Abstract
Neurogenesis plays a critical role in brain physiology and behavioral performance, and defective neurogenesis leads to neurological and psychiatric disorders. Here, we show that PLCβ4 expression is markedly reduced in SENP2-deficient cells and mice, resulting in decreased IP3 formation and altered intracellular calcium homeostasis. PLCβ4 stability is regulated by the SUMO-dependent ubiquitin-mediated proteolytic pathway, which is catalyzed by PIAS2α and RNF4. SUMOylated PLCβ4 is transported to the nucleus through Nup205- and RanBP2-dependent pathways and regulates nuclear signaling. Furthermore, dysregulated calcium homeostasis induced defects in neurogenesis and neuronal viability in SENP2-deficient mice. Finally, SENP2 and PLCβ4 are stimulated by starvation and oxidative stress, which maintain calcium homeostasis regulated neurogenesis. Our findings provide mechanistic insight into the critical roles of SENP2 in the regulation of PLCβ4 SUMOylation, and the involvement of SENP2-PLCβ4 axis in calcium homeostasis regulated neurogenesis under stress.
Subject terms: Sumoylation, Ubiquitylation
Introduction
Neurogenesis is the process by which neurons are generated through the division of neural stem cells and maturation of neural progenitor cells (NPCs) [1]. Adult neurogenesis is regulated and restricted to the subgranular zone of the dentate gyrus in the hippocampus and the subventricular zone in the lateral ventricles of the mammalian brain [2, 3]. Adult neurogenesis is dynamically regulated by extracellular signals and intracellular properties [4, 5]. Increasing evidence has shown that adult neurogenesis plays a critical role in brain repair, olfactory behaviors, and behavioral performance [6, 7].
G protein-coupled receptors (GPCRs) constitute the largest group of integral membrane proteins involved in the activation of intracellular signaling and cellular responses [8]. GPCRs bind to their respective ligands and initiate a broad range of intracellular signaling events, including activation of adenylyl cyclase, phosphodiesterase and phospholipase [9]. Phospholipase C (PLC) is the major enzyme that catalyzes the hydrolysis of phosphatidylinositol 4,5-bisphosphate (PIP2) to yield diacylglycerol and inositol 1,4,5-trisphosphate (IP3). DAG and IP3 activate protein kinase C and release calcium from intracellular stores, respectively [10]. Growing evidence indicates that PLCβ4, one of the major neuronal isoforms of PLC, is abundant in Purkinje cells of the rostral cerebellum and essential for neuronal function [8, 11]. However, whether PLCβ4 is correlated with neurogenesis remain largely unknown.
SUMO (small ubiquitin-like modifier) is a ubiquitin-like protein that can be conjugated to lysine residues of the target protein via E1, E2, and E3 enzymes specific for SUMOylation and a family of SENPs (Sentrin/SUMO-specific proteases) for deSUMOylation [12, 13]. SUMOylation is a versatile regulatory system and has been implicated in multiple biological and pathological processes [14]. There are six SENPs with different substrate specificities, including SENP1-3 and SENP5-7 [13]. SENP2-null embryos do not survive to birth because of a cardiac developmental defect that develops due to the absence of SENP2, which regulates the binding of the polycomb complex to H3K27me3 [15]. SENP2 is also important in regulating myostatin-mediated myogenesis [16] and sudden unexplained death in epilepsy [17–19]. However, the mechanism of the involvement of SENP2 in neurogenesis is largely unknown.
In this study, we show that PLCβ4 is SUMOylated at multiple sites, and SUMOylation regulates the nuclear trafficking of PLCβ4 and mediates its degradation via the ubiquitin-proteasome pathway. In addition, PLCβ4 SUMOylation induces the impairment of intracellular calcium homeostasis, and ultimately impairs normal neurogenesis. Moreover, the SENP2-PLCβ4 axis protects cells from responding to oxidative stress via the regulation of calcium homeostasis. Taken together, our findings reveal the critical role of SENP2 in PLCβ4 deSUMOylation and identify an important function of the SENP2-PLCβ4 axis in the regulation of neurogenesis through calcium homeostasis, with broad implications for the maintenance of cellular homeostasis.
Results
SENP2 deficiency decreases PIP2 hydrolysis and disturbs calcium homeostasis through PLCβ4
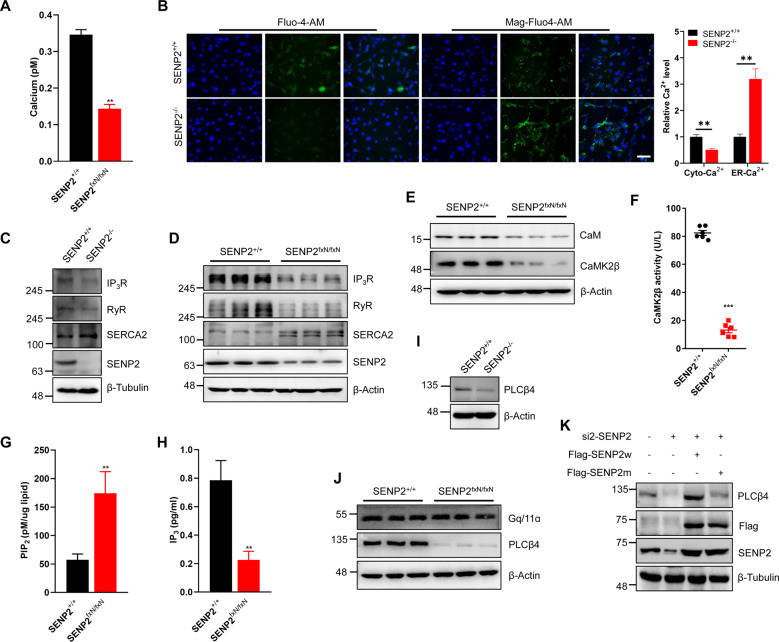
Our previous studies showed that SENP2 deficiency induced hyper-SUMOylation of the Kv7 potassium channel and diminished the M-current [17]. To identify whether calcium channels were affected in SENP2-deficient mice, we measured the intracellular calcium concentration in the brain and found that the it was dramatically decreased in the SENP2-deficient mice (Fig. 1A). Because the ER serves as the largest calcium ion reservoir in the cytosol, we further measured calcium levels using the cytosolic and ER luminal calcium indicators Fluo-4-AM and Mag-Fluo-4-AM in SENP2 MEF cells. The results of both immunofluorescence (IF) staining and flow cytometry revealed that there was a significant decrease in the cytosolic calcium concentration and an increase in the ER calcium concentration in the SENP2−/− MEF cells (Figs. 1B and S1A). We further detected the role of SENP1 and SENP3 on calcium homeostasis, and found that both SENP1 and SENP3 showed no significant effect on calcium homeostasis in SENP1−/− MEF cells or SENP3 knockdown MEF cells (Figs. S1B–S1E). Intracellular calcium homeostasis is mainly regulated by calcium channels and pumps localized in the ER, including IP3R, RyR, and SERCA2. The experiments showed that the transcription levels of IP3R and RyR were decreased in the SENP2-deficient MEF cells and brain (Fig. S1F and S1G); the protein levels of IP3R and RyR were decreased, and the protein level of SERCA2 was increased in the SENP2-deficient MEF cells and brain (Fig. 1C and 1D). We added 1 mM or 2 mM CaCl2 to SENP2+/+ and SENP2−/− MEF cells, and detected the effect of exogenous calcium on calcium homeostasis. However, the exogenous calcium showed no significant effect on the balance of calcium in ER and cytosol (Fig. S1H), suggesting that the calcium gradient between ER and cytosol was generated in a cell autonomous manner due to SENP2 deficiency. Calcium binding is required for the activation of CaM and CaMK2, and the experiments showed that the protein levels of CaM and CaMK2 were decreased in the brain of the SENP2-deficient mice (Fig. 1E). The CaMK2β activity was dramatically decreased in the SENP2-deficient mice (Fig. 1F), indicating that SENP2 deficiency leads to a sustained perturbation of calcium homeostasis.
Fig. 1. SENP2 deficiency affects calcium homeostasis through PLCβ4-regulated ER calcium channels.
A The concentration of calcium in the brain tissue was significantly decreased in the SENP2-deficient mice compared with the wild-type mice. The concentration of calcium in the brain tissue was detected by the ELISA kit (n = 5 mice/group). B SENP2 deficiency induced cytosolic calcium overload and ER luminal calcium depletion in MEF cells. Cytosolic and ER calcium were visualized using the calcium-sensitive fluorescent dyes Fluo-4-AM and Mag-Fluo-4-AM, respectively and observed by fluorescence microscopy (left), and the fluorescence intensity in each group was quantified (right, n = 6 photos/group). The scale bar is 100 µm. C and D Dysregulation of ER-resident calcium channels and pumps in SENP2−/− MEF cells (C) and SENP2-deficient mice (D). Western blot analysis of the cell lysates of the SENP2+/+ and SENP2−/− MEF cells (C) or the brain lysates of the SENP2-deficient and control mice (D) with anti-IP3R, anti-RyR, anti-SERCA2, anti-SENP2, and anti-β-Tubulin antibodies. E The protein levels of CaM and CaMK2β were decreased in the SENP2-deficient mice. Western blot analysis of brain extracts from SENP2-deficient and wild-type mice with anti-CaM, anti-CaMK2β, and anti-β-Actin antibodies. F CaMK2β activity was decreased in the brains of SENP2-deficient mice. The activity of CaMK2β in the brain tissue of the SENP2-deficient and wild-type mice was measured by the ELISA kit (n = 6 mice/group). G and H The PIP2 level was increased (G) and IP3 level was decreased (H) in the SENP2-deficient mice. The concentrations of PIP2 (G) and IP3 (H) in the brain tissue of the SENP2-deficient and wild-type mice were measured by the ELISA kit (n = 6 mice/group). I and J The protein level of PLCβ4 was decreased in the SENP2−/− MEF cells (I) and brain tissue of the SENP2-deficient mice. The cell lysates of the SENP2+/+ and SENP2−/− MEF cells (I) and brain extracts of the SENP2+/+ and SENP2fxN/fxN mice (J) were analyzed by IB with anti-PLCβ4, anti-SENP2, anti-Gq/11α, or anti-β-Actin antibodies. K Wild-type SENP2 increased PLCβ4 expression, while the SENP2 catalytic mutant failed to rescue PLCβ4 expression in the HEK293T cells after SENP2 knockdown. The indicated plasmids were transfected into HEK293T cells, and cell lysates were detected by IB with anti-PLCβ4, anti-Flag, anti-SENP2, or anti-β-Tubulin antibodies.
