcription by Sp1 and Puralpha: evidence for association of Sp1 and Puralpha in brain. J Cell Physiol. 1999;181:160–8. doi: 10.1002/(SICI)1097-4652(199910)181:1<160::AID-JCP17>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 55.Zambrano N, De Renzis S, Minopoli G, Faraonio R, Donini V, Scaloni A, et al. DNAbinding protein Puralpha and transcription factor YY1 function as transcription activators of the neuron-specific FE65 gene promoter. Biochem J. 1997;328:293–300. doi: 10.1042/bj3280293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hsu MT, Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. 1979;280:339–40. doi: 10.1038/280339a0. [DOI] [PubMed] [Google Scholar]
- 57.Sanger H, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly basepaired rod-like structures. Proc Natl Acad Sci USA. 1976;73:3852–6. doi: 10.1073/pnas.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Han D, Li J, Wang H, Su XP, Hou J, Gu Y, et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66:1151–64. doi: 10.1002/hep.29270. [DOI] [PubMed] [Google Scholar]
- 59.Greengard P, Allen PB, Nairn AC. Beyond the dopamine receptor: the DARPP-32/protein phosphatase-1 cascade. Neuron. 1999;23:435–47. doi: 10.1016/s0896-6273(00)80798-9. [DOI] [PubMed] [Google Scholar]
- 60.Xhima K, Markham-Coultes K, Nedev H, Heinen S, Saragovi UH, Hynynen K, et al. Focused ultrasound delivery of a selective TrkA agonist rescues cholinergic function in a mouse model of Alzheimer’s disease. Sci Adv. 2020;6:eaax6646. doi: 10.1126/sciadv.aax6646. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available from the corresponding author upon reasonable request.