Because IP3R is activated by IP3, which is produced by PIP2 hydrolysis, we next measured the intracellular PIP2 and IP3 concentrations in mice brain. As shown, the PIP2 level was increased, and the IP3 level was decreased in the SENP2-deficient mice (Fig. 1G and 1H). PLCβ4 is critical for the regulation of PIP2 hydrolysis, and the experiments showed that the PLCβ4 level was dramatically decreased after SENP2 knockdown in HEK293T cells (Fig. S1I). The PLCβ4 level was also decreased in the SENP2-deficient MEF cells and brain (Fig. 1I and 1J). Further experiments showed that Gq/11α, which activates PLCβ4, was not significantly changed in the brain of the SENP2-deficient mice (Fig. 1J), indicating that PLCβ4, but not its upstream activators, was regulated by SENP2. There was no significant variation in PLCβ4 mRNA levels in the SENP2-deficient MEF cells or brain (Fig. S1J and S1K). Further experiments showed that wild-type SENP2 (SENP2w) enhanced the expression of PLCβ4, whereas the SENP2 catalytic mutant (SENP2m) did not in HEK293T cells (Figs. 1K and S1L), suggesting that PLCβ4 expression is regulated by the catalytic activity of SENP2. Further experiment showed that SENP2, but not SENP1 or SENP3, specifically decreased the protein level of PLCβ4 (Fig. S1M). These results indicate that SENP2 regulates PLCβ4-induced hydrolysis of PIP2, and affects intracellular calcium homeostasis.
PLCβ4 is SUMOylated and SENP2 is a specific protease of PLCβ4
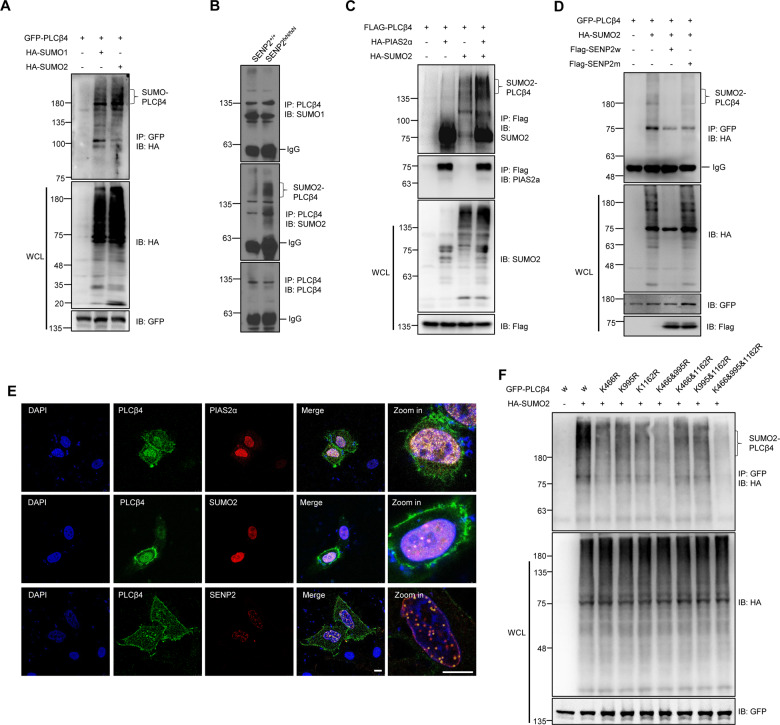
Our results suggest that SENP2 may regulate the expression of PLCβ4 through deSUMOylation (Fig. 1K). The SUMOplot (www.abcepta.com/sumoplot) predicted that PLCβ4 is highly likely to be SUMOylated (Fig. S2A). The western-blot results showed that PLCβ4 was conjugated with exogenous SUMO1 and, to a greater extent, with SUMO2 in HEK293T cells (Fig. 2A). SUMO2-modified, but not SUMO1-modified, bands were highly enriched in the brain of the SENP2-deficient mice (Fig. 2B). Next, the modification of PLCβ4 by endogenous SUMO2 was confirmed by the IP of PLCβ4 from PLCβ4-transfected HEK293T cells (Fig. S2B). Furthermore, the SUMOylation of PLCβ4 was enhanced in SENP2−/− MEF cells (Fig. S2C). PIAS family members are the major SUMO E3 ligases, and the IP assay revealed that PIAS2α was the major SUMO E3 interacting with PLCβ4 (Fig. S2D), and PIAS2α dramatically enhanced the SUMOylation of PLCβ4 (Fig. 2C). These results indicated that PIAS2α is the specific E3 ligase catalyzing PLCβ4 SUMOylation.
Fig. 2. PLCβ4 SUMOylation is modified by SUMO2 and deconjugated by SENP2 in vitro and in vivo.
A PLCβ4 was mainly modified by exogenous SUMO2 in HEK293T cells. The indicated plasmids were transfected into HEK293T cells, and the IP with GFP from cell lysates were detected by IB with anti-HA antibody. The WCL was detected by IB with anti-HA or anti-GFP antibodies. B PLCβ4 was mainly modified by endogenous SUMO2 in the brain tissue of SENP2-deficient mice. The IP with anti-PLCβ4 from the brain lysates of the SENP2-deficient and control mice were detected by IB with anti-SUMO1, anti-SUMO2, or anti-PLCβ4 antibodies. C PIAS2α enhanced the SUMOylation of PLCβ4. The indicated plasmids were transfected into HEK293T cells, and the IP with Flag from cell lysates were detected by IB with anti-SUMO2 or anti-PIAS2α antibodies. The WCL was detected by IB with anti-SUMO2 or anti-Flag antibodies. D SENP2 deconjugates SUMOylated PLCβ4. The indicated plasmids were transfected into HEK293T cells, and the IP with GFP from cell lysates were detected by IB with anti-HA antibody. The WCL was detected by IB with anti-HA, anti-GFP, or anti-Flag antibodies. E Colocalization of PLCβ4 with PIAS2α, SUMO2 and SENP2 in the nucleus. The indicated plasmids were transfected into HEK293T cells, and the cells were harvested for immunocytochemistry with anti-PLCβ4 (green) and anti-PIAS2α, anti-SUMO2 or anti-SENP2 (red) antibodies. DAPI (blue) was used to show nuclei. The scale bar is 5 µm. F K466, K995, and K1162 are the major SUMOylation sites of PLCβ4. Wild-type (w) or mutant GFP-PLCβ4 and HA-SUMO2 were transfected into HEK293T cells, and the IP with GFP from cell lysates were detected by IB with anti-HA antibody. The WCL was detected by IB with anti-HA or anti-GFP antibodies.
SUMOylation is catalyzed by several enzymes and can be reversed by members of the SENP family. The co-IP experiment showed that PLCβ4 strongly binds to SENP2 (Fig. S2E). Furthermore, SENP2w, not SENP2m, deconjugated SUMOylated PLCβ4 (Fig. 2D). IF staining showed that PLCβ4 colocalized with PIAS2α in the nuclear rim, with SUMO2 and SENP2 in the nucleus of HEK293T cells (Figs. 2E and S2F). The bioinformatics analysis of PLCβ4 revealed three potential SUMO-conjugation consensus sites, including K466, K995, and K1162 (Fig. S2A). These predicted SUMO sites are located in the X domain and C-terminus of PLCβ4, respectively (Fig. S2G), and are evolutionarily conserved in different species (Fig. S2H). In mapping experiments, K466, K995, and K1162 were shown to be SUMO-conjugation sites of PLCβ4 (Fig. 2F). These results indicate that PLCβ4 is SUMOylated and SENP2 interacts with PLCβ4 to deconjugate its SUMOylation.
SUMOylation promotes the ubiquitin-mediated degradation of PLCβ4 via the STUBL RNF4
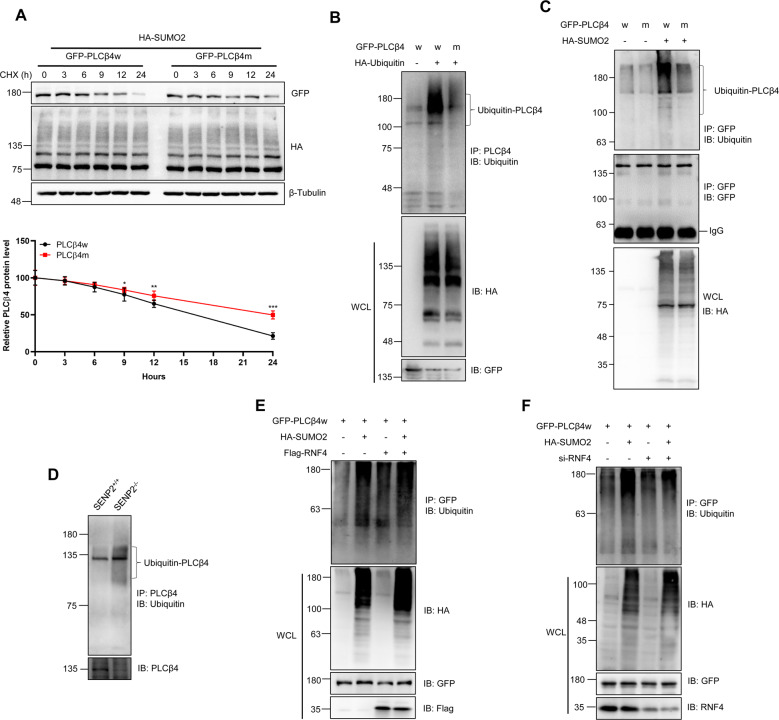
SENP2 deficiency decreased the expression level of PLCβ4 (Fig. 1I and 1J), suggesting that SUMOylation may regulate PLCβ4 stability. We transfected wild-type PLCβ4 (PLCβ4w) or the triple-SUMO site mutant PLCβ4 (PLCβ4m) into HEK293T cells, and harvested proteins after treatment with CHX for different periods of time. As shown, PLCβ4w was degraded much faster than PLCβ4m after 24 h of treatment with CHX (Fig. 3A). SENP2 MEF cells were treated with MG132 or chloroquine to inhibit the ubiquitin or lysosome-dependent degradation pathways, and the results showed that MG132, but not chloroquine, prevented the degradation of PLCβ4 (Fig. S3A). SENP2 knockdown via siRNA also promoted the degradation of PLCβ4, and MG132 treatment prevented its degradation (Fig. S3B). SUMOylation enhanced PLCβ4w ubiquitination, whereas PLCβ4m was poorly ubiquitinated in HEK293T cells (Fig. 3B). We then measured the extent of exogenous ubiquitination of PLCβ4 in HEK293T cells and found that PLCβ4w underwent more ubiquitination than PLCβ4m (Fig. 3C). We further found that PLCβ4 was ubiquitinated by endogenous ubiquitin (Fig. S3C), and ubiquitination of endogenous PLCβ4 was increased in the SENP2−/− MEF cells (Fig. 3D). IF staining showed that PLCβ4 colocalized with ubiquitin in the nuclei of HEK293T cells (Fig. S3D). These results demonstrated that PLCβ4 is SUMOylated and degraded via the ubiquitin-mediated proteasomal degradation pathway.
Fig. 3. SUMOylation contributes to the ubiquitination and degradation of PLCβ4 by the STUBL RNF4.
A SUMOylation decreased the stability of the wild-type PLCβ4 but not the mutant PLCβ4. Wild-type PLCβ4 (w) or the triple-SUMO site mutant (m) and HA-SUMO2 were transfected into HEK293T cells, and the cells were treated with CHX for different periods of time. The cells were harvested, and the cell lysates were detected by IB with anti-GFP, anti-HA, or anti-β-Tubulin antibodies (top). The gray analysis was performed by comparing the PLCβ4 band to the β-Tubulin band (bottom, n = 3 repeats/group). B Exogenous ubiquitin induced the ubiquitination of PLCβ4. Wild type or mutant PLCβ4 and HA-ubiquitin plasmids were transfected into HEK293T cells, and the IP with PLCβ4 from cell lysates were detected by IB with anti-Ubiquitin antibody, and the WCL were detected by IB with anti-HA or anti-GFP antibodies. C SUMOylation of PLCβ4 increased its endogenous ubiquitination. PLCβ4w or PLCβ4m and HA-SUMO2 plasmids were transfected into HEK293T cells, and the IP with GFP from cell lysates were detected by IB with anti-ubiquitin antibody, and the WCL were detected by IB with anti-HA or anti-GFP antibodies. D PLCβ4 was modified by endogenous ubiquitin in SENP2−/− MEF cells. The IP with anti-PLCβ4 from MEF cell lysates were detected by IB with anti-ubiquitin antibody. The WCL was detected by IB with anti- PLCβ4 antibody. E RNF4 enhanced the ubiquitination of PLCβ4. The indicated plasmids were transfected into HEK293T cells, and the IP with anti-GFP from cell lysates were detected by IB with anti-ubiquitin antibody. The WCL was detected by IB with anti-HA, anti-GFP, and anti-Flag antibodies. F Ubiquitination of PLCβ4 was decreased after RNF4 knockdown in HEK293T cells. The indicated plasmids were transfected into HEK293T cells, and the IP with anti-GFP from cell lysates were detected by IB with anti-ubiquitin antibody. The WCL was detected by IB with anti-HA, anti-GFP, and anti-RNF4 antibodies.
SUMOylation of PLCβ4 induced its ubiquitination and degradation, which requires the E3 ubiquitin ligase STUBL (SUMO-targeted ubiquitin ligase). Several ring finger proteins have been reported to have a SIM, such as RNF4 and RNF111 [20], and IF staining showed that PLCβ4 colocalized with RNF4 in the nuclei of HEK293T cells (Fig. S3E). We performed the ubiquitination assay to verify the involvement of RNF4 in the SUMO-triggered ubiquitination of PLCβ4. As shown, both SUMO2 and RNF4 promoted the ubiquitination of PLCβ4 (Fig. 3E). Furthermore, RNF4 knockdown reduced exogenous SUMO-dependent PLCβ4 ubiquitination (Fig. 3F). These data show that RNF4 mediates the ubiquitination of SUMO-modified PLCβ4.
SENP2 regulates the interaction of Nup205 with PLCβ4 for cytoplasm-nucleus transport
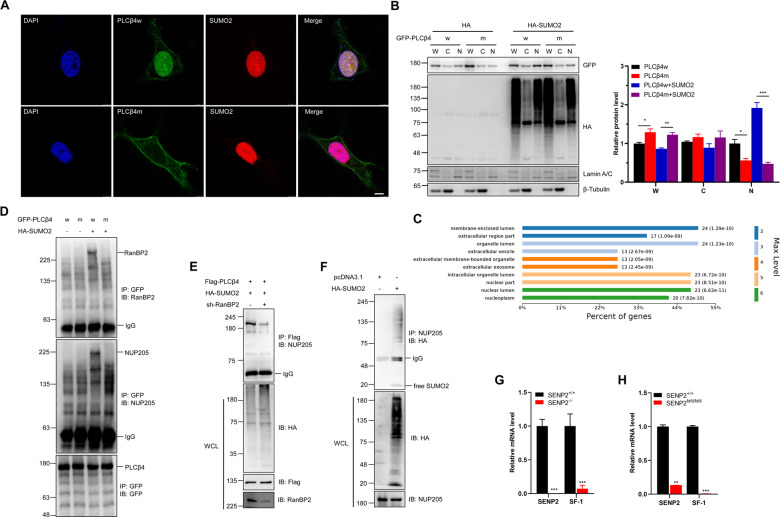
PLCβ4 is a crucial phospholipase and is mainly localized in the cell membrane and cytosol. The IF staining showed that PLCβ4w preferentially colocalized with SUMO2 in the nucleus, whereas PLCβ4m remained in the cytoplasm (Figs. 4A and S4A). Fractionation of transfected HEK293T cells also showed that SUMOylation dramatically enhanced PLCβ4 accumulation in the nucleus (Fig. 4B). These results indicate that hyper-SUMOylation facilitates the nuclear import of PLCβ4 from the cytoplasm.
Fig. 4. SENP2 regulates the interaction of Nup205 with PLCβ4 for cytoplasm-nucleus transport.
A SUMOylation of PLCβ4 promoted its localization in the nucleus. Wild-type or SUMO mutant PLCβ4 and HA-SUMO2 plasmids were transfected into HEK293T cells, and the cells were harvested for immunocytochemistry with anti-PLCβ4 (green) and anti-SUMO2 (red) antibodies. DAPI (blue) was used to show nuclei. The scale bar is 5 µm. B SUMOylation of PLCβ4 promoted its translocation from cytoplasm to the nucleus. The indicated plasmids were transfected into HEK293T cells, and the lysates from the whole cell (W), cytoplasm (C) and nucleus (N) were detected by IB with anti-GFP, anti-HA, anti-Lamin A/C, or anti-β-Tubulin antibodies (left). The gray analysis was performed by GFP band compared to β-Tubulin band (right, n = 3 repeats/group). C Bioinformatics analysis of the top 10 significant different cellular pathways regulated by wild-type PLCβ4. D SUMO mutant PLCβ4 exhibited diminished interaction with RanBP2 and Nup205. The indicated plasmids were transfected into HEK293T cells, and the IP with anti-GFP from cell lysates were detected by IB with anti-RanBP2, anti-Nup205, and anti-GFP antibodies. E RanBP2 is essential for the interaction of PLCβ4 and Nup205. The indicated plasmids were transfected into HEK293T cells, and the IP with anti-Flag from cell lysates were detected by IB with anti-Nup205 antibody. The WCL was detected by IB with anti-HA, anti-Flag, and anti-RanBP2 antibodies. F Nup205 interacted with SUMO2 conjugates. The indicated plasmids were transfected into HEK293T cells, and the IP with anti-Nup205 from cell lysates were detected by IB with anti-HA antibody. The WCL were detected by IB with anti-HA and anti-Nup205 antibodies. G–H The mRNA level of SF-1 was decreased in SENP2−/− MEF cells (G) and the brains of the SENP2-deficient mice (H). The expression levels of SENP2 and SF-1 transcripts in SENP2−/− MEF cells and SENP2-deficient mice were measured by real-time PCR and normalized to control mice (n = 3 mice/group).
To identify the proteins that facilitate nuclear import of PLCβ4, PLCβ4w, or PLCβ4m was transfected into HEK293T cells, and bound proteins were extracted for LC-MS/MS analysis (Fig. S4B). The results showed that PLCβ4w and PLCβ4m bind a large number of different proteins (Fig. S4C), and PLCβ4w binds mostly nuclear lumen and membrane-bound vesicle proteins (Fig. S4D), which is consistent with the GO analysis of different cellular component pathways (Fig. 4C). Among these interacting proteins, binding of the nucleoporin complex NUP205 showed significantly difference between PLCβ4w and PLCβ4m (Fig. S4E). RanBP2 has been implicated in numerous transport pathways as SUMO E3 ligase, and the interaction of RanBP2 and Nup205 with PLCβ4 was significantly decreased after SUMO mutation (Fig. 4D). Furthermore, RanBP2 knockdown dramatically reduced the interaction between PLCβ4 and Nup205 (Fig. 4E). Sequence analysis of NUP205 indicated that the protein encompasses several predicted SIM motifs, which are conserved in various species (Fig. S4F and S4G). The experiment showed that endogenous Nup205 coprecipitated with SUMO2-modified proteins (Fig. 4F). These data indicate that RanBP2 enhances the SUMOylation of PLCβ4 to facilitate its interaction with Nup205 through SIM-SUMO2 interaction.
PIP2 and phosphoinositide-modifying enzymes coexist in the nucleus, and inositol polyphosphate multikinase (IPMK) regulates the transcription of the nuclear receptor steroidogenic factor (SF-1) by directly phosphorylating PIP2 [21]. The mRNA level of SF-1 was measured after SENP2 knockout, and the results showed a significant decrease of SF-1 mRNA in SENP2-deficient MEF cells and brain (Fig. 4G and 4H). These results indicate that PLCβ4 might have similar signaling activity in nuclear and cytoplasmic membranes, regulating PIP2 hydrolysis and reducing the SF-1 transcription mediated by IPMK.
Calcium homeostasis disruption induced by SENP2 deficiency impairs neuronal viability and neurogenesis
We next explored the potential role of SENP2 regulated calcium homeostasis in the neuronal function in mice. As expected, the levels of SENP2 and PLCβ4 were significantly decreased in SENP2-deficient mice (Fig. S5A and S5B). Furthermore, to identify in which cells SENP2 and PLCβ4 were expressed, we performed IF staining with various makers, and the results showed that both SENP2 and PLCβ4 were expressed in mature neurons (Fig. S5C and S5D), immature neurons (Fig. S5E and S5F), and NPCs (Fig. S5G and S5H). Golgi staining revealed dramatically decreased numbers of neurons in the hippocampus in 6-week-old SENP2-deficient mice (Figs. 5A and S5I). Nissl staining revealed a significant decrease in viable neurons and increase in injured neurons in the hippocampus, especially in the DG area, of the SENP2-deficient mice (Fig. 5B). We isolated hippocampal neurons from mice and analyzed their morphology, and the Sholl analysis indicated that SENP2-deficient neurons were shorter with decreased numbers of primary dendrites and less neuronal complexity (Figs. 5C and S5J). These results showed that SENP2 deficiency decreased neuronal viability in mice.
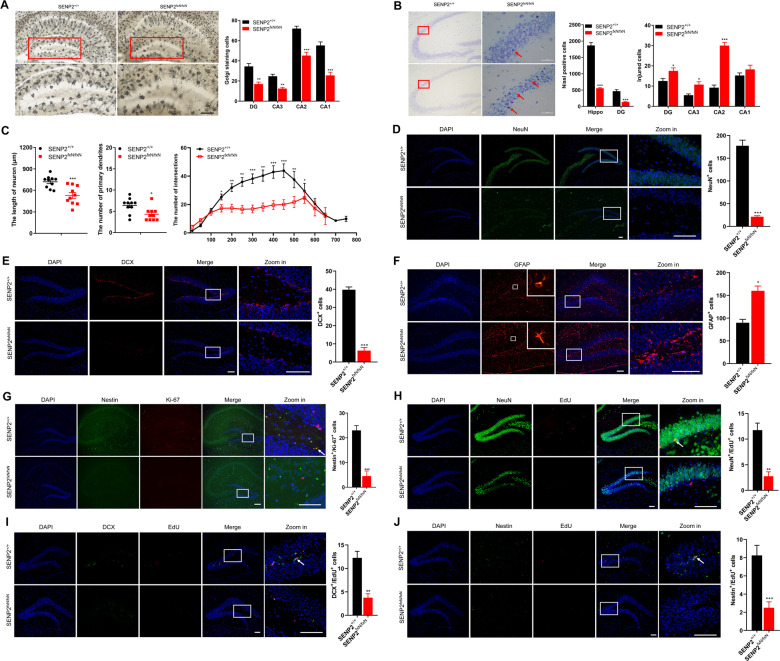
Fig. 5. SENP2-deficient mice exhibit decreased neuronal viability and defective neurogenesis.
A SENP2 deficiency decreased the number of hippocampal neurons. The brains of SENP2-deficient and wild-type mice were sliced for Golgi staining (left). Quantitative analysis of the stained neurons in the DG and CA1-CA3 areas of the hippocampus (right, n = 4 mice/group). The scale bar is 100 μm. B SENP2 deficiency decreased neuronal viability. The brains of SENP2-deficient and wild-type mice were sliced for Nissl staining (left). Quantitative analysis of the stained neurons in the DG and CA1-CA3 areas of the hippocampus (right, n = 4 mice/group). The scale bar is 100 μm. C SENP2 deficiency affected normal neuronal morphology. Hippocampal neurons were cultured for Sholl analysis (n = 10 neurons/group). D SENP2 deficiency decreased the number of mature neurons. The brains of SENP2-deficient and wild-type mice were sliced for immunofluorescent staining with anti-NeuN (green) antibody (left). Quantitative analysis of the stained mature neurons in the hippocampal areas (right, n = 5 mice/group). DAPI (blue) was used to show nuclei. The scale bar is 100 µm. E SENP2 deficiency decreased the number of immature neurons. The brains of SENP2-deficient and wild-type mice were sliced for immunofluorescent staining with anti-DCX (red) antibody. Quantitative analysis of the stained immature neurons in the hippocampal areas (right, n = 5 mice/group). DAPI (blue) was used to show nuclei. The scale bar is 100 µm. F SENP2 deficiency increased the number of astrocytes. The brains of SENP2-deficient and wild-type mice were sliced for immunofluorescent staining with anti-GFAP (red) antibody. Quantitative analysis of the stained astrocytes in the hippocampal areas (right, n = 5 mice/group). DAPI (blue) was used to show nuclei. The scale bar is 100 µm. G SENP2 deficiency inhibited the proliferation of NPCs. The brains of SENP2-deficient and wild type mice were sliced for immunofluorescent staining with anti-Nestin (green) and anti-Ki67 (red) antibodies. Quantitative analysis of the stained NPCs in the hippocampal areas (right, n = 5 mice/group). DAPI (blue) was used to show nuclei, and the arrow was used to indicate the double-positive cells. The scale bar is 100 µm. H–J SENP2 deficiency inhibited neurogenesis. The brains of SENP2-deficient and wild-type mice that had been injected with EdU were sliced for immunofluorescent staining with anti-EdU (red) and anti-NeuN (green, H), anti-DCX (green, I), anti-Nestin (green, J) antibodies. Quantitative analysis of the stained neurons in the hippocampal areas (right, n = 5 mice/group). DAPI (blue) was used to show nuclei, and the arrow was used to indicate the double-positive cells. The scale bar is 100 µm.
Since the depletion of neurons is often due to impaired neurogenesis [22], we explored neurogenesis in 6-week-old SENP2-deficient mice. IF and IHC staining of NeuN indicated that SENP2 deficiency dramatically decreased the number of mature neurons in the brain, especially in the hippocampus (Figs. 5D, S5K, and S5L). In addition, IF staining for DCX indicated that SENP2 deficiency decreased the number of immature neurons in the hippocampus (Fig. 5E). On the other hand, the numbers of two types of gliocytes, astrocytes and microglial cells, were both increased in the hippocampus of the SENP2-deficient mice (Figs. 5F, S5M and S5N). Staining for Nestin and Ki-67 showed that there were significantly decreased number of NPCs, and decreased proliferation of NPCs in the hippocampus of the SENP2-deficient mice (Fig. 5G). BrdU or EdU was injected into 6-week-old mice, and the results showed that SENP2 deficiency significantly inhibited neurogenesis in vivo (Fig. 5H–5J, and S5O). The TUNEL assay indicated that neural apoptosis was not significantly different between the SENP2-deficient and control mice (Fig. S5P). These results indicated that SENP2 deficiency impaired the neurogenesis in hippocampus.
Next, we detected these neuronal markers at postnatal day 10 (p10), and staining for NeuN with DCX or GFAP showed no significant difference between the SENP2-deficient and control mice at p10 (Fig. S5Q and S5R). We further detected neuronal markers in 4-week-old mice, and the results showed that mature neurons were diminished in the hippocampal CA2 area (Fig. S5S), and immature neurons were diminished in the hippocampal DG area of the SENP2-deficient mice (Fig. S5T). Ki67 staining showed less NPC proliferation in the hippocampus of the 4-week-old SENP2-deficient mice than in the wild-type mice (Fig. S5U). These results suggest that SENP2 deficiency affects neurogenesis in a time-dependent manner.
SENP2-regulated neural viability through calcium homeostasis is PLCβ4-dependent
We further identified the roles of PLCβ4 in calcium homeostasis and neural viability affected by SENP2 deficiency. PLCβ4 knockdown decreased the protein levels of IP3R and RyR and increased the protein level of SERCA2 in HT22 and PC12 cells (Figs. 6A, 6B, S6A and S6B), indicating that PLCβ4 knockdown disrupts calcium homeostasis. SENP2 MEF cells were cultured and treated with PLC activator m-3m3-FBS, and the results showed that the PLC activation increased IP3R level and decreased SERCA2 level in SENP2+/+ MEF cells (Fig. 6C). Since there was dramatically decreased PLCβ4 in the SENP2−/− MEF cells, the PLC activator showed no significant effect (Fig. 6C). The SENP2 knockdown similarly diminished the regulatory effect of the PLC activator on calcium homeostasis-related proteins in HT22 cells (Fig. 6D). These results indicate that PLCβ4 plays a critical role in SENP2-mediated calcium homeostasis.
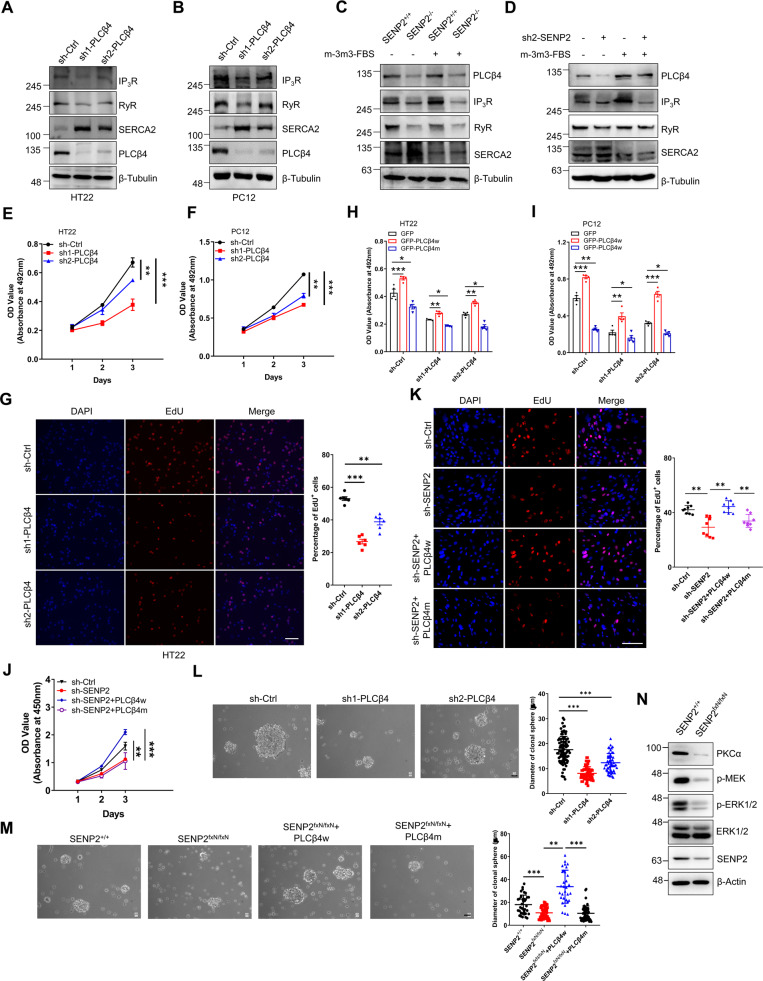
Fig. 6. SENP2 regulation of neural viability through calcium homeostasis is PLCβ4-dependent.
A and B PLCβ4 knockdown affects calcium homeostasis in HT22 (A) and PC12 (B) cells. sh-Ctrl and two sh-PLCβ4 plasmids were packaged and transfected into cells, and the cell lysates were detected by IB with anti-IP3R, anti-RyR, anti-SERCA2, anti-PLCβ4, or anti-β-Tubulin antibodies. C PLC activator m-3m3-FBS increased the expression of calcium channel proteins in SENP2+/+ MEF cells but not in SENP2−/− MEF cells. SENP2+/+ and SENP2−/− MEF cells were treated with DMSO or m-3m3-FBS, and the cell lysates were detected by IB with the indicated antibodies. D The PLC activator m-3m3-FBS had no significant effect on the expression of calcium channel proteins in SENP2 knockdown HT22 cells. sh-ctrl and sh2-SENP2 stable cells were treated with DMSO or m-3m3-FBS, and the cell lysates were detected by IB with indicated antibodies. E and F PLCβ4 knockdown decreased the viability of HT22 (E) and PC12 (F) cells. The viability of PLCβ4-ctrl, sh1-PLCβ4, and sh2-PLCβ4 stably transfected cells was measured by MTT assay (n = 3 repeats/group). G PLCβ4 deficiency inhibited the proliferation of PC12 cells. PC12 cells stably transfected with sh-ctrl, sh1-PLCβ4, or sh2-PLCβ4 were assessed by EdU proliferation assay (left). The EdU fluorescent dye was quantified (right, n = 3 photos/group). DAPI (blue) was used to show nuclei. The scale bar is 100 µm. H and I Wild-type but not SUMO mutant PLCβ rescued the reduced cell viability due to the knockdown of PLCβ4 in PC12 (J) and HT22 (K) cells. GFP, GFP-PLCβ4w, or GFP-PLCβ4m plasmid was transfected into stably knockdown cells, and cell viability was measured via MTT assay (n = 4 repeats/group). J Wild-type but not SUMO mutant PLCβ4 rescued the reduced cell viability in SENP2 knockdown PC12 cells. GFP-PLCβ4w or GFP-PLCβ4m plasmid was transfected into stably knockdown cells, and the cell viability was measured via MTT assay (n = 3 repeats/group). K Wild-type but not SUMO mutant PLCβ4 rescued the reduced cell proliferation in SENP2 knockdown PC12 cells. GFP-PLCβ4w or GFP-PLCβ4m plasmid was transfected into stably knockdown cells, and the cells were assessed by EdU proliferation assay (left). The EdU fluorescent dye was quantified (right, n = 3 photos/group). DAPI (blue) was used to show nuclei. The scale bar is 100 µm. L PLCβ4 knockdown inhibited the neurosphere formation of NPCs. NPCs were isolated and transfected with sh-ctrl or two sh-PLCβ4, and then neurosphere formation was detected (left). The diameter of the clonal spheres was measured and analyzed via ImageJ software (right, n = 51 clones/group). The scale bar is 5 µm. M Wild-type but not SUMO mutant PLCβ4 rescued the neurosphere formation of NPCs. NPCs were isolated and transfected with PLCβ4w or PLCβ4m, and then neurosphere formation was detected (left). The diameter of the clonal spheres was measured and analyzed via ImageJ software (right, n = 50 clones/group). The scale bar is 5 µm. N SENP2 regulated the proliferation of NPCs through PKC-ERK1/2 pathway. NPCs in the hippocampus of wild type or SENP2 deficiency mice were isolated, and the cell lysates were detected by IB with indicated antibodies.
To verify the role of PLCβ4 in neuron viability, PLCβ4 was knocked down in HT22 and PC12 cells, and cell viability was detected by MTT assay and cell counting. The results showed that the cell viability of HT22 and PC12 cells was decreased after PLCβ4 knockdown (Figs. 6E, 6F, S6C and S6D). The EdU proliferation assay showed that the proliferation of HT22 and PC12 cells was largely inhibited after PLCβ4 knockdown (Figs. 6G and S6E). To confirm the critical role of PLCβ4 SUMOylation in neuronal viability, PLCβ4w or PLCβ4m was transfected into PLCβ4 knockdown cells, and the results showed that PLCβ4w, but not PLCβ4m, reversed the decrease in cell viability due to the knockdown of PLCβ4 in HT22 and PC12 cells (Fig. 6H and 6I). The decreased proliferation and viability of SENP2 knockdown PC12 cells were rescued by wild-type, but not SUMO mutant PLCβ4 (Fig. 6J and K). The imbalanced calcium homeostasis in SENP2 knockdown PC12 cells was rescued by PLCβ4 activation and wild-type PLCβ4, but not SUMO mutant PLCβ4 (Fig. S6F). We applied mitochondria-specific calcium probe Rhod-2 to check the calcium of mitochondria, and found that the calcium was significantly overloaded in mitochondria of SENP2−/− MEF cells (Fig. S6G and S6H), indicating that the increased mitochondrial calcium was the major pathway to induce cell death in SENP2−/− MEF cells. To verify the role of PLCβ4 in NPC proliferation, we isolated the primary NPCs and detected neurosphere formation after PLCβ4 knockdown. The results showed that PLCβ4 knockdown inhibited neurosphere formation and the stem cell potential of NPCs (Figs. 6L and S6I). Wild-type but not SUMO mutant PLCβ4 rescued the neurosphere formation and the stem cell potential of SENP2-deficient NPCs (Figs. 6M and S6J).
We further investigated the molecular mechanism how SENP2-PLCβ4 axis regulates the proliferation of NPCs. It was reported that 12-Deoxyphorbols and ELAC (3,12-di-O-acetyl-8-O-tigloilingol) induced proliferation of NPCs via PKC activation [23, 24]. Hypoxia/reoxygenation stimulated proliferation of mouse NPCs through PKC-dependent activation of ERK and Akt [25]. Previous studies showed that ERK activation is both required and sufficient for proliferation of adult neural stem cells [26]. Furthermore, recent report indicated that MiR-140-5p-Prox1 axis regulate the proliferation and differentiation of neural stem cells through the ERK/MAPK signaling pathway [27]. These studies suggested important roles of PKC-ERK in proliferation of NPCs. We isolated NPCs from mice and showed that PKCα, p-MEK, and p-ERK1/2 were dramatically decreased in SENP2-deficient NPCs (Fig. 6N), suggesting that SENP2-PLCβ4 axis might regulate the proliferation of NPCs through PKC-ERK1/2 pathway. Taken together, these results show that PLCβ4 is essential to SENP2 deficiency-induced abnormal calcium homeostasis and decreased neural viability.
The SENP2-PLCβ4 axis acts as a sensor during starvation or oxidative stress
It has been reported that starvation-induced oxidative stress activates protein kinase and neurogenic pathways to mediate neuroprotection [28]. HT22 and PC12 cells were subjected to serum starvation for different periods of time, and the results showed that both SENP2 and PLCβ4 levels were significantly increased in a time-dependent manner (Figs. 7A and S7A), which was followed by a decrease in SUMO1 and SUMO2 modification levels (Fig. S7B and S7C). The levels of IP3R and RyR were enhanced and the level of SERCA2 was diminished after starvation (Fig. 7A). It was reported that starvation could lead to efflux of calcium from the ER to activate the autophagic pathways, and the results showed that the protein levels of Beclin1 and LC3-II were increased after starvation (Fig. 7A). In addition, starvation increased the transcription of SENP2, but not PLCβ4, in HT22, PC12, and SH-SY5Y cells (Figs. 7B and S7D). Starvation inhibited the SUMOylation-mediated ubiquitination of PLCβ4 in HT22 and PC12 cells (Figs. 7C and S7E). Total PLCβ4 and SUMOylated PLCβ4 were highly decreased after SENP2 knockdown even under starvation conditions (Figs. 7D and S7F), indicating that starvation-induced PLCβ4 activation is SENP2-dependent. In addition, SENP2 knockdown diminished the effects of starvation on calcium homeostasis-related protein levels and the rescue of imbalanced calcium homeostasis (Figs. 7D and S7G), indicating an important role for SENP2 and PLCβ4 in the stress response.
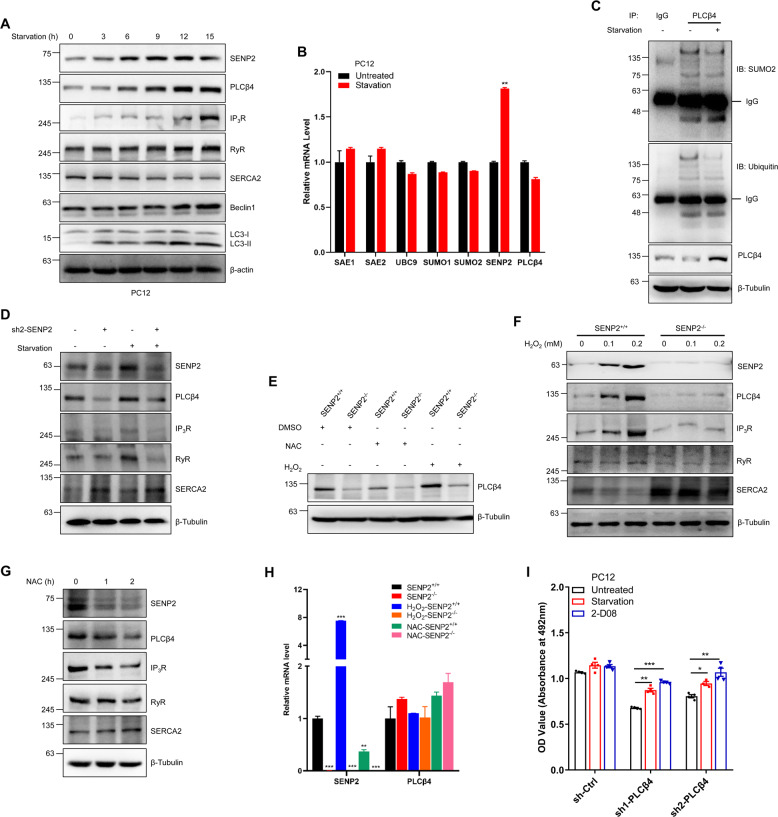
Fig. 7. The SENP2-PLCβ4 axis responds to starvation and oxidative stress.
A Starvation increased SENP2 and PLCβ4 levels, followed by calcium channel proteins, in a time-dependent manner in PC12 cells. PC12 cells were cultured under starvation conditions for different periods of time, and the cell lysates were detected by IB with the indicated antibodies. B Starvation decreased the transcription level of SENP2 in PC12 cells. PC12 cells were treated with serum starvation, and the transcript level of SENP2 and SUMO-related genes were measured by real-time PCR normalized to DMSO control (n = 3 repeats/group). C Starvation decreased the SUMOylation and ubiquitination levels of PLCβ4 in PC12 cells. PC12 cells were starved for 12 h and harvested for IP with IgG or PLCβ4 antibody followed by IB with anti-SUMO2 or anti-ubiquitin antibodies. D SENP2 is essential for starvation-induced calcium homeostasis in PC12 cells. sh-ctrl and sh2-SENP2 stable cells were cultured under starvation conditions for 12 h, and the cell lysates were detected by IB with the indicated antibodies. E SENP2 is essential for oxidative stress-induced expression of PLCβ4. SENP2+/+ and SENP2−/− MEF cells were treated with DMSO, NAC or H2O2 for 30 min, and the cell lysates were detected by IB with the indicated antibodies. F SENP2 and PLCβ4 increased in an H2O2 concentration-dependent manner in SENP2+/+ MEF cells. SENP2+/+ and SENP2−/− MEF cells were treated with different concentrations of H2O2 for 30 min, and the cell lysates were detected by IB with the indicated antibodies. G NAC treatment inhibited SENP2 and PLCβ4-regulated ER channel proteins in a time-dependent manner in PC12 cells. The PC12 cells were treated with NAC for different periods of time, and the cell lysates were detected by IB with the indicated antibodies. H Oxidative stress decreased the transcription level of SENP2, but no significant effect on PLCβ4 in SENP2 MEF cells. SENP2+/+ and SENP2−/− MEF cells were treated with DMSO, NAC, or H2O2 for 30 min, and the transcript levels of SENP2 and PLCβ4 in SENP2−/− MEF cells were measured by real-time PCR normalized to control (n = 3 repeats/group). I Both starvation and SUMO inhibition rescued the viability of PC12 cells. Stably transfected PC12 cells were treated with starvation or 2-D08, and cell viability was measured by MTT assay (n = 4 repeats/group).
The role of the SENP2-PLCβ4 axis under oxidative stress was further defined. SENP2 MEF cells were treated with H2O2 or NAC, and the results showed that H2O2 treatment enhanced the protein level of PLCβ4, whereas NAC treatment diminished the protein level of PLCβ4 (Fig. 7E). H2O2 induced expression of SENP2 and PLCβ4 in a concentration-dependent manner in SENP2+/+ MEF cells, whereas the expression level of PLCβ4 was minimally increased in SENP2−/− MEF cells and was much less than that in SENP2+/+ MEF cells (Fig. 7F), indicating that PLCβ4 is indeed regulated through a SUMO-dependent degradation system, but much less from ROS. The protein levels of IP3R and RyR were increased, and SERCA2 was decreased by H2O2 treatment in a concentration-dependent manner in SENP2+/+ MEF cells, but no response to H2O2 in SENP2−/− MEF cells (Fig. 7F). NAC decreased expression of SENP2 and PLCβ4 in a time-dependent manner in PC12 cells (Fig. 7G). However, the mitochondrial specific antioxidant mitoQ showed no significant effect on the expression levels of IP3R, RyR, and SERCA2 (Fig. S7H). Following decreased SENP2 and PLCβ4, there was enhanced SUMOylation of PLCβ4 (Fig. S7I), and decreased levels of IP3R and RyR and increased levels of SERCA2 (Fig. 7G). In addition, H2O2 increased the transcription of SENP2, whereas NAC decreased the transcription of SENP2 with similar levels of PLCβ4 in SENP2+/+ MEF cells (Fig. 7H). We further verified the role of SENP2-PLCβ4 axis in neuron viability during stress, and the results showed that both starvation and the SUMO inhibitor 2-D08 significantly reversed the decreased cell viability induced by PLCβ4 knockdown in HT22 and PC12 cells (Figs. 7I and S7J). Taken together, these results suggest that SENP2-PLCβ4 axis acts as a specific sensor of starvation and oxidative stress and regulates cell viability through calcium homeostasis.
Discussion
Calcium homeostasis is important for the regulation of a wide variety of cellular processes and is essential for cellular function [29, 30]. SENP2 deficiency induced PLCβ4 degradation, which reduced levels of IP3 and IP3R (Fig. 1). Growing evidence has shown that IP3R and RyR share the same calcium pool, and are functionally coupled [31]. IP3 diffuses into the IP3R channel and activates IP3R on the ER, and subsequently followed by RyR, leading to the uptake of intracellular calcium [32]. SENP2 activates PLCβ4 and promotes the production of IP3, and the activation of IP3R by IP3 recruits neighboring domains of RyR, together leading to calcium release from the ER. SERCA2 usually shows an alteration that is opposite to that of IP3R and RyR to maintain calcium homeostasis in the ER [33, 34]. As shown, the RyR level was decreased, accompanied by increased SERCA2 level (Fig. 1). These results indicated an important role of SENP2-PLCβ4 axis in the maintenance of calcium homeostasis through the IP3-regulated IP3R, RyR and SERCA2.
Neurogenesis is a fine-tuned process that plays important roles during early development and adulthood [35]. Our study demonstrated that SENP2 deficiency has no significant effect on neurogenesis in 10-day-old mice; however, we found fewer mature and immature neurons in 4-week-old SENP2-deficient mice, and substantial defect in neurogenesis in 6-week-old mice (Fig. S5). Six-week-old SENP2-deficient mice showed severe inhibition of neuron proliferation, accompanied by increased numbers of astrocytes and microglia cells, indicating that neurogenesis defects occur via SENP2 deficiency (Fig. 5). These results demonstrate that SENP2 regulates neurogenesis in a time-dependent manner. It is possible that SENP2 deficiency could play a role in neurogenesis in earlier development, such as prior to postnatal day 10; however, the changes were not detectable using immune-histochemical analysis. In our previous publication, SENP2-null embryo died at E10 due to a critical role of SENP2 in the regulation of polycomb repressive complex in gene silencing [15]. Increasing evidence showed that calcium signaling modulates autophagic dynamics [36]. Our studies indicate that the SENP2-PLCβ4 axis plays a critical role in postnatal neurogenesis by maintaining calcium homeostasis, providing mechanistic insights into neurogenesis and development.
A previous study identified that SENP3 was concomitantly induced by ROS and functioned as a suppressor of autophagic flux, forming an intrinsic negative feedback loop in the liver [37]. Our study showed that both mRNA and protein levels of SENP2 were stimulated via starvation or oxidative stress, whereas NAC inhibited the expression of SENP2 (Fig. 7). Oxidative stress exerts a significant effect on hippocampal neurogenesis [38]. The present study showed that SENP2 acts as a unique redox-sensitive SUMO protease in the brain and regulates calcium homeostasis-regulated neurogenesis through PLCβ4 SUMOylation. These findings highlight the critical role of the SENP2-PLCβ4 axis under extracellular stress, showing that it protects cells by maintaining calcium homeostasis regulated neurogenesis.
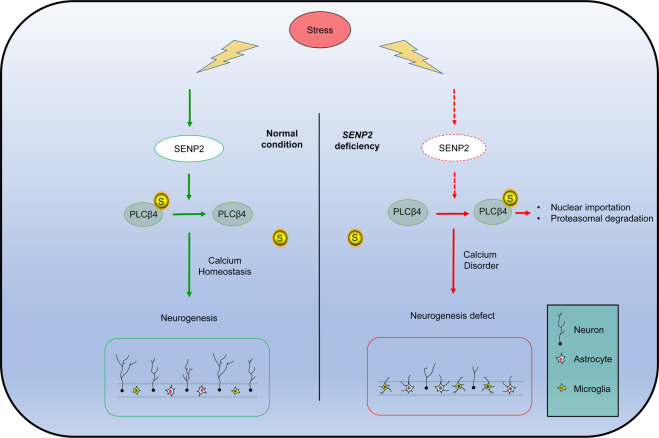
In summary, our study demonstrated a novel role of PLCβ4 SUMOylation in the regulation of neurogenesis via calcium homeostasis under stress (Fig. 8). In SENP2-deficient mice, PLCβ4 is hyper-SUMOylated, which promotes its transport from the membrane and cytoplasm to the nucleus and subsequent degradation. SUMOylated PLCβ4 induces the impairment of intracellular calcium homeostasis and ultimately impaired neurogenesis and neuronal viability, which are novel effects of PTM-mediated regulation of GPCR signaling pathway. In conclusion, our studies indicate that balancing the SUMOylation of PLCβ4 will be a novel intervention for neurological disorders caused by postnatal neurogenesis defects.
Fig. 8. SENP2-PLCβ4 signaling regulates neurogenesis through the maintenance of calcium homeostasis.
Schematic model depicting the role of the SENP2-PLCβ4 axis in the regulation of PLCβ4 stability and calcium homeostasis regulated neurogenesis during stress.
Materials and methods
Cell culture and treatment
HEK293T, HT22, PC12, SH-SY5Y, SENP2+/+ and SENP2−/− MEF cells [16] were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Gibco) containing 10% fetal bovine serum (FBS, Gibco), 100 units/ml penicillin and 100 µg/ml streptomycin at 37 °C in 5% CO2 atmosphere. Cells were transiently transfected using Lipofectamine 3000 (Invitrogen), according to the manufacturer’s instructions. For protein degradation assay, transfected cells were treated with 50 µg/mL cycloheximide (CHX), 10 μM MG132, 100 μM chloroquine for different times, and the whole cell lysates were analyzed by Western blotting. For starvation assay, cells were starved for 12 h in FBS free DMEM medium to mimic calorie restriction. For oxidative stress assay, cells were treated with 100 μM H2O2 or 20 mM NAC for different times. For 2-D08 rescue assay, after seeded for 24 h, the cells were treated with 200 µM 2-D08 for 24 h.
Plasmid construction
Eukaryotic expression plasmids Flag-SENP2 wild type, Flag-SENP2 mutant, HA-SUMO1, HA-SUMO2 and HA-ubiquitin were previously described [15–17]. The Flag-PLCβ4 and pEGFP-PLCβ4 plasmids were constructed by standard PCR-based strategies, and SUMO site mutants were generated using the Quick-change site-directed mutagenesis kit (TianGen). All plasmids were verified by DNA sequencing.
Animal models
SENP2-deficient mice were described previously [17]. All animals were housed at room temperature (22 °C), with a 12 h light-dark cycle with ad libitum access to food and water. Animals were housed 2–5 per cage. All animal care and studies were performed according to protocols “Guide for the Care and Use of Laboratory Animals”, which were approved by the Institutional Animal Care and Use Committee at Shaanxi Normal University, and all manipulations were conducted in consistence with established guidelines. Randomization and blinding were used in animal study. For BrdU/EdU pulse-chase experiments, mice were injected with BrdU or EdU at a dose of 100 mg/kg body weight, twice daily for 5 consecutive days.
RNA knockdown treatment
The siRNA and shRNA oligo-nucleotides targeting SENP2 and PLCβ4 were purchased from Thermo. The si-RNF4 sequence was used to knockdown the RNF4 level [39]. The RanBP2 knockdown plasmid pGFP-V-RS-sh-RanBP2 was kindly provided by Dr. Vallee [40]. RNAi transfections were performed using the Lipofectamine 3000 transfection reagent (Invitrogen).
Generation of the lentiviral system
shRNA knockdown cell lines were generated using a lentiviral system (System Biosciences). Virus was generated in HEK293TN cells by transfecting the pGreen-Puro, packaging (psPAX2), and envelope (pMD2) plasmids. Cell culture media was collected 48 h post transfection and immediately transferred to target cells in the presence of polybrene (Sigma). The transduced cells were selected with puromycin for 48 h. Real-time PCR or Immunoblotting (IB) should be manipulated to detect the efficacy of knockdown.
RNA isolation and real-time PCR
Mice genotyping was described previously [17]. For quantitative analysis of gene expression, total RNA was extracted using the RNeasy protocol (Qiagen) from cultured or transfected cells. RNA was treated with DNase (Promega), and the concentration was determined by measuring absorbance at 260 nm. Equal RNA levels were used to generate complementary DNA using the high-capacity cDNA reverse transcription protocol (Takara). Quantitative real-time PCR was performed using reaction mixtures of cDNA, primers (Supplementary Table S1), and SYBR Green reagent (Takara) with the ABI StepOne system (Perkin-Elmer). PCR was done in triplicate, and standard deviations representing experimental errors were calculated. All data were analyzed using ABI PRISM SDS 2.0 software (Perkin-Elmer). This software, which is coupled to the instrument, allows the determination of the threshold cycle that represents the number of the cycle where the fluorescence intensity is significantly above the background fluorescence intensity.
Cellular sub-fractionation
Subcellular fraction protein was isolated by the kit according to the manual (KeyGen). Briefly, PLCβ4 wild type or sumo mutant and SUMO2 were transfected into HEK293T cells and incubated with MG132 for 4 h before harvest. All cells were washed and lysed in hypotonic buffer with protease inhibitors, and cell lysates were centrifuged. Supernatants were collected as cytoplasmic extracts, and pellets were washed three times and lysed with a high-salt buffer with protease inhibitors, vortexed, and rotated. Cell lysates were centrifuged, and supernatants were collected as nuclear extracts. Equal amounts of cytoplasmic and nuclear extracts were performed for Western blotting.
Western blotting and immunoprecipitation
Whole mouse brain or transfected cells were extracted and subsequently homogenized on ice in lysis buffer with protease inhibitors (Targetmol). Total protein levels were quantified using the BCA assay (Pierce). Equal protein amounts were separated by electrophoresis and transferred to PVDF membranes by electroblotting. Membranes were blocked with 5% nonfat dried milk, incubated overnight with primary antibodies (Supplementary Table S3), washed, incubated with secondary antibody coupled to peroxidase, and protein levels were detected with a chemiluminescence system (Tanon) after additional washing steps. For immunoprecipitation experiments, brain or cell lysates were incubated overnight with antibodies. All incubations were performed at 4 °C with constant agitation. Antibody-bound protein complexes were captured by the addition of protein A/G agarose and incubated for another 2 h. Protein A/G agarose was pelleted by centrifugation, and the immunoprecipitated protein complex was eluted using SDS-PAGE sample buffer and western blotting with antibodies.
Golgi staining
Golgi staining was manipulated using the FD Rapid Golgi Stain kit (FD Neuro Technologies). In brief, the brain tissue of 6-week-old wild type and SENP2-deficient mice were perfused and collected, and then put into a mixture of solution A and B. The mixture of impregnation solution was replaced after 24 h of initial immersion, and then kept in dark for 2 weeks at room temperature, and then incubated in Solution C in dark for 1 week at 4 °C. The brain was sectioned into slices with a vibratome (Leica) and mounted on gelatin-coated cover slips. The sections were stained with premixed Solution D and E for 10 min and rinsed with water and dehydrated with ethanol series, followed by xylene and mounted with DPX (Invitrogen), and then examined by confocal laser scanning microscopy (Zeiss).
Nissl staining
Cresyl violet was used to perform Nissl staining for neuron-specific endoplasmic reticulum granular bodies. Briefly, brain sections were incubated with cresyl violet (Sigma) in sodium acetate buffer for 5 min followed by a brief rinse in water. The staining was differentiated in 1% glacial acetic acid in 95% ethanol until white matter tracts were visible from gray matter. After dehydration with gradient ethanol, the tissue was cleared in xylene and mounted with DPX (Invitrogen), and then examined by confocal laser scanning microscopy (Zeiss).
Hippocampal neuronal culture
Hippocampal neuronal cultures were prepared from 1-day-old SENP2-deficient and wild type control pups [19]. Hippocampi were removed rapidly under stereomicroscopic observation, cut into small pieces, and digested with papain at 37 °C for 15 min. The tissue segments were then transferred to Neurobasal medium (Invitrogen), and triturated with a fire-polished Pasteur pipette in the presence of DNase I. The cells were plated at a density of 2 × 104 per 12 mm glass coverslip precoated with poly-D-lysine (BD) and maintained with Neurobasal medium. Two hours after plating, the medium was removed, and Neurobasal/B27 medium was added. The hippocampal neurons were cultured in Neurobasal supplemented with 2% B27 and 1% Glutamax (Invitrogen) in a humidified atmosphere containing 5% CO2 at 37 °C. Subsequent feeding occurred twice weekly, each time replacing half the volume with medium including FGF2 and AraC. The neurons were used for immunocytochemistry at DIV 14.
Hippocampal NPC culture and neurosphere formation
Isolation of NPC was based on selective expansion from the fetal or adult mice brains [41]. The hippocampus was isolated from embryonic day 18 (E18) pups. After dissection, triturated tissue was plated in NPC culture medium (DMEM/F12 with 2% B27, 1% Glutamax, 100 U/mL Penicillin-Streptomycin, 20 ng/mL EGF, 20 ng/mL b-FGF), which supports the expansion of stem cells but not for other cell types. For neural stem cell neurosphere formation assay, the stable sh-PLCβ4 or sh-ctrl transfected NPC were collected and seeded onto six-well plates and cultured with NPC proliferation medium.
Immunocytochemistry
Cells transfected with indicated plasmids were grown on cover slips. And 48 h later, cells were washed with PBS, fixed with 4% paraformaldehyde (PFA, Sigma) and permeabilized with 0.5% Triton X-100 in PBS and then incubated with antibodies. Nonspecific antibody binding was minimized by treatment with 5% donkey serum in PBS for 30 min at room temperature. Primary antibodies were diluted in 0.1% Triton X-100 and incubated with the cells for 1 h at 37 °C. Cells were washed three times in PBS and then incubated for 1 h at room temperature with secondary antibodies Alexa Fluro 488 or 546 fluorophores (Invitrogen). The cells were then washed three times in PBS and mounted using the anti-fade mounting solution (Dako), and then examined by confocal laser scanning microscopy (Zeiss).
SENP2-deficient and wild type control mice were perfused transcardially with cold PBS and then fixed by 4% PFA. The Brain was then coronally sectioned at a thickness of 10 µm, washed with PBS, and incubated in PBS containing blocking solution (goat serum, Gibco) for 1 h. Primary antibodies were incubated overnight in blocking solution at 4 °C and fluorescence labeled secondary antibodies in blocking solution were incubated for 1 h at room temperature. After three times washing, slices mounted on slides with Prolong Gold Antifade (Invitrogen), and then examined by confocal laser scanning microscope (Zeiss).
For TUNEL assay, paraffin sections were incubated with TUNEL solution for 60 min and alkaline phosphatase antibody for 30 min. The sections were re-stained with hematoxylin. The blue-black nucleus was positive under a light microscope. For BrdU staining, the sectioned slices were washed three times with PBS, incubated in 2 M HCl for 30 min at room temperature, incubated with 1 M sodium borate buffer for 10 min at room temperature, and immediately washed three times with PBS. The steps of blocking and incubation with the primary and secondary antibodies were identical to those described above.
Cell viability assay
Cells were seeded at a density of 3000 cells/well in 96-well plates and incubated in the culture medium. After 24, 48, or 72 h, cell viability was measured using the 3-(4,5-dimethylthiazol-2-yl)−2,5-diphenyltetrazolium bromide (MTT, Sigma) according to the manufacturer’s protocol. Briefly, 15 µL of 5 mg/mL MTT solution was added to 150 µL culture medium. After 4 h incubation at 37 °C, 200 µL dimethyl sulfoxide (DMSO, Sigma) was added to each well. The absorbance was measured using a multimode microplate reader (Thermo) at 490 nm.
For cell counting assay, Cells were seeded at a density of 50,000 cells/well in 24-well plates and incubated in the culture medium. After 24, 48, 72, or 96 h, cells were counted via 0.4% Trypan Blue solution (Sigma-Aldrich) according to the manufacturer’s protocol. In brief, stained cells were non-viable cells, while the un-stained cells were counted and calculated.
For EdU proliferation assay, in the logarithmic growth phase, the cells were seeded in 6-well plates at 1 × 105 cells per well. Edu labeling, red fluorescent staining and Hoechst 33342 nuclear counterstaining were performed according to the EdU Kit (Beyotime biotechnology). EdU-labeled cells (red fluorescently labeled cells) and non-labeled cells were counted, and the percentage of EdU-labeled positive cells was calculated.
Intracellular PIP2, IP3, calcium and CaMK2 activity measurement
PIP2 level was measured by PIP2 mass ELISA kit (Echelon Biosciences) according to the assay protocol. Briefly, the brain tissue of mice was lysed, and acidic lipids were collected and incubated with detection and stop solution, and read absorbance at 450 nm on a plate reader. IP3 level was measured by mouse IP3 ELISA kit (Mybiosource) according to the manual. The brain tissue of mice was homogenized, incubated with Biotin and HRP-avidin antibodies, and finally add stop solution and read at 450 nm on a plate reader. The calcium level was measured by calcium colorimetric assay kit (Sigma) according to the procedure. The brain tissue of mice was homogenized, incubated with calcium assay buffer, and measure the absorbance at 575 nm on a plate reader. CaMK2β activity was measured with human CaMK2β ELISA kit (Abcam) according to the manual instructions. In brief, the brain tissue of mice was rinsed with PBS and homogenized in cell extraction buffer, and incubated with antibody, and record the OD at 450 nm on a plate reader.
Identification of PLCβ4 interacted proteins by mass spectrometric analysis
GFP-PLCβ4 wild type or SUMO mutant and HA-SUMO2 were transfected into HEK293T cells and incubated with MG132 for 4 h before harvest. PLCβ4 interacted proteins were enriched via GFP antibody for SDS-PAGE. The whole bands in two groups were excised and analyzed by Mass Spectrometry. Briefly, the gel bands were dehydrated and digested using sequencing grade-modified trypsin. Eluted peptides were sprayed into Q-Exactive mass spectrometer using a Flex Spray ion source (Thermo). The resulting LC-MS spectra were converted to peak lists using Mascot Distiller and analyzed using Scaffold to probabilistically validate protein identifications.
The cytosolic and ER calcium measurement
The calcium levels of cytosolic and ER were monitored using the specific calcium sensitive fluorescent indicators Fluo-4-AM and Mag-Fluo-4-AM (Invitrogen), respectively. The SENP2 MEF cells were cultured for different treatment, and cells were stained with 2 µM Fluo-4-AM or 2 µM Mag-Fluo-4-AM for 40 min at 37 °C in the dark, followed by wash with HBSS solution. The calcium levels were calculated according to fluorescence images collected by fluorescence microscopy (Nikon) or analyzed by flow cytometry using a FACS Calibur instrument (BD PharMingen).
Quantification and statistical analysis
All data were presented as mean ± S.E.M. or S.D. of at least three separate experiments. All mice data were presented as mean ± S.E.M. or SD of at least five mice in each group. Differences between groups were evaluated by the Student’s t test for two-group comparisons, and one-way ANOVA followed by Dunnett’s or Tukey’s test or two-way ANOVA followed by Bonferroni’s test for multiple comparisons among more than two groups. The variance was similar between the groups that were being statistically compared. Statistical significance was defined as p < 0.05 (*p < 0.05; **p < 0.01, and ***p < 0.001).
Supplementary information
Acknowledgements
We are grateful to Dr. Richard Vallee from Columbia University for kindly providing the sh-RanBP2 plasmid, and we also thank Dr. Xing Li from Shaanxi Normal University for their technical supports and Dr. Yingfang Tian from Shaanxi Normal University for critical reading of the paper.
Author contributions
XC, HMW, and YTQ conceptualized and designed the research. XC, YYQ, and YHZ developed experimental methods, performed most experiments and analyzed the data. XYY, ZCX, YJS, and JKC provided technical supports. HMW and YTQ supervised the study. XC, HMW, and YTQ wrote the paper with editorial input from ETHY. All authors have read and approved the paper.
Funding
This study was supported by grants from the National Natural Science Foundation of China (81671294 and 81870241 to YTQ), and the Fundamental Research Funds for the Central Universities (GK201903066 to HMW).
Data availability
All data are provided in the paper and Supplementary files or are available from the corresponding authors on reasonable request.
Competing interests
The authors declare no competing interests.
Ethical approval
All animal care and studies were performed according to protocols “Guide for the Care and Use of Laboratory Animals”, which were approved by the Institutional Animal Care and Use Committee at Shaanxi Normal University, and all manipulations were conducted in consistence with established guidelines.
Footnotes
Edited by G. Del Sal
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Edward T. H. Yeh, Email: dredyeh@gmail.com
Hongmei Wu, Email: hq8479@snnu.edu.cn.
Yitao Qi, Email: qiyitao@snnu.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41418-021-00857-1.
References
- 1.Fares J, Bou Diab Z, Nabha S, Fares Y. Neurogenesis in the adult hippocampus: history, regulation, and prospective roles. Int J Neurosci. 2019;129:598–611. doi: 10.1080/00207454.2018.1545771. [DOI] [PubMed] [Google Scholar]
- 2.Epp JR, Spritzer MD, Galea LA. Hippocampus-dependent learning promotes survival of new neurons in the dentate gyrus at a specific time during cell maturation. Neuroscience. 2007;149:273–85. doi: 10.1016/j.neuroscience.2007.07.046. [DOI] [PubMed] [Google Scholar]
- 3.Kaplan MS, Hinds JW. Neurogenesis in the adult rat: electron microscopic analysis of light radioautographs. Science. 1977;197:1092–4. doi: 10.1126/science.887941. [DOI] [PubMed] [Google Scholar]
- 4.Zhao C, Deng W, Gage FH. Mechanisms and functional implications of adult neurogenesis. Cell. 2008;132:645–60. doi: 10.1016/j.cell.2008.01.033. [DOI] [PubMed] [Google Scholar]
- 5.Ming GL, Song H. Adult neurogenesis in the mammalian brain: significant answers and significant questions. Neuron. 2011;70:687–702. doi: 10.1016/j.neuron.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moreno-Jimenez EP, Flor-Garcia M, Terreros-Roncal J, Rabano A, Cafini F, Pallas-Bazarra N, et al. Adult hippocampal neurogenesis is abundant in neurologically healthy subjects and drops sharply in patients with Alzheimer’s disease. Nat Med. 2019;25:554–60. doi: 10.1038/s41591-019-0375-9. [DOI] [PubMed] [Google Scholar]
- 7.Takahashi H, Yoshihara S, Tsuboi A. The Functional Role of Olfactory Bulb Granule Cell Subtypes Derived From Embryonic and Postnatal Neurogenesis. Front Mol Neurosci. 2018;11:229. doi: 10.3389/fnmol.2018.00229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kano M, Hashimoto K, Watanabe M, Kurihara H, Offermanns S, Jiang H, et al. Phospholipase cbeta4 is specifically involved in climbing fiber synapse elimination in the developing cerebellum. Proc Natl Acad Sci USA. 1998;95:15724–9. doi: 10.1073/pnas.95.26.15724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gutkind JS. The pathways connecting G protein-coupled receptors to the nucleus through divergent mitogen-activated protein kinase cascades. J Biol Chem. 1998;273:1839–42. doi: 10.1074/jbc.273.4.1839. [DOI] [PubMed] [Google Scholar]
- 10.Exton JH. Regulation of phosphoinositide phospholipases by hormones, neurotransmitters, and other agonists linked to G proteins. Annu Rev Pharm Toxicol. 1996;36:481–509. doi: 10.1146/annurev.pa.36.040196.002405. [DOI] [PubMed] [Google Scholar]
- 11.Watanabe M, Nakamura M, Sato K, Kano M, Simon MI, Inoue Y. Patterns of expression for the mRNA corresponding to the four isoforms of phospholipase Cbeta in mouse brain. Eur J Neurosci. 1998;10:2016–25. doi: 10.1046/j.1460-9568.1998.00213.x. [DOI] [PubMed] [Google Scholar]
- 12.Yeh ET, Gong L, Kamitani T. Ubiquitin-like proteins: new wines in new bottles. Gene. 2000;248:1–14. doi: 10.1016/s0378-1119(00)00139-6. [DOI] [PubMed] [Google Scholar]
- 13.Yeh ET. SUMOylation and De-SUMOylation: wrestling with life’s processes. J Biol Chem. 2009;284:8223–7. doi: 10.1074/jbc.R800050200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang HM, Yeh ETH. SUMO: From Bench to Bedside. Physiol Rev. 2020;100:1599–619. doi: 10.1152/physrev.00025.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kang X, Qi Y, Zuo Y, Wang Q, Zou Y, Schwartz RJ, et al. SUMO-specific protease 2 is essential for suppression of polycomb group protein-mediated gene silencing during embryonic development. Mol cell. 2010;38:191–201. doi: 10.1016/j.molcel.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qi Y, Zuo Y, Yeh ET, Cheng J. An essential role of small ubiquitin-like modifier (SUMO)-specific Protease 2 in myostatin expression and myogenesis. J Biol Chem. 2014;289:3288–93. doi: 10.1074/jbc.M113.518282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qi Y, Wang J, Bomben VC, Li DP, Chen SR, Sun H, et al. Hyper-SUMOylation of the Kv7 potassium channel diminishes the M-current leading to seizures and sudden death. Neuron. 2014;83:1159–71. doi: 10.1016/j.neuron.2014.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu H, Chen X, Cheng J, Qi Y. SUMOylation and potassium channels: links to epilepsy and sudden death. Adv Protein Chem Struct Biol. 2016;103:295–321. doi: 10.1016/bs.apcsb.2015.11.009. [DOI] [PubMed] [Google Scholar]
- 19.Chen X, Zhang S, Huang J, Dong W, Xiao H, Shao H, et al. Hyper-SUMOylation of K(+) Channels in Sudden Unexplained Death in Epilepsy: Isolation and Primary Culture of Dissociated Hippocampal Neurons from Newborn Mice for Subcellular Localization. Methods Mol Biol. 2018;1684:63–71. doi: 10.1007/978-1-4939-7362-0_6. [DOI] [PubMed] [Google Scholar]
- 20.Geoffroy MC, Hay RT. An additional role for SUMO in ubiquitin-mediated proteolysis. Nat Rev Mol Cell Biol. 2009;10:564–8. doi: 10.1038/nrm2707. [DOI] [PubMed] [Google Scholar]
- 21.Blind RD, Suzawa M, Ingraham HA. Direct Modification and Activation of a Nuclear Receptor-PIP2 Complex by the Inositol Lipid Kinase IPMK. Sci Signaling. 2012;5:ra44. doi: 10.1126/scisignal.2003111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anisman H, Merali Z, Hayley S. Neurotransmitter, peptide and cytokine processes in relation to depressive disorder: comorbidity between depression and neurodegenerative disorders. Prog Neurobiol. 2008;85:1–74. doi: 10.1016/j.pneurobio.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 23.Geribaldi-Doldan N, Flores-Giubi E, Murillo-Carretero M, Garcia-Bernal F, Carrasco M, Macias-Sanchez AJ, et al. 12-Deoxyphorbols Promote Adult Neurogenesis by Inducing Neural Progenitor Cell Proliferation via PKC Activation. Int J Neuropsychopharmacol. 2015;19:pyv085. doi: 10.1093/ijnp/pyv085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murillo-Carretero M, Geribaldi-Doldan N, Flores-Giubi E, Garcia-Bernal F, Navarro-Quiroz EA, Carrasco M, et al. ELAC (3,12-di-O-acetyl-8-O-tigloilingol), a plant-derived lathyrane diterpene, induces subventricular zone neural progenitor cell proliferation through PKCbeta activation. Br J Pharm. 2017;174:2373–92. doi: 10.1111/bph.13846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sung SM, Jung DS, Kwon CH, Park JY, Kang SK, Kim YK. Hypoxia/reoxygenation stimulates proliferation through PKC-dependent activation of ERK and Akt in mouse neural progenitor cells. Neurochem Res. 2007;32:1932–9. doi: 10.1007/s11064-007-9390-1. [DOI] [PubMed] [Google Scholar]
- 26.Ma DK, Ponnusamy K, Song MR, Ming GL, Song H. Molecular genetic analysis of FGFR1 signalling reveals distinct roles of MAPK and PLCgamma1 activation for self-renewal of adult neural stem cells. Mol Brain. 2009;2:16. doi: 10.1186/1756-6606-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ding K, Lai Z, Yang G, Zeng L. MiR-140-5p targets Prox1 to regulate the proliferation and differentiation of neural stem cells through the ERK/MAPK signaling pathway. Ann Transl Med. 2021;9:671. doi: 10.21037/atm-21-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharma D, Maslov LN, Singh N, Jaggi AS. Remote ischemic preconditioning-induced neuroprotection in cerebral ischemia-reperfusion injury: preclinical evidence and mechanisms. Eur J Pharmacol. 2020;883:173380. doi: 10.1016/j.ejphar.2020.173380. [DOI] [PubMed] [Google Scholar]
- 29.Berridge MJ, Bootman MD, Roderick HL. Calcium signalling: dynamics, homeostasis and remodelling. Nat Rev Mol Cell Biol. 2003;4:517–29. doi: 10.1038/nrm1155. [DOI] [PubMed] [Google Scholar]
- 30.Berridge MJ, Lipp P, Bootman MD. The versatility and universality of calcium signalling. Nat Rev Mol Cell Biol. 2000;1:11–21. doi: 10.1038/35036035. [DOI] [PubMed] [Google Scholar]
- 31.Janiak R, Wilson SM, Montague S, Hume JR. Heterogeneity of calcium stores and elementary release events in canine pulmonary arterial smooth muscle cells. Am J Physiol Cell Physiol. 2001;280:C22–33. doi: 10.1152/ajpcell.2001.280.1.C22. [DOI] [PubMed] [Google Scholar]
- 32.Berridge MJ. Calcium microdomains: organization and function. Cell Calcium. 2006;40:405–12. doi: 10.1016/j.ceca.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 33.Gordienko DV, Bolton TB. Crosstalk between ryanodine receptors and IP(3) receptors as a factor shaping spontaneous Ca(2+)-release events in rabbit portal vein myocytes. J Physiol. 2002;542:743–62. doi: 10.1113/jphysiol.2001.015966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johny JP, Plank MJ, David T. Importance of Altered Levels of SERCA, IP3R, and RyR in Vascular Smooth Muscle Cell. Biophys J. 2017;112:265–87. doi: 10.1016/j.bpj.2016.11.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jurkowski MP, Bettio L, Emma KW, Patten A, Yau SY, Gil-Mohapel J. Beyond the Hippocampus and the SVZ: Adult Neurogenesis Throughout the Brain. Front Cell Neurosci. 2020;14:576444. doi: 10.3389/fncel.2020.576444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patergnani S, Danese A, Bouhamida E, Aguiari G, Previati M, Pinton P, et al. Various Aspects of Calcium Signaling in the Regulation of Apoptosis, Autophagy, Cell Proliferation, and Cancer. Int J Mol Sci. 2020;21:8323. doi: 10.3390/ijms21218323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu K, Guo C, Lao Y, Yang J, Chen F, Zhao Y, et al. A fine-tuning mechanism underlying self-control for autophagy: deSUMOylation of BECN1 by SENP3. Autophagy. 2020;16:975–90. doi: 10.1080/15548627.2019.1647944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rizk M, Vu J, Zhang Z. Impact of pediatric traumatic brain injury on hippocampal neurogenesis. Neural Regen Res. 2021;16:926–33. doi: 10.4103/1673-5374.297057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tatham MH, Geoffroy MC, Shen L, Plechanovova A, Hattersley N, Jaffray EG, et al. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat Cell Biol. 2008;10:538–46. doi: 10.1038/ncb1716. [DOI] [PubMed] [Google Scholar]
- 40.Baffet AD, Hu DJ, Vallee RB. Cdk1 Activates Pre-mitotic Nuclear Envelope Dynein Recruitment and Apical Nuclear Migration in Neural Stem Cells. Dev Cell. 2015;33:703–16. doi: 10.1016/j.devcel.2015.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johe KK, Hazel TG, Muller T, Dugich-Djordjevic MM, McKay RD. Single factors direct the differentiation of stem cells from the fetal and adult central nervous system. Genes Dev. 1996;10:3129–40. doi: 10.1101/gad.10.24.3129. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are provided in the paper and Supplementary files or are available from the corresponding authors on reasonable request.